Abstract
Purpose:
CDKN2A is the main high-risk melanoma-susceptibility gene, but it has been poorly assessed in Latin America. We sought to analyze CDKN2A and MC1R in patients from Latin America with familial and sporadic multiple primary melanoma (SMP) and compare the data with those for patients from Spain to establish bases for melanoma genetic counseling in Latin America.
Genet Med 18 7, 727–736.
Methods:
CDKN2A and MC1R were sequenced in 186 Latin American patients from Argentina, Brazil, Chile, Mexico, and Uruguay, and in 904 Spanish patients. Clinical and phenotypic data were obtained.
Genet Med 18 7, 727–736.
Results:
Overall, 24 and 14% of melanoma-prone families in Latin America and Spain, respectively, had mutations in CDKN2A. Latin American families had CDKN2A mutations more frequently (P = 0.014) than Spanish ones. Of patients with SMP, 10% of those from Latin America and 8.5% of those from Spain had mutations in CDKN2A (P = 0.623). The most recurrent CDKN2A mutations were c.-34G>T and p.G101W. Latin American patients had fairer hair (P = 0.016) and skin (P < 0.001) and a higher prevalence of MC1R variants (P = 0.003) compared with Spanish patients.
Genet Med 18 7, 727–736.
Conclusion:
The inclusion criteria for genetic counseling of melanoma in Latin America may be the same criteria used in Spain, as suggested in areas with low to medium incidence, SMP with at least two melanomas, or families with at least two cases among first- or second-degree relatives.
Genet Med 18 7, 727–736.
Keywords: CDKN2A, familial, Latin America, melanoma, MC1R
Introduction
Melanoma is the most aggressive of common skin cancers because of its tendency to metastasize. Its incidence is rapidly increasing, especially among Caucasian populations. Melanoma is the second most diagnosed cancer among patients younger than 30 years of age,1 and the 3-year survival rate for patients with metastases is around 15%.2 Identification of individuals at high risk of developing melanoma is necessary since an early diagnosis improves the disease prognosis.3
Melanoma is caused by the interaction of environmental, phenotypic, and genetic factors. The main environmental risk factor for melanoma is sun exposure.4 Individuals with fair skin, red hair, and/or a high nevi count have an increased risk of developing melanoma.5 To date, CDKN2A, which encodes the tumor suppressor proteins p16INK4A and p14ARF, is the major high-risk gene involved in melanoma susceptibility.6 CDKN2A has been widely studied in melanoma patients from the United States, Europe, and Australia.6 The frequency of germline mutations in CDKN2A varies across populations (5–72%) and depends on the selection criteria used.6,7 Haplotype analysis indicates a founder effect for most of the recurrent mutations detected.8 Identification of the prevalence of CDKN2A mutations in patients at high risk for melanoma and the correlation of these mutations with clinical data has been crucial for establishing genetic counseling for melanoma. Melanoma risk may also be modulated by common genetic variants acting as low- to medium-penetrance variants.9 MC1R plays a key role in pigmentation and is responsible for phenotypic characteristics such as hair and skin color and the capacity of response to ultraviolet radiation.10 Several MC1R variants are associated with a moderately increased melanoma risk and also modulate the effect of CDKN2A mutations in carriers.11
Genetic counseling and specific dermatological follow-up may be offered to patients at high risk for melanoma.12 In countries with a low to medium incidence of melanoma, genetic counseling is offered to patients with two primary melanomas and/or to families with two melanoma cases and/or one pancreatic adenocarcinoma and one melanoma in first- or second-degree relatives (the “rule of two”). In countries with a moderate to high incidence of melanoma, however, genetic counseling is offered to patients with three primary melanomas and to families with three cases of melanoma or pancreatic cancer in first- or second-degree relatives (the “rule of three”).13 It has been demonstrated that melanoma genetic counseling has a positive impact on the improvement of total body skin examination and self-examination of the skin in unaffected individuals carrying germline mutations after test reporting, whereas affected carriers maintain high levels of screening adherence.14 Furthermore, after melanoma genetic counseling, unaffected members of high-risk melanoma families report improvements in daily routine sun protection, showing that genetic counseling may motivate sustained improvements in prevention behaviors.15 Thus it is very important for both melanoma patients and unaffected individuals from the family to be included in genetic counseling programs.
Few studies have assessed the prevalence of CDKN2A mutations or MC1R variants and phenotypic characteristics in patients at high risk for melanoma from Latin American countries. CDKN2A mutations have been identified in 13.6% of melanoma-prone families from São Paulo, Brazil,16 whereas one study reported no mutations in Porto Alegre,17 and in a different cohort the mutation frequency was 7%.18 In melanoma-prone families from Uruguay, 5/6 families had CDKN2A mutations.19 Phenotypic and genetic characterization of individuals at high risk for melanoma from Latin America may improve their management and implement genetic counseling in these countries. We present the molecular characterization of CDKN2A and MC1R genes in the largest set of patients at high risk for melanoma from distinct Latin American countries (Argentina, Brazil, Chile, Mexico, and Uruguay), and we compare the data with two sets of Spanish patients at high risk for melanoma to establish bases for genetic counseling in Latin America.
Materials and Methods
The multicenter cross-sectional study included 1,090 patients at high risk for melanoma: 758 patients with familial melanoma (FM) and 332 patients with SMP from Latin American countries and Spain. Because Latin America is a region with a low incidence of melanoma (GLOBOCAN 2012, World Health Organization; http://globocan.iarc.fr), the inclusion criteria followed the rule of two.
Overall, 186 Latin American melanoma patients were recruited from Argentina (n = 10), Chile (n = 28), Mexico (n = 6), Uruguay (n = 25), and Brazil (n = 117), which included two sets of patients: Porto Alegre (Southern Brazil) (n = 58) and São Paulo (southeast region) (n = 59). The contribution of each country to the study resulted in a broad representation of a number of Latin American countries. A set of 904 Spanish patients with melanoma from Barcelona (n = 706) and Valencia (n = 198) also were included using the same selection criteria.
The number of primary melanomas, age at diagnosis, number of melanoma cases in the family, ancestral origin, and phenotypic data (hair and eye color, skin phototype, and nevi count) were recorded by dermatologists for most of the patients. Although the number of missing values was higher in the set of Spanish patients than in the Latin American patients, this did not introduce a bias, and the information recruited was informative for the whole cohort: Spanish patients were recruited consecutively, and missing data were distributed randomly; two different cohorts from Spain where used to minimize the bias due to the data collection procedure; and the variable with the greatest amount of missing data had information from at least 600 Spanish patients. Partial genetic information of the patients with melanoma from Spain and Brazil, and a subset of pedigrees from Uruguay, has been previously reported.16,17,18,19,20,21
The study was approved by the ethical committee of the Hospital Clinic of Barcelona. The patients gave their written, informed consent.
CDKN2A and MC1R molecular screening
Molecular characterization of CDKN2A was performed in all patients. CDKN2A was sequenced in all patients, as previously described.16,18,20,21 MC1R was sequenced as described elsewere.22,23 The MC1R genotype was available from all patients from Argentina and Chile, 57% (33/58) patients from Porto Alegre, Brazil, 92% (54/59) patients from São Paulo, Brazil, 96% (24/25) patients from Uruguay, 59% (419/706) patients from Barcelona, Spain, and 94% (186/198) patients from Valencia, Spain. MC1R genotype data were not available for patients from Mexico.
Statistical analyses
For the statistical analyses, the most common MC1R variants were classified as r variants (not associated with red hair color: p.V60L, p.V92M, p.R163Q) or R variants (associated with red hair color: p.D84E, p.R142H, p.R151C, p.I155T, p.R160W, p.D294H).10
SPSS software version 17.0 (IBM, Chicago, IL) was used. Two-sided Pearson χ2 or Fisher exact tests were used for categorical variables, as applicable. Student's t-test was used for quantitative variables. Adjusted P values were calculated using the Bonferroni correction. The test was considered significant if the P value or adjusted P value (as applicable) was <0.05.
Results
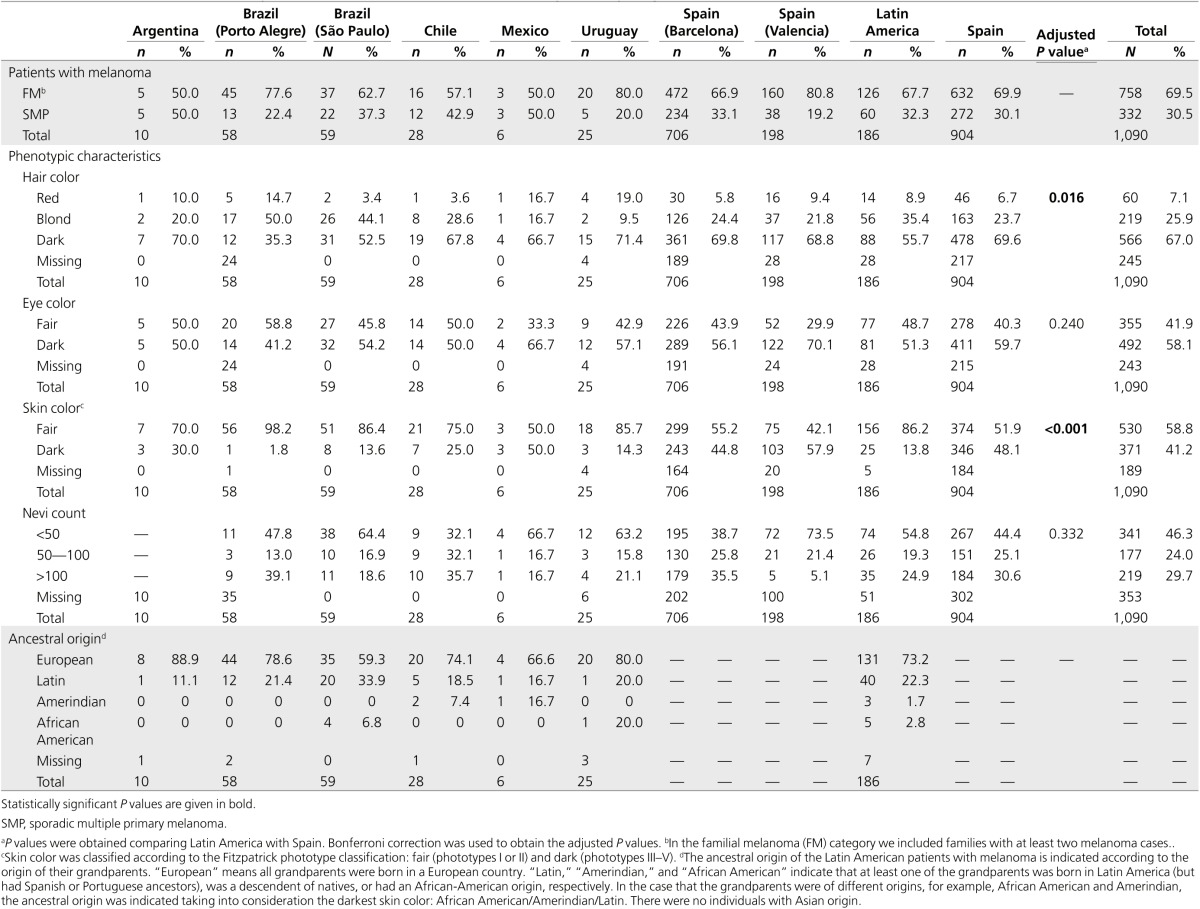
The study included a set of 1,090 patients with melanoma from distinct Latin American countries and Spain. Latin America and Spain had similar frequencies of FM cases (67.7 and 69.9%, respectively) and SMP (32.3 and 30.1%, respectively; P = 0.600), and there were no gender differences (40.3% male and 58.7% female vs. 41.5% male and 58.5% female, respectively; P = 0.806). Since Latin America is a mixed population from European, Native, African and Asian origin as a result of the colonization process and migratory effects,24 we collected information regarding the patients' ancestral origin. The four grandparents of more than 70% of Latin American patients were of European origin. Latin American and Spanish patients differed in pigmentation traits. Latin American patients had fairer hair color (adjusted P = 0.016) and skin phototype (adjusted P < 0.001) than Spanish patients. No differences were observed for nevi count or eye color (Table 1).
Table 1. Characteristics and phenotypic data of patients with melanoma, by country (region).
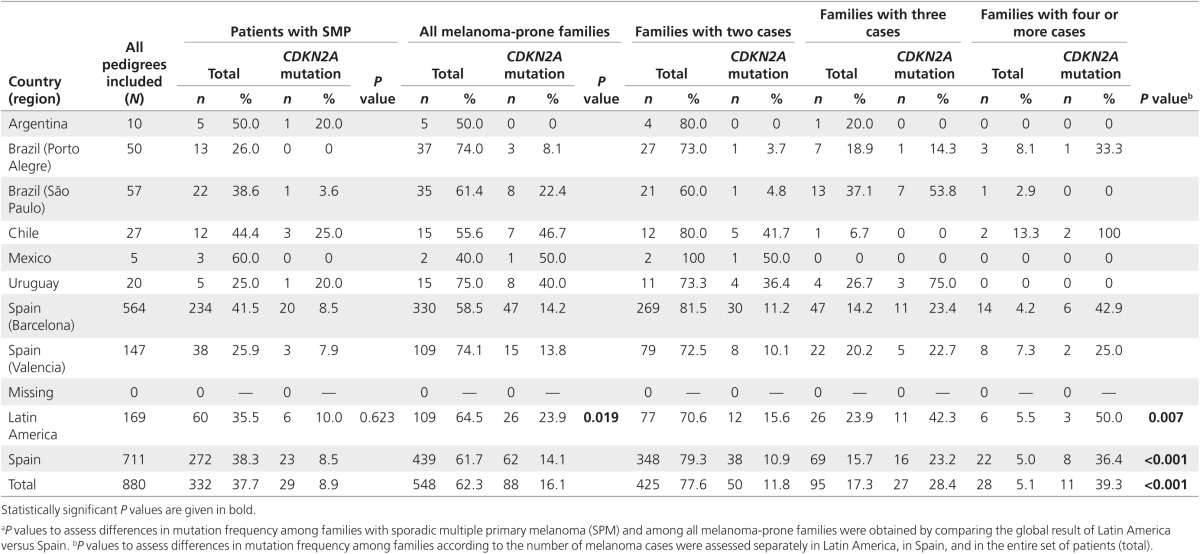
Considering all patients, CDKN2A mutation prevalence was 19% in Latin America and 12% in Spain. CDKN2A mutation frequency in SMP was similar in Latin America (10%) and Spain (8.5%) (P = 0.623). However, the prevalence of CDKN2A mutations in Latin American melanoma-prone families was higher than in Spain (24 and 14%, respectively; P = 0.019). The frequency of mutations varied among countries. Whereas southern Brazil had a low mutation prevalence, Chile and Uruguay showed a high prevalence of mutations in both SMP and FM (Table 2).
Table 2. CDKN2A mutation distribution between families according to the number of melanoma cases by country (region).
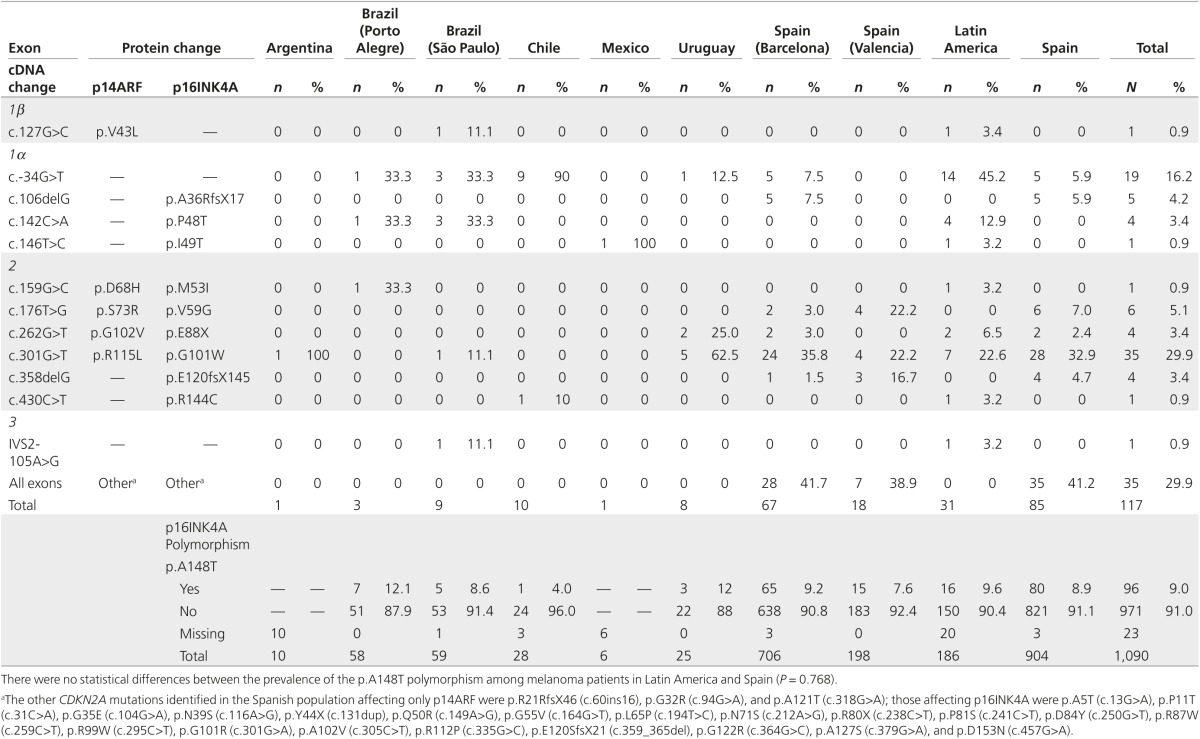
The CDKN2A mutations differed in each country (Table 3). Overall, 74% (23/31) of Latin American CDKN2A mutation carriers had a mutation also found in Spanish patients with melanoma. The most prevalent mutations in Latin America (c.-34G>T and p.G101W (c.301G>T)) were among the most recurrent mutations in Spain, which are p.G101W (33%), p.V59G (c.176T>G) (7%), c.-34G>T (6%), p.A36RfsX17 (c.106delG) (6%), and p.E120fsX145 (c.358delG) (5%) (Table 3). Mutation c.-34G>T was present in 90% of families from Chile, and families from São Paulo (Brazil) and Uruguay with CDKN2A mutations. Mutation p.G101W was present in families from Argentina, São Paulo (Brazil), and Uruguay. The other mutations detected in Latin America were restricted to a few pedigrees.
Table 3. CDKN2A genetic results.
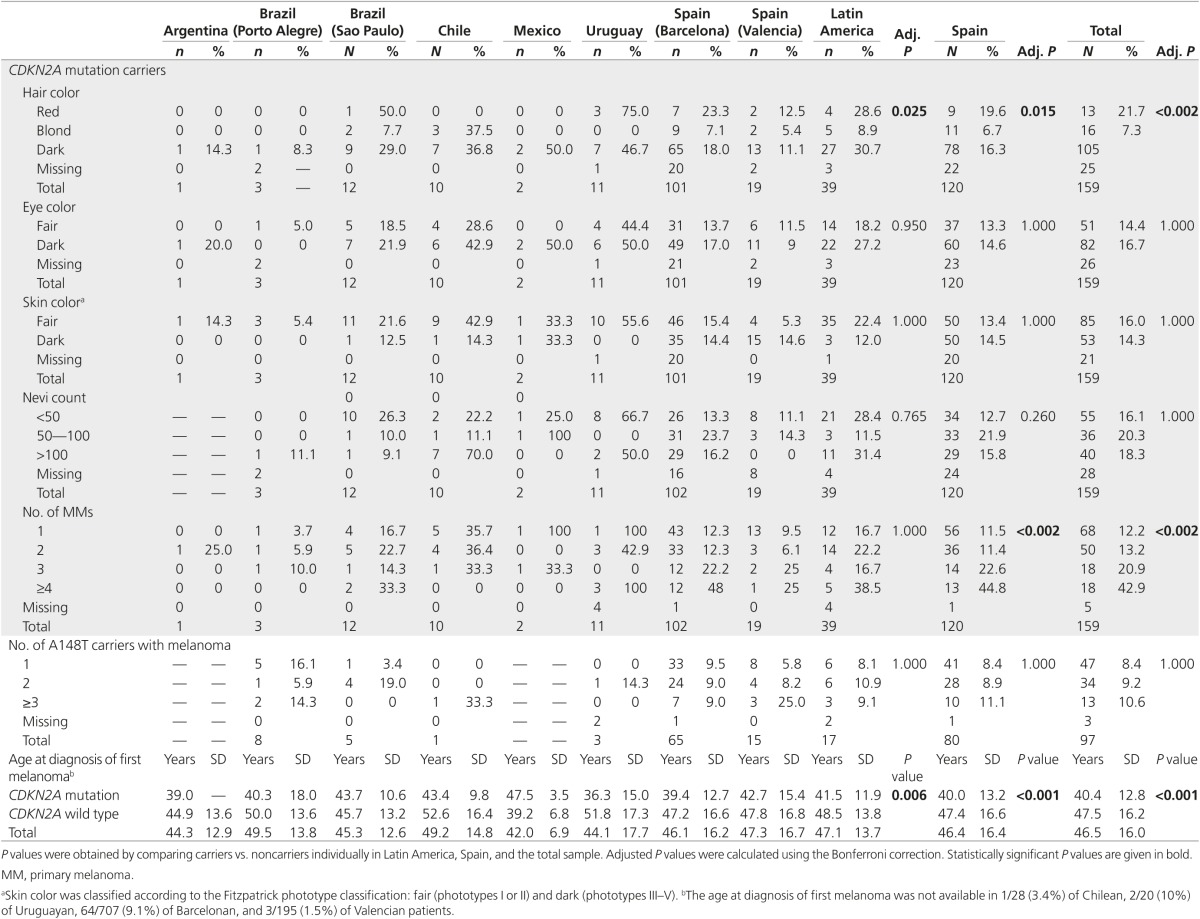
CDKN2A mutations have been previously associated with a lower age at diagnosis, number of primary melanomas, and the number of cases in the family.6 The whole set of patients also showed these associations (Table 4). Latin American patients with melanoma carrying a CDKN2A mutation had an increased number of cases in the family and a lower age at diagnosis, but the number of personal primary melanomas did not reach significance.
Table 4. Clinical and phenotypic characteristics of melanoma patients according to the presence of a CDKN2A mutation, by country.
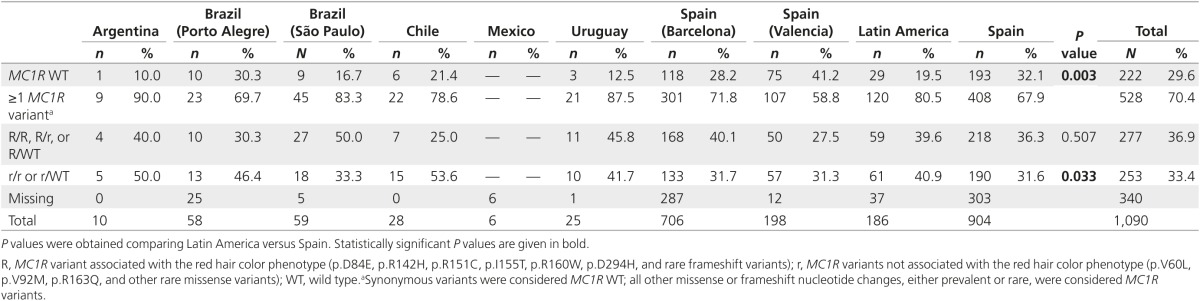
We sequenced MC1R to assess the distribution of MC1R variants across countries (Table 5). We observed differences in the number and type of variants between Latin America and Spain. We detected MC1R variants in 80.5% of Latin American and 67.9% of Spanish patients (P = 0.003), with a similar R variant frequency (39.6 vs. 36.3%, respectively; P = 0.514) but a higher r variant prevalence in Latin America (40.9 vs. 31.6%, respectively; P = 0.033). We analyzed the frequencies of the most common R and r variants, comparing Latin America and Spain (Supplementary Table S1 online). When adjusting using the Bonferroni correction, we found a significantly increased presence of p.R160W (17.4 vs. 7.5%; adjusted P < 0.005) and p.R163Q (14.1 vs. 5.2%; adjusted P < 0.005) in Latin America, but we should take into consideration that all patients carrying the p.R163Q variant in this study were from only three study sites: Brazil (São Paulo), Chile, or Uruguay. The p.D294H variant was more frequent in Spain (5.4 vs. 13.3%; adjusted P = 0.045). The presence of MC1R variants and R variants correlated with phenotype (Supplementary Tables S2 and S3 online).
Table 5. MC1R variant distribution.
Discussion
Latin America has a low incidence of melanoma (GLOBOCAN 2012). The characterization of melanoma genes has allowed other areas with low to medium incidence of melanoma, such as Spain, to recommend genetic counseling for patients with melanoma.12,25 To date, only a few specialized centers in Latin America offer melanoma genetic counseling, and there is little knowledge of the implication of high-risk genes in melanoma susceptibility. This study presents the clinical and molecular characterization of CDKN2A and MC1R in the largest set of Latin American patients at high risk for melanoma.
CDKN2A mutation frequency in melanoma-prone families was higher in Latin America than Spain, using the same selection criteria. By contrast, both areas had similar SMP CDKN2A mutation prevalence, consistent with that reported in other studies (8.2–9%).25,26 The age at diagnosis and number of primary melanomas were associated with the presence of mutations in CDKN2A, as previously reported.6 Otherwise, we did not find associations between CDKN2A mutation and nevi count, suggesting that other genes could play a role in nevogenesis.27,28 Most CDKN2A mutations identified had been previously detected in European or North American patients with melanoma. The most prevalent mutation in Latin America was c.-34G>T. This mutation occurs at a high frequency among unrelated families from Chile, suggesting a possible founder effect. In one family from Chile we detected p.R144C (c.430C>T), previously detected at the germline level in a patient with pancreatic cancer.29 Mutation p.G101W is also frequent in Latin America, as in Mediterranean countries (Italy, France, and Spain)7 where haplotype analysis showed a founder effect.30 We identified four other mutations in Brazil: p.P48T (c.142C>A), previously reported in an Italian population with FM,31 was found in four families, one of them of Italian ancestry, suggesting a possible founder effect32; IVS2-105A>G and p.M53I (c.159G>C), previously reported in melanoma-prone families from the United Kingdom, Australia, and the United States7; and mutation p.V43L (c.127G>C), affecting p14ARF, which has not previously been reported. In Uruguay we detected p.E88X (c.262G>T) in two families, which also was detected in two Spanish pedigrees. In Mexico we identified a mutation in the two probands of one family—p.I49T (c.146T>C)—which was previously reported in a case of FM by Hussussian et al.33 and did not segregate with melanoma in that case. However, functional analysis showed impairment for this variant.34
We detected differences in MC1R variant distribution in our set of patients. Latin American patients with melanoma carry more MC1R variants. These genetic results correlate with the phenotypic data, where Latin American patients with melanoma have fairer skin and hair color. The prevalence of MC1R variants varies between populations.35 In this study, specific variant frequencies differed between Latin American and Spanish patients with melanoma. Latin American patients with melanoma had an increased presence of p.R160W and p.R163Q. However, controls would be needed to assess the melanoma risk associated with carrying these variants in Latin America. p.R160W is associated with an increased risk for melanoma and red hair color.10 By contrast, p.R163Q, which is not associated with pigmentation or tanning response, favors the development of chronic sun exposure melanomas in the Mediterranean population22 and increases the risk for melanoma in areas with high ultraviolet radiation.36 These reports suggest that a possible interaction between p.R163Q and a high ultraviolet radiation dose could favor melanoma development. Most Latin American countries receive a huge amount of ultraviolet radiation compared with northern latitudes; this could explain the increased frequency of SMP and FM with the p.R163Q variant in Latin America, although its frequency in a control Latin American population is unknown.
To date, genetic testing in patients at high risk for melanoma is restricted to CDKN2A and CDK4. More studies of patients wild type for these genes should be conducted to assess the role of other melanoma-susceptibility genes such as MITF, BAP1, TERT, POT1, ACD, and TERF2IF8 for their possible incorporation in melanoma genetic counseling. In this study we demonstrated that CDKN2A germline mutation frequency in melanoma-prone families with at least two melanoma cases is greater in Latin America than Spain (23.9 vs. 14.1%, respectively). Inclusion criteria for genetic testing of melanoma in Spain follow the rule of two.12 Based on the results of this study, the inclusion criteria for genetic counseling for patients with melanoma in Latin America should also follow this rule because it allows the detection of CDKN2A mutations in a significant number of patients, except for southern Brazil, where the rule of three should be used. Genetic testing allows us to identify mutation carriers in families with a high risk of developing the disease. Carriers can be included in specific follow-up programs that allow the detection of melanomas at early stages, which improves the disease prognosis.3,37,38 Digital follow-up with specific dermatologic techniques, including total-body photography and digital dermoscopy, allow early detection of melanomas with a low rate of excision.38 Early melanomas in patients carrying MC1R variants may be difficult to diagnose definitively using dermoscopy, and an integrated approach including clinical history and dermoscopic data should be used when evaluating them.39 Thus, MC1R sequencing could also help to choose the best screening methods. The experience of genetic counseling in Spain over 10 years shows that melanomas can be diagnosed at any time, so the follow-up of individuals at high risk for melanoma should be maintained over time.12
In conclusion, Latin American patients with melanoma and at high risk for melanoma had fair skin and European origin. The mutations found also had been detected in Spanish, European, or North American populations, suggesting that they could have a single origin and that there could be a founder effect. Finally, inclusion criteria for genetic counseling in Latin American patients with melanoma should follow the rule of two: two primary melanomas in an individual or families with at least one invasive melanoma and one or more other diagnoses of melanoma or pancreatic cancer in first- or second-degree relatives.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
The main funding for the study was provided by GenoMEL (contract LSHC-CT-2006–018702) and by the National Cancer Institute of the US National Institutes of Health (CA83115). The research at the Melanoma Unit in Barcelona is partially funded by grants 03/0019, 05/0302, 06/0265, 09/1393, and 12/00840 from Fondo de Investigaciones Sanitarias, Spain; by the CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain; by AGAUR 2014_SGR_603 of the Catalan Government, Spain; and by the European Commission under the 6th Framework Programme. M.P. is the recipient of a PhD Fellowship (PFIS) from Instituto de Salud Carlos III, Spain. F.C. was partially funded by a scholarship (152256/158706) from Consejo Nacional de Ciencia y Tecnología (CONACYT), México. The research at São Paulo, Brazil, was funded by Fundação para o Amparo da Pesquisa do Estado de São Paulo (FAPESP), São Paulo, Brazil (2007/04313-2). The research at Porto Alegre city, Brazil, was funded by the Brazilian Post-Graduation Agency Capes (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). The research in Uruguay was funded by Comisión Honoraria de Lucha Contra el Càncer and Fundación Manuel Pérez, Montevideo, Uruguay. The authors give special thanks to the coordinator of the RO1 grant, David Elder, and those of the FP6 grant, Julia Newton-Bishop and Nelleke Gruis, for their support in the development of the project. The authors also thank their patients and their families, who are the main reason for our studies; the nurses from the Melanoma Unit of Hospital Clínic of Barcelona; Daniel Gabriel, Pablo Iglesias, and Maria E. Moliner for helping to collect patient data; Amanda de Nobrega from São Paulo; Thomas Ruzicka, Carola Berking, and the Department of Dermatology and Allergology of Ludwig Maximilian University, Munich, Germany, for support in performing p.A148T analyses in Brazilian cases from HCPA and Carolina Ribas do Nascimento for performing MC1R tests in HCPA; Lídice Dufrechou from Uruguay and Helena Kruyer for helping with English editing and correction of the manuscript. In addition, the authors thank Victoria Godinez Puig from Mexico for helping to collect patient samples and data, as well as the Biobanco del Instituto Valenciano de Oncología and the A.C. Camargo Biobank.
Statement on prior presentation: part of the results of this study were presented as an Oral Communication in the GenoMEL/BioGenoMEL annual meeting 2014, Valencia, Spain, 7–9 May 2014.
Role of the sponsors: the sponsors had no role in the design and conduct of the study; in the collection, analysis, and interpretation of data; or in the preparation, review, or approval of the manuscript.
Supplementary Material
References
- Bleyer A, Viny A, Barr R. Cancer in 15- to 29-year-olds by primary site. Oncologist 2006;11:590–601. [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009;27:6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Malvehy J. Monitoring patients with multiple nevi. Dermatol Clin 2013;31:565–77, viii. [DOI] [PubMed] [Google Scholar]
- Whiteman DC, Green AC. Melanoma and sun exposure: where are we now? Int J Dermatol 1999;38:481–489. [DOI] [PubMed] [Google Scholar]
- Bertolotto C. Melanoma: from melanocyte to genetic alterations and clinical options. Scientifica (Cairo) 2013;2013:635203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, et al.; Lund Melanoma Study Group; Melanoma Genetics Consortium (GenoMEL). Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007;44:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, et al.; Melanoma Genetics Consortium (GenoMEL). High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006;66:9818–9828. [DOI] [PubMed] [Google Scholar]
- Aoude LG, Wadt KA, Pritchard AL, Hayward NK. Genetics of familial melanoma: 20 years after CDKN2A. Pigment Cell Melanoma Res 2015;28:148–160. [DOI] [PubMed] [Google Scholar]
- Marzuka-Alcalá A, Gabree MJ, Tsao H. Melanoma susceptibility genes and risk assessment. Methods Mol Biol 2014;1102:381–393. [DOI] [PubMed] [Google Scholar]
- Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer 2008;122:2753–2760. [DOI] [PubMed] [Google Scholar]
- Demenais F, Mohamdi H, Chaudru V, et al.; Melanoma Genetics Consortium. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J Natl Cancer Inst 2010;102:1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenas C, Aguilera P, Puig-Butillé JA, Carrera C, Malvehy J, Puig S. Genetic counseling in melanoma. Dermatol Ther 2012;25:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leachman SA, Carucci J, Kohlmann W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009;61:677 e671–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall LG, Taber JM, Leaf SL, Kohlmann W, Leachman SA. Melanoma genetic counseling and test reporting improve screening adherence among unaffected carriers 2 years later. Cancer Epidemiol Biomarkers Prev 2013;22:1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall LG, Taber JM, Kohlmann W, Leaf SL, Leachman SA. Unaffected family members report improvements in daily routine sun protection 2 years following melanoma genetic testing. Genet Med 2014;16:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila AL, Krepischi AC, Moredo LF, et al. Germline CDKN2A mutations in Brazilian patients of hereditary cutaneous melanoma. Fam Cancer 2014;13:645–649. [DOI] [PubMed] [Google Scholar]
- Grazziotin TC, Rey MC, Bica CG, et al. Genetic variations of patients with familial or multiple melanoma in Southern Brazil. J Eur Acad Dermatol Venereol 2013;27:e179–e185. [DOI] [PubMed] [Google Scholar]
- Ashton-Prolla P, Bakos L, Junqueira G Jr, Giugliani R, Azevedo SJ, Hogg D. Clinical and molecular characterization of patients at risk for hereditary melanoma in southern Brazil. J Invest Dermatol 2008;128:421–425. [DOI] [PubMed] [Google Scholar]
- Larre Borges A, Borges AL, Cuéllar F, et al. CDKN2A mutations in melanoma families from Uruguay. Br J Dermatol 2009;161:536–541. [DOI] [PubMed] [Google Scholar]
- Potrony M, Puig-Butillé JA, Aguilera P, et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: implications for genetic counseling. J Am Acad Dermatol 2014;71:888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagore E, Montoro A, Oltra S, et al. Age does not appear to be a major indicator of CDKN2A or CDK4 mutations in melanoma patients in Spain. Melanoma Res 2005;15:555–558. [DOI] [PubMed] [Google Scholar]
- Puig-Butillé JA, Carrera C, Kumar R, et al. Distribution of MC1R variants among melanoma subtypes: p.R163Q is associated with lentigo maligna melanoma in a Mediterranean population. Br J Dermatol 2013;169:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer D, Nagore E, Bermejo JL, et al. Melanocortin receptor 1 variants and melanoma risk: a study of 2 European populations. Int J Cancer 2009;125:1868–1875. [DOI] [PubMed] [Google Scholar]
- Sans M. Admixture studies in Latin America: from the 20th to the 21st century. Hum Biol 2000;72:155–177. [PubMed] [Google Scholar]
- Puig S, Malvehy J, Badenas C, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol 2005;23:3043–3051. [DOI] [PubMed] [Google Scholar]
- Auroy S, Avril MF, Chompret A, et al.; French Hereditary Melanoma Study Group. Sporadic multiple primary melanoma cases: CDKN2A germline mutations with a founder effect. Genes Chromosomes Cancer 2001;32:195–202. [DOI] [PubMed] [Google Scholar]
- Ogbah Z, Badenas C, Harland M, et al. Evaluation of PAX3 genetic variants and nevus number. Pigment Cell Melanoma Res 2013;26:666–676. [DOI] [PubMed] [Google Scholar]
- Ogbah Z, Visa L, Badenas C, et al. Serum 25-hydroxyvitamin D3 levels and vitamin D receptor variants in melanoma patients from the Mediterranean area of Barcelona. BMC Med Genet 2013;14:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiorzo P, Fornarini G, Sciallero S, et al.; Genoa Pancreatic Cancer Study Group. CDKN2A is the main susceptibility gene in Italian pancreatic cancer families. J Med Genet 2012;49:164–170. [DOI] [PubMed] [Google Scholar]
- Ciotti P, Struewing JP, Mantelli M, et al. A single genetic origin for the G101W CDKN2A mutation in 20 melanoma-prone families. Am J Hum Genet 2000;67:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Torre G, Pasini B, Frigerio S, et al. CDKN2A and CDK4 mutation analysis in Italian melanoma-prone families: functional characterization of a novel CDKN2A germ line mutation. Br J Cancer 2001;85:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J, Ramos ES. The P48T germline mutation and polymorphism in the CDKN2A gene of patients with melanoma. Braz J Med Biol Res 2006;39:237–241. [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet 1994;8:15–21. [DOI] [PubMed] [Google Scholar]
- Reymond A, Brent R. p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene 1995;11:1173–1178. [PubMed] [Google Scholar]
- Gerstenblith MR, Goldstein AM, Fargnoli MC, Peris K, Landi MT. Comprehensive evaluation of allele frequency differences of MC1R variants across populations. Hum Mutat 2007;28:495–505. [DOI] [PubMed] [Google Scholar]
- Córdoba-Lanús E, Hernández-Jiménez JG, Medina-Coello C, et al. MC1R gene variants and sporadic malignant melanoma susceptibility in the Canary Islands population. Arch Dermatol Res 2014;306:51–58. [DOI] [PubMed] [Google Scholar]
- Salerni G, Lovatto L, Carrera C, Puig S, Malvehy J. Melanomas detected in a follow-up program compared with melanomas referred to a melanoma unit. Arch Dermatol 2011;147:549–555. [DOI] [PubMed] [Google Scholar]
- Salerni G, Carrera C, Lovatto L, et al. Benefits of total body photography and digital dermatoscopy (“two-step method of digital follow-up”) in the early diagnosis of melanoma in patients at high risk for melanoma. J Am Acad Dermatol 2012;67:e17–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar F, Puig S, Kolm I, et al. Dermoscopic features of melanomas associated with MC1R variants in Spanish CDKN2A mutation carriers. Br J Dermatol 2009;160:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.