Abstract
Much is known about vertebrate DNA methylation and oxidation; however, much less is known about how modified cytosine residues within particular sequences are recognized. Among the known methylated DNA-binding domains, the Cys2-His2 zinc finger (ZnF) protein superfamily is the largest with hundreds of members, each containing tandem ZnFs ranging from 3 to >30 fingers. We have begun to biochemically and structurally characterize these ZnFs not only on their sequence specificity but also on their sensitivity to various DNA modifications. Rather than following published methods of refolding insoluble ZnF arrays, we have expressed and purified soluble forms of ZnFs, ranging in size from a tandem array of two to six ZnFs, from seven different proteins. We also describe a fluorescence polarization assay to measure ZnFs affinity with oligonucleotides containing various modifications and our approaches for cocrystallization of ZnFs with oligonucleotides.
1. INTRODUCTION
The control of gene expression in mammals relies significantly on the modification status of DNA cytosine residues. DNA cytosine modification is a dynamic process catalyzed by specific DNA methyltransferases (DNMTs) that convert cytosine (C) to 5-methylcytosine (abbreviated 5mC or M; Bestor, Laudano, Mattaliano, & Ingram, 1988; Okano, Xie, & Li, 1998), usually within the sequence context of CpG (Bestor et al., 1988; Okano, Bell, Haber, & Li, 1999; Okano et al., 1998) or CpA (Gowher & Jeltsch, 2001; Kubo et al., 2015; Lister et al., 2013, 2009; Ramsahoye et al., 2000; Vlachogiannis et al., 2015). A subset of 5mC may then be oxidized to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the ten-eleven translocation (Tet) dioxygenases in three consecutive Fe(II) and α-ketoglutarate-dependent oxidation reactions (He et al., 2011; Ito et al., 2010, 2011; Tahiliani et al., 2009).
The best-known modified DNA-recognition domains are two that recognize methylated cytosine: methyl-binding domains (MBDs) recognize fully methylated CpG dinucleotides (Dhasarathy & Wade, 2008; Guy, Cheval, Selfridge, & Bird, 2011), and “SET and RING finger-associated” (SRA) domains that bind hemimethylated CpG sites generated transiently by DNA replication (Hashimoto, Horton, Zhang, & Cheng, 2009; Sharif & Koseki, 2011; reviewed in Hashimoto, Zhang, Vertino, & Cheng, 2015; Liu, Zhang, Blumenthal, & Cheng, 2013). Both MBD and SRA domains have been structurally characterized in complexes with 5mC (Arita, Ariyoshi, Tochio, Nakamura, & Shirakawa, 2008; Avvakumov et al., 2008; Hashimoto et al., 2008; Ho et al., 2008; Ohki et al., 2001; Scarsdale, Webb, Ginder, & Williams, 2011).
A third class of mammalian proteins that can recognize methylated DNA is the Cys2-His2 (C2H2) zinc finger (ZnF) proteins, which can preferentially bind to methylated CpG within a longer specific sequence (Sasai, Nakao, & Defossez, 2010). Kaiso is the first known methyl-binding ZnF protein that belongs to the BTB/POZ family (Prokhortchouk et al., 2001), which also includes ZBTB24, whose mutations are associated with immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome (Cerbone et al., 2012; Chouery et al., 2012; de Greef et al., 2011; Nitta et al., 2013), a disease also caused by mutations in a DNA methyl-transferase gene, DNMT3B (Hansen et al., 1999; Okano et al., 1999; Shirohzu et al., 2002; Xu et al., 1999). Recently, ZnF DNA-binding domains from five proteins, Kaiso, Zfp57, Klf4, Egr1, and WT1, have been structurally analyzed in complex with their respective methylated DNA elements (Buck-Koehntop et al., 2012; Hashimoto et al., 2014; Liu et al., 2014; Liu, Toh, Sasaki, Zhang, & Cheng, 2012; Zandarashvili, White, Esadze, & Iwahara, 2015), allowing comparison to other 5mC-binding proteins. In addition, WT1 binds 5caC DNA, as does a mutant Zfp57 (Hashimoto et al., 2014; Liu, Olanrewaju, Zhang, & Cheng, 2013).
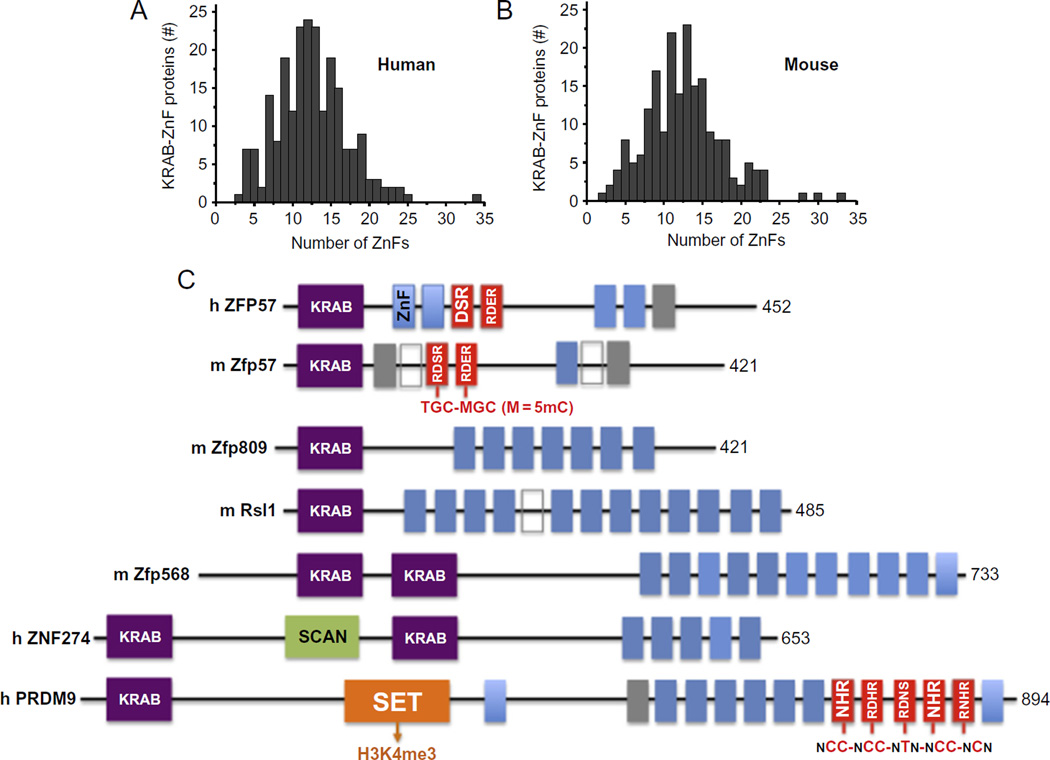
Among the C2H2 ZnF proteins, KRAB-ZnF transcription factors (KRAB-ZnFs) act mostly as chromatin-modulating transcription repressors (Meylan et al., 2011). Of the >300 human or mouse KRAB-ZnF proteins examined, the number of tandem ZnFs ranges from 3 to 35, with a mode of around 11–13 fingers (Liu, Zhang, et al., 2013; Fig. 1A and B). The domain structures of a few examples of mammalian KRAB-ZnF proteins with known biological roles are shown (Fig. 1C). ZFP57 mutations have been found in patients with transient neonatal diabetes (Mackay et al., 2008). Zfp809 restricts retroviral transposition in embryonic stem cells (Wolf & Goff, 2009), and retroviral silencing has been suggested to be the ancestral role of KRAB-ZnFs (Thomas & Schneider, 2011). Regulator of sex limitation (Rsl1) regulates sex- and tissue-specific promoter methylation (Krebs, Schultz, & Robins, 2012). Zfp568 regulates extraembryonic tissue morphogenesis (Garcia-Garcia, Shibata, & Anderson, 2008). Like Zfp57 (Quenneville et al., 2011; Zuo et al., 2012), ZNF274 recruits the histone H3 lysine 9 methyltransferase SETDB1 (SET domain, bifurcated 1; Frietze, O’Geen, Blahnik, Jin, & Farnham, 2010) via the corepressor TRIM28 (tripartite motif-containing 28; also known as KAP1 for Krüppel-associated protein), an essential regulator of genomic imprinting (Messerschmidt et al., 2012). PRDM9 (PR domain zinc finger protein 9), a major determinant of meiotic recombination hotspots, contains a SET domain that methylates histone H3 lysine 4 (Mihola, Trachtulec, Vlcek, Schimenti, & Forejt, 2009). These examples further illustrate the coordinated chromatin controls between DNA methylation and the lysine methylation status of histone H3 (at residues 4 and 9; Cheng & Blumenthal, 2010).
Fig. 1.
KRAB-ZnF proteins. The SysZNF database was examined for Krüppel-associated box (KRAB)-ZnF proteins in human (A) or mouse (B). In each case, the distribution of KRAB-ZnF proteins containing a given number of ZnF repeats is shown. (C) Examples of mammalian KRAB-ZnF proteins with known biologic roles. The size of each protein (in amino acids) is shown on the right. The classic C2H2 ZnF motifs are shown in blue (gray in the print version) boxes and the gray or open boxes indicate degenerate ZnFs that contain mutations affecting zinc coordination. The red (gray in the print version) boxes of mouse Zfp57 and human PRDM9 indicate the structurally characterized ZnFs in complex with their recognition sequences shown below. The amino acids within the red (gray in the print version) box indicate the three or four residues of each ZnF involved in base specific interactions. The SCAN box, a leucine-rich region, was named after SRE-ZBP, CTfin51, AW-1 (ZNF174), Number 18 cDNA (ZnF20) (Williams, Khachigian, Shows, & Collins, 1995). The SET domain was named after Su(var)3–9, Enhancer of zeste, Trithorax (Jenuwein, Laible, Dorn, & Reuter, 1998). Modified from Liu, Y., Zhang, X., Blumenthal, R. M., & Cheng, X. (2013). A common mode of recognition for methylated CpG. Trends in Biochemical Sciences, 38, 177–183.
In the last few years, we have biochemically and structurally characterized mouse Zfp57 (2 ZnFs) (Liu, Olanrewaju, et al., 2013; Liu et al., 2012), mouse Krüppel-like factor 4 (Klf4) (3 ZnFs) (Liu et al., 2014), human early growth response protein (Egr1, also known as Zif268) (3 ZnFs) (Hashimoto et al., 2014), human Wilms tumor protein (WT1) (3 ZnFs) (Hashimoto et al., 2014), human PRDM9 allele-A (5 ZnFs) (Patel, Horton, Wilson, Zhang, & Cheng, 2016), human PRDM9 allele-C (6 ZnFs) (Patel et al., unpublished), and human CTCF (4 ZnFs) (Hashimoto et al., unpublished). Rather than following published methods of refolding insoluble proteins, such as WT1 (Laity, Chung, Dyson, & Wright, 2000) and Egr1/Zif268 (Pavletich & Pabo, 1991), we expressed and purified the tandem array of ZnF DNA-binding domains in soluble form as fusion proteins with glutathione S-transferase (GST). One key to our success in obtaining quality ZnF protein suitable for biochemistry analysis and crystallization is the use of polyethylenimine (PEI), a polymer with repeating unit composed of the amine and two carbon aliphatic spacer, and anion exchange column to completely remove bacterial nucleic acids associated with the ZnF proteins.
2. SOLUBLE EXPRESSION AND PURIFICATION OF ZnF PROTEINS
ZnF cDNA fragments were cloned into the BamHI site of pGEX6p-1 vector (GE Healthcare), leaving five extra residues at the N-terminal, Gly-Pro-Leu-Gly-Ser, after PreScission protease cleavage.
2.1 Expression
Day 1: Inoculate 15 mL of noninducing MDAG media (Studier, 2005) supplemented with 100 µg/mL sterile ampicillin, with a single colony or glycerol freezer stock of Escherichia coli BL21 (DE3) Codon-plus RIL cells containing the expression plasmid. Incubate overnight with shaking at 37°C.
- Day 2: Inoculate 1 L LB medium supplemented with 100 µg/mL ampicillin with 2 mL of the starter culture. Grow at 37°C with shaking at 200 rpm until the A600 reaches ~0.5 when the shaker temperature is reduced to 16°C.Note: Monitor the liquid temperature with a thermometer until it has reached 16°C, which can take 0.5–1.5 h depending on shaker. Add 200 µL of 0.5 M ZnCl2 (to a final concentration of 100 µM) and induce the expression of protein by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to 0.2 mM final concentration. Incubate overnight with shaking at 16°C.
Day 3: Harvest the cells by centrifugation at 3500 rpm for 30 min at 4°C and freeze the pellets at −20°C until purification.
2.2 Purification
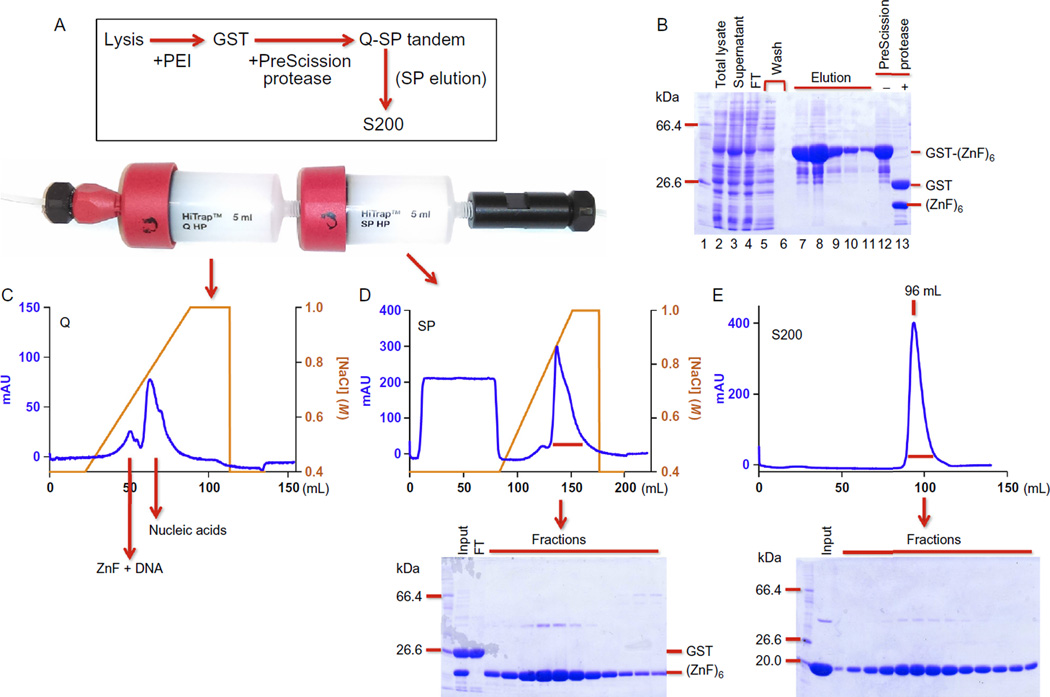
The general scheme involves four-column chromatography (Fig. 2A).
Fig. 2.
One example of ZnF protein purification. (A) The general scheme of four-column chromatography. Inserted is a picture of tandem Q-SP columns. (B) A 12% SDS-PAGE showing total lysate (lane 2), supernatant after PEI treatment (lane 3), flowthrough (FT; lane 4), and washing through the GST column (lanes 5 and 6), five elutions by GSH (lanes 7–11), and before and after PreScission protease cleavage (lanes 12 and 13). Note that more soluble protein appears after PEI treatment (comparing lanes 2 and 3). (C) Elution profile of the HiTrap Q column. (D) Elution profile of the HiTrap SP column and an accompanying SDS-PAGE showing the fractions. (E) Elution profile of a Superdex-200 (16/60 GL) column and an accompanying SDS-PAGE showing the fractions.
Day 1
- Resuspend cells from 6 L culture into 120 mL of lysis buffer [20 mM Tris (pH 7.5), 5% (v/v) glycerol, 25 µM ZnCl2, 0.5mM tris(2-carboxyethyl) phosphine (TCEP), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] containing 250–700 mM NaCl.Note: The salt concentration needs to be individually determined to assure maximum solubility.
Lyse cells by sonication with 1 s on and 2 s off cycles for 8 min in total.
- Treat the lysate with PEI (Sigma—408727) neutralized by HCl to pH 7.Note: Slowly add 6 mL of 2% (w/v) PEI solution drop by drop into the lysate to a final concentration of 0.1% while stirring on an ice bath.
Clear the lysate by centrifugation at 16,500 rpm for 45 min at 4°C.
Load the supernatant onto a Glutathione Sepharose 4B column (GE Healthcare) with 5 mL bed volume equilibrated with 250–700 mM NaCl lysis buffer (see Step 1) at ~0.7 mL/min flow rate. Wash the column with 40 mL of lysis buffer followed by 25 mL of washing buffer containing 100 mM Tris (pH 8.0), 500 mM NaCl, 5% glycerol, 25 µM ZnCl2, and 0.5 mM TCEP. Elute the GST-tagged protein with 50 mL of elution buffer (washing buffer + 20 mM reduced glutathione) into fractions of 10 mL (Fig. 2B).
Remove the GST tag by treating the eluted protein with ~100 µg of PreScission protease (GE Healthcare 27-0843-01 or purified in-house) at 4°C overnight.
Day 2
- Load the protein onto 5 mL HiTrap Q-SP columns connected in tandem (GE Healthcare; Fig. 2A), equilibrated with column buffer (lysis buffer minus PMSF) with 500 mM NaCl. After washing 20 mL with the same buffer, disconnect the Q and SP columns and elute separately with a 50 mL (or 75) linear gradient of 0.5–1 M NaCl (Fig. 2C and D).Note: DNA-free protein flows through the Q column and binds the SP column, while the DNA-containing protein binds to the Q column along with free DNA (Fig. 2C).
Concentrate the protein eluted from the SP column to ~2 mL using a centrifugal concentrator such as Vivaspin. Load onto a Superdex-200 (16/60) column equilibrated with column buffer with 500 mM NaCl. Collect the protein eluted as a single peak (Fig. 2E). Concentrate to about 5 mg/mL and flash freeze with liquid nitrogen and store at −80°C in aliquots. Final yields of the protein range from 10 to 15 mg/6 L culture.
3. FLUORESCENCE POLARIZATION ASSAY FOR ANALYSIS OF DNA BINDING
- Synthesize 5′-FAM (6-carboxyfluorescein)-labeled oligonucleotides containing various cytosine modifications. Only one strand is labeled.Note: Generally, blunt-ended DNA duplex is preferred, although sometimes labeling at one (or two) base overhang at the 5′-end yields better signal (Hashimoto et al., 2014; Liu et al., 2014, 2012). The FAM label may not be compatible with 5caC modification within the same strand due to currently available synthesis chemistry.
Mix twofold serially diluted protein solutions (1–10 µMstarting concentration, 10–15 points) with 1–5 nMfinal concentration of DNA probe in a Corning 3575 plate, using binding buffer of 20 mM Tris–HCl, pH 7.5, 5% glycerol, and 0.5 mM TCEP with varying NaCl concentration (150–300 mM). Incubate the mixture for 10 min at room temperature. Perform at least two duplicate experiments.
- Measure fluorescence polarization at 25°C on a Synergy 4 Microplate Reader (BioTek) using 485/20 nm and 528/20 nm filters for emission and excitation, respectively.Note: The presence of protein should cause no change in fluorescence intensity.
- Calculate the dissociation constants (KD) by fitting the experimental data to the following equation using GraphPad Prism software (version 6.0): [mP]=[maximum mP] × [C] / (KD + [C]) + [baseline mP], then replot the curve using % of saturation calculated as ([mP] − [baseline mP])/([maximum mP] − [baseline mP]), where mP is millipolarization and [C] is protein concentration.Note: The maximum increase of mP observed is protein and/or oligonucleotide dependent, and is most commonly between 50 and 100 mP. Very large mP change (>200) at high protein concentration (>1 µM) often indicates nonspecific binding.
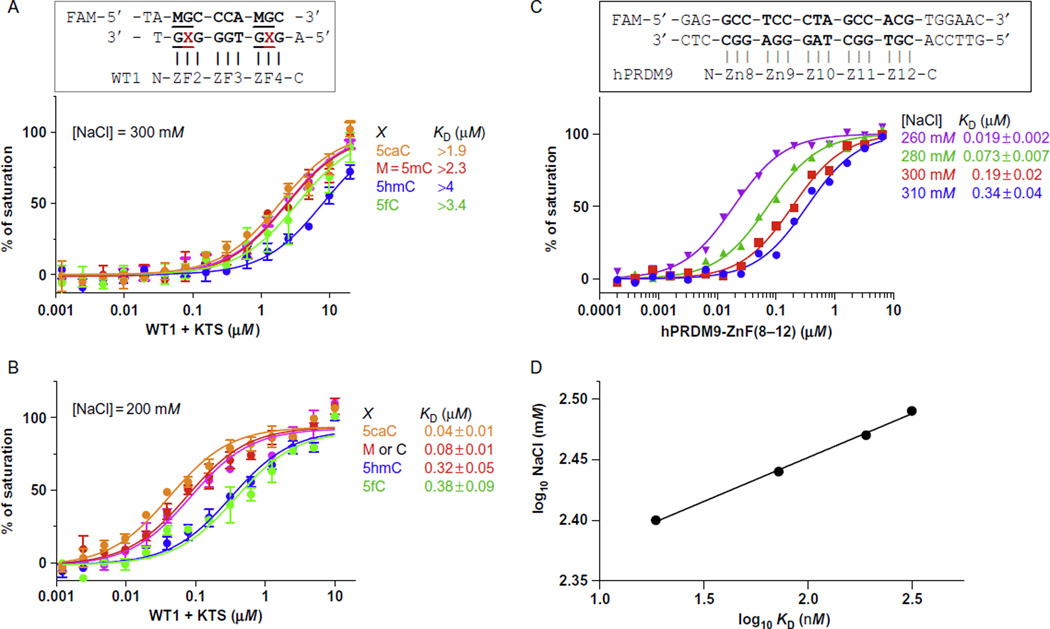
- Effect of NaCl concentration: The KD values are extremely sensitive to the ionic strength of the binding buffer. Fig. 3 illustrated that the WT1 + KTS isoform binds most strongly to 5caC-containing DNA. Affinity is uniformly low in 300 mM NaCl (Fig. 3A) but considerably higher (>10-fold) in 200 mM NaCl (Fig. 3B; Hashimoto et al., 2014). The effect is even more pronounced for human PRDM9 allele-A: increasing NaCl concentration by 20 mM can result in as much as 3-fold increase in KD value (Fig. 3C), and a 50 mM increase of NaCl resulted in an 18-fold reduction in affinity. As documented in previous studies (Jantz & Berg, 2010), the double-logarithmic plot of KD as a function of NaCl concentration is linear (Fig. 3D).Note: Due to extreme salt sensitivity of the assay, different batches of the same buffer formulation can give noticeably different KD values. When possible, use the same batch of buffer for all assays in one study.
Fig. 3.
The effect of NaCl on binding affinity. The DNA-binding affinity of WT1 + KTS isoform is uniformly low in 300 mM NaCl (A) but increases markedly in 200 mM NaCl (B). (C) The DNA-binding affinity (KD) of human PRDM9 allele-A under NaCl concentrations from 260 to 310 mM with 10–20 mM increments. (D) The linear correlation of double-logarithmic plot of KD values and NaCl concentrations. Panel (B): Adopted from Hashimoto, H., Olanrewaju, Y. O., Zheng, Y., Wilson, G. G., Zhang, X., & Cheng, X. (2014). Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes & Development, 28, 2304–2313.
4. CRYSTALLIZATION OF ZnF PROTEINS IN COMPLEX WITH DNA
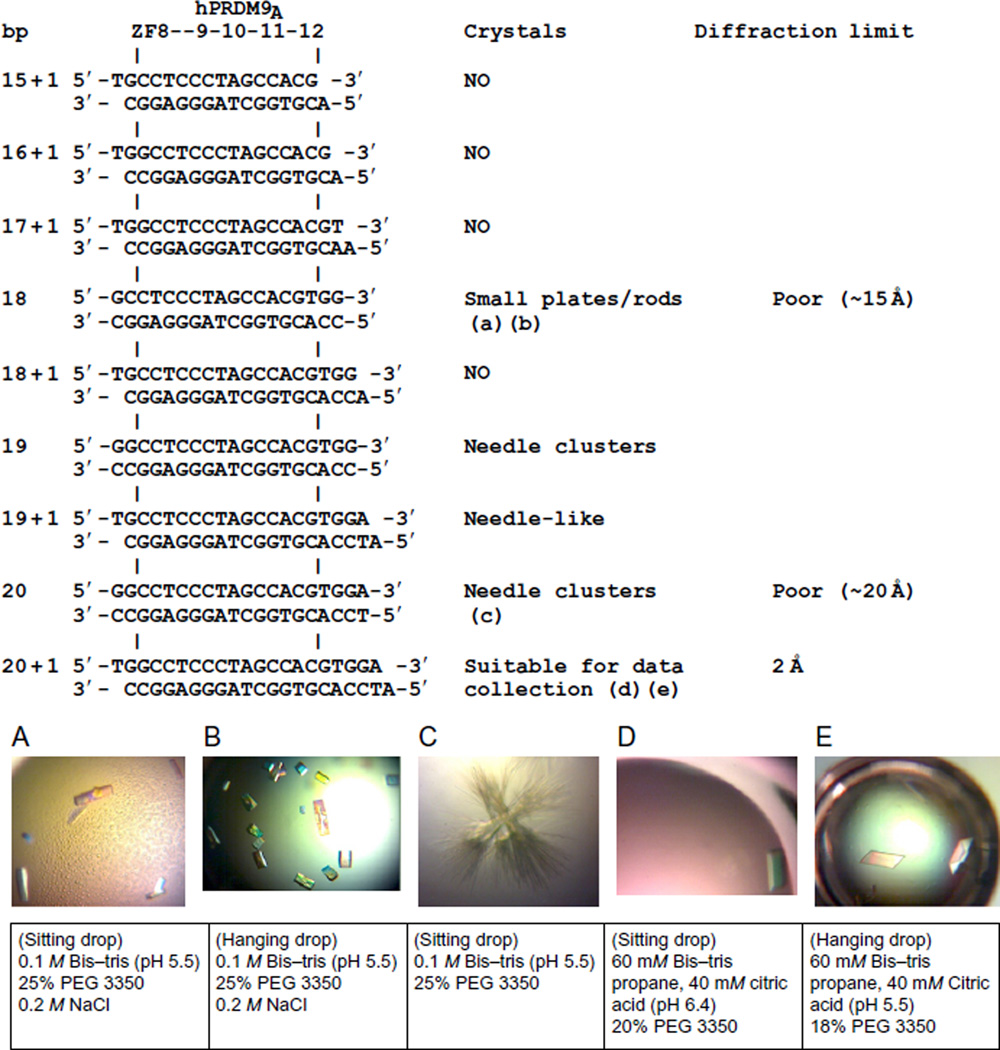
Design of oligonucleotide suitable for cocrystallization: both the length and the ends must be considered. Fig. 4 illustrates the process of obtaining diffraction quality crystals for human PRDM9 allele-A ZnF 8–12 by varying oligonucleotides. We started with a 15 + 1 base pairs (bp) double-stranded oligonucleotide (oligo)—the minimum length required for recognition by five ZnFs—plus a 5′-overhanging thymine or adenine on either strand (Fig. 4). This design was then lengthened 1 or 2 bp at a time to become 16 + 1 (1 bp increase on one end), 17 + 1 (1 bp increase on both ends), 18-, 19-, and 20-bp blunt ends, and 18 + 1, 19 + 1, and 20 + 1 bp with 5′-overhangs. In the end, only the 20 + 1 oligo yielded high-diffraction quality crystals. Alternatively, 3′-overhangings or asymmetric overhanging only on one strand could also be used.
Purification of crude oligonucleotides: Due to cost consideration, we use crude unmodified oligos to screen the length and ends. Resuspend each single-stranded DNA into annealing buffer containing 10 mM Tris (pH 8.0), 50 mM NaCl, and 1 mM EDTA (ethylenediaminetetraacetic acid) to final concentration of ~1 mM. Mix complimentary stands of DNA in equimolar ratio, heat in a boiling water bath that is slowly cooled overnight to room temperature. Load the annealed double-stranded (ds) DNA to a 5 mL HiTrap Q column with 20 mM Tris (pH 8.0) as buffer A and 20 mM Tris (pH 8.0) and 1 M NaCl as buffer B. Collect DNA eluted as a single large peak between 0.55 and 0.65 MNaCl using a linear gradient of NaCl from 0.1 to 1 M. Pool peak fractions and measure DNA concentration by absorbance at 260 nm.
Protein–DNA complex formation by dialysis: Mix ZnF protein with purified dsDNA in 1:1 molar ratio to a final concentration of 25 µM each in buffer containing 20 mM Tris (7.5), 500 mM NaCl, 5% glycerol, 25 µM ZnCl2, and 0.5 mM TCEP. Dialyze the mixture against low salt buffer (150–250 mM NaCl) at 4°C with two 500 mL changes of buffer every 6–8 h. The slow exchange to low salt buffer can potentially reduce nonspecific binding and promote specific complex formation between DNA and protein. [Alternatively, modified oligos are often HPLC or PAGE purified after synthesis, thus can be directly mixed with protein at high concentration (~1 mM).] After dialysis, centrifuge the protein–DNA complex at 4000 rpm for 15 min to remove any precipitate. Concentrate the supernatant to desired concentration and centrifuge at 13,000 rpm for 10 min before crystallization trial.
Fig. 4.
Examples of DNA oligonucleotide sequences used for cocrystallization with human PRDM9 allele-A, crystals observed, and quality of X-ray diffractions. Five examples of crystals and corresponding conditions were shown (A–E).
Acknowledgments
We sincerely thank Yusuf Olatunde Olanrewaju and Yiwei Liu for their early efforts in purifying soluble ZnF proteins. This work was supported by grant from the National Institutes of Health GM049245-22 to X.C. (who is a Georgia Research Alliance Eminent Scholar).
REFERENCES
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. Journal of Molecular Biology. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Buck-Koehntop BA, Stanfield RL, Ekiert DC, Martinez-Yamout MA, Dyson HJ, Wilson IA, et al. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15229–15234. doi: 10.1073/pnas.1213726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbone M, Wang J, Van der Maarel SM, D’Amico A, D’Agostino A, Romano A, et al. Immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome, due to ZBTB24 mutations, presenting with large cerebral cyst. American Journal of Medical Genetics. Part A. 2012;158A:2043–2046. doi: 10.1002/ajmg.a.35486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Coordinated chromatin control: Structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouery E, Abou-Ghoch J, Corbani S, El Ali N, Korban R, Salem N, et al. A novel deletion in ZBTB24 in a Lebanese family with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Clinical Genetics. 2012;82:489–493. doi: 10.1111/j.1399-0004.2011.01783.x. [DOI] [PubMed] [Google Scholar]
- de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. American Journal of Human Genetics. 2011;88:796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasarathy A, Wade PA. The MBD protein family-reading an epigenetic mark? Mutation Research. 2008;647:39–43. doi: 10.1016/j.mrfmmm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3’ ends of ZNF genes. PloS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Shibata M, Anderson KV. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 2008;135:3053–3062. doi: 10.1242/dev.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: The enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. Journal of Molecular Biology. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annual Review of Cell and Developmental Biology. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Cheng X. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics. 2009;4:8–14. doi: 10.4161/epi.4.1.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes & Development. 2014;28:2304–2313. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Zhang X, Vertino PM, Cheng X. The mechanisms of generation, recognition, and erasure of DNA 5-methylcytosine and thymine oxidations. The Journal of Biological Chemistry. 2015;290:20723–20733. doi: 10.1074/jbc.R115.656884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Molecular Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantz D, Berg JM. Probing the DNA-binding affinity and specificity of designed zinc finger proteins. Biophysical Journal. 2010;98:852–860. doi: 10.1016/j.bpj.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cellular and Molecular Life Sciences. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Molecular and Cellular Biology. 2012;32:3732–3742. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, et al. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16:624. doi: 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Chung J, Dyson HJ, Wright PE. Alternative splicing of Wilms’ tumor suppressor protein modulates DNA binding activity through isoform-specific DNA-induced conformational changes. Biochemistry. 2000;39:5341–5348. doi: 10.1021/bi9926678. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Olanrewaju YO, Zhang X, Cheng X. DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 A. Biochemistry. 2013;52:9310–9317. doi: 10.1021/bi401360n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Olanrewaju YO, Zheng Y, Hashimoto H, Blumenthal RM, Zhang X, et al. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Research. 2014;42:4859–4867. doi: 10.1093/nar/gku134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes & Development. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Blumenthal RM, Cheng X. A common mode of recognition for methylated CpG. Trends in Biochemical Sciences. 2013;38:177–183. doi: 10.1016/j.tibs.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nature Genetics. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, et al. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Nitta H, Unoki M, Ichiyanagi K, Kosho T, Shigemura T, Takahashi H, et al. Three novel ZBTB24 mutations identified in Japanese and Cape Verdean type 2 ICF syndrome patients. Journal of Human Genetics. 2013;58:455–460. doi: 10.1038/jhg.2013.56. [DOI] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, et al. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genetics. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Patel A, Horton JR, Wilson GG, Zhang X, Cheng X. Structural basis for human PRDM9 action at recombination hot spots. Genes & Development. 2016;30:257–265. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes & Development. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Molecular Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Research. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsdale JN, Webb HD, Ginder GD, Williams DC., Jr Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Research. 2011;39:6741–6752. doi: 10.1093/nar/gkr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Koseki H. Recruitment of Dnmt1:Roles of the SRA protein Np95 (Uhrf1) and other factors. Progress in Molecular Biology and Translational Science. 2011;101:289–310. doi: 10.1016/B978-0-12-387685-0.00008-1. [DOI] [PubMed] [Google Scholar]
- Shirohzu H, Kubota T, Kumazawa A, Sado T, Chijiwa T, Inagaki K, et al. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. American Journal of Medical Genetics. 2002;112:31–37. doi: 10.1002/ajmg.10658. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expression and Purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Research. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Niederhuth CE, Tuna S, Stathopoulou A, Viiri K, de Rooij DG, et al. The Dnmt3L ADD domain controls cytosine methylation establishment during spermatogenesis. Cell Reports. 2015;10:944–956. doi: 10.1016/j.celrep.2015.01.021. http://dx.doi.org/10.1016/j.celrep.2015.01.021. pii: S2211-1247(15)00034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Khachigian LM, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. The Journal of Biological Chemistry. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Zandarashvili L, White MA, Esadze A, Iwahara J. Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Letters. 2015;589:1748–1753. doi: 10.1016/j.febslet.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. The Journal of Biological Chemistry. 2012;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]