A metabolic gene cluster underlying the known glaucousness loci W1 in wheat and Cer-cqu in barley establishes a novel molecular pathway for β-diketone wax biosynthesis
Abstract
The glaucous appearance of wheat (Triticum aestivum) and barley (Hordeum vulgare) plants, that is the light bluish-gray look of flag leaf, stem, and spike surfaces, results from deposition of cuticular β-diketone wax on their surfaces; this phenotype is associated with high yield, especially under drought conditions. Despite extensive genetic and biochemical characterization, the molecular genetic basis underlying the biosynthesis of β-diketones remains unclear. Here, we discovered that the wheat W1 locus contains a metabolic gene cluster mediating β-diketone biosynthesis. The cluster comprises genes encoding proteins of several families including type-III polyketide synthases, hydrolases, and cytochrome P450s related to known fatty acid hydroxylases. The cluster region was identified in both genetic and physical maps of glaucous and glossy tetraploid wheat, demonstrating entirely different haplotypes in these accessions. Complementary evidence obtained through gene silencing in planta and heterologous expression in bacteria supports a model for a β-diketone biosynthesis pathway involving members of these three protein families. Mutations in homologous genes were identified in the barley eceriferum mutants defective in β-diketone biosynthesis, demonstrating a gene cluster also in the β-diketone biosynthesis Cer-cqu locus in barley. Hence, our findings open new opportunities to breed major cereal crops for surface features that impact yield and stress response.
INTRODUCTION
The extracellular matrix glazing aerial plant organs, the cuticle, serves as an interface with the environment and thus strongly contributes to fitness in unceasingly varying growth conditions. The lipophilic cuticular layer and epicuticular depositions contain complex lipid mixtures “tailored” toward specific roles in development, stress responses, and interactions with other organisms, and thus varying greatly between species, organs, and growth stages. The polyester cutin, a matrix of cross-linked hydroxy C16 and/or C18 fatty acids, forms a major constituent of the cuticle (Yeats and Rose, 2013). The cutin scaffold holds the cuticular waxes, typically complex mixtures of very-long-chain fatty acid (C20-C34) derivatives (Samuels et al., 2008; Yeats and Rose, 2013). The wax compounds common to all plants are formed in three major stages within epidermal cells, starting with the synthesis of C16 fatty acid in plastids, then by further elongation to C20-C34 fatty acids in the endoplasmic reticulum (ER), and finally modification either along a wax alkane-forming pathway or an alcohol-forming pathway (Supplemental Figure 1). Plastidial elongation to C16 depends on the fatty acid synthase (FAS) systems catalyzing four reactions, initially forming a β-ketoacyl-ACP (acyl carrier protein) intermediate and then involving the stepwise conversion of the keto function into CH2. Similarly, the further elongation to very-long-chain fatty acids involves fatty acid elongase systems carrying out analogous reactions, albeit with acyl-CoA intermediates.
In the last decade, major discoveries have greatly advanced our genetic and biochemical understanding of cuticle biosynthesis and assembly, the spatial and temporal transcriptional control of associated metabolic pathways, and, to some extent, structure-function relationships (Yeats and Rose, 2013; Borisjuk et al., 2014; Hen-Avivi et al., 2014). Nevertheless, as most of the work on cuticle biology has been performed in model plant species such as Arabidopsis thaliana, maize (Zea mays), tomato (Solanum lycopersicum), and rice (Oryza sativa), the unique surface features of many other species or families remain poorly understood. In particular, many Poaceae species (including wheat [Triticum spp], barley [Hordeum vulgare], rye [Secale cereale], and oat [Avena sativa]) produce very-long-chain β-diketones with characteristic structures likely formed along pathways different from those leading to more ubiquitous wax compounds (Supplemental Figure 1). Despite many years of research on β-diketone biosynthesis, particularly in barley and wheat (von Wettstein-Knowles, 1976, 1995; von Wettstein-Knowles and Søgaard, 1980; Adamski et al., 2013; Zhang et al., 2013, 2015), the genes and enzymes involved remain to be discovered.
Glaucousness, the light bluish-gray appearance of flag leaf, stem, and spike surfaces due to wax crystals deposited on the surface, is a morphological trait that varies genetically between different plant species. The opposite form is known as glossy (gl), nonglaucous, eceriferum (cer), bloomless (bm), or waxless. Interestingly, the vast majority of the modern wheat (Triticum aestivum) cultivars exhibit a glaucous appearance due to the presence of β-diketones, while both glaucous and glossy phenotypes are found in wild wheat species. Early genetic studies revealed that this trait in hexaploid wheat is predominantly governed by four loci, namely, the wax production loci W1 and W2 and the wax inhibition loci Iw1 and Iw2. W1 and Iw1 reside on the short arm of chromosome 2B (2BS), whereas W2 and Iw2 are located on 2DS (Tsunewaki, 1962; Tsunewaki and Ebana, 1999). Either one of the wax production loci, W1 or W2, is sufficient to produce a glaucous phenotype. The wax inhibitor loci have a dominant epistatic interaction with the wax production loci, and the presence of either Iw1 or Iw2 (or both) suppresses the glaucous phenotype.
Barley is a diploid cereal that produces β-diketones as wax constituents and can therefore serve as an excellent complementary study system together with wheat. Previous genetic work in barley demonstrated that the β-diketone biosynthesis-associated Cer-cqu locus resides in a subtelomeric region on the short arm of chromosome 2H (2HS) (Takahashi et al., 1953; Tsuchiya, 1972; Schondelmaier et al., 1993), in a syntenic position to the wheat W1 locus (Tsunewaki and Ebana, 1999; Varshney et al., 2006). Extensive genetic and biochemical studies in barley proposed a putative β-diketone biosynthesis pathway, which includes three enzymatic steps (von Wettstein- Knowles, 1995, 2012). Two of these were hypothesized to be catalyzed by type-III polyketide synthase (PKS) enzymes, Cer-c and Cer-q. The third step, β-diketone hydroxylation, was predicted to be catalyzed by Cer-u. It remained unclear whether β-diketone biosynthesis in barley is controlled by proteins encoded by three separate but very tightly linked genes (Cer-c, q, and u) or by a single, multifunctional protein with at least three domains (c, q, and u) encoded by a single Cer-cqu gene. The latter scenario was favored (von Wettstein-Knowles and Søgaard, 1981; von Wettstein- Knowles, 1995, 2012). Recently, Schneider et al. (2016) suggested that three different genes rather than one encoding a multifunctional enzyme are involved in the β-diketone biosynthesis pathway in barley.
In this work, we discovered a metabolic gene cluster in wheat and in barley comprising genes that are responsible for β-diketone biosynthesis and encode a PKS, a cytochrome P450, and, unexpectedly, a hydrolase/carboxylesterase. The identification and characterization of the β-diketone biosynthetic route as well as additional results reported here will promote innovative breeding schemes that target plant surface characteristics in major cereal crops.
RESULTS
Analysis of the Glaucousness Trait Using Chromosome-Arm Substitution Lines of Wild Emmer in Common Wheat
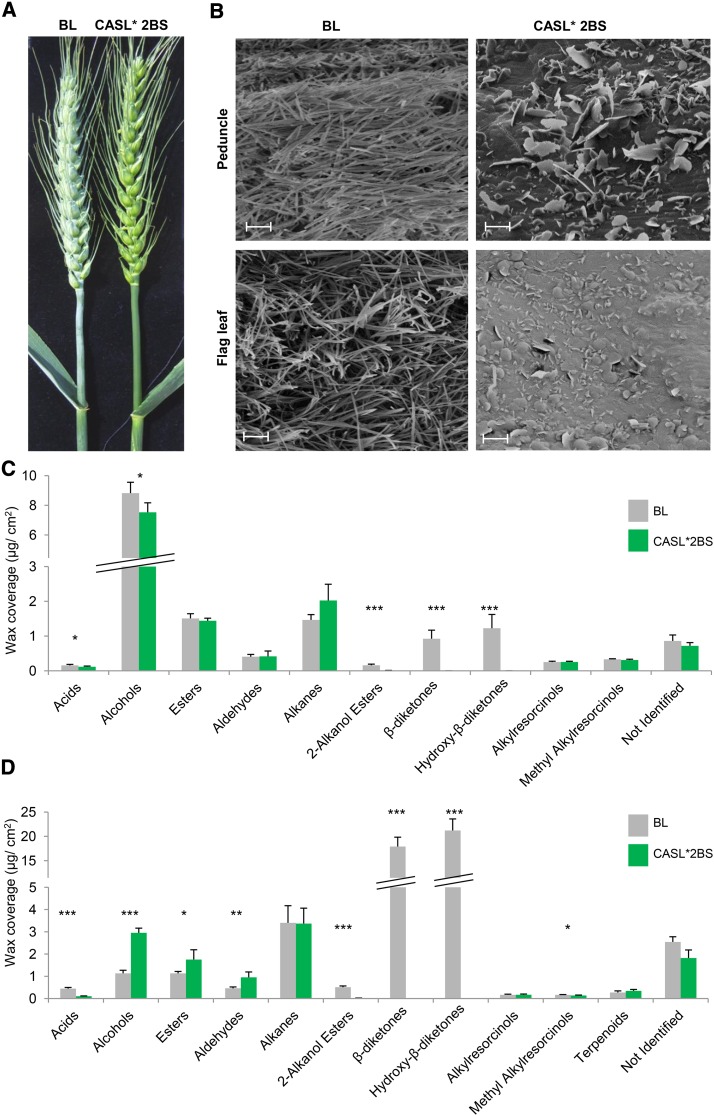
To study β-diketone waxes in wheat, we used a population of lines described by Millet et al. (2013), in which a series of chromosome-arm substitution lines (CASLs) of wild emmer Triticum turgidum ssp dicoccoides accession TTD140 (2n = 4x = 28; genome AABB) were produced in the background of the glaucous common bread wheat T. aestivum cultivar Bethlehem (BL) (2n = 6x = 42; genome AABBDD). One of the most notable observations was the glossy phenotype of CASL*2BS (the short arm of chromosome 2B of wild emmer in the background of common wheat), whereas all the other CASLs had a glaucous appearance. The parental genotype BL and the CASL*2BS were therefore selected to study cuticular wax biosynthesis and composition (for an illustration of lines, see Supplemental Figure 2). The glaucous phenotype of BL (therefore iw1/iw1) and the glossy phenotype of CASL*2BS (carrying Iw1/Iw1 alleles of TTD140; Rong et al., 2000) were noticeably visible in flag leaves, peduncles, leaf sheaths, and spikes (Figure 1A). Cryo-scanning electron microscopy revealed differences in the relative amounts and shape of epicuticular wax crystals between these genotypes (Figure 1B). While the flag leaves and peduncles of BL were covered with tubule-shaped crystals typical for β-diketone-rich wax (Bianchi and Figini, 1986; Jeffree, 2006), the CASL*2BS tissues accumulated platelet-shaped crystals characteristic of alcohol-rich wax (Jeffree, 2006) (Figure 1B). The W1 and Iw1 loci are known to play a role in wax biosynthesis and to be located on 2BS, suggesting that these two loci are the base of the different wax phenotypes.
Figure 1.
Differences in Wax Phenotypes between the Glaucous BL and the Glossy CASL*2BS Wheat Lines.
(A) BL is covered with bluish-gray, waxy material, while CASL*2BS is glossy.
(B) The epicuticular wax covering peduncle and flag leaf (abaxial side) tissues was examined using cryo-scanning electron microscopy. BL and CASL*2BS lines exhibit different wax amounts and crystals shapes. Tubule-shaped crystals predominate on BL, while crystals on CASL*2BS are mostly platelet-shaped. Magnification: 10 k. Bar = 2 µm.
(C) and (D) Chemical compositions of wheat BL and CASL*2BS wax mixtures on flag leaf blade (C) and flag leaf sheath plus peduncle (D) were determined by GC-MS analysis. Values indicate the mean ± sd of n ≥ 4 biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.005.
Surface Wax Analysis Reveals That the Glossy CASL*2BS Plants Lack β-Diketone, Its Hydroxy-β-Diketone Derivatives, and Related 2-Alkanol Esters
To determine the underlying reason for the visible differences in glaucousness between BL and the CASL*2BS, flag leaf blades and flag leaf sheaths plus peduncles of flowering plants were harvested and their cuticular waxes analyzed using gas chromatography with mass spectrometry (GC-MS) and flame ionization detection. CASL*2BS had significantly less total wax, reduced by 20 and 80% in flag leaf blade and flag leaf sheath/peduncle samples, respectively, compared with the equivalent wild-type BL tissues. Wax composition also differed between the genotypes, with β-diketone and hydroxy-β-diketones together making up 13 and 79% of total wax amounts in BL flag leaf blades and sheaths/ peduncles, respectively, while completely absent in the CASL*2BS (Figures 1C and 1D). In addition, only traces of 2-alkanol esters were detected in both CASL*2BS tissues, and free fatty acids were severely depleted in the CASL*2BS leaf sheaths. Conversely, alcohols, esters, and aldehydes accumulated to higher levels in leaf sheaths and peduncle of CASL*2BS, whereas these were low in these tissues of BL. In particular, the marked difference in β-diketone accumulation between the genotypes is in agreement with the accepted impact of β-diketones on glaucousness (Barber and Netting, 1968; Bianchi and Figini, 1986; Adamski et al., 2013; Zhang et al., 2013).
Transcriptome Analysis Implicates Three Genes in β-Diketone Production in Hexaploid Bread Wheat
In order to decipher the β-diketone biosynthesis pathway, transcriptome analysis was conducted in which we compared gene expression in peduncle tissues derived from the BL and CASL*2BS genotypes. Filtering for at least 2-fold changes in expression levels between the two genotypes revealed 628 and 316 transcripts that were more highly expressed in BL or CASL*2BS, respectively. Examination of annotations of these differentially expressed transcripts revealed that one of the genes most highly expressed in BL encodes a putative member of the type-III PKS enzyme family, with significant sequence similarity to curcuminoid synthase in turmeric (Curcuma longa; Ramirez-Ahumada et al., 2006). One of the best-known plant PKSs is chalcone synthase, which catalyzes the formation of naringenin chalcone in the flavonoid biosynthesis pathway (Ferrer et al., 1999; Jez et al., 2000). Interestingly, querying the PKS-like transcript against the available T. aestivum genome (International Wheat Genome Sequencing Consortium, 2014) database revealed the presence of matching genomic sequences on 2BS and 2DS, the chromosomal arms known to carry the W1 and W2 loci, respectively (Tsunewaki and Ebana, 1999; Table 1). Together with the evidence from previous biochemical studies in barley, which implicated type-III PKSs (termed pkKCS) in β-diketone formation (von Wettstein-Knowles, 2012), this PKS-like transcript was a prime candidate to be involved in wheat β-diketone biosynthesis.
Table 1. β-Diketone Biosynthesis Candidate Genes in Wheat and Barley.
| Gene | Transcript No. in Wheat Transcriptome Data | Expression FC BL/CASL*2BSa | Hits in Wheat 2BS Chinese Spring Reference Genome | Hits in Wheat 2DS Chinese Spring Reference Genome | Barley Homolog in 2HS |
|---|---|---|---|---|---|
| DMP (PKS-like) | comp177430 | 4 | Traes_2BS_ 9E10D26DB | Traes_2DS_ B436C6270, Traes_2DS_ 344FD1DE8 | MLOC_59804 |
| DMH (Hydrolase) | comp186033b | 3 | IWGSC_CSS_2BS_scaff_5155033:5108-6160 (not annotated as a gene) | Traes_2DS_ A38359A00 | MLOC_13397 |
| comp166115b | 4 | IWGSC_CSS_2BS_scaff_5155033:6117-6652 (not annotated as a gene) | Traes_2DS_ A38359A00 | ||
| DMC (P450) CYP709J4 | comp178530 | 11 | Traes2_2BS_ 163390FC4 | Not found | MLOC_12151 |
| WES (wax ester synthase)c | comp184322 | 2 | Traes_2BS_ B9F75B638 | Traes_2DS_ 4ABDF6D9C | Not found |
| P450 CYP96B30c | comp188463 | 2.6 | IWGSC_CSS_2BS_ scaff_5201145 dna:scaffold | IWGSC_CSS_2DS_scaff_27654 dna:scaffold | MLOC_71974 |
FC, fold change.
De novo assembly of the wheat transcriptome resulted in two different transcripts matching one gene in both wheat and barley. comp186033 matches the upstream part of the gene and comp166115 matches the downstream part. Together they cover the whole gene in both wheat and the barley homolog MLOC_13397.
Candidate genes based of Zavitan region analysis.
Out of 628 transcripts highly expressed in BL, 139 mapped to chromosome arm 2BS, and two were identified beside the PKS as strong candidates to be associated with β-diketone biosynthesis. The first gene was annotated as a putative carboxylesterase or hydrolase. Carboxylesterases could catalyze the hydrolysis of long-chain fatty acid esters and thioesters, including CoA or ACP esters. The second candidate gene encoded a putative cytochrome P450 protein with similarity to CYP709C1. Members of the 709C1 subfamily of cytochrome P450s have been previously associated with subterminal hydroxylation of long-chain fatty acids, an enzymatic step that is also thought to be required for the formation of hydroxy-β-diketones. This candidate was annotated as CYP709J4 (see Methods). In barley, β-diketone biosynthesis genes were reported to be located on chromosome arm 2HS (Takahashi et al., 1953; Tsuchiya, 1972; Schondelmaier et al., 1993). Consistent with this, all three wheat candidate genes had homologous sequences on barley 2HS (Table 1). The results thus identified these three genes as strong candidates for the W1 locus, and we therefore decided to subject them to further characterization. They were designated as Diketone Metabolism-PKS (DMP), -Hydrolase (DMH), and -CYP450 (DMC) (Table 1; for sequences, see Supplemental File 1).
Molecular Characterization of Chromosome Arm 2BS Demonstrates That the Three Candidate Genes Are Not Found in Lines TTD140 and CASL*2BS
The glaucous phenotype of BL must be an iw1iw2 genotype combined with W1 and/or W2 loci, while CASL*2BS carries the TTD140 Iw1 (Millet et al., 2013) and W1 with unknown genotype instead. The W2 genotype of BL and CASL*2BS was not determined in this study; however, BL and CASL*2BS have the same W2 locus. Therefore, BL and CASL*2BS differ in the Iw1 loci and possibly but not necessarily in their W1 loci. To determine genotypic differences between BL and CASL*2BS, both lines were genotyped with the 90K iSelect SNP Chip (Wang et al., 2014b). This revealed 422 markers that were polymorphic between the genotypes, of which 12 showed no amplification for CASL*2BS but were detected in the BL (referred to as null markers; Supplemental Table 1). Further 90K chip genotyping revealed that the same 12 polymorphic markers were also nulls in the glossy wild emmer wheat T. turgidum ssp dicoccoides accession TTD140 (the 2BS donor in CASL*2BS) but present in the glaucous durum wheat (T. durum 2n = 4x = 28; genome AABB) cultivar Svevo.
The 422 polymorphic markers were ordered along the chromosome arm 2BS of wheat using available genetic maps (Avni et al., 2014; Wang et al., 2014b; Maccaferri et al., 2015) as references. The mapping revealed that the 12 null SNP markers are genetically linked and absent in the glossy genotypes. Importantly, four of these 12 closely linked markers were found to reside inside the DMC gene and another three inside the DMP gene (Supplemental Table 1). The third candidate gene, DMH, is not represented on the 90K iSelect genotyping chip.
To confirm our genotyping results, we attempted to amplify the three candidate genes with specific oligonucleotide pairs from the tetraploid glaucous cultivar Svevo and the glossy TTD140 genotype. While the expected fragments could be amplified from Svevo, no amplification products were observed for TTD140, hence confirming the absence of DMP, DMH, and DMC in TTD140 (Supplemental Figure 3).
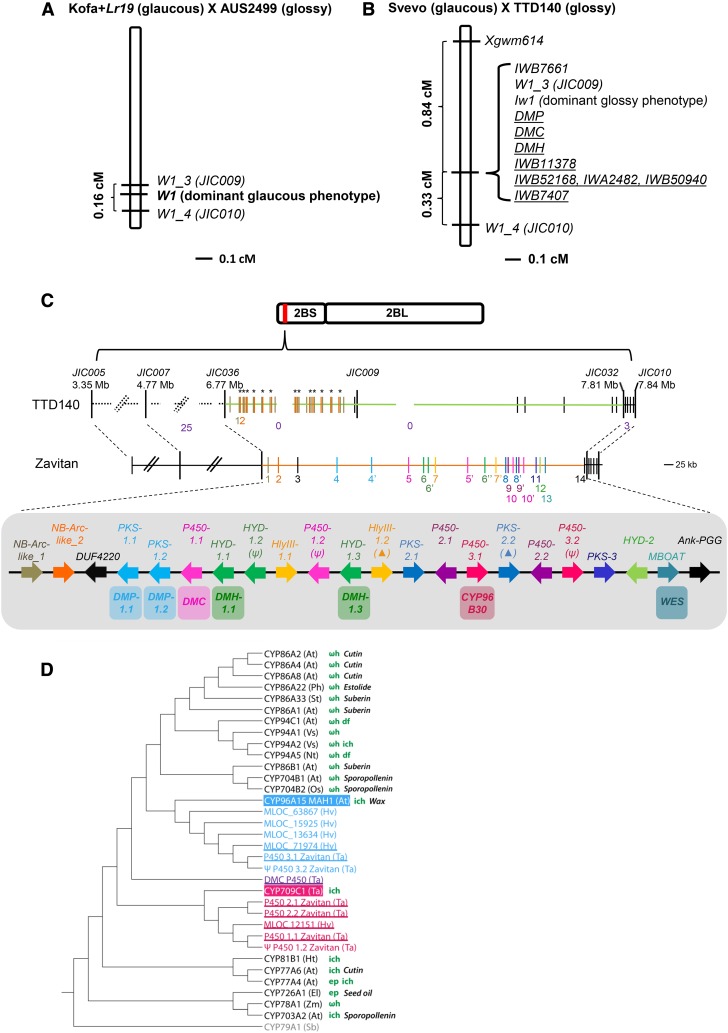
The Fine Map of 2BS in Tetraploid Wheat Reveals Complete Genetic Linkage between the Three β-Diketone Metabolism Candidate Genes, W1, and the Iw1 Loci
Fine genetic mapping of the three candidate genes was performed using segregating populations based on crosses between glaucous and glossy genotypes. One segregating population was developed using the tetraploid accessions Kofa+Lr19 (glaucous) and AUS2499 (glossy). AUS2499 is a mutant derived from a glaucous line. The F1 hybrids were glaucous, confirming that Iw1 is not present in AUS2499 or in Kofa+Lr19. Analysis of 352 F2 progeny revealed the expected 3:1 segregation ratio for a single locus, which was mapped to 2BS. Examination of another 4423 F2 progeny suggested that the dominant locus for β-diketone production designated W1 is confined to a 0.16 cM interval flanked by markers W1_3 and W1_4 (Figure 2A; Supplemental Text 1 and Supplemental Table 2).
Figure 2.
Genetic and Physical Maps of W1/Iw1.
(A) Location of the locus responsible for the dominant glaucous phenotype (W1) based on analysis of 9550 gametes from the AUS2499 × Kofa+Lr19 cross. W1 is contained in a 0.16-cM interval on wheat chromosome 2BS flanked by markers JIC009 and JIC010
(B) The locus responsible for the dominant glossy phenotype (Iw1) on wheat chromosome 2BS is found in a similar location based on analysis of 598 gametes from the Svevo × TTD140 cross. Iw1 is completely linked to JIC009 and to the three candidate wheat genes DMP, DMH, and DMC. Underlined markers represent dominant markers present in Svevo and absent in TTD140.
(C) Wild emmer Zavitan (glaucous) and TTD140 (glossy) physical maps. Gene content of Zavitan across the W1 genetic interval is shown. Out of 22 genes, 15 correspond to either cytochrome P450s (P450; pink), type-III PKS (blue), or hydrolase (HYD; green) genes (including partial genes and pseudogenes). Genes are ordered based on the Zavitan genome and are numbered accordingly on the Zavitan physical map (middle). Duplicated genes with >99% sequence identity are denoted with the same color and with a different numerical suffix. Pseudogenes are denoted with a psi symbol. Partial genes are denoted with a triangle. The Zavitan best homologs for DMP (two duplicated genes DMP-1.1 and DMP-1.2), DMH (two duplicated genes DMH-1.1 and DMH-1.3), and DMC are shown. Among the 22 Zavitan genes, WES and P450-3.1 were annotated as wax ester synthase from the MBOAT family (in turquoise) and CYP96B30 (in dark pink), correspondingly, suggesting a possible involvement in wax biosynthesis. The TTD140 physical map (top) shows a common haplotype with Zavitan in the regions surrounding flanking markers JIC036 and JIC032 (dashed black line). However, apart from the multiple local duplications of the NB-Arc-like genes (genes 1 and 2, duplications are indicated in black asterisks), all other sequences and genes from DUF4220 to Ank_PGG are unique to the Zavitan W1 haplotype (orange line) compared with the TTD140 Iw1 haplotype (light green line). Black vertical lines in the TTD140 physical map are genes exclusive to TTD140. Numbers in purple indicate the number of observed recombinations. A detailed description of the region is provided in Supplemental Text 2 and 3.
(D) Phylogenetic tree of wheat DMC (purple) identified from BL and its homologs from Zavitan accession and from barley, with additional P450s from related clades. TaCYP709C1 and AtCYP96A15 (MAH1) are known to catalyze in-chain hydroxylation and are related by sequence to TaDMC, Zavitan P450(1-3), barley MLOC_12151, and MLOC_71974. Proteins related to TaCYP709C1 and AtCYP96A15 are shown in pink and blue, respectively. Species abbreviations: At, Arabidopsis thaliana; Ph, Petunia hybrida; Sb, Sorghum bicolor; St, Solanum tuberosum; Vs, Vicia sativa; Nt, Nicotiana tabacum; Ta, Triticum aestivum; Hv, Hordeum vulgare; Ht, Helianthus tuberosus; Os, Oryza sativa; El, Euphorbia lagascae; Zm, Zea mays. Known enzymatic activities are shown: ωh, ω-hydroxylation; ich, in-chain hydroxylation; df, diacid formation; ep, epoxidation. Physiological role associated with fatty acid biosynthesis is shown: cutin, cutin synthesis, estolide, estolide synthesis; suberin, suberin synthesis; sporopollenin, sporopollenin synthesis; seed oil, seed oil synthesis; wax, wax synthesis; FA, fatty acid hydroxylation. Genes discussed in this study are underlined.
A second segregating population developed between Svevo (glaucous) and TTD140 (glossy) was subsequently examined. Phenotypic evaluation of F1 hybrids displayed a clear glossy phenotype, supporting previous information regarding the dominant inhibitor (Iw1) on chromosome arm 2BS in TTD140 (Rong et al., 2000). Examination of 299 F2 progeny revealed a single locus with 1:3 (glaucous: glossy) Mendelian segregation ratio. The single locus (corresponding to Iw1) was mapped to 2BS, completely linked to JIC0009. The markers Xgwm614 and JIC0010 were mapped in a distance of 0.84 cM (distal) and 0.33 cM (proximal) to JIC009, respectively. We next genotyped the F2 population with the null markers for DMP, DMH, and DMC and found a complete linkage to JIC0009. Taken together, both W1 and Iw1 are located in very close proximity to JIC0009 and the three candidate genes (Figure 2B; Supplemental Text 1 and Supplemental Table 2).
The Three Candidate β-Diketone Biosynthesis Genes Are Part of a Metabolic Gene Cluster in the Genome of Glaucous Wild Emmer Wheat Accession Zavitan and Are Missing in Glossy Wild Emmer Wheat Accession TTD140
Analysis of the W1/Iw1 interval (between genetic markers JIC036 and JIC032; Figure 2C) in the recently sequenced genome of the glaucous wild emmer wheat accession Zavitan resulted in 22 predicted genes (including partial genes and pseudogenes; see sequences in Supplemental File 1). Six genes encode members of the CYP450 family (P450), five genes encode members of the PKS family, and another four encode members of the esterase/lipase family (hydrolase; HYD), thus accounting for 15 of the 22 genes (Figure 2C). This analysis showed that the three candidate genes are not only located on 2BS but are also tightly clustered together in the W1 wheat locus. The high sequence similarity among the duplicated genes (>99.3%) together with their physical arrangement suggested that a series of independent local duplications have occurred within the Zavitan interval after the wheat-barley divergence (Supplemental Text 2, Supplemental Table 3, and Supplemental Figure 4). A phylogenetic tree of the six P450 proteins from Zavitan with other related P450s showed that these proteins are associated with two similar but distinct P450 clades, namely, that P450-1.1 and P450-1.2 (annotated as CYP709J4 orthologs; see Methods), and P450-2.1 and P450-2.2 (annotated as CYP709J5, partial) are similar to wheat CYP709C1 and that a third pair comprising P450-3.1 and P450-3.2 (annotated as CYP96B30) is related to Arabidopsis MAH1 (CYP96A15; Figure 2D). Both wheat CYP709C1 and Arabidopsis MAH1 catalyze in-chain hydroxylation (Kandel et al., 2005; Greer et al., 2007), and many of the P450s from related clades are known to be involved in fatty acid metabolism (Figure 2D). Surprisingly, in addition to the 15 DMP/H/C homologs, WES (wax ester synthase), belonging to the membrane-bound O-acyl transferase (MBOAT) family, resides in this region (Figure 2C). The β-diketone biosynthesis pathway branch out to 2-alkanol esters (Supplemental Figure 1); therefore, it is likely that the WES gene might be also associated with the β-diketone biosynthesis gene cluster.
Additionally, BAC sequences of the wild emmer TTD140 were employed to analyze the candidate gene regions in a glossy wheat line. Genetic markers JIC007/IWB7407 (distal) and JIC010/CD927782 (proximal) flanking the W1/Iw1 interval were anchored (Figure 2C). While the distal and proximal regions of the interval share ≥96% similarity, the interval between is completely different between TTD140 and Zavitan (over 800 kb in the TTD140 sequence). This is true for both the intergenic space as well as for the gene content, with the exception of two NB-Arc-like genes (NB-Arc-like_1 and _2; Figure 2C; Supplemental Text 3), pointing to haplotype divergence across the W1/Iw1 interval between these two wild emmer accessions.
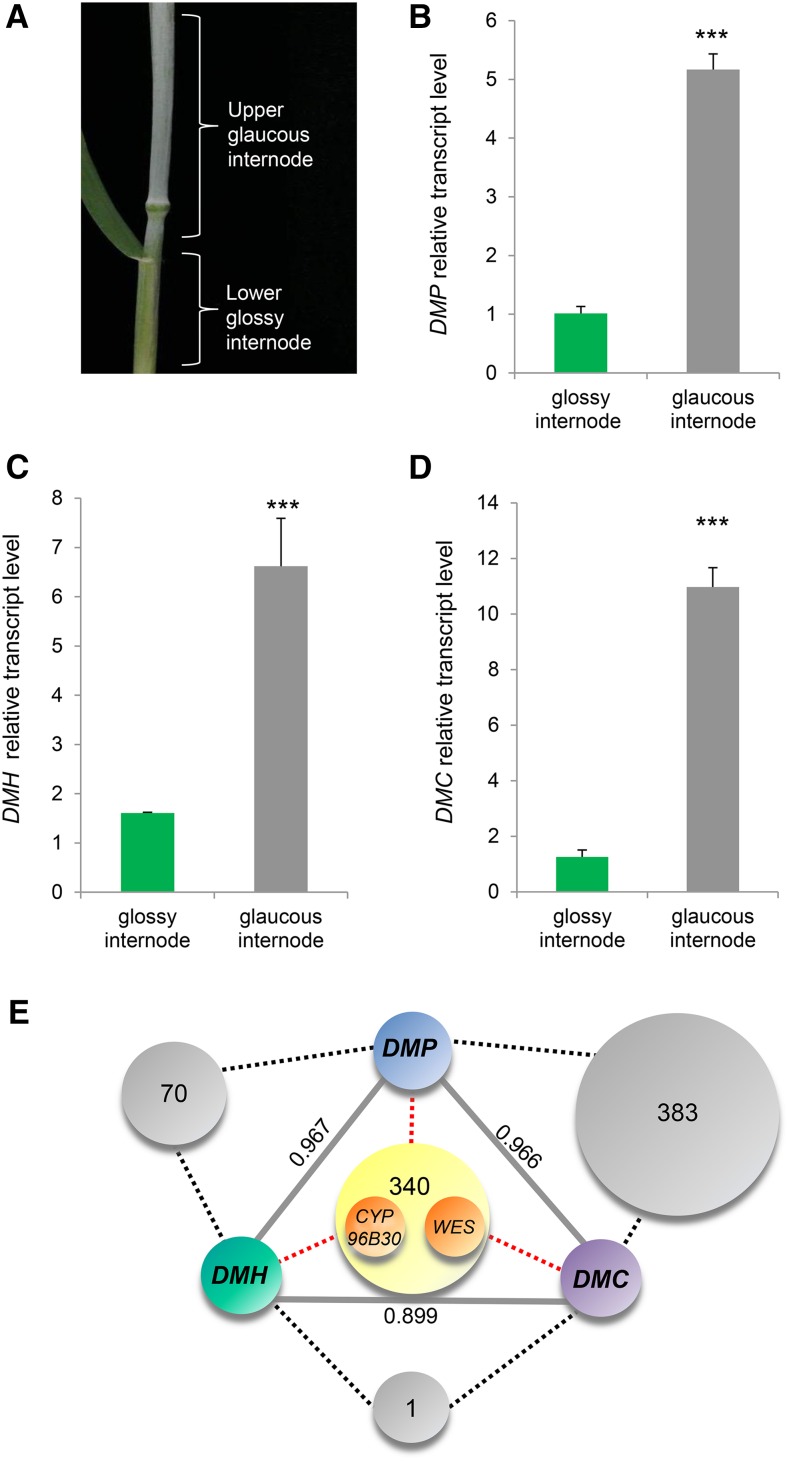
The Candidate β-Diketone Biosynthesis Genes Are Tightly Coexpressed, and Their Expression Corresponds to the β-Diketone Accumulation Pattern
If DMP, DMH, and DMC encode enzymes involved in β-diketone biosynthesis, their expression is expected to correlate with β-diketone accumulation. In glaucous tissues, β-diketones start accumulating toward plant maturation at the uppermost internode, approximately 2 weeks prior to ear emergence. By the time the ear has fully emerged, the flag leaf and the stem become visually glaucous (Troughton and Hall, 1967). Expression of the three candidate genes was examined in BL lower versus upper internodes at a time when the glaucous phenotype becomes visible (Figure 3A). All three genes displayed significantly higher expression in the upper, glaucous internode than in the lower, glossy ones (Figures 3B to 3D). Moreover, these genes exhibited highly significant coexpression between each other (r > 0.85; Figure 3E; Supplemental Data Set 1), further supporting their involvement in wheat β-diketone production.
Figure 3.
The Three β-Diketone Metabolic Gene Cluster Members Are Highly Expressed in Glaucous Tissues and Have Very Similar Expression Patterns.
(A) A wheat tiller in the stem elongation stage in which the upper internode is already glaucous, while the next lower internode is still glossy (Zadoks ∼GS39).
(B) to (D) RT-qPCR analysis of the three β-diketone biosynthesis genes was performed in wheat BL glaucous and glossy internodes. Relative gene expression of DMP (B), DMH (C), and DMC (D), shown as means ± sd of n ≥ 3 biological replicates. ***P < 0.001.
(E) Coexpression analysis, r-value cutoff (unless indicated otherwise in numbers and gray lines) = 0.85. Gray and yellow circles on dashed black and red lines show numbers of genes coexpressed with two or all three β-diketone biosynthesis genes, respectively. WES and CYP96B30 are among the genes that are coexpressed with the DMP, DMH, and DMC genes.
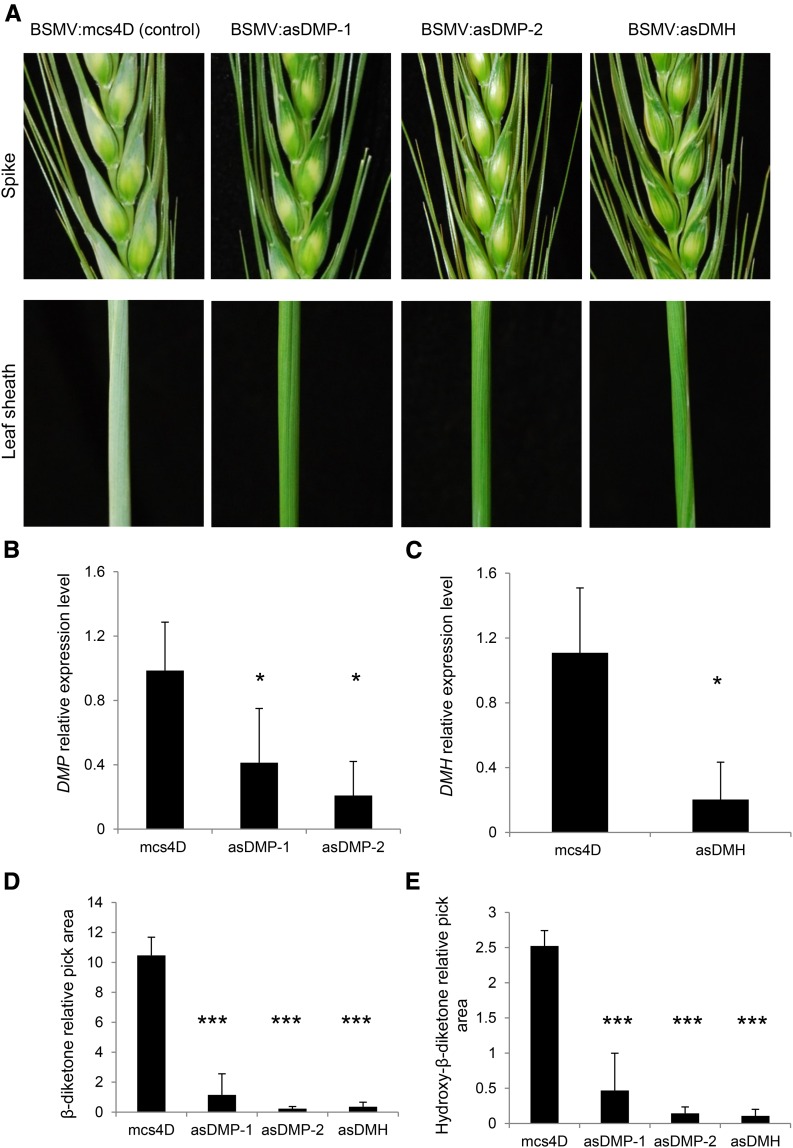
Silencing of DMP and DMH Provides in Planta Evidence for Their Role in the Wheat β-Diketone Biosynthetic Pathway
The DMP and DMH genes were silenced using Barley stripe mosaic virus (BSMV)-mediated virus-induced gene silencing (VIGS) (Scofield et al., 2005; Yuan et al., 2011). Silencing was done in the glaucous wheat cultivar Bobwhite, a genotype that has previously been demonstrated to be amenable to BSMV-VIGS (Scofield et al., 2005; Lee et al., 2015a, 2015b). While we could find a fragment that is predicted to silence DMH specifically, silencing of DMP resulted in a possible confounding effect of off-target silencing of sequence-related PKS gene family members. To mitigate the effects of off-target silencing, we used two independent VIGS constructs designed to target DMP for silencing (BSMV:asDMP-1 and BSMV:asDMP-2; see Methods). Flag leaf sheaths and glumes of newly emerged spikes of BSMV:mcs4D-infected nonsilenced control plants were glaucous, while a glossy phenotype was observed in the flag leaf sheaths and glume tissues of wheat plants inoculated with either BSMV:asDMP-1 or BSMV:asDMP-2. A similar glossy phenotype was observed in plants in which TaDMH was targeted for silencing (BSMV:asDMH; Figure 4A). Flag leaf sheaths and spikes of BSMV:mcs4D-infected control plants were visually similar to uninfected control plants (Supplemental Figure 5A).
Figure 4.
VIGS of DMP and DMH in Wheat Affects β-Diketone Biosynthesis and Results in a Glossy Appearance.
(A) Phenotypes in spikes and leaf sheaths of plants infected with different VIGS constructs: BSMV:mcs4D (control), BSMV:asDMP-1, BSMV:asDMP-2, and BSMV:asDMH.
(B) and (C) Relative expression levels of the targeted genes DMP (B) and DMH (C) in flag leaf sheath.
(D) and (E) Analysis of β-diketones (D) and hydroxy-β-diketones (E) in flag leaf sheaths of plants infected with different VIGS constructs. Relative peak area = compound pick area/internal standard peak area (E). Values indicate the mean ± sd of n ≥ 4 biological replicates. as, antisense. *P < 0.05 and ***P < 0.005.
Expression assays of flag leaf sheath samples harvested from emerging spikes (Zadoks GS 45-47; Zadoks et al., 1974) confirmed that DMP and DMH transcript levels were significantly reduced in BSMV:asDMP-1-, BSMV:asDMP-2-, and BSMV:asDMH-infected plants (Figures 4B and 4C). Scanning electron microscopy revealed significantly fewer epicuticular wax crystals on the flag leaf sheaths and glumes of silenced plants than on the corresponding tissues of control plants (Supplemental Figure 5B). Moreover, β-diketone and hydroxy-β-diketone amounts were significantly reduced in all BSMV:asDMP-1-, BSMV:asDMP-2-, and BSMV:asDMH-infected plants (Figures 4D and 4E), providing further evidence for the role of the DMP and DMH genes in the wheat β-diketone biosynthesis pathway.
A β-Diketone Metabolic Gene Cluster Is Also Present on the 2HS Barley Chromosome Arm
The finding of a β-diketone metabolic gene cluster in the wheat W1 locus prompted us to examine the physical location of the β-diketone biosynthesis genes in barley. The W1 region on wheat 2BS is in synteny with the short arm of barley chromosome 2H (Varshney et al., 2006). Furthermore, it was previously hypothesized that the barley β-diketone biosynthesis gene(s) might represent a metabolic gene cluster located on the barley 2HS chromosome (Cer-c, Cer-q, and Cer-u; von Wettstein-Knowles, 1992). Querying the three β-diketone biosynthesis genes from wheat against the barley genome returned three top hits corresponding to genes MLOC_59804, MLOC_13397, and MLOC_12151 (designated Hv-DMP, Hv-DMH, and Hv-DMC, respectively; Table 1). These genes are located within an interval of <1 Mb on 2HS (the exact cluster size cannot be determined in the current barley genome assembly), indicating a metabolic gene cluster in barley similar to the one in wheat.
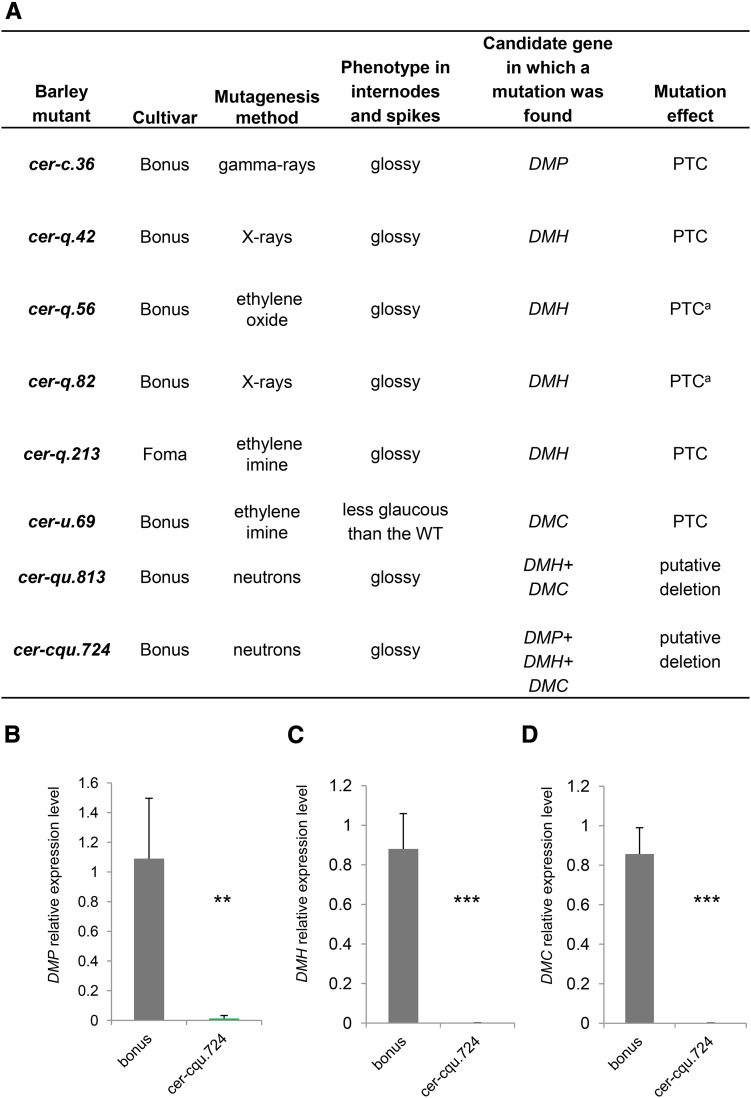
The Barley cer-c, q, and u Single Mutants Possess Mutations in Either One of the Three β-Diketone Metabolic Cluster Genes, Which Are Likely Deleted in Double and Triple Mutants
Hundreds of barley cer mutants (Lundqvist and von Wettstein, 1962, 1971; Lundqvist et al., 1968) are publicly available, including the cer-c, cer-q, and cer-u mutants deficient in β-diketone biosynthesis. To examine if the barley DMP, DMH, and DMC genes are indeed deficient in the previously characterized cer mutants, coding regions of the three cluster genes were amplified from five cer-c, six cer-q, and five cer-u single mutants, three double mutants, and one triple cer-cqu mutant. Of these, one cer-c mutant (cer-c.36) had a premature termination codon (PTC) in the DMP gene, one cer-u mutant (cer-u.69) had a PTC in the DMC gene, and four cer-q mutants (cer-q.42, cer-q.56, cer-q.82, and cer-q.213) had PTCs in the DMH gene. Furthermore, the DMH and DMC genes could not be amplified by PCR from DNA of the double mutant cer-qu.813 using different oligonucleotide pairs targeting the three genes. Finally, the DMP, DMH, and DMC genes could not be amplified in the cer-cqu.724 triple mutant with any of the gene-specific oligonucleotide pairs (Figure 5A; Supplemental Figure 6). Expression analysis of the three genes in cer-cqu.724 supported a deletion of the entire β-diketone gene cluster in this triple mutant (Figures 5B to 5D). In total, eight barley cer mutants deficient in β-diketone biosynthesis were found to bear mutations or deletions of the β-diketone biosynthetic genes, firmly supporting that barley DMP, DMH, and DMC are the genes underlying the cer-c, cer-q, and cer-u phenotypes, respectively.
Figure 5.
Barley cer Mutants Have Mutations in β-Diketone Biosynthesis Genes DMP, DMH, and DMC.
(A) A screen of barley cer mutants revealed eight mutants with either PTC or deletions in the β-diketone genes DMP, DMH, and DMC. acer-q.56 and cer-q.82 have the same mutation in the DMH gene; hence, it cannot be determined if these are two different mutants.
(B) to (D) RT-qPCR analysis of the gene cluster in wild-type bonus and cer-cqu.724 triple mutant supports a deletion in the genetic region comprising DMP, DMH, and DMC. Relative expression is shown for DMP (B), DMH (C), and DMC (D). Values indicate the mean ± sd of n ≥ 3 biological replicates. **P < 0.005 and ***P < 0.001
The Wheat W1 and the Barley Cer-cqu Gene Clusters Are Conserved
The Zavitan reference genome analysis revealed that the W1/Iw1 region contains many gene duplications (Figure 2C). Beside the more recent duplications, which resulted in highly similar genes (i.e., >99.3% identity) and are denoted with a different numerical suffix, there were also probably more ancient duplications, which resulted in less similar genes (i.e., P450-1, P450-2, and P450-3). In order to check whether the whole Zavitan cluster exists in barley, the Zavitan cluster genes were queried against the barley genome (http://www.gramene.org/). Interestingly, we discovered that in addition to the barley homologs of PKS-1, HYD-1, and P450-1 Zavitan genes (MLOC_59804, MLOC_13397, and MLOC_12151, correspondingly), all the other related genes (i.e., PKS-2, PKS-3, HYD-2, P450-2, and P450-3) and even the likely unrelated HlyIII gene that resides inside the Zavitan cluster have hits in the barley cluster region (Table 2). This includes all the highly similar Zavitan genes (denoted with a different numerical suffix; Figure 2C) that had the same hit in barley. Remarkably, the genes that delimit the gene cluster and are not related to β-diketone biosynthesis are also not conserved in the barley cluster. Notably, WES, which is located close to the border or the gene region in Zavitan (Figure 2C), does not have a hit in the gene cluster region in barley. Interestingly, MLOC_11854 is located within the Cer-cqu region, and it encodes a putative BAHD, a family of acyltransferases previously associated with the formation of cuticular lipids esters (Molina and Kosma, 2015).
Table 2. The Wheat Gene Cluster (Accession Zavitan) Is Conserved in the Barley Genome.
| Zavitan Gene(s) | Hit in the Barley Cluster Region |
|---|---|
| NB-Arc-like_1 | No alignment in the cluster region |
| NB-Arc-like_2 | No alignment in the cluster region |
| DUF4220 | No alignment in the cluster region |
| PKS-1 (1.1, 1.2) | MLOC_59804 |
| PKS-2 (2.1, 2.2) | MLOC_59804 |
| PKS-3 | Alignment to an unannotated sequence within the cluster region |
| HYD-1 (1.1, 1.2, 1.3) | MLOC_13397 |
| HYD-2 | MLOC_19415 |
| P450-1 (1.1, 1.2) | MLOC_12151 |
| P450-2 (2.1, 2.2) | MLOC_13649 |
| P450-3 (3.1, 3.2) | MLOC_71974 |
| HlyIII (1.1, 1.2) | Alignment to an unannotated sequence within the cluster region |
| WES | No alignment in the cluster region |
| Ank_PGG | No alignment in the cluster region |
Escherichia coli Cells Expressing Barley DMH Accumulate Predicted Intermediates of the β-Diketone Biosynthesis Pathway
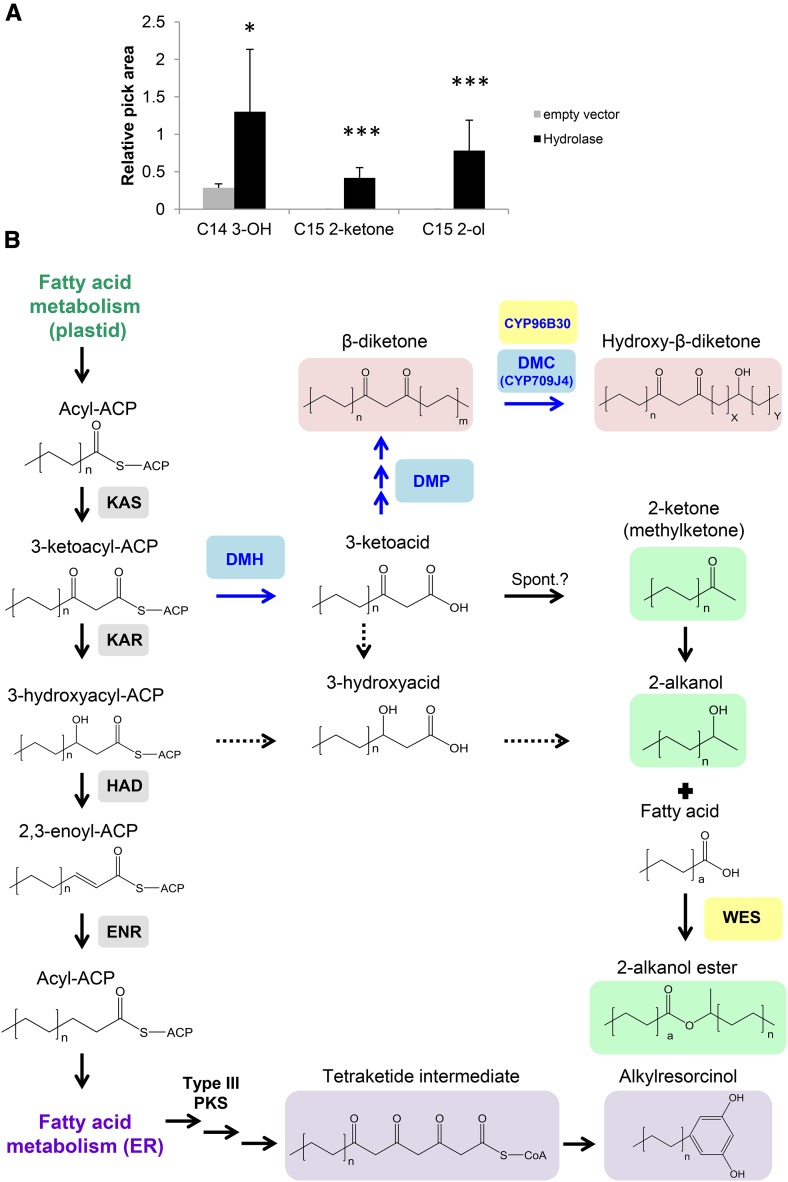
Our findings implicated an esterase/hydrolase enzyme in β-diketone biosynthesis, even though such activity had not been predicted to be part of the pathway before (von Wettstein-Knowles, 2012). E. coli cells expressing the barley DMH gene produced two metabolites that were undetectable in the control, C15 2-ketone, and C15 2-alkanol. Additionally, C14 3-hydroxyacid was present in low concentration in the control and significantly increased in the lipid extract of cells expressing Hv-DMH (Figure 6A; for compound identification, see Supplemental Figure 7). C15 2-ketone is known to arise from spontaneous decarboxylation of the corresponding C16 3-keto fatty acid, either in vivo or during sample preparation and GC-MS analysis (Kornberg et al., 1947). We therefore interpret the ketone as a proxy for the ketoacid and conclude that this acid is the immediate hydrolase product. It seems plausible that both the 3-keto fatty acid and the 2-ketone may have been reduced to 3-hydroxy fatty acid and 2-alkanol by endogenous E. coli enzymes, respectively, thus explaining the other major products identified (Figure 6B). However, it cannot be ruled out that Hv-DMH may have intercepted 3-hydroxy fatty acid elongation intermediates. Since all the major substrate chain lengths occur during plastidial fatty acid synthesis but not during further ER elongation, we suggest that the hydrolase uses 3-ketoacyl-ACP rather than 3-ketoacyl-CoA substrates. DMH may thus be tentatively defined as a medium- and long-chain 3-ketoacyl-ACP thioesterase; however, its exact enzyme activity remains to be determined by detailed biochemical characterization.
Figure 6.
Heterologous Expression of Barley DMH Suggests Its Biochemical Activity and Leads to a Revised β-Diketone Biosynthesis Pathway.
(A) Lipid analysis of DMH-expressing bacteria revealed two compounds not present in the control (C15 2-ketone and C15 2-alkanol). In addition, C14 3-hydroxy acid increased in Hv-DMH-expressing cells. C15 2-ketone is a key intermediate most probably derived from decarboxylation (spontaneous upon GC-MS heating) of C16 3-ketoacid, the DMH reaction product. Values indicate the mean ± sd of n = 5 biological replicates. *P < 0.05 and ***P < 0.005. Relative peak area = compound peak area/internal standard peak area.
(B) Revised β-diketone biosynthesis pathway based on the evidence presented in the current study. Sequential arrows stand for possible sequential enzymatic reactions. Dashed arrows represent reactions that explain the E. coli findings; however, it is not clear whether or not they also occur in planta. DMP, DMH, and DMC are shown in blue. WES and CYP96B30, which were found to be located within the cluster region in the wild emmer wheat Zavitan reference genome, are shown in yellow. Enzymes from the FAS system are shown in gray. Methylketones, 2-alkanols, and 2-alkanol esters that require DMH are shown in light green. β-Diketones and hydroxy-β-diketones that require DMH, DMP, and DMC (DMC only for hydroxy-β-diketones) are shown in light pink. Alkylresorcinol predicted biosynthesis pathway is shown in light purple, based on von Wettstein-Knowles (2012). Spont., spontaneous; a, n, m, x, and y are placeholders to designate chain length variation.
DISCUSSION
The transition from vegetative to reproductive growth in cereals is a critical developmental phase that determines adaptation to a wide range of environments. This process is accompanied by the accumulation of β-diketone waxes on surfaces of various glaucous cereal crops. Indeed, glaucousness has been established as an advantage for yield, especially under dry conditions (Fischer and Wood, 1979; Johnson et al., 1983; Febrero et al., 1998; Merah et al., 2000). Glaucousness in wheat is not only governed by the wax production locus W1 but is influenced by the dominant epistatic effect of Iw1 (in hexaploid wheat also W2 and Iw2). The Iw genes and their inhibition mechanism are still unknown. The Iw genes might be negative regulators, possibly involving RNA silencing similar to the regulation of CER3 in Arabidopsis (Lam et al., 2012, 2015). They could also encode E3 ubiquitin ligases that are specifically involved in β-diketone protein degradation through the proteasome or can inhibit W genes through many other mechanisms. We thus anticipate that the elucidation of genes in the wax production locus W1 reported here will set the stage for elucidating the wax inhibitor Iw1 locus.
Multiple Genes Rather Than a Single One Encoding a Multifunctional Enzyme Are Involved in β-Diketone Biosynthesis in Barley and Wheat
The question whether the barley β-diketone biosynthesis locus is represented by a gene cluster or corresponds to a single multifunctional gene has been a matter of debate in the last three decades. Genetic studies characterizing β-diketone mutants suggested a closely linked location for the three complementation groups of this pathway (Cer-c, Cer-q, and Cer-u) on 2HS of barley (von Wettstein-Knowles and Søgaard, 1980). In a subsequent report, it was shown that the phenotype of double and triple cer mutants could be reverted by additional mutagenesis, suggesting that the Cer-cqu locus most probably corresponds to a multifunctional gene (von Wettstein-Knowles and Søgaard, 1981). Interestingly, among the 434 diketone cer mutants that were generated using seven types of mutagenic treatments, 13 double and triple mutants were found. Eleven out of those 13 double and triple mutants were generated using neutron mutagenesis (von Wettstein-Knowles and Søgaard, 1980). This mutagenesis procedure is known to cause DNA deletions, ranging in size from kilobases to megabases (Li et al., 2001; Men et al., 2002); therefore, revertants are highly unlikely to occur. Indeed, here, we show a correlation between the barley mutant groups (cer-c, q, and u) and mutations in DMP, DMH, and DMC that clearly correspond to a metabolic gene cluster and not to a multifunctional gene. Moreover, deletion mutations in the corresponding genes found in the barley double and triple mutants (i.e., cer-qu.813, and cer-cqu.724) match the finding of a gene cluster. Our results here have been corroborated in a recent report that described the same three clustered genes to be involved β-diketone biosynthesis in barley (Schneider et al., 2016). Using BAC libraries, Schneider et al. (2016) determined that the barley gene cluster has a size of 101 kb. Furthermore, barley mutants were sequenced and homology-based modeling suggested that many of the mutations affect the protein structure or active-site residues.
Genes in the Wheat W1/Iw1 and Barley Cer-cqu Regions Likely Encode Enzymes Involved in the β-Diketone Biosynthesis and Related Branching Pathways
Analysis of the W1/Iw1 interval in the genome of the wild emmer wheat resulted in the detection of 22 predicted genes (including partial genes and pseudogenes). Sequences of 16 of the 22 genes in the cluster region could be associated not only with the biosynthesis of β-diketones but also with additional wax compounds including methylketones, 2-alkanol esters, and maybe even alkylresorcinols (Figure 6). The possible common role of all these compounds is in biotic stress response, acting as bactericides, fungicides, or insecticides (Lin et al., 1987; Reiss, 1989; Chatzivasileiadis and Sabelis, 1997; García et al., 1997; Kozubek and Tyman, 1999). The set of 16 genes belong to four different classes, including cytochrome P450s, type-III polyketide synthase, hydrolases, and the MBOAT family.
The W1/Iw1 region is apparently very dynamic, with recent gene duplications as well as more ancient ones, which likely resulted in diverse gene functions. For example, the hydroxylation reactions leading either to 8-/9- or to 25-hydroxy-β-diketone in different wheat and barley genotypes might be catalyzed by similar hydroxylating enzymes with slightly different substrate and/or product specificity. The different CYP450s found in the wheat cluster and associated (by sequence similarity) with two CYP450 subfamilies (i.e., CYP709C1 [DMC/P450-1.1, P450-2.1, and P450-2.2] and CYP96A15 [P450-3.1]) are both related to proteins mediating midchain hydroxylation and could be catalyzing the synthesis of hydroxy-β-diketones. It is interesting that proteins of both P450 subfamilies (i.e., CYP709C1 and CYP96A15) were found related to several other CYP450 subfamilies whose members are known to be involved with the biosynthesis of plant barriers, including cuticular lipids (wax and cutin), suberin, and sporopollenin. Among the genes in the β-diketone biosynthesis cluster, we also identified a putative WES. As 2-alkanol esters are known to be by-products of the β-diketone pathway (von Wettstein-Knowles, 2012), the involvement of an ester-forming enzyme will be an exciting subject for further investigation. Importantly, both CYP96B30 and WES were found to be highly coexpressed with the three cluster genes DMP, DMH, and DMC. Recently, Zhang et al. (2015) identified the W3 locus, which is linked to W1, and suggested its involvement in β-diketone biosynthesis. Given the complexity of the W1/Iw1 region, W3 might be included in this interval.
Examining the barley cluster region revealed that in addition to the homologs of wheat PKS-1, HYD-1, and P450-1 (barley MLOC_59804, MLOC_13397, and MLOC_12151, respectively), almost all genes in the wheat cluster possess barley homologs in the Cer-cqu region. Genes located at the cluster edges that are likely not related to β-diketone biosynthesis (by homology and annotation) are also not conserved in the barley cluster. This further supports our current notion that duplicated genes within the gene cluster are most likely related to β-diketone biosynthesis since there was a strong evolutionary selection pressure to keep the cluster intact in two different plant species. As mentioned above, the current barley genome assembly does not enable to detect the gene order within the cluster and the exact cluster size. Yet, it is clear that the barley homologs for the Zavitan cluster genes are located within the same genomic region. This is supported by a recent article by Schneider et al. (2016) that found that MLOC_13649, MLOC_71974 (corresponding to Zavitan P450-2 and P450-3), and HlyIII genes are located between two flanking markers of the Cer-cqu locus, together with MLOC_59804, MLOC_13397, and MLOC_12151 (corresponding to Zavitan PKS-1, HYD-1, and P450-1). MLOC_13649 and MLOC_71974 were also found to be deleted in several Cer-cqu mutants. Further investigation of all clustered genes in these two plant species will be subject for future research in order to examine their role in the β-diketone pathway and its branches.
A Revised Model for the β-Diketone Biosynthesis Pathway
Based on our characterization of DMH and the likely reactions catalyzed by the DMP and DMC enzymes, the β-diketone biosynthesis pathway can now be revisited (Figure 6B). We propose that DMH first intercepts keto-fatty acyl-ACP intermediates from the plastidial fatty acid synthase system. The thioester hydrolysis catalyzed by DMH leads to free ketoacids, which are then converted into β-diketone and its hydroxy derivatives by the DMP and DMC enzymes, respectively. Alternatively, the ketoacids may also undergo (either spontaneous or enzymatic) decarboxylation to 2-ketones (methylketones) that serve as precursors for reduction to 2-alkanols and acylation (catalyzed by a WES enzyme) to the 2-alkanol esters.
Activity of DMH as a Hydrolase/Carboxylesterase and the Link to Methylketone Biosynthesis
Two alternative hypotheses have been put forward for the activity of the enzyme encoded by barley Cer-q locus, identified here to be the DMH gene. Originally, it was suggested that the enzyme is localized to the ER, where it functions as a PKS utilizing acyl-CoA substrates to form ketoacyl-CoA intermediates (von Wettstein-Knowles, 2012). The recent discovery that the Cer-q sequence bears similarity with lipases then prompted speculation that the enzyme instead hydrolyses hypothetical glycerolipid intermediates into free acids (Schneider et al., 2016).
However, our experiments testing the DMH activity by heterologous expression in E. coli led to accumulation of C15 2-ketone (C15 methylketone), which is most probably a decarboxylation product of C16 3-ketoacid. The product of the DMH-catalyzed reaction is thus a free ketoacid, rather than the previously assumed acid lacking the keto group. Conversely, this finding strongly suggests that the substrate of the reaction is a ketoacyl compound present in plants as well as E. coli. Finally, based on the sequence similarity with hydrolases and carboxylesterases, we conclude that DMH most likely functions as a thioesterase intercepting common ketoacyl intermediates of fatty acid synthesis. Since the acyl chain lengths involved are known to be generated in plants by the plastidial FAS system, it seems plausible that the ketoacyl-ACP is intercepted between the ketoacyl-ACP synthase and ketoacyl-ACP reductase steps. An alternative interception of a ketoacyl-CoA intermediate of the ER-localized fatty acid elongase system seems unlikely, since they are thought to elongate only acyl chains beyond C16, longer than the chain lengths observed in the E. coli experiment.
Methylketones are known to be side products of the β-diketone biosynthesis pathway (von Wettstein-Knowles, 2012). Previous studies demonstrated that methylketone biosynthesis in Solanum habrochaites involves two enzymes, methylketone synthase 1 (MKS1) and MKS2 (Fridman et al., 2005; Ben-Israel et al., 2009; Yu et al., 2010; Auldridge et al., 2012). While MKS2 (a HotDog fold protein) acts as a thioesterase hydrolyzing (primarily C12 and C14) β-ketoacyl-ACPs to generate the corresponding 3-ketoacids, MKS1 (a α/β-hydrolase fold protein) is a decarboxylase that converts the 3-ketoacids to the corresponding (C11 and C13) 2-ketones (Yu et al., 2010). The tomato study further revealed that decarboxylation occurs either enzymatically (catalyzed by MKS1) or spontaneously, however, with a slow rate (Yu et al., 2010). Taken together, the tomato results thus underpin our conclusion that DMHs (in wheat and in barley) act as thioesterases that hydrolyze β-ketoacyl-ACPs to 3-ketoacids, which are then spontaneously/enzymatically decarboxylated to 2-ketones.
Our wheat wax analyses confirmed earlier results for barley waxes, showing that the amounts of β-diketones are much higher than those of the accompanying 2-alkanol esters, the final products derived from methylketone intermediates. These findings suggest that the overall pathway flux is higher toward β-diketone products then to the 2-ketone intermediates en route to the 2-alkanol ester side products. Therefore, relatively slow, spontaneous decarboxylation of the common ketoacid intermediate may be sufficient to account for the formation of low amounts of 2-alkanol esters. Yet, alternative scenarios for the decarboxylation reaction cannot be excluded at present. On the one hand, a separate, unknown decarboxylase may exist, which might even be encoded within the gene cluster. On the other hand, DMH might act as a bifunctional enzyme having both hydrolase and decarboxylase activities, similar to other enzymes like polyneuridine aldehyde esterase from Rauvolfia serpentina (Dogru et al., 2000; Yang et al., 2009).
Finally, it should also be noted that both tomato proteins involved in formation of 2-ketones (MKS1 and MKS2) localized to the plastid, even though MKS1 lacks a typical transit peptide (Fridman et al., 2005; Yu et al., 2010), as is the case with the wheat and barley DMH proteins. It is worth mentioning that, despite the lack of typical transit peptides, the plant-mPLoc tool (Chou, 2005; Shen and Chou, 2006; Chou and Shen, 2007, 2008, 2010) predicts that the wheat and barley DMHs localize to the chloroplast or the nucleus, but not the ER. Surely, further studies are required in order to fully characterize the DMH enzyme activity and its subcellular localization in both barley and wheat.
Common and Unique Features of the β-Diketone Metabolic Gene Cluster
The β-diketone biosynthesis clusters found here in the wheat W1 and barley Cer-cqu loci join a growing number of plant metabolic gene clusters discovered in the past years (Osbourn, 2010; Boycheva et al., 2014; Nützmann and Osbourn, 2014). These clusters clearly share a few typical features detected in other metabolic gene clusters to date. These include (1) a typical size of a several hundred kilobases (800 kb in wheat), (2) the presence of at least three genes, (3) the involvement of genes encoding a minimum of three types of biosynthetic enzymes, and (4) the tight coexpression of the clustered genes (Nützmann and Osbourn, 2014; Nützmann et al., 2016). Another common feature of plant metabolic gene clusters is the presence of an enzyme catalyzing the first committed step in the pathway. Such enzymes divert the pool of an intermediate metabolite from core/primary pathways into secondary/specialized metabolism. In the case of terpenoid-producing clusters, triterpene and diterpene cyclases draw substrates from the core cytosolic and plastidic isoprenoid pathways, respectively (Qi et al., 2004; Wilderman et al., 2004; Field and Osbourn, 2008; Sawai and Saito, 2011). Other examples include the first committed enzymes in the biosynthesis of Dhurrin, a cyanogenic glycoside from Sorghum bicolor, and the cyclic hydroxamic acid 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one from maize, both utilizing substrates from core amino acid biosynthetic pathways (Frey et al., 1997; Takos et al., 2011). In the case of the β-diketone biosynthesis cluster, DMH likely performs the first committed step intercepting ketoacyl-ACP intermediates from the core FAS system. In terms of enzyme activities, the β-diketone cluster includes for the first time members of the hydrolase/carboxylesterase and PKSs families, the latter often found in bacterial metabolic clusters (Shen, 2003; Wang et al., 2014a).
Boutanaev et al. (2015) proposed that metabolic gene clusters are often located in unstable chromosomal regions (close to the telomere or in highly dynamic regions, i.e., rich in transposable elements). The clustering of genes in highly unstable regions probably reflects the advantage conferred by coinheritance of beneficial gene combinations. Both W1 in wheat and Cer-cqu in barley are located in subtelomeric regions, and W1 is indeed a very dynamic region. Therefore, it seems that through evolution glaucousness was favored and inheritance of the genes as a cluster was established, at least in wheat and barley.
While common in bacterial and fungal clusters of natural product biosynthesis, to date none of the plant metabolic gene clusters appear to contain regulatory genes controlling the corresponding pathway. In a recent example, Cárdenas et al. (2016) demonstrated that an AP2-type transcription factor (GLYCOALKALOID METABOLISM9 [GAME9]) could likely activate genes of the clustered steroidal glycoalkaloid pathway genes such as GAME4. Yet, GAME9 is located on chromosome 1 and hence is not physically associated with the steroidal glycoalkaloid cluster genes situated on chromosomes 7 and 12 in tomato and potato (Solanum tuberosum). The wheat β-diketone cluster could turn out to be the first to contain a regulatory gene, as the Iw1 dominant wax inhibitor resides in the vicinity of the W1 locus and it might encode a negative regulator of the β-diketone pathway.
In this study, we used diverse approaches to shed light on the molecular basis of β-diketone production in wheat and barley. The importance of cuticular lipids including β-diketones to cereal crop performance has been demonstrated several times by now, including the control of the covered/hulled and naked caryopsis phenotype as well as leaf water conservation (Taketa et al., 2008; Chen et al., 2011). Hence, the findings here extend the current toolbox allowing breeding and manipulation of surface traits in major cereal crops for improved biotic and abiotic stress response and ultimately yield.
METHODS
Plant Material
BL and CASL*2BS lines were generously provided by Moshe Feldman (Weizmann Institute of Science). Barley (Hordeum vulgare) accessions were kindly provided by the NordGen gene bank (http://www.nordgen.org/index.php/en). Plants used for scanning electron microscopy imaging, wax analysis, transcriptome libraries, and mutant screens were grown in a greenhouse (22°C during day time and 20°C during the night). Two F2 populations used in this study were (1) tetraploid population derived from a cross between the glossy wild emmer wheat (Triticum turgidum subsp Dicoccoides) accession TTD140 collected from Rosh-Pina, Israel, and glaucous durum wheat Triticum durum cv Svevo; and (2) BL (Triticum aestivum ssp aestivum cv Bethlehem [BL]) crossed with CASL*2BS. The F2 populations were grown during the winter season (November 2013 to May 2014) at the experimental farm of the Hebrew University in Rehovot, Israel. For the VIGS experiments, Nicotiana benthamiana plants for preparation of BSMV sap inoculum and wheat (T. aestivum) cultivar Bobwhite were raised in Levington F2+S Seed and Modular Compost Plus Sand and Rothamsted prescription mix compost (Petersfield Products), respectively, in a controlled environment room with day/night temperature of 23°C/20°C, 60% relative humidity, and a 16-h photoperiod (∼180 μmol m−2 s−1 light intensity).
Scanning Electron Microscopy Imaging
Leaf sheaths and peduncles of plants in Zadoks ∼GS59 were sampled for cryo-scanning electron microscopy. Samples were freshly cut on narrow strips, placed in specially designed cryo-holder, and quickly frozen in liquid nitrogen. The frozen samples were kept in liquid nitrogen and transferred by using a vacuum cryotransfer device (VCT 100; Leica Microsystems) to a freeze-fracture device (BAF 60; Leica Microsystems). Samples were etched for 20 min at −95°C and coated with 6-nm platinum/carbon by double-axis rotary shadowing. Samples were observed at −120°C in an Ultra 55 SEM (Zeiss) using a secondary electron in-lens detector. Cryo-scanning electron microscopy images of glume and flag leaf sheath surfaces of plants infected with VIGS constructs were taken using a JEOL JSM-6360 LVSEM for 500× magnification images. Small (∼5 mm) sections of the flag leaf sheath or glume tissue were cut using a scalpel and mounted on an aluminum stub using a 50:50 mix of Tissue-Tek tissue freezing medium and colloidal graphite. The samples were rapidly frozen in liquid nitrogen and then transferred to the GATAN Alto 2500 cryo-preparation chamber. They were etched at −95°C for 2 min to remove any ice contamination then coated with a layer of gold/palladium (Au/Pd) once the temperature had recovered to −150°C. Samples were then transferred to the scanning electron microscope, which was maintained at −150°C, and imaged using an accelerating voltage of 5 kV.
Preparation of Wax Extracts
Flag leaf blades or leaf sheath together with peduncles were collected from BL and CASL*2BS lines, and surface areas were measured for normalization between samples. Each experiment had ≥4 biological replicates. Samples were submerged in a 20-mL scintillation vial containing 10 mL CHCl3 (Sigma-Aldrich; ≥99%, 0.75% ethanol as stabilizer) and 5 μg of n-tetracosane (Alfa Aesar; ≥99%) as internal standard. Samples were gently shaken at room temperature for 30 s, and then the chloroform was transferred to another vial. A fresh portion of 10 mL of CHCl3 was added, the solution was shaken for another 30 s, and then the two extracts were combined and concentrated under a gentle stream of N2 (Praxair; ≥99.998%), before finally being transferred to 2-mL GC sampler vials. VIGS plant wax extraction was done as above, with few modifications: Leaf sheaths were collected, surface areas were measured, and 4 mL of CHCl3 was used twice for wax extraction. Samples were kept open in a chemical hood for CHCl3 evaporation until dryness. Each sample was redissolved in 1 mL of CHCl3 containing 0.5 μg of n-tetracosane as internal standard, vortexed for 15 s, concentrated to 100 μL under a gentle stream of N2, and transferred to 2-mL GC sampler vials.
Qualitative and Quantitative Analyses of Wax Extracts
BL and CASL*2BS wax samples were derivatized by refluxing in 10 μL N,O-bis(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich; GC grade) and 10 μL pyridine (Sigma-Aldrich; ≥99.8%, anhydrous) at 70°C for 30 min and then removing excess reagents under nitrogen. Once dry, samples were redissolved in 50 μL of CHCl3 for GC analysis. Two GC instruments were used for separation and detection of wax constituents, both equipped with the same type of capillary GC column (6890N; Sigma-Aldrich; HP-1 column, 30-m length, 0.32-mm inner diameter, and 0.1-µm film thickness) using on-column injection and following the same temperature program (2 min at 50°C, ramp 40°C min−1 to 200°C, constant for 2 min, ramp 3°C min−1 to 320°C, constant for 30 min). The first employed helium gas (Praxair; ≥99%) as mobile phase, at a flow rate of 1.4 mL min−1, and was equipped with MS detector (5973N; Agilent; EI-70 eV), serving primarily the purpose of qualitative identification of separated wax compounds. The second used H2 carrier gas (Praxair; ≥99.95%) at 2.0 mL/min and a flame ionization detector for quantification of individual wax homologs based on normalization of peak areas against that of the internal standard. The relative response factors of all cuticular wax classes with respect to the internal standard were approximated to 1.00, in agreement with Riederer and Schneider (1989). VIGS and Escherichia coli samples (concentrated to 100 μL CHCl3) were derivatized by adding 20 μL N,O-bis(trimethylsilyl)trifluoroacetamide and 20 μL pyridine at 70°C for 40 min. A sample volume of 1 μL was injected in splitless mode on a GC-MS system (Agilent 7683 autosampler, 7890A gas chromatograph, and 5975C mass spectrometer). GC was performed (Zb-1ms column; 30-m length, 0.25-mm inner diameter, and 0.50-μm film thickness; Zebron, Phenomenex) with injection temperature 270°C, interface set to 250°C, and the ion source to 200°C. Helium was used a carrier gas at constant flow rate of 1.2 mL min−1. The temperature program was 0.5 min isothermal at 70°C, followed by a 30°C min−1 oven temperature ramp to 210°C and a 5°C min−1 ramp to 330°C, then kept constant during 21 min. Mass spectra were recorded with an m/z 40 to 850 scanning range. The chromatograms and mass spectra were evaluated using MSD Chemstation software (Agilent). E. coli samples were injected as the VIGS samples with the following adaptations: Sample volumes of 1 μL were injected with a split ratio of 50:1. Injection temperature was 250°C, the interface set to 250°C, and the ion source to 200°C. The temperature program was 0.5 min isothermal at 120°C, followed by a 30°C min−1 oven temperature ramp to 200°C, then ramped by 5°C min−1 to 250°C and kept for 21 min. For quantification of β-diketones and hydroxy-β-diketones, the GC peak areas of both the derivatized enol tautomers and the nonderivatized β-diketo forms were added together, as these compounds are not derivatized uniformly. Hydroxy-β-diketones included the 8- and 9-hydroxy isomers.
Expression in E. coli Cells and Lipid Extraction
Barley DMH (MLOC_13397) was cloned into the pET28-TEVH plasmid (Peleg and Unger, 2008) using restriction-free cloning procedure (Unger et al., 2010) to generate the pET28-Hydrolase plasmid. Oligonucleotides are listed in Supplemental Table 4. pET28-Hydrolase and pET28-TEVH (control) were transformed into BL21 E. coli cells. Five colonies of each transformed E. coli were grown in 5 mL of Luria-Bertani (LB) medium supplemented with kanamycin in 37°C overnight. Each sample was diluted 1:100 and grown to OD600 ∼0.6 to 0.8 in 10 mL LB+ kanamycin. Cells were induced with 500 μM IPTG at 15°C overnight. The cells were washed with PBS and kept at −80°C until further use. E. coli cell pellets were resuspended in 1 mL of double-distilled water and transferred to glass vials, and 3.75 mL of chloroform:methanol (1:2) mixture supplemented with 1 μg of n-tetracosane internal standard was added to each sample. Samples were vortexed for 2 h and sonicated for 10 min. To each sample 1.25 mL of chloroform was added and the sample vortexed, and 1.25 mL of double-distilled water was added followed by a second sonication for 10 min. The samples were centrifuged for 5 min at 3000 rpm, and the lower phase was recovered. To extract the remaining lipids, 1.9 mL of chloroform was added to the non-lipid upper phase, following vortex and centrifugation for 5 min at 3000 rpm. The two organic phases were combined in the glass tube and concentrated to 100 μL under a gentle stream of N2. Samples were stored at −20°C until derivatization.
Transcriptome Sequencing, Assembly and Analysis, and RT-qPCR
Wheat BL and CASL*2BS libraries for Illumina high-throughput strand-specific RNA-seq were prepared as follows: Total RNA was extracted with the TRIzol method using standard TRIzol RNA extraction protocol. For each sample, 5 mg of RNA was used for preparation of RNA-seq libraries as described by Zhong et al. (2011) with minor modifications. Briefly, poly(A) RNA was isolated from total RNA using Dynabeads oligo(dT)25 (Invitrogen), fragmented at 94°C for 5 min and eluted. Single-strand cDNA was synthesized using random primers, dNTPs, and SuperScript III (Invitrogen) reverse transcriptase. Second-strand cDNA was generated using DNA polymerase I (Enzymatics) and dATP, dCTP, dGTP, and dUTPs. After end-repair (Enzymatics), dA-tailing with Klenow 3′-5′ (Enzymatics), and adapter ligation (Quick T4 DNA Ligase; New England Biolabs), the second strand (dUTP-containing) was digested by uracil DNA glycosylase (Enzymatics). The remaining first-strand adaptor-ligated cDNA was used for 13 to 15 cycles of PCR enrichment with NEB-Next High-Fidelity PCR Master Mix (New England Biolabs). Sequencing of 12 wheat libraries (BL, CASL*2BS, and F2 progeny of the cross BL × CASL*2BS) was performed on two lanes of HiSeq 2500 using v3 clustering and sequencing reagents; six libraries were multiplexed on each lane (two libraries are used in this study). The sequencing was run using RTA 1.17.20 and analyzed by CASAVA 1.8.2, resulting in 14 to 16 million passed filter 100 base paired end reads per library. Sickle (Joshi and Fass, 2011) was used (in its paired end mode) for trimming the reads. Sequences shorter than 50 bases after trimming were discarded (only 3′ trimming was allowed). The RNA-seq reads were combined and assembled into transcripts using Trinity (version trinityrnaseq-r2013-02-25; Grabherr et al., 2011) with the option -SS_lib_type RF. A total of 396,155 transcripts and 157,340 components were obtained. RSEM (Li and Dewey, 2011) was used for abundance estimation of the components. The obtained transcripts were annotated using Blast2GO (Conesa et al., 2005). The counts were TMM-normalized using edgeR (Robinson and Oshlack, 2010), and TMM-normalized FPKM matrix was calculated using Trinity’s script run_TMM_normalization_write_FPKM_matrix.pl. Two libraries were used in this study- BL and CASL*2BS from peduncle tissue. Hits for the RNA-seq transcripts of barley and wheat genes were received from BLAST at http://www.gramene.org/. For RT-qPCR, RNA was extracted with the TRIzol method using standard TRIzol RNA extraction protocol. DNase treatment and reverse transcription reactions were performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Primers are listed in Supplemental Table 4. Fast SYBR Green reagent and the StepOnePlus instrument (Applied Biosystems) were used for RT-qPCR. Reaction steps were as follows: initial step in the thermal cycler for 20 s at 95°C, followed by PCR amplification for 40 cycles of 3 s at 95°C and 30 s at 59°C, and finally dissociation analysis to confirm the specificity of PCR products. Total reaction volume was 10 µL, containing 2.5 µM of each primer. At least three biological replicate were analyzed for each biological group. Relative transcript levels were calculated according to the ΔΔCt method (Livak and Schmittgen, 2001). Normalization was done by the following reference genes: wheat Actin for RT-qPCR in BL and CASL*2BS internodes, barley Actin for Bonus and cer-cqu.724, and CDC48 for the VIGS experiment (Supplemental Table 4).
CYP450 Nomenclature
The wheat CYP450 proteins in this study were kindly annotated by David R. Nelson (University of Tennessee, Memphis, TN) as follows: wheat DMC (cv BL)-CYP709J4, Zavitan P450-1.1- CYP709J4 ortholog, Zavitan P450-2.1- CYP709J5 (partial), and Zavitan P450-3.1- CYP96B30.
Gene Coexpression Analysis
Wheat transcriptome data (https://urgi.versailles.inra.fr/files/RNASeqWheat/) of five organs during three developmental stages was used for coexpression analysis of the cluster genes. Reads were aligned to the reference transcriptome (created by trinity; see “Transcriptome Sequencing, Assembly and Analysis, and RT-qPCR” above). Expression levels were estimated using RSEM (Li and Dewey, 2011). Cluster genes were used as “baits” for the coexpression analysis. The analysis was performed using the Pearson correlation coefficient with r-value ≥ 0.85.
Development of Molecular Markers on the W1 and the Iw1 Region
PCR markers were developed from selected 90K SNPs showing polymorphism between the parental lines using TaqMan SNP assay (De la Vega et al., 2005) or high-resolution melt methods (Vossen et al., 2009). In addition, three presence/absence markers (wheat DMC, DMP, and DMH) were developed based on results from the RNA-seq experiment described above. Sequence data were obtained from the International Wheat Genome Sequencing Consortium (IWGSC) website and from the wild emmer wheat genome assembly (The International Wild Emmer Wheat Sequencing Consortium; http://wewseq.wix.com/consortium) of the Triticum dicoccoides accession ‘Zavitan’. Specific primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/). Sequence alignments were performed using NCBI-BLAST in order to match the 90K markers to their corresponding genes. BLAST hits were filtered to e-value threshold of 0.0001, >99% identity (sequence similarity/match), and more than 75% of length. Two closely linked EST markers (CD893659; Xu et al., 2015), JIC009 (Adamski et al., 2013), and three SSR markers, Xgwm614 (Röder et al., 1998), Xwmc661, and Xwmc764 (Somers et al., 2004), were also integrated into the current mapping effort. Sequences of all the markers are available in Supplemental Table 2.
Annotation of the W1 Interval and Genotyping with the 90K iSelect SNP Chip
Genomic sequence of chromosome 2B of wild emmer accession Zavitan was annotated between the flanking genetic markers of W1, JIC007, and JIC010. As a first step, the transposable elements were annotated by hand using the TREP database and the nonredundant nucleotide collection hosted on NCBI. The remaining sequence was then searched for open reading frames (ORFs) using the Fgenesh algorithm. The putative genes were translated and compared with the nonredundant protein database of NCBI. In general, ORFs were considered as real genes if their translated protein contained at least one complete protein domain. In addition, the predicted ORFs were tested against the IPK barley BLAST databases “assembly_WGSMorex,” “HC_genes_CDS_Seq,” and “full length cDNA” using BLAST (http://apex.ipk-gatersleben.de/apex/f?p=284:10:32887702692733::NO). The presence of an orthologous locus in barley was not a requirement to call a predicted ORF a real gene. Genotyping procedure for the 90K chip was performed on the parental line DNAs (BL, CASL*2BS, Svevo, and TTD140) as described by Wang et al. (2014b) at the UC Davis Genome Center. Genotypic clusters for each SNP were determined using the manual option of GenomeStudio version 1.9.4 (Illumina), based on the data from all of the described genotypes. For gene sequences, see Supplemental File 1.
Generation of Phylogenetic Trees
Protein sequences were aligned using Muscle web tool (http://www.ebi.ac.uk/Tools/msa/muscle/) with default parameters and Fasta output format. The maximum likelihood phylogenetic tree was created using MEGA6 (for P450 tree in Figure 2D; Tamura et al., 2013) or MEGA7 (for Supplemental Figure 4; Kumar et al., 2016), both with the Jones-Taylor-Thornton matrix based substitution model and a 100 bootstrap replications. P450 proteins in the phylogenetic tree (Figure 2D) included TaDMC, six Zavitan P450s, barley DMC homolog (MLOC_12151), and P450s with known activity (Kandel et al., 2005; Greer et al., 2007; Pinot and Beisson, 2011). The tree was rooted using CYP79A1 from Sorghum bicolor as an outgroup. Text files corresponding to the alignments used can be found in Supplemental Data Sets 2 to 4.
Construction of three BAC Libraries from TTD140
High molecular weight (HMW) DNA was prepared from 20 g of young leaves of T. turgidum accession TTD140 as described by Peterson et al. (2000). Embedded HMW DNA was partially digested with HindIII (New England Biolabs), subjected to two size selection steps, and ligated into pIndigoBAC-5 HindIII-Cloning Ready vector (Epicentre Biotechnologies). Pulsed-field migration programs, electrophoresis buffer, and ligation desalting conditions were performed according to Chalhoub et al. (2004). The first resulting BAC library was named Ttu-B-TTD140. It consisted of 332,160 BAC clones with mean insert size of 126 kb, which represented ∼3.3-fold coverage of total TTD140 genome. This first BAC library was classically arrayed into 865 384-well microtiter plates. High-density colony filters were prepared for this whole genomic BAC library. Labeling and hybridization (with 33P) were performed as described by Gonthier et al. (2010). Positive BAC clones were validated individually by PCR amplification using the primer pairs used for probes synthesis. Two additional BAC libraries named Ttu-B-TTD140-ng (HindIII restriction) and Ttu-B-TD140e-ng (EcoRI restriction) were constructed from the same plant material using a nongridded BAC library protocol based on Isidore et al. (2005) with the following modifications: growth of pooled BAC clones in liquid LB medium, BAC pool DNA amplification by the Whole Genome Genomiphi.v2 phi29 enzyme kit (GE Healthcare) instead of DNA extraction, and use of secondary pooling steps to identify positive clone coordinates with less screening effort after clone picking. HMW DNA extractions and sizing steps were processed as for the first classical BAC library. For Ttu-B-TTD140-ng, 461,540 BAC clones were produced (117 kb average insert size) representing ∼4.5-fold coverage of genome. For Ttu-B-TTD140-ng, 652,427 BAC clones (103 kb average insert size) were produced representing ∼5.6-fold coverage of genome. BAC clones were divided in a total of 1328 pools before overnight growth and DNA amplification. Diluted pools were screened using specific PCR markers previously developed. All these BAC library resources and dedicated screening tools are available upon request (http://cnrgv.toulouse.inra.fr).
BSMV-VIGS of β-Diketone Biosynthesis Genes
The BSMV-VIGS system described by Yuan et al. (2011) comprising three T-DNA binary plasmids, pCaBS-α, pCaBS-β, and pCa-γbLIC, was used in this study. In silico predictions by si-Fi software (siRNA Finder; http://labtools.ipk-gatersleben.de) were used to select the most effective gene-specific fragments for silencing, ranging from 362 to 387 bp in size. According to si-Fi results, it was not possible to identify a region of the wheat DMP gene that would silence DMP alone. In order to mitigate the possible confounding effects of off-target silencing of other PKS family members, two nonoverlapping fragments of the DMP gene were cloned separately to generate two independent VIGS constructs designed to target DMP for silencing (VIGS constructs BSMV:asDMP-1 and BSMV:asDMP-2). DMP-1, DMP-2, and DMH fragments were first amplified from BL cDNA (for primers, see Supplemental Table 4) and cloned into pGEM-T vector (Promega), followed by cloning into pCa-γbLIC in antisense orientation using a ligation-independent cloning strategy. BSMV:mcs4D, which contains noncoding sequence, was included in all VIGS experiments as a nonsilencing virus control. Transformation of the BSMV binary plasmids into Agrobacterium tumefaciens strain GV3101 and agroinfiltration into 3- to 4-week-old N. benthamiana plants were performed as described previously (Lee et al., 2014). Sap from the infiltrated N. benthamiana leaves was used to mechanically inoculate the third fully expanded leaf of 28-d-old wheat plants at the four-leaf stage. Samples for RT-qPCR analysis to determine target gene silencing success were harvested from the flag leaf sheaths of spikes in early boot stage (GS45-47). A minimum of three independent samples per virus treatment, each sample harvested from an individual plant, were analyzed. Flag leaf sheath tissue samples for GC-MS analysis of wax extracts were taken from fully emerged wheat spikes prior to anthesis (corresponding to GS58-59). A minimum of four independent samples per virus treatment were analyzed for each tissue type, with each sample harvested from an individual plant.
Barley cer Mutant Screen
Barley β-diketone cer mutants were ordered from the NordGen bank. The cDNAs corresponding to the cluster genes DMP, DMH, and DMC were amplified in the 20 cer mutants (Figure 5A) and two corresponding wild-type cultivars Bonus and Foma. Oligonucleotides used for amplification are listed in Supplemental Table 4. The cDNAs were amplified using OptiTaq DNA Polymerase (EURX), sequenced, and compared with the relevant wild-type cultivar.
Supplemental Data
Supplemental Figure 1. General scheme of biosynthetic pathways generating different cuticular wax components.
Supplemental Figure 2. Illustration of wheat parental and chromosome arm substitution lines used in this study.
Supplemental Figure 3. Amplification of markers developed for the three candidate genes in glaucous and nonglaucous wheat.
Supplemental Figure 4. Phylogenetic trees of Zavitan proteins.
Supplemental Figure 5. Virus-induced gene silencing of the β-diketone genes reduces wax accumulation and deposition of epicuticular wax crystals.
Supplemental Figure 6. The barley cer-qu.813 double and cer.cqu-724 triple mutants have deletions that include the metabolic gene cluster genes.
Supplemental Figure 7. Compound identification of E. coli expressing HvDMH metabolites.
Supplemental Text 1. Fine genetic map of 2BS W1/Iw1 loci in different segregating populations.
Supplemental Text 2. Analysis of Zavitan W1/Iw1 interval.
Supplemental Text 3. Comparison of the W1/Iw1 interval between the wild emmer accessions TTD140 and Zavitan.
Supplemental Table 1. Linked deleted markers in the TTD140 accession detected by the 90K chip analysis.
Supplemental Table 2. Markers and oligonucleotides used for linkage analysis.
Supplemental Table 3. Homology between Zavitan, TTD140, and the corresponding barley genes.
Supplemental Table 4. Oligonucleotides used in this study.
Supplemental File 1. Gene sequences of β-diketone biosynthesis and those annotated in the genome of the Zavitan accession.
Supplemental Data Set 1. Coexpressed genes using the three β-diketone genes as baits.
Supplemental Data Set 2. Text file of alignments corresponding to the phylogenetic tree of PKS proteins in Supplemental Figure 4.
Supplemental Data Set 3. Text file of alignments corresponding to the phylogenetic tree of hydrolase proteins in Supplemental Figure 4.
Supplemental Data Set 4. Text file of alignments corresponding to the phylogenetic tree of CYP450 proteins in Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Moshe Feldman for kindly providing the wheat lines BL and CASL*2BS used in the study. We thank Rebecca Lauder from Bioimaging at Rothamsted Research, UK, for conducting part of the electron microscopy experiments and Tamar Unger from the Structural Proteomics Unit, faculty of biochemistry at the Weizmann Institute of Science for assistance with protein expression. We thank David R. Nelson for the kind assistance with CYP nomenclature. G.F. is Incumbent of the David and Stacey Cynamon Reaserch Fellow Chair in Genetics and Personalized Medicine. This work was supported by an Israel Science Foundation (ISF) personal grant to A.A. (ISF Grant 646/11) and the European Research Council (SAMIT-FP7 program). We thank the Adelis Foundation, Leona M. and Harry B. Helmsley Charitable Trust, Jeanne and Joseph Nissim Foundation for Life Sciences, Tom and Sondra Rykoff Family Foundation Research, and the Raymond Burton Plant Genome Research Fund for supporting the A.A. lab activity. A.A. is the incumbent of the Peter J. Cohn Professorial Chair. This work was also supported by Natural Sciences and Engineering Research Council Discovery Grant 2208738 (Canada), the Canada Foundation for Innovation Grant 201681, and UK Biotechnology and Biological Sciences Research Council Grants BB/J004588/1, BB/H018824/1, and BB/J00426X/1.
AUTHOR CONTRIBUTIONS
S.H.-A. designed and performed the research and wrote the article. O.S. performed molecular characterization and fine mapping and wrote the article. R.C.R. performed BL and CASL*2BS wax analysis, assisted in GC-MS VIGS experiment analysis, and wrote the article. W.-S.L. performed VIGS assays, assisted in VIGS analysis, and wrote the article. N.M.A. analyzed the physical maps, performed fine mapping, and wrote the article. S.M. assisted with GC-MS operation and analysis. E.A.-S. assisted in the RNA-seq analysis and performed coexpression. M.L. performed coexpression. S.V. and H.B. performed BAC screening and sequencing. G.F. analyzed the RNA-seq data. E.K. conducted the BL and CASL*2BS electron microscopy experiments. G.B.-Z. was involved with generation of the wild emmer wheat reference genome. N.A. assisted in the experiments. C.U. and K.K. designed part of the research and wrote the article. A.A., A.D., and R.J. designed the research and wrote the article.
Glossary
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- PKS
polyketide synthase
- GC-MS
gas chromatography with mass spectrometry
- VIGS
virus-induced gene silencing
- PTC
premature termination codon
- ORF
open reading frame
- HMW
high molecular weight
References
- Adamski N.M., Bush M.S., Simmonds J., Turner A.S., Mugford S.G., Jones A., Findlay K., Pedentchouk N., von Wettstein-Knowles P., Uauy C. (2013). The inhibitor of wax 1 locus (Iw1) prevents formation of β- and OH-β-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. Plant J. 74: 989–1002. [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., Guo Y., Austin M.B., Ramsey J., Fridman E., Pichersky E., Noel J.P. (2012). Emergent decarboxylase activity and attenuation of α/β-hydrolase activity during the evolution of methylketone biosynthesis in tomato. Plant Cell 24: 1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., Zhao R., Pearce S., Jun Y., Uauy C., Tabbita F., Fahima T., Slade A., Dubcovsky J., Distelfeld A. (2014). Functional characterization of GPC-1 genes in hexaploid wheat. Planta 239: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber H.N., Netting A.G. (1968). Chemical genetics of β-diketone formation in wheat. Phytochemistry 7: 2089–2093. [Google Scholar]
- Ben-Israel I., Yu G., Austin M.B., Bhuiyan N., Auldridge M., Nguyen T., Schauvinhold I., Noel J.P., Pichersky E., Fridman E. (2009). Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 151: 1952–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G., Figini M.L. (1986). Epicuticular waxes of glaucous and nonglaucous durum wheat lines. J. Agric. Food Chem. 34: 429–433. [Google Scholar]
- Borisjuk N., Hrmova M., Lopato S. (2014). Transcriptional regulation of cuticle biosynthesis. Biotechnol. Adv. 32: 526–540. [DOI] [PubMed] [Google Scholar]
- Boutanaev A.M., Moses T., Zi J., Nelson D.R., Mugford S.T., Peters R.J., Osbourn A. (2015). Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl. Acad. Sci. USA 112: E81–E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva S., Daviet L., Wolfender J.-L., Fitzpatrick T.B. (2014). The rise of operon-like gene clusters in plants. Trends Plant Sci. 19: 447–459. [DOI] [PubMed] [Google Scholar]
- Chalhoub B., Belcram H., Caboche M. (2004). Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnol. J. 2: 181–188. [DOI] [PubMed] [Google Scholar]
- Chatzivasileiadis E.A., Sabelis M.W. (1997). Toxicity of methyl ketones from tomato trichomes to Tetranychus urticae Koch. Exp. Appl. Acarol. 21: 473–484. [Google Scholar]
- Chen G., et al. (2011). An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc. Natl. Acad. Sci. USA 108: 12354–12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.C. (2005). Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21: 10–19. [DOI] [PubMed] [Google Scholar]
- Chou K.C., Shen H.B. (2007). Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 100: 665–678. [DOI] [PubMed] [Google Scholar]
- Chou K.C., Shen H.B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3: 153–162. [DOI] [PubMed] [Google Scholar]
- Chou K.C., Shen H.B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5: e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cárdenas P.D., et al. (2016). GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 7: 10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Vega F.M., Lazaruk K.D., Rhodes M.D., Wenz M.H. (2005). Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP genotyping assays and the SNPlex genotyping system. Mutat. Res. 573: 111–135. [DOI] [PubMed] [Google Scholar]
- Dogru E., Warzecha H., Seibel F., Haebel S., Lottspeich F., Stöckigt J. (2000). The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the alpha/betahydrolase super family. Eur. J. Biochem. 267: 1397–1406. [DOI] [PubMed] [Google Scholar]
- Febrero A., Fernández S., Molina-Cano J.L., Araus J.L. (1998). Yield, carbon isotope discrimination, canopy reflectance and cuticular conductance of barley isolines of differing glaucousness. J. Exp. Bot. 49: 1575–1581. [Google Scholar]
- Ferrer J.L., Jez J.M., Bowman M.E., Dixon R.A., Noel J.P. (1999). Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6: 775–784. [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification–independent assembly of operon-like gene clusters in different plants. Science 320: 543–547. [DOI] [PubMed] [Google Scholar]
- Fischer R., Wood J. (1979). Drought resistance in spring wheat cultivars. III.* Yield associations with morpho-physiological traits. Aust. J. Agric. Res. 30: 1001–1020. [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699. [DOI] [PubMed] [Google Scholar]
- Fridman E., Wang J., Iijima Y., Froehlich J.E., Gang D.R., Ohlrogge J., Pichersky E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García S., García C., Heinzen H., Moyna P. (1997). Chemical basis of the resistance of barley seeds to pathogenic fungi. Phytochemistry 44: 415–418. [DOI] [PubMed] [Google Scholar]
- Gonthier L., Bellec A., Blassiau C., Prat E., Helmstetter N., Rambaud C., Huss B., Hendriks T., Bergès H., Quillet M.C. (2010). Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Res. Notes 3: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S., Wen M., Bird D., Wu X., Samuels L., Kunst L., Jetter R. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145: 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen-Avivi S., Lashbrooke J., Costa F., Aharoni A. (2014). Scratching the surface: genetic regulation of cuticle assembly in fleshy fruit. J. Exp. Bot. 65: 4653–4664. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Isidore E., Scherrer B., Bellec A., Budin K., Faivre-Rampant P., Waugh R., Keller B., Caboche M., Feuillet C., Chalhoub B. (2005). Direct targeting and rapid isolation of BAC clones spanning a defined chromosome region. Funct. Integr. Genomics 5: 97–103. [DOI] [PubMed] [Google Scholar]
- Jeffree C.E. (2006). The fine structure of the plant cuticle. In Biology of the Plant Cuticle, Riederer M., Muller C., eds (Oxford, UK: Blackwell Publishing), pp. 11–125. [Google Scholar]
- Jez J.M., Ferrer J.-L., Bowman M.E., Dixon R.A., Noel J.P. (2000). Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39: 890–902. [DOI] [PubMed] [Google Scholar]
- Johnson D.A., Richards R.A., Turner N.C. (1983). Yield, water relations, gas exchange, and surface reflectances of near-isogenic wheat lines differing in glaucousness. Crop Sci. 23: 318–325. [Google Scholar]
- Joshi N.A., Fass J.N. (2011). Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.100). Available at https://github.com/najoshi/sickle.
- Kandel S., Morant M., Benveniste I., Blée E., Werck-Reichhart D., Pinot F. (2005). Cloning, functional expression, and characterization of CYP709C1, the first sub-terminal hydroxylase of long chain fatty acid in plants. Induction by chemicals and methyl jasmonate. J. Biol. Chem. 280: 35881–35889. [DOI] [PubMed] [Google Scholar]
- Kornberg A., Ochoa S., Mehler A.H. (1947). Spectrophotometric studies on the decarboxylation of beta-ketoacids. Fed. Proc. 6: 268. [PubMed] [Google Scholar]
- Kozubek A., Tyman J.H. (1999). Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem. Rev. 99: 1–26. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol., http://dx.doi.org/10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Zhao L., Eveleigh N., Yu Y., Chen X., Kunst L. (2015). The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 167: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Zhao L., McFarlane H.E., Aiga M., Lam V., Hooker T.S., Kunst L. (2012). RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 159: 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Rudd J.J., Kanyuka K. (2015a). Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 79: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Rudd J.J., Hammond-Kosack K.E., Kanyuka K. (2014). Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 27: 236–243. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Devonshire B.J., Hammond-Kosack K.E., Rudd J.J., Kanyuka K. (2015b). Deregulation of plant cell death through disruption of chloroplast functionality affects asexual sporulation of Zymoseptoria tritici on wheat. Mol. Plant Microbe Interact. 28: 590–604. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Song Y., Century K., Straight S., Ronald P., Dong X., Lassner M., Zhang Y. (2001). A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 27: 235–242. [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Trumble J.T., Kumamoto J. (1987). Activity of volatile compounds in glandular trichomes ofLycopersicon species against two insect herbivores. J. Chem. Ecol. 13: 837–850. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lundqvist U., von Wettstein D. (1962). Induction of eceriferum mutants in barley by ionizing radiations and chemical mutagens. Hereditas 48: 342–362. [Google Scholar]
- Lundqvist U., von Wettstein D. (1971). Stock list for the eceriferum mutants. Barley Genet. Newsl. 1: 97–102. [Google Scholar]
- Lundqvist U., von Wettstein-Knowles P., von Wettstein D. (1968). Induction of eceriferum mutants in barley by ionizing radiations and chemical mutagens. II. Hereditas 59: 473–504. [Google Scholar]
- Maccaferri M., et al. (2015). A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 13: 648–663. [DOI] [PubMed] [Google Scholar]
- Men A.E., Laniya T.S., Searle I.R., Iturbe-Ormaetxe I., Gresshoff I., Jiang Q., Carroll B.J., Gresshoff P.M. (2002). Fast neutron mutagenesis of soybean (Glycine soja L.) produces a supernodulating mutant containing a large deletion in linkage group H. Genome Lett. 1: 147–155. [Google Scholar]
- Merah O., Deléens E., Souyris I., Monneveux P. (2000). Effect of glaucousness on carbon isotope discrimination and grain yield in durum wheat. J. Agron. Crop Sci. 185: 259–265. [Google Scholar]
- Millet E., Rong J.K., Qualset C.O., McGuire P.E., Bernard M., Sourdille P., Feldman M. (2013). Production of chromosome-arm substitution lines of wild emmer in common wheat. Euphytica 190: 1–17. [Google Scholar]
- Molina I., Kosma D. (2015). Role of HXXXD-motif/BAHD acyltransferases in the biosynthesis of extracellular lipids. Plant Cell Rep. 34: 587–601. [DOI] [PubMed] [Google Scholar]
- Nützmann H.-W., Osbourn A. (2014). Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26: 91–99. [DOI] [PubMed] [Google Scholar]
- Nützmann H.W., Huang A., Osbourn A. (2016). Plant metabolic clusters - from genetics to genomics. New Phytol., http://dx.doi.org/10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. (2010). Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol. 154: 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Y., Unger T. (2008). Application of high-throughput methodologies to the expression of recombinant proteins in E. coli. Methods Mol. Biol. 426: 197–208. [DOI] [PubMed] [Google Scholar]
- Peterson D.G., Tomkins J.P., Frisch D.A., Wing R.A., Paterson A.H. (2000). Construction of plant bacterial artificial chromosome (BAC) libraries: an illustrated guide. Journal of Agricultural Genomics 5: 1–100. [Google Scholar]
- Pinot F., Beisson F. (2011). Cytochrome P450 metabolizing fatty acids in plants: characterization and physiological roles. FEBS J. 278: 195–205. [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ahumada Mdel.C., Timmermann B.N., Gang D.R. (2006). Biosynthesis of curcuminoids and gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): identification of curcuminoid synthase and hydroxycinnamoyl-CoA thioesterases. Phytochemistry 67: 2017–2029. [DOI] [PubMed] [Google Scholar]
- Reiss J. (1989). Influence of Alkylresorcinols from Rye and Related Compounds on the Growth of Food-Borne Molds. (St. Paul, MN: American Association of Cereal Chemists). [Google Scholar]
- Riederer M., Schneider G. (1989). Comparative study of the composition of waxes extracted from isolated leaf cuticles and from whole leaves of Citrus: Evidence for selective extraction. Physiol. Plant. 77: 373–384. [Google Scholar]
- Robinson M.D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J.K., Millet E., Manisterski J., Feldman M. (2000). A new powdery mildew resistance gene: introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 115: 121–126. [Google Scholar]
- Röder M.S., Korzun V., Wendehake K., Plaschke J., Tixier M.H., Leroy P., Ganal M.W. (1998). A microsatellite map of wheat. Genetics 149: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707. [DOI] [PubMed] [Google Scholar]
- Sawai S., Saito K. (2011). Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L.M., Adamski N.M., Christensen C.E., Stuart D.B., Vautrin S., Hansson M., Uauy C., von Wettstein-Knowles P. (2016). The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J. Exp. Bot. 67: 2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schondelmaier J., Fischbeck G., Jahoor A. (1993). Linkage studies between morphological and RFLP markers in the barley genome. Barley Genet. Newsl. 22: 57–62. [Google Scholar]
- Scofield S.R., Huang L., Brandt A.S., Gill B.S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138: 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B. (2003). Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Shen H.B., Chou K.C. (2006). Ensemble classifier for protein fold pattern recognition. Bioinformatics 22: 1717–1722. [DOI] [PubMed] [Google Scholar]
- Somers D.J., Isaac P., Edwards K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Takahashi R., Yamamoto J., Yasuda S., Itano Y. (1953). Inheritance and linkage studies in barley. Ber. Ohara Inst. Landw. Forsch. Kurashiki 10: 29–52. [Google Scholar]
- Taketa S., et al. (2008). Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 105: 4062–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A.M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M.S., Olsen C.E., Sato S., Tabata S., Jørgensen K., Møller B.L., Rook F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68: 273–286. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troughton J., Hall D. (1967). Extracuticular wax and contact angle measurements on wheat (Triticum vulgare L.). Aust. J. Biol. Sci. 20: 509–526. [Google Scholar]
- Tsuchiya T. (1972). Cytogenetics of telotrisomics in barley. Barley Genet. Newsl. 2: 93–98. [DOI] [PubMed] [Google Scholar]
- Tsunewaki K. (1962). Monosomic analysis of synthesized hexaploid wheats. Jpn. J. Genet. 37: 155–168. [Google Scholar]
- Tsunewaki K., Ebana K. (1999). Production of near-isogenic lines of common wheat for glaucousness and genetic basis of this trait. Genes Genet. Syst. 74: 33–41. [Google Scholar]
- Unger T., Jacobovitch Y., Dantes A., Bernheim R., Peleg Y. (2010). Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172: 34–44. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Balyan H.S., Langridge P. (2006). Wheat. In Genome Mapping and Molecular Breeding in Plants, Vol. 1, Cereals and Millet, Kole C., ed (Berlin: Springer), pp. 79–134. [Google Scholar]
- von Wettstein-Knowles P. (1995). Biosynthesis and genetics of waxes. In Waxes: Chemistry, Molecular Biology and Functions, Hamilton R.J., ed (Dundee, UK: The Oily Press), pp. 91–129. [Google Scholar]
- von Wettstein-Knowles P. (1976). Biosynthetic relationships between β-diketones and esterified alkan-2-ols deduced from epicuticular wax of barley mutants. Mol. Gen. Genet. 144: 43–48. [Google Scholar]
- von Wettstein-Knowles P. (1992). Molecular genetics of lipid synthesis in barley. In Barley Genetics, VI: Handelshøjskolens Forlag, Vol. II, Munck L., ed (Copenhagen, Denmark: Munksgaard International Publishers), 753–771. [Google Scholar]
- von Wettstein-Knowles P. (2012). Plant Waxes. (Chichester, UK:John Wiley & Sons; ). [Google Scholar]
- von Wettstein-Knowles P., Søgaard B. (1980). The cer-cqu region in barley: Gene cluster or multifunctional gene. Carlsberg Res. Commun. 45: 125–141. [Google Scholar]
- von Wettstein-Knowles P., Søgaard B. (1981). Genetic evidence that cer-cqu is a cluster gene. In Barley Genetics IV: Proceedings of the 4th International Barley Genetic Symposium. (Edinburgh, UK: Edinburgh University Press; ), pp. 625–630. [Google Scholar]
- Vossen R.H., Aten E., Roos A., den Dunnen J.T. (2009). High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum. Mutat. 30: 860–866. [DOI] [PubMed] [Google Scholar]
- Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. (2014a). Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 111: 9259–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., et al.; International Wheat Genome Sequencing Consortium (2014b). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P.R., Xu M., Jin Y., Coates R.M., Peters R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135: 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Yuan C., Wang J., Fu D., Wu J. (2015). Mapping the glaucousness suppressor Iw1 from wild emmer wheat “PI 481521”. The Crop Journal 3: 37–45. [Google Scholar]
- Yang L., Hill M., Wang M., Panjikar S., Stöckigt J. (2009). Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids. Angew. Chem. Int. Ed. Engl. 48: 5211–5213. [DOI] [PubMed] [Google Scholar]
- Yeats T.H., Rose J.K.C. (2013). The formation and function of plant cuticles. Plant Physiol. 163: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Nguyen T.T., Guo Y., Schauvinhold I., Auldridge M.E., Bhuiyan N., Ben-Israel I., Iijima Y., Fridman E., Noel J.P., Pichersky E. (2010). Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 154: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Li C., Yan L., Jackson A.O., Liu Z., Han C., Yu J., Li D. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks J.C., Chang T.T., Konzak C.F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14: 415–421. [Google Scholar]
- Zhang Z., Wang W., Li W. (2013). Genetic interactions underlying the biosynthesis and inhibition of β-diketones in wheat and their impact on glaucousness and cuticle permeability. PLoS One 8: e54129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wei W., Zhu H., Challa G.S., Bi C., Trick H.N., Li W. (2015). W3 is a new wax locus that is essential for biosynthesis of β-diketone, development of glaucousness, and reduction of cuticle permeability in common wheat. PLoS One 10: e0140524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Joung J.G., Zheng Y., Chen Y.R., Liu B., Shao Y., Xiang J.Z., Fei Z., Giovannoni J.J. (2011). High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011: 940–949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.