Abstract
Transcriptional regulatory networks (TRNs) encode instructions for animal development and physiological responses. Recent advances in genomic technologies and computational modeling have revolutionized our ability to construct models of TRNs. Here, we survey current computational methods for inferring TRN models using genome-scale data. We discuss their advantages and limitations. We summarize representative TRNs constructed using genome-scale data in both normal and disease development. We discuss lessons learned about the structure/function relationship of TRNs, based on examining various large-scale TRN models. Finally, we outline some open questions regarding TRNs, including how to improve model accuracy by integrating complementary data types, how to infer condition-specific TRNs, and how to compare TRNs across conditions and species in order to understand their structure/function relationship.
Introduction
Gene expression can be regulated at multiple steps along the process, including transcriptional initiation and elongation, RNA stability, and accessibility and rate of translation. In this review, we focus on regulation of transcriptional initiation by the action of transcription factors (TFs) and cis-regulatory DNA elements. For the purpose of discussion, we define transcriptional regulatory networks (TRNs) as regulatory interactions among TFs and their target genes. Edges in a TRN thus represent direct interactions between a TF and its target genes. Models of TRNs provide systems-level explanation of developmental and physiological functions. Accurate knowledge about TRNs can benefit a range of basic and applied biomedical researches. It can help us better understand the molecular mechanisms of development and cellular reprogramming, which can lead to better strategies to generate various cell types for regenerative therapies. Mechanisms of diseases that are characterized by dysfunction of TRNs can also be elucidated. Knowledge about TRNs can also guide selection of novel drug targets and development of efficient strategies for cellular engineering.
TRNs are highly complex as reflected by the number of regulatory components and complicated connectivity patterns among the components. They typically exhibit highly dynamic and often nonlinear behaviors in response to external or internal signals. For this reason, computational modeling is an essential component in TRN research.
In recent years, high-throughput technologies have greatly expanded our ability to collect complementary data types that can be used for computational modeling of TRNs. This article reviews recent developments and discusses their implications for future research. We start by reviewing computational methods for inferring TRN models using genome-scale data sets. TRNs inferred using large-scale data tend to have less precision but can be used to infer network components and wiring, which is particularly critical for largely uncharacterized TRNs. We discuss their relative advantages and limitations. Next, we summarize representative TRNs constructed using large-scale approaches in both normal and disease development. We then discuss insights into the structure/function relationship of TRNs, based on examining various large-scale TRN models. Finally we outline some open questions regarding TRNs, including how to integrate heterogeneous data types to improve model accuracy, how to infer condition-specific TRNs, and how to compare TRNs under different conditions and across species to understand their structure/function relationship.
Computational approaches for constructing genome-wide TRN models
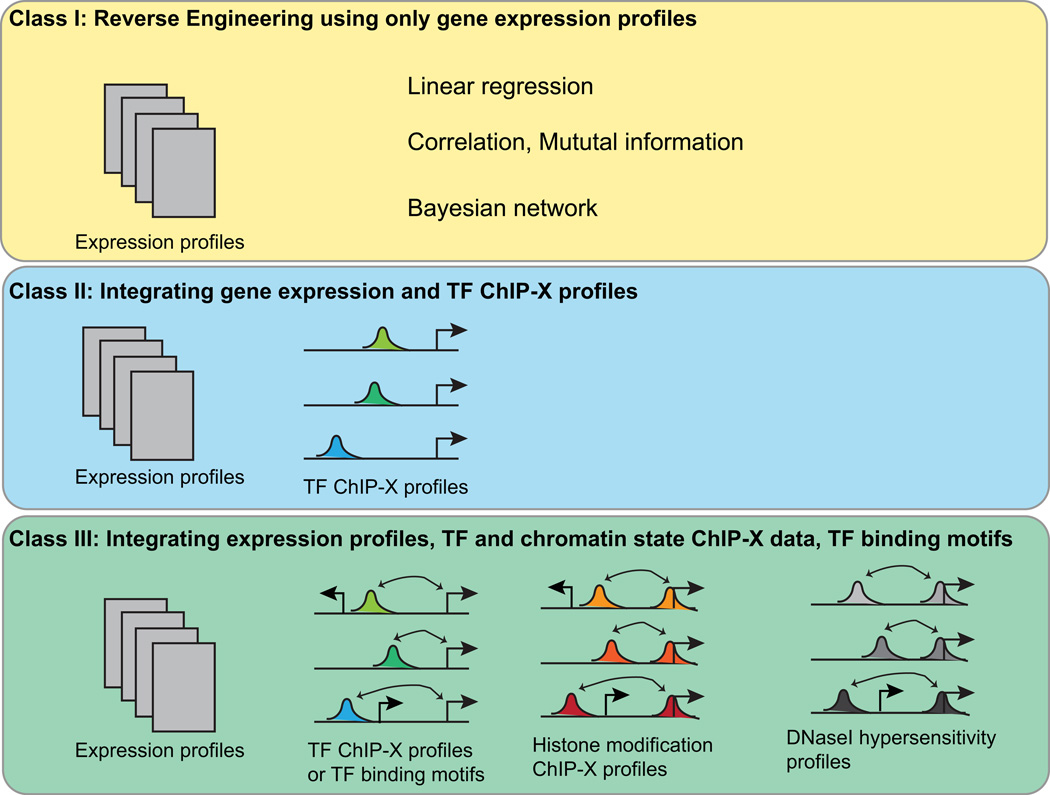
Current approaches to constructing computational models of TRNs can be grouped into three classes based on the type of data used for inference (Figure 1). A list of methods discussed in this review along with their availability is provided in Table 1. The first class utilizes gene expression data as the only input1–8. Because they start with the regulatory output (e.g. expression level), collectively, this class of methods is known as the reverse engineering approach. A number of methods in this class have been developed using various computational frameworks, including linear regression5,9–13, statistical correlation2,3,14–16, and Bayesian network17–21. The basic assumption of regression-based approaches is that the expression levels of the TFs that directly regulate a target gene are the most informative, among all TFs, to predict the expression level of the target gene. When the expression level of a target gene is regressed on the expression levels of TFs, a non-zero regression coefficients indicate statistical dependency which in turn is interpreted as a regulatory interaction between the TF and the gene. Because there are many candidate TFs to consider in a regression, to identify the regulating TFs, a feature selection procedure is typically applied using regularized regression techniques.
Figure 1. Three classes of computational methods for inferring transcriptional regulatory networks.
ChIP-X represents ChIP-ChIP or ChIP-Seq protocol. In the panel representing class III methods, arcs over genome loci and transcription start sites indicate enhancer-promoter links.
Table 1. List of computational methods for modeling transcriptional regulatory networks.
Class I, methods that only use gene expression profiles as their input data; Class II, methods that integrate both gene expression and transcription factor ChIP- X data. Class III, methods that integrate gene expression, transcription factor, and chromatin interaction ChIP-X data. These methods explicitly address a critical subproblem of TRN modeling, i.e. identifying enhancer-promoter interactions.
Correlation-based approaches examine variation in gene expression across different conditions. Variation of gene expression in a large set of conditions provides a means to correlate statistically the expression of a specific TF with the set of expressed genes. The most commonly used correlation measures are Pearson and Spearman correlations. However, they cannot capture nonlinear relationship between two random variables. For this purpose, mutual information has been introduced3,14. Neither correlation measures nor mutual information can distinguish indirect dependencies in a gene-triplet, i.e. dependency between two genes because the expression of both genes is dependent on a third gene. To remove indirectly dependencies, partial correlation coefficient and data processing inequality (DPI) measure have been introduced for correlation- and mutual information-based approaches.
Bayesian networks (BNs) are a class of probabilistic graphical models that represent statistical dependencies among a set of random variables. Formally, a BN is defined by a directed acyclic graph, a set of conditional probability distributions and their parameters, which collectively specify a joint distribution over the set of random variables. In the context of TRN modeling using BNs, nodes in a BN are genes and edges between nodes indicate the conditional dependencies between them. Learning BNs involves two steps: determining the network structure (i.e. structure learning) and estimating the conditional probability values associated with nodes (i.e. parameter learning). The computational challenge underlying BN-based methods is to exhaustively search the space of possible conditional dependencies (i.e. regulatory relationships). Given the numbers of genes and regulatory relationships among them in a typical metazoan species, a full implementation of a BN is intractable. Thus, heuristic approximation methods have been developed that used locally constraint search techniques, making the computational complexity manageable.
The major advantage of the first class of methods is their broad applicability because of the minimal requirement for input data. However, to achieve good performance, these methods typically require a large number of transcriptome profiles, usually at least as many as the number of TFs under study22. However, in most studies, sample size is much smaller than the number of TFs due to high experimental cost. Limited sample size makes the correlations between genes sensitive to noise, and thus highly correlated gene pair needs not imply a true regulatory relationship. A recent study assessed 35 reverse engineering methods using both experimental (E. coli and S. cerevisiae) and computationally simulated data22. The study revealed that no single inference method performs optimally across all data sets. In contrast, integration of predictions from multiple inference methods shows robust and high performance across diverse data sets.
Chromatin immunoprecipitation (ChIP) coupled with high-throughput techniques, such as sequencing or microarray (ChIP-Seq/Chip, hereafter refer to as ChIP-X) can provide genome-wide occupancy information for a given TF. Such data has become increasingly abundant in recent years. Although helpful, the utility of ChIP-X data alone for inferring regulatory interactions is limited because binding events detected by ChIP-X is only necessary but not sufficient for functional regulatory interactions. To address this shortcoming, the second class of methods combines gene expression profiling data with TF ChIP-X data to infer TRNs23–30. These methods fall into two categories in terms of their assumptions and approaches. In the first category, the methods identify subsets of ChIP-X binding sites for which the regulated genes have highly correlated expression profiles, and thus are co-regulated23,28. Methods in the second category use various regression techniques to fit ChIP-X binding data to the observed gene expression profiles in order to infer regulatory interactions24–26,30. Common to these approaches, a linear relationship between gene expression changes and TF binding affinities is assumed.
All class II methods except that by Maienschein-Cline et al. use the gene nearest to a ChIP-X binding site as the candidate regulated gene. This approach works well for compact genomes (e.g. bacteria and yeasts) and promoter-proximal TF binding sites. However, for metazoan species, functional TF binding sites may not reside next to their targets. A number of recent studies using chromosome confirmation capture (3C) based technologies have shown that the majority of enhancers do not target genes closest to them31–33. For this reason, accurate identification of enhancer-promoter interactions becomes a critical first step towards constructing accurate TRNs. Once the enhancer-promoter pairing is predicted, DNA motif analysis of the enhancer sequences is used to infer regulatory interactions between TFs that bind the enhancers and the paired gene promoters. The enormous amount of public data generated by projects such as ENCODE and Epigenomics Roadmap has opened up the door for integrative approaches to constructing TRNs for metazoan species. These integrative approaches represent the third class of methods for modeling TRNs. At the core of these methods is a strategy for linking transcriptional enhancers with target promoters. A general assumption of these methods is that the chromatin states of bona fide enhancers-promoter pairs tend to be correlated across cell/tissue types. Under this general assumption, different genome-wide chromatin state data have been used, including histone modifications34 and chromatin openness (as measured by DNase I hypersensitivity)35. Further development along this line has correlated chromatin state of enhancers with expression profiles of promoters36,37. He et al.38 took this approach further and identified three additional genomic features in addition to chromatin state correlation, including co-expression between promoter and genes encoding transcription factors that occupy the enhancer under consideration, sequence co-evolution between enhancer and promoter. These features when combined with chromatin state correlation were shown to significantly improve the inference accuracy of enhancer-promoter pairs.
Large-scale mapping of TRNs during normal development
In the past several decades, through the effort of individual labs and large research consortia, large-scale TRN models have been constructed for various metazoan species. The TRN controlling sea urchin endomesoderm patterning is the largest and most extensively validated developmental TRN to date39. In C. elegans, TRN for the intestine has been constructed using yeast one-hybrid assay40. At a more global scale, ChIP-Seq was performed for 92 transcription factors spanning 11 developmental stages by the modENCODE consortium. Integration of ChIP-Seq and expression data produced a spatiotemporally resolved TRNs for this species41. In D. melanogasters, dorsal/ventral and anterior/posterior patterning have been studied extensively. TRNs for these two developmental processes have been mapped using ChIP-X assays and computational modeling42–45. Like C. elegans, the modENCODE project also generated TRN models based ChIP-Seq data for 38 fly TFs in different developmental stages and cell types46. In mammalian species, TRNs in various cell types have been constructed by individual labs, including dendritic cell47, macrophage48, embryonic stem cell49–52, hematopoietic stem cell53–55, B cells1, Th17 cells56,57 and T-cell fate specification58. By integrating TF ChIP-Seq data, gene expression profiles, and chromatin modification ChIP-Seq data, the ENCODE and mouse ENCODE projects have also generated TRN models for various human and mouse cells/tissues36,59–61
TRNs in diseased cells
Given the key role of transcriptional regulation in development and cellular homeostasis, it is not surprising that perturbations to TRNs can lead to many diseases. Such perturbations include mutations in regulatory DNA sequences and in transcription factors, co-factors, and chromatin regulators. In order to understand the roles of mutations in pathogenesis, the underlying disease-specific TRNs in which the mutated factors operate need to be defined. Nowadays, the same high throughput technologies used to interrogate TRNs in normal cells are increasingly being applied to study TRNs underlying various human diseases especially cancer62–69. The developing insight is that the same TRNs in normal cells are either rewired or acquire altered activities in diseased cells to promote pathogenesis.
Reverse engineering approaches have been applied to construct TRNs in cancer cells. Carro et al. constructed a glioma-specific TRNs and identified two TFs (C/EBPβ and STAT3) as synergistic master regulators of oncogenic transformation62. Gatta et al.67decipher the oncogenic TRNs controlled by the two TFs, TLX1 and TLX3, in T cell acute lymphoblastic leukemia (T-ALL).
As ChIP-X technology became mature and more sensitive, they have been increasingly used to map TRNs in cancer cells. A recent study revealed that the oncogenic TF TAL1 forms an interconnected autoregulatory loop with two TF partners (RUNX1 and GATA3) in T-ALL. This circuitry contributes to the sustained activation of TAL1-regulated oncogenic program66. Many oncogenic TFs are generated by chromosomal translocation events. A well-known one is the RUNX1/ETO fusion TF, which is generated by the chromosomal translocation t(8;21)70. Using ChIP-Seq and expression profiling, Ptasinska et al.68 showed that the transcriptional program underlying leukemic propagation is regulated by a dynamic equilibrium between the TRNs regulated by the RUNX1/ETO fusion TF and intact RUNX1 complexes. Using the same approach, TRNs in breast cancer64, prostate cancer69, and lung cancer63 have also been studied.
Structure/function relationship of TRNs
Previous effort on mapping TRNs (both small-scale and large-scale) has yielded a large number of TRN models. Analysis of available TRN architectures has revealed the following organizational principles. First, TRNs have a global hierarchical topology36,59,71. TRNs for embryonic development of animal body parts, such as that for specification of endomesoderm in sea urchin39, for specification of gut and mesoderm in C. elegans72, and for specification of eye lens field in zebra fish73,74, tend to have deep hierarchical organization. In comparison, TRNs for terminal fate choice from multipotent stem cells and physiological responses, such as that for specification of erythroid versus myeloid fates bifurcate75,76, T helper versus killer cells diversification77, innate immunity response78, are relatively shallow. The structural difference between embryonic development TRNs and terminal fate TRNs reflects the difference in their functional requirement. Development of the body plan requires a long sequence of progressive decisions that need to be made in different spatiotemporal domains. In contrast, terminal cell fate can be specified with fewer decisions.
Embedded in the global hierarchical topology of TRNs are many over-represented small connectivity patterns, the so-called network motifs79,80. Different types of network motifs are characterized by their different connectivity patterns and distinct functions associated with them. Some of the simplest and most prevalent motifs are feedback loops, either positive or negative. Positive feedback loops are often observed in systems that show switch-like behavior, memory, or bistability. Negative feedback loops are functionally associated with systems that show strong noise resistance to perturbations. A slightly more complex motif is feedforward loop (FFL). This motif consists of three genes: a TF, X, which regulates TF Y, and gene Z, which is regulated by both X and Y. FFL motifs fall into two classes depending on the net sign of the regulatory actions of the two arms of the motif. Coherent FFL motifs have two arms with the same net sign of actions whereas incoherent FFL motifs have two arms with different net signs of action. Coherent FFLs has been shown to filter out brief spurious pulses of signal. Thus, gene Z only respond in the presence of persistent signal that is over its threshold, instead of a brief signal81,82. In incoherent FFLs, the two arms of the FFL act in opposition. They have been shown to generate pulse-like response dynamics by Z after X is activated83. For detailed treatment of other types of network motifs and their structure/function relationship, readers are referred to a number of excellent reviews on this topic80,84,85.
Summary and Outlook
A large number of studies have demonstrated the enormous value that the assembly of TRN models has in understanding normal and disease development. In this review, we only considered networks involving transcription factors and their target genes. However, these networks are part of a much larger and more complex cellular network composed of many other types of molecules and their interactions. For this reason, combining multiple, independently generated observations (such as gene expression, in vivo TF binding and chromatin modification states, protein abundance measure, physical and genetic interactions among genes) to infer network structure can strengthen the resulting models and provide novel insights. Although the majority of current computational methods use only gene expression and/or ChIP-X data as the input to infer TRNs, more integrative methods have been developed in the past few years. For instance, researchers have integrated chromatin modification ChIP-Seq data to construct TRNs (e.g. class III approach). Such integrative approaches will become increasingly powerful as more data becomes available.
Most computational methods and TRN models discussed in this review are global networks. That is, regulatory interactions in these networks are not specific (or not specific enough) to a particular phenotype under study. Such condition-specific-interactions are critical for better understanding the behavior of the network. Advanced methods (both computational and experimental) are needed to allow capturing more nuanced network models.
As models of TRNs start to accumulate rapidly, novel computational methods are needed to allow principled comparisons of TRNs to gain insights into the structure, function and evolution relationship of TRNs. For instance, comparing TRNs of normal and diseased cells will be particularly fruitful for understanding the molecular mechanisms of pathogenesis. Similarly, comparing developmental TRNs across species will provide valuable insights into their evolution and organization principles.
Acknowledgments
B.H. was supported by National Institutes of Health grants HG006130. K.T. was supported by National Institutes of Health grants HG006130, GM104369, GM108716.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review,
Have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 2.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. This paper presents a popular reverse engineering method based on mutual information. The method has been used in a number of successful studies of deregulated TRNs in human cancers.
- 4.Belcastro V, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic acids research. 2011;39:8677–8688. doi: 10.1093/nar/gkr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. This paper presents the first reverse engineering method to identify TRN governing the SOS response in E. coli.
- 6.Bonneau R, et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome biology. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. Reconstructing dynamic regulatory maps. Molecular systems biology. 2007;3:74. doi: 10.1038/msb4100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 9.Della Gatta G, et al. Direct targets of the TRP63 transcription factor revealed by a combination of gene expression profiling and reverse engineering. Genome Res. 2008;18:939–948. doi: 10.1101/gr.073601.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung MK, Tegner J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad Sci U S A. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Someren EP, et al. Least absolute regression network analysis of the murine osteoblast differentiation network. Bioinformatics. 2006;22:477–484. doi: 10.1093/bioinformatics/bti816. [DOI] [PubMed] [Google Scholar]
- 12.Haury AC, Mordelet F, Vera-Licona P, Vert JP. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol. 2012;6:145. doi: 10.1186/1752-0509-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haynes BC, et al. Mapping functional transcription factor networks from gene expression data. Genome Res. 2013;23:1319–1328. doi: 10.1101/gr.150904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 15.Luo W, Hankenson KD, Woolf PJ. Learning transcriptional regulatory networks from high throughput gene expression data using continuous three-way mutual information. BMC Bioinformatics. 2008;9:467. doi: 10.1186/1471-2105-9-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soranzo N, Bianconi G, Altafini C. Comparing association network algorithms for reverse engineering of large-scale gene regulatory networks: synthetic versus real data. Bioinformatics. 2007;23:1640–1647. doi: 10.1093/bioinformatics/btm163. [DOI] [PubMed] [Google Scholar]
- 17. Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. This paper represents one of the first attemp of using Bayesian network to model TRNs using gene expression data in baker's yeast.
- 18.Hartemink AJ, Gifford DK, Jaakkola TS, Young RA. Using graphical models and genomic expression data to statistically validate models of genetic regulatory networks. Pac Symp Biocomput. 2001:422–433. doi: 10.1142/9789814447362_0042. [DOI] [PubMed] [Google Scholar]
- 19.Pe'er D, Regev A, Elidan G, Friedman N. Inferring subnetworks from perturbed expression profiles. Bioinformatics. 2001;17(Suppl 1):S215–S224. doi: 10.1093/bioinformatics/17.suppl_1.s215. [DOI] [PubMed] [Google Scholar]
- 20.Husmeier D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics. 2003;19:2271–2282. doi: 10.1093/bioinformatics/btg313. [DOI] [PubMed] [Google Scholar]
- 21.Zou M, Conzen SD. A new dynamic Bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics. 2005;21:71–79. doi: 10.1093/bioinformatics/bth463. [DOI] [PubMed] [Google Scholar]
- 22. Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. This paper presents a comprehensive evaluation of 35 reverse engineering methods for modeling TRNs. They demonstrated that community network prediction outperforms individual methods.
- 23. Bar-Joseph Z, et al. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 2003;21:1337–1342. doi: 10.1038/nbt890. This paper presents the GRAM algorithm, one of the first algorithm to infer TRNs by integrating gene expression profiles and TF ChIP-Chip data. The method was applied to 106 TF ChIP-Chip data and 500 expression profiles to construct a genome-wide TRN in baker's yeast.
- 24.Gao F, Foat BC, Bussemaker HJ. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulesteix AL, Strimmer K. Predicting transcription factor activities from combined analysis of microarray and ChIP data: a partial least squares approach. Theor Biol Med Model. 2005;2:23. doi: 10.1186/1742-4682-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanguinetti G, Rattray M, Lawrence ND. A probabilistic dynamical model for quantitative inference of the regulatory mechanism of transcription. Bioinformatics. 2006;22:1753–1759. doi: 10.1093/bioinformatics/btl154. [DOI] [PubMed] [Google Scholar]
- 27.Maienschein-Cline M, Zhou J, White KP, Sciammas R, Dinner AR. Discovering transcription factor regulatory targets using gene expression and binding data. Bioinformatics. 2012;28:206–213. doi: 10.1093/bioinformatics/btr628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G, Ji H. ChIPXpress: using publicly available gene expression data to improve ChIP-seq and ChIP-chip target gene ranking. BMC Bioinformatics. 2013;14:188. doi: 10.1186/1471-2105-14-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlebach G, Shamir R. Constructing logical models of gene regulatory networks by integrating transcription factor-DNA interactions with expression data: an entropy-based approach. J Comput Biol. 2012;19:30–41. doi: 10.1089/cmb.2011.0100. [DOI] [PubMed] [Google Scholar]
- 30.Liao JC, et al. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. doi:S0092-8674(11)01517-0 [pii]10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. This study applied chromosome conformation capture carbon copy to interrogate interactions between gene promoters and distal elements in 1% of the human genome. The authors found only ~27% of looping interactions are with the nearest gene, indicating that genomic proximity is a poor predictor for enhancer-promoter interactions.
- 33.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. doi:nature09906 [pii]10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He B, Chen C, Teng L, Tan K. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci U S A. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. This paper presents a state-of-the-art algorithm to predict enhancer-promoter interactions using multiple genomic features. Systematic analysis of enhancer-promoter interactome in 12 human cell types revealed global features of enhancer-promoter communication.
- 39.Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 2009;583:3948–3958. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deplancke B, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. doi:S0092-8674(06)00624-6 [pii] 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 41.Araya CL, et al. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2014;512:400–405. doi: 10.1038/nature13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacArthur S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. doi:gb-2009-10-7-r80 [pii]10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 46.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. doi:nature09990 [pii]10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 49.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. doi:S0092-8674(05)00825-1 [pii]10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. doi:S0092-8674(08)00617-X [pii]10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 53.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. doi:S1934-5909(10)00440-6 [pii]10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 54.Beck D, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 55.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neph S, et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar MS, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 64.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sanda T, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. This study is an excellent example of combining gene expression profile and ChIP-Seq to map deregulated TRN in cancer cell. The authors identified an autoregulatory loop involving TAL-1 and two other TFs. This subnetwork contributes to the sustained activation of TAL1-regulated oncogenic program.
- 67. Della Gatta G, et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med. 2012;18:436–440. doi: 10.1038/nm.2610. This study identified RUNX1 as a tumor suppressor gene in T-ALL. It demonstrates the power of reverse-engineering methods to identify key deregulated regulatory circuits in human cancer.
- 68.Ptasinska A, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8:1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan PY, et al. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32:399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 71. Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. This is an excellent review on structure/function relationship of TRNs for animal development.
- 72.Maduro MF. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim Biophys Acta. 2009;1789:250–260. doi: 10.1016/j.bbagrm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan TM, et al. Developmental gene regulatory networks in the zebrafish embryo. Biochim Biophys Acta. 2009;1789:279–298. doi: 10.1016/j.bbagrm.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 74.Morley RH, et al. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc Natl Acad Sci U S A. 2009;106:3829–3834. doi: 10.1073/pnas.0808382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang P, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci U S A. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rothenberg EV. Decision by committee: new light on the CD4/CD8-lineage choice. Immunol Cell Biol. 2009;87:109–112. doi: 10.1038/icb.2008.100. [DOI] [PubMed] [Google Scholar]
- 78.Amit I, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 80. Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. doi:nrg2102 [pii]10.1038/nrg2102. This is an excellent review on network motifs and their functions, with an emphasis on experimental studies.
- 81.Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 82.Kalir S, Mangan S, Alon U. A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100010. 2005 0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sneppen K, Krishna S, Semsey S. Simplified models of biological networks. Annu Rev Biophys. 2010;39:43–59. doi: 10.1146/annurev.biophys.093008.131241. [DOI] [PubMed] [Google Scholar]
- 85.Lim WA, Lee CM, Tang C. Design principles of regulatory networks: searching for the molecular algorithms of the cell. Mol Cell. 2013;49:202–212. doi: 10.1016/j.molcel.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]