Abstract
Background
Coats' disease is an uncommon form of retinal telangiectasis, and the identification of novel proteins that contribute to the development of Coats' disease is useful for improving treatment efficacy. Proteomic techniques have been used to study many eye diseases; however, few studies have used proteomics to study the development of Coats' disease.
Methods
Isobaric tagging for relative and absolute protein quantification (iTRAQ) was employed to screen differentially expressed proteins (DEPs) in the aqueous humor (AH) between stage 3A patients (n = 8), stage 3B patients (n = 14), stage 4 patients (n = 2) and control patients (n = 20). Differentially co-expressed proteins (DCPs) were present in all three stages of Coats' disease and were considered disease-specific proteins. These proteins were further analyzed using Gene Ontology (GO) functional annotations.
Results
A total of 819 proteins were identified in the AH, 222 of which were significantly differentially expressed (fold change > 2 and P < 0.05) in the samples from at least one stage of Coats' disease. Of the DEPs, 46 were found among all three stages of Coats' disease and the controls; therefore, they were considered Coats' disease-specific proteins (DCPs). A GO classification analysis indicated that the DCPs were closely related to structural molecule activity, cell adhesion molecule binding and receptor binding. Western blotting confirmed the expression levels of haptoglobin and apolipoprotein C-I were significantly up-regulated in Coats’ disease.
Conclusions
The 46 Coats' disease-specific proteins may provide additional insights into the mechanism of Coats' disease and represent potential biomarkers for identifying individuals with Coats' disease.
Introduction
Coats' disease is a form of abnormal telangiectasia that is primarily characterized by aneurysms of retinal vessels and excessive production of yellowish intraretinal and subretinal exudates. Coats’ disease predominantly affects boys or young men [1], can cause retinal detachment and severe visual loss, and has been classified into five stages: stage 1, stage 2, stage 3, stage 4 and stage 5 [2]. Presently, the main therapeutic strategy for Coats' disease is direct coagulation of the abnormal vessels using techniques such as laser photocoagulation [3, 4]. However, direct coagulation eventually increases the subretinal exudates, which promotes secondary retinal detachment [1] as well as complications related to exudative retinal detachment [5]. Thus, this technique is not effective for cases with severe exudative changes [6].
In recent decades, significant efforts have been devoted to investigating the pathogenesis of Coats' disease; however, the cause remains largely unknown. Recently, proteomics have been widely used to obtain abundant information on individual proteins in various ocular diseases, including cataracts and retinopathy [7–10]. In combination with mass spectrometry, isobaric tagging for relative and absolute protein quantification (iTRAQ) has recently been demonstrated to be a sensitive quantitative proteomic method for high-throughput protein identification and quantification [11]. The AH is an intraocular fluid that plays an important role in supplying nutrients and removing metabolic waste from the avascular tissues of the eye [12]. Accumulating evidence suggests that certain proteins in the AH are closely correlated with the mechanisms underlying many eye disorders [13–15]. Studies have shown that vascular endothelial growth factor (VEGF), an important intraocular cytokine, is significantly up-regulated in AH samples from patients with increasingly severe Coats' disease [16–18]. In addition, the levels of nitric oxide in the AH of patients with Coats' disease are elevated, indicating proteins involve in nitric oxide metabolism may affect Coats’ disease development [19]. These proteins, which include antioxidant and immunoregulatory proteins, are essential for regulating homeostasis and may play a crucial role in the pathogenesis of Coats' disease [20].
In this study, we used iTRAQ to conduct comparative proteome profiling of the AH samples between patients with three different stages of Coats’ disease and control patients to identify disease-specific proteins in the AH. Our findings identified potential AH biomarkers that could be used to predict the development of Coats' disease. In addition, our findings may contribute to a better understanding of the molecular events involved in the pathogenesis of Coats' disease.
Materials and Methods
Ethics statement
All patients provided written informed consent prior to participation, and the clinical study was approved by the Medical Ethics Committee of Capital Medical University.
Patient eligibility and recruitment
A total of 44 participants that included 20 controls (CK, senile cataract patients) and 24 patients with Coats' disease were recruited through the medical ethics committee of Capital Medical University in Beijing, China from 2014 to 2015. The participants provided written consent for the donation and use of their aqueous humor samples. All subjects met the inclusion criteria, which included the absence of ocular disease other than Coats' disease and a lack of systemic antimetabolite or immunosuppressant usage. Samples from patients with Coats' disease were classified as stage 3A (n = 8), 3B (n = 14) and 4 (n = 2) according to previously reported classification standards [1]. Unfortunately, no stage 5 patient was found during the sample collection procedure of our investigation. Relative information on the participants, including their gender and age, are listed in Table 1.
Table 1. Summary of the human AH samples from the patients with Coats' disease and the controls without Coats' disease.
| Group/Stage | Specimen | Average age (year) | Gender (F: female, M: male) |
|---|---|---|---|
| control | 20 | 68.15 ± 10.27 | 9 F, 11 M |
| 3A | 8 | 7.38 ± 3.11 | 1 F, 7 M |
| 3B | 14 | 6.93 ± 3.17 | 1 F, 13 M |
| 4 | 2 | 11.5 ± 2.12 | 2 M |
AH collection and processing
The AH samples were obtained from 44 patients using a 26-gauge needle inserted into the anterior chamber of the eye according to previously reported methods [7]. Immediately after collection, the aqueous humor was transferred to dust-free Eppendorf tubes containing a protease inhibitor cocktail. The samples were then centrifuged at 1500 × g for 15 min at 4°C. The supernatant was collected and centrifuged again at 15000 × g for 15 min at 4°C to extract proteins and then stored at −80°C for further analysis.
Proteomic profiling using iTRAQ labeling and LC-MS/MS
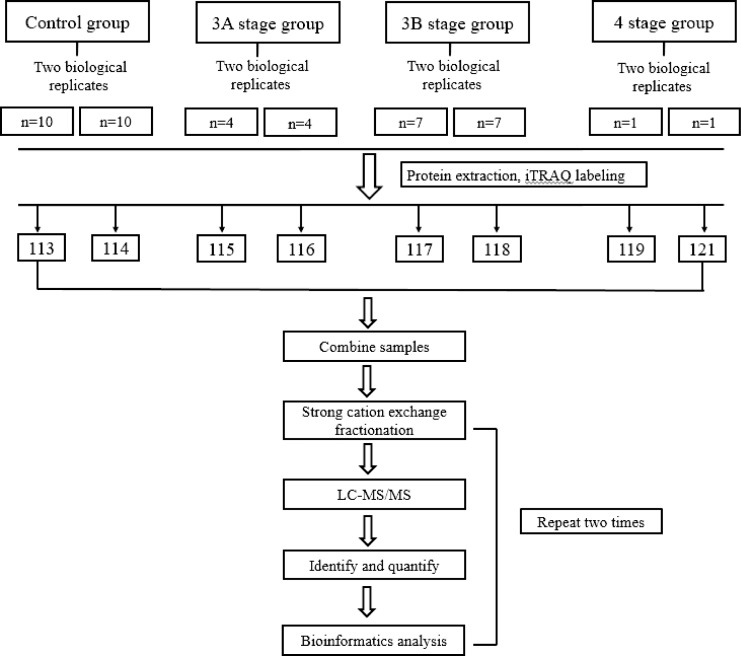
ITRAQ labeling was conducted according to previously reported methods [21]. Briefly, the protein samples from each group were harvested using cold acetone, digested with trypsin overnight at 37°C, and then subjected to iTRAQ labeling. The proteins extracted from the aqueous humor samples were divided into two duplicates and labeled as 113–114 (control), 115–116 (stage 3A), 117–118 (stage 3B) and 119–121 (stage 4) using an iTRAQ Reagent 8-Plex kit (Shanghai AB Sciex Analytical Instrument Trading Co., Shanghai, China) and then vacuum dried. A summary of the workflow performed in the current study is illustrated in Fig 1.
Fig 1. Schematic representation of the workflow of the iTRAQ experiment.
Subsequently, the labeled peptides were separated using strong cation exchange (SCX) chromatography and collected in ten SCX fractions. Each SCX fraction was analyzed on a Triple TOF 5600 system (AB Sciex) in duplicate and expressed as run 1 and run 2. The mass spectra of each iTRAQ labeled sample were obtained in an information-dependent acquisition mode. The mass range was set as 100–2000 m/z for the product ion spectra, and the reporting iTRAQ tags (113 and 114 m/z) were enhanced for quantitation. The ratio of the iTRAQ reporter ion intensities were used to measure the relative amount of the peptides and proteins in each sample according to previously published methods [22, 23]. Database searches were conducted using ProteinPilot Software version 4.5.
Proteins that presented a fold change greater than 2.0 and a p-value less than 0.05 between the patients and CK were considered differentially expressed proteins (DEPs). Hierarchical clustering of the selected probe sets was performed using a Pearson’s correlation analysis for the distance matrix and the Ward's linkage.
Bioinformatics analysis of identified proteins
The DEPs were annotated using the GO database (http://www.geneontology.org/), and the protein classification was performed based on functional annotations using Gene Ontology (GO) terms for cellular components, biological processes and molecular functions. In addition, 46 identified differentially co-expressed proteins (DCPs) were assigned a gene symbol (Table 2) using the Panther database (http://www.pantherdb.org/). We analyzed the DCPs between the patients and CK and calculated a significance value for the GO categories using a cut-off criterion (p < 0.05).
Table 2. DEPs between the patients with stage 3A, 3B and 4 Coats’ disease and the controls (p < 0.05).
| 3A vs. CK | 3B vs. CK | 4 vs. CK | DCPs |
|---|---|---|---|
| 93 | 125 | 146 | 46 |
Western blotting
Western blot analyses were performed on AH samples to validate the results obtained by iTRAQ for some of the proteins that showed significant differences. In brief, equal amounts of sample (20 μg) were loaded on a 10% sodium dodecyl sulfate polyacrylamide gel. The proteins were then transferred into a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, California, USA). After blocking with 2% bovine serum albumin, the membranes were probed with monoclonal antibodies of haptoglobin (sn: ab13429) and apolipoprotein C-I (sn: ab198288) (Abcam, Cambridge, MA, USA) followed by secondary antibody. The signal was then detected by enhanced chemiluminescence using LAS4000 system (GE, Fairfield, Connecticut, USA).
Results
Image acquisition and description of retinal morphology in Coat' disease
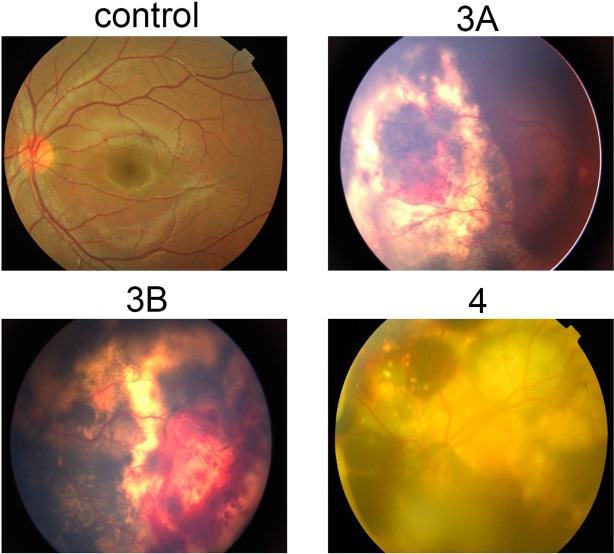
After full dilation of the pupils, retinal photographs were obtained from patients with different stages of Coats' disease and the control participants using a Zeiss FF450+ fundus camera (Zeiss, Germany; and Imedos Systems, Germany). As shown in Fig 2, the control group did not present abnormal vascular changes. However, the fundus examination revealed that stage 3A patients presented with subtotal exudative serous retinal detachment and stage 3B patients presented with total yellow exudative retinal detachment. Compared with stage 3A, additional subretinal fluid and hyperpigmentation were observed in stage 3B. Retinal detachment accompanied by a large number of abnormal expansions of retinal blood vessels and retinal surface bleeding were observed in stage 4 patients (Fig 2).
Fig 2. Representative fundus views of the different stages of Coats' disease, including stage 3A, stage 3B, and stage 4, and the controls.
Proteomic analysis to detect differentially expressed proteins in the aqueous humor
To investigate the mechanisms leading to the development of Coats' disease, a high-throughput quantitative proteomics analysis using iTRAQ labeling was performed to profile the aqueous humor from the patients presenting with three stages of Coats' disease and the control patients. The results of the iTRAQ analysis revealed that the protein profiles of the three stages of Coats' disease were significantly different. Of the total 819 identified proteins, 222 were significantly differentially expressed between the Coats' disease patients and the controls (S1 and S2 Tables). The hierarchical clustering heat map of the 222 DEPs is shown in Fig 3.
Fig 3. Heat map analysis of the differentially expressed proteins between the CK group and three groups of patients with different stages of Coats' disease.
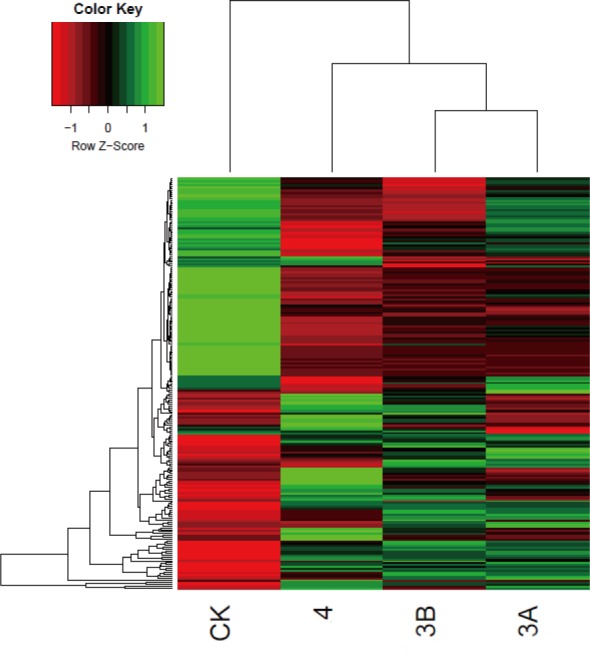
The color scale shown at the top illustrates the relative protein expression level across all the samples. Red represents an expression level lower than the mean, whereas green represents an expression level above the mean.
Proteomic alterations in the aqueous humor of Coats' disease patients
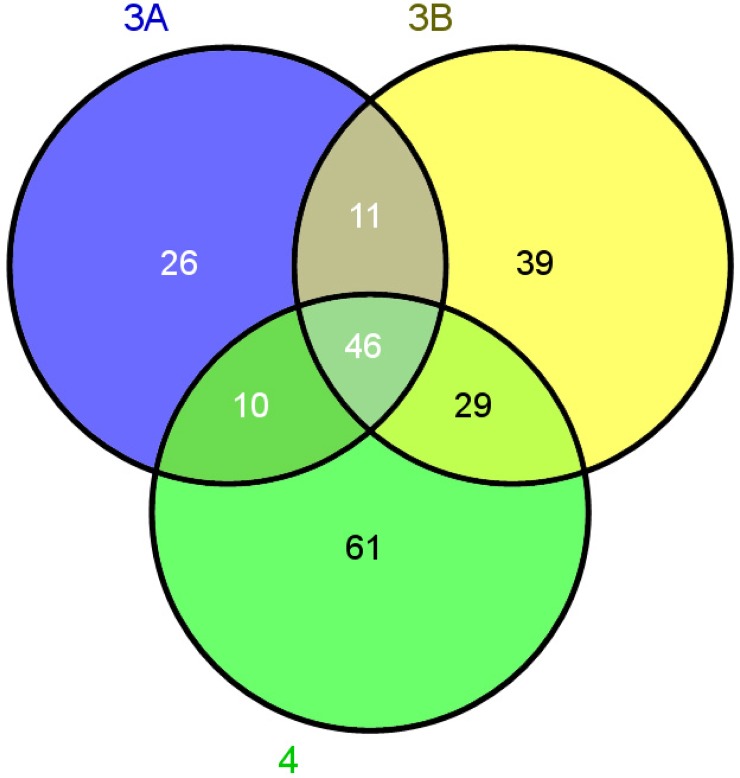
We then identified the proteomic differences between the patients with stage 3A, 3B and 4 of Coats' disease and the CK group. As shown in Table 2, 93 proteins were differently expressed between the stage 3A patients and the CK group. Similarly, we found 125 and 146 DEPs between the stage 3B patients and stage 4 patients and the CK group, respectively. Moreover, a Venn diagram was used to analyze the DCPs between the three stages of Coats' disease and the CK group (Fig 4). The results showed that 46 proteins were common between each stage of Coats' disease and the controls. Among those, 17 proteins were up-regulated and 29 proteins were down-regulated in the patients with Coats' disease (Fig 4 and Table 3).
Fig 4. Venn diagram indicating the differentially co-expressed proteins (DCPs) from the patients with the three stages of Coats' disease.
The numbers in parentheses indicate the total number of DCPs in stage 3A, stage 3B and stage 4, which are represented by purple, yellow and green, respectively.
Table 3. Forty-six DCPs, including 29 down-regulated proteins (log FC < -1) and 17 up-regulated proteins (log FC > 1).
| Gene symbol | Protein Name | Log FC* |
|---|---|---|
| FGB | Fibrinogen beta chain | 4.08 |
| CFH | Complement factor H | 3.14 |
| FN1 | Isoform 10 of Fibronectin | 3.23 |
| ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | 3.31 |
| FGG | Isoform Gamma-A of Fibrinogen gamma chain | 4.27 |
| HP | Haptoglobin | 4.28 |
| IGHA1 | Ig alpha-1 chain C region | 3.17 |
| AFM | Afamin | 3.15 |
| IGHM | Isoform 2 of Ig mu chain C region | 3.65 |
| LUM | Lumican | 2.26 |
| SERPINA7 | Thyroxine-binding globulin | 1.67 |
| VTN | Vitronectin | 2.77 |
| ITIH1 | Inter-alpha-trypsin inhibitor heavy chain H1 | 1.65 |
| APOC1 | Apolipoprotein C-I | 3.96 |
| SAA4 | Serum amyloid A-4 protein | 1.41 |
| F12 | Coagulation factor XII | 1.54 |
| S100A6 | Protein S100-A6 | 1.51 |
| RBP3 | Retinol-binding protein 3 | -4.06 |
| TTR | Transthyretin | -1.58 |
| CTSD | Cathepsin D | -3.14 |
| CST3 | Cystatin-C | -2.12 |
| MYOC | Myocilin | -3.47 |
| GPX3 | Glutathione peroxidase 3 | -2.64 |
| CRYBB1 | Beta-crystallin B1 | -4.14 |
| SEMA7A | Semaphorin-7A | -3.06 |
| CPE | Carboxypeptidase E | -2.77 |
| C1S | Complement C1s subcomponent | -1.64 |
| ENPP2 | Isoform 3 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | -1.89 |
| CRYBB2 | Beta-crystallin B2 | -3.91 |
| CLSTN1 | Isoform 2 of Calsyntenin-1 | -2.06 |
| CRYGS | Beta-crystallin S | -5.42 |
| SOD3 | Extracellular superoxide dismutase [Cu-Zn] | -2.94 |
| B3GNT1 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase | -1.77 |
| FAM3C | Protein FAM3C | -3.18 |
| CDH2 | Cadherin-2 | -2.47 |
| CRYGD | Gamma-crystallin D | -3.64 |
| WIF1 | Wnt inhibitory factor 1 | -2.37 |
| COL9A1 | Collagen alpha-1(IX) chain | -2.42 |
| IGFBP6 | Insulin-like growth factor-binding protein 6 | -1.37 |
| CRYBA1 | Beta-crystallin A3 | -4.23 |
| MFAP4 | Isoform 2 of Microfibril-associated glycoprotein 4 | -2.00 |
| SEMA4B | Semaphorin-4B | -2.14 |
| ASAH1 | Isoform 2 of Acid ceramidase | -2.61 |
| CTSA | Lysosomal protective protein | -1.77 |
| ALDH1A1 | Retinal dehydrogenase 1 | -1.89 |
| IMPG1 | Interphotoreceptor matrix proteoglycan 1 | -3.32 |
* log FC, log2 of the average protein level fold change (FC) in the samples from stage 3A, stage 3B and stage 4 Coats’ disease patients.
Functional analysis of the identified Coats' disease-related proteins
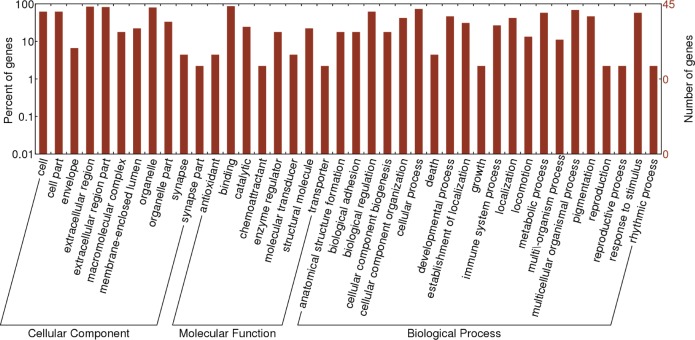
To investigate the functional significance of the DEPs identified in the patients with Coats' disease, 222 DEPs were functionally divided into three groups (cellular components, molecular functions and biological processes) according to the GO annotation analysis (Fig 5). Further analysis (Table 4) revealed that a total of 46 DCPs with a p-value of less than 0.05 were enriched for 6 GO terms. Crystalline lens related proteins, including βB1-crystallin (CRYBB1), βB1-crystallin (CRYBB2), γS-Crystallin (CRYGS) and γD-Crystallin (CRYGD), were found to play a role in structural molecule activity (p = 1.56E-06); myocilin (MYOC) was found to play an important role in protein binding (p = 0.00089) and receptor binding (p = 0.00081); apolipoprotein C-1 (APOC1) was found to be essential for enzyme regulator activity (p = 0.0009); and β-fibrinogen (FGB) was found to be closely associated with most GO terms.
Fig 5. GO annotation and functional classification of the differentially expressed aqueous humor proteins using GO terms for cellular components, molecular functions and biological processes.
Table 4. GO term classification of the DCPs between the patients and CK with p-values < 0.01.
| Term | Biological Process description | Protein number | p-value | Genes |
|---|---|---|---|---|
| GO:0005196 | structural molecule activity | 10 | 1.56E-06 | COL9A1/CRYBA1/CRYBB1/CRYBB2/CRYGD/CRYGS/FGB/FGG/IMPG1/LUM |
| GO:0050839 | Cell adhesion molecule binding | 6 | 2.67E-06 | CPE/FGB/FGG/FN1/VTN/SEMA7A |
| GO:0005102 | Receptor binding | 10 | 0.0008104 | FAM3C/FGB/FGG/FN1/IGHA1/IGHM/MYOC/TTR/VTN/SEMA7A |
| GO:0030234 | Enzyme regulator activity | 8 | 0.0009351 | CST3/ALDH1A1/FN1/APOC1/ITIH1/ITIH4/CTSA/SERPINA7 |
| GO:0070011 | Peptidase activity, acting on L-amino acid peptides | 6 | 0.0024658 | CPE/CTSD/F12/CTSA/RBP3/C1S |
| GO:0008233 | Peptidase activity | 6 | 0.0028979 | CPE/CTSD/F12/CTSA/RBP3/C1S |
Western blot
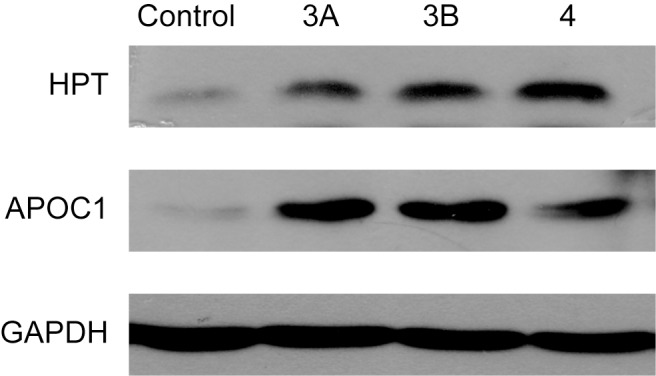
Because of difficulty in acquiring antibodies as well as the limited volume of AH samples, Western blotting was performed for only two of the DCPs, haptoglobin (HPT) and apolipoprotein C-I (APOC1). The results were shonw in Fig 6 and confirmed the up-regulation of these two proteins in Coats’ disease, indicating the potential value of DCPs we identified for further validation and investigation.
Fig 6. Western blotting analyses of 2 DCPs confirmed that the proteins accumulated in three groups of patients.
Discussion
Coats' disease most commonly affects young boys who are often too young to report their symptoms [24]. In our study, 36 patients diagnosed with Coats' disease were classified based on three stages of the disease: stage 3A, stage 3B and stage 4. Most of the patients were between 10 and 15 years of age, which suggests that Coats' disease predominantly affects young males. To identify novel proteins associated with the pathogenesis of Coats' disease, we compared the AH proteins between patients with different stages of Coats' disease and control participants. A heat map analysis showed that the protein profiles from the patients with the three stages of Coats' disease were different from those of the controls, which indicated that the protein content in the AH changes during the development of Coats' disease. Furthermore, we screened 46 DCPs identified between the groups of patients with the three stages of Coats' disease and the controls and found 17 up-regulated proteins and 29 down-regulated proteins in the disease groups compared with the controls. The GO functional enrichment analysis indicated that most of the DCPs were closely correlated with structural molecule activity and molecular functions.
Of the identified DCPs, several crystalline-related proteins, including CRYBB1, CRYBB2, CRYGS and CRYGD, were down-regulated in the AH from patients with Coats' disease. Previous studies have shown that crystalline genes are closely related to lens opacity and microcornea [25]. CRYBB1 accounts for 9% of the total soluble crystalline in the human lens [26, 27] and is a critical protein in the formation of acidic β-crystallin heteromers in the lens. The formation of heteromers is considered important for the maintenance of lens transparency [28]. CRYGS is also important for maintaining lens transparency, and a dysregulation of CRYGS is associated with lamellar cataracts [29, 30]. Therefore, our results suggest that the down-regulation of crystalline-related proteins may play a crucial role in the occurrence of Coats' disease. To the best of our knowledge, vitamin A has diverse biological functions that include sensing light for vision, and the aldehyde form of vitamin A functions as the chromophore for visual pigments in the eye [31, 32]. Plasma RBP is a component of the outer segments of photoreceptors and can mediate cellular vitamin A uptake [33, 34] and prevent the potentially cytotoxic effects of retinoids. In addition, the presence of sufficient quantities of functional RBP3 is critical for photoreceptor survival [35]. The trabecular meshwork (TM) represents the anatomic location with the highest resistance to AH outflow [36]. MYOC, a secreted protein found in human TM, is considered a key regulatory node that governs extracellular matrix turnover homeostasis in the TM [37]. The down-regulation of MYOC promotes AH outflow, which elevates intraocular pressure [38] and may induce the occurrence of Coat's disease. We found that crystalline-related proteins, vitamin A-related proteins and TM skeleton proteins were down-regulated in the AH samples from patients with Coats' disease.
Interestingly, several genes, including APOC1 and FGB, were up-regulated in the AH samples of the patients with Coats' disease (Fig 6). Further analyses indicated that APOC1 is a component of multiple lipoproteins and regulates lipid metabolism and transport [39]. The overexpression of human APOC1 has been demonstrated to produce hyperlipidemia and increase serum lipid levels [40, 41]. Furthermore, APOC1 is closely associated with vascular inflammation and retinitis pigmentosa [42], and reports have indicated that FGB is closely related to elevated plasma fibrinogen levels and ischemic stroke [43, 44].
In summary, our findings revealed that the proteomic composition of AH significantly differed between individuals with Coats' disease and the controls and the proteins identified in this study could serve as potential biomarkers for Coats' disease. In addition, we suggest that the DCPs identified in the patients with Coats' disease contribute to the pathologic changes and complications of Coats' disease. However, further investigations are necessary to explore the exact mechanism by which these DCPs underlie the occurrence of Coats' disease.
Supporting Information
(XLSX)
(XLSX)
Acknowledgments
This study was supported by the Science & Technology Project of Beijing Municipal Science & Technology Commission (code: Z151100001615052), the National Natural Science Foundation of China (No. 81570891), the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (code: ZYLX201307), the National Natural Science Foundation of China (No. 81272981), the Beijing Natural Science Foundation (No. 7151003), the Advanced Health Care Professionals Development Project of Beijing Municipal Health Bureau (No. 2014-2-003) and the Beijing Municipal Administration of Hospitals’ Ascent Plan (Code: DFL20150201). We also thank Dr. Xiang Jin from the Proteomics Center of Chinese Academy of Tropical Agricultural Sciences for kindly help on processing the iTRAQ data.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Science & Technology Project of Beijing Municipal Science & Technology Commission (code: Z151100001615052), the National Natural Science Foundation of China (No. 81570891), the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (code: ZYLX201307), the National Natural Science Foundation of China (No. 81272981), the Beijing Natural Science Foundation (No. 7151003), the Advanced Health Care Professionals Development Project of Beijing Municipal Health Bureau (No. 2014-2-003) and the Beijing Municipal Administration of Hospitals’ Ascent Plan (Code: DFL20150201).
References
- 1.Shields JA, Shields CL, Honavar SG, Demirci H. Clinical variations and complications of Coats disease in 150 cases: the 2000 Sanford Gifford Memorial Lecture. Am J Ophthalmol. 2001; 131(5): 561–571. [DOI] [PubMed] [Google Scholar]
- 2.Shields JA, Shields CL, Honavar SG, Demirci H, Cater J. Classification and management of Coats disease: the 2000 Proctor Lecture. Am J Ophthalmol. 2001; 131(5): 572–583. [DOI] [PubMed] [Google Scholar]
- 3.Theoulakis PE, Halki A, Petropoulos IK, Katsimpris JM. Coats disease in a 14-year-old boy treated with intravitreal ranibizumab and retinal laser photocoagulation. Klin Monbl Augenheilkd. 2012; 229(4): 447–450. 10.1055/s-0031-1299216 [DOI] [PubMed] [Google Scholar]
- 4.Ridley ME, Shields JA, Brown GC, Tasman W. Coats' disease. Evaluation of management. Ophthalmology, 1982; 89(12): 1381–1387. [DOI] [PubMed] [Google Scholar]
- 5.Ogata M, Suzuki T, Nakagawa Y, Hayakawa K, Kawai K. Post-vitrectomy observation of Coats’ disease associated with exudative retinal detachment, successfully treated with long-term silicone oil tamponade. Tokai J Exp Clin Med. 2014; 39(1): 25–28. [PubMed] [Google Scholar]
- 6.Kodama A, Sugioka K, Kusaka S, Matsumoto C, Shimomura Y. Combined treatment for Coats' disease: retinal laser photocoagulation combined with intravitreal bevacizumab injection was effective in two cases. BMC Ophthalmol. 2014; 14: 36 10.1186/1471-2415-14-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang SY, Tsai ML, Wang CY, Chen A, Chou YC, Hsia CW, et al. Proteomic analysis and identification of aqueous humor proteins with a pathophysiological role in diabetic retinopathy. J Proteomics. 2012; 75(10): 2950–2959. 10.1016/j.jprot.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 8.Hadjistilianou T, Giglioni S, Micheli L, Vannoni D, Brogi E, Cevenini G, et al. Analysis of aqueous humour proteins in patients with retinoblastoma. Clin Experiment Ophthalmol. 2012; 40(1): e8–e15. 10.1111/j.1442-9071.2011.02711.x [DOI] [PubMed] [Google Scholar]
- 9.Zhou HY, Yan H, Wang LL, Yan WJ, Shui YB, Beebe DC. Quantitative proteomics analysis by iTRAQ in human nuclear cataracts of different ages and normal lens nuclei. Proteomics Clin Appl. 2015; 9(7–8): 776–786. 10.1002/prca.201400061 [DOI] [PubMed] [Google Scholar]
- 10.Monteiro JP, Santos FM, Rocha AS, Castro-de-Sousa JP, Queiroz JA, Passarinha LA, et al. Vitreous humor in the pathologic scope: insights from proteomic approaches. Proteomics Clin Appl. 2015. 9(1–2): 187–202. 10.1002/prca.201400133 [DOI] [PubMed] [Google Scholar]
- 11.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res. 2006; 5(3): 651–658. [DOI] [PubMed] [Google Scholar]
- 12.To CH, Kong CW, Chan CY, Shahidullah M, Do CW. The mechanism of aqueous humour formation. Clin Exp Optom. 2002; 85(6): 335–349. [PubMed] [Google Scholar]
- 13.Määttä M, Tervahartiala T, Harju M, Airaksinen J, Autio-Harmainen H, Sorsa T. Matrix metalloproteinases and their tissue inhibitors in aqueous humor of patients with primary open-angle glaucoma, exfoliation syndrome, and exfoliation glaucoma. J Glaucoma. 2005; 14(1): 64–69. [DOI] [PubMed] [Google Scholar]
- 14.Ozcan AA, Ozdemir N, Canataroglu A. The aqueous levels of TGF-beta2 in patients with glaucoma. Int Ophthalmol. 2004; 25(1): 19–22. [DOI] [PubMed] [Google Scholar]
- 15.Fleenor DL, Shepard AR, Hellberg PE, Jacobson N, Pang IH, Clark AF. TGFbeta2-induced changes in human trabecular meshwork: implications for intraocular pressure. Invest Ophthalmol Vis Sci. 2006; 47(1): 226–234. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Q, Peng XY, Chen FH, Zhang YP, Wang L, You QS, et al. Vascular endothelial growth factor in Coats' disease. Acta Ophthalmol. 2014; 92(3): e225–228. 10.1111/aos.12158 [DOI] [PubMed] [Google Scholar]
- 17.He YG, Wang H, Zhao B, Lee J, Bahl D, McCluskey J. Elevated vascular endothelial growth factor level in Coats' disease and possible therapeutic role of bevacizumab. Graefes Arch Clin Exp Ophthalmol. 2010; 248(10): 1519–1521. 10.1007/s00417-010-1366-1 [DOI] [PubMed] [Google Scholar]
- 18.Kaul S, Uparker M, Mody K, Walinjkar J, Kothari M, Natarajan S. Intravitreal anti-vascular endothelial growth factor agents as an adjunct in the management of Coats' disease in children. Indian J Ophthalmol. 2010; 58(1): 76–78. 10.4103/0301-4738.58480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Liu ZL. Increased nitric oxide and vascular endothelial growth factor levels in the aqueous humor of patients with coats' disease. J Ocul Pharmacol Ther. 2012; 28(4): 397–401. 10.1089/jop.2011.0168 [DOI] [PubMed] [Google Scholar]
- 20.Elsobky S, Crane AM, Margolis M, Carreon TA, Bhattacharya SK. Review of application of mass spectrometry for analyses of anterior eye proteome. World J Biol Chem. 2014; 5(2): 106–114. 10.4331/wjbc.v5.i2.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu DD, Deng DF, Li X, Wei LL, Li YY, Yang XY, et al. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2014; 14(2–3): 322–331. 10.1002/pmic.201300383 [DOI] [PubMed] [Google Scholar]
- 22.Jin GZ, Li Y, Cong WM, Yu H, Dong H, Shu H, et al. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res. 2011; 10(8): 3418–3428. 10.1021/pr200482t [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Wang G, Angert ER, Wang W, Li W, Zou H. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS One. 2012; 7(2): e30440 10.1371/journal.pone.0030440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris B, Foot B, Mulvihill A. A population-based study of Coats disease in the United Kingdom I: epidemiology and clinical features at diagnosis. Eye (Lond). 2010; 24(12): 1797–1801. [DOI] [PubMed] [Google Scholar]
- 25.Wang KJ, Wang S, Cao NQ, Yan YB, Zhu SQ. A novel mutation in CRYBB1 associated with congenital cataract-microcornea syndrome: the p.Ser129Arg mutation destabilizes the betaB1/betaA3-crystallin heteromer but not the betaB1-crystallin homomer. Hum Mutat. 2011; 32(3): E2050–E2060. 10.1002/humu.21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, et al. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem. 1997; 272(4): 2268–2275. [DOI] [PubMed] [Google Scholar]
- 27.Chan MP, Dolinska M, Sergeev YV, Wingfiled PT, Hejtmancik JF. Association properties of betaB1- and betaA3-crystallins: ability to form heterotetramers. Biochemistry. 2008; 47(42): 11062–11069. 10.1021/bi8012438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajaz MS, Ma Z, Smith DL, Smith JB. Size of human lens beta-crystallin aggregates are distinguished by N-terminal truncation of betaB1. J Biol Chem. 1997; 272(17): 11250–11255. [DOI] [PubMed] [Google Scholar]
- 29.Devi RR, Yao W, Vijayalakshmi P, Sergeev YV, Sundaresan P, Hejtmancik JF. Crystallin gene mutations in Indian families with inherited pediatric cataract. Mol Vis. 2008; 14: 1157–1170. [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, et al. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005; 42(9): 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crouch RK, Chader GJ, Wiggert B, Pepperberg DR. Retinoids and the visual process. Photochem Photobiol. 1996; 64(4): 613–621. [DOI] [PubMed] [Google Scholar]
- 32.Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu Rev Pharmacol Toxicol. 2007; 47: 469–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller J. Interactions of plasma retinol-binding protein with its receptor. Specific binding of bovine and human retinol-binding protein to pigment epithelium cells from bovine eyes. J Biol Chem. 1975; 250(10): 3613–3619. [PubMed] [Google Scholar]
- 34.Rask L, Peterson PA. In vitro uptake of vitamin A from the retinol-binding plasma protein to mucosal epithelial cells from the monkey's small intestine. J Biol Chem. 1976; 251(20): 6360–6366. [PubMed] [Google Scholar]
- 35.Zhu L, Shen W, Zhu M, Coorey NJ, Nguyen AP, Barthelmes D, et al. Anti-retinal antibodies in patients with macular telangiectasia type 2. Invest Ophthalmol Vis Sci. 2013; 54(8): 5675–5683. 10.1167/iovs.13-12050 [DOI] [PubMed] [Google Scholar]
- 36.Seiler T, Wollensak J. The resistance of the trabecular meshwork to aqueous humor outflow. Graefes Arch Clin Exp Ophthalmol. 1985; 223(2): 88–91. [DOI] [PubMed] [Google Scholar]
- 37.Tawara A, Okada Y, Kubota T, Suzuki Y, Taniguchi F, Shirato S, et al. Immunohistochemical localization of MYOC/TIGR protein in the trabecular tissue of normal and glaucomatous eyes. Curr Eye Res. 2000; 21(6): 934–943. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee A, Villarreal G Jr., Rhee DJ. Matricellular proteins in the trabecular meshwork: review and update. J Ocul Pharmacol Ther. 2014; 30(6): 447–463. 10.1089/jop.2014.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skinner NE, Wroblewski MS, Kirihara JA, Nelsestuen GL, Seaquist ER. Sitagliptin Results in a Decrease of Truncated Apolipoprotein C1. Diabetes Ther. 2015; 6(3): 395–401. 10.1007/s13300-015-0123-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berbée JF, van der Hoogt CC, Sundararaman D, Havekes LM, Rensen PC. Severe hypertriglyceridemia in human APOC1 transgenic mice is caused by apoC-I-induced inhibition of LPL. J Lipid Res. 2005; 46(2): 297–306. [DOI] [PubMed] [Google Scholar]
- 41.Muurling M, van den Hoek AM, Mensink RP, Pijl H, Romijn JA, Havekes LM, et al. Overexpression of APOC1 in obob mice leads to hepatic steatosis and severe hepatic insulin resistance. J Lipid Res. 2004; 45(1): 9–16. [DOI] [PubMed] [Google Scholar]
- 42.Avery CL, He Q, North KE, Ambite JL, Boerwinkle E, Fornage M, et al. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet. 2011; 7(10): e1002322 10.1371/journal.pgen.1002322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Li F, Liu G, Cai W, Ling G. [The study of beta-fibrinogen gene—455 G/A, - 148 C/T, 448 G/A polymorphisms and their association with plasma fibrinogen levels]. Zhonghua Xue Ye Xue Za Zhi. 2000; 21(9): 463–465. [PubMed] [Google Scholar]
- 44.Zhao B, Li B, Xing YQ, Xu ZE, Li JL. The beta fibrinogen gene-148C/T polymorphism and its association with plasma fibrinogen's function and level in cerebral infarction. Chinese Journal of Neurology. 2003; 36(6): 450–453. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.