Specific tRNA-derived motifs present in an endogenous dicistronic transcript mediate mRNA transport along the phloem.
Abstract
In plants, protein-coding mRNAs can move via the phloem vasculature to distant tissues, where they may act as non-cell-autonomous signals. Emerging work has identified many phloem-mobile mRNAs, but little is known regarding RNA motifs triggering mobility, the extent of mRNA transport, and the potential of transported mRNAs to be translated into functional proteins after transport. To address these aspects, we produced reporter transcripts harboring tRNA-like structures (TLSs) that were found to be enriched in the phloem stream and in mRNAs moving over chimeric graft junctions. Phenotypic and enzymatic assays on grafted plants indicated that mRNAs harboring a distinctive TLS can move from transgenic roots into wild-type leaves and from transgenic leaves into wild-type flowers or roots; these mRNAs can also be translated into proteins after transport. In addition, we provide evidence that dicistronic mRNA:tRNA transcripts are frequently produced in Arabidopsis thaliana and are enriched in the population of graft-mobile mRNAs. Our results suggest that tRNA-derived sequences with predicted stem-bulge-stem-loop structures are sufficient to mediate mRNA transport and seem to be necessary for the mobility of a large number of endogenous transcripts that can move through graft junctions.
INTRODUCTION
In plants, small interfering RNAs (siRNAs), microRNAs (miRNAs), and mRNAs can move locally from cell to cell via plasmodesmata and can also move over long distances by entering the phloem vasculature. The mobile siRNAs and miRNAs regulate gene expression, affect target mRNAs, and mediate antiviral defense (Ruiz-Medrano et al., 2004; Lough and Lucas, 2006; Kalantidis et al., 2008; Molnar et al., 2010; Melnyk et al., 2011). Distinct mRNAs such as the homeodomain protein-encoding transcripts of potato (Solanum tuberosum) BEL5 and maize (Zea mays) knotted1 also move to other tissues and trigger developmental decisions in targeted cells (Kim et al., 2001; Banerjee et al., 2006).
The molecular mechanisms enabling intercellular mRNA transport and the fate of transported mRNAs in target tissues remain poorly understood. On the one hand, conserved and, thus, predictive mRNA motifs have not been described for known graft-mobile mRNA populations (Guo et al., 2013; Thieme et al., 2015; Yang et al., 2015). On the other hand, recent work in potato showed that the 3′ untranslated region (UTR) of the phloem-mobile transcript BEL5 supports mRNA stability and trafficking into roots, where BEL5 protein initiates tuber formation (Banerjee et al., 2009; Cho et al., 2015).
Viral RNAs can move via the phloem stream in the absence of viral proteins, suggesting that endogenous cellular factors recognize a structural RNA motif and mediate long-distance transport through the phloem (Gopinath and Kao, 2007). Nonconserved viral 3′ UTR sequences, which interact with 5′ UTRs, seem to play a role in facilitating viral RNA cell-to-cell transfer (Lough et al., 2006). Similarly, viroids (infectious, non-protein-coding small RNAs) form specific stem-loop structures not yet identified in other mobile RNAs, allowing them to enter the plant phloem long-distance transport system (Ding, 2009; Takeda et al., 2011).
Many positive-strand RNA viruses harbor conserved stem-loop structures in the 3′ UTR resembling those of canonical tRNAs. Such viral tRNA-like structures (TLSs) seem to play a crucial role in viral replication and infectivity (Dreher et al., 1989; Fechter et al., 2001; Barends et al., 2004). The TLSs are aminoacylated; therefore, the viral clover-like tRNA structures are likely recognized by plant tRNA binding and modifying proteins (Dreher, 2010). Also, viral TLSs recognized by the ribosomal elongation factor eEF1A_GTP form a stable RNA-protein complex repressing viral RNA minus-strand synthesis (Matsuda et al., 2004).
Viral TLS-mediated intercellular or long-distance transport of viral RNAs remains to be shown, but support for the notion that tRNA-related structures might be bona fide RNA mobility motifs for endogenous transcripts was found in the noncoding RNA population of phloem exudate from pumpkin (Cucurbita pepo) (Zhang et al., 2009). In pumpkin phloem exudate, specific subsets of tRNAs are enriched suggesting selective tRNA import from surrounding tissues into conducting phloem vessels. Taken together, the presence of specific tRNAs in the phloem stream, the role of viral TLS, and frequent occurrences of dicistronic poly(A)-mRNA:tRNA transcripts point toward a potential function of tRNA-related sequences in triggering mobility of transcripts. Here, we present evidence that the TLS can trigger mobility of otherwise nonmobile mRNAs and that TLS are significantly enriched in the mobile mRNA populations found in Arabidopsis thaliana.
RESULTS
tRNAMet Fusion Transcripts Move into Flowers
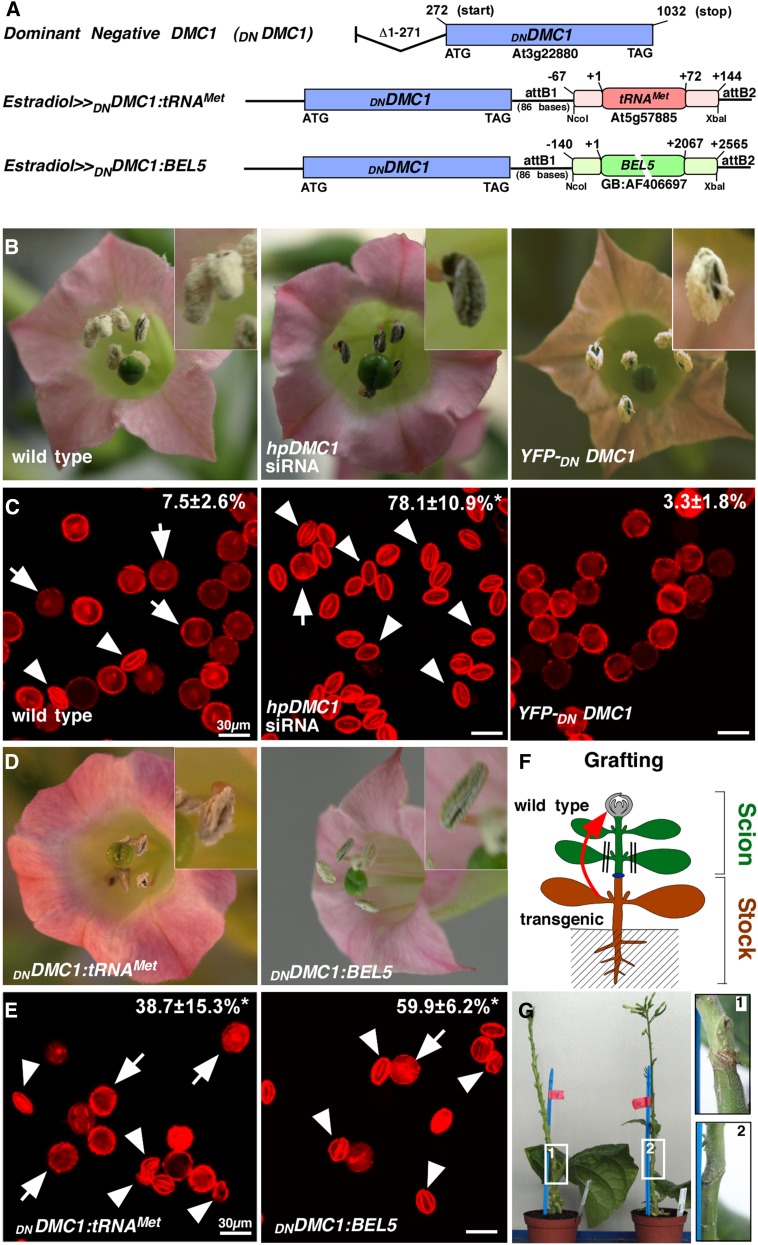
To establish a simple phenotypic scoring system for mRNAs harboring predicted mobility motifs such as TLSs, we used a dominant-negative variant of Arabidopsis DISRUPTION OF MEIOTIC CONTROL1 (DNDMC1) that lacks the N-terminal 92 amino acid residues (Figure 1A) and therefore interferes with the progression of meiosis (Habu et al., 1996). DMC1 is a specific meiotic cell cycle factor and a member of the highly conserved RecA-type recombinase family of DNA-dependent ATPases active during meiosis in sporogenic cells (Doutriaux et al., 1998). Lack of a functional DMC1/RAD51 complex induces achiasmatic meiosis resulting in the formation of anomalously shaped pollen containing an aberrant number of chromosomes and, consequently, is necessary for proper pollen development (Bishop et al., 1992; Zhang et al., 2014). Thus, production of misshaped pollen in anthers and decreased fertility indicate the presence of either DMC1 siRNA (Zhang et al., 2014) (Figures 1B and 1C) or the product of translation of the dominant-negative DNDMC1 mRNA. To implement a reporter system for mRNA mobility, we produced transgenic tobacco (Nicotiana tabacum) lines expressing YFP-DNDMC1 fusion proteins as a fluorescent reporter (Figures 1B and 1C) to test for DMC1 silencing (Zhang et al., 2014) potentially induced by the transgenic DNDMC1 constructs. We also generated lines expressing DNDMC1 mRNA fused to the full-length potato BEL5 transcript, which is known to be mobile (Cho et al., 2015) (DNDMC1:BEL5) as a positive control (Figure 1A). As a negative control, we made lines expressing DNDMC1 mRNA 5′ fused to the vegetative tobacco growth regulator CENTRORADIALIS-like 2 (Amaya et al., 1999; CET2:DNDMC1) (Supplemental Figure 1). Finally, since tRNAMet was detected in the phloem sap of pumpkin (Zhang et al., 2009), we made lines expressing DNDMC1 mRNA fused to full-length tRNAMet (AT5G57885; DNDMC1:tRNAMet; tRNAMet:DNDMC1) (Figure 1A).
Figure 1.
Dominant-Negative DMC1 as a Reporter Construct Causes Male-Sterile Flowers.
(A) Schematic drawing of the DNDMC1 RNA fusion constructs used. Arabidopsis DNDMC1 codes for a truncated protein lacking the N-terminal 92 amino acids and dominantly interferes with meiosis resulting in misshaped pollen and partial male sterility. The DNDMC1 coding sequence was fused to graft-mobile potato BEL5 sequences or phloem tRNAMet at the 3′ UTR to evaluate their potential to trigger DNDMC mRNA transport over graft junctions.
(B) to (E) Fertile anthers of wild-type tobacco plants show regular pollen production with minimal abnormally shaped pollen (2 to 3%), whereas hpDMC1 siRNA transgenic tobacco plants produce high numbers of abnormally shaped pollen and are sterile as previously described (Zhang et al., 2014). YFP-DNDMC1 transgenic plants have normal pollen production similar to the wild type because the N-terminal YFP fusion abolishes the dominant-negative effect of truncated DMC1. Transgenic plants expressing DNDMC1 fused with tRNAMet or BEL5 at the 3′ UTR exhibit increased male sterility.
(C) and (E) Propidium iodide-stained pollen harvested from transgenic plants were imaged by confocal laser scanning microscopy and evaluated by an automatic imaging analysis algorithm to count abnormally shaped pollen (Zhang et al., 2014) indicated by percentage numbers. Arrows indicate normal pollen; arrowheads indicate abnormally shaped pollen. Bars = 30 µm.
(F) and (G) Scheme of performed stem grafts to evaluate transport of mRNA to wild-type flowers.
Independent transgenic plants expressing DNDMC1 mRNA fusion constructs were verified to show a pollen sterility phenotype (Figures 1D and 1E) and used in grafting experiments (Figures 1F and 1G) to evaluate transcript mobility from transgenic source tissue to wild-type flowers. Transgenic lines expressing the YFP-DNDMC1 fusion did not exhibit a dominant-negative effect on endogenous tobacco DMC1 (Figure 1B); also, the fusion transcript was not graft-mobile (Supplemental Figure 1). Thus, these plants could be used to evaluate grafted DNDMC1 transgenic plants for their potential production of mobile siRNAs targeting endogenous DMC1 in wild-type tobacco flowers, triggering sterility (Zhang et al., 2009).
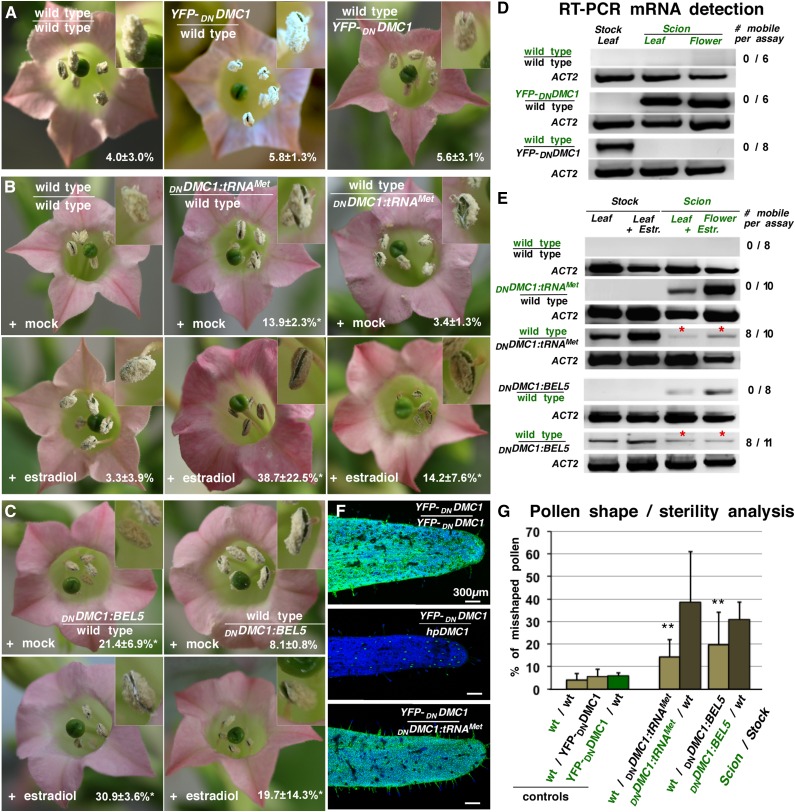
We first confirmed by RT-PCR that DNDMC1 mRNA does not contain the sequences triggering mobility (Supplemental Figure 1) and thus is suitable as a reporter for transcript mobility, producing a pollen phenotype (Figure 2A; Supplemental Figure 1). We next addressed the mobility of the fusion transcripts by grafting DNDMC1:BEL5, DNDMC1:tRNAMet, tRNAMet:DNDMC1, or tobacco CET2:DNDMC1 transgenic plants with wild-type plants and examined pollen sterility and presence of the fusion transcript in the wild-type flowers (Figures 2B to 2G; Supplemental Figure 1). As expected, after induction with estradiol, the DNDMC1:BEL5 and CET2:DNDMC1 scions grafted onto wild-type stocks showed a significantly higher percentage of aberrant pollen formation in their flowers (30.9% ± 7.6% and 51.9% ± 7.8%, respectively) than grafted wild-type plants (4.0% ± 3.0%; Figure 2C; Supplemental Figure 1 and Supplemental Table 1). Confirming previous reports that BEL5 fusion transcripts are mobile (Banerjee et al., 2006), wild-type plants grafted onto DNDMC1:BEL5 stocks produced a significantly higher number of misshapen pollen (19.7% ± 14.3%) than wild-type controls and the presence of fusion transcript was confirmed by RT-PCR in closed wild-type flowers (Figure 2E). Confirming that the DNDMC1 RNA itself does not trigger mobility and that DNDMC1 protein itself is not mobile grafted, CET2:DNDMC1 stock plants did not significantly induce aberrant pollen formation in wild-type flowers (7.8 ± 4.9; Supplemental Figure 1). Thus, DNDMC1 - RNA fusion constructs can be employed as a RNA mobility reporter system by producing a quantifiable pollen phenotype.
Figure 2.
DNDMC1 Fusion Transcript Transport Induces Aberrant Pollen Formation.
(A) Flowers of grafted wild-type stock plants supported by 35Spro:YFP-DNDMC1 transgenic scions and reciprocal grafts are fertile.
(B) Upper panel: Grafted wild-type/wild-type or wild-type/DNDMC1:tRNAMet plants showed normal pollen production when mock treated. Lower panel: Estradiol-induced expression of DNDMC1:tRNAMet in scion or stock plant parts resulted in partially sterile anthers in both transgenic and wild-type flowers. The latter suggests DNDMC1:tRNAMet mRNA transport and expression of the truncated DMC1 protein in wild-type male meiocytes.
(C) Flowers of grafted DNDMC1:BEL5 transgenic plants. Upper panel: Mock-treated wild-type/estradiol>>DNDMC1:BEL5 grafts showed weak male sterility. Lower panel: Flowers of grafted plants treated with estradiol exhibit partial male sterility.
(D) RT-PCR assays on RNAs samples from grafted wild-type tissues revealed that the YFP-DNDMC1 control transcript is not allocated over graft junctions into wild-type stock leaves (n = 6) or scion flowers (n = 8). ACTIN2 (ACT2) specific RT-PCR was used as a positive control.
(E) RT-PCR assays on RNA samples from grafted plants. DNDMC1:tRNAMet and DNDMC1:BEL5 is detected in transgenic and in wild-type scion flowers. Appearance of a specific PCR product in samples from grafted wild-type stock leaves and wild-type flowers (red asterisks) suggests mobility of the DNDMC1:tRNAMet fusion transcript. Number of tested grafted plants is shown on the right.
(F) CLSM images of sepals formed on YFP-DNDMC1 producing scions. Upper panel: YFP-DNDMC1/YFP-DNDMC1 control graft with expected high green fluorescence emitted by YFP-DNDMC1. Middle panel: Control graft with siRNA-producing stock plants (hpDMC1) with expected low YFP fluorescence and distribution in YFP-DNDMC1 flowers (Zhang et al., 2014). Lower panel: YFP-DNDMC1 scion grafted onto DNDMC1:tRNAMet transgenic stock shows similar YFP fluorescence levels as YFP-DNDMC1/YFP-DNDMC1 control grafts. Note that YFP-DNDMC1 fusion protein is detected in all epidermal leaf cells except when grafted with hpDMC1 producing DMC1 siRNA lines. Green, presence of YFP-DNDMC1; blue, plastid autofluorescence. Bar = 300 µm.
(G) Statistical analysis of data from automated detection of misshaped pollen appearing on grafted plants. Misshapen pollen formation was significantly higher on wild-type scions supported by DNDMC1:tRNAMet and DNDMC1:BEL5 stock plants than in control grafts. Asterisks indicate highly significant differences against controls using χ2 test for independence of variables in a contingency table. Biological replicates: n > 8. Error bars indicate sd. For details, see Supplemental Table 1.
Next, to learn whether a phloem-allocated tRNA contains the necessary structural information mediating mRNA movement over long distances, we grafted transgenic plants expressing the 3′ UTR DNDMC1:tRNAMet (Figure 2B) or the 5′ UTR tRNAMet:DNDMC1 (Supplemental Figure 1) fusion construct. Expression was induced by applying estradiol to the transgenic source leaves (stock) or transgenic stem (scion) ∼1.5 weeks after grafting and prior to flower induction. Estradiol-treated grafted plants formed a significantly higher number of misshapen pollen (14.2% ± 7.6%; Supplemental Table 1) compared with control grafts and wild-type plants (Figures 2A and 2B), and RT-PCR assays confirmed the presence of DNDMC1:tRNAMet and tRNAMet:DNDMC1 poly(A) transcripts in wild-type flowers formed on transgenic stock plants (Figure 2E; Supplemental Figure 1).
To exclude the possibility that the grafted chimeric plants produce a mobile DMC1 siRNA that moves into wild-type flower tissues and silences the endogenous DMC1, triggering a pollen sterility phenotype (Zhang et al., 2014), we grafted the DNDMC1:tRNAMet plants with the YFP-DNDMC1 fusion line, which expresses a reporter protein that can be easily detected by fluorescence microscopy. In contrast to the DMC1 siRNA control lines, no systemic siRNA-mediated silencing of the YFP-DNDMC1 reporter construct could be detected in sepals (Figure 2F). Thus, the DNDMC1:tRNAMet fusion transcript does not induce systemic silencing, and the observed defects in pollen formation in grafted plants (Figure 2G) can be attributed to the systemic delivery of the DNDMC1 fusion transcripts. In summary, the presence of the full-length tRNAMet sequence in the 5′ or 3′ UTR triggers transport of the DNDMC1 poly(A) transcript from stock to source leaves and into sporogenic tissues, where it is apparently translated, as it interferes with meiosis in male tissues.
tRNAs Harbor a Signal for Systemic mRNA Movement
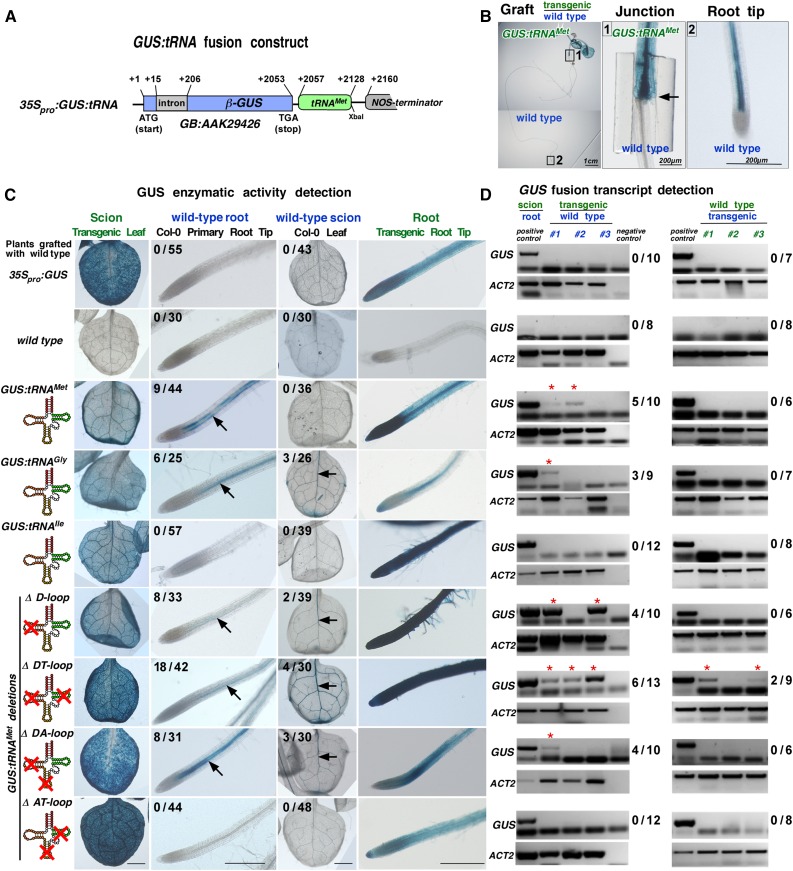
To evaluate whether particular tRNA sequences related to the viral TLS mediate systemic mRNA movement, we used the core sequences of the two phloem-imported tRNAs, tRNAMet (anticodon CAT; 72 bases; TAIR No. AT5G57885) and tRNAGly (anticodon CGG; 74 bases; TAIR No. AT1G71700), and the non-phloem-imported tRNAIle (73 bases; TAIR No. AT3G05835) (Zhang et al., 2009). tRNAGly is also present in the 3′ UTR of the graft-mobile Arabidopsis CHOLINE KINASE1 (CK1) transcript (TAIR No. AT1G71697; Thieme et al., 2015). We fused these three tRNA sequences to the 3′ UTR of the cell-autonomous β-GUS mRNA sequence (Figure 3A; Supplemental Figure 2). To evaluate the mobility of the fusion transcripts, Arabidopsis Col-0 lines expressing 35Spro:GUS or 35Spro:GUS:tRNA fusion constructs were produced and hypocotyl-grafted with Col-0 wild type (shoot or root). Two weeks after grafting, GUS enzyme activity was visualized in situ (Figures 3B and 3C; Supplemental Figure 3). Control grafts with transgenic 35Spro:GUS plants lacking the tRNA sequences in the 3′ UTR showed no GUS activity and GUS mRNA presence in distal wild-type root (n = 0/55 grafts) or leaf (n = 0/43 grafts) tissues, indicating that neither the GUS mRNA nor the GUS protein moves over graft junctions (Figures 3C and 3D). However, GUS activity was detected in phloem-associated cells in wild-type roots after hypocotyl grafting with transgenic scion plants expressing GUS:tRNAMet (n = 9/44 grafts) or GUS:tRNAGly (n = 6/25 grafts). No GUS activity was observed in wild-type roots grafted with plants expressing GUS:tRNAIle (n = 0/57 grafts). RT-PCR assays confirmed the presence of GUS:tRNAMet and GUS:tRNAGly and the absence of GUS:tRNAIle transcripts in wild-type roots after grafting (Figure 3D). Notably, the reverse grafts with transgenic roots and wild-type scions indicate that the shoot-to-root mobile GUS:tRNAMet fusion transcript does not move from root to shoot (n = 0/36 grafts) and that GUS:tRNAGly barely moves from root to shoot (n = 3/26 grafts).
Figure 3.
GUS:tRNA Fusion Transcripts and Mobility in Grafted Arabidopsis.
(A) Schematic drawing of used 35Spro:GUS:tRNA fusion constructs (for sequences of tRNAMet, tRNAGly, tRNAIle, and tRNAMet deletions, see Supplemental Figure 2).
(B) Example of a hypocotyl grafted GUS:tRNAMet/wild-type (Col-0) plant. Blue color indicates presence of GUS activity in the hypocotyl above the graft junction (arrow) and in the wild-type root tip.
(C) GUS activity in leaves and primary root tips detected in GUS:tRNA/wild-type grafts. The numbers indicate the fraction of GUS staining detected in the wild-type root tips or wild-type leaf vasculature (arrows) of plants grafted with the indicated transgenic line. At least three independent transgenic lines were used for each graft combination (for details, see Supplemental Data Set 3, and for additional images of grafted plants, see Supplemental Figure 3).
(D) RT-PCR on poly(A)-RNA samples harvested from grafted plants. Three samples from three to five grafted plants were pooled and tested for presence of GUS transcripts in wild-type tissue (asterisks). Numbers indicate occurrence of GUS poly(A) RNA in the tested wild-type root or wild-type leaves RNA samples. ACT2-specific RT-PCR was used as a positive control confirming mRNA presence in the samples.
To learn whether the whole tRNA sequence or a subsequence is sufficient to mediate mRNA mobility, we used tRNAMet deletion constructs lacking the assigned dihydrouridine (D), anticodon (A), or TψC (T) arm/loop structures and combinations thereof (Figure 3C). Again, plants expressing these GUS mRNA fusion constructs were grafted with the wild type and then tested for GUS activity and presence of the fusion transcript. As indicated by GUS and RT-PCR assays, the ΔD, ΔDT, and ΔDA, but not the ΔAT tRNAMet deletion construct, were sufficient to mediate GUS transport into wild-type roots and, with a very low frequency, to scion leaves (Figures 3C and 3D). Presence of the GUS:ΔDtRNAMet transcript and translation in phloem-associated cells of wild-type roots and leaves suggests that only part of the tRNAMet sequence is required to trigger mobility. This also indicates that A and TψC hairpin-loop sequences have redundant roles in triggering mRNA transport as only deletions of both the Α and TψC hairpin-loop sequences eliminated mobility of the GUS fusion transcript.
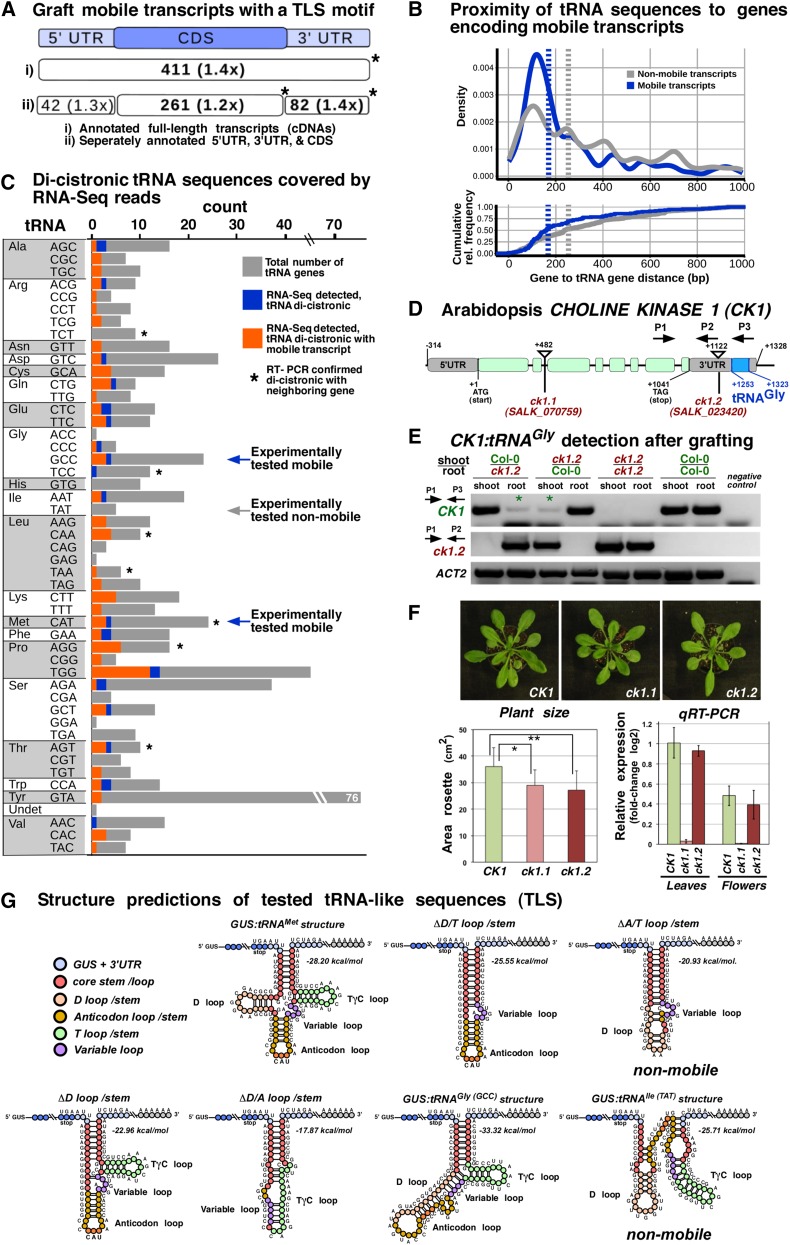
To elucidate whether tRNA sequences or sequences related to viral TLS motifs confer mRNA mobility, we first evaluated whether the endogenous mobile mRNA population found in Arabidopsis is enriched for TLS motifs. We screened the Arabidopsis graft-mobile transcriptome database (n = 3606) (Thieme et al., 2015) for the presence of TLS motifs in the mRNA UTRs and coding sequences (CDSs) (Figure 4A). We performed scans for sequence-independent structure motifs using the established consensus tRNA descriptor (Macke et al., 2001) recognizing the stem-loop arrangements found in most tRNAs (Supplemental Figure 4). The analysis revealed that a significant number of mobile Arabidopsis transcripts (Thieme et al., 2015) (11.4%; n = 411 of 3606) or grapevine (Vitis spp) transcripts (Yang et al., 2015) (7.5%; n = 249 of 3333) harbor a TLS motif in the CDS or 3′ UTR (Figure 4A; Supplemental Data Set 1). Furthermore, annotated tRNA genes were found in closer proximity to genes encoding mobile transcripts than to genes encoding nonmobile transcripts (P < 0.0003; Cohen's D d = 0.313; see Methods). Independent of DNA strand assignment, of all 1125 genes flanked by a tRNA gene, 158 produced mobile RNAs, and of these, 113 are located within 1000 bp of the tRNA gene (34.7% enrichment for mobile transcripts, P = 0.005, Fisher’s exact test; Figure 4B).
Figure 4.
Mobile Arabidopsis mRNAs and Occurrence of tRNA-Like Motifs.
(A) Number of all identified mobile transcripts (n = 3606) with predicted tRNA-like structures found by the default RNAMotif tRNA descriptor, which does not capture the tRNAIle (TAT) (Supplemental Figure 4). Absolute counts and enrichment in relation to transcripts not found in the mobile database are shown. Asterisks indicate significant counts (P < 0.05) according to Fisher’s exact test.
(B) Normalized frequency (estimated density) and cumulative relative frequency (ecdf) of inter-gene distances of tRNA-mRNA tandem gene pairs with the tRNA being located within 1000 nucleotides up- or downstream of genes coding for mobile transcripts (blue) or nonmobile predicted transcripts (gray). Vertical dashed lines indicate medians of shown distributions. Mobile transcript encoding loci in comparison to loci not producing mobile transcript show a significantly closer proximity to tRNA genes (two-sample, two-sided Kolmogorov-Smirnov test, P < 0.0003; Cohen's D d = 0.313).
(C) Number of tRNA genes according to their anticodon, which were detected as poly(A)-RNA:tRNA dicistronic transcripts in RNA-seq data. Orange, distribution of the 94 tRNA genes observed dicistronically; blue, tRNA genes (n = 24) associated with mobile transcripts; gray, TAIR10-annotated tRNA genes; asterisks, tRNA genes with dicistronic transcripts confirmed by RT-PCR; arrows, experimentally tested tRNA fusions.
(D) Schematic diagram of Arabidopsis CK1 gene (AT1G71697) and analyzed insertion mutants. CK1 mRNA exists as a dicistronic poly(A)-tRNA transcript. The ck1.2 mutant harbors a T-DNA insertion between the CK1 stop codon and the annotated tRNAGly (AT1G71700) in the 3′ UTR, resulting in a truncated poly(A) transcript lacking the tRNAGly sequences. RT-PCR primers are indicated as follows: P1 binding to exon 7 of CK1 CDS and P3 binding to tRNAGly sequences were used for wild-type CK1:tRNAGly identification. P1 together with P2, which is stretching the T-DNA left border, and CK1 3′ UTR were used for specific ck1.2 detection.
(E) RT-PCR with the indicated primers revealed that the CK1 poly(A) transcript is present in ck1.2 mutant samples (asterisks) originating from stem-grafted wild-type Col-0 tissue. In the reciprocal wild-type samples, a mutant CK1 poly(A) transcript produced in ck1.2 and lacking the 3′ UTR tRNAGly sequence was not detected.
(F) Phenotype of ck1.1 and ck1.2 and reverse transcription quantitative PCR of transcripts. Wild-type and ck1.2 plants show similarly high levels of CK1 transcript, whereas ck1.1 mutants show very low levels. Rosette area size measurements on adult plants revealed that both ck1.1 and ck1.2 mutants are significantly smaller than the wild type (Student’s t test, mutant versus wild type; P value ck1.1 = 0.006; P value ck1.2 = 0.002; n = 16 plants/line). Error bars indicate sd.
(G) Schematic folding structure of the GUS TLS 3′UTR motifs predicted according to their minimal free energy.
To determine whether these neighboring tRNAs are actually cotranscribed to form dicistronic mRNA-tRNA molecules, we analyzed paired-end RNA-seq data from Arabidopsis (Thieme et al., 2015; Ito et al., 2015) for the presence of poly(A) RNA:tRNA matching sequences and performed RT-PCR assays on selected transcripts (Figure 4C; Supplemental Figure 5 and Supplemental Data Set 2). Both analyses revealed that dicistronic mRNA-tRNA transcripts are produced relatively frequently. In total, 132 dicistronic transcripts spanning protein-coding genes (120 unique loci) and tRNAs (118 unique loci) were supported by RNA-seq data (Supplemental Data Set 2). Of the 120 genes, 27 genes (22.5%) are annotated to produce mobile mRNAs, and of the 118 tRNA genes, 24 tRNA genes were found as dicistronic transcripts in conjunction with a mobile transcript (Figure 4C). Of all mRNA-tRNA tandem sequences within 1000 bp of the respective gene loci, evidence for a dicistronic nature was found 1.6 times more often when the mRNA was annotated as mobile than for nonmobile mRNAs, albeit statistical significance could not be established as the numbers were low (P = 0.1, Fisher’s exact test; Supplemental Data Set 2).
To confirm these findings and to substantiate the notion that tRNAs play a role in transcript mobility, we analyzed insertion mutants of the CK1 gene (TAIR No. AT1G71697), which produces a mobile transcript (Thieme et al., 2015) and expresses an enzyme catalyzing the reaction of choline to phosphatidylcholine (Tasseva et al., 2004). According to the paired-end sequencing data, the tRNAGly core sequence (TAIR No. AT1G71700) is present in the CK1 3′ UTR region forming a dicistronic CK1:tRNAGly transcript (Supplemental Figure 5 and Supplemental Data Set 2). tRNAGly fused to GUS mRNA mediated mobility of the otherwise nonmobile GUS sequence (Figure 3).
To test whether CK1 mRNA mobility depends on the presence of the tRNAGly in the 3′ UTR, we first confirmed and then used two SALK T-DNA insertion lines for grafting experiments: ck1.1 (SALK_070759) and ck1.2 (SALK_023420) (Figure 4D). In ck1.1 mutants, the T-DNA is located within the first intron and the ck1.2 mutants have an insertion between the CK1 stop codon and the annotated tRNAGly sequence. We performed Arabidopsis stem grafting experiments with ck1.2 and the wild type (Col-0) and assayed the presence of wild-type CK1:tRNAGly and truncated ck1.2 mRNA in stock and scion samples via RT-PCR (Figure 4E). Although ck1.2 mutants produce a full-length CK1 poly(A) transcript containing all protein-coding sequences, the truncated transcript lacking the tRNAGly sequence could not be detected in wild-type samples. By contrast, wild-type CK1:tRNAGly transcript was present in both ck1.2 scion and ck1.2 stock tissue samples. This suggests that the CK1:tRNAGly transcript was bidirectionally mobile from stock to scion (Figure 4E), whereas the mutant ck1.2 transcript lacking the tRNAGly sequence was not transported over graft junctions. Thus, the endogenously produced dicistronic CK1:tRNAGly transcript seems to be graft-mobile due to the presence of the 3′ UTR tRNAGly sequence.
As lack of detectable ck1.2 transcript mobility could be a result of low expression levels, we performed quantitative RT-PCR assays to evaluate CK1 expression levels in the two ck1.1 and ck.2 mutants and wild-type plants. Here, only marginal expression could be detected in the ck1.1 mutant, whereas CK1 poly(A)-RNA transcript levels in the ck1.2 mutant were similar to that found in the wild type (Figure 4F). Despite comparably high CK1 transcript levels in wild-type and ck1.2 mutant plants, both the ck1.2 line and the ck1.1 line showed a significant decrease in rosette leaf size compared with the wild type (Figure 4F). This implies that either ck1.2 plants produce less functional CK1 enzyme due to the lack of the 3′ tRNA sequence or that CK1 mRNA presence in expressing cells as well as CK1 mRNA mobility is equivalently essential for normal growth behavior of Arabidopsis.
DISCUSSION
RNAs are arguably the most functionally diverse biological macromolecules found in cells. Their diverse roles are determined by both their complex three-dimensional structure and by their primary sequence. Our study reveals an additional biological role of tRNA sequences in plants. They harbor a motif mediating mRNA transport to distant plant cells. Interestingly, transcript mobility was induced by tRNAMet and tRNAGly, but not by tRNAIle, consistent with the absence of tRNAIle in the pumpkin phloem sap (Zhang et al., 2009). As our results indicate that transcript mobility is mediated by a particular RNA structure, a tRNA motif-scanning algorithm did indeed reveal a significantly high number of identified mobile mRNAs that harbor a TLS motif or are transcribed from genes in close proximity to annotated tRNA genes (Figure 4A; Supplemental Data Set 1), which seem to frequently produce dicistronic poly(A)-RNA:tRNAs (Figure 4C; Supplemental Figure 5 and Supplemental Data Set 2). While the functional role of many mobile mRNAs in distant tissues remains to be elucidated, evidence supports the notion that trafficking of small si/miRNAs and large mRNAs via the phloem plays an important role in regulating plant development (Lucas et al., 2001). A surprisingly high number of mRNAs can be found in phloem exudates (Guo et al., 2013) and move across graft junctions (Thieme et al., 2015; Yang et al., 2015), but no general and easily predictable RNA motif or conserved sequence mediating mobility could be identified in the graft-mobile transcript populations (Calderwood et al., 2016). However, our data suggest that a significant fraction of mobile mRNAs carries a TLS motif potentially mediating mobility across graft junctions.
Highlighting the complexity of the translocation system, mRNA transfer does not strictly follow the source to sink phloem flow as GUS:tRNA fusions not only moved from shoot (source) to root (sink), but also vice versa (Figures 3B and 3C; Supplemental Figure 3). Hence, it is likely that two transport pathways exist for delivering mRNA molecules. One could be based on passive, nonselective delivery from source to sink via the phloem vessels. Obviously, passive delivery such as diffusion depends on transcript stability and abundance. However, another pathway in the form of a targeted and active transport system seems to be in place mediating the delivery of mRNAs from root to shoot. This notion finds support in the observed directional and tissue-specific distribution of graft-mobile mRNAs in Arabidopsis (Thieme et al., 2015). Here, two aspects suggest that a number of mRNAs move in an active and regulated fashion: (1) transport against the phloem flow from root to shoot (sink to source) followed by (2) transfer of distinct mobile mRNAs to specific aboveground tissues such as leaves or flowers. Furthermore, the presence of an active mRNA delivery mechanism is supported by two additional findings presented here: (1) Specific sequences derived from TLSs are sufficient to confer mobility to heterologous mRNAs, and (2) deletion of a TLS in the plant endogenous CK1:tRNAGly dicistronic transcript makes it immobile. This suggests that TLSs or closely related RNA structures mediate transport of a number of graft-mobile transcripts. Here, TLS motifs could provide an evolutionary link between RNA virus transport and mRNA transport. In both systems, TLSs appear to support RNA transport along the phloem vasculature to distant cells. Also, as numerous tRNA genes are dispersed over the genome, it is conceivable that dicistronic gene products are relatively often created by genomic rearrangements. Such randomly created mobile transcripts could complement distant mutant cells harboring nonsense mutations, which seem to occur frequently in higher eukaryotes (McConnell et al., 2013).
In any case, specific RNA transport motifs should interact with plant endogenous RNA binding proteins. This finds support in studies on viroid RNA structures that are specifically necessary for entering specific vascular tissues (Takeda et al., 2011) and in the observed interaction of phloem-delivered mRNAs such as GAI and BEL5 with a phloem-expressed polypyrimidine-tract binding protein (PTB) recognizing poly-cysteine (C)–uracil (U) nucleotide stretches present in the 3′ UTRs of some phloem mobile transcripts (Ham et al., 2009; Cho et al., 2015). However, a predicted RNA sequence related to PTB binding motifs was not found in the tRNA fusion constructs used in this study and thus is unlikely to play the primary role in triggering their movement. A structural analysis revealed that the level of overall base pairings in the 3′ end of Arabidopsis transcripts assigned to be mobile is similar to nonmobile transcripts (two-sample Kolmogorov-Smirnov test, P = 0.011), while the 3′-terminal sequence regions of mobile transcripts form energetically less stable folds (two-sample Kolmogorov-Smirnov test, P < 2.2E-16). This is caused by a significantly lower GC content in the respective region compared with transcripts not found to be mobile (Wilcoxon rank sum test, P < 2.518e-10) (Supplemental Figure 4). Hence, the mobile mRNA population seems to be generally less stable and less likely to bind to PTB proteins.
In general, predicted folds of mobile tRNA variants (Figure 4G) point toward an RNA hairpin motif triggering transport. All tRNA sequences and variants thereof eliciting bidirectional GUS mRNA transfer over graft junctions seem to form a predicted long hairpin motif with a stem (8 to 12 nucleotides)–variable bulge(s)–stem (4 to 7 nucleotides)–variable loop (Figure 4G), which is surprisingly similar to structures formed by precursor miRNAs having predicted stem–short bulge–stem–extensive loop core folds. Interaction of a D stem-loop with a T stem-loop present in mature L-shaped tRNAs seems not to be essential as the D loop deletion actually enhanced the transport activity of GUS fusions. Requirements for TLS-mediated transport seem to be either within the A- or the T-loop and the acceptor stem sequences present in all mobile GUS-tRNA fusion transcripts. Here, it is important to note that not all tRNAs seem to confer mobility to transcripts. The tRNAIle(TAT) core sequence fused to GUS did not trigger GUS transcript mobility over graft junctions. This observation is in line with previous studies in which tRNAIle (TAT) is not or rarely detected in the phloem exudate of pumpkin (Zhang et al., 2009). Also, we did not identify tRNAIle(TAT) dicistronic transcripts in RNA-seq data (Figure 4C; Supplemental Data Set 2) and found evidence that tRNAIle(TAT) has a predicted A- and T-loop folding structure distinct from tRNAGly, tRNAMet, and variants thereof, triggering mobility (Figure 4G).
Another important aspect is that GUS enzyme activity assays and sterility phenotypes detected in distant wild-type tissues corroborate the notion that mobile mRNAs are translated into functional proteins after transport. Interestingly, the observed plant size phenotype in mutants producing a nonmobile CK1 transcript (ck1.2) lacking the dicistronic tRNAGly sequence is equivalent to that observed with the ck1.1 null mutant (Figure 4F). Here, although it seems implausible, CK1 transcript mobility appears to have a similar crucial function as the gene product itself. In any case, we could show that specific tRNA sequences such as tRNAGly, tRNAMet, and tRNAMet-derived sequences trigger transport of otherwise nonmobile transcripts and that a significant number of mobile mRNAs harbor a TLS motif.
METHODS
Plant Material and Growth Conditions
Tobacco (Nicotiana tabacum cv Petite Havana) plants were grown under aseptic conditions on agar-solidified medium containing 30 g L−1 sucrose. Rooted tobacco plants were transferred to soil and grown to maturity under standard greenhouse conditions: relative humidity, 55%; day temperature, 25°C; night temperature, 20°C; diurnal cycle, 16 h light/8 h dark; light intensity, 190 to 600 μE·m−2·s−1; mixed light (ratio 1:1) from metal-halide light (HPIT) and sodium-vapor light (AgroSonT) sources plus sunlight. Arabidopsis thaliana seeds of wild-type (Col-0) and transgenic 35Spro:GUS, ck1.1 (SALK_070759), and ck1.2 (SALK_023420) plants of ecotype Col-0 were used and grown in controlled environmental chambers (light source: mixed fluorescent tubes 50% cold white, 50% warm white) for growth assays or on soil in the greenhouse: relative humidity, 60%; day temperature, 22°C; night temperature, 19°C; diurnal cycle, 16 h light/8 h dark; light intensity, 170 to 200 μE·m−2·s−1; mixed light (ratio 1:1) from metal-halide light (HPIT) and sodium-vapor light (AgroSonT) sources plus sunlight. The SALK lines were obtained from the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003).
Grafting and Estradiol Treatment
Tobacco plants used for grafting experiments were grown 2 to 3 months on soil in the greenhouse. A standard splice grafting procedure was used as previously described (Zhang et al., 2014). In short, plants with the same stem diameter carrying five fully expanded leaves were used as stock and scion material; rootstocks were prepared by removing the apical leaves from the top of the plant and keeping two to three source leaves. Scions were prepared by cutting the stem 3 to 4 cm below the apex and removing the source leaves. A long slanting cut was made on the rootstock stem (∼30 degrees from vertical) with a matching cut at the scion base. The surfaces of both cuts were immediately pressed together and the junction was tightly wrapped with Parafilm. The first week after grafting, the scion was covered with a plastic bag and kept under high humidity. After the graft junction was established, axillary branches and leaves emerging at the stock were removed to enforce apical dominance of the scion. Before flower induction, 5 µM 17-β-estradiol mixed with Lanolin (Sigma-Aldrich) (1000× stock solution: 5 mM 17-β-estradiol in DMSO, stored at −20°C) was applied with soaked tissue paper onto the adaxial side of stock plant leaf surfaces to induce gene expression. The tissue paper was left on the surface to mark the side of induction. After flowers appeared on the scion, part of the induced leaf and emerging first closed flowers were sampled for fusion transcript presence by RT-PCR.
Arabidopsis hypocotyl grafting was performed as described (Thieme et al., 2015). In short, plants were grown vertically on solid 0.5 Murashige and Skoog (MS) medium (1% sucrose) at 22°C with a photoperiod of 8 h light (fluence rate of 100 µmol m−2 s−1). The temperature was increased to 26°C 4 d after germination to reduce adventitious root formation. Six to seven days after germination, seedlings were used for grafting under sterile conditions as described (Thieme et al., 2015). In short, seedlings were cut transversely in the middle of the hypocotyl with a razor blade (Dumont; No. 5), and a silicon collar (NeoTecha; diameter 0.30 × 0.60 mm) was slid over the stock in which the scion was inserted. Grafted plantlets were placed on solid 0.5 MS medium (supplemented with 1% agar and 1% sucrose) and grown at 22°C (8 h light). Appearing adventitious roots were cut every 2 d, and after 2 weeks successfully grafted plants were submitted to histochemical GUS stain assays, or root and shoot material were harvested separately for RT-PCR detection of GUS transcripts. The detailed procedure of Arabidopsis inflorescence stem grafting used for CK1 mobility assays was performed as described (Nisar et al., 2012), and samples were harvested for RNA extraction and RT-PCR detection 1 week after grafting.
Expression Constructs
To produce a dominant-negative Arabidopsis DMC1 with a N-terminal 92-amino acid deletion in Arabidopsis DMC1 (DNDMC1; provided by Rijk Zwaan) transcripts with 3′ UTR and 5′ UTR fusions an expression binary constructs named pRD1 and pRD4 were created based on a pMDC7 backbone (Curtis and Grossniklaus, 2003). The DNDMC1 fragment was introduced 5′ or 3′ of the pMDC7 Gateway cloning cassette, which resulted in a template binary vector used to clone the RNA sequences of BEL5 or tRNAMet between the DNDMC1 open reading frame and promoter or terminator via a Gateway reaction (Figure 1A). Synthetic oligonucleotides were used to produce Gateway Entry clones with the according sequence for Gateway recombination with the binary vector (Supplemental Table 2). The binary vector constructs based on pMDC7 allow estradiol-induced DNDMC1:RNA or RNA:DNDMC1 expression. pEarleyGate104 used for 35Spro:YFP-DNDMC1 expression and the DMC1 siRNA tobacco line (35Spro:BcDMC1 hpRNAi) and its function were previously described (Zhang et al., 2014).
GUS fusion constructs harboring tRNAMet (AUG), tRNAGly (GGC), or tRNAIle (AUA) and tRNAMet (AUG) variants in the 3′ UTR were created by PCR amplification using an NcoI GUS forward primer covering the GUS start codon and by a BstEII GUS reverse primer covering the GUS stop codon and the tRNA sequence. The resulting PCR fragment was amplified again with an unspecific XbaI reverse primer harboring an XbaI site for identification of the cloned fragment. The resulting NcoI-BstEII-digested fragments were cloned into the accordingly digested pCambia1305.1 (Chen et al., 1998), allowing expression of the GUS:tRNA constructs driven by a 35S promoter. All synthetic oligonucleotides used in the PCR reactions are listed in Supplemental Table 2.
RNA Isolation and Reverse Transcription Reactions
Samples were prepared in 1 mL Trizol reagent (Invitrogen) (0.5 mL/100 mg tissue) as described previously (Zhang et al., 2009). After centrifugation (10,000g, 10 min at 4°C), the supernatant (∼1 mL) was transferred to a new RNase-free tube and extracted once with 200 μL and once with 50 μL chloroform. To precipitate the RNA, the supernatant was supplemented with two volumes of 99% isopropanol, 0.1 volumes of 3 M sodium acetate (pH 5.2), and 1 µg of linear acrylamide (Invitrogen) and incubated >1 h at −20°C. After centrifugation (16,000g, 30 min at 4°C), the resulting pellet was washed twice with 80% ethanol, once with 99% ethanol, air dried, and resuspended in 20 μL RNase-free water. To determine RNA quality and concentration, 1 μL of each RNA sample was submitted to agarose gel electrophoresis (2%, agarose, 1× TBE) and quantified using a NanoDrop ND-1000 (Thermo Scientific).
Reverse transcription reaction was performed with 1 unit/μL AMV reverse transcriptase (Promega) with the following modifications: total RNA (∼4 µg) was denatured at 70°C for 10 min in the presence of oligo(dT) primer followed by a 5-min annealing incubation at 37°C prior to the RT reaction, then incubated at 42°C for 1 h, and 72°C for 10 min for deactivation. RT-PCR was conducted under standard PCR conditions with 40 to 45 cycles (Zhang et al., 2014). Oligonucleotides used for RT-PCR are listed in Supplemental Table 2.
Quantitative RT-PCR
Quantitative real-time PCR was performed according to the SYBR Green method in a 5 μL volume using 4 μg total RNA, 2.5 μL SYBR Green Master Mix (Applied Biosystems), and 0.2 μM forward and reverse primers. For each genomic confirmed ck1 mutant, RNA from three to five individual plants was isolated and used. At least three technical replicas were performed. An ABI system sequence detector (Applied Biosystems 7900HT fast real-time PCR) was used with the following regimen of thermal cycling: stage 1, 1 cycle, 2 min at 50°C; stage 2, 1 cycle, 10 min at 95°C; stage 3, 40 cycles, 15 s at 95°C, 1 min 60°C. Dissociation stage was as follows: 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C. Oligonucleotides used for RT-PCR are listed in Supplemental Table 2.
Microscopy and Pollen Shape Analysis
The statistical pollen shape analysis indicating sterility was performed as described previously (Zhang et al., 2014). Tobacco pollen was collected and stained with propidium iodide (0.01 mg/mL; Molecular Probes). To image the shape and size of the pollen, a confocal laser scanning microscope (TCS SP5; Leica Microsystems) was used. The system had the following settings: detection channel 2 (red), 570 to 650 nm; the channel 2 gain (PMT) was set between 500 and 600 V; pinhole, 1.0 airy units; five Z-stacks with 5 to 6 μm were merged and used for the shape recognition algorithm as described (Zhang et al., 2014).
YFP fluorescence was detected as described (Zhang et al., 2014) with the following settings: sequential channel scan mode with a maximum aerial pinhole of 1.5 airy units. To compare the YFP fluorescence intensity between plants, the same settings, such as laser power, gain voltage, pinhole, objective, magnification, and channel/filter wavelengths, were used. Z-stack images were assembled and processed using the Image J software package (NIH): detection channel 1 (green), 535 to 617 nm; detection channel 2 (red), not used; detection channel 3 (blue; chloroplast/plastid autofluorescence), 695 to 765 nm. Channel 1 gain (PMT) was set between 500 and 600 V.
GUS Detection
Histochemical reactions with substrate X-Gluc were performed with plant material incubated in 80% acetone for 20 min at −20°C, washed two times with 50 mM NaPO4 buffer, pH 7.0. The staining solution (1 mM X-Gluc diluted to 25 mg/mL, in 50 mM NaPO4 pH 7.0 buffer, supplemented with 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 0.1% Triton X-100) was vacuum infiltrated for 15 min. The staining reaction was performed at 37°C overnight and stopped by rinsing the tissues three times in 70% ethanol for 1 h. The stained plant material was examined by stereomicroscopy (Leica DFC300). For thin sections, GUS-stained samples were dehydrated in an ethanol series including a fixation step with 20% ethanol, 35% ethanol, 50% ethanol, FAA prepared fresh (50% ethanol, 3.7% formaldehyde, and 5% acetic acid), and 70% ethanol for 30 min each at room temperature. The samples were then embedded in paraffin using the enclosed Leica ASK300S tissue processor and the Leica EG1160 embedding center. The 10- and 20-µm longitudinal and traverse sections were placed on poly-l-lysine-coated slides. After drying the samples overnight at 42°C, the slides were dewaxed twice in Histoclear for 10 min and then incubated twice in 99.8% ethanol for 10 min under constant movement at room temperature. After drying overnight, the cover slips were mounted with Entellan new (Merck Millipore) and examined by an epifluorescence microscope (Olympus BX61).
Bioinformatic Analysis
tRNA Motif Scans
Reference sequences of all protein-encoding genes (available cDNA sequence data associated with all protein-coding Arabidopsis genes, TAIR10 [Lamesch et al., 2012], excluding organellar genomes) were partitioned into distinct sets based on their annotation as mobile or nonmobile as detected in heterografted Arabidopsis accessions or Cuscuta-parasite Arabidopsis-host interactions (Thieme et al., 2015). Subsets were generated for genes common to both mobile sets (n = 486), present in at least one of them (n = 3606), as well as according to the observed movement direction (root-to-shoot, shoot-to-root, and bidirectional). All genes and associated transcripts assigned as nonmobile were used as controls. All sets were filtered for duplicate sequences, and annotated tRNA genes were removed. tRNA sequence data were obtained from the tRNAdb (Jühling et al., 2009). Prior to structure motif scans, each sequence was padded with 50 “N” leading and trailing characters to facilitate the detection of terminally located tRNA structures without asymmetric ends at the tRNA acceptor arm which are required by the default tRNA descriptor. All sets were analyzed by RNAMotif version 3.1.1 (Macke et al., 2001) using the provided tRNA structure descriptor and default parameter settings. Motif enrichment associated with genes encoding mobile transcripts compared with background data was assessed by Fisher’s exact test. Specificity of the searched tRNA-like structure was assessed by permutation scans of the default tRNA descriptor. A total of 20,000 different tRNA descriptors were produced by randomly altering the accepted minimum and maximum lengths limits for the stems and the single-stranded loops in the model (normal distribution using µ = 0, sigma = 5; minimum stem length set to 3 nucleotides). Each descriptor was evaluated against the mobile/nonmobile data by RNAMotif with default settings. Structuredness, i.e., the percentage of base-paired nucleotides and associated energetics, within the 3′ UTR was addressed by excising the 150-nucleotide 3′-terminal sequence portion and subsequent analysis of its predicted secondary structure (RNAfold, default settings).
tRNA-mRNA Tandem Scans
Genes adjacent to tRNA loci were identified according to TAIR10 gene models including protein-coding, noncoding genes, and pseudogenes. Statistical significance of the difference of gene proximity distributions (distances between tRNA genes and mobile versus nonmobile gene neighbors) was estimated by the nonparametric Kolmogorov-Smirnov test; relevance was assessed by the effect size (Cohen's D) based on the mean observed differences and associated standard deviations.
Dicistronic tRNA Analysis
Arabidopsis reference genome (TAIR10) sequence information obtained from TAIR-associated gene model descriptions (gtf version 10.30) was taken from http://plants.ensembl.org. Paired-end RNA-seq data (100-nucleotide reads from both ends) were retrieved from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra): accession numbers SRX853394 (14.1G bases, root sample) and SRX853395 (15.3G bases, shoot sample) (Thieme et al., 2015) as well as DRX014481 (19G bases, root sample) and DRX014482 (32.7G bases, root sample) (Ito et al. 2015). Read data were quality trimmed and Illumina adapter sequences were clipped using Trimmomatic (Lohse et al., 2012) standard settings (ILLUMINACLIP:<adapterfile>:2:40:15, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:36).
Mapping of sequence mate pairs to the Arabidopsis reference genome (TAIR10) was done by STAR v2.5.1 (Dobin et al. 2013) based on Ensembl gene model descriptions. Considering the high number of tRNA genes in the Arabidopsis genome and their similar sequences, reads with multiple alignments were excluded, minimum overhang for gene junctions was set to 10 nucleotides for annotated junctions and 20 nucleotides for unannotated junctions, and maximum number of allowed mismatches per pair was 10 nucleotides (outFilterMultimapNmax 1, alignSJDBoverhangMin 10, alignSJoverhangMin 20, outFilterMismatchNmax 10). Subsequently, all read pairs mapping to chromosomes 1 to 5 with a minimum alignment quality Q≥10 were checked to be intersecting with both tRNA and mRNA gene annotations. Finally, identified 132 dicistronic poly(A)-RNA::tRNA transcripts were grouped by their tRNA gene identity (118 unique tRNA genes; Figure 4C) as well as by the protein-coding gene (120 unique genes) and the assigned transcript mobility. Results were compared with the list of annotated tRNA-mRNA tandems and statistical significance for the observed overlap to the dicistronic transcripts was assessed by Fisher’s exact test.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DMC1 TAIR, AT3G22880; BEL5, GenBank AF406697; CEN2/CET2, GenBank AF145260; tRNAMet, TAIR AT5G57885; tRNAGly, TAIR AT1G71700; tRNAIle, TAIR AT3G05835; ACTIN2 (ACT2), TAIR At3g18780; CK1, TAIR AT1G71697; and GUS, GenBank AKK29426.
Supplemental Data
Supplemental Figure 1. tRNAMet:DNDMC1 movement into flowers and pollen phenotype.
Supplemental Figure 2. tRNA sequences fused to the 3′UTR of GUS.
Supplemental Figure 3. Images of hypocotyl-grafted wild-type/GUS:tRNA.
Supplemental Figure 4. Computational analysis of tRNA-like sequences present in mobile mRNAs.
Supplemental Figure 5. RT-PCR assays confirming the presence of dicistronic poly(A)-RNA:tRNA transcripts in wild-type Arabidopsis flowers and leaves.
Supplemental Table 1. Pollen shape analysis of wild-type, transgenic, and grafted plants.
Supplemental Table 2. Oligonucleotides used in the study.
Supplemental Data Set 1. Distribution of tRNA-like sequence motifs in graft-mobile Arabidopsis and grapevine transcripts.
Supplemental Data Set 2. tRNA proximity to genes, and occurrences of dicistronic poly(A) RNA:tRNA transcripts.
Supplemental Data Set 3. Number of independent transgenic lines used to perform graft experiments.
Supplementary Material
Acknowledgments
We thank Dana Schindelasch and Marina Stratmann (MPI-MPP) for their outstanding technical support. This work was partially funded by MPI-MPP and Rijk Zwaan to F.K.
AUTHOR CONTRIBUTIONS
W.Z. performed grafting experiments, evaluated pollen phenotypes, constructed GUS fusions, analyzed GUS transgenic and CK1 mutants, and performed RT-PCR experiments. G.K. constructed DMC1 fusions and made transgenic tobacco lines. F.A. and W.Z. conducted the pollen shape analysis. N.A. supervised and, supported by W.Z., analyzed ck1 mutant plants and performed ck1 and wild-type grafting experiments. L.Y. performed some Arabidopsis grafts and harvested pollen from grafted tobacco plants. N.W. embedded and analyzed GUS-stained tissue from grafted plants. C.J.T. and D.W. performed the bioinformatic analysis of graft-mobile mRNA sequences data. F.K. outlined the project, suggested experiments, analyzed data, and wrote the manuscript with W.Z., supported by C.J.T., D.W., and N.W.
Glossary
- siRNA
small interfering RNA
- miRNA
microRNA
- UTR
untranslated region
- TLS
tRNA-like structure
- CDS
coding sequence
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Amaya I., Ratcliffe O.J., Bradley D.J. (1999). Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11: 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Lin T., Hannapel D.J. (2009). Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol. 151: 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Chatterjee M., Yu Y., Suh S.G., Miller W.A., Hannapel D.J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S., Rudinger-Thirion J., Florentz C., Giegé R., Pleij C.W., Kraal B. (2004). tRNA-like structure regulates translation of Brome mosaic virus RNA. J. Virol. 78: 4003–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L., Kleckner N. (1992). DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Calderwood A., Kopriva S., Morris R.J. (2016). Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28: 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang S., Beachy R.N., Faucet C. (1998). A protocol for consistent, large scale production of fertile transgenic plants. Plant Cell Rep. 18: 25–31. [Google Scholar]
- Cho S.K., Sharma P., Butler N.M., Kang I.H., Shah S., Rao A.G., Hannapel D.J. (2015). Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction with StBEL5 RNA. J. Exp. Bot. 66: 6835–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47: 105–131. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux M.P., Couteau F., Bergounioux C., White C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257: 283–291. [DOI] [PubMed] [Google Scholar]
- Dreher T.W. (2010). Viral tRNAs and tRNA-like structures. Wiley Interdiscip. Rev. RNA 1: 402–414. [DOI] [PubMed] [Google Scholar]
- Dreher T.W., Rao A.L., Hall T.C. (1989). Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206: 425–438. [DOI] [PubMed] [Google Scholar]
- Fechter P., Rudinger-Thirion J., Florentz C., Giegé R. (2001). Novel features in the tRNA-like world of plant viral RNAs. Cell. Mol. Life Sci. 58: 1547–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath K., Kao C.C. (2007). Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 19: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., et al. (2013). The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45: 51–58. [DOI] [PubMed] [Google Scholar]
- Habu T., Taki T., West A., Nishimune Y., Morita T. (1996). The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 24: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B.K., Brandom J.L., Xoconostle-Cázares B., Ringgold V., Lough T.J., Lucas W.J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., et al. (2015). Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS One 10: e0119724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. (2009). tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37: D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantidis K., Schumacher H.T., Alexiadis T., Helm J.M. (2008). RNA silencing movement in plants. Biol. Cell 100: 13–26. [DOI] [PubMed] [Google Scholar]
- Kim M., Canio W., Kessler S., Sinha N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289. [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Bolger A.M., Nagel A., Fernie A.R., Lunn J.E., Stitt M., Usadel B. (2012). RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40: W622–W627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough T.J., Lucas W.J. (2006). Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57: 203–232. [DOI] [PubMed] [Google Scholar]
- Lough T.J., Lee R.H., Emerson S.J., Forster R.L., Lucas W.J. (2006). Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351: 455–465. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Yoo B.C., Kragler F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2: 849–857. [DOI] [PubMed] [Google Scholar]
- McConnell M.J., Lindberg M.R., Brennand K.J., Piper J.C., Voet T., Cowing-Zitron C., Shumilina S., Lasken R.S., Vermeesch J.R., Hall I.M., Gage F.H. (2013). Mosaic copy number variation in human neurons. Science 342: 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. (2001). RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29: 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D., Yoshinari S., Dreher T.W. (2004). eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321: 47–56. [DOI] [PubMed] [Google Scholar]
- Melnyk C.W., Molnar A., Baulcombe D.C. (2011). Intercellular and systemic movement of RNA silencing signals. EMBO J. 30: 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875. [DOI] [PubMed] [Google Scholar]
- Nisar N., Verma S., Pogson B.J., Cazzonelli C.I. (2012). Inflorescence stem grafting made easy in Arabidopsis. Plant Methods 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cazares B., Kragler F. (2004). The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7: 641–650. [DOI] [PubMed] [Google Scholar]
- Takeda R., Petrov A.I., Leontis N.B., Ding B. (2011). A three-dimensional RNA motif in Potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 23: 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseva G., Richard L., Zachowski A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566: 115–120. [DOI] [PubMed] [Google Scholar]
- Thieme C.J., Rojas-Triana M., Stecyk E., Schudoma C., Zhang W., Yang L., Miñambres M., Walther D., Schulze W.X., Paz-Ares J., Scheible W.-R.D., Kragler F. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Yang Y., Mao L., Jittayasothorn Y., Kang Y., Jiao C., Fei Z., Zhong G.Y. (2015). Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 15: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Sun L., Kragler F. (2009). The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150: 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kollwig G., Stecyk E., Apelt F., Dirks R., Kragler F. (2014). Graft-transmissible movement of inverted-repeat-induced siRNA signals into flowers. Plant J. 80: 106–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.