Abstract
Increasing vulnerability of plants to a variety of stresses such as drought, salt and extreme temperatures poses a global threat to sustained growth and productivity of major crops. Of these stresses, drought represents a considerable threat to plant growth and development. In view of this, developing staple food cultivars with improved drought tolerance emerges as the most sustainable solution toward improving crop productivity in a scenario of climate change. In parallel, unraveling the genetic architecture and the targeted identification of molecular networks using modern “OMICS” analyses, that can underpin drought tolerance mechanisms, is urgently required. Importantly, integrated studies intending to elucidate complex mechanisms can bridge the gap existing in our current knowledge about drought stress tolerance in plants. It is now well established that drought tolerance is regulated by several genes, including transcription factors (TFs) that enable plants to withstand unfavorable conditions, and these remain potential genomic candidates for their wide application in crop breeding. These TFs represent the key molecular switches orchestrating the regulation of plant developmental processes in response to a variety of stresses. The current review aims to offer a deeper understanding of TFs engaged in regulating plant’s response under drought stress and to devise potential strategies to improve plant tolerance against drought.
Keywords: drought, transcription factors, climate change, abiotic stress, crop plants
Introduction
The 21st century agriculture is facing a daunting challenge of attaining nearly up to 70% increase in crop productivity by 2050 (Friedrich, 2015; Joshi et al., 2016a; Wang et al., 2016b). Also, crop productivity needs to witness up to 40% increase by 2015 in view of the cultivated area being increasingly affected by stresses (Pennisi, 2008). Frequent changes in climatic conditions also affect the crop yield (such as flooding after high temperature), which led scientists to undertake research on this aspect in recent time around the world (Nakashima et al., 2014a). Among the various abiotic factors challenging crop production globally, drought stress is increasingly playing a crucial role. Drought is a meteorological term and is commonly defined as a combined interplay of reduced rainfall, decreasing ground water table, limiting water availability with rise in temperature (Singh and Laxmi, 2015; Singh B. et al., 2015). With global climatic vagaries registering escalated frequency, the global drought incidence is likely to swell beyond 20% by the end of this century, especially in Central and South American corn belt, and Central and Western Europic regions (Edmeades, 2013; Singh B. et al., 2015). Similarly in South Asia, where more than 75% of the farmers are dependent on rainfed agriculture, a damage of about US$ 84 billion is predicted owing to the global climate change (Mendelsohn, 2014). Most of the crops are susceptible to drought stress, resulting in more than 50% yield losses (Singh B. et al., 2015). In South and Southeast Asia, drought stress causes about 40% annual loss in productivity, which results in 58% of income loss (Li et al., 2015). Similarly, regional climate change models showed Mediterranean regions and Middle East among the ‘Hot-spots’ of severe drought, directly affecting crop failures and livestock death (Barlow et al., 2015; Saadi et al., 2015). Brazil, which is the second highest producer (40%) of soybean has lost greater than 20% of their productivity because of the occurrence of drought during 2003–2005 (Nakashima et al., 2014b). The uneven distribution of rainfall and ground water shortage often create drought stress conditions in the environment (Luo and Zhang, 2001), which eventually leads to an enormous decrease in the grain yield potential.
Upon exposure to drought, plants manifest multiple impairments including cell injury through reactive oxygen species (ROS) generation and increasing cellular temperature, which result in an increase in the viscosity of cellular contents, alterations in the protein–protein interactions, protein aggregation, and denaturation (Farooq et al., 2008). Cell shrinkage followed by a marked decline in cellular volume becomes evident as an instant symptom caused by dehydration. Besides, greater accumulation of solutes causes toxicity and negatively affects functioning of some enzymes, often leading to reduced photosynthesis and water use efficiency (WUE). Under prolonged dehydration, plants exhibit leaf rolling followed by wilting and bleaching that eventually result in death of the plant (Sahoo et al., 2013). Reproductive stages, i.e., flowering as well as seed development are especially sensitive to drought stress (Samarah et al., 2009a,b,c; Alqudah et al., 2010; Samarah and Alqudah, 2011). A reduction in number of grains per spike, grain filling duration, and dry matter accumulation, leading to decreased grain weight in barley kernels has already been reported earlier (Samarah et al., 2009a; Samarah and Alqudah, 2011). Recently, leaf area, leaf weight as well as leaf growth rate were also reported to hold great relevance while breeding crops grown under Mediterranean environmental conditions such as drought stress (Alqudah and Schnurbusch, 2015; Quan et al., 2016). Substantial progress has already been made to enhance crop productivity using conventional breeding methods. However, development of drought tolerant cultivars using conventional breeding approach is greatly restricted by low heritability of drought tolerance, narrow genetic variation existing in the crop’s exploitable genetic pool, complex and multigenic nature of this trait and high magnitude of environmental interactions (Pardo, 2010; Hill et al., 2013; Jha et al., 2014; Liu J.H. et al., 2014).
Considering global food security, immediate assessment of drought stress impact and innovative techniques to breed tolerant and productive cultivars under dramatically changing climate are essentially warranted (Mishra and Singh, 2010). As a means to provide crop-based answer to burgeoning issues like food insecurity and malnutrition, cereals constitute a vital role as major fraction in the diet, especially in developing nations (Bohra et al., 2015). Rice, wheat and maize are major cereal crops in the world; therefore less water availability exerts profound consequences (Joshi et al., 2011). Development of advanced genotypes with improved stress tolerance and wider adaptability is a simple, cost-effective and eco-friendly approach to cope with drought stressed scenarios (Jha et al., 2014).
Drought tolerance is an outcome of a series of molecular, cellular, and physiological processes including induction/repression of various genes that cause accumulation of various osmolytes, improved antioxidant system, reduced transpiration, inhibited shoot growth, and decreased tillering (Pareek et al., 2010). Besides, the phytohormone abscisic acid (ABA) is reported to be abundant under water-deficit conditions and this in turn causes stomata closure and induces expression of various stress-related genes (Yang et al., 2011). It has been shown that drought inducible gene expression is also governed by ABA-independent regulatory system (Aguado et al., 2014). Plant’s ability to cope with water deficit depends largely on its water status, which changes with environmental conditions (Joshi et al., 2016b). Thus, incorporating drought stress tolerance in crops becomes imperative under changing climatic conditions and it could be a promising approach to meet the global food demand (Todaka et al., 2012). In this context, unraveling the molecular mechanisms that control the perception and transduction of stress signals to initiate adaptive responses is crucial for engineering drought stress tolerance in plants (Ray et al., 2010; Sanchez et al., 2011).
Notable progress has been made toward this end by utilizing modern genetics and functional genomics approaches such as transcriptomics, proteomics and metabolomics and consequently, various drought stress responsive genes have been identified and characterized in crops. These key genes mainly code for proteins that have either metabolic or regulatory roles, such as those involved in detoxification, osmolyte biosynthesis, proteolysis of cellular substrates, water channel, ion transporter, heat shock protein (HSP), and late embryogenesis abundant (LEA) protein (Joshi et al., 2016b). On the other hand, the regulatory class primarily comprises of TFs (AREB, AP2/ERF, NAC, bZIP, MYC, and MYB), signaling protein kinases [mitogen activated protein kinases (MAPK), calcium-dependent protein kinases (CDPK), receptor protein kinases, ribosomal protein kinases, and transcription regulation protein kinases] and protein phosphatases (phosphoesterases and phospholipase), which synchronize signal transduction and expression of genes during stress responses (Wani et al., 2013). Various desiccation-induced genes have been characterized, including those encoding ABA biosynthetic pathway (Sah et al., 2016), osmo-protectants such as proline, trehalose, mannitol, ectoine, glycine betaine providing tolerance to cellular dehydration (Wani and Gosal, 2011; Yao et al., 2011; Varshney et al., 2011), cellular enzymes (Puckette et al., 2007), signaling proteins (Zhu, 2002), and TFs (Mochida et al., 2009; Gosal et al., 2010). Several of these regulatory genes including TFs were found to play essential roles in multiple abiotic stress responses via regulating downstream stress-responsive genes. As illustrated in Figure 1, TFs regulate gene expression through binding to cis-regulatory elements in the promoter region of different stress-related genes (Nuruzzaman et al., 2013; Franco-Zorrilla et al., 2014). Thus, genetic modification of the expression of these regulatory genes can greatly influence plant stress tolerance because they further regulate many downstream stress-responsive genes at a given time (Wang et al., 2016). Among these regulatory genes, stress-responsive TFs have gained widespread attention on account of their significant role in plant stress tolerance (Shao et al., 2015) and allelic variations were examined across Indian wild rice population by researchers in genes with respect to abiotic stresses (Singh et al., 2015a; Mishra et al., 2016). In this article, we elaborate on the key TFs that contribute toward drought stress tolerance, and their networks, cross-talks and potential utilization in crop improvement programs.
FIGURE 1.
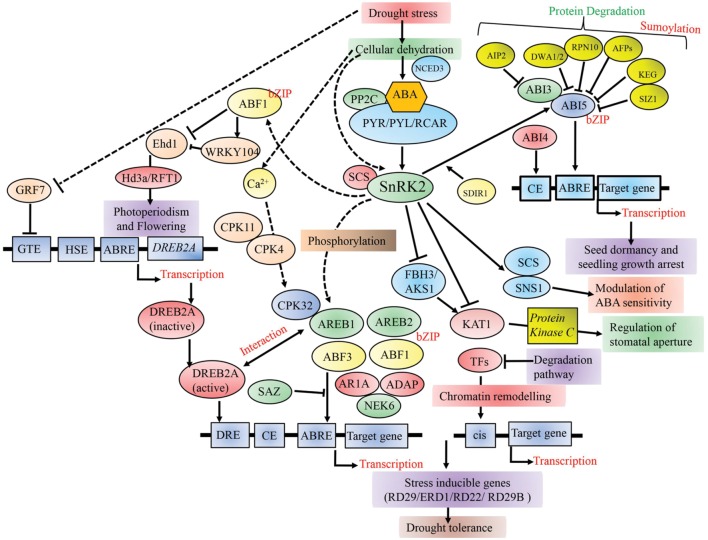
A schematic model of the transcriptional regulation of different TFs playing key roles in cellular dehydration in plants. PP2C-PYR/PYL/RCAR complex positively regulates AREB/ABF-SnRK2 pathway. SnRK2s further modulate other TFs downstream including AREB/ABFs, FBH3/AKS1, and SNS1 during stress as well as seed maturation. Phosphorylated AREB/ABF TFs, AREB1, AREB2, ABF3, and ABF1 bind to the promoter region of target genes and activate their expression in response to dehydration stress. GRF7 suppresses the expression of DREB2A, which is a key TF in ABA-independent gene expression. Broken lines indicate possible roles. PP2C-PYR/PYL/RCAR, pyrabactin resistance1/PYR1-like/regulatory components of ABA receptors; PP2C, protein phosphatase 2C; CE, coupling element; GTE, GRF7-targeting cis element; HSE, heat shock element; TFs, transcription factors.
Molecular Mechanisms Regulating Plant’s Response toward Drought Stress
The response of any crop to drought stress depends primarily on the growth stage and WUE (Pareek et al., 2010). For example, reproductive stage is considered critically susceptible to drought stress in various crops (Moumeni et al., 2015; Farooq et al., 2016). As sessile organisms, plants have evolved several defense mechanisms involving various molecular, physiological and biochemical alterations in response to low soil moisture. Visible symptoms describing drought tolerance include leaf rolling, stay green ability, epicuticular wax deposition, closing of stomata, enhanced root length leading to higher WUE, photochemical quenching, photoinhibition resistance, osmotic adjustment, and membrane stabilization at the cellular level (Khazaei et al., 2013). Leaf rolling, a foremost symptom, constitutes an initial survival mechanism against osmotic stress that can minimize canopy temperature and transpiration rate, thereby improving WUE (Singh B. et al., 2015). Thus, species maintaining higher relative water content (RWC) under osmotic stress are reported to be less susceptible to low water potential, and thus retain their growth and productivity (Joshi and Karan, 2013). It was also observed that crop growth is primarily supported by the soil moisture surrounding plant root system (Tron et al., 2015). Therefore, total root area being directly affected by the decreased soil water content deserves greater attention. It was reported that osmotic stress-induced premature differentiation caused growth inhibition of primary roots and allows outgrowth of lateral roots (Ji et al., 2014). A range of biochemical changes have also shown in response to drought stress, including higher accumulation of osmolytes, ROS, antioxidant species, and antioxidative enzymes (Kholová et al., 2011). These osmolytes are reported to have principal role in maintaining enzyme activity and membrane stability under osmotic stress (Du et al., 2014; Khattab et al., 2014; Singh B. et al., 2015).
Drought stress in plants occurs either due to poor quantity of water, i.e., little rainfall or due to insufficient quality of water, i.e., saline habitats (Buchanan et al., 2015). As a means to survive under water stress conditions, plants respond by alteration in expression of several genes and regulate them through complex transcriptional networks (Singh and Laxmi, 2015). It is therefore imperative to dissect stress regulatory mechanisms and to pinpoint the key regulators that could eventually be harnessed to breed or engineer stress tolerant plants. By analyzing plant genomes and employing modern OMICS tools, including genomics, transcriptomics and proteomics, significant progress has been made in elucidating the stress signaling pathways involved in drought stress response (Liu J.H. et al., 2014). Few of these genes associated with these transcriptional networks have been characterized in various molecular studies and have been found to be key contributors toward drought stress tolerance in transgenic plants (Todaka et al., 2015).
The signaling pathway of any abiotic stress involves certain key steps such as signal perception, transduction, responsiveness, combined with activation of physiological, and metabolic reactions (Pérez-Clemente et al., 2013; Liu J.H. et al., 2014). In this process, plant cells first perceive stress stimulus through sensors or receptors localized mostly at the cell membrane. Then, the receiver signaling molecules activate the intracellular ones through second messengers like calcium ions, inositol phosphate, ROS, cyclic nucleotides (cAMP and cGMP), sugars and nitric oxide. Subsequently, these second messengers initiate the corresponding signaling pathways to transduce the signals (Bhargava and Sawant, 2013). The phosphorylation and dephosphorylation of proteins regulated by protein kinases and phosphatases, respectively, is a significant and effective mechanism in several signal transduction pathways. For example, the MAPKs and CDPKs are well known for their role in drought stress signaling pathways (Huang et al., 2012). At the end of the phosphorylation cascade, TFs are activated or suppressed by protein kinases or phosphatases, and directly regulate the expression of an array of downstream genes by interacting with the specific cis-elements in their promoter region (Danquah et al., 2014). In addition, TFs themselves are regulated at the transcription level by other upstream components (Hirayama and Shinozaki, 2010) and after several modifications at the post-transcription level, such as ubiquitination and sumoylation, they form a complex regulatory network which modulates the expression of stress responsive genes, regulating various physiological and metabolic processes (Mizoi et al., 2013). In the past few decades, considerable research has been carried out toward identification and characterization of different TFs that contribute toward drought stress response.
TFs Can Induce a Range of Stress Responsive Genes in Plants
The multifarious signaling pathways of water stress in plants consist of several proteins, i.e., TFs, enzymes, functional proteins, molecular chaperones, and metabolites (Song et al., 2013). Genetic engineering approaches to mitigate the challenge of drought stress in plants involve the overexpression of these TFs, enzymes and other metabolites (Singh B. et al., 2015). In recent years, a wide range of TF families holding relevance with drought stress response have been identified (Anbazhagan et al., 2015). During the signal transduction, TFs directly regulate the expression of the associated genes via serving as molecular switches. These TFs interact specifically with cis-elements located in the promoter region of the genes they regulate. In plants, a large proportion of genes in the genome (up to 10%) potentially encode TFs (Franco-Zorrilla et al., 2014), which are categorized into different gene families such as AREB, DREB, MYB, WRKY, NAC, and bZIP based on the distinct structure of their DNA-binding domain (Golldack et al., 2011; Jin et al., 2014). In Arabidopsis nearly 6% of the proteome is dedicated to TFs (Rayko et al., 2010). Among these TF genes, several have been reported to respond under drought stress through pathways dependent/independent of ABA. Promising results have been obtained concerning engineering of some TF genes with an aim to improve tolerance against drought stress. In this section, we describe major TF families known to be responsible for drought stress tolerance and discuss the scope for manipulating them in order to obtain transgenic plants with enhanced drought tolerance.
AREB/ABF TFs
Several research groups have elucidated the molecular mechanism related to drought stress transcriptional networks in plants. Under osmotic stress conditions, detailed molecular analyses have found abscisic acid-responsive element binding protein (AREB)/ABFs (ABRE binding factor) as a major transcriptional activator modulating the expression of genes during ABA signaling (Maruyama et al., 2012). ABA-responsive gene expression is controlled by a conserved ABRE (PyACGTGG/TC) cis-element in its promoter region (Figure 1). Studies have revealed the essential role of several ABRE or its combinations with coupling elements (CE) during ABA-responsive genes expression (Fujita et al., 2013; Nakashima and Yamaguchi-Shinozaki, 2013). Structurally, AREB/ABFs consist of four SnRK2 phosphorylation sites containing conserved domains that regulate ABA-dependent gene expression (Fujita et al., 2011, 2013). It was found that vascular tissue specific cells produce ABA and transfer them to target cells (Kuromori et al., 2014). It was also demonstrated that ABA is synthesized within guard cells and vascular tissue specific cells (Bauer et al., 2013). In Arabidopsis, five genes are known to code for NCED (9-cis-epoxy carotenoid dioxygenase), a major enzyme in ABA biosynthesis and expression. Of these, NCED3 showed higher expression under dehydration and was reported to be under the regulation of an AG-box recognition sequence located 2248 bp upstream to its transcriptional start site (Behnam et al., 2013). ABA is perceived by an ABA-bound PYL/PYR/RCARs receptor complex combined with PP2Cs, and subsequently SnRK2s is released (Miyakawa et al., 2013; Nakashima and Yamaguchi-Shinozaki, 2013). The activated SnRKs phosphorylate the downstream proteins like AREB/ABF TFs, which then bind to the ABRE cis-element (PyACGTGG/TC) (Umezawa et al., 2013). In the absence of ABA, PP2Cs inhibit ABA signaling pathway by dephosphorylating SnRK2s. Of the nine AREB/ABF family members, AREB1/ABF2 regulates ABA signaling during drought stress at the vegetative stage (Figure 1). ABA is essential for proper functioning of AREB1, and ABA-dependent phosphorylation regulates its activity (Yoshida et al., 2010). Overexpression of AREB1 was shown to improve drought tolerance in Arabidopsis, rice and soybean (Oh et al., 2005; Barbosa et al., 2013; Yoshida et al., 2015). Recently it was shown that Arabidopsis plants overexpressing wheat transcription factor TaAREB3 have enhanced ABA sensitivity and drought tolerance (Wang et al., 2016a) (Table 1). In primary species, group A PP2Cs have evolved as essential regulators providing osmotic stress tolerance (Komatsu et al., 2013).
Table 1.
Major families of TF genes expressed in response to drought stress in plants.
| Gene Family | Gene (Gene ID) | Identified in crop | Studied crop | Drought/other stress related function | References |
|---|---|---|---|---|---|
| AREB/ABF | AREB1, AREB2/ABF4 and ABF3 | Arabidopsis thaliana | Arabidopsis thaliana | Dehydration/Drought | Fujita et al., 2005; Yoshida et al., 2015 |
| AREB1, AREB2/ABF4 and ABF3 | Arabidopsis thaliana | Arabidopsis thaliana | Osmotic stress | Choi et al., 2000; Kang et al., 2002 | |
| Kobayashi et al., 2008; Lee et al., 2010 | |||||
| ABF2 | Arabidopsis thaliana | Arabidopsis thaliana | Osmotic stress | Kim et al., 2004 | |
| AP2/ERF | SodERF3 | Saccharum officinarum L. cv Ja60-5 | Nicotiana tabacum L. cv. SR1 | Dehydration/Multiple stress | Trujillo et al., 2008 |
| GmERF3 | Glycine max | Nicotiana tabacum | Dehydration/Multiple stress | Zhang et al., 2009 | |
| AtDREB1A | Arabidopsis thaliana | Glycine max | Dehydration | de Paiva Rolla et al., 2014 | |
| OsDREB1F | Oryza sativa | Oryza sativa | Drought/Multiple stress | Wang et al., 2008 | |
| GmDREB2A;2 | Glycine max | Glycine max | Dehydration/Multiple stress response | Mizoi et al., 2013 | |
| DREB1/CBF | Arabidopsis thaliana | Oryza sativa | Dehydration | Paul et al., 2015 | |
| FeDREB1 | Fagopyrum esculentum | Arabidopsis thaliana | Drought/Multiple stress | Fang et al., 2015 | |
| VrDREB2A | Vigna radiata | Arabidopsis thaliana | Drought | Chen et al., 2016 | |
| EaDREB2 | Erianthus arundinaceus | Saccharum spp. hybrid Co 86032 | Drought | Augustine et al., 2015 | |
| AtDREB2A | Arabidopsis thaliana | Glycine max | Drought | Engels et al., 2013 | |
| OsDREB2A | Oryza sativa | Oryza sativa | Drought | Cui et al., 2011 | |
| SsDREB | Suaeda salsa | Nicotiana tabacum | Drought | Zhang X. et al., 2015 | |
| TaDREB2 and TaDREB3 | Triticum aestivum | Triticum aestivum, Hordeum vulgare L. cv. Golden Promise | Drought | Morran et al., 2011 | |
| NAC | SNAC1 | Oryza sativa | Oryza sativa | Drought | Hu et al., 2006 |
| TaNAC2 | Triticum aestivum | Arabidopsis thaliana | Drought/Multiple stress | Mao et al., 2012 | |
| OsNAC5 | Oryza sativa | Oryza sativa | Drought | Song et al., 2011 | |
| OsNAC6 | Oryza sativa | Oryza sativa | Dehydration/Multiple stress | Nakashima et al., 2007 | |
| GmNAC20 | Glycine max | Glycine max | Drought | Hao et al., 2011 | |
| TaNAC69 | Triticum aestivum | Triticum aestivum | Dehydration Tolerance | Xue et al., 2011 | |
| bZIP | GmbZIP44, GmbZIP62, GmbZIP78, GmbZIP132 | Glycine max | Arabidopsis thaliana | Salt and Freezing stress | Liao et al., 2008 |
| GmbZIP1 | Glycine max | Arabidopsis thaliana | Drought | Gao et al., 2011 | |
| OsbZIP23 | Oryza sativa | Oryza sativa | Salinity and Drought tolerance | Xiang et al., 2008 | |
| Wlib19 | Triticum aestivum | Nicotiana tabacum | Abiotic stresses | Kobayashi et al., 2008 | |
| ZnbZIP17 | Zea mays | Zea mays | Drought, Heat, Salt | Jia et al., 2009 | |
| OsAREB1 | Oryza sativa | Arabidopsis thaliana | Drought, Heat | Jin et al., 2010 | |
| SlAREB | Solanum lycopersicum | Solanum lycopersicum, Arabidopsis thaliana | Water deficit and Salt stress | Hsieh et al., 2010 | |
| OsbZIP16 | Oryza sativa | Oryza sativa | Drought | Chen et al., 2012 | |
| DgZFP | Dendronthema grandiform | Nicotiana tabacum | Drought, Salinity, Cold | Liu et al., 2010 | |
| HvDRF1 | Hordeum vulgare | Hordeum vulgare | Drought | Xue and Loveridge, 2004 | |
| Homeodomain-leucine zipper (HD-Zip) proteins | Multiple HD-Zip genes | Glycine max | Glycine max | Drought/Salinity | Chen et al., 2014 |
| Zinc finger proteins | OsTZF1 | Oryza sativa | Oryza sativa | RNA metabolism of stress-responsive genes/Multiple stress response | Jan et al., 2013 |
| DgZFP2 | Dendronthema grandiform | Dendronthema grandiform | Dehydration/Multiple stress | Liu et al., 2012 | |
| TaMYB33 | Triticum aestivum | Arabidopsis thaliana | Drought/Salinity | Qin et al., 2012 | |
| TaPIMP1 | Triticum aestivum | Nicotiana tabacum | Drought/Salinity | Liu et al., 2011 | |
AP2/ERF TFs
APETALA2/Ethylene Response Element binding Factors (AP2/ERF) family covers a large group of plant-specific TFs and is characterized by the presence of a much-conserved AP2/ERF DNA-binding domain (Song et al., 2013). This domain binds with the GCC box, which is a DNA sequence having role in the ethylene-responsive transcription (Rashid et al., 2012). AP2/ERFBP TFs perform diverse roles in plant biological processes, such as cell proliferation, vegetative and reproductive development, plant hormone responses, and abiotic/biotic stress responses (Sharoni et al., 2011; Xu et al., 2011). Genome-wide analyses have resulted in the identification of a multitude of AP2/ERFBP members across several plant species such as 145 in Arabidopsis (Riechmann and Meyerowitz, 1998), 170 in rice (Rashid et al., 2012), 178 in sorghum (Srivastav et al., 2010), 200 in poplar (Zhuang et al., 2008), 291 in Chinese cabbage (Song et al., 2013), 171 in foxtail millet (Lata et al., 2014), and 116 in moso bamboo (Wu et al., 2015). Based on the number and similarity of AP2/ERF domains, the family is further categorized into four major subfamilies: AP2 (Apetala 2), RAV (related to ABI3/VP1), DREB (dehydration-responsive element-binding protein), and ERF (Sharoni et al., 2011; Rashid et al., 2012). Apropos of the plant abiotic and biotic stress response, ERF and DREB subfamilies have been extensively studied. Induced, respectively, by cold and dehydration, DREB1/CBF (with 11 genes in rice) and DREB2 (six genes in rice) function in ABA-independent manner (Srivastav et al., 2010; Nakashima et al., 2014b). The promoter analysis of drought stress regulated genes having ABA-independent expression in Arabidopsis illustrated a cis-element with A/GCCGAC sequence (known as DRE/CRT: Lucas et al., 2011). Functional similarities were observed in the downstream genes of DREB1A and DREB2A by microarray analysis, however, differential expression was established during carbohydrate metabolism in transgenic plants. Four orthologs of CBF/DREB1A in rice viz. OsDREB1A, OsDREB1B, OsDREB1C, and OsDREB1D have already been characterized (Dubouzet et al., 2003). DREB1/CBF TFs specifically interact with the DRE/CRT and regulate the expression of several abiotic stress-responsive genes. Different studies confirmed that DREB1/CBF genes expression is differentially regulated by other genes like ICE1, HOS1, MYB15, SIZ1, PIF7, CAMTA3, and a clock component (Dong et al., 2011; Qin et al., 2011). Plants overexpressing DREB1/CBF showed enhanced expression of several stress responsive genes providing improved tolerance toward drought including tomato, chrysanthemum, potato (Iwaki et al., 2013), soybean (de Paiva Rolla et al., 2014), rice (Datta et al., 2012; Nakashima et al., 2014b; Paul et al., 2015), tobacco (Phuong et al., 2015), sugarcane (Augustine et al., 2015), groundnut (Tiwari et al., 2015), peanut (Bhatnagar-Mathur et al., 2014), and wheat (Shavrukov et al., 2016). In rice, DREB1/CBF-type TFs conferred higher drought tolerance besides expression of cold-responsive genes. Ishizaki et al. (2013) experimentally demonstrated improved survival of NERICA1 (an upland rice cultivar) under drought stress through expressing Arabidopsis DREB1C in the transgenic plants. OsDREB1F is reported to be induced by drought stress and ABA application (Wang et al., 2008). Overexpression of OsDREB1G is also known to promote drought tolerance (Chen et al., 2008). Similarly, constitutive expression of DREB1/CBF3 in transgenic plants led to improved tolerance against salinity, heat and cold (Augustine et al., 2015; Fang et al., 2015; Kidokoro et al., 2015; Chen et al., 2016). Higher accumulation of osmoprotectants, such as free proline and soluble sugars were recorded in the transgenic rice plants overexpressing DREB1A (Ito et al., 2006). More recently, transgenic tobacco with elevated tolerance level against drought and salt was obtained through over-expression of SsCBF4 from succulent halophyte Suaeda salsa (Zhang X. et al., 2015) (Table 1). An affymetrix microarray based analysis of transgenic rice plants facilitated the identification of 404 genes as induced or suppressed by DREB1BI when compared to their respective wild types (Zhuang et al., 2015).
A conserved regulatory mechanism has been established after examining DREB2-type proteins across several crop species such as wheat, maize, rice, barley, and sunflower (Mizoi et al., 2012). Rice harbors six DREB2 family genes (Srivastav et al., 2010) while Arabidopsis contains eight DREB2 genes of which DREB2A and DREB2B rendered higher induction under drought stress (Nakashima et al., 2014b). The constitutive as well as stress inducible expression of OsDREB2A and OsDREB2B in transgenic plants has resulted in enhanced tolerance toward osmotic stress (Cui et al., 2011; Mizoi et al., 2013). Transgenic Arabidopsis plants overexpressing OsDREB2B displayed enhanced expression of DREB2A target genes and improved tolerance toward drought stress (Matsukura et al., 2010). Taken together, these reports suggest that OsDREB2 is a major gene encoding for DREB2-type TF that holds great importance in stress-responsive gene expression. However, in addition to imparting increased drought tolerance, over expression of DREB2A was reported to cause significant growth defects. In contrast, no growth defects were seen in case of Arabidopsis and soybean transgenics where stress-inducible expression of DREB2A led to enhanced drought tolerance (Engels et al., 2013). Under normal condition, DREB2A is regulated by two mechanisms (i) GRF7 (growth-regulating factor7) inhibits expression of DREB2A binding to its short promoter region (Singh and Laxmi, 2015) and (ii) targeted degradation by 26S proteasome mediated proteolysis assisted by DRIP1 (DREB2A- interacting protein1) and DRIP2 protein (Morimoto et al., 2013; Singh and Laxmi, 2015). In other words, conditional expression of DREB2A under drought stress offers a way to minimize the inefficient loss of energy.
Besides DREB1A and DREB2 members, various other AP2/ERF-type and AP2/ERF-like TFs were also characterized in rice with an aim to improve abiotic stress tolerance. Likewise, overexpression of Arabidopsis HARDY gene (a subgroup of A-4 AP2/ERF family) in Trifolium led to enhanced osmotic stress tolerance (Abogadallah et al., 2011). OsDERF1 has been established as a negative modulator in osmotic adjustment by interacting directly with GCC box located in the promoter of OsERF3 (Wan et al., 2011). Furthermore, OsERF3 contains an ethylene-responsive element-binding factor-associated amphiphilic repression (EAR) motif, which transcriptionally represses the ethylene production along with drought tolerance. Mutant EAR lines showed enhanced ethylene emission and drought tolerance as compared to overexpressing as well as wild type plants (Zhang et al., 2013). However, Joo et al. (2013) reported OsERF4a as a positive regulator of plant growth and drought tolerance in rice. In addition, ectopic expression of tomato JERF3 alleviates soluble sugars and proline with increased drought tolerance in rice (Zhang et al., 2010). Similarly, overexpression of another AP2/ERF TF showed increased drought, cold, and salinity stress tolerance in transgenic rice (Table 1).
NAC TFs
The NAC gene family is the largest and plant specific TF family (Puranik et al., 2012; Shao et al., 2015). The NAC acronym is derived from three proteins having a specific NAC domain from petunia, i.e., NAM (no apical meristem) and ATAF1/2 and CUC2 (cup-shaped cotyledon) in Arabidopsis (Shao et al., 2015). Characteristically, a NAC TF comprises of a highly conserved DNA binding NAC domain in the N-terminal region and a variable C-terminal transcriptional regulatory region. The NAC domain is linked with DNA binding, nucleus-oriented localization and formation of homodimers or heterodimers with similar domains, while the C-terminal functions in transcriptional regulation (Puranik et al., 2012). The NAC TFs modulate downstream drought inducible EARLY RESPONSE TO DEHYDRATION1 (ERD1) gene transcription by interacting with NAC recognition sequence (NACRS) having CACG core-DNA binding motif in its promoter (Shao et al., 2015). In rice, SNAC1 TFs were reported to confer drought tolerance through regulating downstream genes OsPP18, a PP2C in ABA independent pathways (You et al., 2014). To date, genome wide characterization has offered numerous putative NAC TFs across different plant species like 204 in Chinese cabbage, 152 in soybean, 152 in maize, 151 in rice, 117 in Arabidopsis and 74 in grape (Shao et al., 2015). In rice, several NAC genes were reported to be induced during early stages of salt and drought stresses (Hong et al., 2016). The role of NAC TFs in drought stress response is further supported by several transcriptome-based analyses undertaken in different crops; examples include 40 NAC genes in rice (Shao et al., 2015), 38 NAC genes in soybean (Le et al., 2011). These stress-responsive NAC genes also showed differential expression patterns such as tissue-specific, developmental stage-specific or stress-specific expression, thereby suggesting their active involvement in the complex signaling networks during plant stress responses. In Arabidopsis, drought tolerance was found to be elevated by overexpressing three genes, i.e., ANACO19, ANACO55, ANACO72. Similarly, overexpression of stress responsive NAC1 (SNAC1) in rice provides tolerance against severe drought stress at the reproductive stage in field conditions without a change in their phenotype or yield penalty (Liu G. et al., 2014). In another study it was found that under drought stress SNAC1 was induced in guard cells and its over-expression reduced transpirational losses due to increased stomatal closure (Singh et al., 2015b). Highlighting the importance of SNAC1 gene, genetic variation of this gene was assessed among Indian wild rice and as a result, discovery of four haplotypes associated with tolerance might be useful for developing drought tolerant crops (Singh et al., 2015b). Similarly, OsNAC5 expression was reported to be induced under drought, cold, ABA, and methyl jasmonate (MeJA) treatments. Overexpressors of OsNAC5 were reported to serve as transcriptional activators that regulate the stress-responsive gene expression (Takasaki et al., 2010). Function of OsNAC5 in drought tolerance was examined by RNA interference (RNAi) and overexpression study in transgenic rice and Arabidopsis (Song et al., 2011). In addition, OsNAC6/SNAC2 is also known to be induced by cold, drought, salinity and ABA (Todaka et al., 2012). Furthermore, overexpressing TaNAC2 and TaNAC29 in Arabidopsis resulted in enhanced drought, salt and cold stress tolerance along with higher transcript levels of stress-responsive genes and improved physiological parameters (Mao et al., 2012; Huang et al., 2015). Contrary to this, OsNAC6 overexpressing rice plants had reduced growth and low yield under normal conditions even though these plants remained tolerant for dehydration and salinity stresses. Based on these studies, OsNAC6 could be established as a potent functional regulator in drought stress response (Todaka et al., 2012). Interestingly, overexpression of HvSNAC1 in barley not only improved drought tolerance but also provided resistance against fungal infection of Ramularia collo-cygni causing leaf spot (Al-Abdallat et al., 2014; McGrann et al., 2015).
Enhanced abiotic stress tolerance in rice caused by overexpressing SNACs was evident when using a root-specific promoter RCc3 (Jeong et al., 2010, 2013; Redillas et al., 2012). Similarly, thicker roots and higher grain yield under drought stress were observed in transgenic rice plants overexpressing OsNAC10 (Jeong et al., 2010), OsNAC5 (Jeong et al., 2013), and OsNAC9/SNAC1 (Redillas et al., 2012) regulated by root specific promoter (Jeong et al., 2010). Furthermore, microarray analysis of these transgenic plants revealed 62 downstream genes to be differentially expressed, which included P450, Zn-finger, HAK5, 2OG-Fe(II), NCED, NAC, KUP3, calcium-transporting ATPase, germin-like protein, and meristem protein (Redillas et al., 2012). Of these, only 17 were found to be upregulated (Jeong et al., 2013). However, as demonstrated by Hao et al. (2011) in soybean GmNAC11 was reported to function as a transcriptional activator, whereas GmNAC20 acted as a mild repressor with C-terminal transcriptional activation activity and its overexpression increases lateral root development in transgenic plants. This study established GmNAC20 as a regulator of stress tolerance, which serves by activating DREB/CBF-COR pathway and lateral root formation by modulating auxin signaling genes (Table 1). Thus, precise manipulation of SNACs with suitable promoter offers a promising way to substantially alter drought stress response in plants (Mei and Lizhong, 2011; Bang et al., 2013; Rusconi et al., 2013; Nakashima et al., 2014b).
bZIP TFs
The basic leucine zipper (bZIP) family contains a conserved bZIP domain, which is composed of a highly basic nuclear localization and DNA binding region at the N-terminus and a leucine-rich motif for dimerization at the C-terminus (Wang et al., 2015). Besides regulating plant growth and development, bZIP TFs remain crucial concerning abiotic stress response such as drought (Llorca et al., 2014). Members of the bZIP TF family have been isolated and characterized in various eukaryotes. Examples include 17 in Saccharomyces, 31 in Caenorhabditis, 55 in grapevine (Liu J. et al., 2014), 75 in Arabidopsis (Jakoby et al., 2002), 89 in rice (Nijhawan et al., 2008), 89 in barley (Pourabed et al., 2015), 92 in sorghum (Wang et al., 2011), 96 in Brachypodium distachyon (Liu and Chu, 2015), 125 in maize (Wei et al., 2012), and 131 in soybean (Liao et al., 2008). Though the physiological role of their homologs with putative zinc finger motif remains unclear, studies on abiotic stress responses have confirmed bZIP TFs to be ABA inducible, i.e., these regulate the expression of stress-related genes in ABA-dependent manner after binding with the promoter region of specific ABRE (Llorca et al., 2014).
Xu et al. (2007) characterized a novel gene ThZF1 from Thellungiella halophila that encodes a functional TF. It contains two conserved Cys-2/His-2 regions with conserved DNA-binding motif similar to the members of Arabidopsis ZFP family. Drought and salinity are reported to induce the transcription of this gene. Ectopic expression of ThZF1 in Arabidopsis mutant azf2 revealed its similarity with Arabidopsis AZF2 in plant development and downstream gene regulation. Likewise, transgenic rice plants overexpressing OsbZIP16 exhibited significantly higher drought tolerance at both seedling and tillering stages (Chen et al., 2012). Under drought stress conditions its downstream drought-inducible genes showed significantly higher expression levels in transgenics than the corresponding wild types. Though OsbZIP16 is reported to be ABA-inducible, overexpression of OsbZIP16 renders transgenic plants more sensitive to ABA. A recent study in Arabidopsis revealed that overexpression of TabZIP60 improved plant’s tolerance against stresses like drought, salt and freezing along with increasing sensitivity to ABA (Zhang L. et al., 2015).
Similarly, OsbZIP23, a close relative of Arabidopsis homologs ABF/AREB, was reported to be a principle regulator of ABA-dependent pathways (Todaka et al., 2015). Rice plants overexpressing OsbZIP23 caused greater ABA sensitivity at both germination and post-germination stages along with increased drought and salt stress tolerance. Microarray analysis showed differential expression of several downstream genes including stress-related TFs, protein kinases, dehydrins and LEA proteins. OsbZIP46 also belong to the subfamily of OsbZIP23 and its constitutively active form OsbZIP46CA1 was developed by mutating OsbZIP46 domain D. Rice plants overexpressing OsbZIP46CA1 showed enhanced drought tolerance. Differentially expressed downstream genes different from those of OsbZIP23 downstream genes were uncovered through a microarray analysis, which in turn suggested independent regulatory mechanisms of OsbZIP46CA1 and OsbZIP23. In contrast, growth of OsbZIP23 or OsbZIP46CA1 overexpressing rice plants is reduced under drought stress (Tang et al., 2012). Furthermore, ABF3 belonging to similar subfamily as OsbZIP46 and OsbZIP23 improved drought tolerance in transgenic Arabidopsis and rice (Todaka et al., 2015). OsbZIP16 and OsbZIP71 are classified into group IV of the rice bZIP subfamily (Todaka et al., 2015). After ABA treatment transgenic rice plants overexpressing OsbZIP16 showed enhanced drought tolerance and reduced plant growth (Chen et al., 2012). However, OsbZIP71 overexpressing rice transgenics with a constitutive promoter or a stress-inducible RD29A promoter showed enhanced drought, salt, and osmotic stress tolerance (Liu J. et al., 2014). Collectively, these reports confirmed the involvement of bZIP TFs in ABA signaling, which could be harnessed for developing better genotypes endowed with drought tolerance (Todaka et al., 2015).
Interactions among Multiple TFs
Drought stress is an unpredictable event and it varies in severity and duration. This results in both general and specific effects on plant growth and development. Thus, plant response toward drought stress is dynamic, which involves multiple stress perception and signal transduction pathways, which may crosstalk at various steps in the pathways (Lenka et al., 2011). For this reason, plants have evolved complex regulatory mehanisms, including metabolic adjustment and gene expression toward physiological and morphological adaptation (Baldoni et al., 2015). Moreover, extensive overlap between signal interactions and adaptation mechanisms can be both synergistic and antagonistic, resulting in positive and negative functional outcomes (Atkinson and Urwin, 2012). This type of regulation allows cell to respond rapidly to changes such as drought stress (Lindemose et al., 2013). TFs are of key importance in these signaling cascades and in generating specificity in stress responses. Interactions among different TFs have made significant advances over recent years and were documented recently by Nakashima et al. (2014a). The TFs control transcription of their downstream genes by interacting with other proteins and binding to a consensus sequence in promoters. Identification of these downstream genes becomes imperative to elucidate molecular machineries of gene activation or repression (Figure 1).
Drought responsive gene promoters contain a DRE/CRT motif where ABA-independent DREB/CBF TF binds and act as a coupling element for ABRE in ABA-dependent gene expression (Singh and Laxmi, 2015). It was already shown that DREB1A/CBF3, DREB2A, and DREB2C proteins interact with AREB/ABF (Lee et al., 2010). Thus, there exists a crosstalk between ABA-dependent and independent signaling and regulatory pathways (Figure 1). Under osmotic stress conditions, AREB/ABF TFs and SnRK2s regulate the transcriptional activation of DREB2A gene, suggesting a complex interaction between DREB and AREB regulatory regions at the transcript and protein level (Kim et al., 2011). Similar interactions were also reported in between AREB/ABFs and NACs. In addition, regulation of ABA-dependent gene expression of ABRE regulons by SNAC TFs was confirmed when Jensen et al. (2013) reported that Arabidopsis SNAC transcription factor ATAF1 directly modulates ABA biosynthetic gene NCED3. By contrast, SNAC gene promoter contains ABRE region (Nakashima et al., 2014b). Under dehydration and osmotic stress, ANAC096 was also found to interact with ABF2/AREB1 and ABF4/AREB2 (Xu et al., 2013). Furthermore, correlation among DREB/CBFs and AP2/ERFs at the transcript levels was also confirmed when Arabidopsis ERF1 was shown to regulate transcript expression of a gene by coupling with two different cis-elements, the GCC box and DRE/CRT during stress response (Cheng et al., 2013). Recently, it was shown that bZIP TFs VEG2 and FLOWERING LOCUS D (FD) interact with nuclear protein CONSTANS (CO) and positively regulate signaling of flowering activator FLOWERING LOCUS T (FT) proteins (Cai et al., 2014; Weller and Ortega, 2015). TFs activate several functional genes encoding wide range of proteins involved in osmolyte production, ROS scavenging and detoxification, macromolecule protection and ubiquitination. As envisaged by Puranik et al. (2012), such versatility in TFs functioning might have evolved to ensure plants’ longevity, survival and reproductive success under environmental stresses. Our current knowledge about these target genes stems mainly from the studies performed using large-scale transcriptome analyses in plants that overexpress TFs. Deeper analyses can better elucidate their roles under drought response.
Future Directions
Here, we highlighted the increasingly important roles of TFs as modern genomic tools to improve plant tolerance toward multiple abiotic stresses. Significant progress has been achieved in deciphering the role of TFs toward tolerance to abiotic stress factors such as drought, and an appreciable number of promising candidates – TF genes have already been identified and validated. Selection of key TFs from such large families and realization of their full potential still remain a strenuous task. Also, development of transgenic plants harboring such TF genes that deliver anticipated level of tolerance in the field conditions still poses a great challenge to the research community. Furthermore, functional redundancy between different TF members might potentially hamper the progress to delineate the functions of an individual member.
The constitutive over-expression of some TF genes may improve the stress tolerance; however, occasional negative effects in transgenic plants such as dwarfing, late flowering and lower yields should also be taken into account while investing research. Furthermore, the complete regulatory mechanism of individual TF encompassing its upstream and downstream co-regulators, as well as their interactions remain largely obscure. A principle concern is also about the selection of plants, as most of the initial experiments are being performed on model crops like Arabidopsis and tobacco. Focus must be shifted to commercial crops like rice, wheat, maize and other cereal crops, which are predominantly affected by drought stress. In comparison to other crops, rice has shown the highest potential under waterlogging conditions (Voesenek and Bailey-Serres, 2013). Although reports are available with improved drought tolerance in transgenic rice under field conditions, further research is required in order to unveil the regulatory mechanism of drought tolerance. Future investigations must focus on pinpointing novel genes that enhance drought tolerance as well as yield. In addition, combination of drought-tolerance with submergence-tolerance in rice is exceptionally useful, as this would allow crop to withstand both drought and waterlogged conditions.
Another attractive approach for discovering novel candidate genes is to intensively examine the mechanisms that describe drought tolerance in extremophiles, i.e., desert plants (Mittler and Blumwald, 2010; Joshi et al., 2015). It is important to mention here that up to 38% of total protein functions are still unknown even in well-analyzed plants and a renewed focus in this aspect might led to the discovery of robust candidate genes. Modification in root architecture is another important factor for improving drought tolerant crops. Transcriptome analysis has shown that drought-responsive TFs like REB1/CBF, DREB2, AREB/ABF, and NAC TFs function under drought response and tolerance. Utilizing a balanced crosstalk between these TFs under osmotic stress, overexpression of TFs may affect their signaling pathways. Identification of multiple stress-responsive TF genes and their prioritization by means of comparing expression patterns deserves attention of the scientific community and pinpointing the commonly regulated genes which have been proposed to be essential for universal stress responses or represent points of cross-talk between signaling pathways (Prasch and Sonnewald, 2015). Genetic manipulation of these multiple stress-responsive TF genes stands to be a powerful approach for improving plant tolerance than addressing each functional gene individually. Furthermore, the field trials are required to critically evaluate the transgenic plants, especially focusing on their growth and tolerance in the whole life period, which unequivocally remains a deciding factor while developing stress-tolerant crops.
Author Contributions
SLS-P and AP conceived the review topic. RJ, BS, and AB drafted the manuscript. SW, ZD, and AL participated in the discussion of topic. All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
RJ acknowledges the Start-Up research grant (Young Scientist) from the Science and Engineering Research Board, Government of India. SLS-P acknowledge the support of research funds in the laboratory from the Department of Science and Technology, Department of Biotechnology, Government of India, and internal grants of ICGEB.
References
- Abogadallah G. M., Nada R. M., Malinowski R., Quick P. (2011). Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta 233 1265–1276. 10.1007/s00425-011-1382-3 [DOI] [PubMed] [Google Scholar]
- Aguado A., Capote N., Romero F., Dodd I. C., Colmenero-Flores J. M. (2014). Physiological and gene expression responses of sunflower (Helianthus annuus L.) plants differ according to irrigation placement. Plant Sci. 227 37–44. 10.1016/j.plantsci.2014.06.009 [DOI] [PubMed] [Google Scholar]
- Al-Abdallat A. M., Ayad J. Y., Elenein J. A., Al Ajlouni Z., Harwood W. A. (2014). Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Mol. Breed. 33 401–414. 10.1007/s11032-013-9958-1 [DOI] [Google Scholar]
- Alqudah A. M., Samarah N. H., Mullen R. E. (2010). “Drought stress effect on crop pollination, seed set, yield and quality,” in Alternative Farming Systems, Biotechnology, Drought Stress and Ecological Fertilization Vol. 6 ed. Lichtfouse E. (New York, NY: Springer; ), 193–213. [Google Scholar]
- Alqudah A. M., Schnurbusch T. (2015). Barley leaf area and leaf growth rates are maximized during the pre-anthesis phase. Agronomy 5 107–129. 10.3390/agronomy5020107 [DOI] [Google Scholar]
- Anbazhagan K., Bhatnagar-Mathur P., Vadez V., Dumbala S. R., Kishor P. B., Sharma K. K. (2015). DREB1A overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 34 199–210. 10.1007/s00299-014-1699-z [DOI] [PubMed] [Google Scholar]
- Atkinson N. J., Urwin P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63 3523–3543. 10.1093/jxb/ers100 [DOI] [PubMed] [Google Scholar]
- Augustine S. M., Ashwin Narayan J., Syamaladevi D. P., Appunu C., Chakravarthi M., Ravichandran V., et al. (2015). Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep. 34 247–263. 10.1007/s00299-014-1704-6 [DOI] [PubMed] [Google Scholar]
- Baldoni E., Genga A., Cominelli E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16 15811–15851. 10.3390/ijms160715811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang S. W., Park S. H., Jeong J. S., Kim Y. S., Jung H., Ha S. H., et al. (2013). Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 237 211–224. 10.1007/s00425-012-1764-1 [DOI] [PubMed] [Google Scholar]
- Barbosa E. G. G., Leite J. P., Marin S. R. R., Marinho J. P., Carvalho J. F. C., Fuganti-Pagliarini R., et al. (2013). Overexpression of the ABA-Dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Rep. 31 719–730. 10.1007/s11105-012-0541-4 [DOI] [Google Scholar]
- Barlow M. B., Zaitchik S., Paz E., Black J. E., Hoell A. (2015). A review of drought in the middle east and southwest asia. J. Climate 10.1175/JCLI-D-13-00692.1 [Epub ahead of print]. [DOI] [Google Scholar]
- Bauer H., Ache P., Lautner S., Fromm J., Hartung W., Al-Rasheid K. A., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23 53–57. 10.1016/j.cub.2012.11.022 [DOI] [PubMed] [Google Scholar]
- Behnam B., Iuchi S., Fujita M., Fujita Y., Takasaki H., Osakabe Y., et al. (2013). Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res. 20 315–324. 10.1093/dnares/dst012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava S., Sawant K. (2013). Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed. 132 21–32. 10.1111/pbr.12004 [DOI] [Google Scholar]
- Bhatnagar-Mathur P., Rao J. S., Vadez V., Dumbala S. R., Rathore A., Yamaguchi-Shinozaki K., et al. (2014). Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33 327–340. 10.1007/s11032-013-9952-7 [DOI] [Google Scholar]
- Bohra A., Sahrawat K. L., Kumar S., Joshi R., Parihar A. K., Singh U., et al. (2015). Genetics- and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J. Appl. Genet. 56 151–161. 10.1007/s13353-014-0268-z [DOI] [PubMed] [Google Scholar]
- Buchanan B. B., Gruissem W., Vickers K., Jones R. L. (2015). Biochemistry and Molecular Biology of Plants. New York, NY: Wiley-Blackwell. [Google Scholar]
- Cai Y., Chen X., Xie K., Xing Q., Wu Y., Li J., et al. (2014). Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 9:e102529 10.1371/journal.pone.0102529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu L., Wang L., Wang S., Cheng X. (2016). VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 129 263–273. 10.1007/s10265-015-0773-0 [DOI] [PubMed] [Google Scholar]
- Chen J. Q., Meng X. P., Zhang Y., Xia M., Wang X. P. (2008). Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 30 2191–2198. 10.1007/s10529-008-9811-5 [DOI] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819 120–128. 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Chen X., Chen Z., Zhao H., Zhao Y., Cheng B., Xiang Y. (2014). Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 9:e87156 10.1371/journal.pone.0087156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Li X., Huang X., Ma T., Liang Y., Ma X., et al. (2013). Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 70 252–260. 10.1016/j.plaphy.2013.05.025 [DOI] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Cui M., Zhang W., Zhang Q., Xu Z., Zhu Z., Duan F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49 1384–1391. 10.1016/j.plaphy.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Danquah A., de Zelicourt A., Colcombet J., Hirt H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32 40–52. 10.1016/j.biotechadv.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Datta K., Baisakh N., Ganguly M., Krishnan S., Yamaguchi Shinozaki K., Datta S. K. (2012). Overexpression of Arabidopsis and rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10 579–586. 10.1111/j.1467-7652.2012.00688.x [DOI] [PubMed] [Google Scholar]
- de Paiva Rolla A. A., de Fátima Corrêa Carvalho J., Fuganti-Pagliarini R., Engels C., do Rio A., Marin S. R., et al. (2014). Phenotyping soybean plants transformed with rd29A:AtDREB1A for drought tolerance in the greenhouse and field. Transgenic Res. 23 75–87. 10.1007/s11248-013-9723-6 [DOI] [PubMed] [Google Scholar]
- Dong M. A., Farré E. M., Thomashow M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7241–7246. 10.1073/pnas.1103741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Shi H., Zhang X., Xu X. (2014). Responses of reactive oxygen scavenging enzymes, proline and malondialdehyde to water deficits among six secondary successional seral species in Loess Plateau. PLoS ONE 9:e98872 10.1371/journal.pone.0098872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. 10.1046/j.1365-313X.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Edmeades G. O. (2013). Progress in Achieving and Delivering Drought Tolerance in Maize- An Update. Ithaca, NY: ISAAA. [Google Scholar]
- Engels C., Fuganti-Pagliarini R., Marin S. R., Marcelino-Guimarães F. C., Oliveira M. C., Kanamori N., et al. (2013). Introduction of the rd29A:AtDREB2A CA gene into soybean (Glycine max L. Merril) and its molecular characterization in leaves and roots during dehydration. Genet. Mol. Biol. 36 556–565. 10.1590/S1415-47572013000400015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Zhang X., Gao J., Wang P., Xu X., Liu Z., et al. (2015). A Buckwheat (Fagopyrum esculentum) DRE-Binding transcription factor gene, FeDREB1 enhances freezing and drought tolerance of transgenic Arabidopsis. Plant Mol. Biol. Rep. 33 1510–1525. 10.1007/s11105-015-0851-4 [DOI] [Google Scholar]
- Farooq M., Basra S. M. A., Wahid A., Cheema Z. A., Cheema M. A., Khaliq A. (2008). Physiological role of exogenously applied glycinebetaine to improve drought tolerance in fine grain aromatic rice (Oryza sativa L.). J. Agron. Crop Sci. 194 325–333. 10.1111/j.1439-037X.2008.00323.x [DOI] [Google Scholar]
- Farooq M., Gogoi N., Barthakur S., Baroowa B., Bharadwaj N., Alghamdi S. S., et al. (2016). Drought stress in grain legumes during reproduction and grain filling. J. Agron. Crop Sci. 10.1111/jac.12169 [DOI] [Google Scholar]
- Franco-Zorrilla J. M., López-Vidriero I., Carrasco J. L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111 2367–2372. 10.1073/pnas.1316278111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T. (2015). A new paradigm for feeding the world in 2050. The sustainable intensification of crop production. Res. Mag. 22:18 10.3389/fpls.2015.00978 [DOI] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M. M., Seki M., et al. (2005). AREB1 is a transcription activator of novel ABREdependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124 509–525. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147 15–27. 10.1111/j.1399-3054.2012.01635.x [DOI] [PubMed] [Google Scholar]
- Gao S. Q., Chen M., Xu Z. S., Zhao C. P., Li L., Xu H. J., et al. (2011). The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 75 537–553. 10.1007/s11103-011-9738-4 [DOI] [PubMed] [Google Scholar]
- Golldack D., Lüking I., Yang O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30 1383–1391. 10.1007/s00299-011-1068-0 [DOI] [PubMed] [Google Scholar]
- Gosal S. S., Wani S. H., Kang M. S. (2010). Biotechnology and crop improvement. J. Crop Improv. 24 153–217. 10.1080/15427520903584555 [DOI] [Google Scholar]
- Hao Y. J., Wei W., Song Q. X., Chen H. W., Zhang Y. Q., Wang F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68 302–313. 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- Hill C. B., Taylor J. D., Edwards J., Mather D., Bacic A., Langridge P., et al. (2013). Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait Loci in bread wheat grown in a water-limited environment. Plant Physiol. 162 1266–1281. 10.1104/pp.113.217851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61 1041–1052. 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- Hong Y., Zhang H., Huang L., Li D., Song F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4 10.3389/fpls.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. H., Li C. W., Su R. C., Cheng C. P., Sanjaya Tsai Y. C., et al. (2010). A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231 1459–1473. 10.1007/s00425-010-1147-4 [DOI] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. T., Ma S. L., Bai L. P., Zhang L., Ma H., Jia P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39 969–987. 10.1007/s11033-011-0823-1 [DOI] [PubMed] [Google Scholar]
- Huang Q., Wang Y., Li B., Chang J., Chen M., Li K., et al. (2015). TaNAC29 a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268 10.1186/s12870-015-0644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T., Maruyama K., Obara M., Fukutani A., Yamaguchi-Shinozaki K., Ito Y., et al. (2013). Expression of Arabidopsis DREB1C improves survival, growth, and yield of upland New Rice for Africa (NERICA) under drought. Mol. Breed. 31 255–264. 10.1007/s11032-012-9785-9 [DOI] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47 141–153. 10.1093/pcp/pci230 [DOI] [PubMed] [Google Scholar]
- Iwaki T., Guo L., Ryals J. A., Yasuda S., Shimazaki T., Kikuchi A., et al. (2013). Metabolic profiling of transgenic potato tubers expressing Arabidopsis dehydration response element-binding protein 1A (DREB1A). J. Agric. Food Chem. 61 893–900. 10.1021/jf304071n [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). bZIP Research Group. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jan A., Maruyama K., Todaka D., Kidokoro S., Abo M., Yoshimura E., et al. (2013). OsTZF1 a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161 1202–1216. 10.1104/pp.112.205385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K., Lindemose S., de Masi F., Reimer J. J., Nielsen M., Perera V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3 321–327. 10.1016/j.fob.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S., Kim Y. S., Baek K. H., Jung H., Ha S. H., Do Choi Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153 185–197. 10.1104/pp.110.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S., Kim Y. S., Redillas M. C., Jang G., Jung H., Bang S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11 101–114. 10.1111/pbi.12011 [DOI] [PubMed] [Google Scholar]
- Jha U. C., Chaturvedi S. K., Bohra A., Basu P. S., Khan M. S., Barh D. (2014). Abiotic stresses, constraints and improvement strategies in chickpea. Plant Breed. 133 163–178. 10.1111/pbr.12150 [DOI] [Google Scholar]
- Ji H., Liu L., Li K., Xie Q., Wang Z., Zhao X., et al. (2014). PEG-mediated osmotic stress induces premature differentiation of the root apical meristem and outgrowth of lateral roots in wheat. J. Exp. Bot. 65 4863–4872. 10.1093/jxb/eru255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Lian Y., Zhu Y., He J., Cao Z., Wang G. (2009). Cloning and characterization of a putative transcription factor induced by abiotic stress in Zea mays. Afr. J. Biotechnol. 8 6764–6771. 10.5897/AJB09.1280 [DOI] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G., Luo J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42 D1182–D1187. 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X. F., Jiong A. S., Peng R. H., Liu J. G., Gao F., Chen J. M., et al. (2010). OsAREB1 an ABRE binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep. 43 34–39. 10.5483/BMBRep.2010.43.1.034 [DOI] [PubMed] [Google Scholar]
- Joo J., Choi H. J., Lee Y. H., Kim Y. K., Song S. I. (2013). A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 238 155–170. 10.1007/s00425-013-1880-6 [DOI] [PubMed] [Google Scholar]
- Joshi R., Karan R. (2013). “Physiological, biochemical and molecular mechanisms of drought tolerance in plants,” in Molecular Approaches in Plant Abiotic Stress, eds Gaur R. K., Sharma P. (Boca Raton, FL: CRC Press; ), 209–231. [Google Scholar]
- Joshi R., Karan R., Singla-Pareek S. L., Pareek A. (2016a). Ectopic expression of Pokkali phosphoglycerate kinase-2 (OsPGK2-P) improves yield in tobacco plants under salinity stress. Plant Cell Rep. 35 27–41. 10.1007/s00299-015-1864-z [DOI] [PubMed] [Google Scholar]
- Joshi R., Singh B., Bohra A., Chinnusamy V. (2016b). “Salt stress signalling pathways: specificity and crosstalk,” in Managing Salinity Tolerance in Plants: Molecular and Genomic Perspectives, eds Wani S. H., Hossain M. A. (Boca Raton, FL: CRC Press; ), 51–78. [Google Scholar]
- Joshi R., Shukla A., Sairam R. K. (2011). In vitro screening of rice genotypes for drought tolerance using polyethylene glycol. Acta Physiol. Plant. 33 2209–2217. 10.1007/s11738-011-0760-6 [DOI] [Google Scholar]
- Joshi R., Ramanarao M. V., Bedre R., Sanchez L., Pilcher W., Zandkarimi H., et al. (2015). “Salt adaptation mechanisms of halophytes: improvement of salt tolerance in crop plants,” in Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives Vol. 2 ed. Pandey G. K. (New York, NY: Springer; ), 243–280. [Google Scholar]
- Kang J. Y., Choi H. I., Im M. Y., Kim S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357. 10.1105/tpc.010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattab H. I., Emam M. A., Emam M. M., Helal N. M., Mohamed M. R. (2014). Effect of selenium and silicon on transcription factors NAC5 and DREB2A involved in drought-responsive gene expression in rice. Biol. Plant. 58 265–273. 10.1007/s10535-014-0391-z [DOI] [Google Scholar]
- Khazaei H., Street K., Bari A., Mackay M., Stoddard F. L. (2013). The FIGS (focused identification of germplasm strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 8:e63107 10.1371/journal.pone.0063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholová J., Hash C. T., Kočová M., Vadez V. (2011). Does a terminal drought tolerance QTL contribute to differences in ROS scavenging enzymes and photosynthetic pigments in pearl millet exposed to drought? Environ. Expt. Bot. 71 99–106. 10.1016/j.envexpbot.2010.11.001 [DOI] [Google Scholar]
- Kidokoro S., Watanabe K., Ohori T., Moriwaki T., Maruyama K., Mizoi J., et al. (2015). Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81 505–518. 10.1111/tpj.12746 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Mizoi J., Yoshida T., Fujita Y., Nakajima J., Ohori T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52 2136–2146. 10.1093/pcp/pcr143 [DOI] [PubMed] [Google Scholar]
- Kim S., Kang J. Y., Cho D. I., Park J. H., Kim S. Y. (2004). ABF2 an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40 75–87. 10.1111/j.1365-313X.2004.02192.x [DOI] [PubMed] [Google Scholar]
- Kobayashi F., Maeta E., Terashima A., Kawaura K., Ogihara Y., Takumi S. (2008). Development of abiotic stress tolerance via bZIP-type transcription factor LIP19 in common wheat. J. Exp. Bot. 59 891–905. 10.1093/jxb/ern014 [DOI] [PubMed] [Google Scholar]
- Komatsu K., Suzuki N., Kuwamura M., Nishikawa Y., Nakatani M., Ohtawa H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4:2219 10.1038/ncomms3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Sugimoto E., Shinozaki K. (2014). Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol. 164 1587–1592. 10.1104/pp.114.235556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Mishra A. K., Muthamilarasan M., Bonthala V. S., Khan Y., Prasad M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 9:e113092 10.1371/journal.pone.0113092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi-Shinozaki K., Shinozaki K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18 263–276. 10.1093/dnares/dsr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Kang J. Y., Park H. J., Kim M. D., Bae M. S., Choi H. I., et al. (2010). DREB2C interacts with ABF2 a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153 716–727. 10.1104/pp.110.154617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka S. K., Katiyar A., Chinnusamy V., Bansal K. C. (2011). Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol. J. 9 315–327. 10.1111/j.1467-7652.2010.00560.x [DOI] [PubMed] [Google Scholar]
- Li T., Angeles O., Radanielson A., MarcaidaIII M., Manalo E. (2015). Drought stress impacts of climate change on rainfed rice in South Asia. Climatic Change 133 709–720. 10.1007/s10584-015-1487-y [DOI] [Google Scholar]
- Liao Y., Zou H. F., Wei W., Hao Y. J., Tian A. G., Huang J., et al. (2008). Soybean GmbZIP44 GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228 225–240. 10.1007/s00425-008-0731-3 [DOI] [PubMed] [Google Scholar]
- Lindemose S., O’Shea C., Jensen M. K., Skriver K. (2013). structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14 5842–5878. 10.3390/ijms14035842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Li X., Jin S., Liu X., Zhu L., Nie Y., et al. (2014). Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 9:e86895 10.1371/journal.pone.0086895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhou X., Dong N., Liu X., Zhang H., Zhang Z. (2011). Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Funct. Integr. Genomics 11 431–443. 10.1007/s10142-011-0228-1 [DOI] [PubMed] [Google Scholar]
- Liu J., Chen N., Chen F., Cai B., Dal Santo S., Tornielli G. B., et al. (2014). Genome wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15:281 10.1186/1471-2164-15-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Peng T., Dai W. (2014). Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 32 303–317. 10.1007/s11105-013-0667-z [DOI] [Google Scholar]
- Liu Q. L., Dong F. L., Xiao F., Wu J., Li Z. J. (2012). Isolation and molecular characterization of DgZFP2: a gene encoding a Cys2/His2-type zinc finger protein in chrysanthemum. Afr. J. Agric. Res. 7 4499–4504. 10.1007/s11033-009-9886-7 [DOI] [Google Scholar]
- Liu Q. L., Xu K. D., Ma N., Zeng L., Zhao L. J. (2010). Isolation and functional characterization of DgZFP: a gene encoding a Cys2/His2-type zinc finger protein in chrysanthemum. Mol. Biol. Rep. 37 1137–1142. 10.1007/s11033-009-9886-7 [DOI] [PubMed] [Google Scholar]
- Liu X., Chu Z. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16:227 10.1186/s12864-015-1457-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca C. M., Potschin M., Zentgraf U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5:169 10.3389/fpls.2014.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S., Durmaz E., Akpinar B. A., Budak H. (2011). The drought response displayed by a DRE-binding protein from Triticum dicoccoides. Plant Physiol. Biochem. 49 346–351. 10.1016/j.plaphy.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Luo L. J., Zhang Q. F. (2001). The status and strategy on drought resistance of rice. Chin. J. Rice Sci. 15 209–214. [Google Scholar]
- Mao X., Zhang H., Qian X., Li A., Zhao G., Jing R. (2012). TaNAC2 a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Expt. Bot. 63 2933–2946. 10.1093/jxb/err462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Todaka D., Mizoi J., Yoshida T., Kidokoro S., Matsukura S., et al. (2012). Identification of cis-acting promoter elements in cold-and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19 37–49. 10.1093/dnares/dsr040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283 185–196. 10.1007/s00438-009-0506-y [DOI] [PubMed] [Google Scholar]
- McGrann G. R. D., Steed A., Burt C., Goddard R., Lachaux C., Bansal A., et al. (2015). Contribution of the drought tolerance-related Stress-responsive NAC1 transcription factor to resistance of barley to Ramularia leaf spot. Mol. Plant Pathol. 16 201–209. 10.1111/mpp.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Lizhong X. (2011). Isolation and characterization of a drought-inducible promoter Oshox24P in rice. J. Huazhong Agric. Univ. 30 525–531. [Google Scholar]
- Mendelsohn R. (2014). The impact of climate change on agriculture in Asia. J. Integ. Agric. 13 660–665. 10.1016/S2095-3119(13)60701-7 [DOI] [Google Scholar]
- Mishra A. K., Singh V. P. (2010). A review of drought concepts. J. Hydrol. 391 202–216. 10.1016/j.jhydrol.2010.07.012 [DOI] [Google Scholar]
- Mishra S., Singh B., Panda K., Singh B. P., Singh N., Misra P., et al. (2016). Association of SNP haplotypes of HKT family genes with salt tolerance in indian wild rice germplasm. Rice 9:15 10.1186/s12284-016-0083-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2010). Genetic engineering for modern agriculture: challenges and perspectives. Ann. Rev. Plant Biol. 61 443–462. 10.1146/annurev-arplant-042809-12116 [DOI] [PubMed] [Google Scholar]
- Miyakawa T., Fujita Y., Yamaguchi-Shinozaki K., Tanokura M. (2013). Structure and function of abscisic acid receptors. Trends Plant Sci. 18 259–266. 10.1016/j.tplants.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Mizoi J., Ohori T., Moriwaki T., Kidokoro S., Todaka D., Maruyama K., et al. (2013). GmDREB2A; 2 a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161 346–361. 10.1104/pp.112.204875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Mochida K. E., Yoshida T. A., Sakurai T. E., Yamaguchi-Shinozaki K. A., Shinozaki K. A., Tran L. S. (2009). In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 16 353–369. 10.1093/dnares/dsp023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K., Mizoi J., Qin F., Kim J. S., Sato H., Osakabe Y., et al. (2013). Stabilization of Arabidopsis DREB2A is required but not sufficient for the induction of target genes under conditions of stress. PLoS ONE 8:e80457 10.1371/journal.pone.0080457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran S., Eini O., Pyvovarenko T., Parent B., Singh R., Ismagul A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9 230–249. 10.1111/j.1467-7652.2010.00547.x [DOI] [PubMed] [Google Scholar]
- Moumeni A., Satoh K., Venuprasad R., Serraj R., Kumar A., Leung H., et al. (2015). Transcriptional profiling of the leaves of near-isogenic rice lines with contrasting drought tolerance at the reproductive stage in response to water deficit. BMC Genomics 16:1110 10.1186/s12864-015-2335-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Jan A., Todaka D., Maruyama K., Goto S., Shinozaki K., et al. (2014a). Comparative functional analysis of six drought-responsive promoters in transgenic rice. Planta 239 47–60. 10.1007/s00425-013-1960-7 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Tran L. S. P., Nguyen D. V., Fujita M., Maruyama K., Todaka D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51 617–630. 10.1111/j.1365-313X.2007.03168.x [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32 959–970. 10.1007/s00299-013-1418-1 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (2014b). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170 10.3389/fpls.2014.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A., Jain M., Tyagi A. K., Khurana J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146 333–350. 10.1104/pp.107.112821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M., Sharoni A. M., Kikuchi S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Song S. I., Kim Y. S., Jang H. J., Kim S. Y., Kim M., et al. (2005). Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138 341–351. 10.1104/pp.104.059147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J. M. (2010). Biotechnology of water and salinity stress tolerance. Curr. Opin. Biotechnol. 21 185–196. 10.1016/j.copbio.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Pareek A., Sopory S. K., Bohnert H. K., Govindjee (2010). Abiotic Stress Adaptation in Plants: Physiolgical, Molecular and Genomic Foundation. Dordrecht: Springer. [Google Scholar]
- Paul S., Gayen D., Datta S. K., Datta K. (2015). Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress. Plant Sci. 234 133–143. 10.1016/j.plantsci.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2008). Plant genetics. The blue revolution, drop by drop, gene by gene. Science 320 171–173. 10.1126/science.320.5873.171 [DOI] [PubMed] [Google Scholar]
- Pérez-Clemente R. M., Vives V., Zandalinas S. I., López-Climent M. F., Muñoz V., Gómez-Cadenas A. (2013). Biotechnological approaches to study plant responses to stress. Biomed Res. Int. 2013:654120 10.1155/2013/654120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuong N. D., Tuteja N., Nghia P. T., Hoi P. X. (2015). Identification and characterization of a stress-inducible gene OsNLI-IF enhancing drought tolerance in transgenic tobacco. Curr. Sci. 109 541–551. [Google Scholar]
- Pourabed E., Golmohamadi F. G., Monfared P. S., Razavi S. M., Shobbar Z. S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57 12–26. 10.1007/s12033-014-9797-2 [DOI] [PubMed] [Google Scholar]
- Prasch C. M., Sonnewald U. (2015). Signaling events in plants: stress factors in combination change the picture. Environ. Expt. Bot. 114 4–14. 10.1016/j.envexpbot.2014.06.020 [DOI] [Google Scholar]
- Puckette M. C., Weng H., Mahalingam R. (2007). Physiological and biochemical responses to acute ozone-induced oxidative stress in Medicago truncatula. Plant Physiol. Biochem. 45 70–79. 10.1016/j.plaphy.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Qin F., Shinozaki K., Yamaguchi-Shinozaki K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52 1569–1582. 10.1093/pcp/pcr106 [DOI] [PubMed] [Google Scholar]
- Qin Y., Wang M., Tian Y., He W., Han L., Xia G. (2012). Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 39 7183–7192. 10.1007/s11033-012-1550-y [DOI] [PubMed] [Google Scholar]
- Quan W., Liu X., Wang H., Chan Z. (2016). Comparative physiological and transcriptional analyses of two contrasting drought tolerant alfalfa varieties. Front. Plant Sci. 6:1256 10.3389/fpls.2015.01256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Guangyuan H., Guangxiao Y., Hussain J., Xu Y. (2012). AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evol. Bioinform. Online. 8 321–355. 10.4137/EBO.S9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Dansana P. K., Bhaskar A., Giri J., Kapoor S., Khurana J. P., et al. (2010). “Emerging trends in functional genomics for stress tolerance in crop plants,” in Plant Stress Biology: From Genomics to Systems Biology, ed. Hirt H. (Weinheim: Wiley-VCH; ), 37–63. [Google Scholar]
- Rayko E., Maumus F., Maheswari U., Jabbari K., Bowler C. (2010). Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188 52–66. 10.1111/j.1469-8137.2010.03371.x [DOI] [PubMed] [Google Scholar]
- Redillas M. C., Jeong J. S., Kim Y. S., Jung H., Bang S. W., Choi Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10 792–805. 10.1111/j.1467-7652.2012.00697.x [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Meyerowitz E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Rusconi F., Simeoni F., Francia P., Cominelli E., Conti L., Riboni M., et al. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64 3361–3371. 10.1093/jxb/ert180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi S., Todorovic M., Tanasijevic L., Pereira L. S., Pizzigalli C., Lionello P. (2015). Climate change and Mediterranean agriculture: impacts on winter wheat and tomato crop evapotranspiration, irrigation requirements and yield. Agric. Water Manag. 147 103–115. 10.1016/j.agwat.2014.05.008 [DOI] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo K. K., Tripathi A. K., Pareek A., Singla-Pareek S. L. (2013). Taming drought stress in rice through genetic engineering of transcription factors and protein kinases. Plant Stress 7 60–72. [Google Scholar]
- Samarah H., Alqudah A. (2011). Effects of late-terminal drought stress on seed germination and vigor of barley (Hordeum vulgare L.). Arch. Agron. Soil Sci. 57 27–32. 10.1080/03650340903191663 [DOI] [Google Scholar]
- Samarah N. H., Alqudah A., Amayreh J., McAndrews G. (2009a). The effect of late-terminal drought stress on yield components of four barley cultivars. J. Agron. Crop Sci. 195 427–441. 10.1111/j.1439-037X.2009.00387.x [DOI] [Google Scholar]
- Samarah N. H., Haddad N., Alqudah A. (2009b). Yield potential evaluation in chickpea genotypes under late terminal drought in relation to the length of reproductive stage. Italian J. Agron. 3 111–117. 10.4081/ija.2009.3.111 [DOI] [Google Scholar]
- Samarah N. H., Mullen R. E., Anderson I. (2009c). Soluble sugar contents, germination and vigor of soybean seeds in response to drought stress. J. New Seeds 10 63–73. 10.1080/15228860902786525 [DOI] [Google Scholar]
- Sanchez D. H., Pieckenstain F. L., Szymanski J., Erban A., Bromke M., Hannah M. A., et al. (2011). Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS ONE 6:e17094 10.1371/journal.pone.0017094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Wang H., Tang X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902 10.3389/fpls.2015.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni A. M., Nuruzzaman M., Satoh K., Shimizu T., Kondoh H., Sasaya T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52 344–360. 10.1093/pcp/pcq196 [DOI] [PubMed] [Google Scholar]
- Shavrukov Y., Baho M., Lopato S., Langridge P. (2016). The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol. J. 14 313–322. 10.1111/pbi.12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Bohra A., Mishra S., Joshi R., Pandey S. (2015). Embracing new-generation ‘omics’ tools to improve drought tolerance in cereal and food-legume crops. Biol. Plant. 59 413–428. 10.1007/s10535-015-0515-0 [DOI] [Google Scholar]
- Singh B. P., Jayaswal P. K., Singh B., Singh P. K., Kumar V., Mishra S., et al. (2015a). Natural allelic diversity in OsDREB1F gene in the Indian wild rice germplasm led to ascertain its association with drought tolerance. Plant Cell Rep. 34 993–1004. 10.1007/s00299-015-1760-6 [DOI] [PubMed] [Google Scholar]
- Singh B. P., Singh B., Kumar V., Singh P. K., Jayaswal P. K., Mishra S., et al. (2015b). Haplotype diversity and association analysis of SNAC1 gene in wild rice germplasm. Indian J. Genet. Plant Breed. 75 157–166. 10.5958/0975-6906.2015.00025.5 [DOI] [Google Scholar]
- Singh D., Laxmi A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895 10.3389/fpls.2015.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S. Y., Chen Y., Chen J., Dai X. Y., Zhang W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234 331–345. 10.1007/s00425-011-1403-2 [DOI] [PubMed] [Google Scholar]
- Song X., Li Y., Hou X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14:573 10.1186/1471-2164-14-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastav A., Mehta S., Lindlof A., Bhargava S. (2010). Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal. Behav. 5 775–784. 10.4161/psb.5.7.11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Kidokoro S., Ito Y., Fujita Y., Shinozaki K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284 173–183. 10.1007/s00438-010-0557-0 [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu M., Gao S., Zhang Z., Zhao X., Zhao C., et al. (2012). Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant. 144 210–224. 10.1111/j.1399-3054.2011.01539.x [DOI] [PubMed] [Google Scholar]
- Tiwari V., Chaturvedi A. K., Mishra A., Jha B. (2015). Introgression of the SbASR-1 gene cloned from a halophyte salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogaea) and acts as a transcription factor. PLoS ONE 10:e0131567 10.1371/journal.pone.0131567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. (2012). Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice 5:6 10.1186/1939-8433-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D., Shinozaki K., Yamaguchi-Shinozaki K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6:84 10.3389/fpls.2015.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tron S., Bodner G., Laio F., Ridolfi L., Leitner D. (2015). Can diversity in root architecture explain plant water use efficiency? A modeling study. Ecol. Model. 312 200–210. 10.1016/j.ecolmodel.2015.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo L. E., Sotolongo M., Menendez C., Ochogava M. E., Coll Y., Hernandez I., et al. (2008). SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol. 49 512–515. 10.1093/pcp/pcn025 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8 10.1126/scisignal.2003509 [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Bansal K. C., Aggarwal P. K., Datta S. K., Craufurd P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16 363–371. 10.1016/j.tplants.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Voesenek L. A., Bailey-Serres J. (2013). Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 16 647–653. 10.1016/j.pbi.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang J., Zhang H., Zhang Z., Quan R., Zhou S., et al. (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 6:e25216 10.1371/journal.pone.0025216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang H., Shao H., Tang X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67 10.3389/fpls.2016.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li Q., Mao X., Li A., Jing R., Wang C., et al. (2016b). Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int. J. Biol. Sci. 12 257–269. 10.7150/ijbs.13538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu X., Zhang X., Smith P., Li L., Filley T. R., et al. (2016a). Size and variability of crop productivity both impacted by CO2 enrichment and warming-A case study of 4 year field experiment in a Chinese paddy. Agric. Ecosys. Environ. 221 40–49. 10.1016/j.agee.2016.01.028 [DOI] [Google Scholar]
- Wang J., Zhou J., Zhang B., Vanitha J., Ramachandran S., Jiang S. Y. (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 53 212–231. 10.1111/j.1744-7909.2010.01017.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67 589–602. 10.1007/s11103-008-9340-6 [DOI] [PubMed] [Google Scholar]
- Wang Z., Cheng K., Wan L., Yan L., Jiang H., Liu S., et al. (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1053 10.1186/s12864-015-2258-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani S. H., Gosal S. S. (2011). Introduction of OsglyII gene into Oryza sativa for increasing salinity tolerance. Biol. Plant. 55 536–540. 10.1007/s10535-011-0082-y [DOI] [Google Scholar]
- Wani S. H., Singh N. B., Devi T. R., Haribhushan A., Jeberson S. M., Malik C. P., et al. (2013). “Engineering abiotic stress tolerance in plants: extricating regulatory gene complex,” in Conventional and Non-Conventional Interventions in Crop Improvement, eds Malik C. P., Sanghera G. S., Wani S. H. (New Delhi: CABI; ), 1–19. [Google Scholar]
- Wei K., Chen J., Wang Y., Chen Y., Chen S., Lin Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19 463–476. 10.1093/dnares/dss026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Ortega R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci. 6:207 10.3389/fpls.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Lv H., Li L., Liu J., Mu S., Li X., et al. (2015). Genome-Wide Analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in moso bamboo (Phyllostachys edulis). PLoS ONE 10:e0126657 10.1371/journal.pone.0126657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tang N., Du H., Ye H., Xiong L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148 1938–1952. 10.1104/pp.108.128199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang X., Chen J. (2007). Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep. 26 497–506. 10.1007/s00299-006-0248-9 [DOI] [PubMed] [Google Scholar]
- Xu Z. S., Chen M., Li L. C., Ma Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53 570–585. 10.1111/j.1744-7909.2011.01062.x [DOI] [PubMed] [Google Scholar]
- Xu Z. Y., Kim S. Y., Kim D. H., Dong T., Park Y., Jin J. B., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25 4708–4724. 10.1105/tpc.113.119099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G.-P., Loveridge C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37 326–339. 10.1046/j.1365-313X.2003.01963.x [DOI] [PubMed] [Google Scholar]
- Xue G. P., Way H. M., Richardson T., Drenth J., Joyce P. A., McIntyre C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4 697–712. 10.1093/mp/ssr013 [DOI] [PubMed] [Google Scholar]
- Yang W., Liu X. D., Chi X. J., Wu C. A., Li Y. Z., Song L. L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233 219–229. 10.1007/s00425-010-1279-6 [DOI] [PubMed] [Google Scholar]
- Yao D., Zhang X., Zhao X., Liu C., Wang C., Zhang Z., et al. (2011). Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics 98 47–55. 10.1016/j.ygeno.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38 35–49. 10.1111/pce.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE–dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- You J., Zong W., Hu H., Li X., Xiao J., Xiong L. (2014). A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166 2100–2114. 10.1104/pp.114.251116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought and diseases in transgenic tobacco. J. Expt. Bot. 60 3781–3796. 10.1093/jxb/erp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu W., Wan L., Li F., Dai L., Li D., et al. (2010). Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res. 19 809–818. 10.1007/s11248-009-9357-x [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Quan R., Pan X., Wan L., Huang R. (2013). EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237 1443–1451. 10.1007/s00425-013-1852-x [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang L., Xia C., Zhao G., Liu J., Jia J., et al. (2015). A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant. 153 538–554. 10.1111/ppl.12261 [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu X., Wu L., Yu G., Wang X., Ma H. (2015). The SsDREB transcription factor from the succulent halophyte Suaeda salsa enhances abiotic stress tolerance in transgenic tobacco. Int. J. Genomics 2015:875497 10.1155/2015/875497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1093/jxb/erp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J., Cai B., Peng R. H., Zhu B., Jin X. F., Xue Y., et al. (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 371 468–474. 10.1016/j.bbrc.2008.04.087 [DOI] [PubMed] [Google Scholar]
- Zhuang J., Wang F., Xu Z. S., Xiong A. S. (2015). Microarray analysis of different expression profiles between wild-type and transgenic rice seedlings overexpression OsDREB1BI gene. Biologia 70 760–770. 10.1515/biolog-20150092 [DOI] [Google Scholar]