Abstract
We discovered six plant extracts that increase yeast chronological lifespan to a significantly greater extent than any of the presently known longevity-extending chemical compounds. One of these extracts is the most potent longevity-extending pharmacological intervention yet described. We show that each of the six plant extracts is a geroprotector which delays the onset and decreases the rate of yeast chronological aging by eliciting a hormetic stress response. We also show that each of these extracts has different effects on cellular processes that define longevity in organisms across phyla. These effects include the following: 1) increased mitochondrial respiration and membrane potential; 2) augmented or reduced concentrations of reactive oxygen species; 3) decreased oxidative damage to cellular proteins, membrane lipids, and mitochondrial and nuclear genomes; 4) enhanced cell resistance to oxidative and thermal stresses; and 5) accelerated degradation of neutral lipids deposited in lipid droplets. Our findings provide new insights into mechanisms through which chemicals extracted from certain plants can slow biological aging.
Keywords: yeast, cellular aging, longevity, plant extracts, aging-delaying chemical compounds
INTRODUCTION
The budding yeast Saccharomyces cerevisiae is a unicellular eukaryote amenable to comprehensive molecular analyses [1–3]. The development of various methods of such analyses for S. cerevisiae has enabled to uncover mechanisms underlying complex biological processes within individual yeast cells and their populations [1, 4, 5]. In addition, S. cerevisiae has relatively short and easy measurable replicative and chronological lifespans [6–13]. Due to these beneficial properties as a model organism for studying mechanisms of aging and longevity, S. cerevisiae has been used for the discovery of genes that slow cellular aging and increase healthy lifespan not only in S. cerevisiae and other yeasts but also in multicellular eukaryotes [6, 7, 9, 11, 14–16]. Furthermore, using S. cerevisiae as a model organism for elucidating mechanisms of cellular aging, several nutrient- and energy-sensing signaling pathways have been revealed; these pathways coordinate an evolutionarily conserved array of longevity-defining cellular processes not only in S. cerevisiae and other yeasts but also in eukaryotes across phyla [9, 11, 17–20]. Moreover, S. cerevisiae has been a model organism employed for uncovering several low molecular weight molecules that slow aging and extend healthy lifespan in various multicellular eukaryotes [10, 21–27]. All these studies employing S. cerevisiae as a model organism have provided evidence that the main features of biological aging have been conserved in the course of evolution [6, 9, 11, 18, 21, 28–31].
Our research is aimed at using S. cerevisiae as a model organism to discover chemical compounds that can slow aging and delay the onset of age-related diseases in evolutionarily distant eukaryotic organisms. Some of such geroprotective compounds have been previously revealed in natural products extracted from certain plants [25, 32, 33]. As a first step towards uncovering novel aging-delaying chemical compounds of plant origin, we conducted a screen for plant extracts (PEs) that can extend yeast chronological lifespan (CLS). Our screen revealed six PEs that increase yeast CLS considerably more efficiently than any of the longevity-extending chemical compounds yet described. We show that each of these PEs decelerates yeast chronological aging and has different effects on several longevity-defining cellular processes.
RESULTS
A screen for PEs that can extend longevity of chronologically aging yeast
We screened a library of PEs for extracts that can increase yeast CLS. This library includes 35 different PEs of known origin and properties (Tables 1 and 2, respectively). To perform the screen for lifespan-extending PEs, we used a robust assay for measuring yeast CLS. This assay was similar to the one described previously [34], but the wild-type strain BY4742 was cultured in the synthetic minimal YNB medium initially containing 2% glucose (instead of the nutrient-rich YEP medium supplemented with 0.5% glucose). Yeast cells cultured on 2% glucose are not limited in calorie supply; these cells age chronologically under so-called non-caloric restriction (non-CR) conditions that accelerate aging in different yeast genetic backgrounds, including BY4742 [6, 10, 11].
Table 1. A list of plant extracts that have been used in this study.
| Abbreviated name | Botanical name | Plant part used | Commercial source |
|---|---|---|---|
| PE1 | Echinacea purpurea | Whole plant | Idunn Technologies |
| PE2 | Astragalus membranaceous | Root | Idunn Technologies |
| PE3 | Rhodiola rosea L. | Root | Idunn Technologies |
| PE4 | Cimicifuga racemosa | Root and rhizome | Idunn Technologies |
| PE5 | Valeriana officinalis L. | Root | Idunn Technologies |
| PE6 | Passiflora incarnate L. | Whole plant | Idunn Technologies |
| PE7 | Polygonum cuspidatum | Root and rhizome | Idunn Technologies |
| PE8 | Ginkgo biloba | Leaf | Idunn Technologies |
| PE9 | Zingiber officinale Roscoe | Rhizome | Idunn Technologies |
| PE10 | Theobroma cacao L. | Cacao nibs | Idunn Technologies |
| PE11 | Camellia sinensis L. Kuntze | Leaf | Idunn Technologies |
| PE12 | Apium graveolens L. | Seed | Idunn Technologies |
| PE13 | Scutellaria baicalensis | Root | Idunn Technologies |
| PE14 | Euterpe oleracea | Fruit | Idunn Technologies |
| PE15 | Withania somnifera | Root and leaf | Idunn Technologies |
| PE16 | Phyllanthus emblica | Fruit | Idunn Technologies |
| PE17 | Camellia sinensis | Leaf | Idunn Technologies |
| PE18 | Pueraria lobata | Root | Idunn Technologies |
| PE19 | Silybum marianum | Seed | Idunn Technologies |
| PE20 | Eleutherococcus senticosus | Root and stem | Idunn Technologies |
| PE21 | Salix alba | Bark | Idunn Technologies |
| PE22 | Glycine max L. | Bean | Idunn Technologies |
| PE24 | Calendula officinalis | Flower | Idunn Technologies |
| PE25 | Salvia miltiorrhiza | Root | Idunn Technologies |
| PE27 | Panax quinquefolium | Root | Idunn Technologies |
| PE28 | Harpagophytum procumbens | Root | Idunn Technologies |
| PE29 | Olea europaea L. | Leaf | Idunn Technologies |
| PE30 | Gentiana lutea | Root | Idunn Technologies |
| PE31 | Piper nigrum | Fruit | Idunn Technologies |
| PE32 | Aesculus hippocastanum | Seed | Idunn Technologies |
| PE33 | Mallus pumila Mill. | Fruit | Idunn Technologies |
| PE34 | Fragaria spp. | Fruit | Idunn Technologies |
| PE35 | Ribes nigrum | Leaf | Idunn Technologies |
| PE36 | Dioscorea opposita | Root | Idunn Technologies |
| PE37 | Cinnamomum verum | Bark | Idunn Technologies |
Table 2. Properties of plant extracts that have been used in this study.
| Abbreviated name | Properties |
|---|---|
| PE1 | Extraction solvent: ethanol (75%)/water (25%). Extract ratio: 4/1. Composition: natural extract, maltodextrin. |
| PE2 | Extraction solvent: denatured ethanol (70%)/water (30%). Extract ratio: 10/1. Composition: natural extract (40–50%), gum arabic (50–60%). |
| PE3 | Extraction solvent: ethanol (60–80%)/water (40–20%). Extract ratio: 15–20/1. Composition: natural extract (80–100%), maltodextrin (0–20%). |
| PE4 | Extract ratio: 6–8/1. Composition: natural extract (28–38%), maltodextrin (60–70%), tricalcium phosphate (0–5%). |
| PE5 | Extraction solvent: denatured ethanol/water. Extract ratio: ∼ 6/1. Composition: natural extract, maltodextrin, silica (0–1%). |
| PE6 | Extraction solvent: water (100%). Extract ratio: 4/1. Composition: natural extract, maltodextrin. |
| PE7 | Extraction solvent: ethanol (80%)/water (20%). Extract ratio: 40/1. Composition: natural extract (90–100%), maltodextrin (0–10%). |
| PE8 | Extraction solvent: ethanol/water. Extract ratio: 50/1. Composition: natural extract. |
| PE9 | Extraction solvent: ethanol/water. Composition: natural extract (96%), gingerols (4%). |
| PE10 | Natural powder/final product ratio: 2–3/1. Composition: natural powder. |
| PE11 | Extraction solvent: ethyl acetate (90%)/water (10%). Extract ratio: 6/1. Composition: natural extract (100%). |
| PE12 | Extraction solvent: ethanol (90%)/water (10%). Extract ratio: 8/1. Composition: natural extract, maltodextrin, modified starch, silica. |
| PE13 | Extraction solvent: ethanol/water. Extract ratio: 4/1. Composition: natural extract. |
| PE14 | Extraction solvent: ethanol/water. Extract ratio: 4/1. Composition: natural extract. |
| PE15 | Extraction solvent: water. Extract ratio: 9/1. Composition: withanolide glycoside conjugates (10%), oligosaccharides (32%), free withanolides (0.5%). |
| PE16 | Extraction solvent: water. Composition: hydrolysable tannins (> 60%), including Emblicanin–A, Emblicanin–B, Punigluconin, Pedunculagin. |
| PE17 | Composition: tea polyphenols (> 90%), including epigallocatechin gallate (> 40%). |
| PE18 | Composition: flavonoids (> 40%), including puerarin. |
| PE19 | Extraction solvent: ethanol/water. Composition: silymarin (> 80%). |
| PE20 | Extraction solvent: water. Composition: eleutheroside B + E (> 0.8%). |
| PE21 | Extraction solvent: ethanol/water. Composition: salicin (> 25%). |
| PE22 | Composition: isoflavones (40%). |
| PE24 | Composition: lutein (> 5%). |
| PE25 | Composition: tanshinones, isotanshinones, cryptotanshinone, isocryptotanshinone, dihydrotanshinone, hydroxytanshinones. |
| PE27 | Composition: ginsenosides (10%, by HPLC–UV), quintozene–free. |
| PE28 | Extraction solvent: ethanol/water. Extract ratio: 40/1. Composition: harpagosides (20%, by HPLC–UV). |
| PE29 | Extraction solvent: ethanol (70%)/water (30%). Extract ratio: 5–10/1. Composition: natural extract, maltodextrin, silica (0.2%). |
| PE30 | Composition: isogentisin (0.04%). |
| PE31 | Extraction solvent: ethanol. Extract ratio: 10/1. Composition: piperine (> 90%). |
| PE32 | Composition: aescin (20%). |
| PE33 | Extraction solvent: ethanol (70%)/water (30%). Extract ratio: 120–130/1. Composition: natural extract (60–70%), maltodextrin (30–40%). |
| PE34 | Extract ratio: 5/1. Composition: natural extract, including polyphenols (> 2%). |
| PE35 | Extraction solvent: water. Composition: polyphenols (15%, by HPLC–UV). |
| PE36 | Composition: diosgenine (> 16%, by HPLC–UV). |
| PE37 | Extraction solvent: water. Composition: polyphenols (25%, by HPLC–UV). |
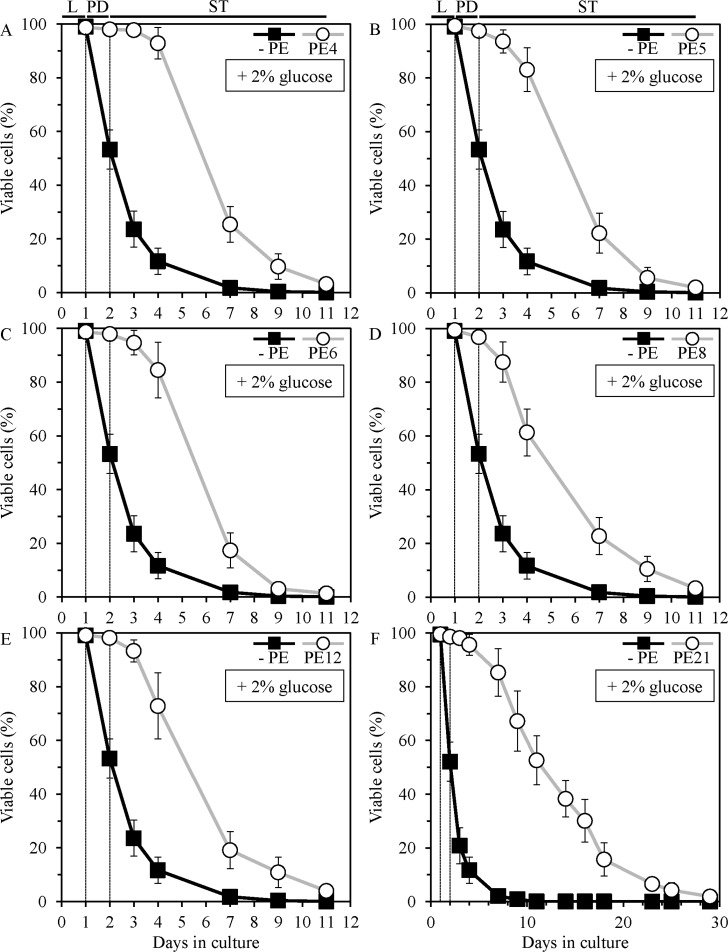
In our screen for longevity-extending PEs, each PE from the library was added to growth medium at the time of cell inoculation at a final concentration in the 0.02% to 1.0% range. Some PEs from the library did not alter the mean and maximum CLS of yeast under non-CR conditions at any concentration examined; among these PEs were PE9, PE13, PE16, PE22, PE28 and PE36 (Figure S2–Figure S5). Many PEs from the library shortened the mean and/or maximum CLS of yeast under non-CR conditions at final concentrations ranging from 0.08% to 1.0%; among these PEs were PE1–PE3, PE7, PE10, PE11, PE14, PE15, PE17–PE20, PE24, PE25, PE27, PE29–PE35 and PE37 (Figure S1–Figure S5). 6 of the 35 PEs from the library significantly increased both the mean and maximum CLS of yeast under non-CR conditions if added at final concentrations ranging from 0.04% to 1.0% (Figure S1–Figure S3). A group of these longevity-extending PEs included the following extracts: 1) 0.5% PE4 from Cimicifuga racemosa (Figure 1A, Figure 3A and 3B, Figure S1); 2) 0.5% PE5 from Valeriana officinalis L. (Figure 1B, Figure 3A and 3B, Figure S1); 3) 1.0% PE6 from Passiflora incarnata L. (Figure 1C, Figure 3A and 3B, Figure S1); 4) 0.3% PE8 from Ginkgo biloba (Figure 1D, Figure 3A and 3B, Figure S1); 5) 0.1% PE12 from Apium graveolens L. (Figure 1E, Figure 3A and 3B, Figure S2); and 6) 0.1% PE21 from Salix alba (Figure 1F, Figure 3A and 3B, Figure S3). None of the six lifespan-prolonging PEs affected growth rates in logarithmic (L) and post-diauxic (PD) phases or impacted the maximum cell density in stationary (ST) phase of yeast cultures under non-CR conditions on 2% glucose (Figure S6). Thus, the observed lifespan extension by each of these PEs is unlikely to be caused by its ability to decrease growth rate or to make yeast more resistant to toxic substances accumulated during culturing in the synthetic minimal YNB medium.
Figure 1. PE4, PE5, PE6, PE8, PE12 and PE21 extend the chronological lifespan (CLS) of yeast grown under non-caloric restriction (non-CR) conditions.
Wild-type (WT) cells were grown in the synthetic minimal YNB medium (0.67% Yeast Nitrogen Base without amino acids) initially containing 2% glucose, in the presence of a PE or in its absence. Survival curves of chronologically aging WT strain cultured with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F) are shown. Data are presented as means ± SEM (n = 21–35). CLS extension was significant for each of the PEs tested (p < 0.05; the p values for comparing survival curves were calculated with the help of the GraphPad Prism statistics software). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
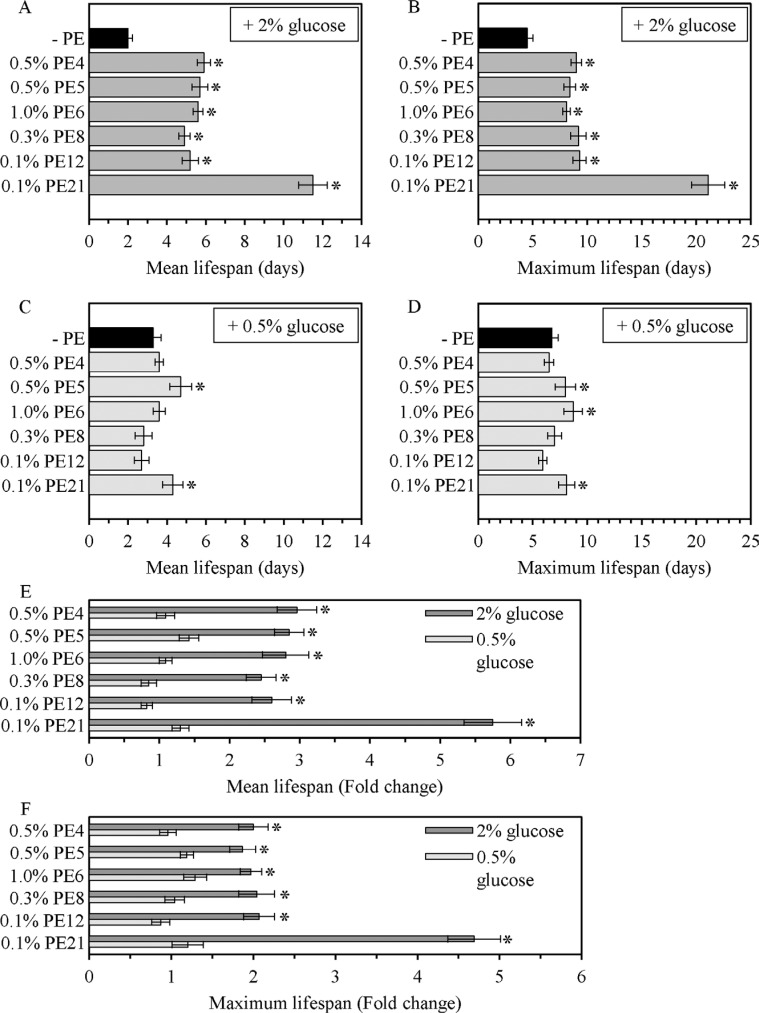
Figure 3. The longevity-extending efficacy under non-CR conditions significantly exceeds that under CR conditions for each of the six lifespan-prolonging PEs.
WT cells were grown in the synthetic minimal YNB medium initially containing 0.5% glucose (CR conditions) or 2% glucose (non-CR conditions), in the presence of a PE or in its absence. The mean (A, C and E) and maximum (B, D and F) lifespans of chronologically aging WT strain cultured under CR (C, D, E and F) or non-CR (A, B, E and F) conditions in the absence of a PE or in the presence of 0.5% PE4, 0.5% PE5, 1% PE6, 0.3% PE8, 0.1% PE12 or 0.1% PE21 are shown; data are presented as means ± SEM (n = 5–7; *p < 0.05). The extent to which each of the PE tested increases the mean and maximum lifespans under non-CR conditions exceeds that under CR conditions (*p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test).
For each of the six lifespan-prolonging PEs, the longevity-extending efficacy under CR conditions is significantly lower than that under non-CR conditions
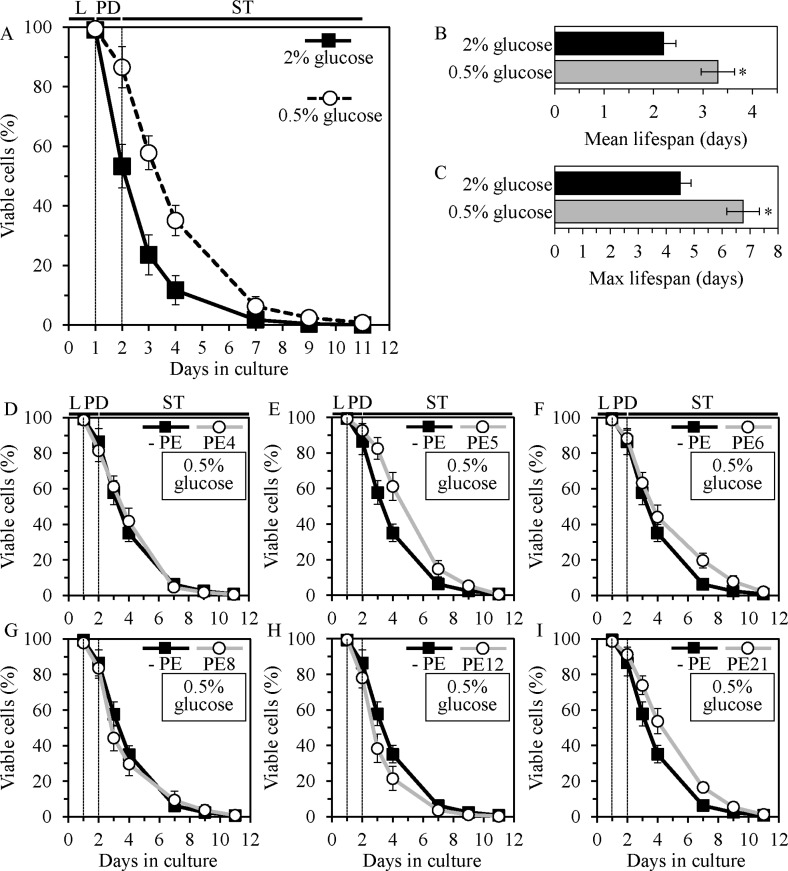
Chronologically aging yeast grown under CR conditions on 0.5% glucose are known to live longer than yeast cultured under non-CR conditions on 2% glucose. Such ability of the CR diet to extend CLS has been reported for yeast cultured in media of various nutrient compositions [6, 10, 11]. We found that, if the CR diet is administered by culturing yeast in the YNB medium initially containing 0.5% glucose, it significantly increases both the mean and maximum CLS of S. cerevisiae (Figure 2A–2C). We discovered that 0.5% PE5 and 0.1% PE21 (but not 0.5% PE4, 1.0% PE6, 0.3% PE8 or 0.1% PE12) extend the mean CLS of yeast grown under CR conditions (Figure 2D–2I, Figure 3C). We also revealed that 0.5% PE5, 1.0% PE6 and 0.1% PE21 (but not 0.5% PE4, 0.3% PE8 or 0.1% PE12) extend the maximum CLS of yeast grown under CR conditions (Figure 2D–2I, Figure 3D). Akin to their effects under non-CR conditions, none of the six lifespan-prolonging PEs influenced growth rates in L and PD phases or altered the maximum cell density in ST phase of yeast cultures under CR conditions on 0.5% glucose (Figure S7). Importantly, each of the six lifespan-prolonging PEs extended both the mean and maximum CLS of yeast cultures under non-CR conditions on 2% glucose to a significantly higher extent than those of yeast under CR at 0.5% glucose (Figure 3A–3F). This observation indicates that each of these PEs could mimic the longevity-extending effect of CR.
Figure 2. PE5 and PE21, but not PE4, PE6, PE8 or PE12, extend the CLS of yeast grown under CR conditions.
WT cells were grown in the synthetic minimal YNB medium initially containing 0.5% glucose (CR conditions) or 2% glucose (non-CR conditions), in the presence of a PE or in its absence. Survival curves (A), the mean (B) and maximum (C) lifespans of chronologically aging WT strain cultured under CR or non-CR conditions in the absence of a PE are shown; data are presented as means ± SEM (n = 5–7). CR caused significant extension of CLS (A) (p < 0.05; the p values for comparing survival curves were calculated with the help of the GraphPad Prism statistics software). CR extended both the mean (B) and maximum (C) lifespans (*p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Survival curves of chronologically aging WT strain cultured under CR on 0.5% glucose with or without 0.5% PE4 (D), 0.5% PE5 (E), 1% PE6 (F), 0.3% PE8 (G), 0.1% PE12 (H) or 0.1% PE21 (I) are shown; data are presented as means ± SEM (n = 5–7). CLS extension under CR on 0.5% glucose was significant for PE5 and PE21 (p < 0.05; the p values for comparing survival curves were calculated with the help of the GraphPad Prism statistics software). CLS extension under CR on 0.5% glucose was not significant for PE4, PE6, PE8 and PE12. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Each of the six longevity-extending PEs is a geroprotector which delays the onset and slows the progression of yeast chronological aging by eliciting a hormetic stress response
As we found, PE4, PE5, PE6, PE8, PE12 and PE21 greatly extend the mean CLS of yeast cultured under non-CR conditions (Figure 1A, Figure 3A and 3B, Figure S1–S3). Mean lifespan is believed to be directly proportional to the survival rates of organisms in the population during development and maturity stages of organismal aging; mean lifespan is likely to be under control of certain extrinsic (environmental) factors [35–38]. Thus, it is conceivable that PE4, PE5, PE6, PE8, PE12 and PE21 decrease the extrinsic rate of yeast chronological aging prior to cell entry into quiescence or senescence.
Furthermore, we revealed that PE4, PE5, PE6, PE8, PE12 and PE21 also substantially increase the maximum CLS of yeast grown under non-CR conditions (Figure 1A, Figure 3A and 3B, Figure S1–S3). Maximum lifespan is believed to reflect the duration of “healthy” life period (i.e. healthspan) during quiescence/senescence stage of organismal aging; maximum lifespan is likely to be controlled by certain intrinsic (cellular and organismal) longevity modifiers [7, 35–37, 39, 40]. One could therefore conclude that PE4, PE5, PE6, PE8, PE12 and PE21 also decrease the intrinsic rate of yeast chronological aging after cell entry into quiescence or senescence.
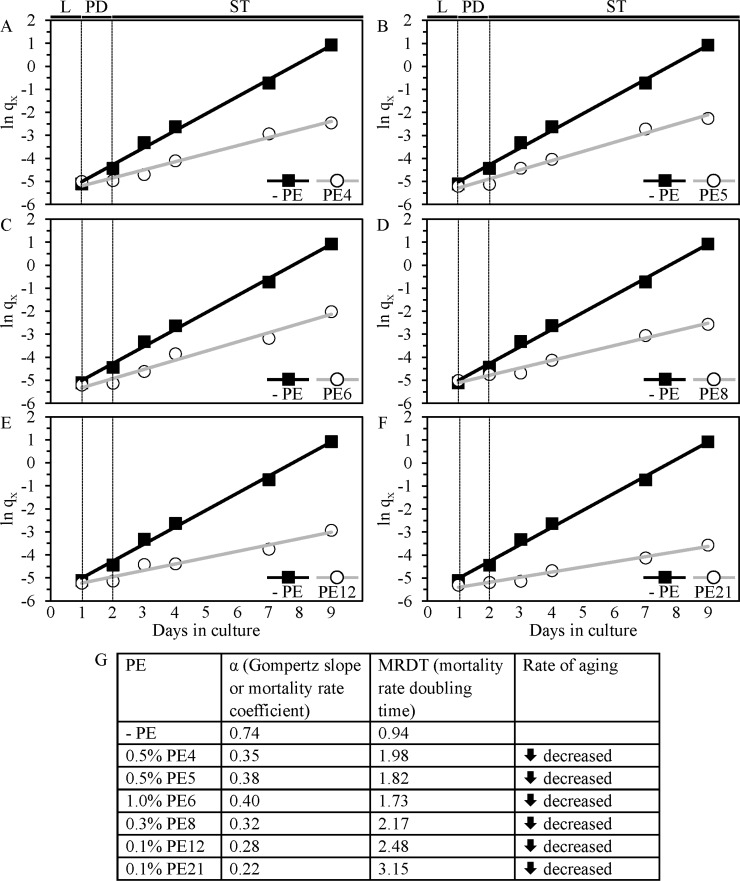
Our analysis of the Gompertz mortality function further validated the above conclusion that PE4, PE5, PE6, PE8, PE12 and PE21 significantly reduce the rate of yeast chronological aging. Indeed, we found that each of these longevity-extending PEs causes a substantial decrease in slope of the Gompertz mortality rate (also known as mortality rate coefficient α) and a considerable increase in the mortality rate doubling time (MRDT) (Figure 4A–4G). Such changes in the values of α and MRDT are characteristic of interventions that decrease the rate of progression through the process of biological aging [37, 41–44].
Figure 4. Analysis of the Gompertz mortality function indicates that PE4, PE5, PE6, PE8, PE12 and PE21 significantly decrease the rate of chronological aging in yeast.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. Survival curves shown in Figure 1A–1F were used to calculate the age-specific mortality rates (qx) of chronologically aging WT yeast populations cultured with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F). Each of these longevity-extending PEs caused a substantial decrease in slope of the Gompertz mortality rate (also known as mortality rate coefficient α) and a considerable increase in the mortality rate doubling time (MRDT) (G). The values of qx, α and MRDT were calculated as described in Materials and methods. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Noteworthy, our analyses of how different concentrations of PE4, PE5, PE6, PE8, PE12 and PE21 impact yeast longevity under non-CR conditions revealed that each of them causes a so-called “hormetic” stress response in chronologically aging yeast with respect to longevity. Indeed, the dose-response curves (i.e. the curves that reflect relationships between PE concentrations and mean or maximum CLS) for PE4, PE5, PE8, PE12 and PE21 were inverted U-shaped, whereas the dose-response curve for PE6 was J-shaped (Figure S1–S3). Such nonlinear and biphasic dose-response curves denote a hormetic kind of stress response, in which 1) lower (hormetic) concentrations of a chemical compound increase the survival of a cell or an organism by stimulating biological processes that allow to maintain cellular or organismal stress at a level which is below a threshold of toxicity; while 2) higher concentrations of this chemical compound decrease the survival of a cell or an organism by creating stress which exceeds such threshold [25, 45–48].
Each of the six lifespan-extending PEs alters the age-related chronology of longevity-defining traits of mitochondrial functionality
We hypothesized that PE4, PE5, PE6, PE8, PE12 and PE21 slow yeast chronological aging by influencing certain cellular processes. We sought to identify these longevity-defining processes. Certain aspects of mitochondrial functionality (such as mitochondrial respiration, mitochondrial membrane potential and mitochondrial reactive oxygen species [ROS] homeostasis) are known to define the rate of chronological aging in yeast [6, 11, 23, 34, 52, 55–67]. We therefore assessed how PE4, PE5, PE6, PE8, PE12 and PE21 impact these longevity-defining processes in chronologically aging yeast cultures under non-CR conditions on 2% glucose.
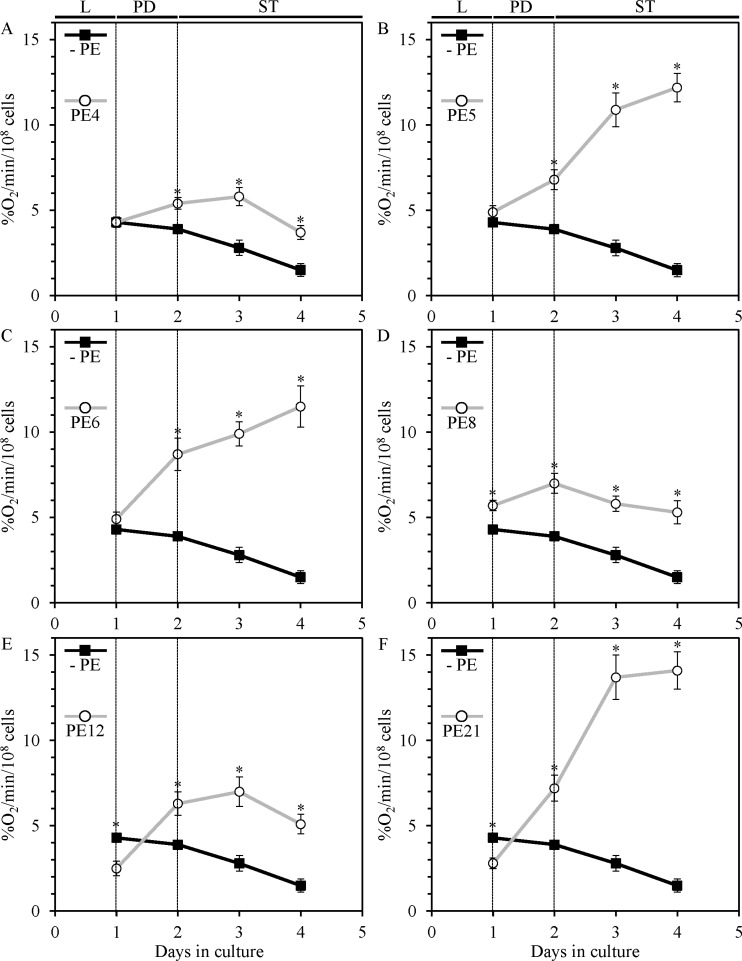
We found that each of the six lifespan-extending PEs stimulates coupled mitochondrial respiration, which was monitored by measuring the rate of oxygen consumption by yeast cells. PE4, PE8 and PE12 decreased the extent to which such respiration declined in ST-phase cultures (Figure 5A, 5D and 5E), whereas PE5, PE6 and PE21 considerably increased the rate of mitochondrial respiration in yeast during PD and ST growth phases (Figure 5B, 5C and 5F).
Figure 5. PE4, PE5, PE6, PE8, PE12 and PE21 alter the age-related chronology of mitochondrial oxygen consumption by yeast grown under non-CR conditions.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. A polarographic assay was used to measure oxygen uptake by live yeast cells, as described in Materials and methods. Age-dependent changes in the rate of mitochondrial oxygen consumption by chronologically aging WT strain cultured under non-CR conditions on 2% glucose with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F) are shown; data are presented as means ± SEM (n = 7–9; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
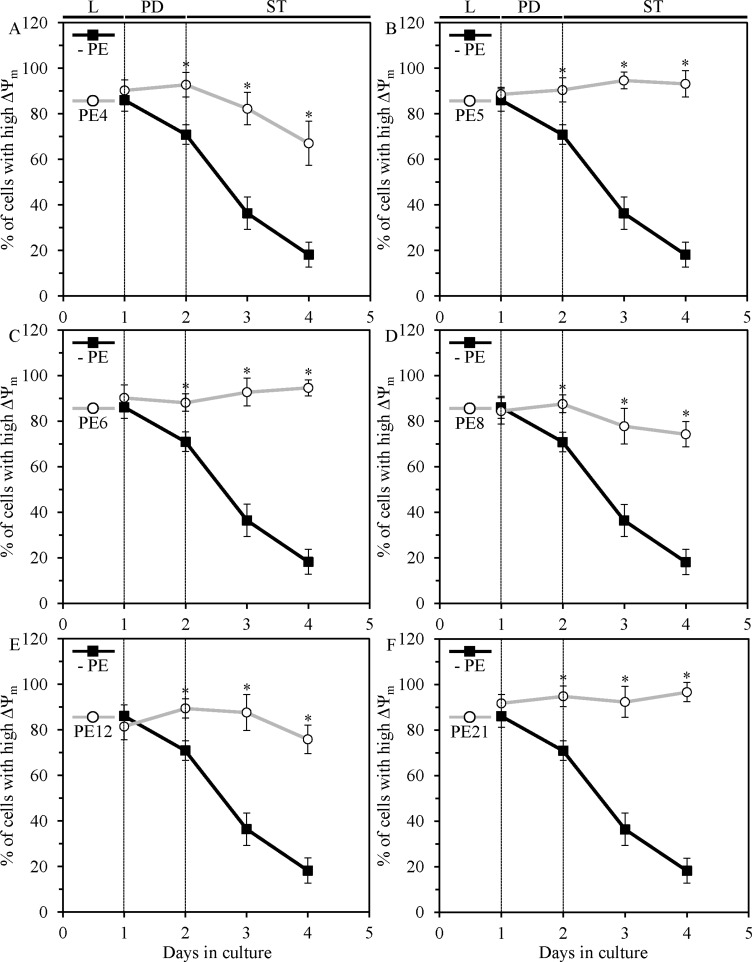
We also found that each of the six lifespan-extending PEs sustains healthy populations of functional mitochondria that exhibit high mitochondrial membrane potential (ΔΨm). PE4, PE8 and PE12 substantially reduced the extent to which ΔΨm declined during PD and ST growth phases (Figure 6A, 6D and 6E; Figure S8 and S9), whereas PE5, PE6 and PE21 completely prevented such decline (Figure 6B, 6C and 6F; Figure S8 and S9).
Figure 6. PE4, PE5, PE6, PE8, PE12 and PE21 sustain healthy populations of functional mitochondria that exhibit high mitochondrial membrane potential (ΔΨm) in chronologically aging yeast grown under non-CR conditions.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. ΔΨm was measured in live yeast by fluorescence microscopy of Rhodamine 123 staining, as described in Materials and methods. Age-dependent changes in the percentage of WT cells displaying high ΔΨm in chronologically aging yeast cultures under non-CR conditions on 2% glucose with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F) are shown; data are presented as means ± SEM (n = 3–4; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
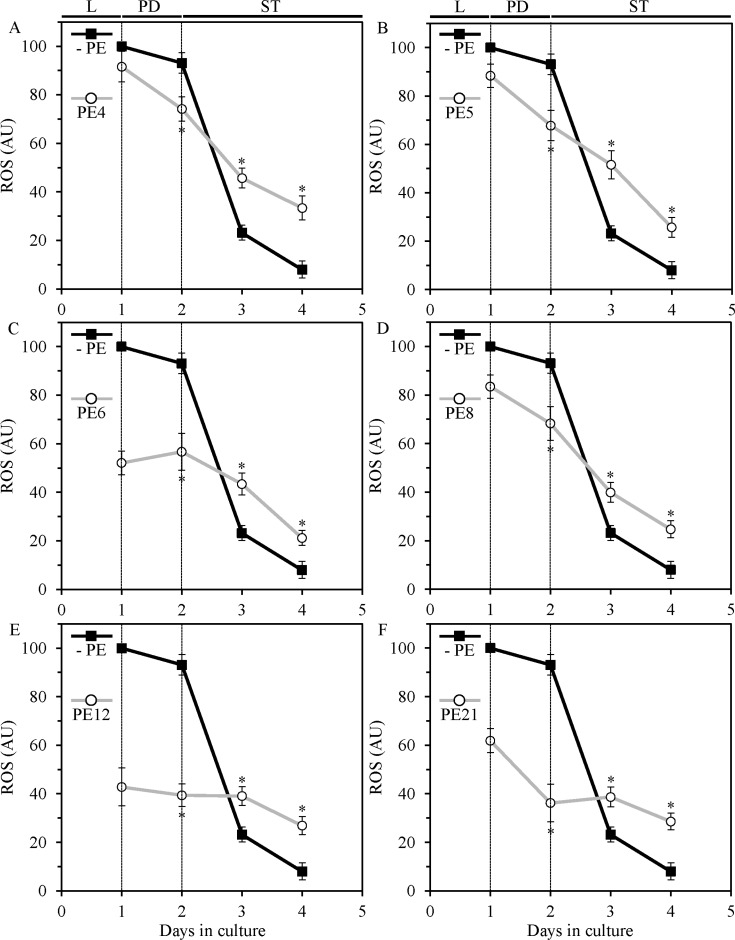
PE4, PE5, PE6, PE8, PE12 and PE21 also caused significant changes in the age-related chronology of intracellular ROS, which in yeast and other organisms are known to be formed mainly as by-products of mitochondrial respiration [68, 69]. Each of these PEs decreased the extent to which the intracellular concentration of mitochondrially generated ROS declined during PD and ST growth phases (Figure 7A–7F). On days 3 and 4 of culturing, ROS concentrations in yeast grown with PE4, PE5, PE6, PE8, PE12 or PE21 exceeded those in yeast grown without it (Figure 7A–7F).
Figure 7. In yeast grown under non-CR conditions, PE4, PE5, PE6, PE8, PE12 and PE21 alter the patterns of age-related changes in intracellular reactive oxygen species (ROS) known to be generated mainly as by-products of mitochondrial respiration.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. The intracellular concentrations of ROS were measured in live yeast by fluorescence microscopy of dihydrorhodamine 123 staining, as described in Materials and methods. Age-dependent changes in ROS concentrations within chronologically aging WT cells cultured under non-CR conditions on 2% glucose with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F) are shown; data are presented as means ± SEM (n = 3–4; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
The six lifespan-extending PEs differently influence the extent of an age-related oxidative damage to cellular proteins, membrane lipids, mitochondrial and nuclear genomes
A body of evidence supports the following view on the relationships between cellular ROS, oxidative molecular damage and aging in organisms across phyla: 1) if cellular concentrations of ROS exceed a threshold of toxicity, ROS cause oxidative damage to proteins, lipids and DNA; 2) oxidative damage to each kind of these macromolecules accumulates with age; and 3) cumulative oxidative damage to the different kinds of macromolecules is one of the major causes of aging [53, 54, 69–75]. We therefore examined how PE4, PE5, PE6, PE8, PE12 and PE21 impact the extent of oxidative damage to proteins, lipids and DNA in chronologically aging yeast cultured under non-CR conditions on 2% glucose.
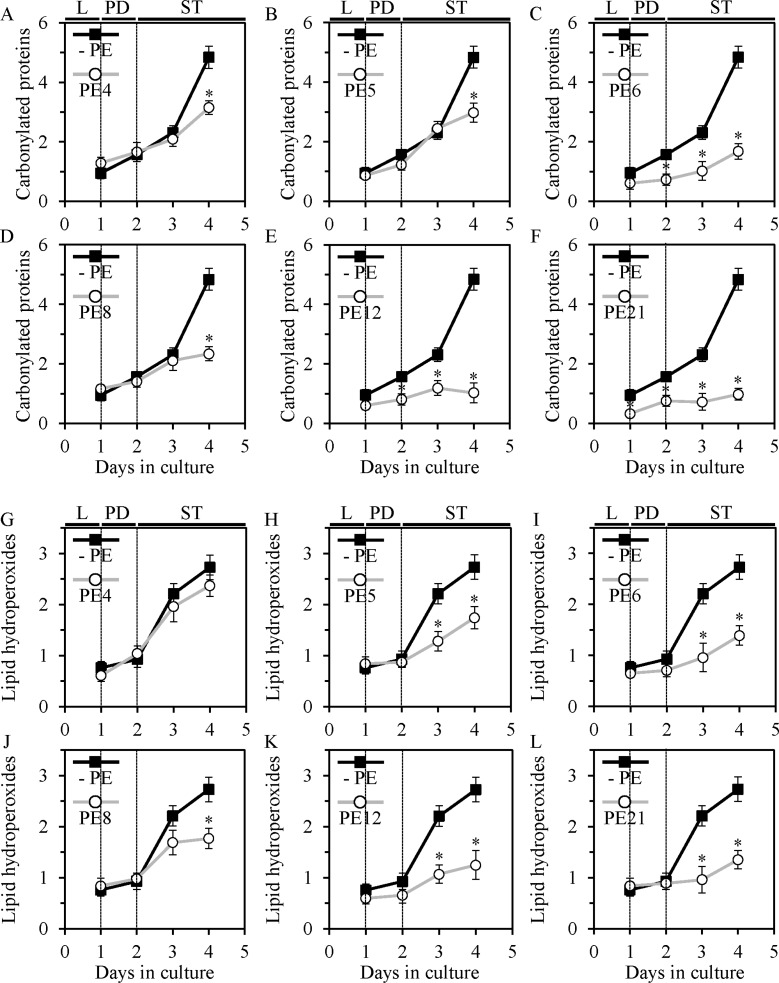
We found that each of the six lifespan-extending PEs delays an age-dependent rise in the extent of oxidative damage to cellular proteins. PE6, PE12 and PE21 reduced oxidative carbonylation of proteins in yeast cells progressing through the entire ST phase (Figure 8C, 8E and 8F). PE4, PE5 and PE8 elicited such inhibitory effect on oxidative protein damage only later in ST phase, on day 4 of culturing (Figure 8A, 8B and 8D).
Figure 8. PE4, PE5, PE6, PE8, PE12 and PE21 delay an age-dependent rise in the extent of oxidative damage to cellular proteins in chronologically aging yeast grown under non-CR conditions. PE5, PE6, PE8, PE12 and PE21, but not PE4, have similar effects on the extent of oxidative damage to membrane lipids.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. Carbonylated cellular proteins (A–F) and oxidatively damaged membrane lipids (G–L) were determined as described in Materials and methods. Age-dependent changes in the concentrations of these oxidatively damaged macromolecules within chronologically aging WT cells cultured under non-CR conditions on 2% glucose with or without 0.5% PE4 (A and G), 0.5% PE5 (B and H), 1% PE6 (C and I), 0.3% PE8 (D and J), 0.1% PE12 (E and K) or 0.1% PE21 (F and L) are shown; data are presented as means ± SEM (n = 2–4; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Furthermore, PE5, PE6, PE8, PE12 and PE21 (but not PE4) caused a significant reduction in the levels of oxidatively damaged membrane lipids; such reduction was observed late in ST phase, on days 3 and/or 4 of culturing (Figure 8H–8L).
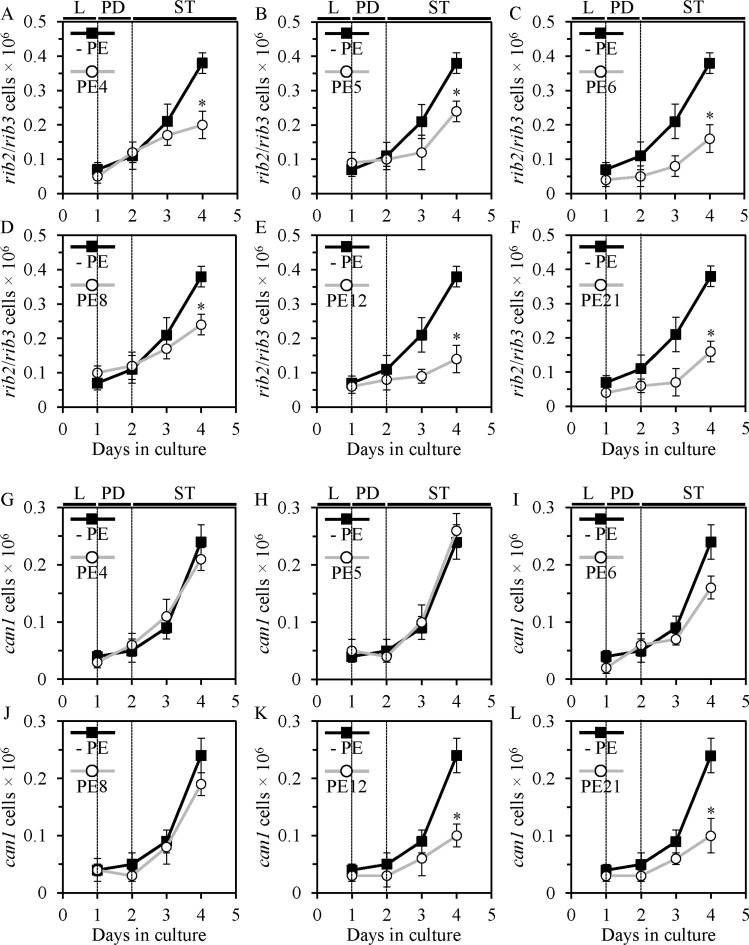
Moreover, PE4, PE5, PE6, PE8, PE12 and PE21 decreased the frequencies of spontaneous point mutations in the RIB2 and RIB3 genes of mitochondrial DNA (mtDNA) (Figure 9A–9F) - likely due to a reduced extent of oxidative damage to mtDNA in yeast cells exposed to any of these PEs. Such inhibitory effects of the six lifespan-extending PEs on oxidative damage to mtDNA was observed late in ST phase, on day 4 of culturing.
Figure 9. PE4, PE5, PE6, PE8, PE12 and PE21 slow down an age-dependent rise in the frequency of spontaneous point mutations in the rib2 and rib3 loci of mitochondrial DNA (mtDNA) in chronologically aging yeast grown under non-CR conditions. PE12 and PE21, but not PE4, PE5, PE6 or PE8, have similar effects on the frequency of spontaneous point mutations in the CAN1 gene of nuclear DNA (nDNA).
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. The frequency of spontaneous point mutations in the rib2 and rib3 loci of mtDNA (A–F), as well as the frequency of spontaneous point mutations in the CAN1 gene of nDNA (G–L), were measured as described in Materials and methods. Age-dependent changes in the frequencies of these mtDNA and nDNA mutations in chronologically aging WT cells cultured under non-CR conditions on 2% glucose with or without 0.5% PE4 (A and G), 0.5% PE5 (B and H), 1% PE6 (C and I), 0.3% PE8 (D and J), 0.1% PE12 (E and K) or 0.1% PE21 (F and L) are shown; data are presented as means ± SEM (n = 3–5; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
We also revealed that PE12 and PE21 (but not PE4, PE5, PE6 or PE8) caused a significant reduction in the frequencies of spontaneous point mutations in the CAN1 gene of nuclear DNA (nDNA) (Figure 9K and 9L) - possibly due to a decreased degree of oxidative damage to nDNA in yeast cells grown in the presence of PE12 or PE21. Such inhibitory effects of PE12 or PE21 on oxidative damage to nDNA was also seen late in ST phase, on day 4 of culturing.
The six lifespan-extending PEs differently influence the resistance of chronologically aging yeast to chronic oxidative and thermal stresses
A body of evidence implies that the development of resistance to chronic (long-term) oxidative and/or thermal stresses can extend longevity in organisms across phyla, including yeast [6, 9, 10, 11, 34, 46–48, 68, 76–80]. We therefore assessed how PE4, PE5, PE6, PE8, PE12 and PE21 influence the abilities of chronologically aging yeast cultured under non-CR conditions to resist chronic oxidative and thermal stresses.
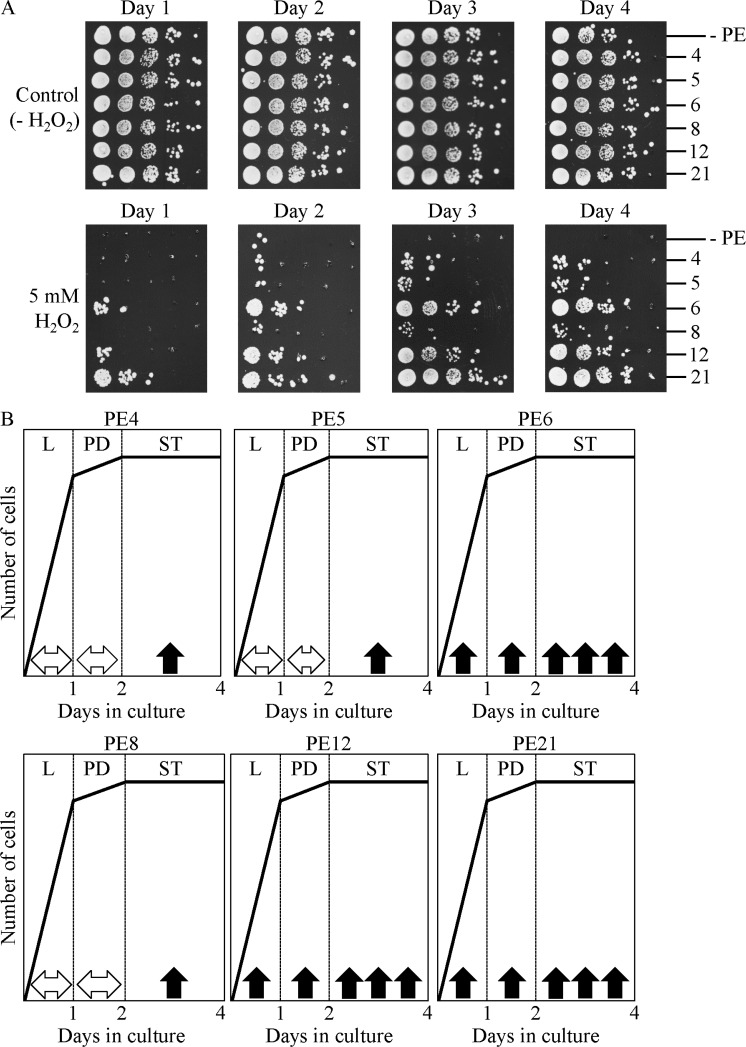
Chronic oxidative stress was administered by recovering yeast cells progressing through L, PD or ST phases of growth/culturing in liquid YNB medium initially containing 2% glucose, spotting these cells on solid YEP medium with 2% glucose and 5 mM hydrogen peroxide, and incubating them for 3 days. We found that PE6, PE12 and PE21 significantly increase cell resistance to chronic oxidative stress in yeast cultures progressing through L, PD and ST phases (Figure 10A and 10B). PE4, PE5 and PE8 enhanced the ability of cells to resist chronic oxidative stress only in yeast cultures progressing through ST phase, but did not alter such ability during L and PD phases (Figure 10A and 10B).
Figure 10. PE4, PE5, PE6, PE8, PE12 and PE21 enhance the ability of chronologically aging yeast grown under non-CR conditions to resist chronic oxidative stress.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. (A) Spot assays for monitoring oxidative stress resistance were performed as described in Materials and methods. Serial 10-fold dilutions of cells recovered at different days of culturing were spotted on plates with solid YEP medium containing 2% glucose as carbon source, with or without 5 mM hydrogen peroxide. All pictures were taken after a 3-d incubation at 30°C. (B) A model for how 0.5% PE4, 0.5% PE5, 1% PE6, 0.3% PE8, 0.1% PE12 and 0.1% PE21 influence the resistance of yeast to chronic oxidative stress during logarithmic (L), post-diauxic (PD) or stationary (ST) phases of growth.  or
or  Denote unaltered or enhanced, respectively, cell resistance to chronic oxidative stress during a particular phase of growth. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Denote unaltered or enhanced, respectively, cell resistance to chronic oxidative stress during a particular phase of growth. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
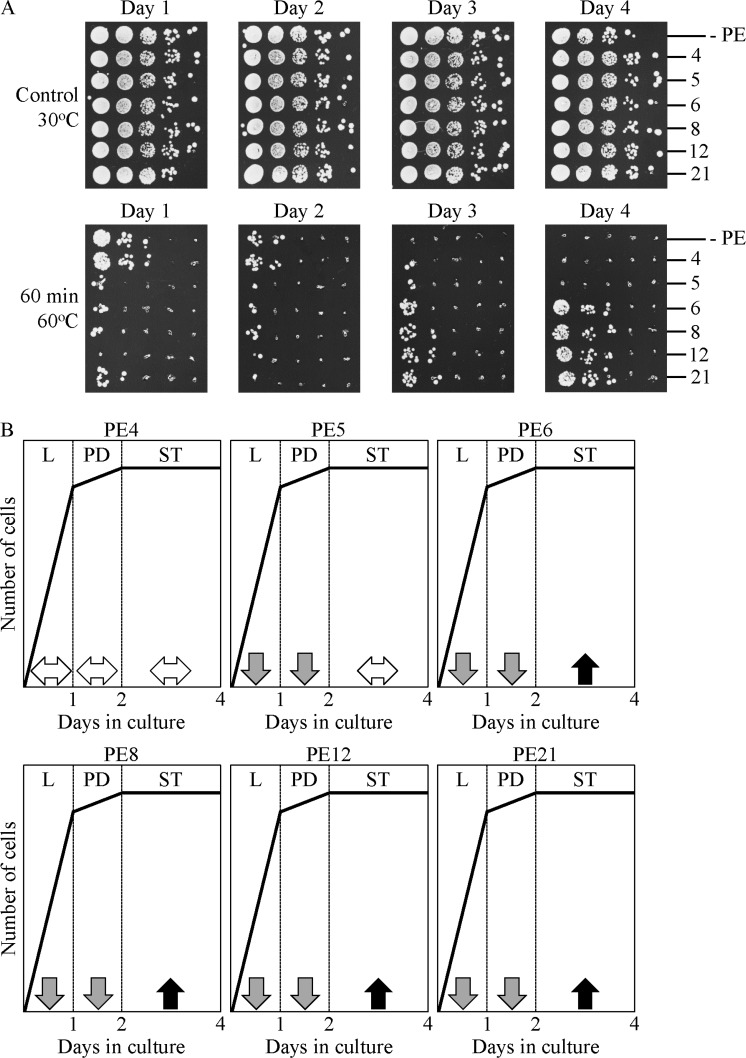
Chronic thermal stress was administered by recovering yeast cells progressing through L, PD or ST phases of growth/culturing in liquid YNB medium initially containing 2% glucose, spotting these cells on solid YEP medium with 2% glucose and incubating at 60°C for 60 min, and then transferring plates with these cells to 30°C and incubating at this temperature for 3 days. We found that PE6, PE8, PE12 and PE21 increase cell resistance to chronic thermal stress only in yeast cultures progressing through ST phase (Figure 11A and 11B). In contrast, each of these four lifespan-extending PEs weakened the ability of cells to resist chronic thermal stress during L and PD phases (Figure 11A and 11B). Furthermore, neither PE4 nor PE5 altered cell resistance to chronic thermal stress in yeast cultures progressing through ST phase (Figure 11A and 11B). PE4 did not affect the ability of cells to resist chronic thermal stress also during L and PD phases, whereas PE5 reduced such ability in yeast cultures progressing through these two phases (Figure 11A and 11B).
Figure 11. PE4, PE5, PE6, PE8, PE12 and PE21 exhibit different effects on the ability of chronologically aging yeast grown under non-CR conditions to resist chronic thermal stress.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. (A) Spot assays for monitoring thermal stress resistance were performed as described in Materials and methods. Serial 10-fold dilutions of cells recovered at different days of culturing were spotted on plates with solid YEP medium containing 2% glucose as carbon source. Plates were initially incubated at 30°C (control) or 60°C for 60 min, and were then transferred to 30°C. All pictures were taken after a 3-d incubation at 30°C. (B) A model for how 0.5% PE4, 0.5% PE5, 1% PE6, 0.3% PE8, 0.1% PE12 and 0.1% PE21 influence the resistance of yeast to chronic thermal stress during logarithmic (L), post-diauxic (PD) or stationary (ST) phases of growth.  ,
,  or
or  Denote unaltered, reduced or enhanced, respectively, cell resistance to chronic thermal stress during a particular phase of growth. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Denote unaltered, reduced or enhanced, respectively, cell resistance to chronic thermal stress during a particular phase of growth. Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
Each of the six lifespan-extending PEs causes rapid degradation of neutral lipids deposited in lipid droplets
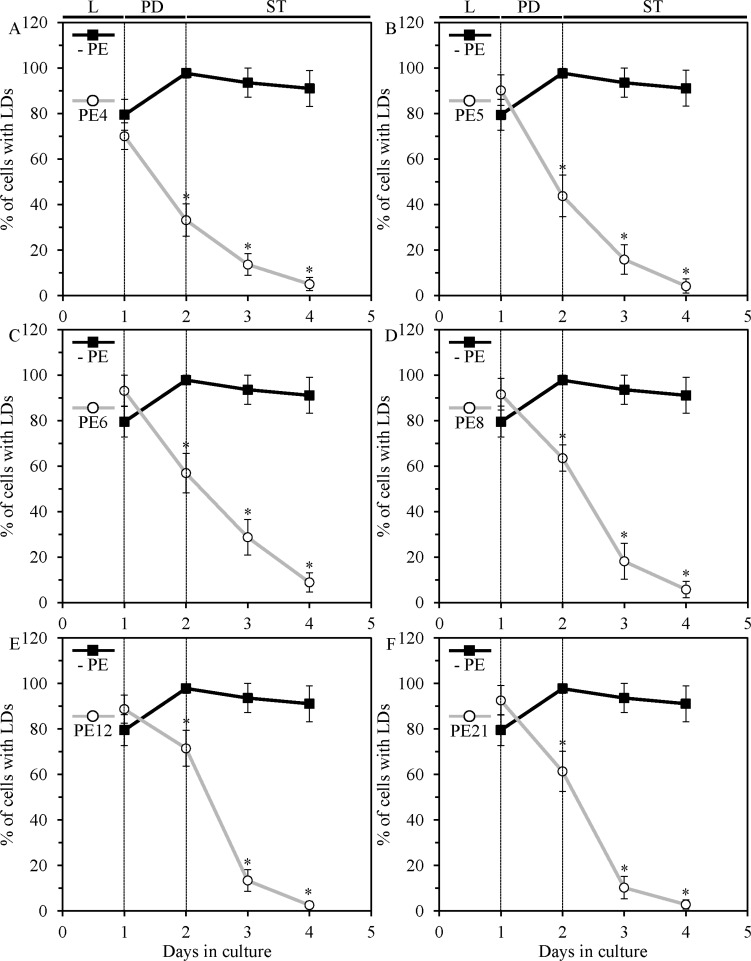
Triacylglycerols and steryl esters are uncharged (and therefore are called “neutral” or “nonpolar”) classes of lipids that can be found in cells of all eukaryotic organisms [81–83]. After being initially synthesized in the endoplasmic reticulum and then deposited in lipid droplets (LDs), these two highly hydrophobic lipids can undergo lipolytic degradation to provide substrates for the synthesis of phospholipids and sphingolipids [82, 84–87]. Emergent evidence supports the view that the biosynthesis, storage and lipolysis of neutral lipids are longevity assurance processes; importantly, it has been shown that these processes can be controlled by certain dietary and pharmacological interventions known to delay aging in various eukaryotes, including yeast [6, 23, 34, 87–108]. We therefore used live-cell fluorescence microscopy to examine how PE4, PE5, PE6, PE8, PE12 and PE21 influence the age-related dynamics of changes in the intracellular concentration of neutral lipids confined to LDs in chronologically aging yeast grown under non-CR conditions.
We found that each of the six lifespan-extending PEs elicits rapid age-related decline in the number of yeast cells exhibiting LDs (Figures 12A–12F; Figure S10 and S11). In contrast, no significant changes in the number of cells with LDs were seen in yeast progressing through L, PD and ST phases of culturing in medium without a PE (Figure 12A–12F; Figures S10 and S11). These findings demonstrate that in chronologically aging yeast grown under non-CR conditions, each of the six lifespan-extending PEs causes rapid lipolytic degradation of neutral lipids stored in LDs.
Figure 12. PE4, PE5, PE6, PE8, PE12 and PE21 induce rapid consumption of neutral lipids deposited in lipid droplets (LDs) of chronologically aging yeast grown under non-CR conditions.
WT cells were grown in the synthetic minimal YNB medium initially containing 2% glucose, in the presence of a PE or in its absence. Neutral lipids deposited in LDs were measured in live yeast by fluorescence microscopy of BODIPY 493/503 staining, as described in Materials and methods. Age-dependent changes in the percentage of WT cells exhibiting LDs in chronologically aging yeast cultures under non-CR conditions on 2% glucose with or without 0.5% PE4 (A), 0.5% PE5 (B), 1% PE6 (C), 0.3% PE8 (D), 0.1% PE12 (E) or 0.1% PE21 (F) are shown; data are presented as means ± SEM (n = 3–4; *p < 0.05; the p values for comparing the means of two groups were calculated with the help of the GraphPad Prism statistics software using an unpaired two-tailed t test). Abbreviations: Logarithmic (L), post-diauxic (PD) or stationary (ST) growth phase.
DISCUSSION
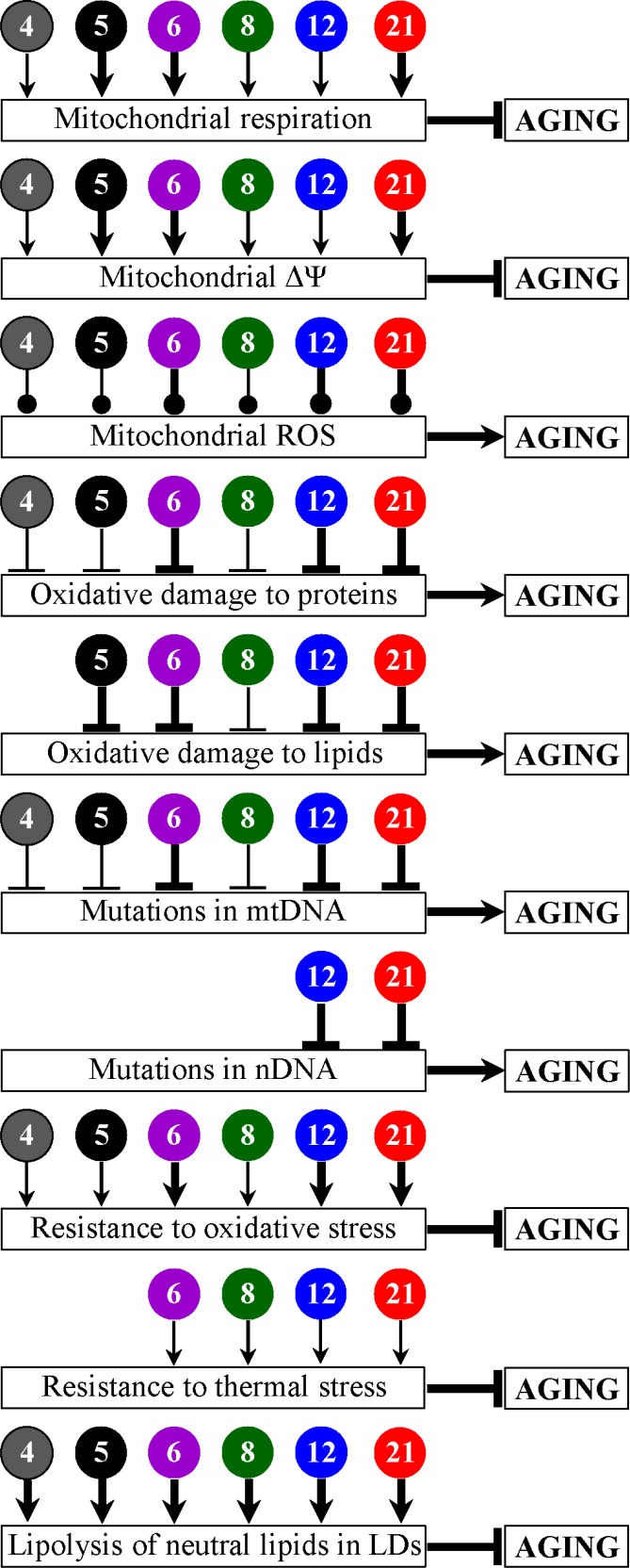
In this study, we performed a screen for PEs capable of extending longevity of the chronologically aging yeast S. cerevisiae. Our screen revealed six PEs (which we call PE4, PE5, PE6, PE8, PE12 and PE21) that can significantly increase yeast CLS. We demonstrated that each of these PEs is a geroprotector which delays the onset and slows the progression of yeast chronological aging by eliciting a hormetic stress response. We provided evidence that each of these geroprotective PEs has different effects on cellular processes known to define longevity in organisms across phyla. Such effects include the following: 1) amplified mitochondrial respiration and membrane potential; 2) increased or decreased concentrations of ROS; 3) reduced oxidative damage to cellular proteins, membrane lipids, and mitochondrial and nuclear genomes; 4) enhanced cell resistance to oxidative and thermal stresses; and 5) accelerated degradation of neutral lipids deposited in LDs (Figure 13). These findings provide important new insights into mechanisms through which some chemical compounds of plant origin can slow biological aging.
Figure 13. PE4, PE5, PE6, PE8, PE12 and PE21 delay yeast chronological aging and have different effects on several longevity-defining cellular processes.
Arrows pointing at boxes with the terms of longevity-defining cellular processes denote activation of these processes, T bars denote inhibition of these processes, whereas lines with filled circles denote change in the age-related chronology of intracellular ROS. The thickness of such arrows, T bars and lines with filled circles correlates with the extent to which a PE activates, inhibits or alters the age-related chronology (respectively) of a particular longevity-defining cellular process. Arrows and T bars pointing at boxes with the term “AGING” denote acceleration or deceleration (respectively) of yeast chronological aging.
Each of the six longevity-extending PEs increases lifespan more efficiently than any lifespan-prolonging chemical compound currently known
Our findings imply that the efficiency of longevity extension by PE4, PE5, PE6, PE8, PE12 or PE21 greatly exceeds that for any of the 42 chemical compounds known to increase lifespan in yeasts, filamentous fungi, nematodes, fruit flies, daphnias, mosquitoes, honey bees, fishes, mammals and cultured human cells (Table S1). Indeed, under non-CR conditions these longevity-extending PEs increase the mean and maximum CLS of yeast by 145%–475% and 80%–369%, respectively (Figure 3A and 3B; Table S1); the corresponding rows in Table S1 are highlighted in yellow. In contrast, any of the 42 currently known lifespan-extending chemical compounds has been shown to extend cellular and/or organismal lifespan in evolutionarily distant eukaryotes much less efficiently, within the 5% to 75% range (Table S1) [references 1–27, 30–57, 59–61 for Table S1]. Only two chemical compounds, spermidine under non-CR conditions and lithocholic acid under CR-conditions, have been reported to exhibit the lifespan-extending efficiencies that are comparable to those for PE4, PE5, PE6, PE8 and PE12 (Table S1) [references 28, 29, 58 for Table S1]. Specifically, both these pharmacological interventions were demonstrated to increase the RLS and/or CLS of yeast and human peripheral blood mononuclear cells by 83%–200%; the corresponding rows in Table S1 are highlighted in green. Of note, PE21 appears to be the most potent longevity-extending pharmacological intervention presently known. It increases the mean and maximum CLS of yeast by 475% and 369%, respectively (Figure 3A and 3B; Table S1).
Future perspectives
In the future, it would be important to further explore the following key aspects of the mechanisms through which each of the six longevity-extending PEs slows biological aging.
First, it is intriguing to identify the individual chemical compounds responsible for the ability of each of these PEs to delay the onset and decrease the rate of yeast chronological aging. Such identification is already underway in our laboratory; of note, it is conceivable that only some combinations of certain chemicals composing these PEs (but not individual chemical compounds per se) can be responsible for their extremely high efficiencies as aging-delaying interventions.
Second, it is interesting to elucidate how genetic interventions that impair any of the few nutrient- and energy-sensing signaling pathways known to define longevity of chronologically aging yeast [6, 10, 11, 23] influence the extent to which each of the six longevity-extending PEs can slow aging. These studies may allow to identify protein components of the longevity-defining signaling pathways that are targeted by each of the PEs. These studies may also reveal that certain combinations of these PEs and genetically impaired components of pro-aging signaling pathways exhibit additive or synergistic effects on the efficiencies of lifespan and healthspan extensions.
Third, it is important to investigate how various combinations of the six longevity-extending PEs with each other and with presently known aging-delaying chemical compounds alter the extent of CLS extension in yeast. These studies may identify such combinations of various pharmacological interventions that impose substantial additive or synergistic effects on the efficiencies with which organismal lifespan and healthspan can be prolonged.
Fourth, our ongoing studies indicate that the six longevity-extending PEs also extend longevities of other eukaryotic model organisms, delay the onset of age-related diseases and/or exhibit anti-tumor effects. In this regard, it needs to be mentioned that genetic, dietary and pharmacological interventions known to delay aging in yeast and other eukaryotes have been shown to selectively kill cultured human cancer cells and/or decrease the incidence of cancer [29, 88, 109–120]. The challenge for the future is to define mechanisms through which the six geroprotective PEs prolong healthy lifespan and decelerate tumorigenesis.
MATERIALS AND METHODS
Yeast strains, media and growth conditions
The wild-type strain Saccharomyces cerevisiae BY4742 (MATa his3D1 leu2D0 lys2D0 ura3D0) from Thermo Scientific/Open Biosystems was grown in a synthetic minimal YNB medium (0.67% Yeast Nitrogen Base without amino acids) initially containing 2% or 0.5% glucose and supplemented with 20 mg/l histidine, 30 mg/l leucine, 30 mg/l lysine and 20 mg/l uracil. Cells were cultured at 30°C with rotational shaking at 200 rpm in Erlenmeyer flasks at a “flask volume/medium volume” ratio of 5:1.
CLS assay
A sample of cells was taken from a culture at a certain day following cell inoculation and PE addition into the medium. A fraction of the sample was diluted in order to determine the total number of cells using a hemacytometer. Another fraction of the cell sample was diluted and serial dilutions of cells were plated in duplicate onto YEP (1% yeast extract, 2% peptone) plates containing 2% glucose as carbon source. After 2 d of incubation at 30°C, the number of colony forming units (CFU) per plate was counted. The number of CFU was defined as the number of viable cells in a sample. For each culture, the percentage of viable cells was calculated as follows: (number of viable cells per ml/total number of cells per ml) × 100. The percentage of viable cells in mid-logarithmic growth phase was set at 100%.
A screen for PEs that can extend yeast CLS
CLS analysis in the presence of various PEs was performed as described above. A 20% stock solution of each PE in ethanol was made on the day of adding this PE to cell cultures. For each PE, the stock solution was added to growth medium with 2% glucose immediately following cell inoculation into the medium. The final concentration of each PE in the medium was 0.02%, 0.04%, 0.06%, 0.08%, 0.1%, 0.3%, 0.5% or 1.0%.
Miscellaneous procedures
The age-specific mortality rate (qx) [37, 42], Gompertz slope or mortality rate coefficient (α) [41, 42], and mortality rate doubling time (MRDT) [41, 42] were calculated as previously described. Oxygen consumption assay for monitoring mitochondrial respiration [34], mitochondrial membrane potential measurement in live yeast [34], ROS measurement in live yeast [121], BODIPY 493/503 staining for examining neutral lipids deposited in LDs [122], fluorescence microscopy [34], quantitative assays for oxidatively damaged proteins and membrane lipids [64], measurements of the frequencies of spontaneous mutations in mitochondrial and nuclear DNA [123], and plating assays for the analysis of resistance to oxidative and thermal stresses [123] have been described elsewhere.
Statistical analysis
Statistical analysis was performed using Microsoft Excel's (2010) Analysis ToolPack-VBA. All data are presented as mean ± SEM. The p values for comparing the means of two groups (using an unpaired two-tailed t test) and survival curves (using a two-tailed t test) were calculated with the help of the GraphPad Prism statistics software.
SUPPLEMENTARY MATERIAL FIGURES AND TABLE
Acknowledgments
We acknowledge the Centre for Microscopy and Cellular Imaging at Concordia University for outstanding services.
Footnotes
CONFLICTS OF INTEREST
Vicky Lutchman, Younes Medkour, Eugenie Samson, Anthony Arlia-Ciommo, Pamela Dakik, Berly Cortes, Rachel Feldman, Sadaf Mohtashami, Mélissa McAuley Marisa Chancharoen, Belise Rukundo and Vladimir I. Titorenko have no conflicts of interest to declare. Éric Simard is the president of Idunn Technologies Inc.
FUNDING
This study was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada and Concordia University Chair Fund. E.S. was supported by the Concordia Undergraduate Research Award. A.A.-C. was supported by the Fonds québécois de la recherche sur la nature et les technologies Master's Research Scholarship Award. P.D. was supported by the Concordia University Graduate Fellowship Award. V.I.T. is a Concordia University Research Chair in Genomics, Cell Biology and Aging.
REFERENCES
- 1.Botstein D, Fink GR. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldmann H, editor. Yeast: Molecular and Cell Biology. 2. Wiley-Blackwell; Weinheim, Germany: 2012. p. 464. [Google Scholar]
- 3.Lachance M-A. eLS. Chichester, West Sussex, UK: John Wiley & Sons; 2010. Yeasts; pp. 1–12. [Google Scholar]
- 4.Smith JS, Burke DJ, editors. Springer; New York, USA: 2014. Yeast Genetics: Methods and Protocols; p. 378. [Google Scholar]
- 5.Weissman J, Guthrie C, Fink GR, editors. Guide to Yeast Genetics: Functional Genomics, Proteomics, and Other Systems Analysis. Academic Press; Burlington, USA: 2010. p. 892. [Google Scholar]
- 6.Arlia-Ciommo A, Leonov A, Piano A, Svistkova V, Titorenko VI. Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microbial Cell. 2014a;1:164–178. doi: 10.15698/mic2014.06.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arlia-Ciommo A, Piano A, Leonov A, Svistkova V, Titorenko VI. Quasi-programmed aging of budding yeast: a trade-off between programmed processes of cell proliferation, differentiation, stress response, survival and death defines yeast lifespan. Cell Cycle. 2014b;13:3336–3349. doi: 10.4161/15384101.2014.965063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denoth Lippuner A, Julou T, Barral Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev. 2014;38:300–325. doi: 10.1111/1574-6976.12060. [DOI] [PubMed] [Google Scholar]
- 9.Fontana L, Partridge L, Longo VD. Extending healthy life span - from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyström T, Liu B. Protein quality control in time and space - links to cellular aging. FEMS Yeast Res. 2014;14:40–48. doi: 10.1111/1567-1364.12095. [DOI] [PubMed] [Google Scholar]
- 13.Váchová L, Cáp M, Palková Z. Yeast colonies: a model for studies of aging, environmental adaptation, and longevity. Oxid Med Cell Longev. 2012;2012:601836. doi: 10.1155/2012/601836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longo VD, Fabrizio P. Chronological aging in Saccharomyces cerevisiae. Subcell Biochem. 2012;57:101–121. doi: 10.1007/978-94-007-2561-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piper PW. Maximising the yeast chronological lifespan. Subcell Biochem. 2012;57:145–159. doi: 10.1007/978-94-007-2561-4_7. [DOI] [PubMed] [Google Scholar]
- 16.Sutphin GL, Olsen BA, Kennedy BK, Kaeberlein M. Genome-wide analysis of yeast aging. Subcell Biochem. 2012;57:251–289. doi: 10.1007/978-94-007-2561-4_12. [DOI] [PubMed] [Google Scholar]
- 17.Evans DS, Kapahi P, Hsueh WC, Kockel L. TOR signaling never gets old: aging, longevity and TORC1 activity. Ageing Res Rev. 2011;10:225–227. doi: 10.1016/j.arr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jazwinski SM. The retrograde response and other pathways of interorganelle communication in yeast replicative aging. Subcell Biochem. 2012;57:79–100. doi: 10.1007/978-94-007-2561-4_4. [DOI] [PubMed] [Google Scholar]
- 20.Jazwinski SM. The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta. 2013;1833:400–409. doi: 10.1016/j.bbamcr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Cabo R, Carmona-Gutierrez D, Bernier M, Hall MN, Madeo F. The search for antiaging interventions: from elixirs to fasting regimens. Cell. 2014;157:1515–1526. doi: 10.1016/j.cell.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg AA, Richard VR, Kyryakov P, Bourque SD, Beach A, Burstein MT, Glebov A, Koupaki O, Boukh-Viner T, Gregg C, Juneau M, English AM, Thomas DY, et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging (Albany NY) 2010a;2:393–414. doi: 10.18632/aging.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbard BP, Sinclair DA. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol Sci. 2014;35:146–154. doi: 10.1016/j.tips.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonov A, Arlia-Ciommo A, Piano A, Svistkova V, Lutchman V, Medkour Y, Titorenko VI. Longevity extension by phytochemicals. Molecules. 2015;20:6544–6572. doi: 10.3390/molecules20046544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011;3:716–732. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinclair DA, Guarente L. Small-molecule allosteric activators of sirtuins. Annu Rev Pharmacol Toxicol. 2014;54:363–380. doi: 10.1146/annurev-pharmtox-010611-134657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M. The biology of aging: citizen scientists and their pets as a bridge between research on model organisms and human subjects. Vet Pathol. 2015 doi: 10.1177/0300985815591082. pii:0300985815591082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of Aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitt JN, Kaeberlein M. Why is aging conserved and what can we do about it? PLoS Biol. 2015;13:e1002131. doi: 10.1371/journal.pbio.1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argyropoulou A, Aligiannis N, Trougakos IP, Skaltsounis AL. Natural compounds with anti-ageing activity. Nat Prod Rep. 2013;30:1412–1437. doi: 10.1039/c3np70031c. [DOI] [PubMed] [Google Scholar]
- 33.Menendez JA, Joven J, Aragonès G, Barrajón-Catalán E, Beltrán-Debón R, Borrás-Linares I, Camps J, Corominas-Faja B, Cufí S, Fernández-Arroyo S, Garcia-Heredia A, Hernández-Aguilera A, Herranz-López M, et al. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: a new family of gerosuppressant agents. Cell Cycle. 2013;12:555–578. doi: 10.4161/cc.23756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldberg AA, Bourque SD, Kyryakov P, Gregg C, Boukh-Viner T, Beach A, Burstein MT, Machkalyan G, Richard V, Rampersad S, Cyr D, Milijevic S, Titorenko VI. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp Gerontol. 2009a;44:555–571. doi: 10.1016/j.exger.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Abrams PA. Evolutionary biology: mortality and lifespan. Nature. 2004;431:1048–1049. doi: 10.1038/4311048a. [DOI] [PubMed] [Google Scholar]
- 36.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–747. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 37.McDonald RB. Biology of aging. Garland Science, Taylor & Francis Group, LLC; 2014. pp. 1–54. Chapters 1 and 2. [Google Scholar]
- 38.Medkour Y, Svistkova V, Titorenko VI. Cell-non-autonomous mechanisms underlying cellular and organismal aging. Int Rev Cell Mol Biol. 2016;321 doi: 10.1016/bs.ircmb.2015.09.003. in press. [DOI] [PubMed] [Google Scholar]
- 39.Blagosklonny MV. Answering theultimate question “what is the proximal cause of aging?”. Aging (Albany NY) 2012;4:861–877. doi: 10.18632/aging.100525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fries JF, Bruce B, Chakravarty E. Compression of morbidity 1980–2011: a focused review of paradigms and progress. J Aging Res. 2011;2011:261702. doi: 10.4061/2011/261702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Magalhães JP, Cabral JA, Magalhães D. The influence of genes on the aging process of mice: a statistical assessment of the genetics of aging. Genetics. 2005;169:265–274. doi: 10.1534/genetics.104.032292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finch CE. Longevity, senescence, and the genome. University of Chicago Press; Chicago: 1990. [Google Scholar]
- 43.Gavrilov LA, Gavrilova NS. The biology of life span: a quantitative approach. New York, New York/Chur, Switzerland: Harwood Academic; 1991. [Google Scholar]
- 44.Lashmanova E, Proshkina E, Zhikrivetskaya S, Shevchenko O, Marusich E, Leonov S, Melerzanov A, Zhavoronkov A, Moskalev A. Fucoxanthin increases lifespan of Drosophila melanogaster and Caenorhabditis elegans. Pharmacol Res. 2015;100:228–241. doi: 10.1016/j.phrs.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Burstein MT, Beach A, Richard VR, Koupaki O, Gomez-Perez A, Goldberg AA, Kyryakov P, Bourque SD, Glebov A, Titorenko VI. Interspecies chemical signals released into the environment may create xenohormetic, hormetic and cytostatic selective forces that drive the ecosystemic evolution of longevity regulation mechanisms. Dose Response. 2012a;10:75–82. doi: 10.2203/dose-response.11-011.Titorenko. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calabrese V, Cornelius C, Dinkova-Kostova AT, Iavicoli I, Di Paola R, Koverech A, Cuzzocrea S, Rizzarelli E, Calabrese EJ. Cellular stress responses, hormetic phytochemicals and vitagenes in aging and longevity. Biochim Biophys Acta. 2012;1822:753–783. doi: 10.1016/j.bbadis.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Calabrese EJ, Mattson MP. Hormesis provides a generalized quantitative estimate of biological plasticity. J Cell Commun Signal. 2011;5:25–38. doi: 10.1007/s12079-011-0119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg AA, Kyryakov P, Bourque SD, Titorenko VI. Xenohormetic, hormetic and cytostatic selective forces driving longevity at the ecosystemic level. Aging (Albany NY) 2010b;2:461–470. doi: 10.18632/aging.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beach A, Leonov A, Arlia-Ciommo A, Svistkova V, Lutchman V, Titorenko VI. Mechanisms by which different functional states of mitochondria define yeast longevity. Int J Mol Sci. 2015a;16:5528–5554. doi: 10.3390/ijms16035528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagliarini DJ, Rutter J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 2013;27:2615–2627. doi: 10.1101/gad.229724.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruetenik A, Barrientos A. Dietary restriction, mitochondrial function and aging: from yeast to humans. Biochim Biophys Acta. 2015;1847:1434–1447. doi: 10.1016/j.bbabio.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shadel GS, Horvath TL. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Hekimi S. Mitochondrial dysfunction and longevity in animals: Untangling the knot. 2015;350:1204–1207. doi: 10.1126/science.aac4357. [DOI] [PubMed] [Google Scholar]
- 55.Beach A, Richard VR, Leonov A, Burstein MT, Bourque SD, Koupaki O, Juneau M, Feldman R, Iouk T, Titorenko VI. Mitochondrial membrane lipidome defines yeast longevity. Aging (Albany NY) 2013;5:551–574. doi: 10.18632/aging.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beach A, Richard VR, Bourque S, Boukh-Viner T, Kyryakov P, Gomez-Perez A, Arlia-Ciommo A, Feldman R, Leonov A, Piano A, Svistkova V, Titorenko VI. Lithocholic bile acid accumulated in yeast mitochondria orchestrates a development of an anti-aging cellular pattern by causing age-related changes in cellular proteome. Cell Cycle. 2015b;14:1643–1656. doi: 10.1080/15384101.2015.1026493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burstein MT, Titorenko VI. A mitochondrially targeted compound delays aging in yeast through a mechanism linking mitochondrial membrane lipid metabolism to mitochondrial redox biology. Redox Biol. 2014;2:305–307. doi: 10.1016/j.redox.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leonov A, Titorenko VI. A network of interorganellar communications underlies cellular aging. IUBMB Life. 2013;65:665–674. doi: 10.1002/iub.1183. [DOI] [PubMed] [Google Scholar]
- 60.Martins D, Titorenko VI, English AM. Cells with impaired mitochondrial H2O2 sensing generate less •OH radicals and live longer. Antioxid Redox Signal. 2014;21:1490–1503. doi: 10.1089/ars.2013.5575. [DOI] [PubMed] [Google Scholar]
- 61.Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY) 2009;1:131–145. doi: 10.18632/aging.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richard VR, Leonov A, Beach A, Burstein MT, Koupaki O, Gomez-Perez A, Levy S, Pluska L, Mattie S, Rafeh R, Iouk T, Sheibani S, Greenwood M, et al. Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis. Aging (Albany NY) 2013;5:234–269. doi: 10.18632/aging.100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroeder EA, Shadel GS. Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan. Mech Ageing Dev. 2014;135:41–49. doi: 10.1016/j.mad.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Titorenko VI, Terlecky SR. Peroxisome metabolism and cellular aging. Traffic. 2011;12:252–259. doi: 10.1111/j.1600-0854.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 69.Ristow M, Schmeisser K. Mitohormesis: Promoting health and lifespan by increased levels of reactive oxygen species (ROS) Dose Response. 2014;12:288–341. doi: 10.2203/dose-response.13-035.Ristow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beach A, Burstein MT, Richard VR, Leonov A, Levy S, Titorenko VI. Integration of peroxisomes into an endomembrane system that governs cellular aging. Front Physiol. 2012;3:283. doi: 10.3389/fphys.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct. 2012;2012:646354. doi: 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gladyshev VN. The origin of aging: imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet. 2013;29:506–512. doi: 10.1016/j.tig.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gladyshev VN. The free radical theory of aging is dead. Long live the damage theory! Antioxid Redox Signal. 2014;20:727–731. doi: 10.1089/ars.2013.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calabrese V, Cornelius C, Cuzzocrea S, Iavicoli I, Rizzarelli E, Calabrese EJ. Hormesis, cellular stress response and vitagenes as critical determinants in aging and longevity. Mol Aspects Med. 2011;32:279–304. doi: 10.1016/j.mam.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 77.D'Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 78.Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 79.Veal E, Day A. Hydrogen peroxide as a signaling molecule. Antioxid Redox Signal. 2011;15:147–151. doi: 10.1089/ars.2011.3968. [DOI] [PubMed] [Google Scholar]
- 80.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koch B, Schmidt C, Daum G. Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol Rev. 2014;38:892–915. doi: 10.1111/1574-6976.12069. [DOI] [PubMed] [Google Scholar]
- 82.Kohlwein SD, Veenhuis M, van der Klei IJ. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat - store ‘em up or burn ‘em down. Genetics. 2013;193:1–50. doi: 10.1534/genetics.112.143362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klug L, Daum G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014;14:369–388. doi: 10.1111/1567-1364.12141. [DOI] [PubMed] [Google Scholar]
- 85.Ohsaki Y, Suzuki M, Fujimoto T. Open questions in lipid droplet biology. Chem Biol. 2014;21:86–96. doi: 10.1016/j.chembiol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 86.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang CW. Lipid droplet dynamics in budding yeast. Cell Mol Life Sci. 2015;72:2677–2695. doi: 10.1007/s00018-015-1903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arlia-Ciommo A, Piano A, Svistkova V, Mohtashami S, Titorenko VI. Mechanisms underlying the anti-aging and anti-tumor effects of lithocholic bile acid. Int J Mol Sci. 2014c;15:16522–16543. doi: 10.3390/ijms150916522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beach A, Titorenko VI. In search of housekeeping pathways that regulate longevity. Cell Cycle. 2011;10:3042–3044. doi: 10.4161/cc.10.18.16947. [DOI] [PubMed] [Google Scholar]
- 90.Beach A, Titorenko VI. Essential roles of peroxisomally produced and metabolized biomolecules in regulating yeast longevity. Subcell Biochem. 2013;69:153–167. doi: 10.1007/978-94-007-6889-5_9. [DOI] [PubMed] [Google Scholar]
- 91.Blüher M. Fat tissue and long life. Obes Facts. 2008;1:176–182. doi: 10.1159/000145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 93.Chiu CH, Lin WD, Huang SY, Lee YH. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970–1975. doi: 10.1101/gad.1213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goldberg AA, Bourque SD, Kyryakov P, Boukh-Viner T, Gregg C, Beach A, Burstein MT, Machkalyan G, Richard V, Rampersad S, Titorenko VI. A novel function of lipid droplets in regulating longevity. Biochem Soc Trans. 2009b;37:1050–1055. doi: 10.1042/BST0371050. [DOI] [PubMed] [Google Scholar]
- 95.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 97.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 98.Krahmer N, Farese RV, Jr., Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- 100.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richard VR, Beach A, Piano A, Leonov A, Feldman R, Burstein MT, Kyryakov P, Gomez-Perez A, Arlia-Ciommo A, Baptista S, Campbell C, Goncharov D, Pannu S, et al. Mechanism of liponecrosis, a distinct mode of programmed cell death. Cell Cycle. 2014a;13:3707–3726. doi: 10.4161/15384101.2014.965003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richard VR, Bourque SD, Titorenko VI. Metabolomic and lipidomic analyses of chronologically aging yeast. Methods Mol Biol. 2014b;1205:359–373. doi: 10.1007/978-1-4939-1363-3_21. [DOI] [PubMed] [Google Scholar]
- 104.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 105.Sheibani S, Richard VR, Beach A, Leonov A, Feldman R, Khelghatybana L, Piano A, Greenwood M, Vali H, Titorenko VI. Macromitophagy, neutral lipids synthesis and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell Cycle. 2014;13:138–147. doi: 10.4161/cc.26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–R481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS - lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arlia-Ciommo A, Svistkova V, Mohtashami S, Titorenko VI. A novel approach to the discovery of anti-tumor pharmaceuticals: searching for activators of liponecrosis. Oncotarget. 2016 doi: 10.18632/oncotarget.6440. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blagosklonny MV. Aging and immortality: quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle. 2006;5:2087–2102. doi: 10.4161/cc.5.18.3288. [DOI] [PubMed] [Google Scholar]
- 111.Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging (Albany NY) 2009;1:357–362. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goldberg AA, Beach A, Davies GF, Harkness TA, Leblanc A, Titorenko VI. Lithocholic bile acid selectively kills neuroblastoma cells, while sparing normal neuronal cells. Oncotarget. 2011;2:761–782. doi: 10.18632/oncotarget.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goldberg AA, Titorenko VI, Beach A, Sanderson JT. Bile acids induce apoptosis selectively in androgen-dependent and -independent prostate cancer cells. PeerJ. 2013;1:e122. doi: 10.7717/peerj.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 116.Kaeberlein M. Longevity and aging. F1000Prime Rep. 2013;5:5. doi: 10.12703/P5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22:R741–R752. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 118.Partridge L. Intervening in ageing to prevent the diseases of ageing. Trends Endocrinol Metab. 2014;25:555–557. doi: 10.1016/j.tem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 119.Piano A, Titorenko VI. The Intricate interplay between mechanisms underlying aging and cancer. Aging Dis. 2014;6:56–75. doi: 10.14336/AD.2014.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kyryakov P, Beach A, Richard VR, Burstein MT, Leonov A, Levy S, Titorenko VI. Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front Physiol. 2012;3:256. doi: 10.3389/fphys.2012.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bascom RA, Chan H, Rachubinski RA. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants. Mol Biol Cell. 2003;14:939–957. doi: 10.1091/mbc.E02-10-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Burstein MT, Kyryakov P, Beach A, Richard VR, Koupaki O, Gomez-Perez A, Leonov A, Levy S, Noohi F, Titorenko VI. Lithocholic acid extends longevity of chronologically aging yeast only if added at certain critical periods of their lifespan. Cell Cycle. 2012b;11:3443–3462. doi: 10.4161/cc.21754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.