Abstract
The genus Pan is the closest genus to our own and it includes two species, Pan paniscus (bonobos) and Pan troglodytes (chimpanzees). The later is constituted by four subspecies, all highly endangered. The study of the Pan genera has been incessantly complicated by the intricate relationship among subspecies and the statistical limitations imposed by the reduced number of samples or genomic markers analyzed. Here, we present a new method to reconstruct complete mitochondrial genomes (mitogenomes) from whole genome shotgun (WGS) datasets, mtArchitect, showing that its reconstructions are highly accurate and consistent with long-range PCR mitogenomes. We used this approach to build the mitochondrial genomes of 20 newly sequenced samples which, together with available genomes, allowed us to analyze the hitherto most complete Pan mitochondrial genome dataset including 156 chimpanzee and 44 bonobo individuals, with a proportional contribution from all chimpanzee subspecies. We estimated the separation time between chimpanzees and bonobos around 1.15 million years ago (Mya) [0.81–1.49]. Further, we found that under the most probable genealogical model the two clades of chimpanzees, Western + Nigeria-Cameroon and Central + Eastern, separated at 0.59 Mya [0.41–0.78] with further internal separations at 0.32 Mya [0.22–0.43] and 0.16 Mya [0.17–0.34], respectively. Finally, for a subset of our samples, we compared nuclear versus mitochondrial genomes and we found that chimpanzee subspecies have different patterns of nuclear and mitochondrial diversity, which could be a result of either processes affecting the mitochondrial genome, such as hitchhiking or background selection, or a result of population dynamics.
Keywords: genome diversity, chimpanzee, bonobo, bioinformatics, next-generation sequencing, mtArchitect
Introduction
The genus Pan consists of the two living species most closely related to humans: the chimpanzee (Pan troglodytes, P. t.) and the bonobo (Pan paniscus, P. p.). The study of their population structure and diversity allows us to infer their evolutionary history, which can be taken as a frame of reference for a better understanding of our own (Marques-Bonet et al. 2009; Prado-Martinez et al. 2013; Rogers and Gibbs 2014). Currently, four subspecies are recognized within chimpanzees and they are frequently denoted by their geographical ranges (fig. 1A): (i) P. t. verus is the western chimpanzee, found in Senegal, Guinea-Bissau, Guinea, Sierra Leone, Liberia, Mali, and Ivory Coast; (ii) P. t. troglodytes is the central subspecies, spread across Congo, Gabon, Central African Republic, Equatorial Guinea, and Cameroon; (iii) P. t. schweinfurthii, also known as the eastern chimpanzee, is located in Tanzania, Burundi, Rwanda, Uganda, and the Democratic Republic of the Congo (Stone et al. 2010); and (iv) P. t. ellioti, is the Nigeria-Cameroon chimpanzee, and although its establishment has been very controversial (Becquet et al. 2007), the latest nuclear genome (Bowden et al. 2012; Prado-Martinez et al. 2013) as well as Y chromosome studies (Hallast et al. 2016) have found clear evidence supporting its distinct identity. In contrast, bonobos are not classified in subspecies but three clades have been described based on mitochondrial DNA analysis (Zsurka et al. 2010) and they are found exclusively in the Democratic Republic of the Congo, separated from chimpanzees by the Congo River.
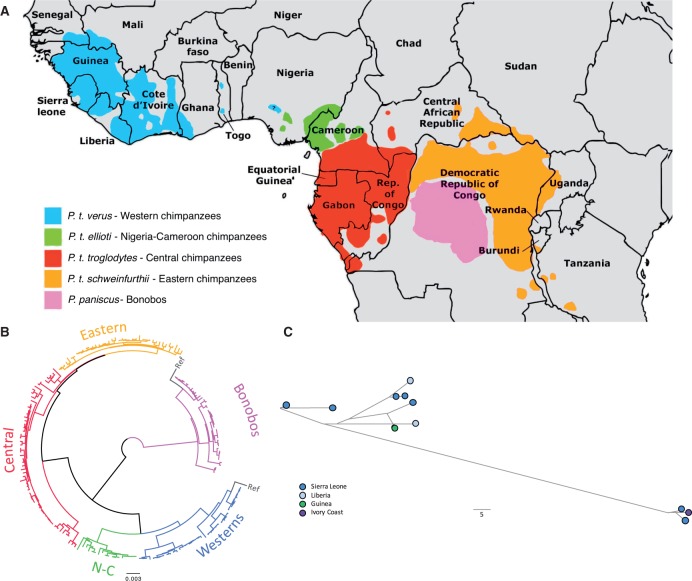
Fig. 1.—
(A) Geographical distribution of the genus Pan species and subspecies. (B) Neighbor-joining bootstrap consensus tree of the mitogenome of the 200 samples. The scale is in changes per base. The bootstrap values for each of the five clusters are all 100. (C) Median joining network of 11 P. t. verus indicating their geographical origin. Branch lengths are proportional to the amount of differences, symbolizing phylogenetic relations.
Mitochondrial DNA has been widely used in evolutionary studies due to its high mutation rate (Lynch et al. 2006) and its absence of recombination; as such it represents a simple and highly informative genetic marker in natural populations over short time frames (Galtier et al. 2009). Mitogenomes have suffered a strong selective pressure towards losing their genes, which have migrated to the nuclear genome, where they can be protected from the accumulation of deleterious mutations, or Muller’s ratchet, by sexual reproduction and recombination (Wallace 2007). Along with the 13 oxidative phosphorylation (OXPHOS) genes, whose protein products are organized in five (I–V) complexes, mammalian mitochondrial genomes maintain 12S and 16S rRNAs, 22 tRNAs, and what is called the D-loop, or control region, which contains the promoters for both strands and two hyper variable regions broadly used in phylogenetic research (Wise et al. 1997; Eriksson et al. 2004).
Over the past years, several studies have characterized the diversity and phylogeny of the genus Pan and other great apes (Stone et al. 2010; Zsurka et al. 2010; Bjork et al. 2011; Fisher et al. 2011; Prado-Martinez et al. 2013; Hvilsom et al. 2014). However, the limited number of samples, the non-homogenous access to the whole spectrum of subspecies diversity and most importantly, the non-matched information for mitochondrial and nuclear genome sequences, has hindered a proper assessment of the evolutionary processes affecting Pan mitogenomes. In fact, the study with the highest total number of individuals was Hvilsom et al. (2014) with 88 individuals, none of them with nuclear genome information. Only Fisher et al. (2011) contained nuclear sequences but this comparison was restricted to 150 kilobases (kbs).
Recent technological developments have made it much simpler to generate full genome datasets in humans (The 1000 Genomes Project Consortium 2015) and closely related species (Prado-Martinez et al. 2013, Carbone et al. 2014). This has motivated the development of methods to reconstruct mitochondrial sequences from WGS data, with MITObim (Hahn et al. 2013) being the most powerful and recently published methodology. This software uses modules of the MIRA sequence assembler (Chevreux et al. 1999) to map the reads to a reference sequence based on conserved regions. The resulting contigs are subsequently used to retrieve overlapping reads and incorporate them in an iterative way to extend and complete the sequence. Although this approach proved useful and has a good accuracy rate (more than 99.5% for publication data and 99.38% for our data), it has limitations such as the inclusion of nuclear mitochondrial regions (NUMTs) into the final sequence or lower mapping efficiency in the start and end of the linearized mitogenome. In order to overcome these limitations, we developed a different approach, which we called mtArchitect (http://www.biologiaevolutiva.org/tmarques/mtarchitect/).
Considering both published and new mtArchitect reconstructed mitogenomes, this work gathers the largest dataset of complete mitochondrial sequences in the Pan genus, including 200 samples. We analyzed and compared mitochondrial and nuclear genomes, trying to infer the evolutionary forces and demographic changes that shaped the mitochondrial genome of the genus Pan.
Materials and Methods
Dataset
A new set of 20 complete chimpanzee genomes was sequenced to a mean coverage of 25× from peripheral blood on an Illumina HiSeq 2000 sequencing platform (supplementary table 1, Supplementary Material online) and their mitogenome was reconstructed with our new computational approach, mtArchitect (Genbank access numbers KU308529-KU308548) (fig. 2). In addition, we added de novo mitochondrial genome reconstructions from whole genome data of 32 individuals (Prado-Martinez et al. 2013) (Genbank access nos. KX211928–KX211959) and we merged data from previously published articles (Stone et al. 2010; Zsurka et al. 2010; Bjork et al. 2011; Fisher et al. 2011; Hvilsom et al. 2014). To avoid the presence of redundant sequences from relatives in our dataset, we looked for matrilineally related individuals in the studbooks (Carlsen and Jongh 2007; Ross 2009) and eliminated the sample with the lowest quality of each corresponding pair. Because many samples did not have any family information, we aligned each subset against the rest using BLAST (Camacho et al. 2009). As identical sequences most likely come from the same individual or from closely related individuals we removed the inferior of each pair.
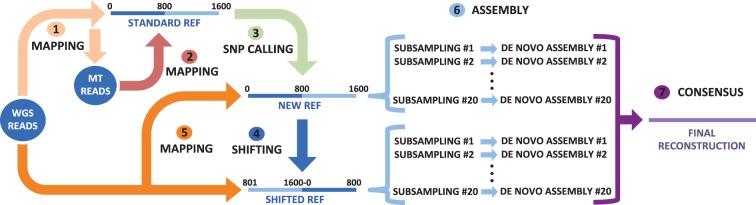
Fig. 2.—
mtArchitect overview. (1) Whole genome sequencing reads are mapped to a standard mitochondrial reference sequence with low stringency parameters to retrieve mitochondrial reads. (2) and (3) After these reads are mapped with regular parameters to the reference, SNPs are called and incorporated into the reference, creating a new specific sequence. This step is iterative in order that the newly incorporated SNPs favour the mapping of more reads at each iteration. (4) The final modified reference start is shifted 8 kb so that the highly polymorphic D-loop is centred and more reads covering it can be retrieved. (5) All whole genome-sequencing reads are mapped to both modified references. (6) The final set of reads is subsampled up to 150× and a de novo assembly is performed 20 times for each modified reference. (7) The final sequence is constructed from the consensus of the 40 assemblies.
The final dataset of complete mitogenomes (18 P. t. ellioti, 40 P. t. schweinfurthii, 59 P. t. troglodytes, 38 P. t. verus, 43 P. paniscus, and the NCBI references for both chimpanzee and bonobo (table 1 and supplementary table 1, Supplementary Material online)) was aligned to the chimpanzee NCBI reference sequence (NC001643) using MAFFT 7.130b (Katoh and Standley 2013). Neighbor joining trees of whole mitochondrial genomes, coding regions and D-loops were constructed using MAFFT with 1,000 bootstrap repetitions and were edited with FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). The similarity of the trees was assessed by calculating the linear correlation coefficient (r) between the distance matrices obtained for each alignment with ClustalW2 (Larkin et al. 2007).
Table 1.
Summary table of the individuals included in this study. Data were gathered from six published articles. Phase I samples are 5 unpublished mitochondrial newly reconstructed sequences from Prado-Martinez et al. (2013) and Phase II are the 20 newly sequenced, unpublished and reconstructed samples. NCBI samples are chimpanzee and bonobo references (NC001643 and NC001644) and Jenny (X93335), a P. t. verus from Arnason et al. (1996)
| Subspecies | Source |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stone (2010) | Zsurka (2010) | Bjork et al. (2011) | Fischer (2011) | Prado-Martinez (2013) | Hvilsom (2014) | Phase I | Phase II | NCBI | TOTAL | |
| Pan paniscus | — | 7 | — | 20 | 12 | 4 | — | — | 1 | 44 |
| P. t. ellioti | 1 | — | 3 | 4 | 5 | — | 5 | — | — | 18 |
| P. t. schweinfurthii | 1 | — | 6 | 17 | 6 | 5 | — | 5 | — | 40 |
| P. t. troglodytes | 1 | — | 12 | 16 | 4 | 21 | — | 5 | — | 59 |
| P. t. verus | 4 | — | 1 | 13 | 5 | 9 | — | 5 | 2 | 39 |
| Total | 7 | 7 | 22 | 70 | 32 | 39 | 5 | 15 | 3 | 200 |
Algorithm Overview
We have developed a new approach to reconstruct the complete mitochondrial genomes from whole genome sequencing data (fig. 2). The reconstruction is based on a two-stage read capture and assembly approach.
In the first stage, a sample-specific modified mitochondrial sequence is created. To do this, we first determine a pool of mitochondrial reads by mapping the whole genome paired-end data to a mitochondrial reference genome with Burrows–Wheeler Alignment tool (BWA) (Li and Durbin 2009). This baiting step is run with low stringency parameters (bwa mem –A 4) and paired-end reads with only one of the reads mapped are also retrieved and included in the set using SAMtools (Li et al. 2009) (samtools view –f 4 –F 8), which allows the capture of a wide range of reads. Once the mitochondrial read pool is defined, an iterative process is performed to capture more distant reads. The collected reads are mapped to the mitochondrial reference genome (bwa mem). Variants are then called (samtools mpileup –L 1,000 –d 1,000 –C50) and applied to the reference sequence with Vcftools (Danecek et al. 2011) (vcf-consensus function). This way, only well-supported variants are introduced into the reference, preventing a non-ending cycle of variants being called at the same position as well as the inclusion of NUMTs into the final sequence. This iterative strategy maximizes the number of captured reads in highly variable regions because the incorporation of variants allows the mapping of previously unmapped reads, generating a new reference that represents the sequenced mitochondrial genome much more accurately than the initial reference. Finally, in order to increase the number of captured reads at the ends of this reference and taking advantage of the circularity of the mitochondrial genome, WGS reads are mapped (bwa mem) to a further-modified mitochondrial sequence obtained by shifting the start 8 kbs towards the middle of the mitogenome. From the set of reads captured up to this step, only high-quality paired-end reads are kept by retaining read pairs only if both reads are mapped, properly paired and with a mapping quality higher than 50 (samtools view –f 2 –q 50). Moreover, paired-end pairs with at least one read having a median Phred quality score lower than 32 are discarded.
In the second stage, the final set of reads is sub-sampled to have at most 150-fold depth of coverage and Hapsembler 1.1 is used to construct contigs (Donmez et al. 2011) (-p Illumina -t 4 -d no –PHRED_OFFSET 33 –MIN_CONTIG_SIZE 1000 –EPSILON 0.05). This random sub-sampling and contig construction is performed 20 times on both the standard and the shifted-origin sample-specific reference sequences. In this way, the random representation of reads that could lead to problems such as the incorporation of NUMTs into the assembly is solved, as the NUMT reads are expected to be less than 1% of retrieved reads. At each of the 40 de novo assemblies, the contigs are oriented using local alignments to the corresponding reference (BLAST) and joined using MAFFT 7.130b. N’s are incorporated in existing gaps and those sites remaining unresolved due to differences in overlapping contigs. Finally, the reconstructed mitogenome is built from the consensus of the 40 de novo assemblies.
Comparisons Between mtArchitect, PCR, and MITObim
In order to test the accuracy of our method we compared mtArchitect reconstructions with 10 mitochondrial sequences obtained by long-range PCR, eight from Hvilsom et al. (2014) and two from Fischer et al. (2011). We reconstructed the mitogenomes of the same five bonobos and five chimpanzees with mtArchitect and aligned them to their corresponding PCR sequence using ClustalW2. All the mitochondrial genomes of these 10 individuals were also reconstructed using MITObim 1.7 standard commands (Mitobim.pl –start 0 –end 10) and –quick option, given the existence of reference sequences for both bonobo and chimpanzee.
Genetic Diversity
Genetic diversity analyses were performed using our full dataset and were compared with a similar dataset of Homo sapiens mitochondrial sequences (200 African and 200 European individuals) randomly obtained from MitoTool (Fan and Yao 2011). Intraspecific and interspecific nucleotide pairwise distances (nucleotide diversity, π, Nei 1987) of complete mitogenomes were calculated using MEGA 6.06 (Tamura et al. 2013). To compare mitochondrial with nuclear variation, chromosome 21 pairwise distances were also calculated with data from Prado-Martinez et al. (2013), using R (R Development Core Team 2014). The mitochondrial multifasta alignment was divided in gene sequences using Jalview 2 (Waterhouse et al. 2009), maintaining the reading frame. Interpopulational ϕst values, the number of segregating sites (S), Tajima’s D (Tajima 1989), and Fu’s Fs (Fu 1997) were obtained for each gene and for the whole mitogenome of each subset using Arlequin (Excoffier and Lischer 2010). The πA/πS indexes were calculated with DNAsp 5.10.01 (Librado and Rozas 2009) and a phylogenetic network of 11 P. t. verus was obtained with Network (Bandelt et al. 1995).
Demographic History
Two alternative models of the genealogical relationships among species and subspecies of the genus Pan were tested through Approximate Bayesian Computation (ABC) (Beaumont et al. 2002; for a review see e.g. Bertorelle et al. 2010). Under Model 1 an initial divergence event separating the Pan paniscus/Pan troglodytes clades is followed by the divergence of the P. t. verus/P. t. ellioti clade from the P. t. troglodytes/P. t. schweinfurthii one. Subsequently, two divergence events separate the subspecies within each clade. Model 2 is also characterized by a first divergence event separating Pan paniscus from all Pan troglodytes subspecies, but in the second divergence event P. t. verus detaches first from all others, followed by P. t. ellioti and eventually by the separation of P. t. troglodytes from P. t. schweinfurthii. According to previous studies (Stone et al. 2010; Bjork et al. 2011; Prado et al. 2013), in Model 1 the divergence between P. t. verus and P. t. ellioti was set to precede the one separating P. t. troglodytes and P. t. schweinfurthii (supplementary fig. 1, Supplementary Material online).
For this analysis, the control regions of the 198 samples were used, removing missing data, gaps and length polymorphisms to focus on potentially neutrally evolving sites. We generated 500,000 gene genealogies for each model by coalescent simulation with ABCtoolbox (Wegmann et al. 2010). The prior distributions were all uniform (population sizes between 250 and 100,000 individuals; mutation rates between 1×10−7 and 5×10−5 per site per generation for the whole mitochondrial sequence; first population split between 50 and 50,000 generations ago; following splits separated from the previous one by a minimum of 10 to a maximum of 50,000 generations. The number of segregating sites (S), the number of different haplotypes (k), haplotype diversity (H), Tajima’s D, intra- and inter-mean number of pairwise differences (MNPD), and pairwise Fst were calculated for the observed and simulated data. The summary statistics were selected for model choice by applying Kolmogorov–Smirnov (KS) test, as implemented in R, to each summary statistic at 10 random subsets of 10,000 simulations from each model. They were ranked based on the resulting P value and the first 10 summary statistics overlapping among the 10 random subsets were chosen. To calculate each model’s posterior probability we used a weighted multinomial logistic regression (LR) (Beaumont 2008) using R scripts from https://code.google.com/archive/p/popabc/, modified by S.G. and A.B. To evaluate the stability of the posterior probabilities, we fixed different thresholds of retained simulations (supplementary fig. 1, Supplementary Material online). Type I Error and the rate of true and false positives were estimated by generating 1,000 pseudo-observed datasets according to each model analyzed, with parameter values randomly chosen from the corresponding prior distributions.
Further, we inferred the demographic history by Bayesian Markov Chain Monte Carlo (MCMC) with BEAST 2 v.2.1.3 (Bouckaert et al. 2014). Alignments with four partitions (coding region sites 1 + 2, coding region sites 3, rRNAs and the D-loop; as found most appropriate by Endicott and Ho 2008) excluding all gaps and ambiguous nucleotides, were used to infer the divergence times between the lineages, changes in their effective population sizes (Ne), and the mutation rates (μ) of each partition. The substitution model for each subspecies was chosen with PartitionFinder (Lanfear et al. 2014), and the model was optimized with Path Sampler by changing one parameter each run. The previously estimated divergence time between chimpanzees and bonobos, 1.2 million years ago (Hvilsom et al. 2014; Prado et al. 2013) with a standard deviation of 0.17, was used as a root time calibration, allowing the estimation of absolute divergence times. To calculate the Ne, the generation time used was 24.63 years (Langergraber et al. 2012). Gene ND6 was not excluded from the final analysis as test runs showed that its inclusion or exclusion does not affect the results significantly. Also, despite being the only coding gene on the light strand of the mitochondrial genome, both synonymous and non-synonymous mutation rates at this gene are similar to those of other mitochondrial coding genes (Saccone et al. 1999). The MCMC length necessary to obtain effective sample sizes bigger than 200 was from 10 to 30 million steps. Every chain was run twice independently to check the consistency and the results were summarized with Tracer v1.6 (Rambaut et al. 2013).
Results
The quality of mtArchitect reconstructions was assessed by comparing our computational reconstructions to sequences obtained by applying PCR-based procedures to a subset of 10 individuals. This comparison showed a mean identity of 99.96% (fewer than three errors per mitochondrial genome), whereas MITObim had an identity of 99.41% (more than 90 errors). Even though our method performs better than MITObim for the more complicated D-loop region (99.74% vs. 95.65%), this remains a major source of errors, constituting the 62% of the mistakes (supplementary table 2 and supplementary fig. 2, Supplementary Material online), seemingly by way of the elevated concentration of variants. We also found that mtArchitect performs well even when the only available reference mitogenome is highly divergent from the species of interest. Our analyses suggest that mtArchitect reconstructions are accurate when using a reference with up to 13% divergence (supplementary fig. 3, Supplementary Material online).
We used mtArchitect to reconstruct the mitochondrial genomes of 20 newly sequenced chimpanzee individuals. After joining them with sequences from previous studies, we selected a total of 200 samples (table 1 and supplementary table 1, Supplementary Material online). The phylogenetic topologies of the complete mitochondrial genome, coding regions and D-loop neighbour joining trees are very similar (fig. 1B and supplementary fig. 4, Supplementary Material online), particularly the complete and coding trees, which maintain almost all the substructure, with a linear correlation coefficient of 0.99 between their distance matrices. The D-loop tree differs in individual and small clusters connections (r = 0.90 with the coding tree). All trees show the chimpanzee and bonobo separation, the subdivision of chimpanzees in four subspecies (providing strong evidence for the split of the Nigeria–Cameroon chimpanzee from all other subspecies) as well as the presence of three distinct clades within bonobos (Zsurka et al. 2010). These clustering patterns in the chimpanzee support the two-clade distribution (western vs. east) as previously suggested (Prado-Martinez et al. 2013). Surprisingly, the chimpanzee and the bonobo NCBI references cluster with different subgroups in the coding and D-loop trees. Because they are the only samples presenting this pattern, this suggests that the complete reference sequences might have been constructed from more than one individual, as mentioned by Zsurka et al. (2010) for bonobos. As this can bias diversity estimations, references were not included in subsequent analyses.
The available geographic location of some of the samples allowed us to explore to which extent we could observe stratification by geographic origin. We found no correlation between mitochondrial genotypes and geographical origin within chimpanzee subspecies (exemplified in Western chimpanzees, fig. 1C). This result is perhaps the product of maternal inheritance of the mitochondrial genome, its lack of recombination and the social organization of the genus Pan: in both species, males typically spend their entire lives in their natal communities, and females commonly transfer to neighbouring groups during adolescence (Gerloff et al. 1999; Mitani et al. 2002). This social structure would naturally lead to a larger spread of mitochondrial haplogroups within the subspecies than for the nuclear genome.
We then compared two divergence models by ABC. Model 1, in which, after the separation of chimps from bonobos, the first divergence event separated a Central-Western clade from a Central-Eastern clade, received the strongest support, having a posterior probability of 76%. Considering a probability threshold of 0.5, we found that the type I Error was always ≤ 0.23 (supplementary table 3, Supplementary Material online). Thus, the models appear to be reasonably well recognized as the probability to identify the correct model is approximately three-fold as large as the probability of Type I error, even when the probability threshold is 0.5. When higher probability thresholds were considered, the increase of the Type I Error probability is not due to an incorrect attribution of the model but to the fact that it was not possible to assign support to either model. Model 1 appears roughly three-fold as likely as the alternative Model 2 as a description of Pan demographic history.
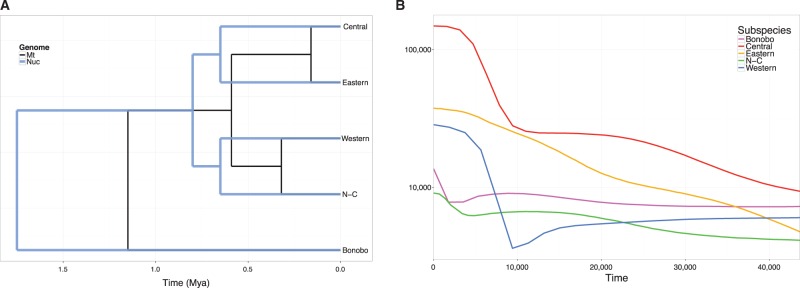
The demographic history estimated via ABC was confirmed by BEAST 2. The demographic parameters estimated with BEAST 2 are consistent among runs, resulting in a divergence time of 1.15 million years ago (Mya) between the ancestor of the four chimpanzee subspecies and bonobos and 0.59 Mya between central + eastern and western + Nigeria-Cameroon branches. As in previous studies (Prado et al. 2013; Hvilsom et al. 2014), the divergence between Nigeria-Cameroon and western chimpanzees appears to be older (0.32 Mya) than the divergence of central and eastern subspecies (0.16 Mya) (fig. 3A). The effective population size (Ne) estimated from the mitochondrial genomes (fig. 3B) show that bonobos, and West-African chimpanzees (P. t. verus and P. t. ellioti), have a more stable recent demographic history, whereas the eastern and the central chimpanzee have experienced an increase in their effective population sizes. Eastern chimpanzees have experienced a moderate expansion, whereas central chimpanzees have a more explosive expansion, with a fold change in Ne of 7 and 20, respectively.
Fig. 3.—
(A) Nuclear and mitochondrial divergence times. Nuclear divergence times are represented by the blue tree and the mitochondrial times by the black tree. (B) Inferred historical effective population size (Ne) of each population.
To investigate possible evolutionary constraints over mitochondrial genes we calculated mitochondrial genes πA/πS ratios grouped by OXPHOS complex to facilitate the interpretation of the results (supplementary table 4 and supplementary fig. 5, Supplementary Material online). We found evidence of strong purifying selection acting over most of the complexes in Pan (median of 0.122). Homo has similar πA/πS values for complexes III and IV, but it shows evidence of a relaxation of selective constraint in complexes I and V (median πA/πS is around four-fold higher than in Pan). Also, the low πA/πS ratio of complex III in western chimpanzee (0.029) stands out (median of the other subspecies is 0.175), suggesting a very strong purifying selection unique to this subspecies. Moreover, consistent with BEAST 2 results, Tajima’s D and Fu’s Fs test statistics rejected neutrality for eastern and central chimpanzee mitochondrial genome, irrespectively of whether the D-loop sequence was, or was not, considered (table 2), but this was not always the case for Nigeria-Cameroon. To gain insight into the evolutionary processes, these tests were also individually performed for each gene of each subset (supplementary table 5). This way, eastern and central chimpanzees have several genes with significantly negative Tajima’s D (P value < 0.05) and all of their genes show significantly negative Fu’s Fs values, whereas there are no significant tests for any other subset.
Table 2.
Diversity statistics. For each (sub)species (D-loop, coding and complete mitochondrial genome) we report the number of individuals (N), the number of haplotypes (k), the number of segregating sites (S), Tajima’s D (D) and Fu’s Fs statistics, mitochondrial diversity (π) (with its standard deviation (SD)), haplotype diversity (H) and mean number of pairwise differences (MNPD)
| N | k | S | D | D P value | Fs | Fs P value | π (SD) | H (SD) | MNPD (SD) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dloop | P. t. schweinfurthii | 40 | 36 | 101 | −1.88708 | 0.0121 | −22.71477 | 0 | 0.010992 (0.005653) | 0.9949 (0.0069) | 11.508974 (5.328178) |
| P. t. troglodytes | 59 | 53 | 153 | −0.98407 | 0.1628 | −23.71742 | 0.0002 | 0.022666 (0.011210) | 0.9930 (0.0061) | 23.731151 (10.582534) | |
| P. t. ellioti | 18 | 15 | 68 | −1.07001 | 0.1318 | −2.30847 | 0.1431 | 0.014027 (0.007368) | 0.9739 (0.0293) | 14.686275 (6.898381) | |
| P. t. verus | 38 | 33 | 110 | 0.63672 | 0.7951 | −6.16683 | 0.0399 | 0.029312 (0.014543) | 0.9929 (0.0077) | 30.689900 (13.703112) | |
| P. paniscus | 43 | 28 | 85 | 0.23429 | 0.6736 | −2.14633 | 0.26 | 0.019982 (0.009988) | 0.9723 (0.0121) | 20.921373 (9.417391) | |
| Coding region | P. t. schweinfurthii | 40 | 38 | 287 | −2.12846 | 0.0028 | −13.45385 | 0.0017 | 0.001889 (0.000937) | 0.9974 (0.0063) | 28.971795 (12.939033) |
| P. t. troglodytes | 59 | 57 | 550 | −1.80945 | 0.0111 | −16.48661 | 0.0011 | 0.003816 (0.001853) | 0.9988 (0.0034) | 58.515488 (25.629206) | |
| P. t.ellioti | 18 | 18 | 161 | −1.34843 | 0.0691 | −4.08519 | 0.0321 | 0.002080 (0.001065) | 1.0000 (0.0185) | 31.895425 (14.605480) | |
| P. t. verus | 38 | 34 | 275 | 0.77392 | 0.837 | −1.69194 | 0.2553 | 0.005148 (0.002517) | 0.9900 (0.0105) | 78.944523 (34.744269) | |
| P. paniscus | 43 | 36 | 331 | −0.19018 | 0.4855 | −0.93566 | 0.3832 | 0.004732 (0.002308) | 0.9900 (0.0078) | 72.569214 (31.879795) | |
| Complete mitogenome | P. t. schweinfurthii | 40 | 38 | 388 | −2.08028 | 0.0044 | −9.6645 | 0.007 | 0.002471 (0.001217) | 0.9974 (0.0063) | 40.480769 (17.952297) |
| P. t. troglodytes | 59 | 57 | 703 | −1.63536 | 0.023 | −11.66109 | 0.006 | 0.005020 (0.002430) | 0.9988 (0.0034) | 82.246639 (35.893197) | |
| P. t. ellioti | 18 | 18 | 229 | −1.27599 | 0.0887 | −2.88198 | 0.061 | 0.002843 (0.001446) | 1.0000 (0.0185) | 46.581699 (21.180277) | |
| P. t. verus | 38 | 36 | 385 | 0.73975 | 0.8305 | −2.39312 | 0.153 | 0.006692 (0.003264) | 0.9972 (0.0068) | 109.634424 (48.125356) | |
| P. paniscus | 43 | 40 | 416 | −0.1024 | 0.5271 | −3.1576 | 0.122 | 0.005707 (0.002778) | 0.9967 (0.0059) | 93.490587 (40.977571) |
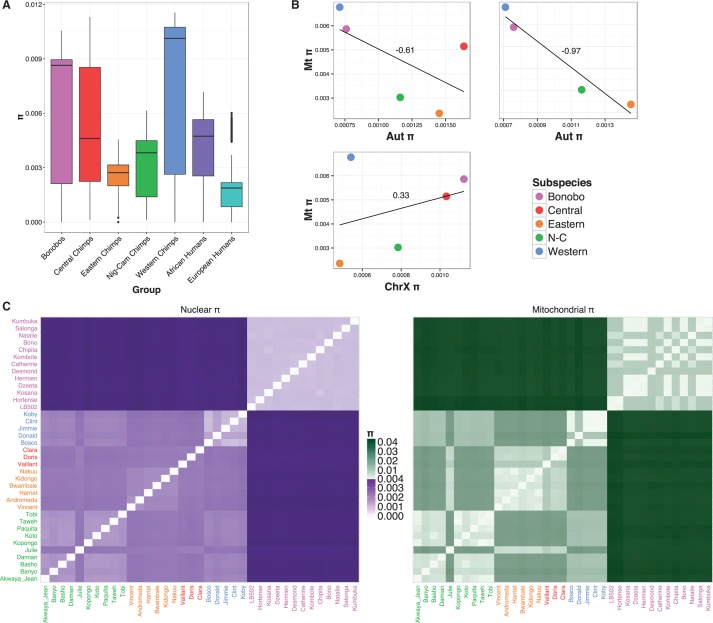
Nucleotide diversity, π, shows a wide diversity in all Pan members, all of them being more diverse than European humans (fig. 4A), whereas African human populations fall within their variability. Bonobos and western chimpanzees show the most diverse mitogenomes and eastern and Nigeria-Cameroon chimpanzees have about half their diversity, as previously reported (Stone et al. 2010; Fischer et al. 2011). The advantage of our approach is that we can compare for the first time genome-wide diversity with mitochondrial diversity. The difference in mitochondrial and nuclear π heat map scales (fig. 4C) illustrates the lower diversity of the nuclear genome, which is most likely a result of its lower mutation rate and the smaller proportion of neutrally evolving sites in mitochondrial genomes. In nuclear DNA, we observe a range from 0.08% of mean variation within bonobos up to 0.41% between them and western chimpanzees, whereas the mitochondrial genome ranges from a 0.3% of intraspecific changes in eastern chimpanzees to a 4.05% between bonobos and central chimpanzees, resulting in an average range of 50–675 differences per mitochondrial genome. The comparison of nuclear versus mitochondrial diversities (first panel in fig. 4B) shows that bonobos and western chimpanzees have the most diverse mitochondrial genome but the least diverse nuclear genome. Eastern and the Nigeria-Cameroon chimpanzees have the opposite pattern, that is, low mitochondrial diversities and high nuclear diversities. This is evidenced by the low correlation coefficient between mitochondrial and nuclear π (r=−0.61). This seems to be caused mostly by the central chimpanzees, because excluding them, both variables present a high and negative correlation (r=−0.97). To explore if these differences were caused by female demographic history being different from that of males, we looked at X chromosome diversity (third panel in fig. 4B) and we found that it poorly correlates with mitochondrial diversity (r = 0.33), being western chimpanzees the outlier group in this case.
Fig. 4.—
(A) Nucleotide diversity of each (sub)species, African human samples and European human samples. Coloured boxes span from the first to the third quartile and the segment inside them is the median. The vertical lines mark the maximum and minimum values (excluding outliers) and the outliers are represented by dots. (B) Correlation of mitochondrial (Mt) diversity (π) with autosomal (Aut) and X chromosome (ChrX) diversity. (C) Heat maps of intraspecific and interspecific pairwise nucleotide diversity at the mitochondrial and nuclear genome calculated for the samples from Prado-Martinez et al. (2013). Note that the scale values of mitogenomes are 10-fold the values of nuclear data.
Further, all interpopulational φst values were smaller than 0.005 (table 3), illustrating that most of the diversity resides within each subspecies rather than between them (table 4).
Table 3.
Inter sub(species) ϕst values. The ϕst value for each pair of (sub)species is shown
| P. paniscus | P. t. ellioti | P. t. schweinfurthii | P. t. troglodytes | P. t. verus | |
|---|---|---|---|---|---|
| P. paniscus | 0 | — | — | — | — |
| P. t. ellioti | 0.00328 | 0 | — | — | — |
| P. t. schweinfurthii | 0.00439** | 0.00126 | 0 | — | — |
| P. t. troglodytes | 0.00374*** | 0.00061 | 0.0018 | 0 | — |
| P. t. verus | 0.0046** | 0.00147 | 0.00264* | 0.002* | 0 |
Significance: *P < 0.05; **P < 0.01; ***P < 0.001.
Table 4.
Mitochondrial mutation rates (substitutions/site/year). The mean and 95% highest posterior density (HPD) intervals of mitochondrial mutation rates obtained in this study, Endicott and Ho (2008) and Hvilsom et al. (2014) are shown for the mitogenome and its four partitions: The D-loop, concatenated ribosomal RNA (rRNA) sequences, first and second positions of gene coding codons (PC1 + 2) and the third position of gene coding codons (PC3)
| This study |
Endicott and Ho (2008) |
Hvilsom et al. (2014) |
||||
|---|---|---|---|---|---|---|
| Mean | 95% HPD | Mean | 95% HPD | Mean | 95% HPD | |
| Whole | 2.49×10−8 | [1.75×10−8–3.32×10−8] | 9.66×10−8 | [7.35×10−8–1.16×10−7] | 2.69 × 10−8 | 1.73 × 10−8–3.84 × 10−8 |
| D-loop | 6.27×10−8 | [3.83×10−8–9.14×10−8] | 3.02×10−7 | [2.23×10−7–3.73×10−7] | 2.09 × 10−7 | 1.08 × 10−7–3.29 × 10−7 |
| rRNA | 4.58×10−9 | [2.56×10−9–6.9×10−9] | 2.21×10−8 | [1.13×10−8–3.24×10−8] | 1.29 × 10−8 | 7.04 ×10−9–2.05 × 10−8 |
| PC1 + 2 | 1.15×10−8 | [7.71×10−9–1.56×10−8] | 1.11×10−8 | [7.23×10−9–1.53×10−8] | 9.38 × 10−9 | 5.37 × 10−9–1.51 × 10−8 |
| PC3 | 5×10−8 | [3.47×10−8–6.8×10−8] | 5.09×10−8 | [3.44×10−8–6.80×10−8] | 4.83 × 10−8 | 2.63 × 10−8–7.51 × 10−8 |
Discussion
Traditionally, mitochondrial genomes have been sequenced by long-range PCR, but today next-generation sequencing (NGS) technologies have created the need for a new technique to accurately reconstruct mitochondrial genomes from whole genome sequencing reads. The problem of reconstructing a mitochondrial sequence using a closely related reference is the need for flexibility in mapping to highly variable regions whereas at the same time, ensuring some stringency so that no false variants are incorporated, particularly those arising from NUMTs. Here, we validate a new pipeline, mtArchitect, which reconstructs sequences with higher precision than other published methods. The main advantage of our approach is the combination of low stringency mapping with a de novo assembly strategy. This allows mtArchitect to incorporate all the existing variability, when introducing very few mistakes. Furthermore, the D-loop, the most variable region, is usually situated at one end of the reference and moving it to the centre markedly helps resolve local variation. Also, avoiding the incorporation of NUMTs into the final sequence, a problem that arises when using WGS data, can be achieved only by incorporating the most common allele at potentially heterozygous positions. Given that the ratio of mitochondrial to nuclear DNA is usually higher than 100 to 1 this allows us to use a majority rule in allele selection. Further, some of the differences between mtArchitect reconstructions and the PCR sequences are shared with MITObim. We cannot assess whether these discrepancies manifest PCR mistakes or sequencing or reconstructing errors.
The application of our algorithm revealed interesting features of the chimpanzee mitogenome. First, the NCBI references cluster with different subgroups depending on which part of the mitochondrial genome is used to construct the tree, the coding region or the D-loop. Hence, there is reason to believe the NCBI reference was generated from more than one individual as previously reported for bonobos (Zsurka et al. 2010). It would be advisable to use other sequences as NCBI references. We suggest our reconstructions of Clint and Bono (KX211957 and KX211936) as the new mitogenome reference sequences for chimpanzee and bonobo, respectively. Currently, there is only one NCBI reference for chimpanzees, a western chimpanzee, but in light of the divergence between subspecies, which reaches 2.16% between central and western chimpanzees, it would be recommendable to establish a mitochondrial reference for each Pan troglodytes subspecies, including P. t. ellioti, the Nigeria-Cameroon subspecies. The bootstrap value in this study (100 in complete and coding trees; 95 in D-loops tree) provides support for the P. t. ellioti subspecies designation, as do well-established conclusions based on nuclear data (Bowden et al. 2012; Prado et al. 2013).
The divergence time between bonobos and chimpanzees (P. paniscus and P. troglodytes) has been estimated using varying types of genetic data. From nuclear data, it has been inferred to be from 1.56 Mya using microsatellites (Becquet et al. 2007) to ≈1.8 Mya from multilocus polymorphisms (Prado-Martinez et al. 2013). From the non-recombining portion of the Y chromosome (NRY), it was estimated to be 1.8 Mya (Stone et al. 2002). Recently, it has also been inferred from the male-specific region of the Y chromosome (MSY) at 1.57 Mya (Oetjens et al. 2015). Here we estimated the divergence time between bonobos and chimpanzees from mitogenomes to be 1.15 [0.81–1.49] which is more recent than the nuclear estimates. This may be explained by the smaller effective population size of mitochondrial genomes (¾ the nuclear Ne) and the saturation of variants produced by its higher mutation rate resulting in a compression of the basal branches. As for the divergence between the subspecies, the ABC analysis showed that the most probable model for chimpanzee sub-speciation is one in which the Central-Western and Central-Eastern clades separate first, followed by the split between the western subspecies, P. t. verus and P. t. ellioti, and ending with the division of the eastern clade into P. t. troglodytes and P. t. schweinfurthii. This result is in agreement with the MCMC analysis with BEAST 2, that estimated the dates of these events to be 0.59 Mya [0.41–0.78], 0.32 Mya [0.22–0.43], and 0.16 Mya [0.17–0.34]. This dates are very close to the ones obtained in Hvilsom et al. (2014) but are more recent than others, which estimated the divergence of bonobos and chimpanzees from 1.94 to 2.1 (Stone et al. 2010; Bjork et al. 2011; Oetjens et al. 2015). This difference could be due to the selection of different calibration points. Whereas these studies used the Homo-Pan divergence, we used the divergence between bonobos and chimpanzees as the calibration point, alike in Hvilsom et al. (2014). With respect to the variation in effective population size over time, our analysis with BEAST 2 indicates the occurrence of a recent expansion of central and eastern populations. This conclusion is also supported by the observed excess of rare variants illustrated by the significantly negative Tajima’s D and Fu’s Fs values for these same subspecies. The poor correlation between X chromosome and mitochondrial π discards the possibility of female population dynamics accounting for these patterns. The question of whether these signals are a result of processes affecting only the mitochondria or affecting the whole populations is rather complex. Previous analyses of nuclear data also suggest that the eastern subspecies has indeed expanded (Wegmann and Excoffier 2010; Prado-Martinez et al. 2013), perhaps by a factor of 8. In contrast, the population expansion exhibited at the nuclear genome by central chimpanzees is not as strong. As the recombination and slower mutation rate make the nuclear genome less sensitive to recent population dynamics, perhaps the absence of that signal is a product of the more limited power in nuclear genome studies. Nevertheless, given the absence of recombination, mitogenomes are more prone to be subject of hitchhiking or background selection, and as these processes produce an excess of rare variants that would result in the signals we observe, they cannot be discarded as explanatory variables for chimpanzee mitochondrial diversity. Further analyses combining mitochondrial and nuclear sequences, including explicit tests of phylogenetic and demographic models, are needed to cast light upon these phenomena.
Supplementary Material
Acknowledgments
Centre de Regulació Genomica/Centro Nacional de Analisis Genomico (CRG-CNAG), Baldiri Reixach 4-8, 08023 Barcelona, Spain/SGR AGAUR/Maria de Maeztu. The authors thank Luca Betti for drawing the map of the Chimpanzees distribution, Stefania Rubbini for helping with the dataset management, T.M-B is supported by ICREA (www.icrea.cat), EMBO YIP (www.embo.org) 2013, MINECO BFU2014-55090-P (FEDER), BFU2015-7116-ERC and BFU2015-6215-ERCU01 MH106874 grant, Fundacio Zoo Barcelona and Secretaria d Universitats i Recerca del Departament d Economia i Coneixement de la Generalitat de Catalunya. S.T. was supported by the Young Researcher Fellowship (5×1000 Year 2010 and Unicredit 2013 Funds) from the University of Ferrara.
Literature Cited
- Arnason U, Xu X, Gullberg A. 1996. Comparison between the complete mitochondrial DNA sequences of Homo and the common chimpanzee based on nonchimeric sequences. J. Mol. Evol. 42(2):145–152. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. 1995. Mitochondrial portraits of human populations using median networks. Genetics 141(2):743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA. 2008. Joint determination of topology, divergence time, and immigration in population trees In: Renfrew C, Matsumura S, Forster P, editor. Simulation, genetics and human prehistory. McDonald Institute Monographs 134; Cambridge: McDonald Institute for Archaeological Research. p. 1541. [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162(4):2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C, et al. 2007. Genetic structure of chimpanzee populations. PLoS Genetics 3(4):e66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Benazzo A, Mona S. 2010. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol. Ecol. 19(13):2609–2625. [DOI] [PubMed] [Google Scholar]
- Bjork A, et al. 2011. Evolutionary history of chimpanzees inferred from complete mitochondrial genomes. Mol. Biol. Evol. 28(1):615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10(4):e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden R, et al. 2012. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genetics 8(3):e1002504.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:(1), 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, et al. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature 513(7517):195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen F, de Jongh T. 2007. European studbook for the chimpanzee Pan troglodytes. Copenhagen Zoo. Roskildevej 38 p. 1–278. [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information German Conference on Bioinformatics. p. 45–56. [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez N, Brudno M. (2011). Hapsembler: an assembler for highly polymorphic genomes In: Research in Computational Molecular Biology. Berlin, Heidelberg: Springer; p. 38–52. [Google Scholar]
- Endicott P, Ho SY. 2008. A Bayesian evaluation of human mitochondrial substitution rates. Am J Human Genet. 82(4):895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Hohmann G, Boesch C, Vigilant L. 2004. Rivers influence the population genetic structure of bonobos (Pan paniscus). Mol. Ecol. 13(11):3425–3435. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Fan L, Yao YG. 2011. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 11(2):351–356. [DOI] [PubMed] [Google Scholar]
- Fischer A, et al. 2011. Bonobos fall within the genomic variation of chimpanzees. PLoS One 6(6):e21605.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Nabholz B, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18(22):4541–4550. [DOI] [PubMed] [Google Scholar]
- Gerloff U, et al. 1999. Intracommunity relationships, dispersal pattern and paternity success in a wild living community of bonobos (Pan paniscus) determined from DNA analysis of faecal samples. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 266(1424):1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. 2001. Is the population size of a species relevant to its evolution? Evolution 55(11):2161–2169. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, et al. 2016. Great-ape Y-Chromosome and mitochondrial DNA phylogenies reflect sub-species structure and patterns of mating and dispersal. Genome Res. Gr-198754.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, Carlsen F, Heller R, Jaffré N, Siegismund HR. 2014. Contrasting demographic histories of the neighboring bonobo and chimpanzee. Primates 55(1):101–112. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 14(1):82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber KE, et al. 2012. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. 109(39):15716–15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311(5768):1727–1730. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Ryder OA, Eichler EE. 2009. Sequencing primate genomes: what have we learned? Annu. Rev. Genomics Human Genetics 10:355–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani JC, Watts DP, Muller MN. 2002. Recent developments in the study of wild chimpanzee behavior. Evol. Anthropol.: Iss. News Rev. 11(1):9–25. [Google Scholar]
- Nei M. (1987). Molecular evolutionary genetics. Columbia: Columbia University Press. [Google Scholar]
- Oetjens M, Shen F, Zou Z, Kidd J. 2015. Inferring chimpanzee Y chromosome history and amplicon diversity from whole genome sequencing. bioRxiv 029702. [Google Scholar]
- Prado-Martinez J, et al. 2013. Great ape genetic diversity and population history. Nature 499(7459):471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Available from: http://www.R-project.org/. [Google Scholar]
- Rambaut A, Drummond AJ, Suchard M. 2013. Tracer v1. 6.
- Rogers J, Gibbs RA. 2014. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat. Rev. Genetics 15(5):347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SR. (2009). North American regional chimpanzee studbook (Pan troglodytes). Chicago, IL, USA: Lincoln Park Zoo. [Google Scholar]
- Saccone C, et al. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238 (1999):195–209. [DOI] [PubMed] [Google Scholar]
- Stone AC, Griffiths RC, Zegura SL, Hammer MF. 2002. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc. Natl. Acad. Sci. 99(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone AC, et al. 2010. More reliable estimates of divergence times in Panusing complete mtDNA sequences and accounting for population structure. Philos. Trans. R. Soc. B: Biol. Sci. 365(1556):3277–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76:781–821. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, et al. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Excoffier L. 2010. Bayesian inference of the demographic history of chimpanzees. Mol. Biol. Evol. 27(6):1425–1435. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinf. 11(1):116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise CA, Sraml M, Rubinsztein DC, Easteal S. 1997. Comparative nuclear and mitochondrial genome diversity in humans and chimpanzees. Mol. Biol. Evol. 14(7):707–716. [DOI] [PubMed] [Google Scholar]
- Zsurka G, et al. 2010. Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans. BMC Evol. Biol. 10(1):270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.