In vivo PIF1 targeting to specific promoter sites is determined by PIF1-interacting transcription factors and their binding to G-box coupling sequence elements.
Abstract
The bHLH transcription factor PHYTOCHROME INTERACTING FACTOR1 (PIF1) binds G-box elements in vitro and inhibits light-dependent germination in Arabidopsis thaliana. A previous genome-wide analysis of PIF1 targeting indicated that PIF1 binds 748 sites in imbibed seeds, only 59% of which possess G-box elements. This suggests the G-box is not the sole determinant of PIF1 targeting. The targeting of PIF1 to specific sites could be stabilized by PIF1-interacting transcription factors (PTFs) that bind other nearby sequence elements. Here, we report PIF1 targeting sites are enriched with not only G-boxes but also with other hexameric sequence elements we named G-box coupling elements (GCEs). One of these GCEs possesses an ACGT core and serves as a binding site for group A bZIP transcription factors, including ABSCISIC ACID INSENSITIVE5 (ABI5), which inhibits seed germination in abscisic acid signaling. PIF1 interacts with ABI5 and other group A bZIP transcription factors and together they target a subset of PIF1 binding sites in vivo. In vitro single-molecule fluorescence imaging confirms that ABI5 facilitates PIF1 binding to DNA fragments possessing multiple G-boxes or the GCE alone. Thus, we show in vivo PIF1 targeting to specific binding sites is determined by its interaction with PTFs and their binding to GCEs.
INTRODUCTION
The seven PHYTOCHROME INTERACTING FACTORs (PIF1, PIF3, PIF4, PIF5, PIF6, PIF7, and PIF8) are bHLH transcription factors that belong to Arabidopsis (Arabidopsis thaliana) bHLH subgroup 15 along with eight other bHLH proteins including LONG HYPOCOTYL IN FAR-RED (HFR1) (Toledo-Ortiz et al., 2003; Leivar and Quail, 2011). PIFs are a well-conserved protein family important in regulating various light responses downstream of phytochromes; they inhibit seed germination (Oh et al., 2004), promote hypocotyl negative gravitropism (Oh et al., 2004; Shin et al., 2009; Kim et al., 2011), inhibit seedling photomorphogenesis (Ni et al., 1998; Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004), promote shade avoidance responses (Lorrain et al., 2008; Leivar et al., 2012; Li et al., 2012), and promote leaf senescence (Sakuraba et al., 2014). Some of these responses require individual PIFs, while others require the coordinate activity of multiple PIFs (Leivar et al., 2008a; Shin et al., 2009; Jeong and Choi, 2013). In addition to light responses, PIFs also integrate hormonal and environmental signals to promote hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008; Koini et al., 2009; Franklin et al., 2011; Zhong et al., 2012; Oh et al., 2014).
The regulation of seed germination by PIF1 illustrates how PIFs regulate light responses downstream of phytochromes. In imbibed seeds, PIF1 inhibits seed germination in the dark both by coordinating hormone signaling and by repressing cell wall loosening (Oh et al., 2006, 2007, 2009). PIF1 activates abscisic acid (ABA), auxin, and jasmonate signaling, which all inhibit seed germination. It also represses gibberellic acid (GA) and brassinosteroid signaling, which promote seed germination (Oh et al., 2009). For ABA signaling, PIF1 indirectly activates the expression of ABA biosynthetic genes (e.g., ABA1 and NCED6), indirectly represses the expression of the ABA catabolic gene CYP707A2 (Oh et al., 2007), and directly activates the expression of ABA positive signaling component genes (e.g., ABSCISIC ACID INSENSITIVE3 [ABI3] and ABI5) (Oh et al., 2009). For GA signaling, PIF1 indirectly represses the expression of GA biosynthetic genes (e.g., GA3ox1 and GA3ox2), indirectly activates the expression of the GA catabolic gene GA2ox2, and directly activates the expression of GA negative signaling component genes (e.g., GIBBERELLIC ACID INSENSITIVE [GAI], REPRESSOR OF GA [RGA], and BOTRYTIS SUSCEPTIBLE1 INTERACTOR [BOI]) (Oh et al., 2007; Park et al., 2013). When phytochromes are activated by light, they enter the nucleus to interact with PIF1 (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999; Huq et al., 2004; Oh et al., 2004). Once there, the activated phytochromes inhibit PIF1 both by reducing its ability to bind DNA (Park et al., 2012) and by promoting its degradation (Shen et al., 2005; Oh et al., 2006). This loss of PIF1 frees seeds to germinate. Similar transcriptional regulation of cell wall loosening genes, hormone biosynthetic genes, and hormone signaling genes by other PIFs has been observed in hypocotyl elongation and shade avoidance responses (Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013; Pfeiffer et al., 2014).
PIFs bind to G-box elements (CACGTG) in vitro and in vivo. The first evidence that PIFs bind to G-box elements came from a random binding site selection procedure using PIF3 (Martínez-García et al., 2000). Subsequently, electrophoretic mobility shift assays also demonstrated in vitro binding of other PIFs (i.e., PIF1/PIL5, PIF3, PIF4, PIF5/PIL6, and PIF7) to G-box elements (Huq and Quail, 2002; Toledo-Ortiz et al., 2003; Huq et al., 2004; Oh et al., 2007; Shin et al., 2007; Leivar et al., 2008b; Hornitschek et al., 2009). Chromatin immunoprecipitation (ChIP) assays have also confirmed in vivo binding of PIFs to promoters containing G-box elements (Oh et al., 2007, 2009, 2012; Shin et al., 2007; Hornitschek et al., 2012; Zhang et al., 2013). In addition to G-box elements, PIFs also bind weakly in vitro to an E-box element called the PIF binding E-box (PBE; CACATG) (Shin et al., 2007; Kim et al., 2008; Zhang et al., 2013; Pfeiffer et al., 2014). This PBE is also known as the Hormone Up at Dawn (HUD) (Michael et al., 2008) and MYC-responsive element (Abe et al., 1997).
However, genome-wide PIF binding analyses indicate that PIFs also target promoters that lack G-box elements. For PIF1, 748 binding sites in imbibed seeds have been identified using the ChIP microarray technique (ChIP-chip; Oh et al., 2009). While 438 (59%) of these binding sites possess G-box elements, 310 (41%) do not. Of the 1064 PIF3 binding sites in etiolated seedlings identified by the ChIP-seq, only 531 (50%) possess G-box elements (Zhang et al., 2013; Pfeiffer et al., 2014). Similarly, G-boxes are present in only 29% of PIF4 binding sites and 46% of PIF5 binding sites (Hornitschek et al., 2012; Pfeiffer et al., 2014). Overall, 29 to ∼59% of the known PIF binding sites possess G-box elements. In addition, since the Arabidopsis genome contains 14,545 G-box elements, only a fraction of them actually bind PIFs. Clearly, other factors must direct the binding of PIFs, not only to specific G-box elements, but also to the PIF-regulated promoters that lack G-boxes entirely.
In fact, the binding of transcription factors to loci containing only a small subset of core promoter sequences and even to loci lacking core promoter sequences is rather common in vivo. CIRCADIAN CLOCK ASSOCIATED1 (CCA1), a circadian transcriptional repressor in Arabidopsis that binds in vitro to evening elements (EEs), targets only a fraction of the EEs across the genome in vivo (Alabadí et al., 2001; Nagel et al., 2015). CCA1 also targets loci that lack EEs but contain G-boxes or other sequence elements (Nagel et al., 2015; Kamioka et al., 2016). A few mechanisms explaining this in vivo transcription factor binding specificity have been discovered. Sequences flanking core promoter elements, for example, can influence the in vivo binding of transcription factors. Two closely related yeast bHLH transcription factors, Cbf1 and Tye7, bind G-boxes in vitro (MacIsaac et al., 2006; Zhu et al., 2009). In vivo, however, the binding of Cbf1 and Tye7 to different targets throughout the genome can be explained partly by sequences flanking the core G-box element (Harbison et al., 2004; Gordân et al., 2013). In vivo transcription factor binding is also influenced by the binding of other interacting transcription factors (Kazemian et al., 2013). When alone, Isl1, a homeodomain transcription factor that regulates motor neuron development, binds to TAAKKR sequences (K = G or T, R+A, or G). The heterodimerization of Isl1 with Lhx3, another homeodomain transcription factor that regulates spinal motor neuron development (Mazzoni et al., 2013), alters this binding specificity. Similarly, MyoD, a bHLH transcription factor that regulates muscle differentiation, binds E-boxes (CANNTG) when alone. MyoD heterodimers with the bHLH transcription factor E, on the other hand, bind VCASCTGT sequences (V = not T, S = G or C) (Cao et al., 2010). In eukaryotes, the local chromatin environment can also influence the specificity of transcription factor targeting. To this point, it remains unclear what determines the in vivo specificity of PIF binding. We report here that in vivo binding of PIF1 is partly determined by the presence of G-boxes and partly by PIF1-interacting transcription factors that bind G-box coupling elements.
RESULTS
In Vivo PIF1 Targeting: A Hypothesis
Using the ChIP-chip technique, we previously reported 748 PIF1 binding sites (PBSs) in imbibed Arabidopsis seeds (Oh et al., 2009). We will refer to the 438 (59%) of these that possess at least one G-box element as PBS-Gs and to the remaining 310 (41%) that lack G-box elements as PBS-Ns (PBS with no G-box). In our ChIP-chip data, we defined PIF1 binding peaks as peaks appearing with six consecutive probes whose binding intensities fell in the top 1% of all probe binding intensities. We defined a PBS as the 500 bp surrounding each PIF1 binding peak. For comparison, we defined nonbinding sites (NBSs) as stretches of 500 bp whose signal intensities for the six consecutive probes fell in the bottom 30% of all probe signal intensities. We then classified NBSs as either NBS-G or NBS-N depending on the presence of a G-box element. Given that the Arabidopsis genome contains 14,545 G-boxes and that we identified 748 PBSs, something must distinguish PBS-Gs from NBS-Gs and PBS-Ns from NBS-Ns to direct proper PIF binding.
We first pursued the features that distinguish PBS-Gs from NBS-Gs. One simple hypothesis is that PBS-Gs might possess conserved flanking sequences outside the core G-box. We therefore examined G-boxes in PBS-Gs and NBS-Gs but were unable to identify any such sequences.
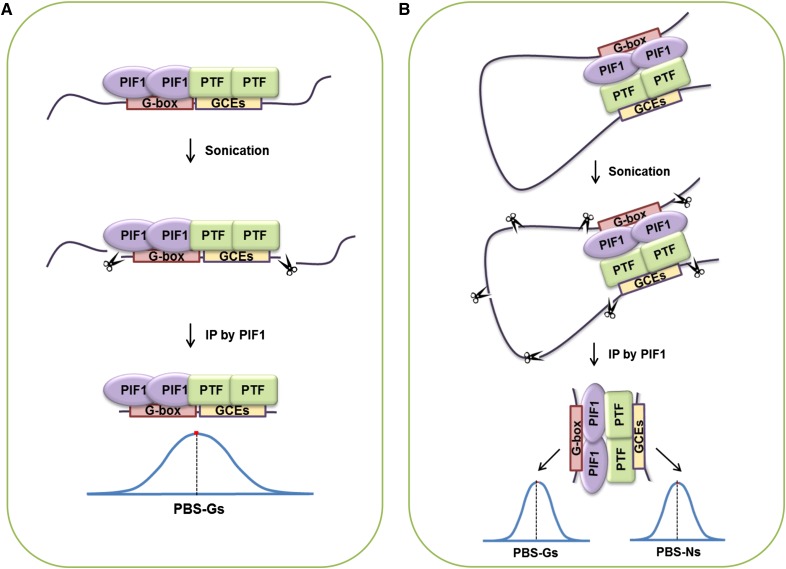
Alternatively, PIF1 may bind stably in vivo to specific G-box elements only in the presence of PIF1-interacting transcription factors (PTFs) that bridge PIF1 and other nearby sequence elements that are enriched at PBSs. We will refer to these PTF binding sequences as G-box coupling elements (GCEs) (Figure 1A). Simultaneous binding of PIF1 to a G-box and a nearby GCE-bound PTF would increase the binding affinity of PIF1 to that specific G-box. PTFs and GCEs may also explain PBS-Ns (Figure 1B) with PIF1 bound to a promoter containing a G-box looping to interact with a PTF binding a GCE. In a ChIP assay, such DNA loops would produce PBS-Ns if they were sheared and the GCEs were precipitated by PIF1 because of their interaction with PTFs (Figure 1B).
Figure 1.
Possible Mechanisms Determining in Vivo PIF1 Binding Specificity.
(A) PIF1 binding specific G-boxes in the Arabidopsis genome via interaction with PTFs that themselves bind GCEs.
(B) Looping interactions between PIF1 and GCE binding PTFs may explain the appearance of PBS-Ns in ChIP results.
In Vivo PIF1 Binding Sites Are Enriched with Multiple G-Boxes and G-Box Coupling Sequence Elements
The hypothesis above depends on the enrichment of GCEs within PBSs in a pattern similar to that of G-box elements. We identified hexameric sequence elements enriched in PBSs while using the two-sample Kolmogorov-Smirnov (KS) test to exclude hexamers whose distribution differs from that of G-box elements. Table 1, which is sorted by enrichment P value, lists the top 10 hexamers enriched in PBS-Gs and PBS-Ns.
Table 1. Top 10 Hexameric Sequence Elements Enriched in PBSs.
| Rank | Motif | Exp.a | Obs.b | Ratioc | P Value (Enrichment)d | P Value (Distribution)e |
|---|---|---|---|---|---|---|
| PBS-Gs | ||||||
| 1 | CACGTG | 52.69 | 1442 | 27.37 | 0 | 1 |
| 2 | CGCGTG/CACGCG | 15.54 | 145 | 9.33 | 1.36E-86 | 0.554 |
| 3 | GCGCGT/ACGCGC | 11.58 | 121 | 10.45 | 6.14E-78 | 0.369 |
| 4 | CGCGCG | 4.72 | 58 | 12.28 | 5.25E-42 | 0.0236 |
| 5 | CACGTC/GACGTG | 36.27 | 133 | 3.67 | 4.22E-35 | 0.7293 |
| 6 | CGTGGC/GCCACG | 29.04 | 116 | 3.99 | 4.83E-34 | 0.0140 |
| 7 | TGACGT/ACGTCA | 50.88 | 156 | 3.07 | 2.58E-32 | 0.115 |
| 8 | GGGTCC/GGACCC | 23.61 | 99 | 4.19 | 6.81E-31 | 0.744 |
| 9 | GCGTGA/TCACGC | 28.81 | 109 | 3.78 | 3.55E-30 | 0.865 |
| 10 | ACGCGT | 19.32 | 88 | 4.56 | 4.05E-30 | 0.0181 |
| PBS-Ns | ||||||
| 1 | CGCGTG/CACGCG | 11.00 | 83 | 7.55 | 1.28E-43 | 0.072 |
| 2 | TGACGT/ACGTCA | 36.01 | 136 | 3.78 | 3.93E-37 | 0.035 |
| 3 | GCGCGT/ACGCGC | 8.19 | 65 | 7.93 | 8.97E-36 | 0.145 |
| 4 | CACGTC/GACGTG | 25.67 | 87 | 3.39 | 1.97E-21 | 0.01 |
| 5 | CACATG/CATGTG | 79.28 | 172 | 2.17 | 1.42E-19 | 0.601 |
| 6 | ACGCGT | 13.67 | 54 | 3.95 | 1.43E-16 | 0.401 |
| 7 | TGTCGT/ACGACA | 44.02 | 104 | 2.36 | 1.10E-14 | 0.016 |
| 8 | CGTCAT/ATGACG | 39.66 | 94 | 2.37 | 1.64E-13 | 0.053 |
| 9 | GACGTC | 23.77 | 66 | 2.78 | 8.84E-13 | 0.482 |
| 10 | CCACGT/ACGTGG | 31.78 | 79 | 2.49 | 1.35E-12 | 0.127 |
Motifs expected in the Arabidopsis genome.
Motifs observed in PBS-Gs or PBS-Ns.
Ratio of Exp. to Obs.
P value for motif enrichment (via binomial tests).
P value for comparing the distribution of G-boxes with each motif (via two-sample KS tests). P values ≤ 0.01 indicate the motifs whose distribution is significantly different from that of G-boxes.
PBS-Gs are enriched with multiple G-box elements, four ACGC-containing elements, and two ACGT-containing elements (Table 1). We named the ACGC-containing elements GCE1 and the ACGT-containing elements GCE2 (Table 2). We noticed that PBS-Gs tend to contain multiple G-boxes (Figure 2A). Of the 438 PBS-Gs, 204 (46.6%) contain more than two G-boxes. Only 4.3% of the NBS-Gs have more than two G-boxes. It is possible that these multiple G-boxes serve to recruit additional PIF1 molecules, thus increasing their chance of immunoprecipitation by PIF1 in the ChIP assay. Alternatively, the prototypical G-box (CACGTG) may be a special case of the ACGT-containing GCE2.
Table 2. G-Box Coupling Elements Found in PBSs.
| Name | Sequence Elements |
|---|---|
| G-box | CACGTG |
| GCE1 | ACGC-containing elements |
| GCE2 | ACGT-containing elements |
| GCE3 | CACATG |
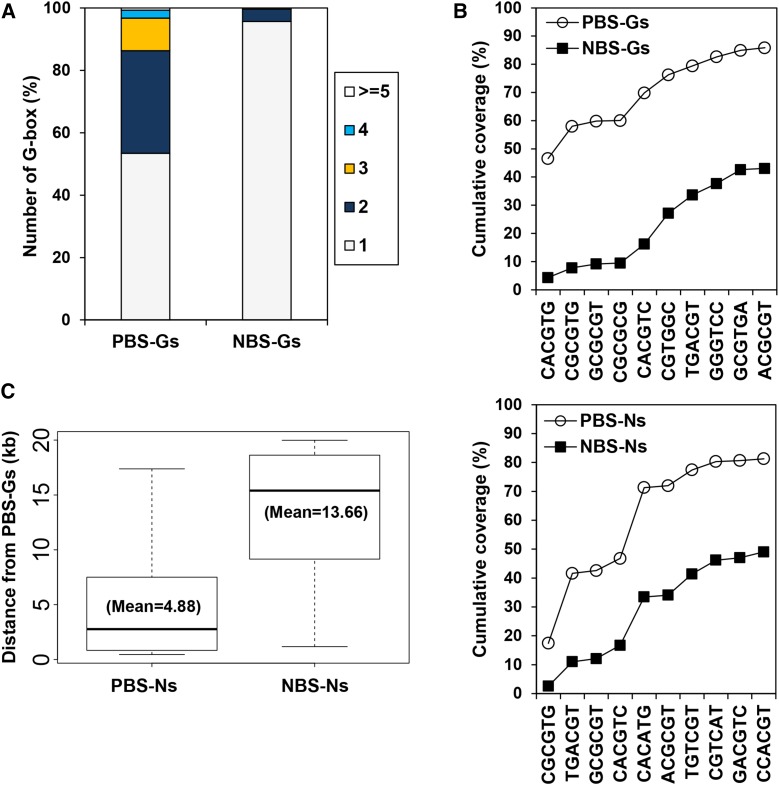
Figure 2.
In Vivo PIF1 Binding Sites Are Enriched with Multiple G-Boxes and G-Box Coupling Sequence Elements.
(A) Quantifying the percentage of PBS-Gs that contain multiple G-boxes.
(B) Many PBSs contain multiple G-boxes and GCEs. Cumulative percentage of PBSs and NBSs possessing each hexamer. The first G-box sequence in the upper graph indicates additional G-boxes in PBS-Gs.
(C) PBS-Ns are closer to PBS-Gs than NBS-Ns to PBS-Gs. The distance between each and the closest PBS-G within a 20-kb window is plotted in a standard box plot.
Again, 46.6% of PBS-Gs contain multiple G-boxes (Figure 2A). Of the remaining PBS-Gs, an additional 31.7% contain at least one GCE1 or GCE2. This means 78.3% of PBS-Gs have either multiple G-boxes or a single G-box plus at least one GCE1 or GCE2 (Figure 2B). Only 30.2% of NBS-Gs follow this pattern. This significant presence of GCEs in PBS-Gs supports the hypothesis that in vivo PIF1 binding is concentrated at PBS-Gs enriched with multiple G-boxes and/or GCEs.
Our hypothesis further predicts that similar coupling elements should appear in the PBS-Ns. Indeed, we identified similar hexameric elements enriched in PBS-Ns. These include three GCE1s, four GCE2s, and a GCE3 (CACATG) (Tables 1 and 2). GCE3 is identical to the sequence element previously known as HUD, which was overrepresented in promoters of morning-specific phytohormone genes (Michael et al., 2008) or PBEs shown to bind weakly to PIFs in vitro (Shin et al., 2007; Kim et al., 2008; Zhang et al., 2013; Pfeiffer et al., 2014). Thus, GCE3 could serve as a direct PIF1 binding sequence or could act as another coupling element that binds PTFs. Similar to what we observed with PBS-Gs, 74.5% of PBS-Ns contain GCE1s, GCE2s, or GCE3s, whereas only 37.7% of NBS-Ns follow this pattern (Figure 2B). This is consistent with the hypothesis that PIF1 preferentially binds PBS-Ns enriched with GCEs. None of the three GCEs are preferentially associated with transcriptionally activated or repressed PIF1 target genes in imbibed seeds (Supplemental Table 1).
If PBS-Ns can interact with PBS-Gs via PIF1-PTF interactions, PBS-Ns should appear in association with PBS-Gs more often than NBS-Ns appear in association with PBS-Gs. Thus, we calculated the average distance from PBS-Ns to PBS-Gs within a 20-kb sliding window. Indeed, while 97 of 310 PBS-Ns (∼31%) are located within 20 kb from the nearest PBS-G, only 417 of 28,301 of NBS-Ns (∼1%) are located within 20 kb from the nearest PBS-G. The average distance from a PBS-N to the nearest PBS-G is 4.88 kb, whereas the average distance from an NBS-N to the nearest PBS-G is 13.66 kb (Figure 2C). This fact that PBS-Ns are located significantly closer to PBS-Gs lends further support to the hypothesis that PBS-Ns are associated with PBS-Gs in vivo.
T-DNA Insertion within or between PBSs Disrupts in Vivo PIF Targeting
If GCEs are important for in vivo PIF1 targeting to PBSs, the insertion of a T-DNA between the G-box and a nearby GCE should disrupt PIF1 targeting. To examine this possibility, we isolated mutants containing T-DNA insertions in PBSs. Since the T-DNAs used in this study are longer than 5 kb and include two different genes and various sequence elements (Sessions et al., 2002; Rosso et al., 2003), they may be able to decouple G-boxes and GCEs when inserted between them.
The PIL2/PIF6 promoter contains three G-boxes that are important for PIF-dependent gene expression. We identified one T-DNA insertion (sail_158_H03) between two of these G-boxes (Figure 3A). We will refer this sail_158_H03 line as the pil2-sl158 mutant. We next asked whether PIF1 binds the PIL2 promoter in the pil2-sl158 mutant by performing a ChIP assay in imbibed seeds using an antibody to the endogenous PIF1. We compared the enrichment of the two DNA fragments on either side of the T-DNA insertion site with the enrichment of a fragment 1.6 kb upstream of the PIL2 transcription start site. We found PIF1 enriches PIL2 promoter fragments in pil2-sl158 mutant seeds less than half as much as in wild-type seeds (Figure 3C). On the other hand, PIF1 enriches the HFR1 and BOI promoter fragments equally well in pil2-sl158 mutant and wild-type seeds (Supplemental Figure 1). This reduced enrichment is not due to a reduction in PIF1 protein (Figure 3D). Since it is possible this reduced PIF1 binding is secondary to an increase in heterochromatin formation around the T-DNA inserted in the PIL2 promoter, we measured the levels of the heterochromatin marker H3K27me3 (Liu et al., 2010). A second ChIP assay using an H3K27me3 antibody showed similar enrichment of PIL2 promoter fragments in wild-type and pil2-sl158 mutant seeds (Figure 3E). Together, these results indicate the physical separation of G-boxes within a PBS by T-DNA insertion can disrupt in vivo PIF1 targeting.
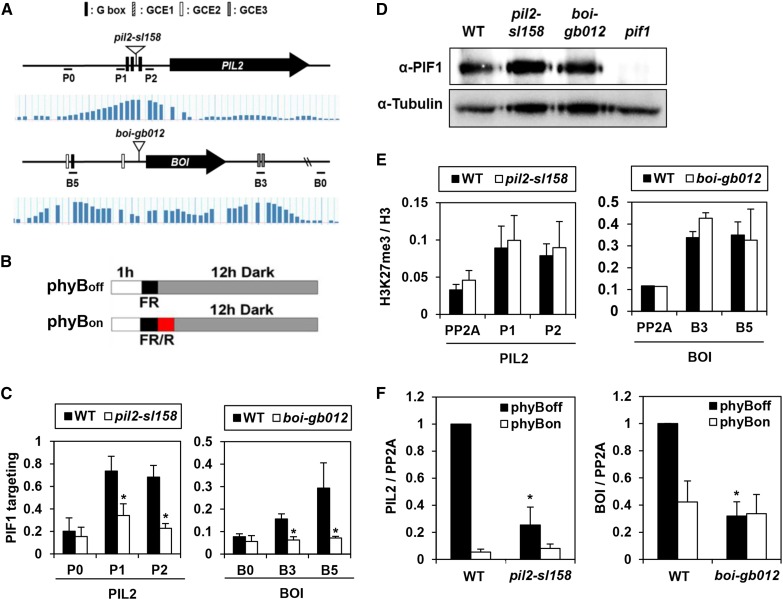
Figure 3.
T-DNA Insertion within or between PBSs Disrupts PIF1 Targeting.
(A) The PIL2 and BOI loci (upper panel), PIF1 binding signals from ChIP-chip assays (lower panel), sequence elements in PBSs, T-DNA insertion sites, and ChIP-PCR amplicons. The vertical bars in the lower panel indicate PIF1 binding intensities for each site in previously reported ChIP-chip assays. The inverted open triangles indicate T-DNA insertion sites in the pil2-sl158 and boi-gb012 mutants. P0, P1, P2, B0, B5, and B3 indicate ChIP amplicons.
(B) Diagrams explaining the phyBon and phyBoff conditions. Seeds are sterilized, irradiated with either a far-red pulse (phyBoff) or a red pulse followed by a far-red pulse (phyBon), and then incubated in the dark for 12 h before sampling. Red fluence rate, 11.5 µmol m−2 s−1; far-red fluence rate, 2.59 µmol m−2 s−1.
(C) Reduced PBS enrichment by PIF1 in T-DNA insertion mutant imbibed phyBoff seeds. Asterisks indicate statistical differences (P < 0.05, Student’s t test). Error bars indicate sd (n = 3 biological replicates).
(D) Wild-type and two T-DNA insertion mutant imbibed phyBoff seeds have similar PIF1 protein levels.
(E) PIL2 and BOI loci in imbibed phyBoff seeds show similar H3K27me3 levels irrespective of T-DNA insertions. This ChIP assay used an anti-H3K27me3 antibody and an anti-H3 antibody. The y axis indicates H3K27me3 enrichment relative to H3 enrichment. Error bars indicate sd (n = 2 biological replicates).
(F) T-DNA insertion mutants in phyBon and phyBoff conditions show reduced expression of PIL2 and BOI mRNAs. Asterisks indicate statistical differences (P < 0.05, Student’s t test). Error bars indicate sd (n = 3 biological replicates).
The BOI gene is flanked by two PBSs separated by ∼3 kb (Figure 3A). The 5′ PBS is a PBS-G, containing a G-box and a GCE2, whereas the 3′ PBS is a PBS-N, containing only two GCE3s. We identified a T-DNA (gabi_012H08) inserted between these two PBSs that increases the distance between them by 5 kb. We will refer to this T-DNA insertion as boi-gb012. Again using ChIP, we found that PIF1 enriches both of these PBSs less than a half as much in boi-gb012 seeds as in wild-type seeds (Figure 3C). In contrast, PIF1 enriches the HFR1 and PIL2 promoter fragments equally well both in boi-gb012 mutant and wild-type seeds (Supplemental Figure 1). As with the pil2-sl158 mutant, this reduced enrichment can neither be attributed to reduced levels of PIF1 protein nor to increased heterochromatin formation at the BOI locus (Figures 3D and 3E). Thus, T-DNA insertion between a PBS-G and a PBS-N also disrupts in vivo PIF1 targeting.
Since PIF1 is the primary transcription factor responsible for activating PIL2 and BOI expression in imbibed seeds, T-DNA insertions that inhibit PIF1 targeting to these loci should reduce the expression of these genes. We therefore compared the expression of PIL2 and BOI in phyBoff and phyBon conditions (Figure 3B) in wild-type and T-DNA insertion mutant seeds. In the phyBoff condition, PIFs are active leading to high levels of both PIL2 and BOI in wild-type seeds. In the phyBon condition, PIFs are inactive and PIL2 and BOI levels are expectedly low. In contrast, the T-DNA insertion mutant seeds (pil2-sl158 and boi-gb012) express low levels of PIL2 and BOI, respectively, in both the phyBoff and phyBon conditions (Figure 3F). Thus, T-DNA insertion between G-boxes and GCEs not only inhibits PIF1 binding but also reduces light-dependent PIF1 target gene expression.
ABI5 Targets a Subset of PBSs Possessing G-Box and GCE2
We noticed that four of the ACGC-containing GCE1s (i.e., CACGCG, ACGCGC, TCACGC, and ACGCGT) bear a resemblance to the FAR-RED ELONGATED HYPOCOTYLS3/FAR-RED IMPAIRED RESPONSE1 (FHY3/FAR1) binding site (FBS; CACGCGC) (Lin et al., 2007; Li et al., 2011). One study showed that FHY3 interacts with PIF1 to regulate chlorophyll biosynthesis (Tang et al., 2012). Under our experimental conditions, we too found that PIF1 interacts with FHY3 (Supplemental Figure 2). Interestingly, we also found that FHY3 binds in vitro to both the FBS and to a GCE1 (CACGCGA) (Supplemental Figure 2). This suggests FHY3/FAR1 may be a PTF that links PIF1 to GCE1s. We also noticed that G-boxes and GCE2s share a common core sequence (ACGT) that may serve as a binding site for bZIPs like ELONGATED HYPOCOTYL5 (HY5) and ABI5 (Chattopadhyay et al., 1998; Carles et al., 2002; Kim et al., 2002; Lee et al., 2007; Song et al., 2008; Zhang et al., 2011; Lee et al., 2012). Previous studies have shown that PIF1 interacts with HY5 (Chen et al., 2013) and that the genome-wide binding sites of HY5 overlap significantly with PIF1 binding sites (Lee et al., 2007; Zhang et al., 2011). However, since both the fhy3 and hy5 mutants do not show strong phyB-dependent germination phenotypes (Supplemental Figure 3), we reasoned that there must be other bZIPs that serve as PTFs regulating seed germination. The group A bZIPs, including ABI5 and the ABA RESPONSIVE ELEMENTS BINDING FACTORs, are known to inhibit seed germination under ABA signaling (Kang et al., 2002; Lopez-Molina et al., 2002; Finkelstein et al., 2005) and are therefore candidate PTFs. Consistent with this hypothesis, PIF1 and ABA signaling coregulates many genes in imbibed seeds (Supplemental Figure 4).
We next cloned two group A bZIPs (ABI5 and ENHANCED EM LEVEL [EEL]) and performed electrophoretic mobility shift assays (EMSAs) to determine if they bind GCE2s. Indeed, both ABI5 and EEL bind biotin-labeled G-box and GCE2 (Figure 4A) and their binding can be competed out by identical but unlabeled DNA fragments. Unlike our results with the ACGT-containing G-box and GCE2, ABI5 and EEL do not bind strongly to GCE1 and GCE3.
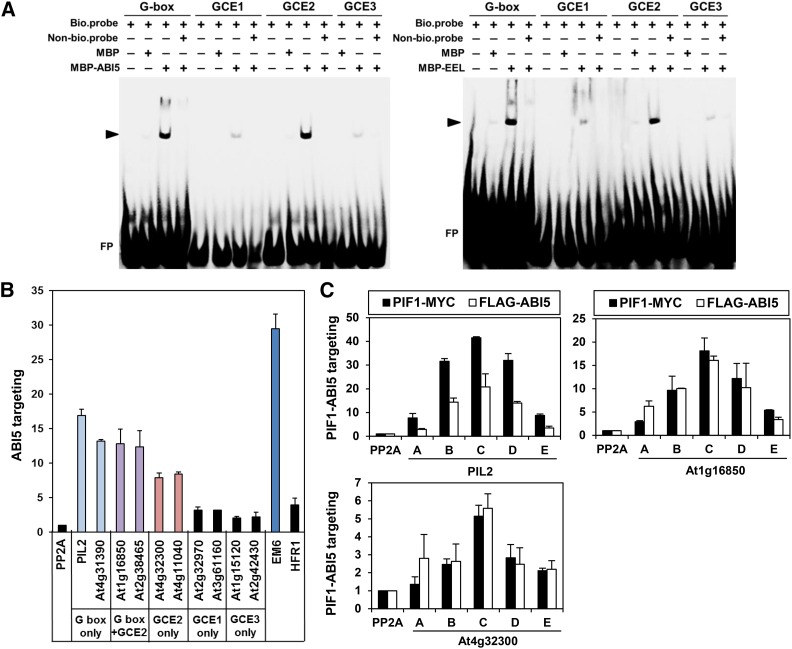
Figure 4.
ABI5 Targets a Subset of PBSs Possessing GCE2s.
(A) EMSA demonstrating the binding of group A bZIPs (ABI5 and EEL) to a G-box and a GCE2. MBP-tagged ABI5 and EEL proteins and biotinylated double-stranded oligomers (bio.probe) were used for this assay. Nonbiotinylated double-stranded oligomers (Non-bio.probe) were used at 100× the concentration of the biotinylated double-stranded oligomers. The triangles indicate the shifted bands (ABI5, left panel; EEL, right panel). FP indicates free, unbound DNA probes. The sources for each probe sequence are as follows: G-box, the PIL2 promoter; GCE1, the At3g18080 promoter; GCE2, the At1g16850 promoter; GCE3, the At1g17830 promoter.
(B) ABI5-mediated enrichment in imbibed phyBoff seeds of PBSs possessing a G-box, a GCE2, or a G-box and a GCE2. Gene names on the x axis indicate amplified promoter fragments covering either PIF1 binding peaks, an ABI5 binding site (Em6), or nonbinding control sites within the body of the PP2A gene (Supplemental Figure 5). Stable transgenic plant seeds expressing FLAG-tagged ABI5 and anti-FLAG antibody were used for this ChIP assay. Error bars indicate sd (n = 2 biological replicates).
(C) ABI5 and PIF1 show similar enrichment profiles at three selected PBSs in imbibed phyBoff seeds. The letters on the x axis indicate ChIP-PCR amplicons in each promoter shown in Supplemental Figure 5. Stable transgenic plant seeds expressing either MYC-tagged PIF1 (PIF1-MYC) or FLAG-tagged ABI5 (FLAG-ABI5) and anti-MYC or FLAG antibodies were used for this ChIP assay. Error bars indicate sd (n = 2 biological replicates).
Next, we performed a ChIP assay using a transgenic line expressing FLAG-tagged ABI5 in imbibed seeds. ABI5 is known to bind EARLY METHIONINE-LABELED6 (EM6) promoter (Nakamura et al., 2001; Carles et al., 2002; Lopez-Molina et al., 2002). Consistent with this, ABI5 strongly enriches a fragment of the EM6 promoter that possesses an ACGT-containing G-box and a GCE2 (Figure 4B; Supplemental Figure 5). In the same ChIP assay, ABI5 also strongly enriches PBSs with multiple G-boxes (PIL2 and At4g31390), PBSs with both a G-box and a GCE2 (At1g16850 and At2g38465), and PBSs with only GCE2s (At4g32300 and At4g11040) (Figure 4B). In further confirmation that ABI5 and PIF1 target similar promoter regions, ABI5 and PIF1 produce identical enrichment profiles for three selected promoters (PIL2, At1g16850, and At4g32300) (Figure 4C). Unlike with the ACGT-containing G-boxes and GCE2s but consistent with EMSA data, ABI5 did not strongly enrich PBSs with either GCE1s (At2g32970 and At3g61160) or GCE3s (At1g15120 and At2g42430) alone (Figure 4C). Oddly, ABI5 does not enrich the PBS in the HFR1 promoter even though it possesses a G-box and a GCE2 (Figure 4C). Taken together, ABI5 preferentially targets a subset of PBSs that possess ACGT-containing G-box and/or GCE2s.
Group A bZIPs Interact with PIF1 Protein
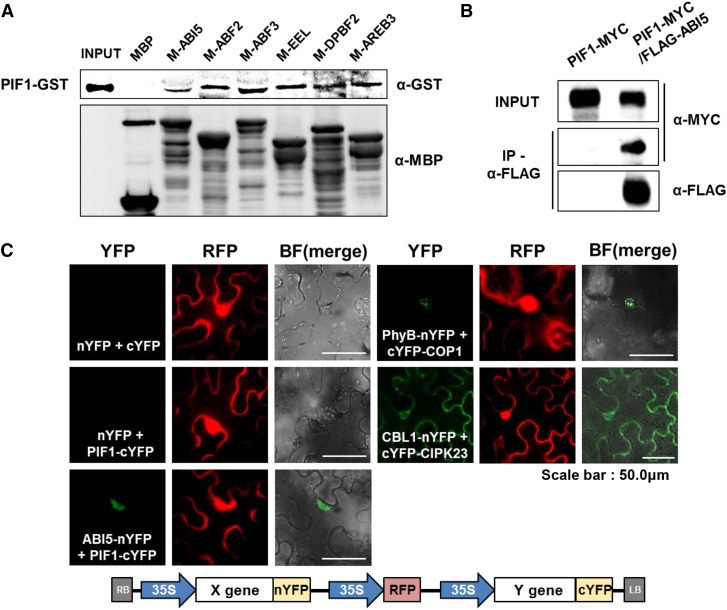
If the group A bZIPs are legitimate PTFs, they must interact with PIF1 protein. We purified six recombinant MBP-fused group A bZIP proteins including ABI5 and found that they are all able to pull down PIF1 protein, while MBP alone cannot (Figure 5A). We also found that an anti-FLAG antibody can pull down PIF1-MYC from lysates of stable transgenic plants expressing both PIF1-MYC and FLAG-ABI5, but not from lysates of transgenic plants expressing PIF1-MYC alone (Figure 5B). Clearly, ABI5 interacts with PIF1 both in vitro and in vivo. Still, we used a bimolecular fluorescence complementation (BiFC) assay to obtain further confirmation of the in vivo interaction between PIF1 and ABI5. After infiltrating split YFP constructs into Nicotiana benthamiana leaves, we examined them under fluorescence microscopy. As expected, the split YFPs (nYFP and cYFP) alone produce no YFP fluorescence, but the positive controls CBL1-nYFP with cYFP-CIPK23 and PHYB-nYFP with cYFP-COP1 produce the expected YFP signals at the cell membrane and speckled in the nucleus, respectively (Grefen and Blatt, 2012). In the same assay, we found that ABI5-nYFP with PIF1-cYFP produce diffuse YFP fluorescence in the nucleus (Figure 5C), whereas nYFP alone with PIF1-cYFP does not. This confirms the interaction of PIF1 and ABI5 in vivo.
Figure 5.
Group A bZIP Proteins Interact with PIF1 Protein.
(A) An in vitro binding assay showing the interaction between PIF1 protein and group A bZIP proteins. PIF1-GST was pulled down by resin-bound MBP-bZIP proteins and detected by an anti-GST antibody (upper panel). Resin-bound MBP-bZIPs was eluted and detected by an anti-MBP antibody (lower panel).
(B) A coimmunoprecipitation assay demonstrating an in vivo interaction between PIF1 and ABI5. Stable transgenic plants expressing MYC-tagged PIF1 either alone (PIF1-MYC) or with FLAG-tagged ABI5 (PIF1-MYC/FLAG-ABI5) were used. An anti-FLAG antibody was used to immunoprecipitate FLAG-ABI5 and the coimmunoprecipitated PIF1-MYC was detected with an anti-MYC antibody.
(C) A BiFC assay demonstrating the interaction between PIF1 and ABI5. Split YFP vectors with PIF1-cYFP and ABI5-nYFP were agroinfiltrated into N. benthamiana leaf cells and observed 3 d later. nYFP and cYFP indicate N-terminal and C-terminal split YFP, respectively. YFP and RFP indicate individual channel images while BF(merge) indicates the merged YFP/bright-field image. We used the RFP signal (35S-driven RFP in the vector diagram below) to identify infiltrated cells. PhyB-nYFP/cYFP-COP1 and CBL1-nYFP/cYFP-CIPK23 were used as positive controls for nuclear and cytosolic interactions, respectively.
Group A bZIPs Assist PIF1 to Target Specific Sites in Vivo
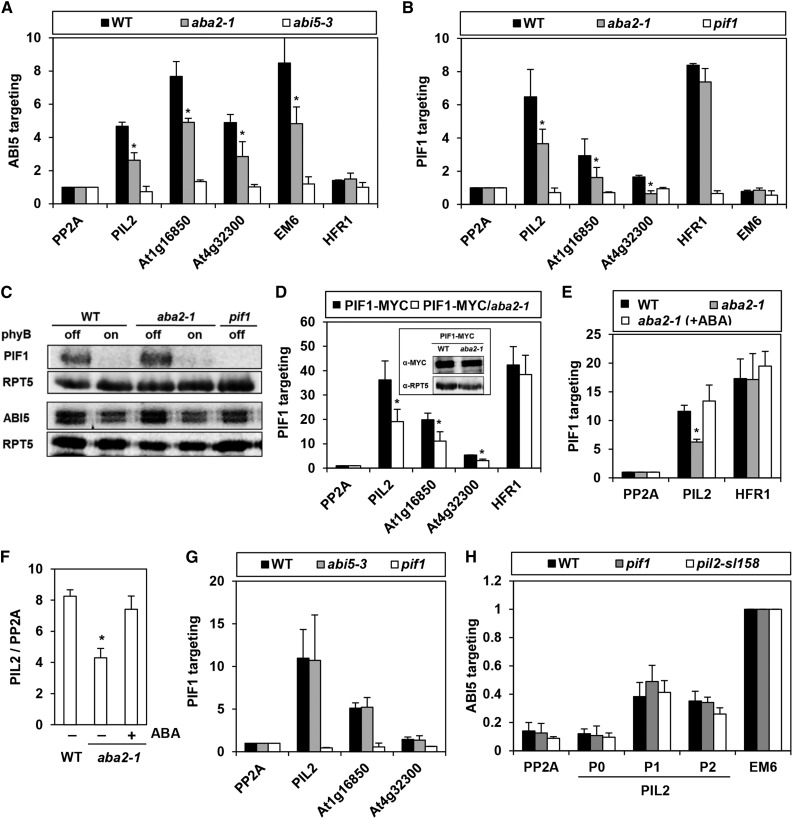
If the group A bZIP are legitimate PTFs, PIF1 targeting to a subset of PBSs should be disrupted in their absence. We therefore performed a ChIP assay using a PIF1-specific antibody in imbibed aba2 mutant seeds. These seeds cannot synthesize ABA because of a mutation in an ABA synthetic xanthoxin dehydrogenase (Schwartz et al., 1997). Since ABA signaling enhances DNA binding of ABI5 and presumably other group A bZIPs (Uno et al., 2000; Lopez-Molina et al., 2001; Lopez-Molina et al., 2002), we used the aba2 mutant as a mild proxy for a general group A bZIP mutant. Consistent with this, although ABI5 strongly enriches EM6 promoter fragments in wild-type seeds, in the aba2 mutant seeds, the level of enrichment falls by more than 50% (Figure 6A). In the same ChIP assay, the ABI5-mediated enrichment of three PBSs that possess multiple G-boxes, a single G-box accompanied by a GCE2, and GCE2s alone is also much lower in the aba2 mutant. This indicates ABI5 targeting to PBSs is also enhanced by ABA signaling. Thus, if PIF1 requires group A bZIPs in vivo to bind specific sites, PIF1’s enrichment of cotargeted PBSs should also be reduced in the aba2 mutant. Indeed, PIF1’s enrichment of PIL2 and other cotargeted fragments is lower in aba2 mutant seeds than in wild-type seeds, while its enrichment of the HFR1 promoter is unaffected by the aba2 mutation (Figure 6B). This reduced enrichment is not an artifact of reduced PIF1 protein levels in the aba2 mutant (Figure 6C). For further proof, we performed another ChIP assay on imbibed seeds of a 35S-driven PIF1-MYC overexpression line and found a similar reduction in cotargeted fragment enrichment in the aba2 mutant. Again, we found no enrichment of HFR1 promoter fragments (Figure 6D). Exogenous ABA treatment restores PIF1 binding to the PIL2 promoter (Figure 6E) and increases PIL2 mRNA levels in aba2 mutant seeds (Figure 6F). In the abi5 single mutant, on the other hand, PIF1 enriches PIL2 or other cotargeted fragments equally well (Figure 6G). Since ABI5 binds the PIL2 promoter even in the presence of the pif1 and pil2-sl158 mutations, ABI5 must target the PIL2 promoter either independently of PIF1 or depending on other PIF family members (Figure 6H). Together, our results suggest the group A bZIPs facilitate PIF1 binding to specific target sites in vivo.
Figure 6.
PIF1 Targeting to a Subset of PBSs Is Compromised in the aba2 Mutant.
(A) Reduced enrichment of promoter fragments by ABI5 in the aba2 mutant. This ChIP assay used an anti-ABI5 antibody in imbibed phyBoff seeds. Gene names on the x axis indicate the amplified promoter fragments that cover either PIF1 binding peaks (PIL2, At1g16850, and At4g32300), the ABI5 binding site (Em6), or a nonbinding site (PP2A and HFR1). Asterisks indicate statistical differences (P < 0.05, Student’s t test). Error bars indicate sd (n = 2 biological replicates).
(B) Reduced enrichment of ABI5-targeting PBSs (PIL2, At1g16850, and At4g32300) by PIF1 in the aba2 mutant. This ChIP assay used an anti-PIF1 antibody in imbibed phyBoff seeds. PP2A and Em6 were used as nonbinding controls of PIF1. Asterisks indicate statistical differences (P < 0.05, Student’s t test). Error bars indicate sd (n = 2 biological replicates).
(C) Immunoblot analysis using anti-PIF1 and anti-ABI5 antibodies shows that the wild type, aba2, and pif1 mutant imbibed phyBoff and phyBon seeds have similar levels of PIF1 and ABI5 proteins.
(D) Reduced enrichment of ABI5-targeting PBSs (PIL2, At1g16850, and At4g32300) by PIF1-MYC in the aba2 mutant. This ChIP assay used an anti-MYC antibody in imbibed phyBoff seeds. Stable transgenic plant seeds expressing MYC-tagged PIF1 (PIF1-MYC) were used. The inset shows that the wild-type and aba2 mutant backgrounds both produce similar levels of 35S-driven PIF1-MYC. Asterisks indicate statistical differences (P < 0.05, Student’s t test). Error bars indicate sd (n = 2 biological replicates).
(E) A ChIP assay showing exogenous ABA treatment (10 μM) rescues the reduced PIF1 binding to the PIL2 promoter in the aba2 mutant. This ChIP assay used an anti-PIF1 antibody on imbibed phyBoff seeds. For the ABA treatment, aba2 seeds were imbibed in media containing 10 μM ABA. The asterisk indicates a statistical difference (P < 0.05, Student’s t test). Error bars indicate sd (n = 2 biological replicates).
(F) PIL2 transcript levels in wild-type and aba2 mutant seeds in either the presence or absence of exogenous ABA (10 μM). The asterisk indicates a statistical difference (P < 0.05, Student’s t test). Error bars indicate sd (n = 2 biological replicates).
(G) Similar targeting of PIF1 to PBSs (PIL2, At1g16850, and At4g32300) in both wild-type and abi5-3 mutant seeds. This ChIP assay used an anti-PIF1 antibody on imbibed phyBoff seeds. Error bars indicate sd (n = 2 biological replicates).
(H) Similar enrichment of PIL2 promoter by ABI5 in both pif1 and pil2-sl158 mutant seeds. This ChIP assay used an anti-ABI5 antibody in imbibed phyBoff seeds. P0, P1, P2, Em6, and PP2A indicate ChIP amplicons shown in Figure 3A. Error bars indicate sd (n = 3 biological replicates).
ABI5 Enhances Binding of PIF1 to PBSs
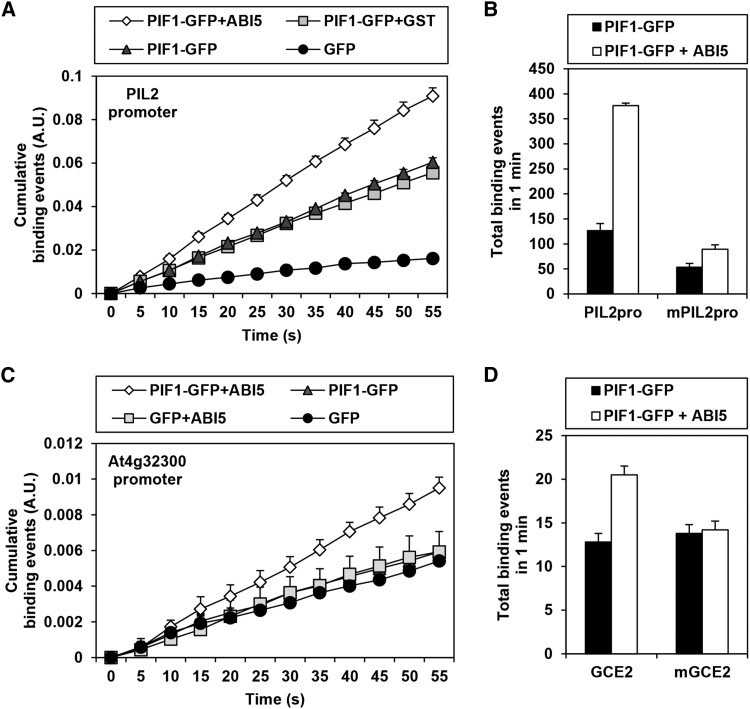
According to the hypothesis being tested by this study, PTFs bridge PIF1 and nearby GCEs, thereby enhancing the binding of PIF1 to PBSs. To examine the feasibility of this part of the hypothesis, we performed real-time single-molecule fluorescence imaging (Lee et al., 2013). We immobilized biotinylated PIL2 promoter fragments, which possess three G-boxes, on a slide. We then used total internal reflection fluorescence (TIRF) microscopy to observe real time approach events of PIF1-GFP at the slide surface in the presence or absence of ABI5. We counted GFP fluorescence spikes as binding events of PIF1-GFP to the immobilized promoter fragments and charted cumulative binding events over time. As expected, PIF1-GFP binding events occurred significantly more often than binding events for GFP alone because PIF1-GFP binds the immobilized PIL2 promoter fragments. Consistent with a role for ABI5 as a PTF, PIF1-GFP binding events occurred even more often in the presence of ABI5 but not GST (Figure 7A; Supplemental Figure 6). The G-boxes of the PIL2 promoter are necessary for the binding as the frequency of PIF1-GFP binding events is significantly reduced when they are absent (Figure 7B). We also asked whether ABI5 can enhance the binding of PIF1-GFP to the At4g32300 promoter fragment, which possesses only one GCE2. According to our EMSA results, ABI5 but not PIF1 binds GCE2s (Figure 4A; Supplemental Figure 7). Consistent with this, PIF1-GFP only binds the At4g32300 promoter fragment in the presence of ABI5 (Figure 7C) and the binding of PIF1-GFP is lost if the GCE2 is mutated in the single-molecule imaging assay (Figure 7D). Together, our data support a function for ABI5 as a PTF that stabilizes the binding of PIF1 to a subset of PBSs.
Figure 7.
ABI5 Enhances PIF1 Binding to PBSs.
(A) Real-time single-molecule fluorescence imaging analysis shows that ABI5 enhances PIF1 binding to a PIL2 promoter fragment containing three G-boxes. Promoter fragments are immobilized on a slide and binding events are detected by TIRF as GFP molecules approach the surface. Error bars indicate se (n = 3 locations).
(B) The enhancement of PIF1 binding to the PIL2 promoter fragment by ABI5 requires intact G-boxes. PIL2pro indicates immobilized PIL2 promoter fragments containing three intact G-boxes, while mPIL2pro indicates the same fragment with three mutated G-boxes. Error bars indicate se (n = 3 locations).
(C) ABI5 enhances PIF1 binding to an At4g32300 promoter fragment containing a GCE2. Error bars indicate se (n = 3 locations).
(D) The enhancement of PIF1 binding to the At4g32300 promoter fragment by ABI5 requires an intact GCE2. GCE2 indicates immobilized promoter fragments containing an intact GCE2, while mGCE2 indicates the same fragments with a mutated GCE2. Error bars indicate se (n = 20 locations).
DISCUSSION
PIFs bind to G-box elements in vitro, but genome-wide binding analyses using either the ChIP-chip or ChIP-seq techniques reveal a pattern in which in vivo PIF targeting is restricted to a group of promoters, some of which contain G-boxes while others do not (Martínez-García et al., 2000; Huq and Quail, 2002; Toledo-Ortiz et al., 2003; Huq et al., 2004; Oh et al., 2007, 2009, 2012; Shin et al., 2007; Leivar et al., 2008b; Hornitschek et al., 2009, 2012; Zhang et al., 2013; Pfeiffer et al., 2014). In our previous report, we found that PIF1 binds 748 sites in imbibed seeds, only 438 of which possess at least one G-box (Oh et al., 2009). In another study, PIF1 binds 3016 sites in dark-grown seedlings, only 1199 of which sites possess at least one G-box (Pfeiffer et al., 2014). This study is our attempt to determine both how PIF binding is restricted to a subset of the 14,545 G-boxes in the Arabidopsis genome and how PIFs bind promoters that do not contain G-boxes. We show that PIF1 binding sites with and without G-boxes are enriched with sequences we call GCEs (Table 1). Rather than binding PIF1 directly, we expected that these GCEs bind PTFs that themselves mediate the interaction with PIF1. We found that ABI5 and other group A bZIP proteins bind ACGT-containing GCEs and interact with PIF1 protein both in vitro and in vivo (Figure 5). These sorts of interactions would stabilize the targeting of PIF1 to specific sites in vivo.
Consistent with this hypothesis, we have shown that the separation of promoter elements by T-DNA insertion disrupts the in vivo targeting of PIF1 to these PBSs (Figure 3). We have also shown that ABI5 enhances the targeting of PIF1 to a subset of PBSs and, via single molecule fluorescence imaging (Figure 7), that ABI5 strengthens PIF1 binding to PBSs. Therefore, our working model is as follows: PBSs are enriched with G-boxes and GCEs. PIF1 binds directly to G-boxes, while PTFs bind directly to GCEs. Since the PTFs interact with PIF1, their presence stabilizes the binding of PIF1 to PBSs in vivo (Figure 1).
There are precedents for this sort of cooperative binding of two transcription factors to shared target sites in plants. The MADS box proteins, which are important for floral organ and ovule development, are a classic example (Theissen and Saedler, 2001). During ovule development, two MADS box proteins, SEPALLATA3 (SEP3) and SEEDSTICK (STK), heterodimerize. Then, they presumably form a tetrameric complex that binds CArG boxes in the VERDANDI (VDD) promoter (Mendes et al., 2013). Of the three CArG boxes in the VDD promoter, the SEP3-STK complex binds preferentially to CArG box1 and box3, which are separated by a 437-bp loop. When one of the three CArG boxes is mutated, the SEP3-STK complex binds the remaining two CArG boxes. If two of the three CArG boxes are mutated, however, SEP3-STK binding is abolished. This indicates the interaction between SEP3 and STK with at least two of their cognate binding sites is necessary for in vivo targeting of these proteins to the VDD promoter. This is very similar to what we have proposed for PIF1-PTF interactions and G-box-GCE targeting. Studies have also shown that PIF1 interacts with ABI3, BZR1, FHY3, HY5, TOC1, and VQ29 (Yamashino et al., 2003; Park et al., 2011; Oh et al., 2012; Tang et al., 2012; Chen et al., 2013; Li et al., 2014). PIF3 interacts with HY5 and TOC1 (Yamashino et al., 2003; Chen et al., 2013), and PIF4 interacts with AUXIN RESPONSE FACTOR6 (ARF6), BZR1, CO, LEC1, and TOC1 (Yamashino et al., 2003; Oh et al., 2014; Huang et al., 2015; Fernandez et al., 2016). The interactions between PIFs and other transcription factors mean multiple transcription factors target the same promoters with additive, synergistic, or antagonistic effects on target gene expression. Some interactions even permit cooperative binding. While PIF4 enhances the binding of ARF6 to the SAUR15 promoter (Oh et al., 2014), ABI3 does not affect PIF1 binding to the SOMNUS promoter (Park et al., 2011). HY5 does not affect PIF3 binding to the F3H promoter (Shin et al., 2007), but HY5 does induce cooperative of PIF3 to NDB2 and other ROS-related gene promoters (Chen et al., 2013). HY5 also competes in vitro with PIF3 for binding to G-boxes (Toledo-Ortiz et al., 2014). Further experiments will be required to determine the extent to which other interacting transcription factors also act as PTFs that affect PIF targeting in vivo.
The direct binding of ABI5 to a subset of PIF1-dependent promoters suggests that PIF1 regulates a subset of its target genes via a coherent feed-forward loop. In imbibed seeds, PIF1 either directly or indirectly activates the expression of six group A bZIP transcription factors (i.e., ABI5, ABF4, AREB3, GBF4, EEL, and bZIP13) (Oh et al., 2009), which then target and regulate a subset of PIF1-dependent promoters. In other words, both PIF1 and the PIF1-dependent group A bZIPs regulate a subset of PIF1-dependent genes. Signaling pathways that include such coherent feed-forward loops are highly robust (Le and Kwon, 2013), and this is not the only such PIF1-dependent loop in imbibed seeds. PIF1 also activates the genes necessary for ABA biosynthesis, increasing the level of ABA in imbibed seeds (Oh et al., 2007, 2009). ABA signaling, in turn, induces the posttranslational activation of group A bZIPs, completing another feed-forward loop (Lopez-Molina et al., 2001; Fujii et al., 2007; Piskurewicz et al., 2008; Nakashima et al., 2009; Sirichandra et al., 2010). PIF1 also reduces the level of GA in imbibed seeds and directly activates the expression of two DELLAs (GAI and RGA), increasing the level of all DELLA proteins, including RGL2 (Oh et al., 2006, 2007, 2009). Since DELLAs also increase the level of ABA, PIF1’s regulation of GA and the DELLAs completes a third feed-forward loop (Piskurewicz et al., 2008, 2009). Since ABA and GA levels depend on environmental conditions (e.g., temperature, salt, and water availability), the coherent feed-forward loops we have identified that link PIF1, GA, ABA, the DELLAs, and the group A bZIPs form a focal point in the integration of the environmental conditions that regulate the germination of imbibed seeds (Olszewski et al., 2002; Nambara and Marion-Poll, 2005).
Though our main analysis is limited to the targeting of PIF1 in imbibed seeds, PIF1 targeting in seedlings is also probably facilitated by similar PTF/GCE interactions. Indeed, a hexamer analysis indicates that both G-box and non-G-box PIF1 binding sites in seedlings are also enriched with GCE1, GCE2, and GCE3 sequences (Supplemental Data Set 1). Similarly, the targeting of other PIFs may also be facilitated by PTF/GCE interactions. Similar hexamer analyses on other PIFs binding sites uncovered enriched sequences similar to the GCEs identified in this study (Supplemental Data Set 1). The top 10 enriched hexamers among G-box-containing PIF5 binding sites are additional G-boxes, three GCE1s, and three GCE2s. G-box-containing PIF3 and PIF4 binding sites are also enriched with these hexamers, suggesting that the in vivo targeting of other PIFs is facilitated by PTF/GCE interactions. The non-G-box binding sites of PIF3, PIF4, and PIF5 are also enriched with GCE1, GCE2, and GCE3, but only GCE3 appears in the top ten lists. Also included in the top 10 list, however, are hexamers containing an ATGGG core (Supplemental Data Set 1), suggesting that these elements play more dominant roles in targeting the other PIFs to non-G-box binding sites in seedlings. It is interesting to note that the ATGGG sequence is identical to a core sequence of the PHYTOCHROME A (PHYA) promoter proposed to mediate the light-induced repression of PHYA expression (Bruce et al., 1991; Dehesh et al., 1994). Genome-wide binding assays have also shown that PIFs target a region of the PHYA promoter that contains the ATGGG sequences (Oh et al., 2009, 2012; Hornitschek et al., 2012; Pfeiffer et al., 2014). It is not yet clear which transcription factors bind these ATGGG-containing sequence elements, but it seems likely that they will also act as PTFs.
Our hexamer analyses identified enriched sequences other than the GCEs we have already discussed that may also be physiologically meaningful. For example, TGTCGT is the 7th most enriched hexamer in non-G-box PIF1 binding sites in imbibed seeds (Table 1). This sequence shares an AuxRE core (TGTC), which is the binding site for the ARF transcription factors (Ulmasov et al., 1999; Boer et al., 2014). TGTC-containing sequences are enriched in the binding sites for other PIFs, but they do not appear in the top 10 lists (Supplemental Data Set 1). The TGTC-containing sequence CATGTC, for example, is enriched 1.93-fold in PIF4 non-G-box binding sites (Supplemental Data Set 1). A recent study showed that ARF6 interacts with PIF4 to promote hypocotyl elongation (Oh et al., 2014). It is unclear if ARFs serve as PTFs facilitating PIF targeting to specific sites, but the direct ARF6-PIF4 interaction and the enrichment of hexamer sequences that share the core AuxRE suggest as much (Oh et al., 2014). In addition, the ABI3 binding RY motif (CATGCA) also appears on one of the lower ranking lists of enriched sequences (Supplemental Data Set 1), and ABI3 is known to interact with PIF1 (Mönke et al., 2004; Park et al., 2011). We expect that a more thorough analysis of the rest of the minor enriched sequences will reveal various combinations of coupling elements and interacting transcription factors.
Finally, it should be noted that in vivo transcription factor targeting is determined not only by the binding of the transcription factors to coupling elements but also by means of higher-level chromatin structures. In vivo, eukaryotic DNA is packed into chromatin, arranged in repeating nucleosomal units by histone proteins. Transcription factor targeting can be affected by the degree of chromatin compaction, which differs from region to region depending on things like DNA sequence, DNA methylation, histone modifications, and transcriptional status (Kouzarides, 2007; Li et al., 2007; Pfluger and Wagner, 2007; Liu et al., 2010). Regions of heterochromatin, which have high levels of H3K27me3, are highly compact and less accessible to many transcription factors and so are characterized by low transcriptional activity. Regions of euchromatin, on the other hand, have low levels of H3K27me3, are less compact, and are more accessible, so they are characterized by high transcriptional activity (Huisinga et al., 2006; Grewal and Jia, 2007; Liu et al., 2010). Even within euchromatic regions, nucleosome-free regions allowing even greater transcription factor accessibility (Bi et al., 2004; Oki and Kamakaka, 2005). These nucleosome-free regions are actively regulated by chromatin remodeling complexes and can thus change with changing conditions (Bi et al., 2004; Oki and Kamakaka, 2005). Studies have shown that light alters the chromatin status of both light-induced and -repressed loci (Crosio et al., 2000; Tessadori et al., 2009; van Zanten et al., 2010; Barneche et al., 2014). Thus, although it is currently unclear whether this light-dependent chromatin modification requires or induces PIF binding, it is possible.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana plants used in this study were in the Col-0 background and grown at 22 to 24°C in a growth room with a 16-h-light/8-h-dark cycle. T-DNA insertion mutants were obtained from the Arabidopsis stock centers (TAIR and NASC): pil2-sl158 (sail_158_H03), boi-gb012 (gabi_012H08), and fhy3 (Salk_002711). The abi5-3 mutant was kindly provided by Ruth Finkelstein (Finkelstein and Lynch, 2000). The pif1 aba2 double mutant was generated by combining the pif1 (pil5-1) and aba2-1 mutants. 35S-driven PIF1-MYC and ABI5-FLAG transgenic lines correspond to the previously described PIL5-OX3 (Oh et al., 2007) and ABI5-FLAG1 (Lim et al., 2013) lines, respectively.
Seed Light Treatment
Seeds were surface-sterilized and plated on 0.6% phytoagar without sucrose. Within 1 h after the start of surface sterilization, phytochromes were either inactivated (phyBoff condition) or activated (phyBon) by monochromatic light treatment and incubated in the dark for 12 h for molecular analysis and 4 d for germination assays. For the phyBoff condition, plated seeds were irradiated with far-red light (2.59 µmol m−2 s−1) for 5 min. For the phyBon condition, plated seeds were irradiated with red light (11.5 µmol m−2 s−1) for 5 min followed by the far-red pulse.
Gene and Protein Expression Analysis
Seeds were incubated in the dark for 12 h after light irradiation (phyBoff or phyBon condition) and collected for gene or protein expression analysis. For gene expression analysis, total RNAs were extracted using the Spectrum plant total RNA kit (Sigma-Aldrich). The transcript levels of interest were determined by real-time PCR using specific primer sets (Supplemental Table 2) and normalized with that of PP2A. For protein analysis, seeds were frozen in liquid nitrogen and ground very fine using a mortar and pestle. Total proteins were extracted with denaturing buffer (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M urea, pH 8.0) and the debris was removed by centrifugation. The extracted proteins were further denatured by boiling at 100°C for 5 min. The protein levels were detected by immunoblot analysis using an anti-PIF1 antibody, anti-ABI5 antibody, anti-MYC antibody (2276S, lot number 24; Cell Signaling), anti-FLAG antibody (F7425; Sigma-Aldrich), anti-α-tubulin antibody (T5168; Sigma-Aldrich), or anti-RPT5 antibody (Biomol).
ChIP and EMSA
ChIP assays were performed as described by Oh et al. (2007). Seeds were incubated in the dark for 12 h after light irradiation (phyBoff) and then cross-linked in 1% formaldehyde under vacuum for 1 h in the dark. The seeds were collected and finely ground in liquid nitrogen. The chromatin complexes were isolated and sonicated with a Bioruptor (30 s on/30 s off cycles and high-power output) to obtain 200- to 600-bp DNA fragments. Protein-DNA complexes were immunoprecipitated using the same antibodies as for the protein expression analysis. The cross-linking was then reversed and the enrichment of DNA fragments was determined by real-time PCR using specific primer sets (Supplemental Table 2).
EMSA assays were performed as described by Oh et al. (2007) using recombinant proteins (MBP-fused ABI5 and EEL, GST-fused N-terminal FHY3 [200 amino acids], and 6X-HIS fused PIF1) and biotinylated probes (Supplemental Table 2). Biotinylated probes were used at a concentration of 15 to 20 ng/μL and nonbiotinylated probes at 5 to 100 times that of the biotinylated probes. Each protein was mixed with probes in binding buffer (10 mM Tris, 50 mM KCl, and 1 mM DTT, pH 7.5) with 100 ng/μL poly (dI⋅dC), 0.05% Nonidet P-40, and 5 mM MgCl2. The buffers for GST-FHY3N and PIF1-HIS also included 2.5% glycerol, 0.25 mM EDTA, and 100 µM ZnCl2. The bound complexes were resolved on a 6% polyacrylamide gel and transferred to a nylon membrane (Amersham Hybond N+) before being UV-cross-linked at 120 mJ/cm2. The results were detected by a chemiluminescent nucleic acid detection module kit (Pierce).
In Vitro Binding Assay
GST-fused PIF1 was purified by cloning the PIF1 cDNA into a pET42 vector (Novagene). MBP-fused group A bZIPs (ABI5, ABF2, ABF3, EEL, DPBF2, and AREB3) were purified by cloning cDNAs into a pMAL-c2X vector (New England Biolabs). For in vitro binding assays, each resin-bound MBP-fused protein was incubated with purified GST-PIF1 in a binding buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 10% glycerol, 50 µM MG132, and protease inhibitor cocktail) for 4 h at 4°C. After the incubation, MBP beads were collected by centrifugation and washed three times with 500 μL binding buffer. Bound proteins were eluted by boiling with SDS sample buffer and detected by immunoblot analysis using an anti-GST antibody (sc-138; Santa Cruz Biotechnology) and an anti-MBP antibody (sc-809; Santa Cruz Biotechnology).
In Vivo Binding Assay
Four-day-old dark-grown PIF1-MYC and PIF1-MYC/ABI5-FLAG seedlings were sampled and ground fine in liquid nitrogen. To make cell extracts, two grams of the ground samples were solubilized in 2 mL lysis buffer (50 mM Tris-HCl, pH 7.5, 75 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 5% glycerol, 1 mM PMSF, 50 µM MG132, and protease inhibitor cocktail), sonicated, and centrifuged at 20,000g at 4°C for 10 min to remove debris. FLAG-tagged ABI5 in the cell extracts was precipitated with anti-FLAG antibody-conjugated resin (F2426; Sigma-Aldrich). Coprecipitated PIF1-MYC was detected by immunoblot analysis using an anti-MYC antibody.
BiFC Assay
ABI5 and PIF1 were cloned into pBiFCt-2in1-CC (Grefen and Blatt, 2012), while PHYB, COP1, CBL1, and CIPK23 were cloned into pBiFCt-2in1-CN vectors. Each construct was transformed into Nicotiana benthamiana leaf epidermal cells by agroinfiltration. Three days after the infiltration, YFP fluorescence signals were observed with an Olympus BX51 fluorescence microscope. Since pBiFCt-2in1 vectors possess 35S-driven RFP as a transfection control, YFP signals were observed only in epidermal cells emitting RFP fluorescence.
Real-Time Single-Molecule Fluorescence Imaging Analysis
HIS-fused PIF1-GFP was purified by cloning PIF1-GFP into the pET29a vector. ABI5 was also purified by cloning into the pET29a vector. Biotinylated PIL2 promoter fragments (426 bp) were generated by amplification using a biotinylated primer set (Supplemental Table 2). For the At4g32300 promoter fragments, a biotinylated double-stranded oligomer (53 bp) covering GCE2 was synthesized. Imaging chambers were constructed on a quartz slide cleaned with piranha solution. The surface of the slide was coated with methoxypolyethylene glycol:biotin-PEG (Laysan Bio) (100:1 [mol/mol]) and 0.1 mg/mL neutravidin (Invitrogen) was attached for 5 min. DNA molecules were immobilized using the neutravidin-biotin interaction for 5 min, and 1 nM PIF1-GFP with 1 μM ABI5-His or GST-His protein was used for single-molecule imaging. TIRF microscopy, which excites fluorescent proteins within 100 nm of the surface, was used to visualize GFP using 473-nm lasers. An electron-multiplying CCD (iXon DU897v; Andor Technology) was used to produce videos of GFP binding events. The 7×7-pixel gaussian mask fitting was used to determine single-molecule signals and total intensity (Thompson et al., 2002). To distinguish GFP binding and unbinding events, fluorescence intensity steps were detected from the recorded intensity traces using a model-independent nonuniform step detection method (Kalafut and Visscher, 2008). Signals were measured from at least three different locations. Two independent experimental data sets using independently purified recombinant proteins and independently prepared slides are presented in Figure 7 and Supplemental Figure 6.
Hexamer Analysis and KS Test
Overrepresented hexamers were identified using the Oligo-analysis method (van Helden et al., 1998). Oligo-analysis calculates the probability of observing a given oligonucleotide using a binomial distribution model. We compared the occurrences of overrepresented hexamers in PBSs and the whole Arabidopsis genome. PBSs were defined as 500-bp sequences surrounding PIF1 binding peaks, as described by Oh et al. (2009). For PIF3, PIF4, and PIF5, and seedling PIF1 binding sites where ChIP-seq instead of ChIP-chip methods were used, PBSs were defined as 200 bp surrounding the PIF binding peak. To exclude false positives, we used two-sample KS tests implemented in R programming language. The null hypothesis of these two-sample KS tests is that the distribution in the two samples is the same. Low P values for a specific motif in the KS tests mean that the distribution of that motif is different from the distribution of G-boxes, suggesting that it is likely a false positive. We defined the threshold for valid motifs P ≤ 0.01.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PIF1 (AT2G20180), ABI5 (AT2G36270), ABF2 (AT1G45249), ABF3 (AT4G34000), EEL (AT2G41070), DPBF2 (AT3G44460), AREB3 (AT3G56850), ABI3 (AT3G24650), SOM (AT1G03790), ATAF1 (AT1G01720), FHY3 (AT3G22170), PIL2 (AT3G62090), BOI (AT4G19700), HY5 (AT5G11260), HFR1 (AT1G02340), EM6 (AT2G40170), ABA2 (AT1G52340), PHYB (AT2G18790), COP1 (AT2G32950), CIPK23 (AT1G30270), CBL1 (AT4G17615), PP2A (AT1G13320), At4g31390, At1g16850, At2g38465, At4g32300, At4g11040, At2g32970, At3g61160, At1g15120, and At2g42430. ChIP-chip and ChIP-seq data used in this article are GSE14450 (seed PIF1), GSE39217 (PIF3), GSE43286 (seedling PIF1 and PIF4), and GSE35062 (PIF5). Microarray data used in this article are GSE15700 (wild type/aba2-1) and GSE14374 (wild type/pif1).
Supplemental Data
Supplemental Figure 1. T-DNA insertions do not affect PIF1 binding to independent loci.
Supplemental Figure 2. FHY3 protein interacts with PIF1 and binds GCE1s.
Supplemental Figure 3. pif1 mutant but not fhy3 and hy5 mutant seeds germinate in the phyBoff condition.
Supplemental Figure 4. PIF1 and ABA coregulate a subset of genes in imbibed seeds.
Supplemental Figure 5. Diagrams showing various loci (upper panels), PIF1 binding signals from ChIP-chip assays (lower panels), PBS sequence elements, and ChIP-PCR amplicons.
Supplemental Figure 6. Another data set showing ABI5 enhances PIF1 binding to PBSs from Figure 7.
Supplemental Figure 7. PIF1 binds to the G-box but not GCEs in vitro.
Supplemental Table 1. Classification of PIF1 target genes based on GCEs and expressional direction.
Supplemental Table 2. List of primers
Supplemental Data Set 1. Hexamer analyses of PIFs binding sites.
Supplementary Material
Acknowledgments
We thank TAIR and NASC for providing information and mutant seeds. This work was supported in part by grants from the National Research Foundation of Korea (2015R1A2A1A05001091 and 2011-0031955) and the Rural Development Administration (SSAC-PJ011073) to G.C.
AUTHOR CONTRIBUTIONS
Ju.K., H.K., J.P., T.Y., and G.C. designed experiments. Ju.K., H.K., J.P., W.K., J.Y., N.L., and Ja.K. performed experiments. Ju.K. and G.C. wrote the article.
Glossary
- ABA
abscisic acid
- GA
gibberellic acid
- PBE
PIF binding E-box
- ChIP
chromatin immunoprecipitation
- EE
evening element
- PBS
PIF1 binding site
- NBS
nonbinding site
- PTF
PIF1-interacting transcription factor
- GCE
G-box coupling element
- KS
Kolmogorov-Smirnov
- EMSA
electrophoretic mobility shift assay
- BiFC
bimolecular fluorescence complementation
- TIRF
total internal reflection fluorescence
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Barneche F., Malapeira J., Mas P. (2014). The impact of chromatin dynamics on plant light responses and circadian clock function. J. Exp. Bot. 65: 2895–2913. [DOI] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J.J., Zou Y. (2004). Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24: 2118–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., Weijers D., Coll M. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Bruce W.B., Deng X.W., Quail P.H. (1991). A negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J. 10: 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., Gentleman R.C., Tapscott S.J. (2010). Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C., Bies-Etheve N., Aspart L., Léon-Kloosterziel K.M., Koornneef M., Echeverria M., Delseny M. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30: 373–383. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Ang L.H., Puente P., Deng X.W., Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., Lin R. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C., Cermakian N., Allis C.D., Sassone-Corsi P. (2000). Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 3: 1241–1247. [DOI] [PubMed] [Google Scholar]
- Dehesh K., Franci C., Sharrock R.A., Somers D.E., Welsch J.A., Quail P.H. (1994). The Arabidopsis phytochrome A gene has multiple transcription start sites and a promoter sequence motif homologous to the repressor element of monocot phytochrome A genes. Photochem. Photobiol. 59: 379–384. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez V., Takahashi Y., LeGourrierec J., Coupland G. (2016). Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J., http://dx.doi.org/10.1111/tpj.13183. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S., Lynch T.J., Thomas T.L., Rock C.D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59: 253–267. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086. [DOI] [PubMed] [Google Scholar]
- Gordân R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L. (2013). Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Reports 3: 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2012). A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314. [DOI] [PubMed] [Google Scholar]
- Grewal S.I.S., Jia S. (2007). Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., et al. (2004). Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., Lopez-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang M., Hu Y., Liu X., Li Y., Hou X. (2015). Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27: 3099–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K.L., Brower-Toland B., Elgin S.C. (2006). The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma 115: 110–122. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafut B., Visscher K. (2008). An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput. Phys. Commun. 179: 716–723. [Google Scholar]
- Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemian M., Pham H., Wolfe S.A., Brodsky M.H., Sinha S. (2013). Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 41: 8237–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Shin J., Lee S.H., Kweon H.S., Maloof J.N., Choi G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 108: 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Ma J., Perret P., Li Z., Thomas T.L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Le D.H., Kwon Y.K. (2013). A coherent feedforward loop design principle to sustain robustness of biological networks. Bioinformatics 29: 630–637. [DOI] [PubMed] [Google Scholar]
- Lee H.W., Ryu J.Y., Yoo J., Choi B., Kim K., Yoon T.Y. (2013). Real-time single-molecule coimmunoprecipitation of weak protein-protein interactions. Nat. Protoc. 8: 2045–2060. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26: 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008a). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008b). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Li G., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622. [DOI] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jing Y., Li J., Xu G., Lin R. (2014). Arabidopsis VQ MOTIF-CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME-INTERACTING FACTOR1. Plant Physiol. 164: 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., Kim J., Kang H., Kim D.H., Kawakami N., Choi G. (2013). ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E. (2006). An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- Mazzoni E.O., Mahony S., Closser M., Morrison C.A., Nedelec S., Williams D.J., An D., Gifford D.K., Wichterle H. (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16: 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes M.A., Guerra R.F., Berns M.C., Manzo C., Masiero S., Finzi L., Kater M.M., Colombo L. (2013). MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 25: 2560–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G., Altschmied L., Tewes A., Reidt W., Mock H.P., Bäumlein H., Conrad U. (2004). Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219: 158–166. [DOI] [PubMed] [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M., Kamakaka R.T. (2005). Barrier function at HMR. Mol. Cell 19: 707–716. [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Nguyen K.T., Park E., Jeon J.S., Choi G. (2013). DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J., Wagner D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28: 2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Nagatani A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10: 859–868. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.Y., Kim J., Paek N.C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5: 4636. [DOI] [PubMed] [Google Scholar]
- Schwartz S.H., Léon-Kloosterziel K.M., Koornneef M., Zeevaart J.A. (1997). Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994. [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Davanture M., Turk B.E., Zivy M., Valot B., Leung J., Merlot S. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One 5: e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., et al. (2008). DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol. 146: 1862–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F., et al. (2009). Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471. [DOI] [PubMed] [Google Scholar]
- Thompson R.E., Larson D.R., Webb W.W. (2002). Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82: 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M., Halliday K.J. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19: 309–319. [DOI] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97: 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., André B., Collado-Vides J. (1998). Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281: 827–842. [DOI] [PubMed] [Google Scholar]
- van Zanten M., Tessadori F., McLoughlin F., Smith R., Millenaar F.F., van Driel R., Voesenek L.A.C.J., Peeters A.J.M., Fransz P. (2010). Photoreceptors CRYTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol. 154: 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. (2003). A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629. [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., et al. (2009). High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.