Abstract
The simplest forms of mutations, base substitutions, typically have negative consequences, aside from their existential role in evolution and fitness. Hypermutations, mutations on steroids, occurring at frequencies of 10−2 – 10−4 per base pair, straddle a domain between fitness and death, depending on the presence or absence of regulatory constraints. We portray two facets of hypermutation, one in Escherichia coli involving DNA polymerase V (pol V), the other in humans, involving activation-induced deoxycytidine deaminase (AID). Pol V is induced as part of the DNA-damage-induced SOS regulon, and is responsible for generating the lion’s share of mutations when catalyzing translesion DNA synthesis (TLS). Four regulatory mechanisms, temporal, internal, conformational, and spatial, activate pol V to copy damaged DNA and then deactivate it. On the flip side of the coin, SOS-induced pols V, IV and II mutate undamaged DNA, thus providing genetic diversity heightening long-term survival and evolutionary fitness. Fitness in humans is principally the domain of a remarkably versatile immune system marked by somatic hypermutations (SHM) in immunoglobulin variable (IgV) regions that ensure antibody (Ab) diversity. AID initiates SHM by deaminating C → U, favoring hot WRC (W = A/T, R = A/G) motifs. Since there are large numbers of trinucleotide motif targets throughout IgV, AID must exercise considerable catalytic restraint to avoid attacking such sites repeatedly, which would otherwise compromise diversity. Processive, random, and inefficient AID-catalyzed dC deamination simulates salient features of SHM, yet generates B-cell lymphomas when working at the wrong time in the wrong place.
Keywords: Hypermutation, SOS mutagenesis, DNA polymerase, RecA* nucleoprotein filament, translesion DNA synthesis, AID, immunological diversity
THE IMPACT OF DNA POLYMERASE FIDELITY ON SPONTANEOUS MUTATION
In the beginning, Watson & Crick wrote: “it seems plausible to us that a spontaneous mutation, which we imagine to be a change in the sequence of bases, is due to a base occurring very occasionally in one of the less likely tautomeric forms, at the moment when the complementary chain is being formed” [Watson and Crick 1953b]. Along with deciphering the structure of DNA, this remarkably intuitive insight on an atomic model for how the simplest base substitution mutations might arise predated by three years the discovery of DNA polymerase (pol) in Escherichia coli by A. Kornberg and colleagues in 1956 [Bessman et al., 1956]. In I. R. Lehman’s superb J. Biol. Chem. Reflections article [Lehman 2003], he asks, “Was it possible that the DNA polymerase was performing the template-directed replication proposed by Watson and Crick for their double-stranded structure of DNA [Watson and Crick 1953a]?” The stunning result (quoting Lehman) was that DNA synthesis by the E. coli pol represents replication of a DNA template [Lehman et al., 1958]. This brief historical comment pretty much reflected the state of affairs in 1965, namely that DNA pol performs template directed DNA synthesis save for when a rare base mispair might occur stochastically in the pol active site. In other words, the polymerase has virtually no choice in base selection during DNA synthesis.
In 1965, however, J. Speyer identified a T4 bacteriophage mutant that exhibited strongly elevated spontaneous and base analog-induced mutation rates, ~ 100-fold in a T4-rIIB reporter gene [Speyer 1965]. This mutator mutation mapped in gene 43, which encodes the T4 pol [De Waard et al., 1965]. In 1968, J. Drake identified gp43 T4 pol mutants with similarly dramatic opposite behaviors, having strongly reduced spontaneous and induced mutation rates in rIIB [Drake and Allen 1968]. Soon thereafter, biochemical studies by M. J. Bessman [Muzyczka et al., 1972] and N. Nossal [Gillin and Nossal 1976], revealed that mutator and antimutator phenotypes could be explained by an ability to edit nucleotide misinsertions via a 3′-exonuclease proofreading activity present in T4 pol. Antimutator alleles were highly efficient editors, mutators poor editors, with wild type in between [reviewed in Goodman and Fygenson 1998; Schaaper 1998, focused on antimutators]. And finally, to square the circle, a pol-associated 3′-exonuclease was first observed as a supposed contaminant of E. coli pol I, which Lehman tried to eliminate, of course to no avail [Lehman 2003], and which D. Brutlag and A. Kornberg subsequently identified as having an editing function [Brutlag and Kornberg 1972]. Therefore, while on the one hand, pols act predominantly as DNA template-directed catalysts, they can also strongly influence the fidelity of DNA synthesis, which can profoundly influence the fitness and well-being of all organisms from bacterial viruses to humans.
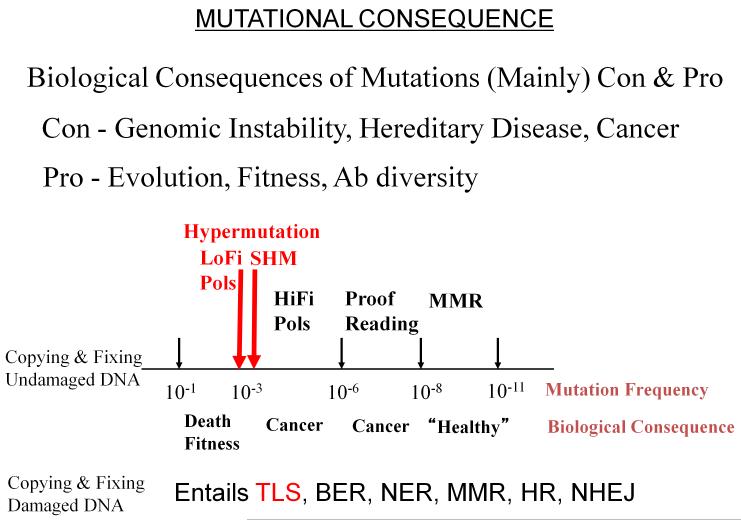
High fidelity DNA pols involved in chromosomal replication, E. coli pol III or human pols δ and ε, typically misinsert deoxynucleotides ~ 10−3 to 10−6 per bp, depending on the specific type of mismatch, e.g., G•T, A•G, and surrounding sequence context. 3′-exo proofreading may reduce the mutational load by about 100-fold. Therefore, even the best scenario for a pol-regulated mutation frequency of about 10−8 per bp would lead to poor biological consequences in human chromosomal DNA containing 3 billion base pairs. An additional 1000-fold reduction of errors with post replication mismatch repair (MMR) is essential to minimize the chance of human disease (Fig. 1).
Fig. 1.
Sketch depicting approximate ranges of mutation frequencies accompanied by potential biological consequences. Hypermutation (~ 10−3 – 10−4 mutations/base pair) is the germane range for our review, which focuses on low fidelity (LoFi) E. coli DNA polymerase V (pol V), which catalyzes error-prone translesion DNA synthesis (TLS) responsible for SOS mutagenesis, and on human activation-induced deoxycytidine deaminase (AID), responsible for initiating somatic hypermutation (SHM) in immunoglobulin variable (IgV) regions on the path toward antibody (Ab) diversity in humans. DNA repair pathways indicated are BER (base excision repair), NER (nucleotide excision repair), MMR (post-replication mismatch repair), HR (homologous recombination), NHEJ (non-homologous end-joining).
Figure 1 also depicts a high-end hypermutation frequency range, ~10−2 - 10−3. Proteins responsible for hypermutation need to be tightly regulated to avoid biological chaos. We’ve indicated two types of hypermutation, one type involves copying DNA with low fidelity DNA polymerases present in prokaryotes and eukaryotes [reviewed in Goodman and Woodgate 2013; Ohmori et al., 2001], another involves somatic hypermutation (SHM), which occurs in immunoglobulin variable region (IgV) DNA, and is responsible for generating antibody (Ab) diversity [reviewed in Di Noia and Neuberger 2007; Peled et al., 2008]. Figure 1 further indicates error-free DNA repair pathways, base excision repair (BER), nucleotide excision repair (NER), post-replication mismatch repair (MMR), homologous recombination (HR) and non-homologous end-joining (NHEJ), along with an error-prone pathway, mentioned above, that uses LoFi translesion synthesis (TLS) pols to enforce a brute-force copying of damaged DNA template bases.
This review will discuss our studies on the biochemical basis of hypermutation, centered on two key enzymes, E. coli pol V involved in TLS and human activation-induced deoxycytidine deaminase (AID) required to initiate SHM. Each enzyme plays an indispensible role in generating hypermutation.
SOS REGULON
Concurrent with W-C, another notable discovery in 1953 was J. Weigle’s observation that λ bacteriophage inactivated by UV radiation and unable to lyse E. coli was reactivated following UV irradiation of the bacterial host [Weigle and Dulbecco 1953]. Weigle’s observation was the impetus for the work of E. Witkin and M. Radman in initiating the field of SOS error-prone DNA repair [reviewed in Schlacher and Goodman 2007]. In 1967, E. Witkin showed that Weigle-reactivation could be attributed to the induction of E. coli proteins that repaired UV damaged bacterial DNA, while at the same time repairing the phage DNA thus adventitiously restoring λ infectivity [Witkin 1967]. Phage reactivation was accompanied by a large increase in λ mutagenesis, referred to as Weigle mutagenesis [Weigle and Dulbecco 1953]. The connection between negatively regulated DNA repair and DNA damaged-induced mutagenesis was embodied in M. Radman’s SOS error-prone repair hypothesis [Radman 1974].
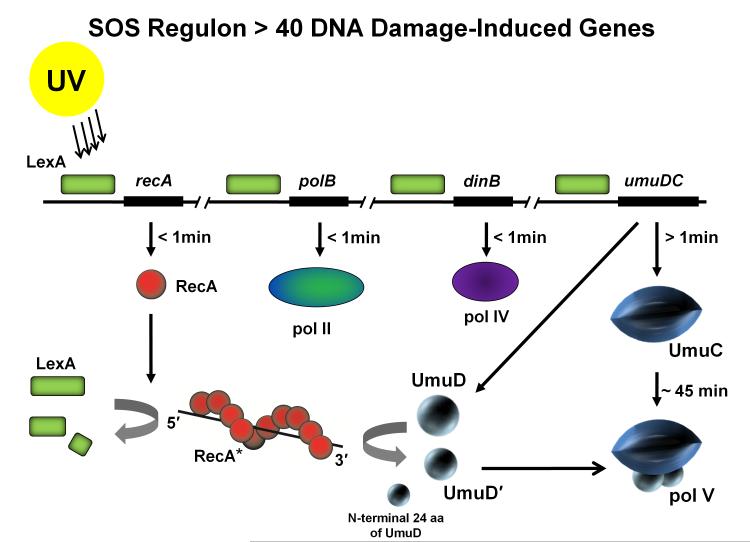
The SOS regulon in E. coli is induced in response to chromosomal damage and is composed of more than 40 genes principally engaged in repair, replication, recombination and the regulation of cell division [Courcelle et al., 2001]. Temporal regulation of gene expression results from the variable affinity binding of LexA repressor to consensus operator sequences for each of the genes in the regulon. Weakly bound genes (e.g., recA) are turned on rapidly following DNA damage (≤ 1min), tightly bound genes (e.g., umuDC operon) are induced somewhat later. When free in solution, LexA is destroyed by binding to a RecA nucleoprotein filament (RecA*), which acts as a coprotease to cleave and thereby inactivate the repressor [Little 1984; Little and Mount 1982; Yu and Egelman 1993] (Fig. 2). Three of the SOS genes encode pols II, IV, and V. Pols II and IV are present in the cell at substantial constitutive concentrations, and are then elevated to much higher levels almost immediately upon SOS induction (Fig. 2) [Goodman 2002; Goodman and Woodgate 2013]. In contrast, pol V cannot be detected in undamaged cells (< 7 molecules/cell) and remains below detectable levels in the cytosol until around 45 min after UV irradiation [Sommer et al., 1998]. The very late appearance of pol V is indicative of its favored status among the SOS pols with respect to catalyzing TLS. SOS mutagenesis induced either by UV radiation or by numerous chemicals that cause DNA damage is virtually absent in cells lacking pol V [Kato and Shinoura 1977; Woodgate and Sedgwick 1992]. Indeed, the umuDC operon encoding pol V (UmuD'2C) was identified and named based on the absence of mutations above spontaneous background levels in UV irradiated cells that contain mutations in either of the UV mutagenesis genes umuC or umuD [Kato and Shinoura 1977; Steinborn 1978]. The lengthy delay in the induction of pol V provides a wide window of opportunity for error-free BER, NER and HR, prior to resorting to pol V-catalyzed error-prone TLS to rescue replication forks stalled at unrepaired template damage sites. Evidently, neither pol II nor pol IV can substitute effectively for pol V, at least with regard to UV- and a variety of chemical-induced TLS.
Fig. 2.
Induction of the SOS response in E. coli. The LexA repressor (green box) binds to a 20 base pair consensus sequence in the operator region of the SOS genes suppressing their expression. Upon UV irradiation, expression of RecA (red spheres) is induced after DNA damage occurs. RecA forms a nucleoprotein filament on ssDNA (RecA*) and functions as a coprotease to cleave and thereby inactivate the LexA repressor. The umuDC operon is fully derepressed ~15 min after DNA damage, however, due to rapid proteolysis of the Umu proteins, the UmuD'2C complexes themselves do not accumulate until ~ 45 min after damage. RecA* facilitates cleavage of UmuD (15 kDa) to UmuD' (12 kDa) removing the first 24 aa from the N–terminus of UmuD. Two molecules of UmuD' (UmuD'2) form a physical complex with UmuC (48kDa) forming pol V (72 kDa).
TLS IS MEDIATED BY POL V AND RECA*
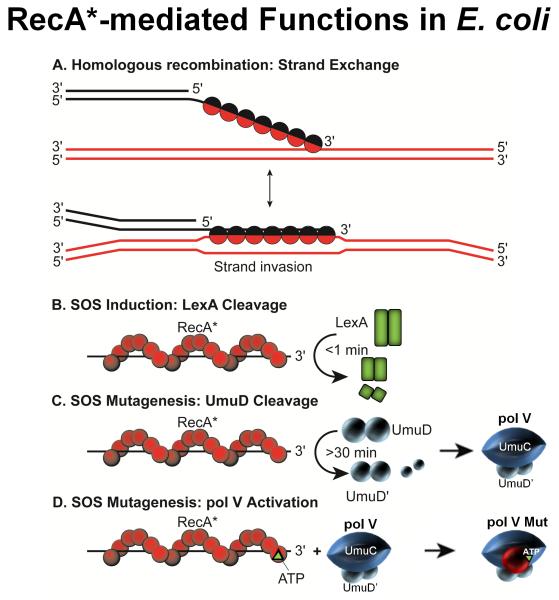
Grasping TLS requires establishing how the interactions between pol V and RecA* required for SOS mutagenesis can be understood biochemically. This turned out to be a complex problem owing to RecA’s ubiquitous roles in the cell and to pol V’s complicated and recalcitrant biochemical properties. RecA in the form of a nucleoprotein filament RecA*, has four distinct cellular roles (Fig. 3). Its principal task is to catalyze strand exchange during homologous recombination (Fig. 3A). It also has two coproteolytic roles, one to cleave LexA to turn on SOS (Figs. 1 and 3B), and another to cleave UmuD → UmuD' to form pol V (UmuD'2C) (Figs. 1 and 3C). A 4th role for RecA* is its direct involvement in SOS mutagenesis (Fig. 3D). In establishing a direct mutagenic function for RecA*, R. Devoret and G. Walker identified a RecA(S117F) mutant (originally named RecA1730) that rendered cells UV non-mutable, while retaining its other three roles [Dutreix et al., 1989]. Pol V offered several biochemical challenges, the first being that the insolubility of UmuC in aqueous solution [Woodgate et al., 1989] frustrated the early attempts by Hatch Echols, Roger Woodgate, MFG and colleagues to reconstitute TLS using purified proteins, UmuC, UmuD'2, RecA*, and pol III holoenzyme (pol III HE) [Rajagopalan et al., 1992].
Fig. 3.
The four cellular functions of the RecA protein in E. coli. A: RecA protein is involved in many aspects of recombinational DNA repair, and promotes a variety of DNA strand exchange reactions in the context of this function. One key type of reaction is shown – DNA strand invasion. In this process, RecA forms a filament (RecA*) on the 3′ end of a single-stranded DNA, aligns that DNA with its complement in a duplex DNA, and pairs the bound DNA with the complementary strand of the duplex (displacing the other duplex strand). B: The autocatalytic cleavage of LexA repressor occurs only in the presence of RecA* filaments. Since these filaments generally do not form except when the number of single strand DNA gaps is increased as a result of excessive DNA damage, LexA cleavage and the accompanying SOS system induction generally occur only in response to such damage. In the presence of RecA* filaments, LexA cleavage occurs rapidly. C: RecA* is required for the autocatalytic cleavage of the UmuD protein to form active UmuD'. This reaction is much slower than LexA cleavage and occurs >30mins after DNA damage. D: A RecA monomer and a molecule of ATP are transferred to pol V from the 3′-proximal end of a RecA* filament to form the active pol V Mut.
Following Hatch’s untimely passing in 1993, in collaboration with Roger, we showed that a tight complex is formed between UmuC and UmuD'2, which is soluble in solution [Bruck et al., 1996]. While this observation was a key step in successfully reconstituting TLS using UmuD'2C, RecA*, and pol III HE to copy DNA containing a site-directed abasic lesion, an enigmatic issue was that a more robust TLS reaction occurred without pol III HE [Tang et al., 1998]. Not only that, but increasing concentrations of pol III HE competitively inhibited TLS. We commented on the likelihood that UmuD'2C might be a new type of error-prone DNA polymerase, which turned out to be the case, where E. coli pol V = UmuD'2C [Tang et al., 1998], with the UmuC subunit harboring the pol active site [Reuven et al., 1999; Tang et al., 1999]. Even so, pol V is for all intents and purposes is virtually dead for TLS or when copying undamaged DNA templates unless RecA* is present in the reaction.
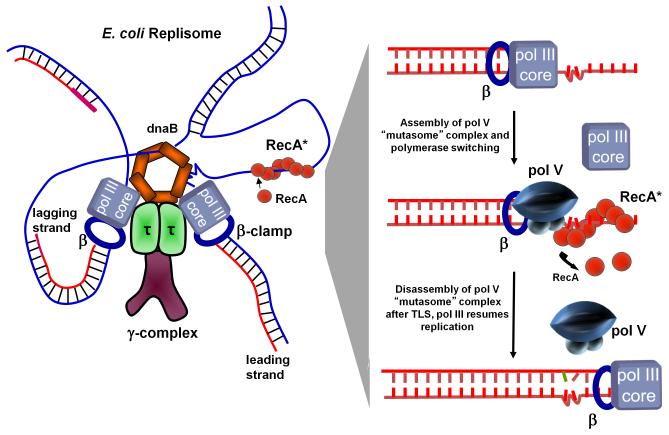
A series of longstanding TLS models beginning with the benchmark 1985 Bridges-Woodgate model [Bridges and Woodgate 1985], and subsequent models that incorporated new biochemical discoveries [Echols and Goodman 1990; Fujii et al., 2004; Maor-Shoshani et al., 2000; Pham et al., 2001], assumed that RecA* had to assemble in cis, i.e., on the damaged template strand being copied, proximal to a pol III replication fork arrested at the site of a DNA template lesion (Fig. 4). In the pol V version of the model, pol III HE blocked by a leading strand lesion (Fig. 4, sketch at left) is replaced on the β-processivity clamp with pol V, which carries out TLS, and is replaced, in turn, by pol III after the lesion has been copied past (Fig. 4, sketch at right). I have referred to this as the Willie Sutton RecA* pol V cis-activation model for TLS. When asked why he robbed banks, Willie Sutton, ca, 1920’s, famously replied because that’s where the money is. So it seemingly makes excellent sense to locate RecA* in cis on the template strand being copied, because that’s where the lesion is.
Fig. 4.
TLS cis-activation model. A model of an E. coli replisome stalled at a lesion site is depicted in the left panel and individual TLS steps are shown in the right panel. DNA pol III stalls at the lesion site, DnaB helicase unwinds dsDNA ahead of the blocked fork resulting in regions of ssDNA where RecA* assembles in cis. The cis-activation model requires polymerase switching. DNA pol V replaces pol III core on the β clamp and bypasses lesions with concurrent 3′ → 5′ displacement of RecA* formed ahead of the lesion by pol V. Following TLS, pol III core replaces pol V to continue rapid replication of undamaged DNA downstream of the lesion.
THE ACTIVE FORM OF POL V IS POL V MUT (UmuD'2C-RecA-ATP)
In 2006, there were two unresolved enigmatic questions in SOS mutagenesis: 1) Why is RecA* required for pol V-catalyzed DNA synthesis? 2) What is the active form of pol V?
It is important to point out that even if RecA* were to act in cis to activate pol V, the model (Fig. 4, sketch at right) provides no insight into the molecular interactions between RecA* and pol V required for TLS. In 2006, an experiment conceived and performed by Kathi Schlacher, a graduate student in the lab, demonstrated pol V-catalyzed TLS with RecA* acting in trans, thus inserting a fly in the cis filament ointment. She used pol V to copy a short hairpin p/t DNA with a 3 nt template overhang. Since the footprint of a single RecA monomer is 3 nt, there’s simply no room to assemble RecA* in cis on the template strand. Instead, ~100% of the template was copied in the presence of RecA* formed on a separate ssDNA. Therefore, RecA* acting in trans enables the replication of DNA by pol V [Schlacher et al., 2006].
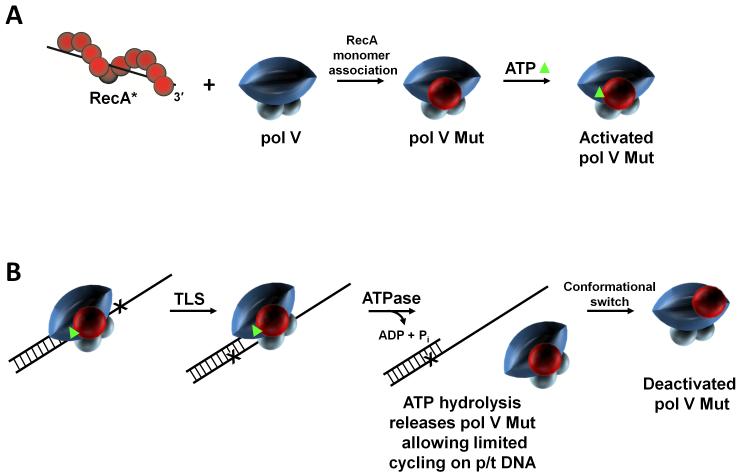
The two questions concerning the requirement for RecA* and the identity of the activated form of pol V were resolved by observing that pol V was modified by incubating with transRecA* (Fig. 5A). Attachment of RecA* covalently to beads permitted the complete separation of transRecA* beads by centrifugation from pol V following incubation [Erdem et al., 2014; Jiang et al., 2009]. The analysis of the post-incubated polymerase by MALS (multiangle light scattering) showed the presence of two light scattering peaks, one at 113 kDa and another at 73 kDa, corresponding to the molecular mass of pol V (UmuD'2C). SDS-PAGE electrophoresis showed that the heavier (113 kDa) peak was composed of UmuD'2C-RecA-ATP in equimolar UmuD'2:UmuC:RecA:ATP stoichiometric ratios [Jiang et al., 2009]. Therefore, the answers to the two questions are that RecA* is required to transfer a RecA monomer from its 3′-proximal tip to pol V, and that the activated mutasomal form of pol V, pol V Mut = UmuD'2C-RecA-ATP (Fig. 5A).
Fig. 5.
RecA and ATP regulation of pol V Mut activity. Pol V activity is regulated in multiple steps. (A) First, a RecA subunit is transferred to pol V from the 3′-proximal tip of RecA* to form pol V Mut. Pol V Mut is activated by binding a molecule of ATP (green triangle). (B) Activated pol V Mut associates with p/t DNA and DNA synthesis proceeds until ATP is hydrolyzed. Pol V Mut is a unique DNA-dependent ATPase, where a single ATP hydrolytic event leads to enzyme dissociation from p/t DNA (internal regulation), followed by limited cycling on p/t DNA. After DNA synthesis, pol V Mut becomes deactivated but can be reactivated by exposure to a new RecA*. We have proposed a conformational regulatory mechanism in which activated and deactivated forms of pol V Mut are determined by the location of RecA on pol V.
4D REGULATION OF POL V MUT: TEMPORAL, INTERNAL, CONFORMATIONAL, AND SPATIAL
Once it has been formed, pol V Mut performs TLS in the absence of RecA* [Jiang et al., 2009]. When copying undamaged DNA, pol V Mut has fidelity on the order of 1 nt misincorporation per 100 to 1000 bp [Tang et al., 2000], which is roughly about 100-fold lower than the replicative pol III HE [Bloom et al., 1997]. It’s this highly error-prone behavior that implies a need for tight regulation. One means of regulation is temporal, in which the UmuD'2C complexes themselves do not accumulate until ~ 45 min after DNA damage, thereby providing a window in time for BER, NER, and MMR to repair the damage (Figs. 1 and 2). In the absence of temporal regulation, a RecAE38K mutant (formerly called RecA730), described by J. Sweasy and Witkin [Sweasy et al., 1990], constitutive for SOS induction, generates a 100-fold increase in mutation in the absence of UV.
A second form of regulation involving ATP and ATP hydrolysis, is what we’ve referred to as internal, that appears to act as a clock to limit pol V Mut processivity. When we had determined that a single ATP molecule was present as an integral part of the pol V Mut complex [Jiang et al., 2009], we had no idea what, if any, its role might be. We had initially assumed that since ATP binds to RecA and is required for RecA* nucleoprotein assembly, and ATP hydrolysis for RecA* disassembly, that the nucleotide cofactor had just come along for the ride during the transfer of RecA from transRecA* to pol V, and remained bound to RecA in the activated complex [Jiang et al., 2009]. However, once we succeeded in removing ATP while retaining RecA, we found that the mutasomal complex composed of UmuD'2C-RecA was catalytically dead, because it could not bind to p/t DNA [Erdem et al., 2014]. The re-addition of ATP restored full catalytic activity. Therefore, the presence of ATP in pol V Mut is required for binding to a 3′-primer terminus (Fig. 5B). Although pol V alone binds ssDNA [Bruck et al., 1996; Rehrauer et al., 1998], ATP and RecA are necessary for catalytically productive binding [Erdem et al., 2014].
But a greater surprise was to follow. Pol V Mut contains an intrinsic DNA-dependent ATPase activity [Erdem et al., 2014]. This ATPase activity is distinct from the long-established canonical Walker B or A motif required for activity of RecA [Walker et al., 1982 and reviewed in Lusetti and Cox 2002], because pol V Mut retains its intrinsic ATPase activity when formed using an ATPase-deficient RecAK72R mutant [Erdem et al., 2014]. A measurement of the rate of pol V Mut dissociation from p/t DNA coincides with an independent measurement of the rate of ATP hydrolysis, which implies that a single ATP turnover releases pol V Mut from a 3′-primer end (Fig. 5B). We speculate that the intrinsic DNA-dependent ATPase provides a way to limit the length of time that the low fidelity polymerase remains bound during DNA synthesis. This can be viewed as an internal clock mechanism to restrict pol V Mut processivity to the vicinity of a lesion, thus limiting the extent of mutagenesis on undamaged DNA downstream of TLS. The clock molecular mechanism remains an open question; hydrolysis might be coupled to the number of nt incorporated, to DNA synthesis rate, or it might be uncoupled behaving stochastically in time, which is our currently favored hypothesis.
A third regulation mode is conformational. Generically, DNA polymerases bind to p/t DNA, incorporate variable numbers of deoxynucleotides depending on individual catalytic rate and processivity, and following pol-p/t DNA dissociation, can cycle to a different DNA, presumably ad infinitum. That seems not be the case for pol V Mut, which exists in either of two states, activated or deactivated [Jiang et al., 2009]. When activated, pol V Mut exhibits limited cycling between DNA substrates and then deactivates. Subsequent exposure to transRecA* reactivates pol V Mut [Jiang et al., 2009]. The deactivated form of pol V Mut retains RecA in the polymerase complex. Reactivation occurs by incubation with transRecA*, i.e., pol V Mutdeactivated + transRecA* → pol V Mutactivated, with a new RecA replacing the old RecA in the complex [Jiang et al., 2009]. A mass spectroscopic analysis of pol V Mut identifies a RecA surface that is in close proximity to amino acid residues on two distinct surfaces of UmuC of pol V, separated by roughly 50 Å [Gruber et al., 2015]. One of these is uniquely prominent in the activated form of pol V Mut [Gruber et al., 2015]. A different cross-linked surface on UmuC occurring in an inactive form of pol V Mut is predicted to interact with the replicative β-clamp [Boudsocq et al., 2002], suggesting perhaps that a conformational switch of RecA on UmuC might be needed for pol V Mut to bind to β. We have hypothesized a toggle switch mechanism to convert between activated and deactivated forms of pol V Mut [Gruber et al., 2015; Jiang et al., 2009]. The two different location of RecA relative to UmuC in the activated and deactivated forms of pol V Mut is sketched in Figure 5B.
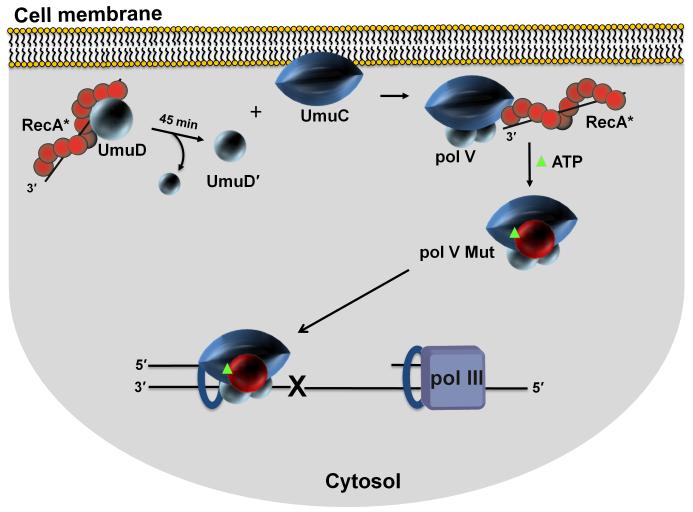
In 2015, thanks to the power of single-molecule fluorescence microscopy, a new spatial regulation mode for pol V was revealed by visualizing UmuC inside living cells in space and time [Robinson et al., 2015](Fig. 6). Replacement of UmuC on the chromosome with a fluorescent UmuC-mKate2 showed that UmuC was synthesized at around 45 min following UV irradiation, but it is not immediately activated. Instead, UmuC is sequestered on the inner cell membrane. Its release into the cytosol requires RecA*-mediated cleavage of UmuD → UmuD', since UmuC remains bound to the membrane in cells containing a non-cleavable umuD(K97A) mutant. The inability to observe UmuC in the cytosol unless it’s converted UmuD'2C provides visual affirmation of the early biochemical observations that UmuC, by itself, was insoluble in aqueous solution [Woodgate et al., 1989], but that it can be obtained as a soluble UmuD'2C complex [Tang et al., 1998].
Fig. 6.
Spatial regulation of pol V Mut activity in vivo. Sketch depicting cell membrane localization of UmuC. Single molecule live cell fluorescence imaging data indicate that after an initial delay post UV irradiation, UmuC is synthesized and localized on the inner cell membrane. UmuC is released into the cytosol only after it interacts with UmuD'2 [Robinson et al., 2015]. As RecA and UmuD are found in or near the membrane, additional steps in the formation of pol V Mut (UmuD'2C-RecA-ATP) may occur in proximity to the membrane. Sequestration of UmuC at the membrane provides additional levels of pol V Mut regulation. A delay between the production of UmuC and the formation of cytosolic pol V Mut provides time for error-free DNA repair to occur before mutagenic pol V Mut lesion bypass is necessary. Infrequent co-localization of pol V Mut with replisomes is consistent with pol III reinitiating synthesis downstream of a lesion, leaving behind an ssDNA gap. Pol V Mut can then bind to a β-clamp left behind by a departing replisome and catalyze TLS.
In vivo mutagenesis assays show that the fluorescent-tagged UmuC is active for spontaneous and damage-induced mutagenesis. However, following UV irradiation, foci of mutagenically active pol V Mut do not co-localize with pol III-replisomes, visualized as DnaX (pol III HE τ subunit)-YPet, suggesting that TLS may be occurring at lesions skipped over by Pol III (Fig. 6). As part of the pol V RecA*-cis-activation model, it had been tacitly assumed that the exchange of pol III with pol V occurred proximal to a lesion (Fig. 4). The observation that pol V Mut might gain access to a lesion without having to displace pol III core at the site of lesion, while perhaps counterintuitive, is nonetheless consistent with a replication-restart model involving lesion skipping by the replisome proposed by J. Yeeles and K. Marians [Yeeles and Marians 2013]. In contrast, in SOS constitutive RecAE38K cells, UmuC is synthesized in the absence of UV and is released immediately in the cytosol [Robinson et al., 2015]. Here the RecAE38K form of pol V Mut (UmuD'2C-RecAE38K-ATP) does co-localize with the replisome in the absence of UV [Robinson et al., 2015]. It seems likely that the 100-fold increase in SOS untargeted mutations reflects the ability of the constitutively expressed pol V MutE38K to gain access to a transiently arrested replication fork, or perhaps to even one that’s moving somewhat fitfully, and displace pol III core on the β-clamp, thus catalyzing error-prone DNA synthesis on undamaged template DNA.
In essence, four mechanisms have been identified that limit the amount of time that the low fidelity pol V has to access DNA: 1) transcriptional and posttranslational regulation that initially keep the intracellular levels of pol V to a mutationally tolerable minimum; 2) an intrinsic ATPase function that limits pol processivity on p/t DNA; 3) conformational switching that moves RecA relative to UmuC to go from activated to deactivated forms of the polymerase; 4) spatial regulation via transient sequestration of UmuC on the membrane, which prevents pol V from entering the cytosol until needed for TLS.
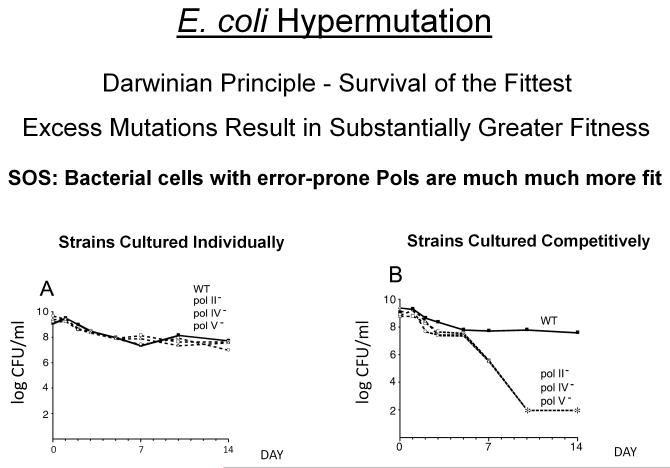
While pol V appears to play a dominant role in TLS-generated SOS mutation, pol V along with pols II and IV have a much more evenhanded responsibility to introduce mutations in E. coli, to help rescue cells in times of stress. Figure 7 provides a striking example showing that SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness [Yeiser et al., 2002]. Wild type (WT) cells containing all of the SOS pols or cells lacking one SOS pol (pol II−, pol IV−, or pol V−) is cultured either individually (Fig. 7A) or with each pol deletion mutant growing in the presence of the WT strain (Fig. 7B). When grown separately over a long period (e.g., 14 days), there’s no discernible difference in individual strain survival patterns (Fig. 7A). When grown together, there’s also not much difference in survival patters for about 6 days, but major differences in strain survival are clearly observed by day 7 and by day 10 the WT strain has basically taken over the population. Therefore, mutants lacking even just one SOS pol suffer a severe reduction in fitness when competing with WT for diminishing nutritional supplies. More generally, different combinations of SOS pols can be used to establish distinct growth phase-dependent hierarchies of pol mutant strain competitiveness, with pol II conferring a significant physiological advantage during periods of rapid growth, and hypermutation generated by pols IV and V making the largest contribution to evolutionary fitness during long-term stationary phase [Corzett et al., 2013].
Fig. 7.
Long-term survival patterns of wild type and SOS polymerase mutant cells. A: Composite of survival patterns when cells are grown alone in batch culture. B: Composite of survival patterns of each polymerase-deficient (pol−) strain when grown in competition with wild-type cells. Solid line with filled boxes, wild type; dashed lines are mutant strains: open squares, pol II−; open circles, pol IV−; open triangles, pol V- mutants. CFU/ml, colony-forming units per ml. Asterisks correspond to cell titers below the limit of detection of 100 CFU/ml.
BIOCHEMICAL BASIS OF AID IN INITIATING SHM
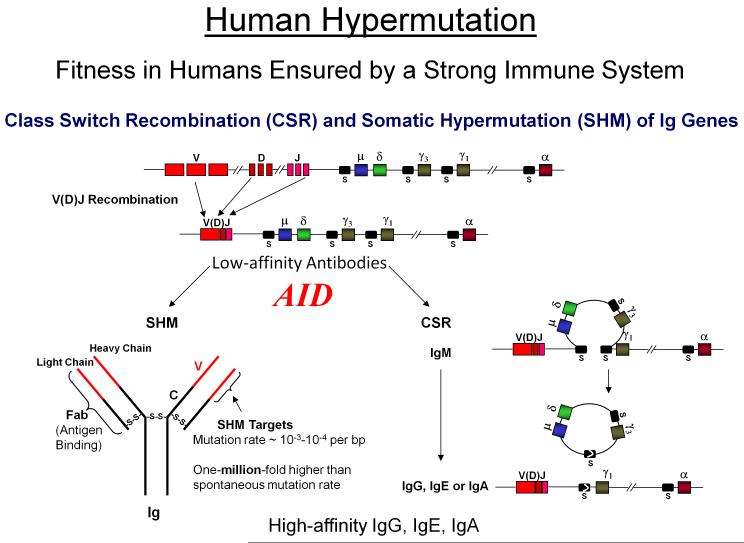
In response to the question of why don’t you die when I sneeze, humans have a robust immune response generating an almost unlimited diversity of antibodies (Abs) that bind invading antigens and bring about their elimination. The generation of low-affinity Abs through V(D)J recombination is required for survival. The generation of high-affinity Abs, while not required for survival is essential for quality of survival, i.e., optimal fitness (Fig. 8) [reviewed in Di Noia and Neuberger 2007; Peled et al., 2008]. AID plays the key role ensuring immunological diversity by initiating SHM and class-switch recombination (CSR) in B cells. The hypermutation frequency in IgV of ~ 10−3 – 10−4 per bp is roughly a million times greater than somatic cell mutation frequencies (Fig. 8).
Fig. 8. Steps involved in Ab diversification.
V(D)J recombination, mediated by RAG proteins, is responsible for generating a repertoire of low affinity Abs required for viability. Low affinity Ab-producing genes undergo SHM and CSR which require C →U deamination by AID at V(D)J and switch (S) regions. AID actions at V(D)J regions generate SHM rates of 10−3 – 10−4 per bp, which are roughly a million-fold higher than spontaneous somatic cell mutation rates. AID targeting to the donor switch (Sμ) and a downstream acceptor S (shown in this case Sγ1) initiates CSR which joins the V(D)J exon with one of the appropriate downstream constant C regions converting IgM to other Ab isotypes IgG, IgE, or IgA. By generating virtually unlimited types of high affinity Abs, SHM and CSR while not necessary for survival are essential for quality of life.
AID performs a seemingly simple task, the deamination of C→U on ssDNA [Bransteitter et al., 2003], but for this to occur in the right place at the right time in the cell requires a highly complex pattern of choreographed interactions between AID and many other proteins. These interactions target AID to transcriptionally active immunoglobulin variable (IgV) regions during SHM and to transcribed R-loops in switch (IgS) regions during CSR [reviewed in Pavri and Nussenzweig 2011]. Each process involves a sizable number of AID-targeting proteins identified mainly through ChiP studies.
AID initiates both processes, SHM and CSR, by deaminating C most often in WRC hot motifs (W = A/T, R = A/G) [Pham et al., 2003]. In the case of SHM in IgV regions, the fate of the nascent U•G mispair is either to be replicated properly by incorporation of A opposite U generating transition mutations at deaminated C sites, or alternatively, the mispair may be subjected to mismatch repair (MMR) or to base excision repair (BER). Filling in of repair tracts by the error-prone pol η (a human Y-family homolog of E. coli pol V), results in transition and transversion mutations at A and T sites. In the case of CSR, deamination of C→U in the numerous WRC hot motifs present in IgS regions produces double-strand DNA breaks. NHEJ-mediated recombination then coverts, i.e., switches, IgM to IgG, IgE and IgA isotypes (Fig. 8) [reviewed in Stavnezer et al., 2008]. The availability of tractable biochemical reconstitution systems using purified proteins to dissect the interactions of AID with its many protein partners in SHM and CSR remains a distant goal. Nonetheless, AID acting by itself on ssDNA reveals basic biological properties of SHM [Pham et al., 2003].
A biochemical approach that has provided a broad, yet detailed, perspective of AID acting on ssDNA was obtained by incubating AID with bacteriophage M13 dsDNA containing a lacZ reporter sequence in a gapped ssDNA region, allowing the detection of AID-catalyzed C→U deaminations, detected as C→T mutations in lacZ [Pham et al., 2003]. The experimental conditions, e.g., AID/DNA substrate ratios, were designed to ensure that the mutations were generated by the action of a single AID molecule, as shown by absence of mutations on 98% of the target DNA (M13 blue plaques). The mutated DNA (M13 white plaques) typically contained a broad distribution of anywhere from 5 – 70 mutations per clone. The mutations were distributed randomly along the DNA, but also included many examples of clusters containing 2 to 10 closely spaced mutations. Therefore, AID is highly processive. The spectra revealed that AID-catalyzed deaminations occur preferentially in WRC hot motifs in accord with SMH spectra observed in vivo [Pham et al., 2003]. But beyond the identification of hot motifs, the mutational spectra obtained in vitro agree remarkably well with the mutability index measured in human IgV DNA in vivo, for hot and cold motifs [Pham et al., 2003]. The mutability index is defined as the frequency of observed C target base mutations compared to the expected unbiased mutation frequency [Shapiro et al., 2002] (Table 1).
Table I.
Comparison Of Mutability Index For Hot And Cold Sequence Motifs For AID Deamination In Vitro And SHM In Vivo*
| Motif | AID in vitro |
SHM in vivo** |
|---|---|---|
| Hot spots | ||
| AAC | 1.72 | 2.00 (3.41) |
| AGC | 1.45 | 2.58 (3.38) |
| TAC | 2.07 | 2.24 (2.89) |
| TGC | 2.15 | 0.71 (2.09) |
| WRC (Average) | 1.85 ± 0.26 | 1.88 ± 0.59 |
| Cold spots | ||
| CCC | 0.61 | 0.07 (0.25) |
| CTC | 0.64 | 0.26 (0.35) |
| GCC | 0.05 | 0.29 (0.28) |
| GTC | 0.28 | 0.41 (0.16) |
| SYC (Average) | 0.40 ± 0.25 | 0.26 ± 0.10 |
The Mutability Index is defined as the number of times a given oligonucleotide within a segment of DNA contains a mutation, divided by the number of times the oligonucleotide would be expected to be mutated for a mechanism with no sequence bias. In vivo data are for trinucleotides 5′-NNC and also for complementary (5′-GNN).
Recently, we have expanded upon the lacZ reporter assay by incorporating in frame mutational cassettes containing different combinations of motifs, e.g., homogeneous (AGC)n hot motifs, alternating (AACAGC)n hot-hot' motifs, alternating (AACGTC)n hot-non-hot motifs, composed of variable numbers of motifs, typically of length n ~ 30. In this expanded system, white or blue plaques still report on the presence or absence of mutations in lacZ, but what’s new is that the distribution of mutations is being scored in each mutational cassette. The mutational patterns in the cassettes exhibit the same basic features observed for lacZ; these include random distributions of mutations including individually mutated sites interspersed with mutational clusters separated by non-mutated DNA regions [Pham et al., 2011] (see, e.g., Fig. 9A).
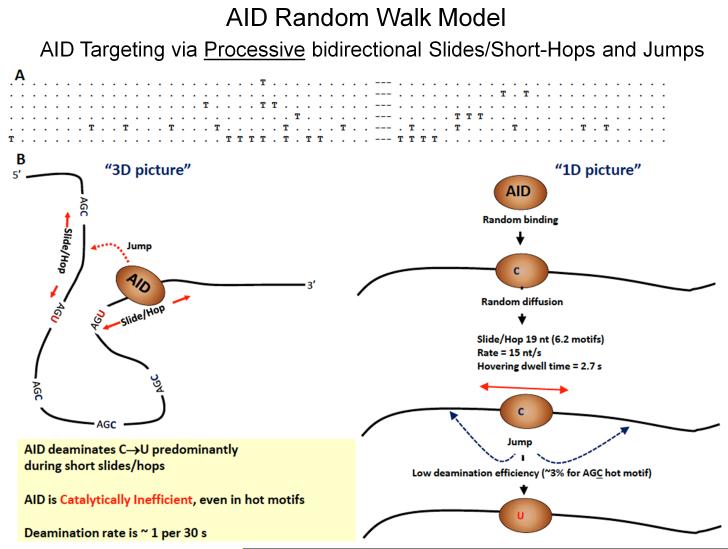
Fig. 9. Random walk model depicting AID scanning and catalyzing haphazard, inefficient deamination of C to U on ssDNA.
A: Representative mutated clones on a cassette containing 32 and 24 consecutive AGC motifs separated by a 9 nt spacer embedded in-frame in a lacZ reporter sequence in an ssDNA gapped region of bacteriophage M13 dsDNA (see text for description). Each T denotes a deaminated WRC motif; each dot denotes a non-deaminated WRC motif; dashes denote 9 nt spacer region. B: AID binds at a random location on ssDNA and scans in random bidirectional short slides/hops. AID only rarely deaminates C → U, at about 3% efficiency even for preferred AGC hot motifs, thus ensuring mutational diversity. The deamination efficiency, slide/hop rates and distances, deamination motif dwell times are predicted from a random-walk mathematical model by an analysis of the types of clonal mutational distributions shown above in A [Mak et al., 2013]. Random deamination clusters can when AID slides repeatedly over the same region of ssDNA. These types of random clustered mutations (termed kataegis) in WRC motifs have been observed as B-cell lymphoma mutational signatures [Qian et al., 2014].
We formulated a random-walk mathematical model to analyze AID scanning ssDNA [Mak et al., 2013; Mak et al., 2015]. The model was used to analyze the catalytic properties of AID, its deamination rates and efficiencies, along with the motional properties of AID, its scanning rates, distances and motif dwell times. The model predictions based on the mutational cassette data are shown in Figure 9B [Mak et al., 2013; Mak et al., 2015]. The most important property of AID is its remarkable catalytic inefficiency. A deamination efficiency of ~ 3% in AGC motifs means that for every 100 encounters between AID and one of its hottest motifs, the conversion of C → U takes place just 3 times. In contrast to AID, scanning enzymes like uracil glycosylase [Porecha and Stivers 2008] or FAPY glycosylase [Blainey et al., 2006] are searching for aberrant target bases, U or 8OxoG, that may be present once in about 106 – 107 bp, a needle-in-a-haystack. To be effective, the glycosylases must remove the errant bases efficiently, ~ 70% or greater [Porecha and Stivers 2008], whereas AID, on the other hand, encounters numerous target needles. Therefore, from a teleological perspective, the poor catalytic behavior of AID, low deamination rate (~1/46 s) and efficiency (~3%) (Fig. 9B) make sense, since inefficient and haphazard catalysis in IgV offers an optimum way to generate Ab diversity.
AID deaminations in favored motifs are sensitive to surrounding sequence context. We’ve observed, for example, that hot AAC motifs were deaminated at more than 2-fold lower rates when located next to GAC motifs [Pham et al., 2011]. The mathematical analysis, which explicitly includes intertwined catalysis and diffusion mechanisms provided an excellent fit to the data [Mak et al., 2015]. An in vivo example suggesting that DNA sequence context information has been programmed into the evolved catalytic behavior of AID is shown by a strong correlation between IgV-region mutations observed in a MMR-BER-defective mouse in vivo and AID deaminations catalyzed in vitro [MacCarthy et al., 2009]. The in vitro experiment entailed incubating AID with either IgV non-transcribed or transcribed strand ssDNA sequences fused downstream of a lacZ reporter sequence. The in vitro and in vivo mutation profiles were highly correlated for the non-transcribed, i.e., ssDNA strand, but not for the transcribed strand, showing that mutation process entails considerable complexity beyond motif targeting.
From the standpoint of human fitness, AID acting at the wrong time in the wrong place is responsible for generating B-cell lymphoma [Pasqualucci et al., 2008; Pasqualucci et al., 2001]. Notably, lymphoma mutational signatures are characterized as random clusters of mutations in WRC hot motifs, termed kataegis [Qian et al., 2014]. The disease clustering pattern is consistent with the processive, stochastic action of AID scanning ssDNA [Pham et al., 2003].
LOOKING JUST AHEAD
Leonard Mlodinow’s first-rate book entitled “The Drunkard’s Walk: How Randomness Rules our Lives” [Mlodinow 2008] provides an apt metaphor for AID, where haphazard mutations in IgV provide a rapid and trustworthy way to cope with exposure to unexpected and unfamiliar hazards. For us, in the near-term, we want to rigorously test the predictions from our random-walk enzymes’ model [Mak et al., 2013; Mak et al., 2015]. Using spatial distributions of mutations distributed along the ssDNA as input to the model, the calculations then specify, i.e., predict, the deamination rates and efficiencies and the displacement rates, distances and motif dwell times that best fit the mutational data. An initial foray into comparing theory with experiment was our recent TIRF-FRET study that visualized the motion of AID scanning ssDNA at single-molecule resolution. The data showed AID moving in random bidirectional short slides, on average ~ 6 motifs, over the entire ssDNA (72 nt) molecule while remaining bound to the same ssDNA molecule for ~ 5 min [Senavirathne et al., 2015]. These processive scanning characteristics of AID agree with the model [Senavirathne et al., 2015].
The real power of the model resides in its potential for predicting AID scanning coupled to catalysis in any DNA sequence, in other words, the effect of sequence context on deamination rates and efficiencies. The TIRF-FRET data indicate that superimposed on the random motion of AID over the entire ssDNA, there appears to be more restricted random movements localized to the vicinity of hot motifs [Senavirathne et al., 2015]. This type of quasi-localized scanning, a sort of hovering motion, is consistent with the model prediction that AID dwells in the vicinity of hot motif sites for about 3 s. The next step would be to study the real-time motion of AID at high resolution, varying the types and arrangement of motifs, in different sequence contexts. Long stretched DNA would best be used, which could then also include IgV, IgS and c-myc sequences.
We will use Damon Runyon’s observation that all life is 6 to 5 against (Damon Runyon in “A Nice Price”, c.a. 1933), as a closing metaphor for our ongoing studies with pol V. Given the unexpected twists in the pol V story it’s a fool’s errand to try to anticipate what’s next, but we’ll give it a go anyway. First, single molecule microscopy might be able to resolve the postulated RecA-UmuC toggle switching provided that a pol V Mut containing a dual fluorescent label (Cy3-UmuC and Cy5-RecA) were to remain active. Second, pol V Mut’s intrinsic ATPase appears unique, lacking Walker A or B motifs. RecA*’s ATPase seems not germane because pol V Mut assembled with an ATPase-deficient RecA K72R mutant, pol V Mut E38K/K72R, has an avid DNA-dependent ATPase [Erdem et al., 2014]. Thus, a near-term challenge is to identify the interacting surfaces between RecA and UmuD'2C that form this new type of hybrid ATPase active site. Recently, we have used desthobiotin-ATP crosslinking [Patricelli et al., 2007] and tandem mass spectrometry to identify a lysine residue in UmuC (K403) that contributes to ATP binding-initiated DNA synthesis and to ATPase activity. It’s a small step in the right direction.
Apart from TLS, pol V Mut has an existential role in bacterial fitness and evolution, as do its two companion SOS-induced pols II and IV (Fig. 7) [Corzett et al., 2013; Yeiser et al., 2002]. Intuitively, one might expect that mass action is responsible for determining the competition between TLS pols for binding to the β-sliding clamp vacated by pol III core. Although SOS-induced cytosolic concentrations of pol II and, even more so, pol IV dwarf those of pol V, yet it’s pol V that’s responsible for virtually all of the UV-induced mutagenesis. Despite the much higher levels of pols II and IV, somehow pol V manages to dominate error-prone TLS, at least at UV lesions and at many different sites of chemically induced DNA damage. Live-cell imaging is ideally suited to visualize when and under what conditions (e.g., ± UV) any of the SOS polymerases co-localize with pol III, and further to determine whether the presence or absence of each SOS polymerase alters the competition for binding to DNA. The power of super-resolution microscopy comes into full play because individual cells can be studied as part of an ensemble of cells. Here, as before [Robinson et al., 2015], live cell imaging can provide a missing link between genetics, biochemistry and cell physiology.
As expressed far more articulately by Mlodinow [Mlodinow 2008] and Runyon, the randomness provided by hypermutation, be it programmed, as for AID, or less programmed, as for pol V Mut, is necessary to provide fitness for large and small forms of life. Haphazard mutations are thankfully available to deal with unexpected hazards.
2015 EMGS AWARD MOTIVATION
The Environmental Mutagenesis and Genomics Society conferred this award to Dr. Myron F. Goodman in recognition of his multiple seminal discoveries concerning molecular mechanisms responsible for accurate and inaccurate DNA synthesis that have contributed to advancements in understanding of mutagenesis, carcinogenesis, heritable birth defects, AIDS, and the development of the immune system. His work firmly established the paradigm that when DNA replication is blocked by lesions in DNA, a set of specialized error-prone DNA polymerases are recruited to the lesion and the action of family Y polymerases is principally responsible for the mutations caused by the lesions. Dr. Goodman has also contributed a number of innovations toward improved and more precise methods for studies of pro-mutagenic activities by the DNA synthesis enzymes and co-factors.
ACKNOWLEDGMENTS
I am indebted to past and present students and postdoctoral associates who bestowed invaluable intellectual support for the SOS-pol V studies. There were so many important USC contributors, but with apologies I can acknowledge only a few, Phuong Pham, Menjia Tang, Kathi Schlacher, Qingfei Jiang, Aysen Erdem, Malgorzata Jaszczur, Jeff Bertram; our collaborators, Kevin McEntee, Roger Woodgate, Michael Cox, Antoine van Oijen, Andrew Robinson, with special affection and thanks to Hatch Echols, and my general appreciation to Evelyn Witkin, Graham Walker, Raymond Devoret, and John Petruska. For the AID studies, again Phuong Pham, Ronda Bransteitter, Linda Chelico, Peter Calabrese, Lars Pedersen, Samir Afif, Chi Mak, David Rueda with huge gratitude to Matty Scharff. I express my sincere gratitude to Moishe Bessman for his friendship, support and inspiration spanning 50 years and counting. Many thanks to Paul B. Wolfe, NIGMS; Dan Shaughnessy, NIEHS; Dick Pelroy, NCI, each of whom were instrumental in providing material support, generous advice and intellectual encouragement.
Grant sponsor: National Institutes of Health, National Institute of Environmental Health Sciences
Grant numbers: GM21422; ES012259; ES013192
Footnotes
This paper was presented at 2015 Environmental Mutagenesis & Genomics Society Award lecture in New Orleans, Louisiana.
REFERENCES
- Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–198. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom LB, Chen X, Fygenson DK, Turner J, O#x0027;Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Ling H, Yang W, Woodgate R. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amst) 2002;1:343–358. doi: 10.1016/s1568-7864(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R. The two-step model of bacterial UV mutagenesis. Mutat Res. 1985;150:133–139. doi: 10.1016/0027-5107(85)90110-1. [DOI] [PubMed] [Google Scholar]
- Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD'C complex from Escherichia coli. Cooperative binding of UmuD'C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- Corzett CH, Goodman MF, Finkel SE. Competitive fitness during feast and famine: how SOS DNA polymerases influence physiology and evolution in Escherichia coli. Genetics. 2013;194:409–420. doi: 10.1534/genetics.113.151837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Waard A, Paul AV, Lehman IR. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci U S A. 1965;54:1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Drake JW, Allen EF. Antimutagenic DNA polymerases of bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1968;33:339–344. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H, Goodman MF. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990;236:301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- Erdem AL, Jaszczur M, Bertram JG, Woodgate R, Cox MM, Goodman MF. DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent ATPase. Elife. 2014;3:e02384. doi: 10.7554/eLife.02384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Gasser V, Fuchs RP. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Gillin FD, Nossal NG. Control of mutation frequency by bacteriophage T4 DNA polymerase. II. Accuracy of nucleotide selection by the L88 mutator, CB120 antimutator, and wild type phage T4 DNA polymerases. J Biol Chem. 1976;251:5225–5232. [PubMed] [Google Scholar]
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Goodman MF, Fygenson KD. DNA polymerase fidelity: from genetics toward a biochemical understanding. Genetics. 1998;148:1475–1482. doi: 10.1093/genetics/148.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AJ, Erdem AL, Sabat G, Karata K, Jaszczur MM, Vo DD, Olsen TM, Woodgate R, Goodman MF, Cox MM. A RecA protein surface required for activation of DNA polymerase V. PLoS Genet. 2015;11:e1005066. doi: 10.1371/journal.pgen.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Lehman IR. Discovery of DNA polymerase. J Biol Chem. 2003;278:34733–34738. doi: 10.1074/jbc.X300002200. [DOI] [PubMed] [Google Scholar]
- Lehman IR, Zimmerman SB, Adler J, Bessman MJ, Simms ES, Kornberg A. Enzymatic Synthesis of Deoxyribonucleic Acid. V. Chemical Composition of Enzymatically Synthesized Deoxyribonucleic Acid. Proc Natl Acad Sci U S A. 1958;44:1191–1196. doi: 10.1073/pnas.44.12.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- MacCarthy T, Kalis SL, Roa S, Pham P, Goodman MF, Scharff MD, Bergman A. V-region mutation in vitro, in vivo, and in silico reveal the importance of the enzymatic properties of AID and the sequence environment. Proc Natl Acad Sci U S A. 2009;106:8629–8634. doi: 10.1073/pnas.0903803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak CH, Pham P, Afif SA, Goodman MF. A Mathematical Model for Scanning and Catalysis on Single-stranded DNA, Illustrated with Activation-induced Deoxycytidine Deaminase. J Biol Chem. 2013;288:29786–29795. doi: 10.1074/jbc.M113.506550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak CH, Pham P, Afif SA, Goodman MF. Random-walk enzymes. Phys Rev E Stat Nonlin Soft Matter Phys. 2015;92:032717. doi: 10.1103/PhysRevE.92.032717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Shoshani A, Reuven NB, Tomer G, Livneh Z. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc Natl Acad Sci U S A. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodinow L. The Drunkard’s Walk: How Randomness Rules our Lives. Pantheon Books; 2008. [Google Scholar]
- Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. others. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Pham P, Bertram JG, O#x0027;Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Pham P, Calabrese P, Park SJ, Goodman MF. Analysis of a single-stranded DNA-scanning process in which activation-induced deoxycytidine deaminase (AID) deaminates C to U haphazardly and inefficiently to ensure mutational diversity. J Biol Chem. 2011;286:24931–24942. doi: 10.1074/jbc.M111.241208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, Liang G, Tang Z, Mathe E, Benner C. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In: FS L. Prakash, Miller MW, Lawrence CW, Tabor HW., editors. Molecular and environmental aspects of mutagenesis. Charles C. Thomas; Springfield, Ill: 1974. pp. 128–142. [Google Scholar]
- Rajagopalan M, Lu C, Woodgate R, O#x0027;Donnell M, Goodman MF, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD', and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci U S A. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer WM, Bruck I, Woodgate R, Goodman MF, Kowalczykowski SC. Modulation of RecA nucleoprotein function by the mutagenic UmuD'C protein complex. J Biol Chem. 1998;273:32384–32387. doi: 10.1074/jbc.273.49.32384. [DOI] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Robinson A, McDonald JP, Caldas VE, Patel M, Wood EA, Punter CM, Ghodke H, Cox MM, Woodgate R, Goodman MF. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 2015;11:e1005482. doi: 10.1371/journal.pgen.1005482. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Antimutator mutants in bacteriophage T4 and Escherichia coli. Genetics. 1998;148:1579–1585. doi: 10.1093/genetics/148.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- Senavirathne G, Bertram JG, Jaszczur M, Chaurasiya KR, Pham P, Mak CH, Goodman MF, Rueda D. Activation-induced deoxycytidine deaminase (AID) co-transcriptional scanning at single-molecule resolution. Nat Commun. 2015;6:10209. doi: 10.1038/ncomms10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro GS, Aviszus K, Murphy J, Wysocki LJ. Evolution of Ig DNA Sequence to Target Specific Base Positions Within Codons for Somatic Hypermutation. J. Immunol. 2002;168:2302–2306. doi: 10.4049/jimmunol.168.5.2302. [DOI] [PubMed] [Google Scholar]
- Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD'C interaction. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- Speyer JF. Mutagenic DNA polymerase. Biochem Biophys Res Commun. 1965;21:6–8. doi: 10.1016/0006-291x(65)90417-1. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O#x0027;Donnell M, Goodman MF. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD'2C mutagenic complex and RecA protein. Proc Natl Acad Sci U S A. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O#x0027;Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O#x0027;Donnell M, Woodgate R, Goodman MF. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953a;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953b;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- Weigle JJ, Dulbecco R. Induction of mutations in bacteriophage T3 by ultraviolet light. Experientia. 1953;9:372–373. doi: 10.1007/BF02167637. [DOI] [PubMed] [Google Scholar]
- Witkin EM. The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction. Proc Natl Acad Sci U S A. 1967;57:1275–1279. doi: 10.1073/pnas.57.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD'. Proc Natl Acad Sci U S A. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R, Sedgwick SG. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ. Dynamics of leading-strand lesion skipping by the replisome. Mol Cell. 2013;52:855–865. doi: 10.1016/j.molcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci U S A. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Egelman EH. The LexA repressor binds within the deep helical groove of the activated RecA filament. J Mol Biol. 1993;231:29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]