ABSTRACT
The ability of Staphylococcus aureus to resist host innate immunity augments the severity and pervasiveness of its pathogenesis. Nitric oxide (NO˙) is an innate immune radical that is critical for the efficient clearance of a wide range of microbial pathogens. Exposure of microbes to NO˙ typically results in growth inhibition and induction of stress regulons. S. aureus, however, induces a metabolic state in response to NO˙ that allows for continued replication and precludes stress regulon induction. The regulatory factors mediating this distinctive response remain largely undefined. Here, we employ a targeted transposon screen and transcriptomics to identify and characterize five regulons essential for NO˙ resistance in S. aureus: three virulence regulons not formerly associated with NO˙ resistance, SarA, CodY, and Rot, as well as two regulons with established roles, Fur and SrrAB. We provide new insights into the contributions of Fur and SrrAB during NO˙ stress and show that the S. aureus ΔsarA mutant, the most sensitive of the newly identified mutants, exhibits metabolic dysfunction and widespread transcriptional dysregulation following NO˙ exposure. Altogether, our results broadly characterize the regulatory requirements for NO˙ resistance in S. aureus and suggest an intriguing overlap between the regulation of NO˙ resistance and virulence in this well-adapted human pathogen.
IMPORTANCE The prolific human pathogen Staphylococcus aureus is uniquely capable of resisting the antimicrobial radical nitric oxide (NO˙), a crucial component of the innate immune response. However, a complete understanding of how S. aureus regulates an effective response to NO˙ is lacking. Here, we implicate three central virulence regulators, SarA, CodY, and Rot, as major players in the S. aureus NO˙ response. Additionally, we elaborate on the contribution of two regulators, SrrAB and Fur, already known to play a crucial role in S. aureus NO˙ resistance. Our study sheds light on a unique facet of S. aureus pathogenicity and demonstrates that the transcriptional response of S. aureus to NO˙ is highly pleiotropic and intrinsically tied to metabolism and virulence regulation.
INTRODUCTION
The versatile Gram-positive pathogen Staphylococcus aureus is the leading cause of skin and soft tissue infections (SSTIs) in the United States but can also cause more severe illnesses, including pneumonia, osteomyelitis, endocarditis, and bacteremia (1–5). The ability of S. aureus to infect virtually every tissue of the body can be partially attributed to its extensive capacity for subverting host immunity. One unique defense of S. aureus against the immune system is resistance to nitric oxide (NO˙), a membrane-permeable radical and broad-spectrum innate immune effector required for the clearance of bacterial, viral, and fungal pathogens (6). The ability of S. aureus to continue growth in the presence of NO˙ at concentrations that are inhibitory to other bacteria, including closely related staphylococci, contributes to its invasiveness and pathogenic success. For example, the NO˙-sensitive species Staphylococcus epidermidis is rapidly cleared from model SSTIs in wild-type (WT) mice but is able to achieve high bacterial density in mice deficient in NO˙ production (i.e., lacking an inducible nitric oxide synthase [iNOS]) (7, 8). In contrast, S. aureus is less affected by the presence of iNOS in multiple-infection models.
NO˙ and other reactive nitrogen species (RNS) are formed by the oxidation of NO˙, target heme, iron-sulfur clusters, redox-active thiols, lipids, and DNA (9–13). Thus, bacteria exposed to NO˙ encounter a variety of biochemical and metabolic stresses, such as DNA damage, altered metal homeostasis, and disrupted metabolic enzymes, including components of the respiratory chain. In Bacillus subtilis, Escherichia coli, and other NO˙-sensitive bacterial species, NO˙ strongly induces general and oxidative stress response regulons (i.e., SigB, PerR, Spx, OhrR, OxyR, and SoxR) (14–18). In contrast, there is no evidence of increased SigB-dependent transcription or oxidative stress regulon induction (PerR) in S. aureus following NO˙ exposure, potentially reflecting the significant NO˙ resistance of S. aureus compared to these other species (6, 14). Instead, S. aureus evokes a unique program of genetic regulation, resulting in the induction of a distinct metabolic state that can support NO˙-resistant growth (6, 19, 20).
When high-level NO˙ disrupts respiration in S. aureus cells, rising NADH levels inactivate the transcriptional repressor Rex, resulting in the derepression of two lactate dehydrogenases, ddh and ldh1, of which ldh1 is unique to S. aureus (6, 21). In conjunction with a third lactate dehydrogenase, ldh2, these enzymes allow nonrespiring S. aureus to perform heterolactic fermentation and maintain redox homeostasis until NO˙ levels subside sufficiently for respiration to resume (19). Whereas a Δldh1 Δldh2 dehydrogenase mutant is extremely sensitive to NO˙ and avirulent in a mouse bacteremia model, individual lactate dehydrogenase mutants are only moderately sensitive (19). This redundancy underscores the evolutionary importance of NO˙ resistance to the pathogenic success of S. aureus but also complicates the identification of individual genes required for NO˙ resistance.
Respiration-limiting conditions imposed by NO˙ also stimulate the SrrAB two-component system, which controls genes involved in maximizing respiratory capacity, repairing iron-sulfur clusters, and supporting anaerobic metabolism (22, 23). Like its homolog ResDE in B. subtilis, SrrAB is required for maximal expression of hmp, encoding a conserved flavohemoprotein that detoxifies NO˙ to nitrate (6, 23, 24). Despite the fact that cells without SrrAB activity retain residual Hmp-mediated NO˙ detoxification, ΔsrrAB mutants are much more sensitive to NO˙ and attenuated for virulence in a mouse model of bacteremia than Δhmp mutants (6). These data imply that in addition to hmp, other SrrAB-regulated genes are important for S. aureus NO˙ resistance. However, of the remaining SrrAB-controlled genes, only qoxABCD, encoding an aa3-type terminal oxidase, has thus far been shown to contribute to S. aureus NO˙ resistance (20, 23). Consequently, a complete understanding of how SrrAB contributes to S. aureus NO˙ resistance is still lacking.
Another regulon commonly stimulated by NO˙ exposure is that controlled by the ferric uptake regulator (Fur). This regulator represses the expression of genes involved in iron acquisition, so long as its ferrous iron corepressor is abundant (25, 26). Given that NO˙ can nitrosylate fur-bound ferrous iron and render the transcription factor inactive, the Fur regulon is highly induced by NO˙ in S. aureus (6, 27). S. aureus Δfur mutants are highly sensitive to NO˙, yet the precise role of Fur in S. aureus NO˙ resistance has not been fully explained (6, 15, 18, 28–30).
Several components of the known S. aureus NO˙ response are controlled by as-yet-unidentified regulatory mechanisms, suggesting that additional NO˙-responsive regulators exist beyond those currently characterized. For example, while SrrAB is required for maximal hmp expression, hmp is still NO˙ inducible in an srrAB mutant (6). In many Firmicutes and most beta- and gammaproteobacteria, hmp transcription is repressed by the Rrf2 family transcription factor NsrR, which responds to NO˙ via a redox active iron-sulfur cluster to derepress hmp (31–33). However, S. aureus entirely lacks an NsR homolog, suggesting that other NO˙-responsive regulators affect hmp transcription in S. aureus. The regulation of ldh1 is likewise incompletely understood; full ldh1 induction requires S. aureus growth on glucose through an unknown mode of transcriptional regulation that is independent of CcpA (34). There are also numerous other S. aureus genes that respond differentially to NO˙ but do not fall into the characterized SrrAB, Fur, or Rex regulon (6, 14).
Considering the complex and redundant nature of NO˙ resistance and the fact that the most severely NO˙-sensitive mutants identified thus far are regulators (Fur and SrrAB), we reasoned that the identification of key regulons important during NO˙ stress is a viable strategy for expanding our understanding of S. aureus NO˙ resistance. In the present study, we combine a comprehensive screen of regulator transposon mutants with transcriptomics to broadly define the regulatory requirements for an effective S. aureus response to NO˙. In addition to SrrAB and Fur, we identified three regulators not previously associated with NO˙ resistance: SarA, CodY, and Rot. Most notably, the sarA mutant is as sensitive to NO˙ as the srrAB mutant and exhibits global gene dysregulation and a unique aberrant metabolism following NO˙ exposure. Additionally, SarA, CodY, and Rot are all important virulence regulators, highlighting the integration of NO˙ resistance and virulence gene regulation in S. aureus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material. The Δrot::Spr, ΔsarA::Spr, ΔargGH::Err, and ΔguaAB ΔpbuX Δxpt::Err mutants were created via allelic exchange using previously described methods (20). The ΔsrrAB::Err and Δfur::Kmr mutants were reconstructed directly in S. aureus Newman and COL, using the previously constructed allelic exchange vectors pTR43 and pTR46, respectively (6). Mutations were subsequently transduced back into the original strain background (using ϕ80 for transduction from COL or Newman and ϕ11 for transduction from LAC or JE2) to reduce the possibility of secondary mutations developed during the mutant-making process. Bursa aurealis transposon mutants from the Nebraska Library were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA). The LAC Δaur ΔsspAB ΔscpA Δspl::Er (protease-null) mutant was provided by Lindsey Shaw (University of South Florida, Tampa, FL) (35). The LAC and COL sarA::Kmr mutants were generated by ϕ11-mediated transduction from a previously constructed mutant (36). The Newman and LAC ΔcodY::Tcr mutants were constructed by ϕ80-mediated transduction from a previously constructed mutant provided by Christiane Wolz (University of Tübingen, Germany) (37). Complementation of srrAB was carried out by cloning either cydAB or qoxABCD into the NdeI site of pOS1-plgt (38), downstream of the constitutive lgt promoter, using a Gibson assembly (NEB).
S. aureus strains were cultured in tryptic soy broth (TSB), Luria-Bertani broth (LB), LB with 0.5% glucose and 0.05 M Tris at pH 7.4 (LBGT), or chemically defined medium (PN medium) with 0.5% glucose. Growth curves were carried out in 200-μl cultures within a 96-well plate. Change in absorbance (650 nm) was measured at 15-min intervals by a Tecan Infinite M200 Pro microplate reader programmed for a series of 96 cycles of 1-mm orbital shaking for 830 s, followed by 1-mm linear shaking for 30 s. Growth curves were inoculated from overnight cultures grown in TSB, washed with phosphate-buffered saline (PBS), and diluted to an optical density at 660 nm (OD660) of 0.01. The NO˙ donors used in this study were 2,2′-(hydroxynitrosohydrazono)bis-ethanimine (DETA)-NO or a mixture of NOC-12 and diethylamine nitric oxide (DEA-NO), each resuspended in 0.01 N NaOH.
Measurement of NO˙ sensitivity during lag phase.
Growth curves were performed in the 96-well plate format as described above with NO˙ added at the time of inoculation. Due to the differential NO˙ sensitivities of various S. aureus strain backgrounds, medium type and NO˙ concentration were used in the following combinations for each strain background: JE2, LB medium, 5 mM DETA-NO; Newman, LBGT medium, 5 mM DETA-NO; COL, LBGT medium, 10 mM DETA-NO; and LAC, LBGT medium, 10 mM DETA-NO. We selected LB broth for the initial screen because it contains less heme (an NO˙-scavenging agent) than TSB and brain heart infusion (BHI) agar (data not shown). For growth curve experiments performed to replicate the S. aureus COL ΔsrrAB, ΔqoxABCD Δhmp, and ΔcydAB Δhmp mutant genotypes reported by Kinkel et al. (23), our growth conditions were modified to match those reported in their study (i.e., PN medium, 1 mM DEA-NO, and 5 mM NOC-12 added at the time of inoculation, with a starting OD660 of 0.033). These conditions were maintained for the ΔsrrAB mutant complementation experiments.
The NO˙ sensitivity of each strain was determined based on the extent to which NO˙ extended its lag time. Lag time is defined as the time until an OD650 of 0.2 (representing entry into early exponential phase) is reached. The following equation was used to determine the amount by which NO˙ extended the lag time of each mutant compared to that of the WT (where t is equal to the lag time of NO˙-exposed [tNO˙] or aerobically cultured [taerobic] cells): (tNO˙ − taerobic)mutant/(tNO˙ − taerobic)WT. All mutant lag times were normalized to the lag time of the WT on the same day to account for experimental variability in DETA-NO strength. The viability of Newman mutants exposed to NO˙ in this assay was determined by serial dilution and plating of cells at the time of inoculation and after 8 h of NO˙ exposure.
Measurement of peroxide sensitivity.
Newman mutants grown to an OD660 of 0.2 in LBGT were exposed to 20 mM peroxide and shaken at 37° in 1-ml culture volumes. Just before the addition of peroxide and again following 1 h of peroxide exposure, cells were serially diluted and plated to determine the change in viability.
Iron chelation experiments.
S. aureus Newman WT and Δfur mutant strains were grown in the 96-well plate growth curve format (described above) in LBGT either with or without the addition of 2 mM 2,2′-dipyridyl, a divalent cation chelator. To enhance NO˙-mediated killing of the Δfur mutant (compared to the moderate killing observed when DETA-NO is added at the time of inoculation), cells were exposed to an NO˙ mixture of 10 mM NOC-12 and 1 mM DEA-NO at an OD650 of 0.15. Viability was determined just before the addition of NO˙ and at 4 h following NO˙ exposure by serial dilution and plating.
Hydroxyl radical measurement.
S. aureus Newman WT and the Δfur mutant were grown from a starting OD650 of 0.04 in 96-well plates in LBGT with 20 μM hydroxyphenyl fluorescein (Thermo Fisher Scientific). After reaching an OD650 of 0.25 (after ~3.5 h), bacteria were washed once in PBS and then resuspended in fresh LBGT, and an NO˙ mixture (10 mM NOC-12, 1 mM DEA-NO) or 1 mM peroxide was added to select wells. After 2 h, bacteria were resuspended in PBS, the OD650 was remeasured, and then cells were transferred to black-bottom plates for measurement of hydroxyphenyl fluorescein (HPF) fluorescence (excitation, 490 nm; emission, 520 nm) on a Tecan M200 plate reader.
Exponential-phase NO˙ exposure and characterization of the fermentative/respiratory phases of growth.
S. aureus COL cells were grown in the 96-well plate growth curve format (described above) in PNG (0.5% glucose) either aerobically or with a NO˙ mixture (1 mM DEA-NO, 10 mM NOC-12) added at an OD650 of 0.15. For both aerobic and NO˙-exposed cultures, the time point at which cultures reached an OD650 of 0.15 was designated t0. Growth rates were calculated at 15-min intervals using the equation Δln(Abs650)/Δt (in hours). The fermentative growth rate is the average growth rate over a 3-h period (corresponding to h 1 to 4) following the addition of NO˙. The respiratory growth rate is the average growth rate over a 45-min period corresponding to the peak growth reached between 4 and 12 h following the addition of NO˙. The aerobic growth rate is the average growth rate over the 45-min period corresponding to peak logarithmic growth in cultures without the addition of NO˙ donor.
Intracellular ATP concentrations were measured using the BacTiter-Glo microbial cell viability assay (Promega), according to the manufacturer's instructions. ATP levels were normalized to the OD650 value for each time point. Aerobic ATP levels were measured at the time of the maximum growth rate (approximately t1), and NO˙ ATP levels were measured at 2-h intervals following the addition of NO˙.
For measurement of excreted metabolites, culture aliquots were removed at 1-h intervals, heat inactivated at 70° for 5 min, and the supernatants were collected. l-Lactic acid and acetic acid concentrations were enzymatically determined using commercially available kits (Roche Yellow line kits; R-Biopharm AG, Darmstadt, Germany).
RNA-Seq.
S. aureus COL WT and ΔsarA mutant strains were grown in PNG and exposed to 4 mM DETA-NO at an OD660 of 0.4. RNA was harvested after 1 h of incubation using a Qiagen RNeasy kit with the modified protocol described by Carroll et al. (39). RNA was also collected from WT cultures grown aerobically (without addition of NO˙) at an OD660 of 0.5. Sample preparation for transcriptome sequencing (RNA-Seq) and sequencing using the Ion Personal Genome Machine (PGM) system were conducted as described previously (39, 40). Briefly, an Agilent 2100 Bioanalyzer in combination with the Agilent RNA 6000 nano kit were used to quantify RNA, equimolar amounts from three independent replicates were pooled for each sample, and MICROBExpress (Life Technologies) and Ribo-Zero (Epicentre) kits were used to remove rRNA, which was confirmed via the Agilent 2100 Bioanalyzer (Agilent RNA 6000 nano kit). RNA-Seq was conducted using the Ion total RNA-Seq kit version 2 to prepare samples, the Ion PGM template OT2 200 kit to generate template-positive Ion Sphere Particles, and the Ion 318 Chip version 2 for sequencing with the Ion PGM sequencing 200 kit version 2. Data were analyzed using CLC Genomics Workbench and were normalized via the quantile normalization approach (41). Reads were mapped to the S. aureus COL genome. Reads per kilobase per million mapped reads (RKPM) were designated the expression values, and genes with RPKM of <10 for both conditions in a given comparison (i.e., WT cells grown aerobically versus WT cells exposed to NO˙) were excluded from analysis. Genes with ≥2-fold change in expression were further analyzed by STRING version 10.0 (42) and GoMiner (43) to identify groups of related genes with changed expression.
qRT-PCR.
For verification of the RNA-Seq results, quantitative real-time PCR (qRT-PCR) was performed on RNA extracts collected from aerobically grown and NO˙-exposed S. aureus COL cultures independently of those used for RNA-Seq analysis. The SensiFAST SYBR & fluorescein one-step kit (Bioline) was used, according to the manufacturer's instructions, to analyze 50 ng of total RNA per reaction on a MyiQ single-color real-time PCR detection system (Bio-Rad). The primers used are listed in Table S1 in the supplemental material.
RESULTS
Five regulators are essential for full NO˙ resistance in multiple S. aureus strains.
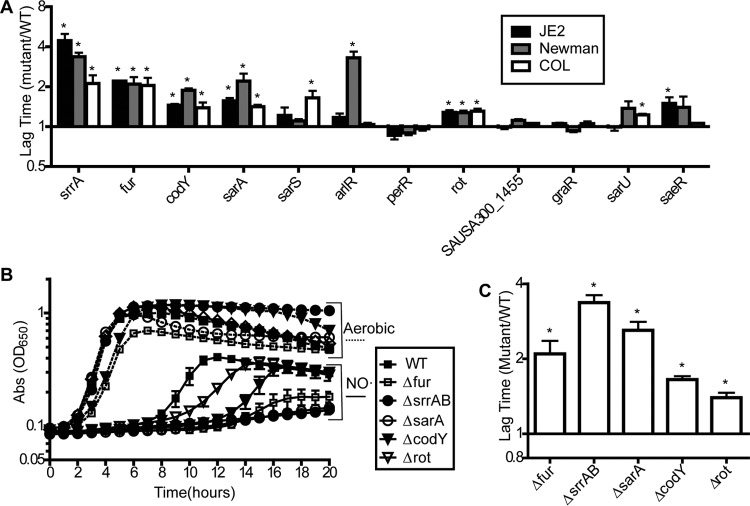
To identify regulatory proteins essential for NO˙ resistance in S. aureus, we screened a library of transposon mutants with insertions in every nonessential predicted transcriptional regulator for signs of NO˙ sensitivity (mutants obtained from the Nebraska Transposon Mutant Library). In total, we screened 115 regulatory transposon mutants comprising 110 transcriptional regulators (approximately 80% of all predicted transcriptional regulators in S. aureus), three sigma factors, and a Ser/Thr kinase/phosphatase enzyme pair known to modulate the functions of multiple regulatory proteins (44–48). To screen for NO˙ sensitivity, we compared the normalized lag time (see Materials and Methods) of the regulatory mutants to that of WT S. aureus following treatment with NO˙ at the time of inoculation. WT cells typically exhibit an ∼2-h lag in buffered LB medium supplemented with glucose; however, NO˙ donors, such as DETA-NO, can extend the lag to ∼5 h (see Fig. S1 in the supplemental material). Mutants with a significantly extended lag time specifically upon NO˙ exposure were considered NO˙ sensitive. Using this lag time readout, we identified 28 transposon mutants that were significantly more sensitive to NO˙ than the WT strain JE2 (see Data set S1 in the supplemental material). To verify that NO˙ sensitivity was associated with the transposon insertion and not a secondary mutation, we transduced the top 12 most sensitive mutants back into the WT JE2 strain background. We were primarily interested in regulators important for NO˙ resistance across multiple strain backgrounds, so we additionally transduced each of the 12 mutations into S. aureus strains COL and Newman.
Following transduction, five regulator mutants were significantly sensitive to NO˙ in all three strain backgrounds: srrAB, fur, sarA, codY, and rot (Fig. 1A). We observed interesting strain differences in NO˙ sensitivity for several other mutants (arlR, sarS, sarU, and saeR), but for this paper, we focus on the five regulators with significant sensitivity in all three backgrounds. We next constructed deletion mutants of each of these five regulators in S. aureus Newman to use for subsequent experiments and to further confirm phenotypes. Each deletion mutant exhibited a NO˙-susceptible phenotype comparable to its corresponding transposon mutant (Fig. 1B and C).
FIG 1.
Five regulators are required for full NO˙ resistance in multiple strains of S. aureus (SA). (A) Mutations in S. aureus transposon mutants identified as sensitive to NO˙ in the initial screen were transduced back into S. aureus JE2 as well as into S. aureus COL and Newman. The transduced strains were screened for NO˙ sensitivity when exposed to 5 mM DETA-NO at inoculation. The lag time (time to OD650 of 0.2) for each NO˙-exposed strain was normalized to the lag time for the strain under aerobic conditions. Graphs depict the ratio of mutant to WT lag times (n = 3; error bars show the standard error of the mean [SEM]). (B) Mutants with deletions in the five most broadly NO˙-sensitive regulators constructed in S. aureus Newman were grown in LBGT either with or without the addition of 5 mM DETA-NO (n = 3; error bars show SEM). (C) Mutant-to-WT lag time ratios of the NO˙-exposed Newman strains (depicted in panel B) normalized to their respective aerobic lag times (n = 3; error bars show SEM). Significance was calculated using Student's two-sided t test to compare the normalized mutant lag time to the normalized WT lag time (*, significantly different from the WT; P ≤ 0.05).
Increased intracellular free iron sensitizes the Δfur mutant to NO˙.
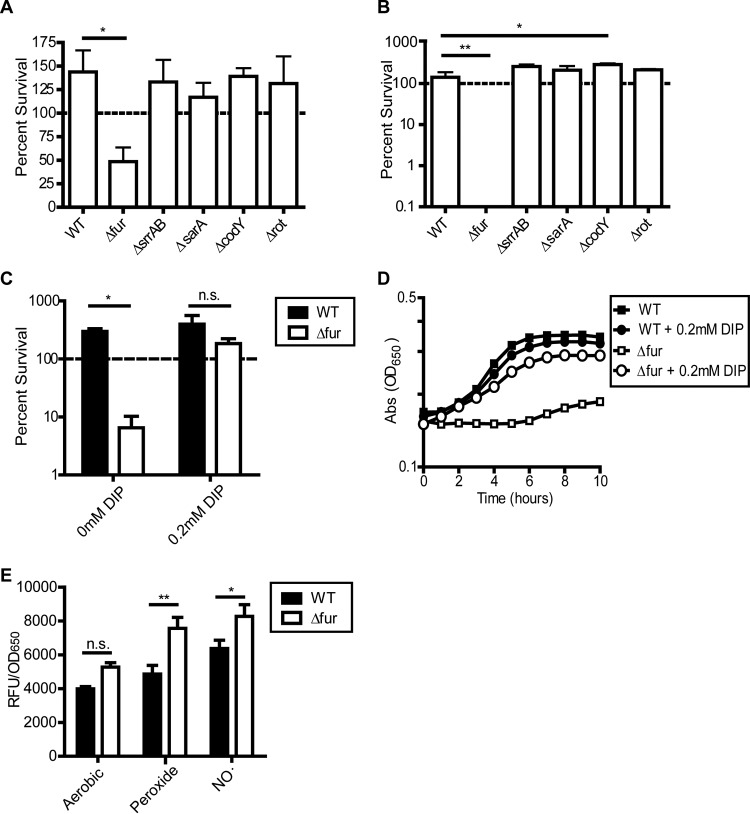
NO˙ generally exerts bacteriostatic effects on sensitive bacteria, with S. aureus being the primary exception, given its ability to continue replicating in the presence of this immune radical. Consistent with this, NO˙ was bacteriostatic for most of the identified sensitive regulatory mutants, with the exception of the Δfur mutant. NO˙ exposure at the time of inoculation reduced the viability of the Δfur mutant by ∼60% in 8 h (Fig. 2A), which likely resulted in its extended lag time.
FIG 2.
The Δfur single mutant is susceptible to killing by NO˙ and is rescued by iron chelation. (A) Percent survival of S. aureus Newman regulator mutants 8 h after exposure to 5 mM DETA-NO in LBGT medium (n = 3; error bars show SEM). (B) Percent survival of S. aureus Newman regulator mutants 1 h after exposure to 20 mM peroxide in LBGT medium (n = 3; error bars show SEM). (C and D) Percent survival (C) and growth (D) of WT Newman and Δfur strains 4 h after exposure to a mixture of 10 mM NOC-12 and 1 mM DEA-NO in LBGT medium with or without 200 μM 2,2′-dipyridyl (DIP). NO˙ was added at an OD650 of 0.15 (t0) (n = 3; error bars show SEM). (E) Hydroxyl radical formation was measured by detecting the fluorescence of hydroxyphenyl fluorescein (HPF) dye in the WT and the Δfur mutant following 2 h of aerobic growth, exposure to a 1 mM peroxide, or exposure to a NO˙ mixture (10 mM NOC-12, 1 mM DEA-NO). Relative fluorescence units (RFU) are normalized to the OD650 (data were pooled from two separate experiments for n of 4; error bars show SEM). Significance for panels C and E was determined by a two-way analysis of variance test (ANOVA) with a post hoc Bonferroni multiple-comparison test (*, P ≤ 0.05; **, P ≤ 0.01). n.s., nonsignificant.
One feature of NO˙ toxicity is the potentiation of reactive oxygen species (ROS)-mediated bacterial killing (49). High-level NO˙ disrupts cellular respiration, which can lead to the buildup of cellular reductants capable of reducing iron to its ferrous state (Fe2+) (50, 51). Free Fe2+ then catalyzes the Fenton reaction, whereby the highly reactive hydroxyl radical (˙OH) is generated from peroxide. NO˙-mediated respiration inhibition also enhances endogenous ROS generation via increased univalent reduction of oxygen resulting from the excessive buildup of electrons in the respiratory chain (52, 53). Because increased sensitivity to oxidative stress might be a mechanism for enhanced NO˙ susceptibility, we tested all five NO˙-sensitive regulatory mutants for sensitivity to peroxide. We found that only the Δfur mutant exhibited enhanced susceptibility to killing by peroxide (Fig. 2B). This result suggests that most NO˙-sensitive regulatory mutants exhibit defects that are specific to NO˙, except for the Δfur mutant, which is highly sensitive to ROS as well.
Fur, the master regulator of bacterial iron homeostasis, has been described as essential for ROS tolerance as well as NO˙ resistance in S. aureus and many other bacterial species (26, 54, 55). A Δfur mutant constitutively expresses iron acquisition genes due to an inability to sense iron abundance. The resultant increase in intracellular free iron potentiates Fenton chemistry (i.e., the generation of the highly reactive hydroxyl radical) under conditions of oxidative stress, resulting in the observed sensitivity to peroxide (54). To determine whether NO˙ sensitivity in the Δfur mutant is also due to excess intracellular iron, we exposed the Δfur mutant to NO˙ in the presence of the divalent cation chelator 2,2′-dipyridyl (Fig. 2C and D). Iron chelation rescued both the survival and growth of the Δfur mutant in the presence of NO˙, indicating that free iron accumulation is the primary cause of the Δfur mutant NO˙ sensitivity as well as ROS sensitivity.
To determine whether hydroxyl radical generation might play a role in the NO˙ sensitivity of the Δfur mutant, we used the dye hydroxyphenyl fluorescein (HPF), which fluoresces green following oxidation by ˙OH (Fig. 2E). Relative to the WT, the Δfur mutant exhibited enhanced fluorescence suggestive of excessive ROS production even during aerobic growth, and the situation worsened following exposure to peroxide and NO˙. Importantly, while NO˙ exposure increased HPF fluorescence for both the WT and the Δfur mutant, levels of fluorescence remained higher in the Δfur mutant, indicating a possible role for ROS in its NO˙ sensitivity. These data should be interpreted with caution, because HPF can also be oxidized by peroxynitrite (a product of the reaction of NO˙ and superoxide), oxidized metals, and an unknown product(s) in dying cells (56, 57). However, the greater HPF fluorescence observed in the Δfur mutant under all conditions is likely attributable to excess iron potentiating ROS generation, regardless of what species are contributing to dye oxidation.
In general, regulators required for NO˙ resistance do not affect the expression of known NO˙ resistance determinants.
We next investigated whether any of the five identified regulators are required for the NO˙-mediated induction of hmp and ldh1, genes known to be critical to S. aureus NO˙ resistance. We used reporter plasmids harboring a transcriptional fusion of gfp to either the hmp or ldh1 promoter to determine the level of NO˙-mediated induction in each regulatory mutant. The Δfur, ΔsarA, and Δrot mutants normally induced ldh1, and the ΔsrrAB and ΔcodY mutants actually showed increased ldh1 induction compared to the WT (see Fig. S2A in the supplemental material). Only the ΔsrrAB mutant exhibited moderate defects in hmp induction upon NO˙ exposure, a phenomenon well established in previous literature (see Fig. S2B). Thus, the NO˙ sensitivity of these regulatory mutants, aside from the ΔsrrAB mutant, cannot be attributed to reduced transcriptional induction of previously characterized NO˙ resistance determinants.
SrrAB is essential for the respiratory phase of S. aureus NO˙-resistant growth.
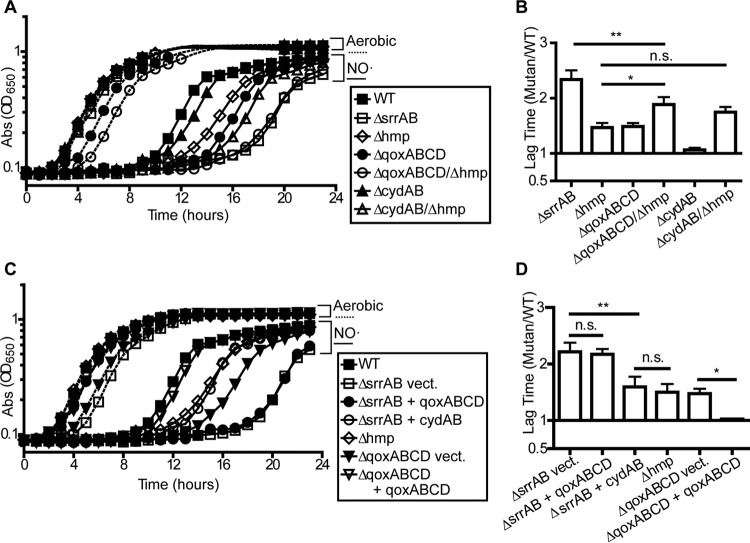
It has been suggested that the contribution of SrrAB to NO˙ resistance in S. aureus is 2-fold. First, SrrAB is essential for full induction of the flavohemoprotein Hmp. This enzyme detoxifies NO˙ to nitrate, thereby reducing intracellular NO˙ levels such that respiration can resume (58). Reduced Hmp production in a ΔsrrAB mutant presumably extends the period of time during which NO˙ levels surpass the threshold that limits respiration. Second, NO˙-mediated induction of respiratory cytochromes is also SrrAB dependent (23). Increasing the levels of the aa3 and bd respiratory cytochromes (encoded by qoxABCD and cydAB, respectively) may aid in overcoming the inhibition of respiration imposed by NO˙. In fact, it has been reported that combining the Δhmp and ΔqoxABCD mutations fully recapitulates the severe NO˙ sensitivity of a ΔsrrAB mutant (23). We replicated this finding and found that the Δhmp ΔqoxABCD double mutant indeed phenocopies the ΔsrrAB mutant during NO˙ stress (Fig. 3A and B). However, the Δhmp ΔqoxABCD mutant also has a defect during normal aerobic cultivation compared to the ΔsrrAB mutant. Thus, the ratiometric comparison of lag times indicates that the ΔqoxABCD Δhmp mutant is still significantly less sensitive to NO˙ than the ΔsrrAB mutant (Fig. 3B). Additionally, whereas the ΔcydAB single mutant is not sensitive to NO˙, we found that combining the Δhmp and ΔcydAB mutations leads to worsened NO˙ sensitivity compared to that with the Δhmp mutation alone alone (Fig. 3A and B). Together, these data suggest that induction of both respiratory cytochromes by SrrAB serves to overcome respiration inhibition during NO˙ stress.
FIG 3.
SrrAB is required for induction of both terminal oxidases during NO˙ stress. (A and C) S. aureus COL strains were grown in PNG either without or with 5 mM NOC-12 and 1 mM DEA-NO (n = 3; graphs show representative growth curves). (B and D) The ratio of mutant to WT lag times in the presence of NO˙, normalized to aerobic lag times, was calculated for the strains and growth conditions depicted in panels A and C, respectively (n = 4 for panel B, and n = 3 for panel D; error bars show SEM). Significance was determined by one-way ANOVA with a post hoc Student-Newman-Keuls test (*, P ≤ 0.05; **, P ≤ 0.01). vect., vector.
To test the relative importance of SrrAB-mediated induction of each respiratory cytochrome during NO˙ stress, we constitutively expressed either qoxABCD or cydAB in the ΔsrrAB mutant. We found that overexpression of cydAB significantly rescued the NO˙ sensitivity of the ΔsrrAB mutant to a level comparable to that of the Δhmp single mutant, suggesting that the NO˙ sensitivity of the ΔsrrAB mutant can indeed be partially attributed to reduced respiratory capacity (Fig. 3B and C). Unexpectedly, however, overexpression of qoxABCD had no effect on the NO˙ sensitivity of the ΔsrrAB mutant, even though expression complemented the ΔqoxABCD single mutant (indicating functionality of the complement vector) (Fig. 3C and D). This observation can be attributed to the fact that SrrAB is also required for the induction of CtaAB, which catalyzes the conversion of b-type heme molecules to the a-type required for QoxABCD function (23, 59). Taken together, these data suggest that the network of genes induced by SrrAB in response to NO˙ is complex but ultimately contributes to overcoming the respiration inhibition induced by high NO˙ levels. This is accomplished in part by maximizing NO˙ detoxification (via the hmp gene) and cytochrome production (via qoxABCD and cydAB).
ΔsrrAB and ΔsarA mutants exhibit defects specifically during the respiratory phase of S. aureus NO˙ resistance.
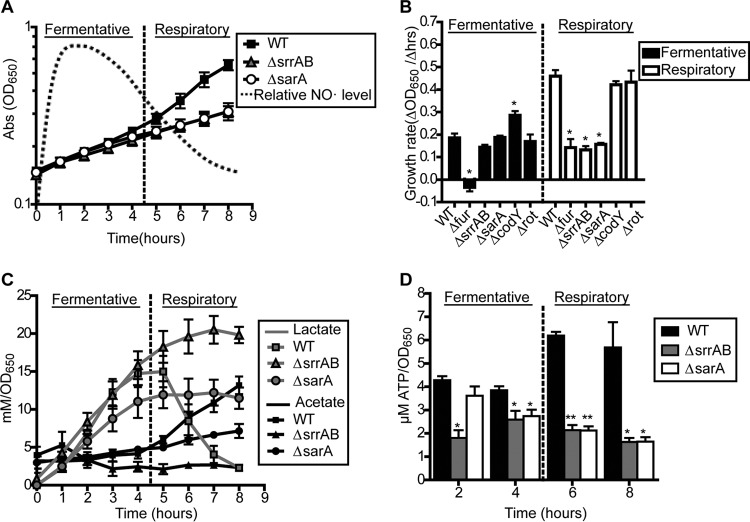
To further explore the role of SrrAB as well as the other regulators in overcoming respiration inhibition following NO˙ stress, we used an assay in which NO˙ donors are added to aerobically growing S. aureus in early exponential phase rather than lag phase. Following the addition of donor in this assay, NO˙ levels rapidly increase to the extent that respiration becomes inhibited, and then they subsequently decrease (due to NO˙ escape and enzymatic detoxification) to a level at which respiration can resume. We are able to visualize the shift from fermentative growth (high NO˙) back to respiratory growth (low NO˙) in distinct phases in this assay. The fermentative phase of NO˙-resistant growth lasts for ∼4 h and is easily visualized by a reduced growth rate and a diversion of glycolytic carbon from acetate to lactate (Fig. 4A and C). Conversely, resumption of respiratory growth is marked by an increased growth rate, reassimilation of extracellular lactate, and renewed production of acetate (Fig. 4A to C). In the resumption of respiratory growth phase, NO˙ is still present but at a level below that required for complete respiration inhibition (data not shown).
FIG 4.
ΔsrrAB and ΔsarA mutants exhibit reduced growth rates associated with aberrant metabolisms and reduced energy charge following exponential-phase NO˙ exposure. (A) Two phases of growth are observed following the addition of a NO˙ mixture (10 mM NOC-12 and 1 mM DEA-NO) to WT COL during early exponential phase (OD650, 0.15): an initial fermentative phase characterized by linear growth rate and lactate production while NO˙ levels are high and a subsequent respiratory phase characterized by resumption of exponential growth and respiration-dependent lactate consumption as NO˙ levels decrease (n = 3). (B) Fermentative and respiratory growth rates of S. aureus COL regulator mutants following addition of the NO˙ mixture at early exponential phase (OD650, 0.15) (n = 3). Significance was calculated using Student's two-sided t test (*, significantly different from the WT; P ≤ 0.05). (C and D) Lactate and acetate production (C) and ATP levels (D) were measured periodically following exposure of S. aureus COL strains to the NO˙ mixture. Significance was calculated using Student's two-sided t test (asterisks indicate significant difference from the WT; *, P ≤ 0.05; **, P ≤ 0.01).
Consistent with our findings outlined above, the ΔsrrAB mutant has no growth defect during the fermentative phase but fails to transition into the respiratory phase (Fig. 4B). That is, the ΔsrrAB mutant never resumes acetogenesis or reassimilates lactate (Fig. 4C). Consequently, ΔsrrAB mutants exhibit drastically reduced energy charge (ATP levels) compared to WT cells during the period in which the WT has successfully entered the respiratory phase (Fig. 4C). Although the ΔcodY and Δrot mutants are significantly sensitive to NO˙ when exposed at the time of inoculation, they do not exhibit growth rate defects when NO˙ is added to exponential-phase cells, suggesting that their NO˙ sensitivity is specific to stationary- or lag-phase cells (Fig. 4B). In contrast, the ΔsarA mutant behaves similarly to the ΔsrrAB mutant in that it exhibits growth defects specifically in the respiratory phase but replicates at WT rates during the fermentative growth phase (Fig. 4A and B). However, unlike the ΔsrrAB mutant, the ΔsarA mutant resumes acetogenesis at the appropriate time, albeit at reduced levels, but fails to reassimilate lactate during the respiratory phase (Fig. 4C). Accordingly, the ΔsarA mutant exhibits reduced energy charge (ATP levels) primarily during the respiratory phase (Fig. 4D). This reduction in acetogenesis and ATP levels in the ΔsarA mutant is a specific effect of NO˙ exposure since it is not apparent during aerobic growth (see Fig. S3 in the supplemental material). Taken together, these data indicate that SarA is also required for an effective switch from NO˙-resistant fermentative growth to NO˙-resistant respiratory growth, but its role in this process is distinct from that of SrrAB.
sarA mutant exhibits global gene dysregulation following exponential-phase NO˙ exposure.
Because the SarA regulon has not been studied in the context of NO˙ stress, we wanted to identify transcriptional differences in the ΔsarA mutant during NO˙ exposure that might account for its respiratory-phase growth defect. Previous studies of the S. aureus response to NO˙ were performed with very low NO˙ concentrations because the unique degree of S. aureus NO˙ resistance had not yet been appreciated (6, 14). We reasoned that exposure to a higher NO˙ dose, more representative of that encountered during infection, might result in a more robust transcriptional response. Consequently, before examining the ΔsarA mutant, we first wanted to extend our characterization of genes that differentially respond to NO˙ in WT cells. Therefore, we performed RNA-Seq on WT S. aureus COL either grown aerobically or exposed for 1 h to 4 mM DETA-NO, an elevated dose compared to that used in previous studies. NO˙ induced a massive transcriptional response, with 347 genes induced and 322 genes repressed in NO˙-exposed compared to aerobically grown cells (see Data set S2 in the supplemental material). We used qRT-PCR to verify the RNA-Seq results for a subset of these genes (see Fig. S4 in the supplemental material). While many genes found to be induced by NO˙ in a previous study were also upregulated in our study, including much of the SrrAB and Rex regulons (hmp, qox, cyd, scdA, pflAB, ldh1, and ddh), we observed an additional 322 genes induced by NO˙ that were not previously reported (6). Furthermore, whereas the previous study found that approximately 45% of the genes induced by NO˙ belonged to the Fur regulon, we did not observe an appreciable derepression of the Fur regulon following NO˙ exposure. This likely reflects the differences in the NO˙ donors used between the two experiments.
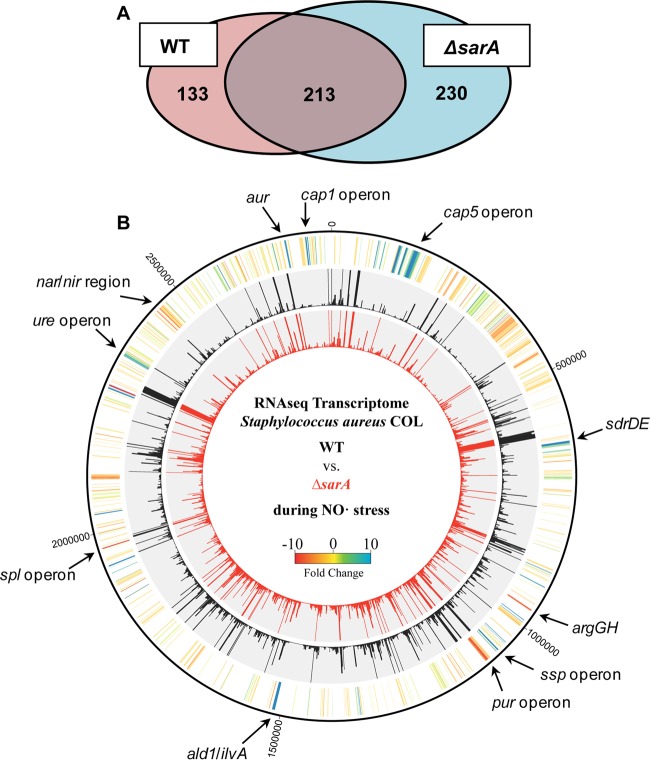
We next performed RNA-Seq on ΔsarA mutant cells exposed to NO˙ and then compared transcriptional response to that of the WT NO˙-exposed cells. Of the 347 genes induced by NO˙ in WT cells, 133 were underexpressed in the ΔsarA mutant NO˙-exposed cells (Fig. 5; see also Data set S3 in the supplemental material). We used STRING version 10 (42), which groups genes based on both physical and functional protein-protein interactions, to distinguish clusters of related genes that are NO˙ inducible in the WT but underexpressed in a ΔsarA mutant. The largest clusters included genes related to nitrate metabolism and biosynthesis of purines, heme, arginine, and histidine (see Fig. S5A in the supplemental material). To investigate whether any of these pathways are essential for S. aureus NO˙ resistance, we made selected mutants in several of these groups (ΔguaAB ΔpbuX Δxpt triple mutant for purine biosynthesis; ΔnirBD mutant, ΔnarGH ΔnirBD double mutant, and ΔnarK mutant for nitrate metabolism; and ΔargGH mutant for arginine biosynthesis) and screened for NO˙ sensitivity (see Fig. S6 in the supplemental material). Individually, only the ΔguaAB ΔpbuX Δxpt mutant exhibited sensitivity to NO˙. However, supplementation with guanine fully rescued the ΔguaAB ΔpbuX Δxpt mutant NO˙ sensitivity but had no effect on the ΔsarA mutant. These results suggest that the inability to fully induce purine biosynthesis, nitrate metabolism, or arginine biosynthesis alone does not explain the NO˙ sensitivity of the ΔsarA mutant.
FIG 5.
The ΔsarA mutant exhibits global gene dysregulation following NO˙ exposure. RNA-Seq was performed on S. aureus COL WT and ΔsarA mutant strains grown in PNG and exposed to 4 mM DETA-NO for 1 h at an OD660 of 0.4. (A) Venn diagram of genes induced by NO˙ in the WT and the ΔsarA mutant. Genes induced in the WT were designated by a >2-fold increase in expression in NO˙-exposed WT cells compared to aerobically grown WT cells. NO˙-exposed WT expression levels were compared to NO˙-exposed ΔsarA mutant expression levels, and genes with >2-fold-lower expression in the ΔsarA mutant were said to be induced in the WT only. Genes with >2-fold-greater expression in NO˙-exposed ΔsarA mutant cells compared to NO˙-exposed WT cells are said to be induced in the ΔsarA mutant only. (B) Genomic map showing transcription profiles of NO˙-exposed WT and ΔsarA mutant strains. The middle (black) circle depicts RPKM values for NO˙-exposed WT cells, and the inner (red) circle depicts RPKM values for the NO˙-exposed ΔsarA mutant. The outer (yellow) circle is a heat map showing fold changes in expression between the WT and the ΔsarA mutant.
We also found that 230 genes were overexpressed in NO˙-exposed ΔsarA cells compared to NO˙-exposed WT cells. STRING analysis identified capsule biosynthesis, ureases, and Na+/H+ transport as the largest functional groups of genes overexpressed in the ΔsarA mutant during NO˙ stress (see Fig. S5B in the supplemental material). Additionally, a gene ontology analysis using GoMiner (43) identified genes in the category of pathogenesis, including proteases (spl, ssp, and aur) and toxins, as the most highly enriched category of genes that are overexpressed in the ΔsarA mutant, consistent with previous ΔsarA mutant transcriptomics studies (60–63).
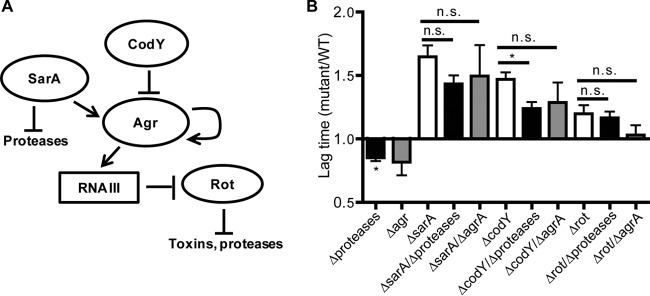
Interestingly, the ΔsarA mutant is not the only NO˙-sensitive mutant we identified that overproduces proteases; the ΔcodY and Δrot mutants have also been characterized as overproducers of at least several of the 10 extracellular proteases secreted by S. aureus (64–69). SarA and Rot directly repress protease transcription, whereas CodY indirectly represses protease transcription by repressing agrA (Fig. 6A). Because of these connections, we wondered whether protease overproduction and/or effects on agr activity might play a role in the NO˙ sensitivity of these mutants. To test this, we obtained a protease-null S. aureus mutant in strain LAC. We constructed codY, rot, and sarA mutations in the protease-null mutant background, as well as in an agrA mutant background, and screened for NO˙ sensitivity. Although abolishing protease production did diminish the lag time of the ΔcodY and ΔsarA mutants under NO˙ stress, it also significantly diminished the lag time of WT LAC under NO˙ stress to the same extent (Fig. 6B). Similarly, mutation of agrA trended toward reducing the lag time of the ΔsarA, ΔcodY, and Δrot mutants but also trended toward a comparably shortened lag for the WT (Fig. 6B). These data suggest that although secreted proteases and agr activity appear to be counterproductive to NO˙ resistance, the NO˙ sensitivity of the ΔsarA, ΔcodY, or Δrot mutant cannot be explained by protease overproduction or regulatory effects on agrA activity.
FIG 6.
Increased protease secretion and/or effects on agr activity do not explain the NO˙ sensitivity of the ΔsarA, ΔcodY, and Δrot mutants. (A) Summary of known regulatory interactions among SarA, CodY, and Rot, protease production, and the agr system of S. aureus. (B) Ratio of mutant to WT lag times of S. aureus LAC mutants grown in LBGT with the addition of 10 mM DETA-NO at the time of inoculation. The lag time (time to OD650 of 0.2) for each NO˙-exposed strain was normalized to its aerobic lag time (n = 7 for the protease-null [Δprotease] background, and n = 3 for the agr background; error bars show SEM). For comparison to the WT, significance was calculated using two-sided Student's t test (*, P ≤ 0.05). For comparisons between strains, significance was determined by one-way ANOVA with a post hoc Student-Newman-Keuls test (*, P ≤ 0.05).
DISCUSSION
S. aureus is unique among pathogens in that it can continue to replicate in the presence of high levels of NO˙. Correspondingly, the transcriptional response of S. aureus to NO˙ is quite distinct from that of other bacteria. However, the complete network of genetic regulation mediating the S. aureus NO˙ response remains poorly understood. In the present study, we further define the regulatory requirements for S. aureus growth during nitrosative stress by identifying five transcription factors required for full NO˙ resistance: SrrAB, Fur, CodY, SarA, and Rot. Our findings elaborate upon the requirement for Fur and SrrAB during NO˙ stress, implicate several known virulence factor regulators in NO˙ resistance, and provide an initial foray into the metabolic and transcriptional roles of the most NO˙-sensitive mutant newly identified in our study, the ΔsarA mutant.
Our results indicate that excess intracellular iron mediates NO˙ toxicity in the Δfur mutant, an effect that is likely attributable to increased Fenton chemistry and elevated ROS generation (Fig. 2). However, the intracellular labile iron pool can also react with NO˙ directly to form dinitrosyl iron complexes (DNICs), which are the foremost cause of protein S-nitrosylation in NO˙-exposed cells (70). DNICs can alter the activities of many metabolic enzymes and have widespread pathophysiological effects. Because chelation of the labile iron pool would reduce both Fenton chemistry and DNIC formation, we cannot distinguish from our results which of these plays a more prominent role in the NO˙ toxicity experienced by the Δfur mutant. Whichever chemistry results in the observed NO˙-sensitive phenotype of the Δfur mutant, both reaction mechanisms are enhanced upon overloading of cytosolic free iron, a situation that results from genetic inactivation of fur. Interestingly, the RNA-Seq results presented in this study differ from those in a previous report in that we did not observe derepression of the Fur regulon upon NO˙ exposure (6). This could be explained by the fact that the previous microarray study used the NO˙ donor S-nitroso-N-acetyl-dl-penicillamine (SNAP). This NO˙ donor favors a transnitrosation reaction in which an NO+ equivalent directly nitrosylates Fur-bound ferrous iron, inactivating the transcription factor and resulting in massive derepression of the Fur regulon. In the present study, we used an authentic NO˙ donor (DETA-NO) less prone to catalyzing this type of chemistry.
This study confirms that the SrrAB two-component system is essential for NO˙ resistance due to its role in the maximal expression of hmp. Additionally, we highlight another major role of SrrAB during NO˙ stress in maximizing terminal oxidase expression to overcome the inhibitory effects of NO˙ on respiration. Respiration is inhibited when NO˙ competitively binds the heme cofactors of terminal oxidases (19). By increasing the total number of terminal oxidases per cell, SrrAB induction of Qox and Cyd would effectively raise the threshold of NO˙ required to wholly inhibit respiration, allowing Hmp to more quickly reduce NO˙ to respiration-permissive levels. Indeed, the SrrAB-dependent induction of both CydAB and QoxABCD contributes to NO˙ resistance in that combining either the ΔcydAB or the ΔqoxABCD mutation with the Δhmp mutation exacerbated NO˙ sensitivity (Fig. 3). The reason that the mutant carrying the ΔqoxABCD mutation alone had a NO˙ phenotype and the ΔcydAB mutant did not likely stems from differences in the expression levels of QoxABCD and CydAB. In fact, although both oxidases are induced by NO˙, the relative transcript abundance of qoxABCD remains >10-fold higher than that of cydAB (see Data set S2 in the supplemental material). Moreover, overexpression of cydAB improves the NO˙ sensitivity of the ΔsrrAB mutant, supporting the idea that induction of either of the respiratory cytochromes is an important function of the SrrAB regulon during NO˙ stress. However, we cannot rule out that differential affinities of the two terminal oxidases for NO˙ may also contribute to the increased importance of Cyd when Hmp induction is lacking.
On the other hand, the failure of qoxABCD overexpression to rescue the ΔsrrAB mutant following NO˙ exposure suggests that other SrrAB-regulated genes, aside from hmp and qoxABCD, are required for the NO˙-resistant growth of S. aureus. In fact, it has been reported that ctaAB, which encodes enzymes required for the biosynthesis of heme A, is SrrAB regulated (23, 59). Given that cytochrome aa3 (Qox) requires heme A for activity but cytochrome bd (Cyd) does not, the loss of ctaB induction in the ΔsrrAB mutant might preclude the restoration of Qox function even with the overexpression of qoxABCD. Thus, although Qox and Cyd are both important for SrrAB-mediated maximization of respiratory flux, other genes within the regulon are also required. Altogether, we have established that the contribution of SrrAB to NO˙ resistance in S. aureus involves the induction of NO˙ detoxification and the maximization of respiratory capacity. Both serve to reestablish respiration, a source of energy generation far superior to substrate-level phosphorylation (via glycolytic lactic acid fermentation), to NO˙-stressed cells.
In addition to SrrAB, we found that the global regulator SarA is required for S. aureus NO˙-resistant metabolism. Like the ΔsrrAB mutant, the ΔsarA mutant exhibits a major defect in transitioning to the respiratory phase of NO˙-resistant growth, as indicated by a reduced growth rate, inability to reassimilate lactate, and reduced ATP levels (Fig. 4). Although the ΔsrrAB and ΔsarA mutants exhibit similar NO˙-sensitive phenotypes, our RNA-Seq results for the ΔsarA mutant and published transcriptomics data for ΔsrrAB mutants suggests that the two do not regulate each other (23, 59). The identification of specific SarA-regulated genes required for NO˙ resistance is complicated by the large effect of SarA on overall transcript abundance following NO˙ stress (>350 genes), a result consistent with SarA functioning as a DNA- and RNA-binding architectural protein rather than a prototypical transcription factor (62, 71). This proposed role for SarA is based on the observations that SarA has an unusually high copy number (50,000), a large number of predicted binding sites in the chromosome (>1,000), the ability to bind mRNA and influence turnover, and the ability to bend DNA to affect topology and promoter accessibility (62, 72–77). It has been suggested that the extensive DNA binding of SarA may play a role in protecting DNA from oxidative damage (76). However, we did not observe increased susceptibility of the ΔsarA mutant to peroxide, which is substantially more damaging to DNA than NO˙ alone (Fig. 2B).
Many of the genes differentially expressed in the ΔsarA mutant following NO˙ exposure include mRNAs previously shown to have differential stability in a ΔsarA mutant during various growth phases (e.g., those stabilized in a sarA-dependent manner, argGH, guaB, purDFHLM, and hemD; and those destabilized in a sarA-dependent manner: ureABCDEFG, cap5MOP, nuc, and sspBC) (61, 62). Interestingly, the NO˙-exposed sarA mutant had moderately lower transcript levels of at least three genes required for full NO˙ resistance (hmp, qoxABCD, and ldh1) (see Data set S3 in the supplemental material). Previous mRNA stability studies showed these transcripts to be among those stabilized in a sarA-dependent manner (61, 62), which might explain why we did not observe reduced gfp transcription from the hmp and ldh1 promoters in the ΔsarA mutant (see Fig. S2 in the supplemental material). Individual overexpression of hmp or qoxABCD in the ΔsarA mutant failed to rescue its respiratory-phase NO˙ sensitivity, suggesting that reduced transcript abundance of either one of these genes does not alone explain the ΔsarA genotype (data not shown). However, this does not rule out the possibility that a collective destabilization of multiple transcripts required for NO˙ resistance plays a role in the NO˙ sensitivity of the ΔsarA mutant.
Alternatively, given the central role of SarA in modulating both DNA topology and RNA stability, the requirement for SarA during NO˙ stress may be for global transcriptional homeostasis rather than specific regulation of a subset of genes during NO˙ stress. Interestingly, modulation of RNA stability is an important component of the bacterial stress response in many species; it provides a posttranscriptional mechanism for influencing protein production and, potentially, for removal of damaged RNAs (78–81). Given that S. aureus modulates mRNA stability in response to stressful conditions, including temperature and pH shock, it is possible that the modulation of mRNA turnover is a key response to NO˙ stress (82, 83). Regardless, due to the potentially global function of SarA in S. aureus, we expect that further high-throughput methods, such as genetic interaction mapping (e.g., performing transposon sequencing [Tn-Seq] in a sarA mutant background), may be necessary to more specifically define the requirement for SarA during NO˙ stress.
Notably, both our RNA-Seq data and previously published microarray data indicate that genes belonging to the CodY regulon are further repressed during NO˙ stress (ilvABCDE and dapABD), reinforcing the importance of CodY when NO˙ is present (6). The reasons for the importance of CodY and Rot during NO˙ stress remain a subject of ongoing investigation in our lab. While eliminating protease production did not rescue the NO˙ sensitivity of the ΔcodY, Δrot, or ΔsarA mutant, the observation that the deletion of proteases generally improved the NO˙ resistance of all strains, including the WT, is intriguing. Interestingly, protease deletion did not affect the sensitivity of cells exposed to NO˙ during early exponential phase, indicating that the negative effect of proteases on NO˙ susceptibility is specific to late-stationary-/lag-phase cells (data not shown). The deletion of proteases may increase the capacity of stationary-phase S. aureus cells to aggregate via cell-surface proteins and thus might provide enhanced physical protection when these aggregated cells are used to inoculate cultures in the presence of NO˙ (84, 85).
Collectively, the results of our study provide a broad characterization of the regulatory requirements for NO˙ resistance in S. aureus. The five regulators identified as essential for NO˙ resistance, Fur, SrrAB, SarA, CodY, and Rot, are all pleiotropic transcription factors that modulate virulence and appropriate adaptation to the extremely diverse ambient conditions inside a host, of which NO˙ is a major component. These regulators do not exist solely or even primarily to respond to NO˙, however, suggesting that NO˙ resistance is extremely multifaceted and deeply integrated into complex networks of virulence and metabolism regulation in S. aureus. These findings advance our understanding of a highly unique adaptation that contributes to S. aureus pathogenesis, and they suggest that NO˙ resistance likely evolved alongside and as a component of S. aureus pathogenicity rather than as a separate independent trait.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NIH grant from the Institute of Allergy and Infectious Diseases (grant 5-R01-AI093613), a Pew Biomedical Scholars award (grant A12-0105), and an American Heart Association predoctoral fellowship (grant 13PRE19830003).
Funding Statement
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00229-16.
REFERENCES
- 1.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core Surveillance (ABCs) MRSA Investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Fowler VG, Miró JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey GR, Spelman D, Bradley SF, Barsic B, Pappas PA, Anstrom KJ, Wray D, Fortes CQ, Anguera I, Athan E, Jones P, van der Meer JTM, Elliott TSJ, Levine DP, Bayer AS, ICE Investigators. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 3.Moet GJ, Jones RN, Biedenbach DJ, Stilwell MG, Fritsche TR. 2007. Contemporary causes of skin and soft tissue infections in North America, Latin America, and Europe: report from the SENTRY Antimicrobial Surveillance Program (1998–2004). Diagn Microbiol Infect Dis 57:7–13. doi: 10.1016/j.diagmicrobio.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Diekema DJ, Pfaller MA, Jones RN, Doern GV, Kugler KC, Beach ML, Sader HS. 2000. Trends in antimicrobial susceptibility of bacterial pathogens isolated from patients with bloodstream infections in the USA, Canada and Latin America. Int J Antimicrob Agents 13:257–271. doi: 10.1016/S0924-8579(99)00131-4. [DOI] [PubMed] [Google Scholar]
- 5.Jensen AG, Espersen F, Skinhøj P, Rosdahl VT, Frimodt-Møller N. 1997. Increasing frequency of vertebral osteomyelitis following Staphylococcus aureus bacteraemia in Denmark 1980–1990. J Infection 34:113–118. doi: 10.1016/S0163-4453(97)92395-1. [DOI] [PubMed] [Google Scholar]
- 6.Richardson AR, Dunman PM, Fang FC. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol 61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 7.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler J, Breitbach K, Renner C, Heitsch A-K, Bast A, van Rooijen N, Vogelgesang S, Steinmetz I. 2011. NADPH-oxidase but not inducible nitric oxide synthase contributes to resistance in a murine Staphylococcus aureus Newman pneumonia model. Microbes Infect 13:914–922. doi: 10.1016/j.micinf.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Wink DA, Mitchell JB. 1998. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med 25:434–456. doi: 10.1016/S0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 10.Schopfer F, Baker PR, Freeman BA. 2003. NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem Sci 28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Radi R, Beckman JS, Bush KM, Freeman BA. 1991. Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide. Arch Biochem Biophys 288:481–487. doi: 10.1016/0003-9861(91)90224-7. [DOI] [PubMed] [Google Scholar]
- 12.Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest 99:2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS. 1991. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 14.Hochgrafe F, Wolf C, Fuchs S, Liebeke M, Lalk M, Engelmann S, Hecker M. 2008. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 190:4997–5008. doi: 10.1128/JB.01846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A 101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schapiro JM, Libby SJ, Fang FC. 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc Natl Acad Sci U S A 100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunoshiba T, DeRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci U S A 90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore CM, Nakano MM, Wang T, Ye RW, Helmann JD. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J Bacteriol 186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 20.Fuller JR, Vitko NP, Perkowski EF, Scott E, Khatri D, Spontak JS, Thurlow LR, Richardson AR. 2011. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front Cell Infect Microbiol 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagels M, Fuchs S, Pané-Farré J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, Wachenfeldt von Liebeke CM, Lalk M, Sander G, Eiff von Proctor CRA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 76:1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MKR. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. doi: 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- 23.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4(6):e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano MM. 2002. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J Bacteriol 184:1783–1787. doi: 10.1128/JB.184.6.1783-1787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong A, Singh VK, Cabrera G, Jayaswal RK. 2000. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 146:659–668. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh MJ, Ingham E, Foster SJ. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Autreaux B, Touati D, Bersch B, Latour J-M, Michaud-Soret I. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci U S A 99:16619–16624. doi: 10.1073/pnas.252591299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu W, Zhou Y, Shao C, Sun Y, Zhang Q, Chen C, Jia J. 2009. Helicobacter pylori proteins response to nitric oxide stress. J Microbiol 47:486–493. doi: 10.1007/s12275-008-0266-0. [DOI] [PubMed] [Google Scholar]
- 29.Yuhara S, Komatsu H, Goto H, Ohtsubo Y, Nagata Y, Tsuda M. 2008. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 154:1763–1774. doi: 10.1099/mic.0.2007/015537-0. [DOI] [PubMed] [Google Scholar]
- 30.Husain M, Jones-Carson J, Liu L, Song M, Saah JR, Troxell B, Mendoza M, Hassan H, Vazquez-Torres A. 2014. Ferric uptake regulator-dependent antinitrosative defenses in Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun 82:333–340. doi: 10.1128/IAI.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, Hutchings MI. 2008. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker NP, Le Brun NE, Dixon R, Hutchings MI. 2010. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol 18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Crooke AK, Fuller JR, Obrist MW, Tomkovich SE, Vitko NP, Richardson AR. 2013. CcpA-independent glucose regulation of lactate dehydrogenase 1 in Staphylococcus aureus. PLoS One 8:e54293. doi: 10.1371/journal.pone.0054293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wormann ME, Reichmann NT, Malone CL, Horswill AR, Grundling A. 2011. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol 193:5279–5291. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan PF, Foster SJ. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469–2479. [DOI] [PubMed] [Google Scholar]
- 37.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll RK, Weiss A, Shaw LN. 2015. RNA-sequencing of Staphylococcus aureus messenger RNA. Methods Mol Biol 1373:131–141. [DOI] [PubMed] [Google Scholar]
- 40.Weiss A, Ibarra JA, Paoletti J, Carroll RK, Shaw LN. 2014. The δ subunit of RNA polymerase guides promoter selectivity and virulence in Staphylococcus aureus. Infect Immun 82:1424–1435. doi: 10.1128/IAI.01508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN. 2005. High-throughput GoMiner, an “industrial-strength” integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of common variable immune deficiency (CVID). BMC Bioinformatics 6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibarra JA, Pérez-Rueda E, Carroll RK, Shaw LN. 2013. Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14:126–126. doi: 10.1186/1471-2164-14-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravcheev DA, Best AA, Tintle N, Dejongh M, Osterman AL, Novichkov PS, Rodionov DA. 2011. Inference of the transcriptional regulatory network in Staphylococcus aureus by integration of experimental and genomics-based evidence. J Bacteriol 193:3228–3240. doi: 10.1128/JB.00350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiba J, Hartmann T, Cluzel M-E, Cohen-Gonsaud M, Delolme F, Bischoff M, Molle V. 2012. A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J Biol Chem 287:43607–43619. doi: 10.1074/jbc.M112.418913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cluzel M-E, Zanella-Cléon I, Cozzone AJ, Fütterer K, Duclos B, Molle V. 2010. The Staphylococcus aureus autoinducer-2 synthase LuxS is regulated by Ser/Thr phosphorylation. J Bacteriol 192:6295–6301. doi: 10.1128/JB.00853-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun F, Ding Y, Ji Q, Liang Z, Deng X, Wong CCL, Yi C, Zhang L, Xie S, Alvarez S, Hicks LM, Luo C, Jiang H, Lan L, He C. 2012. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci U S A 109:15461–15466. doi: 10.1073/pnas.1205952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacelli R, Wink DA, Cook JA, Krishna MC, DeGraff W, Friedman N, Tsokos M, Samuni A, Mitchell JB. 1995. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J Exp Med 182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodmansee AN, Imlay JA. 2003. A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol Microbiol 49:11–22. doi: 10.1046/j.1365-2958.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- 51.Woodmansee AN, Imlay JA. 2002. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J Biol Chem 277:34055–34066. doi: 10.1074/jbc.M203977200. [DOI] [PubMed] [Google Scholar]
- 52.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 53.Messner KR, Imlay JA. 1999. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem 274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 54.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol 177:2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 58.Gonçalves VL, Nobre LS, Vicente JB, Teixeira M, Saraiva LM. 2006. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett 580:1817–1821. doi: 10.1016/j.febslet.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 59.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, Wasserman GA, Chen J, Shopsin B, Gilmore MS, Skaar EP, Cassat JE. 2015. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, Hurlburt B, Smeltzer M, Overbeek R, Disz T, Dunman PM. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J Bacteriol 188:2593–2603. doi: 10.1128/JB.188.7.2593-2603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrison JM, Anderson KL, Beenken KE, Smeltzer MS, Dunman PM. 2012. The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front Cell Infect Microbiol 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang S-J, Rice KC, Bayles KW, Smeltzer MS. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 64.Mootz JM, Benson MA, Heim CE, Crosby HA, Kavanaugh JS, Dunman PM, Kielian T, Torres VJ, Horswill AR. 2015. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol Microbiol 96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One 3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, Gupta RK, Lee CY, Beenken KE, Smeltzer MS. 2015. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 4:436–451. doi: 10.1002/mbo3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bosworth CA, Toledo JC, Zmijewski JW, Li Q, Lancaster JR Jr. 2009. Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci U S A 106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schumacher MA, Hurlburt BK, Brennan RG. 2001. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature 409:215–219. doi: 10.1038/35051623. [DOI] [PubMed] [Google Scholar]
- 72.Sterba KM, Mackintosh SG, Blevins JS, Hurlburt BK, Smeltzer MS. 2003. Characterization of Staphylococcus aureus SarA binding sites. J Bacteriol 185:4410–4417. doi: 10.1128/JB.185.15.4410-4417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morfeldt E, Tegmark K, Arvidson S. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol 21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Manna AC, Pan C-H, Kriksunov IA, Thiel DJ, Cheung AL, Zhang G. 2006. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc Natl Acad Sci U S A 103:2392–2397. doi: 10.1073/pnas.0510439103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujimoto DF, Brunskill EW, Bayles KW. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J Bacteriol 182:4822–4828. doi: 10.1128/JB.182.17.4822-4828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, Smeltzer MS, Hurlburt BK. 2009. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol 74:1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. 2011. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol 193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen C, Deutscher MP. 2005. Elevation of RNase R in response to multiple stress conditions. J Biol Chem 280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 79.Cheng ZF, Deutscher MP. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci U S A 100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morrison JM, Dunman PM. 2011. The modulation of Staphylococcus aureus mRNA turnover. Future Microbiol 6:1141–1150. doi: 10.2217/fmb.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson KL, Dunman PM. 2009. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int J Microbiol 2009:5254911–5254915. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anderson KL, Roux CM, Olson MW, Luong TT, Lee CY, Olson R, Dunman PM. 2010. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol Med Microbiol 60:208–250. doi: 10.1111/j.1574-695X.2010.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. 2012. PLoS One: planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS One 7:e41075. doi: 10.1371/journal.pone.0041075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.