Abstract
Telomeres are DNA-protein structures that cap linear chromosomes and are essential for maintaining genomic stability and cell phenotype. We identified a novel human telomere-associated protein, TIN2, by interaction cloning using the telomeric DNA-binding-protein TRF1 as a bait. TIN2 interacted with TRF1 in vitro and in cells, and co-localized with TRF1 in nuclei and metaphase chromosomes. A mutant TIN2 that lacks amino-terminal sequences effects elongated human telomeres in a telomerase-dependent manner. Our findings suggest that TRF1 is insufficient for control of telomere length in human cells, and that TIN2 is an essential mediator of TRF1 function.
Introduction
Telomeres consist of several thousand copies of a repetitive DNA sequence (TTAGGG in vertebrates) and an unknown number of proteins. The telomeric nucleoprotein structure is essential for preventing chromosome fusions and genomic instability1. Telomeres also influence gene expression. In lower eukaryotes, genes located near telomeres are silenced, and proteins that mediate this silencing can alter gene expression at non-telomeric loci2–4. In higher eukaryotes, shortening of telomeres causes changes in cell phenotype5. The ability of telomeres to prevent genomic instability and alter gene expression depends on their length and the proteins that associate with them.
Telomere length, or the terminal restriction fragment (TRF), is 15–20 kb in the human germ line and early embryonic cells, and is maintained in part by the enzyme telomerase6–8. In the absence of telomerase, each round of DNA replication leaves 50–200 bp of unreplicated DNA at the 3′ end. Telomerase adds telomeric repeats to this 3′ overhang, thereby replenishing the telomeres. Most human cells do not express telomerase, and thus lose telomeric DNA with each division. Once the TRF reaches 5–7 kb, cells enter an irreversible state of arrested growth and altered function, termed replicative senescence9–11.
Telomerase alone does not ensure proper regulation of telomere length. Ectopic expression of telomerase prevents telomere erosion and senescence in some, but not all, human cells12–14. In addition, some cells, such as stimulated T lymphocytes, transiently express telomerase, but their telomeres shorten nonetheless15,16. Many tumour cells express telomerase, but maintain TRFs that are longer or shorter than 5–7 kb (ref. 17), and some maintain telomeres without telomerase (presumably by recombination18). Studies in lower eukaryotes suggest that telomere-associated proteins control whether and how telomerase gains access to the 3′ terminus6,7,19.
Lower eukaryotes such as Saccharomyces cerevisiae maintain telomeres by balancing elongation by telomerase and shortening by exonuclease activity. This equilibrium is controlled in part by the double-stranded, telomeric DNA-binding-protein Rap1p. Rap1p negatively regulates telomere length and maintains chromosome stability and telomeric silencing20,21. At least two Rap1p binding proteins, Rif1p and Rif2p, are important for Rap1p function22. Rap1p also binds components of the SIR protein complex, which regulate silencing at telomeric and non-telomeric loci4,23. The Cdc13 and Stn1 proteins associate with the telomeric 3′ overhang, and also negatively regulate telomere length24,25.
Three genes encoding human telomere-associated proteins have been cloned. The first, TERF1 (ref. 26), may be a functional homologue of RAP1. TERF1 encodes two proteins, TRF1 (ref. 26) and PIN2 (derived by alternative splicing27), that bind double-stranded telomeric DNA and negatively regulate telomere length28. TRF1 also promotes parallel pairing of telomeric DNA (ref. 29). A second gene, TERF2, encodes TRF2, which is structurally similar to TRF1. TRF2 prevents chromosome fusion30 and mediates formation of the terminal telomeric t loop31. The third gene, TNKS, encodes the protein tankyrase, which binds TRF1 and has poly-ADP ribosylase activity32. Here we describe TINF2 (also known as TIN2), a novel human gene that encodes a protein (TIN2) that binds TRF1, localizes to telomeres and is essential for proper regulation of telomere length.
Results
Identification of TIN2
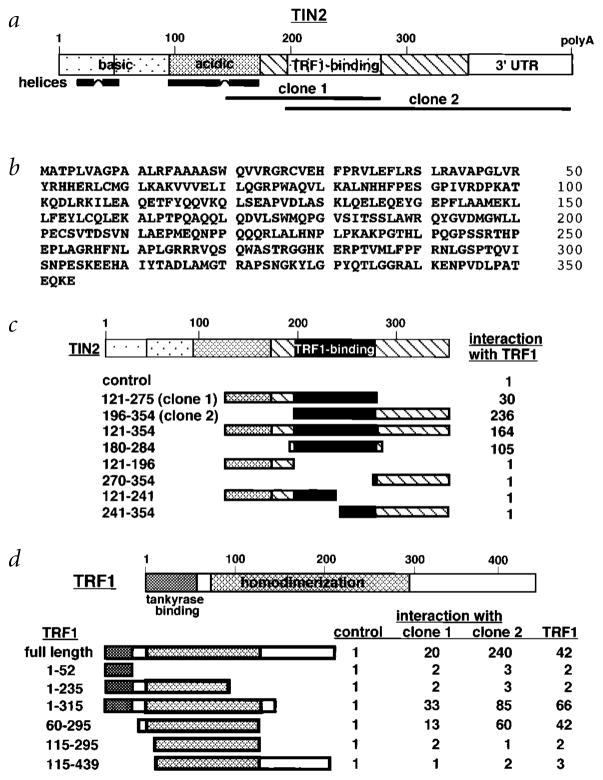
We screened a human fibroblast yeast two-hybrid cDNA library using the TERF1 cDNA fused to the GAL4 binding domain33. Positive clones contained 0.4-kb (clone 1) or 1.0-kb (clone 2) inserts that overlapped in sequence (Fig. 1a). The 1-kb insert had a polyadenylation site and was used to clone the ORF by 5′-RACE. The longest cDNA contained a 1,062-bp ORF, flanked by 5′ (263 bp) and 3′ (870 bp) UTRs. The ORF encoded a 354–amino-acid protein (MW 39,752 daltons; pI=9.5) that we named TRF1-interacting nuclear protein 2 (TIN2; Fig. 1b). TIN2 shares no homology with known genes or proteins. It has few structural motifs, aside from two highly basic regions (aa 1–45, pI 12.8; aa 45–90, pI 10.4) and an adjacent acidic region (aa 90–170, pI 4.3) that have the potential to form α-helices (Fig. 1a).
Fig. 1.
Sequence and structural characteristics of human TIN2. a, Structural features of TIN2. Shown are regions corresponding to the cDNA inserts recovered from the two-hybrid screen (clone 1, aa 147–275; clone 2, aa 196–354), the basic and acidic regions, potential helical structures and TRF1-binding domain. b, Deduced amino acid sequence of TIN2. c, TIN2 domains that interact with TRF1. We transformed TINF2 cDNA fragments (encoding the indicated amino acids) in pGAD-424 into yeast with pGBT9 containing TERF1 cDNA, and assessed interaction by a luminescent β-galactosidase assay. Control luminescence (interaction of pGAD-424 with pGBT9-TRF1) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination. c, TRF1 domains that interact with TIN2. Depicted is TRF1, showing the tankyrase-binding and homodimerization domains. We transformed TERF1 cDNA fragments (encoding the indicated amino acids) in pGBT9 into yeast with pGAD10 containing no insert (control), TINF2 clone 1, TINF2 clone 2 or full-length TERF1 cDNA, and assessed interaction by luminescent β-galactosidase assay. Control luminescence (interaction with insertless pGAD10) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination.
The sequence overlap of clones 1 and 2 suggested that TIN2 interacts with TRF1 through a domain that lies between amino acids 196 and 275 (Fig. 1a). Analysis of additional TINF2 fragments in yeast confirmed the importance of this region for interaction with TRF1 (Fig. 1c). To map the region in TRF1 that interacts with TIN2, we tested TERF1 fragments for the ability to interact with clones 1 and 2 in yeast. TRF1 interacted with TIN2 via a domain within the TRF1 homodimerization region (Fig. 1d). There was no overlap between the region in TRF1 that binds tankyrase32 and that which binds TIN2 (Fig. 1d).
Interaction with TRF1 in vitro and in cells
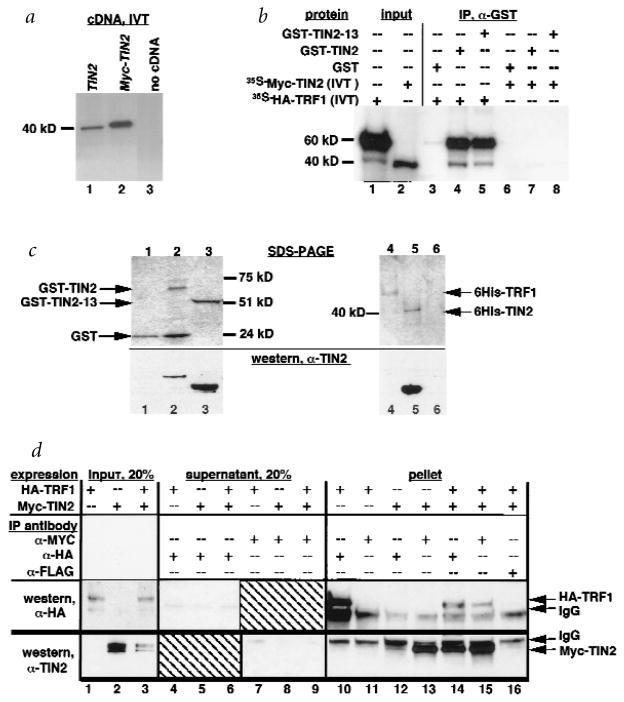
To verify the TIN2–TRF1 interaction, and facilitate further analyses, we prepared several reagents. First, we confirmed by in vitro translation that TINF2 cDNA directs the synthesis of a protein of approximately 40 kD (Fig. 2a, lane 1). Second, we tagged TIN2 with a 13–amino-acid Myc epitope (Myc–TIN2), and TRF1 with a 10–amino-acid haemagglutinin epitope (HA–TRF1). Myc–TINF2 cDNA (lacking the 5′ UTR) directed the synthesis of a protein that migrated more slowly than unmodified TIN2 (Fig. 2a, lane 2). The HA–TRF1 cDNA directed the synthesis of a major protein with an apparent molecular weight of 60 kD (ref. 26), and a minor species of approximately 40 kD that may be a degradation product (Fig. 2b, lane 1). We also produced recombinant glutathione-S-transferase (GST) fusion proteins in which GST was linked to the TIN2 N terminus, and 6-histidine (6His)-tagged TIN2 and TRF1. We confirmed protein purity and identity by SDS–PAGE and western blot using an affinity-purified rabbit polyclonal antibody raised against TIN2 amino acids 121–354 (anti-TIN2). Anti-TIN2 antibody detected single proteins of the expected sizes on western blots of GST–TIN2 (Fig. 2c, lane 2), a GST–TIN2 N-terminal deletion mutant (GST–TIN2-13; Fig. 2c, lane 3), 6His–TIN2 (Fig. 2c, lane 5) and human cell lysates (Fig. 3). Anti-TIN2 antibody did not cross-react with GST (Fig. 2c, lane 1), 6His–TRF1 (Fig. 2c, lane 4) or other cell proteins (Fig. 3), confirming its specificity.
Fig. 2.
TIN2 interacts with TRF1 in vitro and in cells. a, Translation products of TINF2 and Myc–TINF2 cDNAs. We transcribed and translated with 35S-methionine the TINF2 and Myc–TINF2 cDNAs in vitro, and analysed the translation products by SDS–PAGE. Lane 1, TINF2 cDNA; lane 2, Myc–TINF2 cDNA (lacking the 5′ UTR); lane 3, no cDNA control. b, TIN2 binds TRF1, but not TIN2, in vitro. We generated radiola-belled Myc–TIN2 and HA–TRF1 proteins by in vitro translation, and analysed 2 μl of the reactions by SDS–PAGE (lanes 1,2). In parallel, we incubated 5 μl of the reactions with 20 ng of GST (lanes 3,6), GST–TIN2 (lanes 4,7) or GST–TIN2-13 (lanes 5,8), immunoprecipitated the GST complexes, eluted proteins in the immune complexes into SDS–PAGE sample buffer and analysed 50% of the eluate by SDS–PAGE. c, Recombinant proteins and anti-TIN2 antibody. We expressed GST (lane 1), GST–TIN2 (lane 2), and GST–TIN2-13 (lane 3) in Escherichia coli, and 6His–TRF1 (lane 4), 6His–TIN2 (lane 5) and 6His–control (6His plus 36-bp vector sequence; lane 6) proteins using baculovirus and insect cells, purified them from cell lysates by glutathione (GST proteins) or nickel (6His proteins) chromatography, and analysed them by SDS–PAGE (top) and western blot (bottom) using affinity-purified anti-TIN2. d, TIN2 and TRF1 interact in cells. We prepared pre-cleared lysates from HT1080 cells that overexpress HA–TRF1, Myc–TIN2 or both (expression), subjected 20% of each lysate to SDS–PAGE (lanes 1-3), immunoprecipitated the remaining 80% with mouse monoclonal anti-HA, anti-Myc or anti-FLAG (control) antibodies (IP antibody), and collected immune complexes on protein A–Sepharose beads. We analysed 20% of each depleted supernatant by SDS–PAGE (lanes 4–9). We released proteins in the immune complexes into SDS sample buffer, subjected them to SDS–PAGE (lanes 10–16), and analysed them by western blot using affinity-purified rabbit polyclonal anti-HA (top; western, anti-HA) or anti-TIN2 (bottom; western, anti-TIN2) antibodies. Indicated are the positions of HA–TRF1, Myc–TIN2 and cross-reacting IgG heavy chains.
Fig. 3.
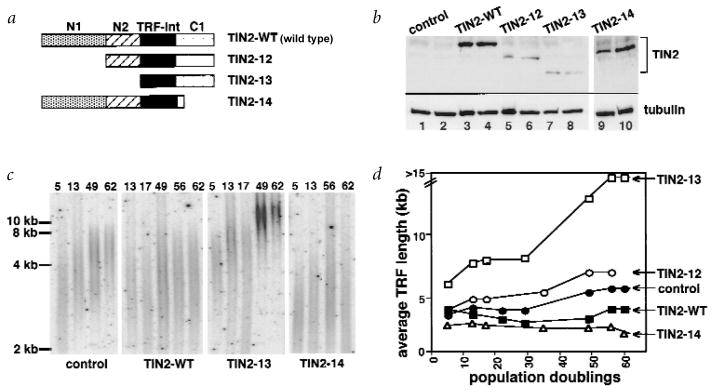
Truncated TIN2 proteins extend telomere length. a, TIN2 proteins used in this experiment. Shown are the N-terminal regions N1 (aa 1–120) and N2 (aa 120–196), TRF1-interaction domain (TRF-int) and C-terminal domain (C1). b, TIN2 expression. We infected HT1080 cells with control or the indicated Myc–TIN2-expressing retroviruses, and analysed cell lysates prepared 3–6 PD (lanes 3,5,7,9) and 60 PD (lanes 4,6,8,10) after selection by western blot, using anti-TIN2 or anti-tubulin (control) antibodies. c,d, Effects on TRF length. We infected HT1080 cells with control or the indicated Myc–TIN2-expressing retroviruses, selected for virus-expressing cells and permitted the cells to proliferate for the PD number indicated above each lane before DNA was isolated and analysed for TRF length. c, Hybridization from one experiment. d, Average intensity of the peak hybridization signal versus PD number from two or three independent experiments.
We incubated radiolabelled HA–TRF1 and Myc–TIN2, synthesized in vitro (Fig. 2b, lanes 1,2), with GST or GST–TIN2. We then immunoprecipitated GST-containing complexes, and identified associated radiolabelled proteins by SDS–PAGE. GST–TIN2 (Fig. 2b, lane 4), but not GST (lane 3), co-precipitated the major (60 kD) and minor (40 kD) HA–TRF1 species. Approximately 40% of the input HA–TRF1 precipitated with GST–TIN2, whereas less than 1% precipitated with GST. Neither GST–TIN2 nor GST (Fig. 2b, lanes 6,7) co-precipitated Myc–TIN2, suggesting that TIN2 does not interact with itself.
To test for interaction in cells, we expressed Myc–TIN2 and HA–TRF1 in human HT1080 fibrosarcoma cells using amphotropic retro-viruses. After selection for infected cells, we prepared cell lysates for immunoprecipitation with anti-Myc, anti-HA or anti-FLAG (control) antibodies. We identified proteins in the immune complexes and depleted supernatants by western blot using anti-HA or anti-TIN2 antibodies. In lysates from cells expressing both proteins (Fig. 2d, lanes 14–16), but not cells expressing either protein alone (Fig. 2d, lanes 10–13), anti-HA antibody precipitated Myc–TIN2 (Fig. 2d, lane 14) and anti-Myc antibody precipitated HA–TRF1 (Fig. 2d, lane 15). Approximately 80% of the HA–TRF1 and Myc–TIN2 co-precipitated (Fig. 2d, lanes 10–16 versus lanes 1–3 and 4–9).
We therefore conclude that TIN2 interacts with TRF1 in yeast, in vitro and in human cells. It seems that TIN2 does not form homotypic complexes. This was true in yeast (data not shown) and in vitro (Fig. 2b), although it is possible that the Gal4 and GST moieties interfered with TIN2 homodimerization.
TIN2 localizes to human telomeres
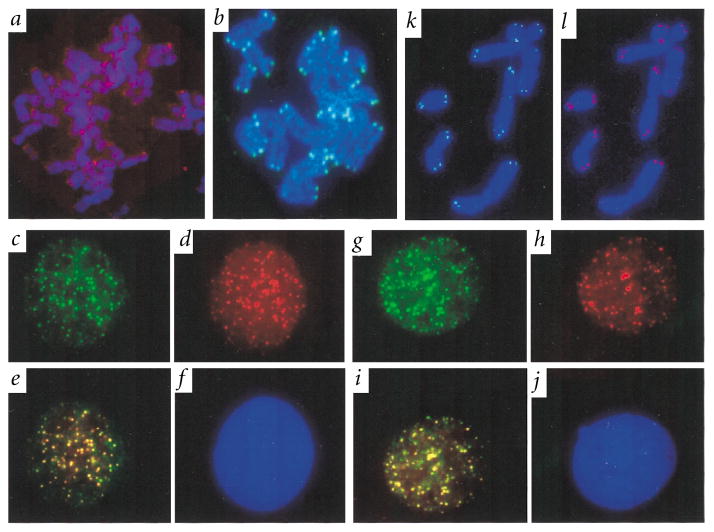
We determined the subcellular localization of endogenous TIN2 and retrovirally expressed Myc–TIN2 and HA–TRF1 by immunofluorescence. Endogenous TIN2, like endogenous TRF1 (ref. 26), is not highly expressed (Fig. 3), and was therefore difficult to visualize by immunostaining. Nonetheless, anti-TIN2 antibody detected endogenous TIN2 as weak but distinct immunostaining at the telomeres of human metaphase chromosomes (Fig. 4a). Anti-Myc antibody detected retrovirally expressed Myc–TIN2 as strong immunostaining at each discernible telomere (Fig. 4b); HA–TRF1 showed identical telomeric localization (Fig. 4k). Anti-Myc antibody also localized Myc–TIN2 to telomeric DNA, detected by a fluorescent DNA probe, in both interphase nuclei and metaphase chromosomes (data not shown), as reported for TRF1 (ref. 26).
Fig. 4.
TIN2 subcellular localization. We fixed HT1080 cells, uninfected or expressing HA–TRF1, Myc–TIN2 or Myc–TIN2-13 retroviruses, while proliferating, or after treatment with colcemid to obtain metaphase chromosomes, stained them with anti-HA, anti-Myc or anti-TIN2 antibodies and applied secondary antibodies (FITC (green fluorescence)- or Texas Red (red fluorescence)-conjugated anti-rabbit or anti-mouse IgG). We visualized DNA by DAPI staining (blue fluorescence), and photographed the cells and chromosomes using a digital camera, merging the images where indicated. a, Metaphase chromosomes from uninfected cell stained with anti-TIN2 (endogenous TIN2) antibody. b, Meta-phase chromosomes from Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. c, Interphase nucleus of an HA–TRF-1/Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. d, Inter-phase nucleus of the same HA–TRF-1/Myc–TIN2-expressing cell stained with anti-HA (retroviral TRF1) antibody. e, Co-localization of HA–TRF1 and Myc–TIN2 in nucleus shown in (c) and (d) (merged image). f, DAPI staining of nucleus shown in (c–e). g, Inter-phase nucleus of another HA–TRF-1/Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. h, Interphase nucleus of the same HA–TRF-1/Myc–TIN2-expressing cells stained with anti-HA (retroviral TRF1) antibody. i, Co-localization of HA–TRF1/Myc–TIN2 in nucleus shown in (g) and (h) (merged image). j, DAPI staining of nucleus shown in (g–i). k, Metaphase chromosomes from HA–TRF1/Myc–TIN2-13–expressing cell stained with anti-HA antibody (telomeric localization of TRF1 in the presence of TIN2-13). l, Metaphase chromosomes from HA–TRF1/Myc–TIN2-13–expressing cells stained with anti-Myc antibody (telomeric localization of TIN2-13).
In the interphase nuclei of HT1080 cells that expressed both Myc–TIN2 and HA–TRF1, anti-HA antibody localized HA–TRF1 to small, randomly distributed foci26 (Fig. 4d,h). Most cells contained more than 80 such foci, near the expected number of 92 telomeres. Control-infected cells showed no anti-HA staining (data not shown). Anti-Myc antibody localized Myc–TIN2 to similar nuclear foci (Fig. 4c,g), more than 70% of which co-localized with HA–TRF1 (Fig. 4e,i). Foci that were not positive for both proteins may indicate that they do not invariably co-localize during interphase. Because the antibodies did not always stain with equal intensity (Fig. 4c–j), the merged images likely underestimate the degree of TIN2 and TRF1 co-localization. Our results indicate that TIN2 co-localizes with human telomeres and TRF1.
TINF2 expression pattern
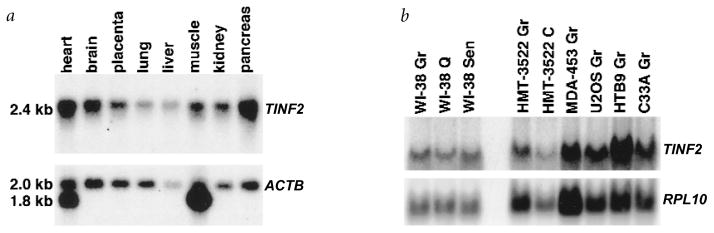
TINF2 cDNA detected a single 2.4-kb mRNA on northern blots of poly(A)+ RNA from human heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas (Fig. 5a), and total RNA from cultured fibroblasts (WI-38 (Fig. 5b); IMR-90 fetal lung, HCA2 neonatal foreskin (data not shown)). Proliferating, quiescent and senescent fibroblasts expressed similar levels of TINF2 mRNA (Fig. 5b), as did several immortal or tumorigenic cell lines (HMT-3522 non-tumorigenic breast epithelial cells, MDA-453 breast carcinoma, U2OS osteosarcoma, HTB9 bladder carcinoma, C33A (Fig. 5b) and HeLa (data not shown) cervical carcinoma). TINF2 expression was similar in non-tumorigenic breast cells, whether proliferating or confluent, and aggressive breast cancer cells (Fig. 5b). Thus, a wide variety of human tissues and cell types expressed TINF2, and expression did not vary with growth state, immortalization or transformation.
Fig. 5.
Expression pattern of TINF2 mRNA. a, Expression in human tissues. We analysed RNA from the indicated human tissues by northern blot to detect the TINF2 and ACTB (β-actin) mRNAs. Indicated are the 2.4-kb TINF2, 2.0-kb ACTB and 1.8-kb cross-hybridizing cardiac and skeletal muscle actin mRNAs. b, Expression in human cells. We analysed RNA from the indicated cell cultures by northern blot to detect the TINF2 and RPL10 (control37) mRNAs.
TIN2 mutants that extend telomeres in telomerase-positive cells
To characterize the function of TIN2, we created three Myc–TIN2 mutants, all of which retained the TRF1-binding domain (Fig. 3a): TIN2-12, an N-terminal deletion of amino acids 1–120; TIN2-13, an N-terminal deletion of amino acids 1–196; and TIN2-14, a carboxy-terminal deletion of amino acids 276–354.
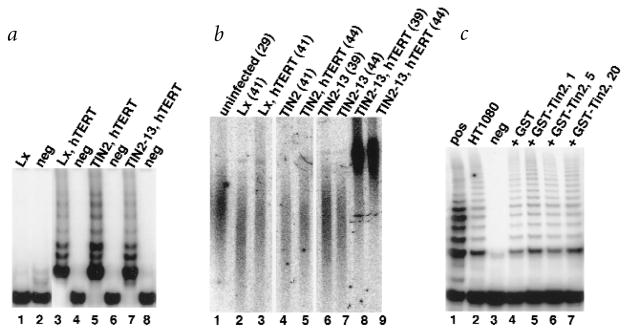
We expressed wild-type or mutant proteins in HT1080 cells using retroviral vectors. Western-blot analysis showed that the viruses expressed high levels of TIN2-WT and TIN2-14 (C-terminal deletion; more than tenfold the level of endogenous TIN2; Fig. 3b, lanes 1,2 versus lanes 3,4,9,10). The viruses expressed lower levels of TIN2-12 (small N-terminal deletion) and TIN2-13 (large N-terminal deletion; two- to fourfold the endogenous level; Fig. 3b, lanes 5–8). Expression from the retroviruses was stable over at least 60 population doublings (PD; Fig. 3b, lanes 3,5,7,9 versus lanes 4,6,8,10), and had no effect on cell growth or morphology (data not shown). There was, however, an effect on telomere length.
HT1080 cells express telomerase and maintain relatively short telomeres26,28 (3–5 kb average TRF over >60 PD; Fig. 3c,d). Cells expressing TIN2-WT maintained an average TRF of 3 kb (Fig. 3c,d), suggesting that TIN2 overexpression either has no effect or slightly shortens the telomeres, similar to the effect of overexpressing TRF1 (ref. 28). TIN2-14 (C-terminal deletion) slightly shortened the TRF to approximately 2 kb (Fig. 3c,d). By contrast, TIN2-12, which lacks 120 N-terminal amino acids, increased the TRF to 6–7 kb (Fig. 3c,d). TIN2-13, which lacks 196 N-terminal amino acids, increased the TRF to more than 15 kb, beyond the resolution of the gel (Fig. 3c,d). The first evidence of telomere elongation by TIN2-13 was apparent within 5 PD, and telomere elongation continued progressively over 40–50 PD (Fig. 3d).
Telomere elongation by TIN2-13 was dependent on telomerase. We expressed TIN2-WT or TIN2-13 in normal human fibroblasts (WI-38), which do not express telomerase (Fig. 6a, lane 1) and senesce after approximately 50 PD. Neither protein induced telomerase activity (data not shown), altered replicative lifespan (data not shown), nor telomere length (Fig. 6b, lane 2 versus 4,6,7). This was not the case when the cells were rendered telomerase-positive by a retrovirus carrying hTERT, the catalytic subunit of human telomerase34,35. hTERT induced telomerase activity (Fig. 6a, lane 3), retarded telomere shortening (Fig. 6b, compare lanes 1–3) and extended replicative lifespan (data not shown). Co-expression of hTERT and TIN2-WT had no effect or slightly shortened the TRF over 10–15 PD (Fig. 6b, lane 3 versus 5). Co-expression of hTERT and TIN2-13 increased the TRF to more than 10 kb (Fig. 6b, lane 3 versus 8,9) over the same interval. This increase persisted for at least 25 PD (data not shown).
Fig. 6.
Telomerase dependence. a, Telomerase activity. We infected WI-38 (lanes 1,2) or hTERT (hT)-expressing WI-38 (lanes 3–8) cells with control (Lx, lanes 1–4), TIN2 (lanes 5,6) or TIN2-13 (lanes 7,8) retroviruses, selected for virus-expressing cells and prepared cell lysates. We analysed cell lysate volumes equivalent to equal numbers of cells for telomerase activity by TRAP assay. neg, extracts heated to 85 °C before assay. b, TRF length. We infected WI-38 cells at PD 29 (lane 1) with pBabe control (lanes 2,4,6,7) or hTERT-expressing (lanes 3,5,8,9) virus. After five to six PD, we superinfected the cells with viruses expressing LXSN control (Lx; lanes 2,3), TIN2 (lanes 4,5) or TIN2-13 (lanes 6–9). We isolated DNA at the indicated PD levels, and analysed the DNA for TRF length. c, TIN2 does not inhibit telomerase activity in vitro. We prepared extracts from HT1080 cells (lane 2) and mixed equal aliquots with 20 ng GST (lane 4), or 1 (lane 5), 5 (lane 6) or 20 (lane 7) ng GST–TIN2. We incubated the extracts for 10 min at 4 °C before assaying for telomerase activity. pos, positive extract from the assay kit; neg, extract heated to 85 °C.
These results suggest that wild-type TIN2 negatively regulates telomere elongation by telomerase, and that TIN2-13 (and to a lesser extent TIN2-12) interferes with this function in a dominant-negative fashion. TIN2 very likely regulates telomere length by an indirect effect on telomerase. An HA-tagged hTERT protein36, when transiently expressed in Myc–TIN2-expressing cells, did not immunoprecipitate with Myc–TIN2 (data not shown). Neither GST–TIN2 (Fig. 6c, lanes 5–7), GST–TIN2-13 nor 6His–TIN2 (data not shown) affected telomerase (TRAP) activity when added to cell lysates.
Tin2-13 does not displace TRF1 from telomeres
The effect of TIN2-13 resembled that of DNA-binding-deficient TRF1, which elongated telomeres in a dominant-negative fashion28. TIN2-13, like wild-type TIN2, was capable of binding TRF1 (but not TIN2) in vitro (Fig. 2b, lanes 5,8), raising the possibility that TIN2-13 extends telomeres by displacing TRF1 from telomeric DNA.
To test this, we determined the localization of TRF1 in the presence of TIN2-13. We infected HT1080 cells with HA–TRF1 and Myc–TIN2-13–expressing retrovirus, and prepared meta-phase chromosomes after five days, before substantial elongation occurred. This avoided the complication that might arise if TRF1, after initial displacement, reassociated with the elongated telomeres. Under these conditions, TRF1 was detectable only at the telomeres (Fig. 4k), and the same was true for TIN2-13 (Fig. 4l). HA–TRF1 and Myc–TIN2-13 remained co-localized on metaphase chromosomes, and also during interphase, after more than 40 PD (data not shown). These findings suggest that TIN2-13 does not displace TRF1 from the telomere.
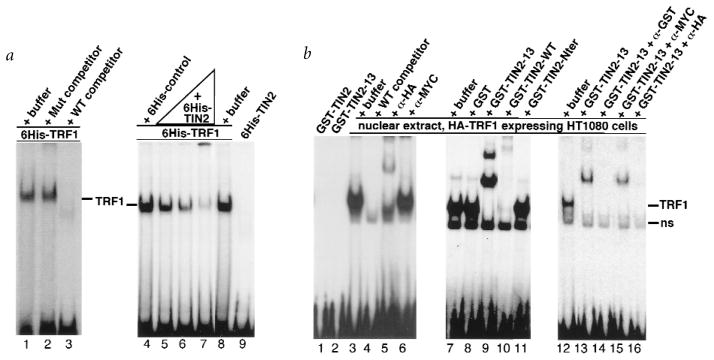
We obtained additional evidence that TIN2-13 does not displace TRF1 from electrophoretic mobility shift assays (EMSA). Neither 6His–TIN2 (Fig. 7a, lane 9), GST–TIN2 (Fig. 7b, lane 1) nor GST–TIN2-13 (Fig. 7b, lane 2) bound the double-stranded telomeric probe. We tested the proteins over a range of concentrations (10–150 ng/EMSA; data not shown), but consistently failed to detect DNA-binding activity. The proteins were active because they altered probe mobility in the presence of TRF1 (Fig. 7a, lanes 5–7; Fig. 7b, lanes 9,10,13). Control proteins did not alter the mobility of the TRF1–DNA complex (Fig. 7a, lane 4; Fig. 7b, lanes 8,11). Control proteins included the TIN2 N-terminal 196 amino acids (lacking the TRF1-binding domain; Fig. 7b, lane 11), con-firming that the N-terminal 196 amino acids are not involved in the interaction with TRF1. Although DNA-binding may require a modification that was not present in these proteins, or unidentified nuclear proteins may stimulate TIN2-DNA binding in vivo, the simplest interpretation is that TIN2 does not bind telomeric DNA directly. As expected26, TRF1 demonstrated DNA binding, whether provided as a 6His-tagged protein (Fig. 7a, lanes 1–8) or nuclear extract from HA–TRF1-expressing cells (Fig. 7b, lanes 3–16). TRF1-DNA binding was specific because the labelled band was abolished by excess of unlabelled wild-type, but not mutant, probe (Fig. 7a, lanes 1–3), and was disrupted by anti-HA antibody but not an irrelevant (anti-Myc) antibody (Fig. 7b, lanes 3–6).
Fig. 7.
TIN2-13 does not displace TRF1. a, 6His–TRF1 and 6His–TIN2 DNA-binding activity. We incubated recombinant proteins with a double-stranded TTAGGG6 probe, and analysed protein-DNA complexes by EMSA. Lane 1, 6-His–TRF1 (150 ng) analysed alone; lane 2, 6-His–TRF1 (150 ng) plus 100-fold excess unlabelled mutant [TTAGGC]7 probe (Mut competitor); lane 3, 6-His–TRF1 (150 ng) plus 100-fold excess unlabelled wild-type [TTAGGG]7 probe (WT competitor; lane 3); lane 4, 150 ng 6His–TRF1 plus an equal volume of 6His–control protein (6His plus 36 bp vector sequence, expressed and purified identically to 6His–TRF1 and 6His–TIN2); lanes 5–8, 150 ng 6His–TRF1 plus 10, 40, 150 or 0 ng 6His–TIN2; lane 9, 150 ng 6His–TIN2 alone. The TRF1-specific band is indicated. b, GST–TIN2 and GST–TIN2-13 binding activity, and interaction with TRF1. We incubated recombinant proteins, without or with nuclear extract (NE) from HA–TRF1-expressing HT1080 cells or antibodies, with a double-stranded TTAGGG13 probe, and analysed protein-DNA complexes by EMSA. Lane 1, 20 ng GST–TIN2 alone; lane 2, 20 ng GST–TIN2-13 alone; lane 3, NE alone; lane 4, NE plus 100-fold excess unlabelled [TTAGGG]7 (WT competitor); lane 5, NE plus 0.2 μg anti-HA antibody; lane 6, NE plus 0.2 μg anti-Myc (control) antibody; lanes 7–11, NE plus 20 ng GST, GST–TIN2-13, GST–TIN2-WT or TIN2 aa 1–196 (lacking the TRF1-binding domain) fused to GST (GST–TIN2-Nter); lane 12, NE; lane 13, NE plus 20 ng GST–TIN2-13; lane 14, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-GST; lane 15, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-Myc (control); lane 16, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-HA. The TRF1-specific band, and a non-specific band (ns) present in some of the gels, is indicated.
To determine how TIN2-13 interacts with TRF1 bound to DNA, we added GST–TIN2-13 or GST–TIN2 to nuclear extracts from HA–TRF1-expressing cells. GST–TIN2-13 shifted the TRF1 complex into a major and minor larger complex (Fig. 7b, lanes 9). Anti-GST and anti-HA (Fig. 7b, lanes 14,16) antibodies, but not an irrelevant antibody (anti-Myc; Fig. 7b, lane 15), disrupted the large complexes, indicating that they contained both HA–TRF1 and GST–TIN2-13. We obtained similar results when 6His-TRF1 was used in place of nuclear extracts (data not shown). The results support the conclusion that TIN2-13 binds TRF1 when it is bound to DNA, and that TIN2-13 does not displace TRF1 from telomeric DNA. Wild-type TIN2, whether provided as 6His–TIN2 (Fig. 7a, lanes 5–7) or GST–TIN2 (Fig. 7b, lane 10), disrupted the TRF1 complex. The interaction between TIN2 and TRF1 bound to telomeric DNA is currently under study, but preliminary results suggest that TIN2 promotes formation of a large, multimeric complex containing the probe, TRF1 and TIN2 (S.-h.K., unpublished data).
Discussion
TIN2 binds TRF1 and appears to function in the control of telomere length. TIN2 proteins with a truncated N terminus (TIN2-13 and, to a lesser extent, TIN2-12) extended telomeres, and did so at expression levels that only modestly exceeded that of endogenous wild-type protein. Thus, TIN2-13 (and TIN2-12) had properties of a dominant-negative mutant. Our findings suggest that wild-type TIN2 is a negative regulator of telomere length, and essential for proper regulation of telomere length in human cells.
The effect of TIN2-13 on telomere length was similar to that of a DNA-binding-deficient TRF1 mutant, which also elongated telomeres in a dominant-negative fashion28. Our results indicate that TRF1 binding alone is insufficient for proper regulation of telomere length in human cells. TIN2-13 did not displace TRF1 from telomeric DNA in vitro, and TRF1 remained at the telomeres in the presence of TIN2-13. Nonetheless, telomere length control was lost in the presence of TIN2-13. We propose that TRF1 recruits TIN2 to the telomere, where TIN2 acts to dampen telomere elongation by telomerase.
How might TIN2 regulate telomere length? TIN2 did not inhibit telomerase activity in vitro or interact directly with the catalytic component, yet telomere elongation by TIN2-13 was strictly telomerase-dependent. These findings suggest that TIN2 does not limit telomere length by suppressing the recombination pathway that is thought to elongate telomeres in telomerase-negative tumour cells18. TIN2, like TRF1, is widely and constitutively expressed, suggesting that these proteins act together to counterbalance telomere elongation by telomerase. TIN2 mutants that retain TRF1-binding but lack N-terminal sequences (120 or 196 aa) increased telomere length. This indicates two possible mechanisms by which TIN2 may act, both of which require the TRF1-binding domain (because TIN2 cannot bind the telomere directly) and the N terminus (which is missing in TIN2-13). First, TIN2 may recruit proteins to the telomere that inhibit telomerase. Thus, it is possible that telomerase inhibitors cannot be recruited to the telomere when TIN2 proteins with a deleted N terminus are bound, resulting in unregulated telomere extension. Alternatively, TIN2 may promote a compact telomeric structure that limits telomerase access to its substrate, the 3′ telomeric terminus. TRF1 has been shown to promote the parallel pairing of telomeric DNA tracts29. TIN2-binding may stimulate this activity of TRF1. In the absence of the TIN2 N terminus, the terminal telomeric junction may have a more open structure, thereby providing telomerase greater access to the 3′ overhang. Our preliminary data suggesting that TIN2 stimulates the telomeric pairing activity of TRF1 favour the latter possibility. Recently, mammalian telomeres, lacking all proteins except TRF2, were shown to form lasso-like t-loop structures in which the 3′ overhang was proposed to invade the DNA duplex at the junction between the lasso circle and tail31. The structure of the t loop junction is not known, but was proposed to limit telomerase access to the 3′ overhang. TIN2, in concert with TRF1, may stabilize the t loop. For example, the t-loop circle may form a coil in the presence of TIN2 and TRF1. Alternatively, TIN2 may promote anti-parallel telomeric DNA pairing, which would compress the circle.
We do not yet know precisely how TIN2 interacts with TRF1. Because TIN2 bound TRF1 near the TRF1 homodimerization domain, TIN2 binding may alter the ability of TRF1 to dimerize with other TRF1 molecules on the same DNA strand, or even on adjacent DNA strands, which would favour the pairing or clustering of telomeric DNA tracts. Alternatively, TIN2 may induce a conformational change in TRF1, which might increase its ability to form parallel telomeric tracts, allow it to form anti-parallel tracts, or alter other properties of TRF1 or TRF1-associated proteins such as tankyrase. Finally, the TIN2 N terminus may recruit other, as-yet-unidentified nuclear proteins to the telomere, which in turn may regulate the activity of telomerase or other proteins important for telomere structure or function.
TRF1 has structural similarity, albeit no sequence similarity, to Rap1p, the double-stranded, telomeric DNA-binding protein that regulates telomere length and gene silencing in S. cerevisiae4,20,21. Rap1p associates with a several yeast proteins, including Rif1p, Rif2p and the silencing proteins Sir3p and Sir4p (refs 22,23). TIN2 has no sequence or structural similarity to these Rap1p-binding proteins, but may be functionally similar to RIF proteins, which, together with Rap1p, control telomere length. Whether TIN2 has a role in controlling gene expression or telomeric silencing remains to be determined.
Methods
Interaction cloning in yeast
We cloned the TERF1 cDNA from a human fibroblast cDNA library37 using the published sequence26 and PCR, and subcloned it into the two-hybrid33 vector pGBT9 (Clontech). We generated a random/poly-dT-primed cDNA library in pGAD-10 (Clontech) using RNA from WI-38 cells (70% senescent, 30% proliferating) and kits (Stratagene and Clontech), and transformed DNA from 106 bacterial transformants into yeast strains HF7C (Clontech) and PJ69-4A (ref. 38) expressing pGBT9-TRF1. Four colonies grew on selective media containing 3-aminotriazole (10–30 mM) and expressed the lacZ (β-galactosidase) reporter. They contained 0.4-kb (clone 1) and 1.0-kb (clone 2) inserts that overlapped in sequence. We cloned full-length TINF2 using a 5′-RACE kit and human fibroblast Marathon library (Clontech), cloned the RACE products into pGEM-TA (Promega) and verified the sequence. We cloned the TERF1 cDNA into pGAD-10, and TINF2 and TERF1 cDNA fragments, generated by PCR, into pGAD-424 and pGBT9. We transformed the vectors into yeast (Y190, Clontech), and, after selection, measured β-galactosidase using a luminescent assay kit (Tropix), normalizing for cell number.
Vectors and recombinant proteins
We generated Myc–TINF2 (wild type or mutant) and HA–TRF1 cDNAs by PCR to add epitope tags, and cloned them into pBluescriptII-SK (Stratagene) or pLXSN (ref. 39). We cloned hTERT cDNA (ref. 35) into pBabe-puro (ref. 40), and TINF2 cDNAs into pGex-4X-1. We expressed GST proteins in Escherichia coli and purified them by glutathione-affinity chromatography using a kit (Pharmacia). We produced baculoviruses expressing 6His-TRF1 or 6His-TIN2, or 6His-vector sequence (6His-control) in Sf9 cells, purified the virally expressed proteins by Ni+2-chelation chromatography using a kit (Pharmigen), and assessed protein purity by SDS–PAGE.
In vitro transcription, translation and immunoprecipitation
We transcribed Myc–TINF2 (in pGEM-TA) and HA–TRF1 (in BlueScript II-SK) cDNAs in vitro, translated the transcripts using a rabbit reticulocyte lysate kit (Promega) and 35S-methionine, separated the translation products by 4–15% PAGE, and visualized them by autoradiography. We incubated GST or GST fusion proteins (60 ng) with translation reaction (5 μl) for 2 h at 4 °C, and added anti-GST antibody (1 μg, Santa Cruz Biotechnology) for 1 h. Alternatively, we incubated translation reactions (5 μl), alone or mixed, with anti-Myc (1 μg; Invitrogen) or anti-HA (Boehringer) antibodies for 1 h at 4 °C. We collected the immune complexes on protein A-Sepharose beads and analysed them by SDS–PAGE and autoradiography.
Cell culture
We cultured WI-38 cells and made them quiescent or senescent as described41,42. We cultured HT1080, U2OS, HTB9, C33A (from the American Type Culture Collection) and MDA-452 (from R. Lupu) as described for WI-38 cells, and HMT-3522 (from M. Bissell) in serum-free medium43.
Northern- and western-blot analyses
We performed northern-blot analysis as described41, hybridizing membranes with poly(A)+ RNA (2 μg) from human tissues (Clontech), or total RNA (30 μg) from cultured cells, to a TINF2 (clone 2) probe and rehybridizing with β-actin or RPL10 probes. We performed western analysis as described37, using enhanced chemiluminescence (Amersham) and autoradiography, and rabbit polyclonal anti-TIN2 antibody to detect TIN2 and mouse monoclonal anti-tubulin antibody (Calbiochem) to detect tubulin (control).
Retroviruses
We produced amphotropic retroviruses by transient transfection using cells and vectors44 (CellGenesys). We collected culture medium containing virus, froze and thawed it, and assayed it for reverse transcriptase (RT). We infected proliferating cells with equivalent RT units, and selected pLXSN-infected cells for 5 d in 400 μg/ml G-418 (maintained in 100 μg/ml G-418), and pBabe-infected cells for 7 d in 0.75 μg/ml puromycin (maintained without puromycin).
Antibody production and immunolocalization
We used TIN2 aa 121–354 fused to GST (GST–TIN2121–354) to produce polyclonal antiserum in rabbits using a commercial service (Babco). We affinity-purified the antibodies using membrane-immobilized GST–TIN2121–354 as described45, and tested them on western blots of GST and GST–TIN2, or control and Myc–TIN2-expressing cell lysates. We immunostained cells as described46. Briefly, we stained cells grown on cover slips with mouse monoclonal anti-HA (Boehringer), affinity-purified rabbit polyclonal anti-HA (Santa Cruz Biotechnology), mouse monoclonal anti-Myc (Boehringer) or affinity-purified rabbit polyclonal anti-TIN2 antibodies for 1 h at RT, washed and applied secondary antibodies of goat anti-mouse or goat anti-rabbit IgG conjugated to FITC or Texas Red (Vector Laboratories). To prepare metaphase chromosomes, we cultured cells in colcemid (0.1 μg/ml) for 3 h, lysed them in hypotonic buffer, and deposited metaphase chromosomes by centrifugation onto poly-lysine–coated slides. We mounted cells and chromosomes in medium containing DAPI (Vector Laboratories).
Telomere length and telomerase measurements
We isolated DNA, digested it with HinfI and RsaI, analysed it by Southern blot using a (TTAGGG)3 probe as described9, and quantified the hybridization signals using a phosphorimager and ImageQuant. We determined telomerase activity by the telomere repeat amplification protocol (TRAP) using a kit (Oncor/Intergen).
Electrophoretic mobility shift assays (EMSA)
We prepared nuclear extracts as described41, and dialysed them against HEPES (20 mM, pH 7.9), KCl (100 mM), DTT (0.5 mM) and PMSF (0.5 mM). We excised [TTAGGG]6 and [TTAGGG]13 from BlueScriptII-SK, labelled the fragments using Klenow or PCR and gel-purified them. We performed EMSA as described47 in 20 μl containing GST (0–20 μg) or GST-fusion proteins or nuclear extract protein (6–8 μg), using a 30 min incubation at RT and 5% PAGE run with 1×Tris-borate-EDTA buffer.
Acknowledgments
We thank P. James for yeast PJ69-4A and sharing strains before publication; R. Weinberg for hTERT cDNA; and M. Bissell and R. Lupu for HMT-3522 and MDA-453 cells. Supported by research (AG09909) and training (AG00266) grants from the National Institute on Aging, a fellowship (2F1B-0026) from the University of California Breast Cancer Research Program and research grant from the Ellison Medical Foundation, under contract DE-AC03-76SF00098 from the U.S. Department of Energy.
Footnotes
GenBank accession number:TINF2, AF195512.
References
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1995;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 3.Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, function in silencing, cell cycle progression and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 4.Marchand S, Buck SW, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by RAP1 protein. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. The biology of replicative senescence. Eur J Cancer. 1997;33:703–709. doi: 10.1016/S0959-8049(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 6.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 7.Lingner J, Cech TR. Telomerase and chromosome end maintenance. Curr Opin Genet Dev. 1998;8:226–232. doi: 10.1016/s0959-437x(98)80145-7. [DOI] [PubMed] [Google Scholar]
- 8.Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 9.Harley CB, Futcher AB, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 10.Shay JW, Wright WE. Defining the molecular mechanisms of human cell immortalization. Biochim Biophys Acta. 1991;1071:1–7. doi: 10.1016/0304-419x(91)90003-4. [DOI] [PubMed] [Google Scholar]
- 11.Campisi J, Dimri GP, Hara E. Control of replicative senescence. In: Schneider E, Rowe J, editors. Handbook of the Biology of Aging. Academic; New York: 1996. pp. 121–149. [Google Scholar]
- 12.Bodnar AG, et al. Extension of life span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 13.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 14.Kiyono T, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 15.Bodnar AG, Kim NW, Effros RB, Chiu CP. Mechanism of telomerase induction during T cell activation. Exp Cell Res. 1996;228:58–64. doi: 10.1006/excr.1996.0299. [DOI] [PubMed] [Google Scholar]
- 16.Buchkovich KJ, Greider CW. Telomerase regulation during entry into the cell cycle in normal human T cells. Mol Biol Cell. 1996;7:1443–1454. doi: 10.1091/mbc.7.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 18.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 19.Shore D. Telomerase and telomere-binding proteins: controlling the end game. Trends Biochem Sci. 1997;22:233–235. doi: 10.1016/s0968-0004(97)01082-7. [DOI] [PubMed] [Google Scholar]
- 20.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 21.Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wotton D, Shore D. A novel RAP1p-interacting factor, RIF2p, cooperates with RIF1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 23.Cockell M, et al. The carboxy termini of SIR4 and RAP1 affect SIR3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandin N, Reed SI, Charbonneau M. STN1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with CDC13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 25.Nugent CI, Hughes TR, Lue NF, Lundblad V. CDC13p: a single-strand telomeric DNA binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 26.Chong L, et al. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 27.Shen M, Haggblom C, Vogt M, Hunter T, Lu KP. Characterization and cell cycle regulation of the related telomeric proteins PIN2 and TRF1 suggest a role in mitosis. Proc Natl Acad Sci USA. 1997;94:13618–13623. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 29.Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- 30.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end to end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 31.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 32.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly (ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 33.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 35.Counter CM, et al. Dissociation among in vitro telomerase activity telomere maintenance and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is upregulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 37.Dimri GP, Testori A, Acosta M, Campisi J. Replicative senescence, aging and growth regulatory transcription factors. Biol Signals. 1996;5:154–162. doi: 10.1159/000109185. [DOI] [PubMed] [Google Scholar]
- 38.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–988. [PMC free article] [PubMed] [Google Scholar]
- 40.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimri GP, Hara E, Campisi J. Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J Biol Chem. 1994;269:16180–16186. [PubMed] [Google Scholar]
- 42.Dimri GP, et al. A novel biomarker identifies senescent human cells in culture and aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briand P, Petersen OW, van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in a chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 44.Finer MH, Dull TJ, Qin L, Farson D, Roberts MR. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 45.Sambrook J, Fritch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor Press; New York: 1989. [Google Scholar]
- 46.Compton DA, Yen TJ, Cleveland DW. Identification of a novel centromere/kinetochore-associated protein using monoclonal antibodies generated against human mitotic chromosome scaffolds. J Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;12:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]