Abstract
Translation of genetic information into functional proteins is critical for all cellular life. Accurate protein synthesis relies on proper aminoacylation of transfer RNAs (tRNAs) and decoding of mRNAs by the ribosome with the use of aminoacyl-tRNAs. Mistranslation can lead to pathologic consequences. All cells contain elaborate quality control mechanisms in translation, although translational fidelity may be regulated by various factors such as nutrient limitation or reactive oxygen species. Translation fidelity is maintained via the accuracy of tRNA aminoacylation by the aminoacyl-tRNA synthetases and matching of the mRNA codon with the tRNA anticodon by the ribosome. Stringent substrate discrimination and proofreading are critical in aminoacylating tRNAs with their cognate amino acid to maintain high accuracy of translation. Although the composition of the cellular proteome generally adheres to the genetic code, accumulating evidence indicates that cells can also deliberately mistranslate; they synthesize mutant proteins that deviate from the genetic code in response to stress or environmental changes. Mistranslation with tRNA charged with noncognate amino acids can expand the proteome to enhance stress response and help adaptation. Here, we review current knowledge on mistranslation through tRNA misacylation and describe advances in our understanding of translational control in the regulation of stress response and human diseases.
Keywords: tRNA, protein synthesis, translation fidelity, stress response, adaptation methionine, molecular regulation, nutrigenomics
Introduction
Protein synthesis is a vital process in all organisms, and proper control of protein synthesis is crucial for cellular physiology. Protein synthesis requires an adequate supply of amino acids. Approximately one-half of the 20 amino acids are essential components of the mammalian diet, and nonessential amino acids can be generated from other compounds (1). Faithful translation of the genetic code during protein synthesis is critical to the growth, development, and function of living organisms. Aminoacyl-transfer RNA (tRNA)4 synthetases (aaRSs) define the genetic code by correctly pairing amino acids with their cognate tRNAs; they are the first enzymes in the translation pathway that are required to maintain high translational fidelity. Translation fidelity is commonly cited as between 10−5 and 10−3 per codon, depending on the measurement method and the codon context (2–4).
These error frequencies are typically interpreted as the tolerance threshold of the translation machinery. The composition of the cellular proteome is commonly thought to strictly adhere to the genetic code. However, high-fidelity translation is not performed in the cell at all times. Accumulating evidence indicates that cells can deliberately synthesize mutant protein molecules that deviate from the genetic code when cells are under stressed conditions or when they undergo environmental adaptation. The deliberate synthesis of mutant proteins leads to the hypothesis that some mistranslated proteins could be useful in cellular stress response and adaptation (5–7).
Amino acid substitution at a codon that codes for a different amino acid than that dictated by the genetic code is termed mistranslation. The mechanism of mistranslation can either be because of aberrant or alternate aaRS activity or ribosomal decoding errors. One of the first descriptions of systematic, generalized codon ambiguity through tRNA was in the fungal pathogen Candida albicans (8, 9). The Candida genus of yeast contains a tRNA coded in the genome that is charged with Ser but reads the CUG codon, which codes for Leu in all non-Candida organisms. All Candida species that contain this tRNASer(CAG) gene reprogrammed their genomes so that the CUG codon is read as Ser rather than Leu (9). In C. albicans, 95–97% of this tRNASer(CAG) is charged with Ser, but 3–5% with Leu, leading to widespread mistranslation at CUG codons. Increasing mistranslation was accompanied with massive genomewide rearrangements (10, 11).
The inherent, ambiguous decoding of the CUG codons and the resulting proteomic and phenotypic diversity may facilitate this opportunistic pathogen to evade the adaptive immune system of the host (6). Some members of the mycoplasma family have evolved with either absent or degenerate editing domains in their leucyl-tRNA synthetase and phenylalanyl-tRNA synthetase and possibly threonyl-tRNA synthetase (ThrRS) (12–14). These editing domain-deficient synthetases mischarge tRNAs at higher amounts, and the corresponding mistranslated peptides are also detected. The physiologic basis for the generation of mistranslated proteins is unclear; it has been suggested that mistranslation in the mycoplasma parasites could enable specific host-pathogen interactions beneficial to mycoplasma (12).
We described the discovery of Met mistranslation in mammals (15) and yeast (16). In mammals, Met mistranslation is mediated through mismethionylation of non-methionyl-tRNAs and the use of these mismethionylated tRNAs by the ribosome. The amount of mismethionylated tRNAs is regulated by an elevated amount of reactive oxygen species (ROSs) generated under oxidative stress and mediated through phosphorylation of the methionyl-tRNA synthetases (MetRSs) by the extracellular signal-regulated kinase (ERK) kinase (17). Because Met residues in proteins are known to scavenger ROSs and protect proteins against oxidative damage (18, 19), Met mistranslation can benefit cells through the contribution of mistranslated proteins to oxidative stress response (15, 17). So far, only 2 studies on natural, regulated mistranslation in mammals have been published (15, 17). This paucity of published work is not because translation in mammals is intrinsically more accurate than in microorganisms. Rather, it reflects the timing of these discoveries (first in 2009) and the technical challenges in investigating mistranslation.
Accumulating evidence suggests that errors in protein translation are far more common than previously thought. In the past, mistranslation was viewed differently than in previous decades, given that more mistranslation cases are observed in many species. Here, we describe some advances in our understanding of translational control that focuses on tRNA aminoacylation, highlight the potential roles of beneficial mistranslation in cellular stress response, and offer perspectives on future studies.
Quality Control of tRNA Aminoacylation
How accurately the genetic code is translated depends strongly on the accurate synthesis of aminoacyl tRNAs. The charging of aminoacyl tRNA is a 2-step process. In the first step, aaRSs catalyze the synthesis of activated amino acids to generate aminoacyl-adenylates. Noncognate amino acids that vary substantially in size, shape, charge, and hydrophobicity will be excluded from respective binding pockets of aaRSs. Inclusion of a noncognate amino acid in the binding pocket may lead to its activation and subsequent tRNA misacylation.
A number of reviews have been written about how aaRS clears cognate tRNAs mischarged with noncognate amino acids, a process called editing (20–22), so only a brief description is provided here. Quality control by aaRS editing includes a double-sieve mechanism; the first sieve is the active site of the aaRS, which determines the specificity for the cognate tRNA and amino acid, and the second sieve is an editing or proofreading activity to clear either activated near-cognate amino acids or mischarged tRNAs.
Some aaRSs have inherently low error rates, whereas other aaRSs may incorporate incorrect amino acids first, and then the mischarged tRNAs are deacylated by the appended editing domains or by additional proteins that deacylate mischarged tRNAs after they are released from the aaRS (23). For instance, in Mycoplasma mobile, leucyl-tRNA synthetase variant missing the editing domain can mischarge tRNALeu with Met, Ile, and Val, which causes mistranslation in the parasite (13). The mycoplasma phenylalanyl-tRNA synthetase lacks a canonical editing activity, relying instead on discrimination against the noncognate amino acid by kinetic proofreading, leading to a higher misacylating rate. Of the 20 canonical aaRSs, one-half are absolutely selective against noncognate amino acids, whereas the other one-half include aaRSs of Met, Ile, Leu, Val, Ala, Lys, Gln, Pro, Phe, and Thr and are less selective (24, 25).
A second way for an aaRS to misacylate tRNA is by attaching its cognate amino acid to noncognate tRNA. This involves a diagonally opposite situation from editing whereby the aaRS attaches a noncognate amino acid to its cognate tRNA. The only well-known case for this mechanism so far is the tRNA misacylation with Met in which the MetRS catalyzes methionylation of non-Met-tRNAs in vivo and in vitro. The Escherichia coli MetRS can specifically misacylate the E. coli tRNAThr(CGT) and tRNAArg(CCT) in vitro (26). The yeast MetRS is part of a 3-protein complex, and this complex mismethionylates a large number of yeast tRNAs (16, 27). The mammalian MetRS is part of an 11-protein complex, but the MetRS alone can misacylate tRNALys in vitro (17). In vivo, the aminoacylation fidelity of the mammalian MetRS is controlled by ERK phosphorylation of 2 specific Ser residues; the phosphorylated MetRS misacylates a large number of mammalian tRNAs (15, 17). We do not yet understand how the fidelity of MetRS of mismethionylating tRNAs is controlled at the molecular level.
Mistranslation through tRNA Misacylation in Cells
Other factors that affect the quality control of aaRSs include a low ratio of cognate to noncognate amino acids, owing to nutrient limitation and stress conditions. Misincorporation of amino acids in recombinant proteins has been reported in mammalian cells and yeast (15–17, 28), some clearly because of nutritional stress via amino acid starvation in culture (29). Oxidative stress has been reported as a main cause of mistranslation. Many aaRSs have specific thiol groups that are targeted by oxidative stress. For example, hydrogen peroxide oxidizes a critical ThrRS editing site Cys residue to sulfinic acid which reduces the translational fidelity in E. coli by causing ThrRS to mischarge tRNAThr with Ser (30). Bacteria may use ThrRS editing to sense the oxidant amounts in the environment (31). Oxidation of amino acids by reactive species such as hydroxyl radicals and superoxide anions can result in alteration of amino acid structure, such as the addition of a hydroxyl group, creating potential substrates for tRNA misacylation (32). Oxidative stress-induced tRNA misacylation by MetRS was recently demonstrated in mammalian cells. In unstressed cells, mismethionylated tRNAs are present at ∼1% amount compared with correctly charged Met-tRNAs. On oxidative stress, the amounts of misacylated Met-tRNAs increase up to 10-fold (15). The mismethionylated tRNAs are used in translation as demonstrated by their same utilization kinetics as the cognate tRNAs, the incorporation of mismethionylated tRNAs into proteins by gel electrophoresis and MS. Mammalian tRNA mismethionylation is regulated through phosphorylation of Ser209 and Ser825 of MetRS by ERK kinase under ROS-induced stressed conditions; phosphorylated MetRS shows increased affinity for noncognate tRNAs and lower affinity for cognate tRNAMet (17). This controlled inaccuracy of MetRS serves as a defense mechanism against ROS-mediated damage at the cost of translational fidelity. Redox reactions within proteins have critical physiologic roles in the cell. In addition to the sulfur-containing amino acids Met and Cys, several other amino acids are also prone to being oxidized (33). Protein modification and oxidative damage are extensively characterized in eukaryotes and strongly associated with human diseases (34–36).
Mistranslation and Nutrient Dependence in Mammals
Regulated mistranslation in mammals was only demonstrated so far through ROSs, and the nicotinamide adenine dinucleotide (phosphate) reduced form oxidases (NOXs) play a role in the induction of tRNA misacylation with Met (15). Many forms of ROS generation and the NOX protein activities are well documented to be related to dietary intakes such as the abundance of proteins, carbohydrates, and fat (37). ROS is also known to trigger the activation of the Ras–mitogen-activated protein ERK kinase (MEK)–ERK signal transduction pathway that ultimately leads to the known mechanism of generating the low-fidelity form of the human MetRSs (17).
The genetic code contains the 20 canonical amino acids found in all organisms plus selenocysteine and pyrrolysine encoded in some genomes (38, 39). In addition to these amino acids, protein synthesis can use many other naturally occurring nonproteinogenic amino acids (NPAs). The potential use of NPAs during translation depends in part on their ability to be charged by aaRSs (22, 32). NPAs occur widely in nature and are well-characterized by-products and/or intermediates of biosynthetic pathways. The accumulation of NPAs in the cell can occur in a number of ways, for example, through degradation of proteins that contain post-translational modifications (40). Degradation of modified protein residues introduces NPAs into the pool of free amino acids in the cell which may increase aaRS to mischarge tRNA.
The NPA m-Tyr is detected in eukaryotic and bacterial proteomes and is one of the products of canonical aromatic amino acid oxidation (32). Recently, it was discovered that the NPA β-aminomethylalanine (BMAA) is mistranslated at Ser codons in human tissues (41). Ingestion of BMAA is associated with an increased risk of neurodegenerative disorders such as amyotrophic lateral sclerosis, Alzheimer disease, and parkinsonism (42). Media supplementation with Ser drastically reduces the degree of this mistranslation, suggesting that noncognate BMAA competition with cognate Ser for tRNA synthetase for Ser may be the source of the errors in protein synthesis. It is conceivable that inducing specific changes in amino acid pools to chemically modify the selectivity of aaRSs may be used to combat many diseases with minimal deleterious effects (43, 44). For instance, Liu et al. (45) developed a general approach that allows unnatural amino acids with diverse physicochemical and biological properties to be genetically encoded in mammalian cells. A mutant E. coli aaRS is first evolved in yeast to selectively aminoacylate its tRNA with 6 unnatural amino acids, including p-methoxyphenylalanine, p-acetylphenylalanine, p-benzoylphenylalanine, p-iodophenylalanine, p-azidophenylalanine, and p-propargyloxyphenylalanine. This mutant aaRS together with a suppressor tRNA is then used to site-specifically incorporate the unnatural amino acid into a protein in Chinese hamster ovary cells. This method facilitated the introduction of biological probes into proteins for cellular studies and may ultimately facilitate the synthesis of therapeutic proteins that contain unnatural amino acids in mammalian cells (46).
Approaches to Analyze Amino Acid Misincorporation in Proteins
Although the occurrence of regulated mistranslation is no longer surprising, the tools for studying its biological relevance are still in their infancy. We developed a microarray-based method to detect and quantify tRNA misacylation in cells which led to the discovery of regulated tRNA misacylation in mammalian cells (15). The method relies on pulse-labeling cells with the use of radioactive amino acids such as 35S-Met, followed by total RNA extraction and hybridization to microarrays printed with tRNA complementary oligonucleotide DNA probes. Although powerful, this method only analyzes mistranslation at the level of tRNA misacylation. How these misacylated tRNAs are selectively used by the ribosome remains unclear.
Traditional measurement of mistranslation is performed indirectly by quantifying amino acid substitutions in exogenously expressed reporters such as luciferase or fluorescent proteins (47–49), in which critical residues of the reporter proteins are mutated and mistranslation of the codon of interest restores the wild-type protein. For example, TagRFP fluorescent protein reporters were constructed for the study of Met mistranslation in mammalian cells by mutating the codon for Met in the amino acid triad that forms the fluorophore to the codons for Lys, His, or Gly (17). These mutant proteins are not fluorescent themselves, but they are converted back to the fluorescent wild-type protein through Met mistranslation. Reporter protein activity is quantified and residue-specific mistranslation is inferred as mistranslation activity. Although useful, a drawback of the reporter-based approach includes detection limits that may not be sufficiently high for low-frequency amino acid substitution events in the cell. Reporters are generally only useful for studying specific amino acid substitutions at one chosen codon at a time, thus limiting the scope to a case-by-case analysis in a specific sequence context (43).
In our opinion, the greatest challenge in studying mistranslation is quantitative measurement of amino acid substitutions which are generally of low frequency and present in the background of a large excess of wild-type protein molecules. Developments in the analytic power afforded by sensitive MS and bioinformatic tools should substantially facilitate the study of mistranslation. MS can be used to directly detect and quantify low-level mistranslation in a codon-specific manner. In particular, LC-electrospray ionization MS/MS, coupled with the use of multiple reaction monitoring modes, can be the new technique of choice (28, 43, 50, 51). Multiple reaction monitoring modes allows for greater detection of low-frequency mistranslation with higher resolution and sensitivity, providing effective tools to examine comparative proteome perturbations caused by changes in the cellular environment.
With the development of modern proteomic combinations of MS, intact mass measurements have become the most common methods for the detection of amino acid misincorporation. Generally, the peptide mass map is at the heart of the analysis with MS/MS spectra of peaks individually assigned to amino acid sequence with the use of error-tolerant matching of spectra to protein sequence in protein identification tools such as Mascot (29, 52). These kinds of analyses have been able to identify mistranslated peptides present at much lower amounts (as low as 10−5) than previous studies. For instance, application of the method to proteins expressed in E. coli easily detected 16 different misincorporations in a set of 6 proteins in 9 different sites (53).
Benefit of Making Mistranslated Proteins
Organisms use many mechanisms to respond to stress and adapt to their environments. Some cells have lost their reliance on particular steps in translation quality control, indicating that, although quality control is crucial for optimal growth under normal conditions, there are certain situations in which translational mistakes can be tolerated (2, 54). E. coli mutants that mistranslate Asp, Glu, or Cys at >10% of the time do not exhibit a growth defect under standard laboratory conditions (55). Candida species can tolerate up to 28% misincorporation of Leu at designated Ser codons without important ill effects (56, 57).
Several lines of evidence suggest that increased amounts of mistranslation can even be advantageous under some stress conditions. The increased rate of Ser misincorporation at CUG codons in Saccharomyces cerevisiae enhances their ability to prevent challenges from toxic agents, including cadmium, arsenate, and hydrogen peroxide (5). CUG codon mistranslation is maintained during the evolution of C. albicans because of its potential to generate cell surface variability, which substantially alters fungus-host interactions (56, 57). In Acinetobacter baylyi, genetic mutation in tRNA synthetase for Ile that allows for mischarging of tRNAIle with Val confers an increased growth rate compared with the wild-type bacteria under conditions of limiting Ile and excess Val (58). Further analysis showed that Val may compensate for the limited availability of Ile to enable protein synthesis to proceed efficiently under such conditions. Mistranslation may therefore enhance the competitiveness of A. baylyi under amino acid starvation. In mycobacteria, increasing amino acid misincorporation frequency results in phenotypic resistance to rifampicin, but decreasing mistranslation produces increased susceptibility to the rifampicin (49). This raises the possibility that errors in protein translation play a role in adapting to stress through increasing proteomic diversity, and altering translational fidelity represents a unique form of environmental adaptation in mycobacteria. In other mycoplasma species, aaRSs have natural mutations in their editing domains, resulting in the inactivation of their editing activities. In vivo mistranslation was detected by MS of the parasite proteome (12). It was suggested that this type of mycoplasma mistranslation evolved as a mechanism for antigen diversity to escape host defense systems. In E. coli, cells with 40% Met residues substituted by norleucine die more rapidly than control cells when exposed to hypochlorite, hydrogen peroxide, or ionizing radiation (59). Global misincorporation of Met into protein may serve a protective function against oxidative stress (18, 19).
Mistranslation can also benefit mammalian cells. Mammalian cells contain many noncognate tRNAs mischarged with Met at amounts that are dynamically regulated (15). The baseline misacylation for Met in unstressed cells is ∼1% (the amount of cognate Met-tRNAMet is set to 100%), whereas Met misacylation can rise up to ∼13% when cells are infected with several viruses or exposed to Toll-like receptor ligands poly-IC, CpG oligonucleotides, and lipopolysaccharides, indicating that mistranslation constitutes an immune response in cells. Induced tRNA misacylation can also be triggered by exposure to oxidative chemicals such as hydrogen peroxide or arsenite. Induction of misacylation is inhibited by inhibitor of NOXs. These results indicate that ROS is the cellular trigger of tRNA misacylation with Met. Subsequent studies show that Met mistranslation is indeed a useful mechanism that contributes beneficially to oxidative stress response (17).
We examined 5 other amino acids, Cys, Ile, Val, Phe, and Tyr for tRNA misacylation in HeLa cells and detected no misacylation for these 5 amino acids (15). Whether tRNA misacylation and hence mistranslation also occurs with the 14 other amino acids in mammalian cells remains to be determined.
At the amount of codon selectivity for Met mistranslation in mammals, tRNALys(CTT), which decodes AAG codons, seems to be mismethionylated at higher levels than tRNALys(TTT), which decodes AAA codons, suggesting that Lys-to-Met substitutions may occur more frequently at AAG than at AAA codons (15). Other such tRNA-mismethionylation-based codon preferences may include preferred Arg-to-Met substitutions at CGU/CGC/CGA codons compared with CGG, AGA/AGG codons. Whether these codon substitution preferences truly exist at the proteome amount remains to be determined.
On the basis of protein structure, some mutant proteins produced through the mistranslation pathway must be functionally more useful than others, depending on the specific location of the amino acid substitution. Clearly, some mutant proteins generated by mistranslation also harm cells, for example, through aggregation. In the case of Met mistranslation, we have proposed that Met-substituted mutant proteins may protect against oxidation better than the wild-type protein because the extra Met residue may be strategically localized. This beneficial hypothesis is based on the unique chemical and cellular properties of Met residue in proteins that can absorb ROS molecules through oxidation but is reduced by peptidyl-Met sulfoxide reductases (60).
Mistranslation through aaRS Mutations and Diseases
aaRS-related mistranslation can benefit cells but can also be toxic to cells. For instance, chromosomal mutations of tRNA synthetase for Ala leading to Ser-for-Ala mistranslation cause severe neuropathologies in mice (61, 62). Several aaRS mutations are linked to various human diseases (63, 64), although the detailed mechanisms on how these mutations lead to diseases remain unclear. Misfolded proteins are associated with several pathologic conditions, including neurodegeneration. At the cellular level, aaRS mutations can reduce cell proliferation and can promote cell death (65). These findings suggest that the pathologic consequences of diminished tRNA synthetase editing activity depend on the cell type and the extent of editing disruption (66). This is supported by findings that certain mutations in human tRNA synthetases for Gly and Tyr which do not possess editing function cause neurodegenerations known as the Charcot-Marie-Tooth disease (67).
Future Perspectives of Adaptive Mistranslation
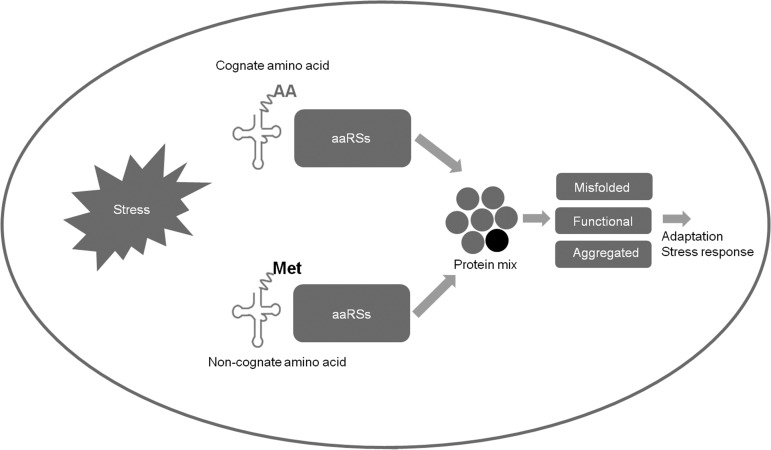
In general, cells minimize errors during protein synthesis by using extensive quality control mechanisms to ensure that the genetic code is translated with high fidelity. However, despite the widespread use of quality control and proofreading, one should not assume that aminoacylation errors always lead to protein unfolding and aggregation (7, 22). An emerging view suggests that cells place varying demands on the quality control machinery under different growth conditions with diverse outcomes (Figure 1). For example, during exponential growth in E. coli and yeast, certain aaRS editing activities are dispensable, in contrast to strict quality control in responding to temperature, antibiotic, or starvation stress (54). Cells are able to respond to amino acid deprivation stress through multiple systems, for example, mammalian target of rapamycin complex 1, an evolutionarily conserved Ser/Thr kinase that senses signals from amino acid availability and the energy status of the cell (68, 69). Quality control can be applied to different degrees in response to changing environmental conditions in between these 2 ranges of states. Although translational errors were generally categorized as deleterious in the past, sufficient evidence suggests useful applications of controlled mistranslation in stress response and adaptation. Documentations of amino acid misincorporation in cellular proteomes are limited at this point. This is changing because more sensitive methods for analysis of these low-abundance events are being developed. Further investigations of misincorporation in protein products compared with native proteins are required. Under certain nutrient limitation conditions, mistranslation with the use of nonstandard amino acids to synthesize protein variants is an intriguing prospect for new drug development that may improve human health. A main future challenge is to develop improved techniques to better understand how internal and external factors modulate translational fidelity and how this, in turn, affects the biology of the cell.
FIGURE 1.
Mistranslation can be beneficial in response to stress. Under stress conditions or in organisms with editing-defective aaRS, aaRS could misacylate noncognate amino acid which can be used in translation to make mutant proteins. Mutant proteins derived from mistranslation can misfold, aggregate, or perform functional roles to respond to stress and help adapt to environmental changes. AA, amino acid; aaRS, aminoacyl-transfer RNA synthetase.
Acknowledgments
Both authors read and approved the final manuscript.
Footnotes
Abbreviations used: aaRS, aminoacyl-tRNA synthetase; BMAA, β-aminomethylalanine; ERK, extracellular signal-regulated kinase; MetRS, methionyl-tRNA synthetase; NOX, nicotinamide adenine dinucleotide (phosphate), reduced form oxidase; NPA, nonproteinogenic amino acid; ROS, reactive oxygen species; ThrRS, threonyl-tRNA synthetase; tRNA, transfer RNA.
References
- 1.Proud CG. Control of the translational machinery by amino acids. Am J Clin Nutr 2014;99:231S–6S. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Soll D. Quality control mechanisms during translation. Science 1999;286:1893–7. [DOI] [PubMed] [Google Scholar]
- 3.Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 2010;16:1797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 2009;10:715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos MA, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol Microbiol 1999;31:937–47. [DOI] [PubMed] [Google Scholar]
- 6.Moura GR, Carreto LC, Santos MA. Genetic code ambiguity: an unexpected source of proteome innovation and phenotypic diversity. Curr Opin Microbiol 2009;12:631–7. [DOI] [PubMed] [Google Scholar]
- 7.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev Genet 2013;47:121–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T, Ueda T, Ohama T, Osawa S, Watanabe K. The gene for serine tRNA having anticodon sequence CAG in a pathogenic yeast, Candida albicans. Nucleic Acids Res 1993;21:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res 1995;23:1481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos MA, Keith G, Tuite MF. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J 1993;12:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes AC, Miranda I, Silva RM, Moura GR, Thomas B, Akoulitchev A, Santos MA. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol 2007;8:R206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Boniecki MT, Jaffe JD, Imai BS, Yau PM, Luthey-Schulten ZA, Martinis SA. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA 2011;108:9378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Palencia A, Lukk T, Li Z, Luthey-Schulten ZA, Cusack S, Martinis SA, Boniecki MT. Leucyl-tRNA synthetase editing domain functions as a molecular rheostat to control codon ambiguity in Mycoplasma pathogens. Proc Natl Acad Sci USA 2013;110:3817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadavalli SS, Ibba M. Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res 2013;41:1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 2009;462:522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiltrout E, Goodenbour JM, Frechin M, Pan T. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res 2012;40:10494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Kim DG, Kim BG, Yang WS, Hong J, Kang T, Oh YS, Kim KR, Han BW, Hwang BJ, et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation against oxidative stresses. J Cell Sci 2014;127:4234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA 1996;93:15036–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol Cell Biochem 2002;234–235:3–9. [PubMed] [Google Scholar]
- 20.Hellmann RA, Martinis SA. Defects in transient tRNA translocation bypass tRNA synthetase quality control mechanisms. J Biol Chem 2009;284:11478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minajigi A, Deng B, Francklyn CS. Fidelity escape by the unnatural amino acid beta-hydroxynorvaline: an efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth. Biochemistry 2011;50:1101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullwinkle T, Lazazzera B, Ibba M. Quality control and infiltration of translation by amino acids outside of the genetic code. Annu Rev Genet 2014;48:149–66. [DOI] [PubMed] [Google Scholar]
- 23.So BR, An S, Kumar S, Das M, Turner DA, Hadad CM, Musier-Forsyth K. Substrate-mediated fidelity mechanism ensures accurate decoding of proline codons. J Biol Chem 2011;286:31810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakubowski H. Quality control in tRNA charging. Wiley Interdiscip Rev RNA 2012;3:295–310. [DOI] [PubMed] [Google Scholar]
- 25.Lo WS, Gardiner E, Xu Z, Lau CF, Wang F, Zhou JJ, Mendlein JD, Nangle LA, Chiang KP, Yang XL, et al. Human tRNA synthetase catalytic nulls with diverse functions. Science 2014;345:328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones TE, Alexander RW, Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc Natl Acad Sci USA 2011;108:6933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015;162:1286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu XC, Borisov OV, Alvarez M, Michels DA, Wang YJ, Ling V. Identification of codon-specific serine to asparagine mistranslation in recombinant monoclonal antibodies by high-resolution mass spectrometry. Anal Chem 2009;81:9282–90. [DOI] [PubMed] [Google Scholar]
- 29.Harris RP, Kilby PM. Amino acid misincorporation in recombinant biopharmaceutical products. Curr Opin Biotechnol 2014;30:45–50. [DOI] [PubMed] [Google Scholar]
- 30.Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA 2010;107:4028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Fan Y, Ling J. Mechanism of oxidant-induced mistranslation by threonyl-tRNA synthetase. Nucleic Acids Res 2014;42:6523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullwinkle TJ, Reynolds NM, Raina M, Moghal A, Matsa E, Rajkovic A, Kayadibi H, Fazlollahi F, Ryan C, Howitz N, et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife 2014;3:e02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob C, Giles GI, Giles NM, Sies H. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew Chem Int Ed Engl 2003;42:4742–58. [DOI] [PubMed] [Google Scholar]
- 34.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 1997;272:20313–6. [DOI] [PubMed] [Google Scholar]
- 35.Stadtman ER. Protein oxidation and aging. Free Radic Res 2006;40:1250–8. [DOI] [PubMed] [Google Scholar]
- 36.Sultana R, Butterfield DA. Oxidative modification of brain proteins in Alzheimer’s disease: perspective on future studies based on results of redox proteomics studies. J Alzheimers Dis 2013;33 Suppl 1:S243–51. [DOI] [PubMed] [Google Scholar]
- 37.Görlach A, Dimova EY, Petry A, Martinez-Ruiz A, Hernansanz-Agustin P, Rolo AP, Palmeira CM, Kietzmann T. Reactive oxygen species, nutrition, hypoxia and diseases: problems solved? Redox Biol 2015;6:372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Böck A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci 1991;16:463–7. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 2002;296:1459–62. [DOI] [PubMed] [Google Scholar]
- 40.Wells-Knecht MC, Huggins TG, Dyer DG, Thorpe SR, Baynes JW. Oxidized amino acids in lens protein with age. Measurement of o-tyrosine and dityrosine in the aging human lens. J Biol Chem 1993;268:12348–52. [PubMed] [Google Scholar]
- 41.Glover WB, Mash DC, Murch SJ. The natural non-protein amino acid N-beta-methylamino-L-alanine (BMAA) is incorporated into protein during synthesis. Amino Acids 2014;46:2553–9. [DOI] [PubMed] [Google Scholar]
- 42.Chiu AS, Gehringer MM, Welch JH, Neilan BA. Does alpha-amino-beta-methylaminopropionic acid (BMAA) play a role in neurodegeneration? Int J Environ Res Public Health 2011;8:3728–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moghal A, Mohler K, Ibba M. Mistranslation of the genetic code. FEBS Lett 2014;588:4305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren W, Truong TM, Ai HW. Study of the binding energies between unnatural amino acids and engineered orthogonal tyrosyl-tRNA synthetases. Sci Rep 2015;5:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Brock A, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods 2007;4:239–44. [DOI] [PubMed] [Google Scholar]
- 46.Ewalt KL, Schimmel P. Activation of angiogenic signaling pathways by two human tRNA synthetases. Biochemistry 2002;41:13344–9. [DOI] [PubMed] [Google Scholar]
- 47.Ilegems E, Pick HM, Vogel H. Monitoring mis-acylated tRNA suppression efficiency in mammalian cells via EGFP fluorescence recovery. Nucleic Acids Res 2002;30:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 2007;13:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Javid B, Sorrentino F, Toosky M, Zheng W, Pinkham JT, Jain N, Pan M, Deighan P, Rubin EJ. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci USA 2014;111:1132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo D, Gao A, Michels DA, Feeney L, Eng M, Chan B, Laird MW, Zhang B, Yu XC, Joly J, et al. Mechanisms of unintended amino acid sequence changes in recombinant monoclonal antibodies expressed in Chinese Hamster Ovary (CHO) cells. Biotechnol Bioeng 2010;107:163–71. [DOI] [PubMed] [Google Scholar]
- 51.Feeney L, Carvalhal V, Yu XC, Chan B, Michels DA, Wang YJ, Shen A, Ressl J, Dusel B, Laird MW. Eliminating tyrosine sequence variants in CHO cell lines producing recombinant monoclonal antibodies. Biotechnol Bioeng 2013;110:1087–97. [DOI] [PubMed] [Google Scholar]
- 52.Koenig T, Menze BH, Kirchner M, Monigatti F, Parker KC, Patterson T, Steen JJ, Hamprecht FA, Steen H. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J Proteome Res 2008;7:3708–17. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Shah B, Bondarenko PV. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry 2013;52:8165–76. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol 2010;8:849–56. [DOI] [PubMed] [Google Scholar]
- 55.Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Soll D. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA 2008;105:16502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miranda I, Silva-Dias A, Rocha R, Teixeira-Santos R, Coelho C, Goncalves T, Santos MA, Pina-Vaz C, Solis NV, Filler SG, et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio 2013;4:e00285–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sárkány Z, Silva A, Pereira PJ, Macedo-Ribeiro S. Ser or Leu: structural snapshots of mistranslation in Candida albicans. Front Mol Biosci 2014;1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bacher JM, Waas WF, Metzgar D, de Crecy-Lagard V, Schimmel P. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J Bacteriol 2007;189:6494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J 2009;23:464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA 2001;98:12920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006;443:50–5. [DOI] [PubMed] [Google Scholar]
- 62.Schimmel P. Mistranslation and its control by tRNA synthetases. Philos Trans R Soc Lond B Biol Sci 2011;366:2965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol 2006;13:1091–100. [DOI] [PubMed] [Google Scholar]
- 64.Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci 2008;17:1643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J, Bergert M, Walther A, Suter B. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat Commun 2014;5:5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA 2014;111:17570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Latour P, Thauvin-Robinet C, Baudelet-Mery C, Soichot P, Cusin V, Faivre L, Locatelli MC, Mayencon M, Sarcey A, Broussolle E, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2010;86:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B, Qian SB. Translational regulation in nutrigenomics. Adv Nutr 2011;2:511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]