Werner syndrome (WS) is a human autosomal recessive disorder that causes the premature appearance of a partial array of disorders characteristic of old age1,2. These disorders include atherosclerosis, cancer, type 2 diabetes, osteoporosis, cataracts, wrinkled skin and grey hair, among other ailments. Cells cultured from WS subjects have a shortened replicative life span3,4 and elevated rates of chromosome translocations, large deletions and homologous recombination5,6. The gene defective in WS, WRN, encodes a large RecQ-like DNA helicase7 of 1432 aa. Defects in another human RecQ-like helicase, BLM, result in Bloom’s syndrome8 (BS), a genetic disorder that is quite different from WS. BS is manifested by short stature, neoplasia, immunodeficiency and high risk of cancer. Cells from BS subjects show an increase in sister chromatid exchanges.
DNA helicases can function in replication, repair, recombination, transcription or RNA processing. As WRN and BLM share no obvious homology outside the helicase domain, the non-helicase domains probably determine in which process each RecQ-like helicase participates, which provides the basis for the disparate cellular and organismal phenotypes that result from defects in these proteins.
Statistical sequence analyses showed subtle but significant similarities between WRN and several 3′→5′ exonucleases9,10. To test the prediction that WRN is an exonuclease, we produced tagged recombinant wild-type and mutant WRN proteins. Two mutants had amino-acid substitutions at either position 82 (D82A) or 84 (E84A), two of the five residues predicted to be critical for exonuclease activity9,10. A third mutant had a substitution at position 577 (K577M), which abolished WRN helicase activity11. The fourth mutant was an N-terminal fragment (aa 1–333; N333) containing the putative exonuclease domain, but lacking the helicase domain. A tagged 36-aa vector-derived peptide served as a negative control (mock).
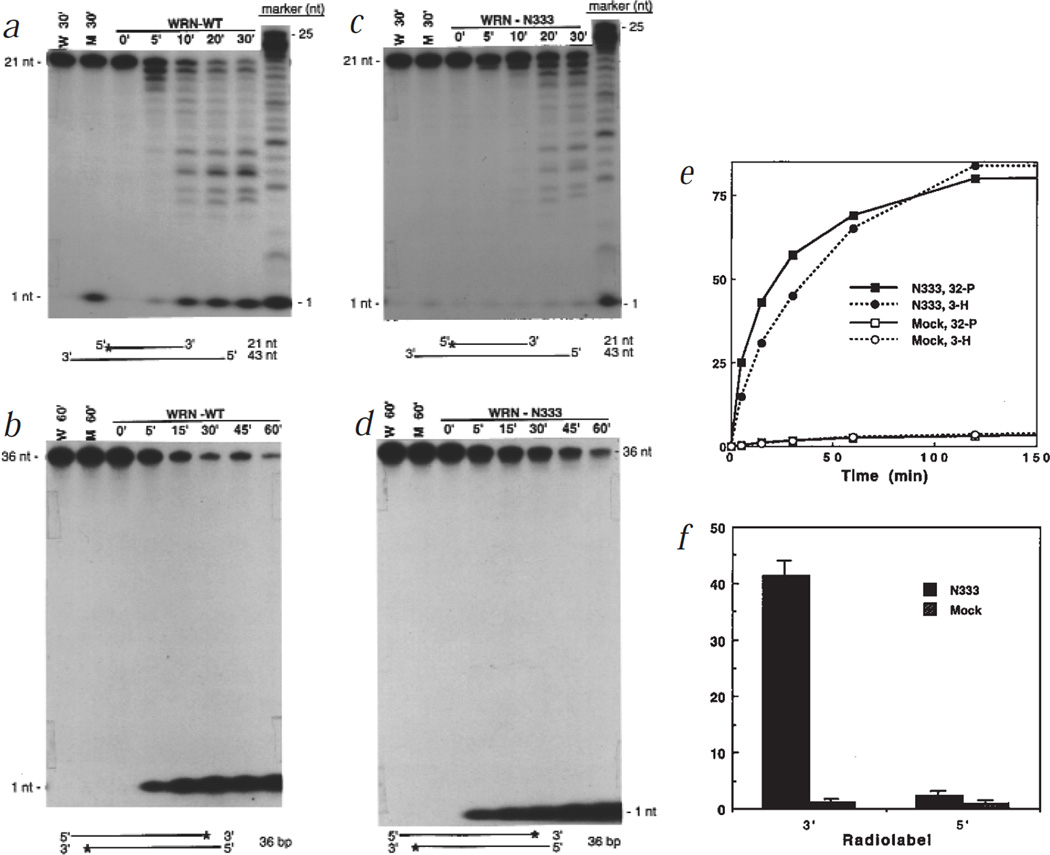
Purified WRN and mock proteins were incubated with doubled-stranded DNA substrates. Wild-type WRN degraded a 5′ labelled substrate to a series of smaller, labelled products (Fig. 1a), and a 3′ labelled substrate to a single labelled product that migrated as a mononucleotide (Fig. 1b). Thus, WRN degraded DNA with 3′→5′ directionality. Although mock and full-length WRN preparations contained low levels of a contaminating 5′→3′ exonuclease, as shown by release of the 5′ label as a mononucleotide (Figs 1a,2b), 3′→5′ degradation was entirely dependent on WRN.
Fig. 1.
Exonuclease activity of wild-type WRN and the N333 fragment. 6×his-tagged proteins were purified from baculovirus-infected insect cells using either nuclear (WRN, D82A, E84A, K577M, mock control) or cytosolic (N333, mock control) extracts. WRN, N-333, or mock proteins (10 ng, a,b; 5 ng, c,d) were incubated with 10,000 cpm of either a partial DNA duplex containing a 21-nt 5′ 32P-labelled fragment annealed to an unlabelled 43-nt fragment (a,c) or a 36-bp DNA fragment 32P-labelled at the 3′ end (b,d) in nuclease assay buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 5 mM DTT, 0.1 mg/ml BSA) at 37 °C for the indicated intervals. Products were analysed on 20% polyacrylamide-8 M urea denaturing gels and visualized by autoradiography. M, mock; W, no protein addition. e, N333 or mock proteins (20 ng) were incubated with a 374-bp DNA fragment labelled with 32P at the 3′ end and 3H at internal thymidine residues for the indicated intervals. The products were precipitated with 70% ethanol, and ethanol-soluble radioactivity was determined by scintillation counting. The activity is presented as percentage of input label (10,000 cpm 32P; 6,000 cpm 3H) that was soluble in 70% ethanol. f, Equal amounts of 6×his-affinity purified N333 and mock proteins were resolved on a 8% non-denaturing polyacrylamide gel. The region containing N333 and a comparable region in the mock lane were excised, and the proteins eluted into nuclease buffer. The eluted N333 (20 ng) and mock proteins were incubated with 10,000 cpm of DNA substrates labelled to similar specific activities with 32P at either the 5′ or 3′ end (a,b, respectively) for 1 h, and the activity was determined as in (e).
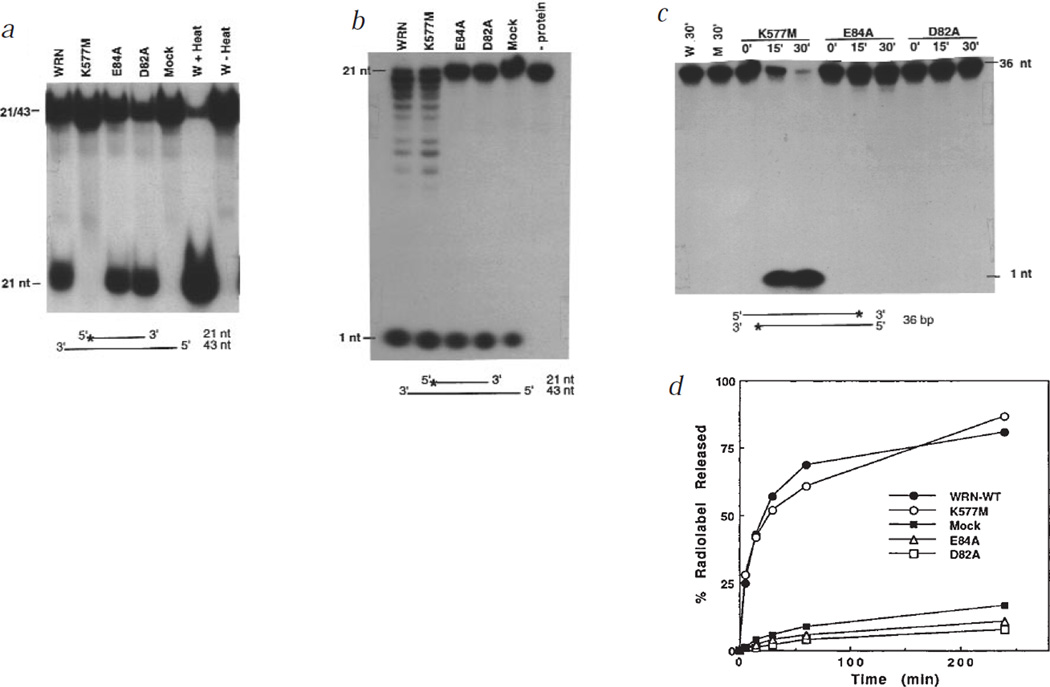
Fig. 2.
Helicase and exonuclease activities of wild-type and mutant WRN proteins. a, WRN, K577M, E84A, D82A or mock proteins (10 ng) were assayed for helicase activity by incubating 5′ 32P-labelled DNA substrate (0.4 pmol; Fig. 1a) in helicase assay buffer (40 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 5 mM DTT, 1 mM ATP, and 0.1 mg/ml BSA) at 37 °C for 10 min. Products were resolved on a 10% non-denaturing polyacrylamide gel and visualized by autoradiography. Controls were no protein addition (W), and heating (+ Heat) or not heating (− Heat) the sample to 95 °C before loading. The displaced species co-migrated with the 21-nt fragment. The same proteins (10 ng) were assayed for nuclease activity by incubating for 1 h (b) or 0–30 min (c) with 10,000 cpm of 5′- (b) or 3′-labelled (c) DNA substrates, and analysing the products as described (Fig. 1a,b). d, Proteins (5 ng) were incubated with 10,000 cpm of the substrate used in (c) for the indicated intervals. The activity was determined as in Fig. 1e.
WRN exonuclease activity resided in the N terminus. N333, which was essentially free of contaminating 5′→3′ exonuclease, degraded 5′ and 3′ labelled substrates similarly to full-length WRN (Fig. 1c,d). When incubated with a 374-bp DNA fragment labelled at the 3′ end with 32P, and internally with 3H, N333 released most of both labels (Fig. 1e). Thus, the WRN exonuclease is capable of substantial DNA degradation. Consistent with 3′→5′ directionality, N333 released 32P from 3′ ends more rapidly than 3H from internal residues. In addition, gel-purified N333, which lacked contaminating nuclease activities, efficiently removed the 3′, but not the 5′, label when incubated with DNA substrates labelled at either the 3′ or the 5′ end (Fig. 1f).
Genetic evidence for WRN exonuclease activity was obtained by introducing point mutations at critical amino acids in the exonuclease domain (D82A and E84A). These mutants retained the wild-type level of helicase activity (Fig. 2a), but had little or no 3′→5′ exonuclease activity, using either a 5′ (Fig. 2b) or 3′ (Fig. 2c) 32P-labelled substrate, and were indistinguishable from mock protein in this regard (Fig. 2d). The K577M mutant, in contrast, was devoid of helicase activity (Fig. 2a), as expected, but had 3′→5′ exonuclease activity comparable to that of wild-type WRN (Fig. 2b–d).
Our data indicate that WRN is indeed a 3′→5′ exonuclease. This activity resides in the N terminus, and is physically and functionally separable from the helicase activity. The identification of an exonuclease activity in WRN clearly distinguishes it from other human RecQ-like helicases, and may help explain the differences between WS and BS.
What are the functions of the WRN exonuclease in vivo? It may participate in recombination and DNA repair. Exonucleases are integral components of some recombination pathways12, and WRN appears to have a role in recombination5,6,13. The finding that WS cells are hypersensitive to the DNA damaging agent 4-nitroquinoline-1-oxide14 suggests a role for WRN in DNA repair. Finally, WRN is homologous to FFA-1 (replication focus-forming activity 1) in Xenopus laevis15, raising the possibility that WRN may also be involved in DNA replication. In this context, the WRN exonuclease may provide 3′→5′ proofreading function to DNA polymerases that lack such activity. Whatever the case, an understanding of the functions of WRN exonuclease and their relationships to the other functions of WRN will lead to new insights into the molecular and cellular basis for WS and a subset of age-associated pathologies.
Acknowledgments
We thank S. Linn for valuable discussions and H. Asahara for technical advice. This work was supported by the National Institute on Ageing (grants AG11658 (J.C.); AG05776 (S.H.); AG14446 (J.O.)) and Department of Energy under contract DE-AC0376SF00098 to the University of California.
References
- 1.Martin GM. Birth Defects. 1978;14:5–39. [PubMed] [Google Scholar]
- 2.Goto M. Mech. Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 3.Martin GM, Sprague CA, Epstein CJ. Lab. Invest. 1970;23:86–92. [PubMed] [Google Scholar]
- 4.Salk D, Bryant E, Hoehn H, Johnston P, Martin GM. Adv. Exp. Bio. Med. 1985;190:305–311. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- 5.Fukuchi K, Martin GM, Monnat RJ. Proc. Natl Acad. Sci. USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng RZ, Murano S, Kurz B, Shmookler-Reis RJ. Mutat. Res. 1990;237:259–269. doi: 10.1016/0921-8734(90)90008-f. [DOI] [PubMed] [Google Scholar]
- 7.Yu CE, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 8.German J. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 9.Main IS. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mushegian AR, Bassett DE, Jr, Boguski MS, Bork P, Koonin EV. Proc. Natl Acad. Sci. USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray MD, et al. Nature Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 12.Linn SM, Lloyd RS, Roberts RT. Nucleases. New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 13.Yamagata K, et al. Proc. Natl Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogburn CE, et al. Hum. Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Chen C-Y, Kobayashi R, Newport J. Nature Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]