Abstract
Seeking to assess the representative and instructive value of an engineered mouse model of pancreatic neuroendocrine tumors (PanNET) for its cognate human cancer, we profiled and compared mRNA and miRNA transcriptomes of tumors from both. Mouse PanNET tumors could be classified into two distinctive subtypes, well-differentiated islet/insulinoma tumors (IT) and poorly differentiated tumors associated with liver metastases, dubbed metastasis-like primary (MLP). Human PanNETs were independently classified into these same two subtypes, along with a third, specific gene mutation–enriched subtype. The MLP subtypes in human and mouse were similar to liver metastases in terms of miRNA and mRNA transcriptome profiles and signature genes. The human/mouse MLP subtypes also similarly expressed genes known to regulate early pancreas development, whereas the IT subtypes expressed genes characteristic of mature islet cells, suggesting different tumorigenesis pathways. In addition, these subtypes exhibit distinct metabolic profiles marked by differential pyruvate metabolism, substantiating the significance of their separate identities.
SIGNIFICANCE
This study involves a comprehensive cross-species integrated analysis of multi-omics profiles and histology to stratify PanNETs into subtypes with distinctive characteristics. We provide support for the RIP1-TAG2 mouse model as representative of its cognate human cancer with prospects to better understand PanNET heterogeneity and consider future applications of personalized cancer therapy.
INTRODUCTION
Genetically engineered mouse models of human cancer have fueled progress in understanding mechanisms of tumor development and progression in different organs, induced by various driving oncogenes and/or loss of tumor suppressors (1). Among these, the RIP1-TAG2 (RT2) mouse model, in which pancreatic neuroendocrine tumors (PanNET) are induced by expression of the SV40 T-antigen oncogenes in insulin-producing islet β cells (2, 3), has proved a valuable prototype for studying the stepwise progression of multistage tumorigenesis. For example, this model has revealed the angiogenic switch (4), the importance of attenuating apoptosis (5), and the determinants of progression to an invasive growth phenotype (6–9). The RT2 model has also proved to be tractable for preclinical trials of targeted therapies. For example, preclinical trials of angiogenesis inhibitors targeting the VEGF signaling pathway (10–15) predicted efficacy and incentivized clinical trials (16) that led to the approval of sunitinib for treating human PanNET. These results notwithstanding, a persistent question concerns the extent to which these tumors, induced by a viral oncogene that abrogates the p53 and RB tumor suppressors, represent human PanNET. This study has sought to address the question via independent profiling followed by comparative analysis (cross-filtering) of the mRNA and miRNA transcriptomes of tumors from the mouse model and from human patients.
Human PanNET can be categorized based on the World Health Organization (WHO)’s classification into comparatively benign, well-differentiated neuroendocrine tumors (WD-NET, also known as NET Grade G1, with 0% to 2% Ki67 cellular proliferation marker-based immunolabeling, or G2, with 3% to 20% Ki67 labeling) that can be functional or nonfunctional, secreting in the former case insulin (insulinoma) or other islet cell hormones. Aggressive, poorly differentiated neuroendocrine carcinomas (PD-NEC or NEC G3, with >20% Ki67 labeling), on the other hand, are mostly nonfunctional and are defined by loss or marked downregulation of the islet cell hormone genes that define their origins (17–21). However, it is increasingly being recognized that not all G3 neuroendocrine neoplasms are poorly differentiated. In fact, a proportion of G3 aggressive tumors display well-differentiated morphology (22).
Although the majority of the PanNETs are sporadic, a certain fraction represents familial hereditary disease that results from inactivating mutations in the multiple endocrine neoplasia type 1 (MEN1) gene (23, 24). Notably, exome sequencing of human PanNET has confirmed somatic inactivation of MEN1 in about 40% of cases and revealed a spectrum of somatic mutations in genes associated with chromatin remodeling (DAXX, encoding death-domain– associated protein, and ATRX, encoding α-thalassemia/mental retardation syndrome X-linked protein) and in negative regulators of the PI3K–mTOR pathway (TSC2, encoding tuberous sclerosis 2 protein, and PTEN, encoding a lipid phosphatase that modulates PI3K signaling; refs. 23, 24). However, these tumor genome analyses have not provided clarity into the underlying determinants that correspond to the varying degrees of malignancy observed in human PanNET.
The RT2 model develops PanNETs of varying malignancy, including both adenomatous (encapsulated tumors with well-defined margins and less invasion into surrounding exocrine pancreas) islet/insulinoma tumors (IT) and invasive carcinomas (IC2), the latter being defined in part by loss of E-cadherin expression (6) and desmosomes (25). Expression of the SV40 large T-antigen hybrid oncogene in the approximately 400 pancreatic islets elicits a highly synchronous tumorigenesis pathway: Hyperplastic/dysplastic islets begin arising at 3 weeks of age, of which about 25% switch on angiogenesis by 6 to 9 weeks, followed by the formation of PanNET; at end stage of 14 to 16 weeks, every mouse has 2 to 10 solid tumors (2, 3). This synchronicity might have predicted homogeneity in histologic and genetic phenotypes, in contrast with the bona fide human cancer. Interestingly, however, there is evidence for genetic and phenotypic heterogeneity. First, genome profiling by array comparative genomic hybridization (CGH) has revealed that developing PanNETs in this model acquire a range of chromosomal aberrations (26, 27). Second, profiling of the miRNA transcriptome during the stereotypical stages of tumorigenesis in this pathway revealed, in addition to stepwise changes in miRNA expression during progression, the existence of two distinctive tumor subtypes, defined by markedly distinct miRNA signatures. The majority of tumors profiled fell into a class that was similar to the earlier tumor development stages (hyperplastic/dysplastic islets and angiogenic islets) and were designated as canonical IT. The remaining primary tumors clustered together with a very distinct miRNA profile and had clear similarity to a small sample set of liver metastases. This latter set, defined by its miRNA signature as a distinct subtype, was dubbed “metastasis-like primary” [or met-like primary (MLP); ref. 28 ]. Herein, we establish the existence of human miRNA PanNET subtypes that resemble those seen in mouse by characterizing the human and mouse PanNET transcriptomes. We further use this information to define associated phenotypic characteristics that are suggestive of differing metabolism and distinct cellular phenotypes or tumorigenesis pathways of PanNETs.
RESULTS
miRNA Profiling of Human PanNET Reveals Distinctive Molecular Subtypes Similar to Mouse PanNET
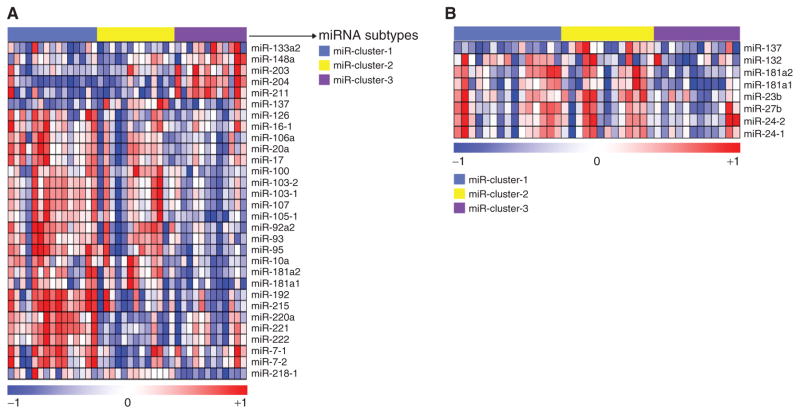
Given the identification of PanNET subtypes in the mouse model by miRNA profiling (28), we began by similarly analyzing miRNA profiles from 40 human PanNETs, including liver or lymph node (LN) metastases, designated as our core clinical miRNA dataset (from a previous report; ref. 29). We chose 88 variable miRNAs that have SD greater than 0.5 across samples and asked whether the tumors could be clustered into distinctive subtypes using the consensus clustering-based non-negative matrix factorization (NMF) method (30), as we have described previously (31–32). The human PanNETs clustered into three distinct subsets with an NMF-based cophenetic coefficient (cc)—indicative of a robust number of clusters (Supplementary Fig. S1A)—being highest for k = 3. These clusters were named miR-cluster-1, miR-cluster-2, and miR-cluster-3. To identify subtype-specific miRNA signatures, we performed Significance Analysis of Microarrays (SAM; ref. 33), as described in our previous publications (31, 32). We identified a 30-miRNA signature (PanNETassignermiR) that was differentially expressed across the three subtypes (Fig. 1A; Supplementary Table S1A). The three miRNA PanNET subtypes were subsequently validated with an independent human PanNET miRNA profiling dataset (n = 50; miRNA validation dataset), applying the PanNETassignermiR signatures and NMF analysis (Supplementary Fig. S1B and S1C and Supplementary Table S1B).
Figure 1.
miRNA expression subtypes in human PanNET and their similarity to mouse MLP PanNET. Heatmaps of human PanNET samples showing (A) three miRNA expression subtypes as defined using NMF and (B) the expression of previously identified RT2 mouse MLP-specific miRNAs (from Olson and colleagues; ref. 28) in the three human PanNET subtypes defined in A. In both cases, the columns represent individual human tumors. In A, the rows indicate 30 differentially expressed miRNAs from 40 samples. In the rainbow bar below the heatmap, red indicates elevated expression, blue decreased, and white no change. (Supplementary Table S1A also lists the miRNAs in the same order as the columns in part A.) The miRNAs shown in A and Supplementary Table S1A constitute a signature, designated as the “PanNETassigner-miR signature” (genes were selected using SAM analysis with a delta value, Δ = 1.3 and a false discovery rate, FDR < 0.05).
Next, we cross-filtered the mouse and human miRNA profiles to assess possible similarities between the two miRNA subtypes seen in the mouse (28) and the three miRNA clusters found in human PanNET. We performed this analysis by mapping the mouse MLP signature miRNAs from Olson and colleagues (28) onto the human PanNET data. Indeed, eight mouse MLP- and metastases-specific miRNAs (miR-137, -132, -181a2, -181a1, -23b, -27b, -24-1, and -24-2) were highly expressed in the majority of the samples in miR-cluster-1 and -2. Among these eight miRNAs, miR-137, the most highly enriched miRNA in the mouse MLP subtype, was highly expressed in the human miR-cluster-2 (Fig. 1B). Given the implication that the mouse model was in fact recapitulating some of the heterogeneity seen in human disease, according to this metric, we went on to profile the mRNA transcriptomes of both human and mouse PanNETs, as well as the premalignant stages in the mouse model.
mRNA Profiling Confirms the Existence of Molecular Subtypes in Human PanNET
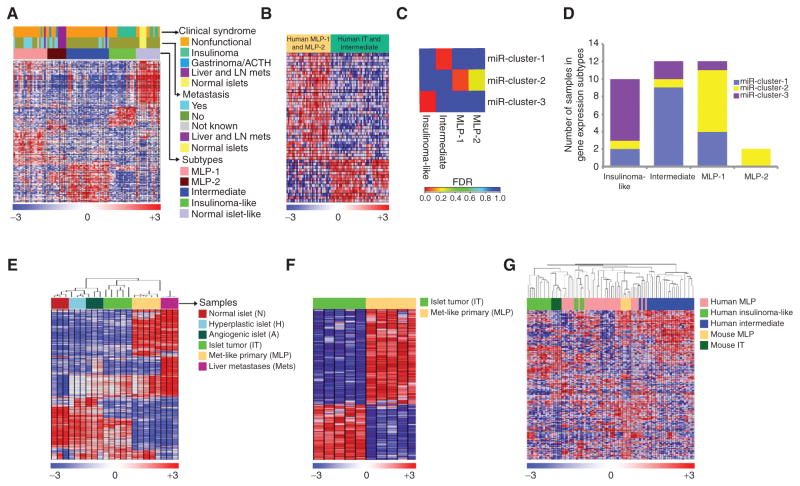
Human mRNA transcriptomes (n = 86; a core clinical gene expression dataset that primarily included nonfunctional PanNETs, insulinomas, and normal pancreatic islet samples) obtained from a previous study (34) were analyzed using NMF consensus clustering analysis. This dataset includes four normal islets, seven liver and lymph node metastases, and 75 primary tumors with or without associated metastasis. This dataset was analyzed after selecting variable genes that were defined to have SD > 0.8, which revealed five clusters of PanNET (cc highest for k = 5; Fig. 2A; Supplementary Fig. S2A; and Supplementary Table S1C). One of these clusters consisted in large part (28%) of normal human islet samples and was therefore termed “normal islet-like.” This cluster was not considered further as a tumor subtype in our current study. Among the four other subtypes, the one termed “islet/insulinoma tumors” consisted primarily of less-aggressive insulinomas (n = 16) that expressed insulinoma-associated genes, such as INS, IAPP, and INSM1. None of these tumors from this subtype were associated with metastases.
Figure 2.
Gene expression subtypes and cross-species analysis of human and mouse PanNETs. A, heatmap of human PanNET samples showing subtypes defined by gene expression differences, and the association of these subtypes with distant metastasis and clinical syndrome. The subtypes are defined by sets of genes—signatures—that are listed in the same order in Supplementary Table S1C and labeled as “PanNETassignermRNA signatures” (genes were selected using SAM analysis with Δ = 11.1 and FDR < 0.05). ACTH represents ectopic adrenocorticotropic hormonesecreting tumor. B, the heatmap shows genes (listed in Supplementary Table S1D) that were commonly expressed/repressed in human MLP-1 and MLP-2 subtypes and inversely expressed in tumors of the other two subtypes (genes were selected using SAM analysis with Δ = 1.7 and FDR < 0.05). C, enrichment of miRNA subtype samples in gene expression subtype samples was assessed using the hypergeometric test (red represents a significant enrichment of samples between miRNA and gene expression subtypes with FDR < 0.05). D, a bar plot displays the total number of miRNA subtype samples in each gene expression subtype. E and F, heatmaps of RT2 mouse PanNET samples showing gene expression profile differences. E, the distinct stages of tumorigenesis in the RT2 mouse model are shown, including IT, MLP, and liver metastasis (mets) samples. Genes (908 genes selected with SD > 0.8, and highest variable probe across samples in the cases where multiple probes per gene were present) and samples are listed in the same order in Supplementary Table S1F. F, mRNAs that clearly delineate the two subtypes—IT and MLP—are shown. The rows indicate 394 differentially expressed genes (genes were selected using SAM analysis with Δ = 0.6 and FDR < 0.05) among the profiled genes of 10 samples. These genes define “m-PanNETassigner-mRNA” signature, with the individual genes listed in the same order in Supplementary Table S1G. G, heatmap showing cross-species analysis and association of human and mouse PanNET subtypes using ComBat-merged gene expression profiles of RT2 mouse (n = 10) and patient (n = 72) PanNET samples. In all the heatmaps, the rows indicate differentially expressed genes (Supplementary Table S1H) among the profiled gene expression data. In the rainbow bar beneath the heatmaps, red indicates elevated expression, blue decreased, and white no change.
Two of the other subtypes consisted primarily of samples (n = 31) categorized as nonfunctional that were associated (in more than 50% of primary tumor samples) with metastasis. As such, we termed these groups MLP-1 and MLP-2 using nomenclature from our work classifying tumors from the RT2 mouse model (28). In general, MLP-1 and MLP-2 share signatures that are enriched for genes associated with fibroblasts/stroma, stem cells, and hypoxia (Fig. 2B; additional information about these genes is available in Supplementary Table S1D). Although these subtypes have subtle differences in their constitution (e.g., 45% of the MLP-2 samples were from liver/LN metastases, in contrast with only 5% of the MLP-1 subtype samples; Fig. 2A), on the whole, there were more similarities than differences. As such, MLP-1 and MLP-2 were subsequently considered as a single MLP PanNET subtype, thereby reducing the total number of human PanNET transcriptome subtypes to three. Three primary PanNETs with functional symptoms of insulinoma, gastrinoma, and Cushing’s syndrome [secretes ectopic adrenocorticotropic hormone (ACTH)] were classified as MLP-1 and MLP-2, respectively. These tumors were metastatic and hence probably denote that rarely functional PanNETs with malignant phenotype can be classified into the MLP subtype. The third distinctive subtype was predominantly composed of nonfunctional PanNETs (n = 25), of which approximately 16% of primary tumors were associated with distant metastasis (Fig. 2A). In addition, this subtype shared many genes with the IT subtype while also being moderately associated with metastasis. As such, it was provisionally named the “intermediate subtype.”
To validate the robustness of these aforementioned subtypes in other PanNET datasets, we derived gene signatures (PanNETassigner-mRNA; 221 genes) specific to each of the subtypes using SAM (Fig. 2A; Supplementary Table S1C). We then used another PanNET dataset (n = 29; gene expression validation dataset) and mapped the PanNETassigner-mRNA signatures onto the new dataset and performed NMF analysis (Supplementary Fig. S2B and S2C and Supplementary Table S1E). Concordantly, in this validation dataset, we observed all five (including normal-like and two MLP) mRNA expression subtypes.
Comparison of miRNA and mRNA Subtypes in Human PanNET
Next, we sought to associate the human PanNET mRNA and miRNA transcriptome subtypes using the matched samples (n = 36) from the core clinical gene (mRNA) expression dataset and the miRNA dataset. Interestingly, each of the three miRNA subtypes was significantly enriched in one of the three mRNA transcriptome subtypes (Fig. 2C and D). Tumors from miR-cluster-1 were significantly enriched in the intermediate subtype; tumors from miR-cluster-2 were enriched in the MLP-1 sub-subtype; and finally, miR-cluster-3 were enriched in the IT subtype (Fig. 2C and D). Due to low sample size in the MLP-2 subtype, it is not significantly associated with any of the miRNA subtypes. However, both of the MLP-2 samples belong to miR-cluster-2 (Fig. 2D). Given the regulatory nature of miRNA for mRNA, concordance between miRNA and mRNA subtypes is not unexpected, but serves to further validate the existence of distinct subtypes of human PanNET.
Transcriptome Subtypes of Mouse PanNET and Its Neoplastic Stages
Having established the existence of three human PanNET subtypes, we next investigated the mRNA expression profiles from the progressive stages of disease in RT2 mice, including pools of normal (n = 3), hyperplastic/dysplastic (n = 3), and angiogenic (n = 3) islets, as well as individual tumors (n = 10) and liver macrometastases (mets; n = 3). As expected, hierarchical clustering (genes selected with SD > 0.8 across samples and average linkage method applied) grouped these samples according to the stage of disease progression, with all of the normal, hyperplastic, and angiogenic islets grouped into one main cluster, whereas tumors and mets grouped into a second main cluster (Fig. 2E; Supplementary Table S1F). The tumor samples were further split into two distinct clusters, one of which shared similarities with the metastasis samples. Based on our current understanding, we expected these tumors to be the MLPs, with the remaining tumors being ITs. In an effort to identify distinguishing characteristics between these mouse tumor subtypes, we performed differential gene expression analysis using the SAM analysis and identified 394 genes (m-PanNET-assignermRNA) that were differentially expressed (Fig. 2F; Supplementary Table S1G). These data indicate that both human PanNET and RT2 tumors can be classified into distinct subtypes based on their miRNA or mRNA expression profiles.
Cross-Species Analysis of Human and Mouse PanNET
Next, we sought to further investigate the similarities between the mouse and human PanNET subtypes, this time at the mRNA level. To this end, we first compared mRNA expression profiles of the distinct subtypes of human Pan-NET and RT2 tumors. Human PanNET mRNA expression profiles from the MLP, IT, and intermediate subtypes and mouse PanNET mRNA expression profiles from the MLP and IT subtypes were combined and analyzed using two different batch correction methods after selecting those genes that had an SD > 0.8 from each dataset. First, we applied the ComBat (“combatting batch effects when combining batches of gene expression microarray data”) method (35) directly to the SD-selected datasets to remove batch effects, followed by hierarchical clustering. Second, we applied distance weighted discrimination (DWD; ref. 36) analysis after normalizing the data (described in the Methods section below), followed by hierarchical clustering to validate the results from ComBat. Both of these analyses revealed that the majority of mouse and human MLPs clustered together and that the mouse and human ITs clustered together. None of the mouse tumors clustered with human tumors of the intermediate subtype (Fig. 2G; Supplementary Table S1H for ComBat analysis; Supplementary Fig. S3 and Supplementary Table S1I for DWD analysis). In Fig. 2G, six human MLP tumors clustered more closely than most MLPs with the human and mouse ITs, and these are suspected to represent a subset of human MLPs with increased IT-specific genes (similar to the subset referred to below as insulin-high MLP mouse and human tumors). Similarly, four human ITs clustered more closely with the majority of the human and mouse MLPs, again suggesting that on rare occasions ITs can be aggressive and metastatic. In sum, this cross-species analysis clearly demonstrates concordance between the two human and mouse PanNET subtypes—IT and MLP.
Mutational Associations in PanNET Subtypes
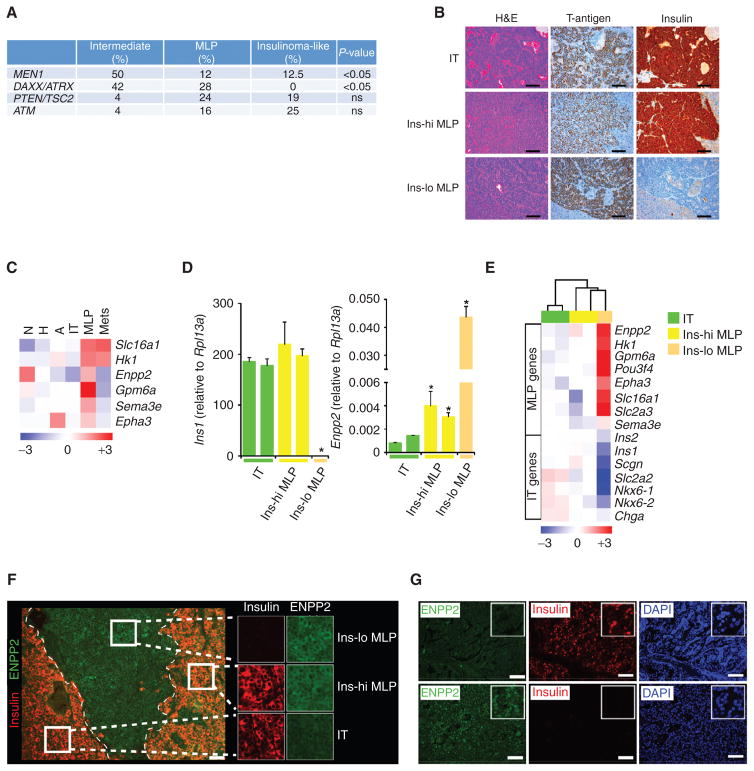
The intermediate human subtype is not observed in the RT2 mouse model (Fig. 2G). One possibility is that this subtype has a molecular pathogenesis different from the concomitant loss of function in the TP53 and RB tumor suppressor pathways (as is suspected for this and many forms of human NET), but rather through the loss of the MEN1 tumor suppressor. In order to address this hypothesis, we performed targeted next-generation DNA sequencing using 65 human tumor specimens representing all three subtypes. Indeed, we observed that 50% (n = 24) of the intermediate tumors were MEN1 mutant, whereas less than 13% of tumors from each of the other two subtypes had mutations in MEN1 (P < 0.05; Fisher exact test; Fig. 3A; Supplementary Table S2). These data provide a possible explanation for the absence of intermediate subtype tumors among those samples from the mouse model and provide insight into the etiology of intermediate subtype human PanNET tumors. As such, we have renamed this subtype as “MEN1-like” to reflect this association. We infer that Men1 mutations are not occurring in either IT or MLP subtypes in the RT2 mouse model of PanNET, as evidenced by the lack of detectable copy-number losses by CGH on mouse chromosome 19 where Men1 resides (26, 27). Notably, most MEN1-related human PanNETs have reported loss of heterozygosity detectable by CGH (37).
Figure 3.
Mutation changes and biomarker expression in human and mouse PanNET subtypes. A, a summary of MEN1, DAXX/ATRX, mTOR pathway (PTEN/TSC2) and ATM gene mutations in human PanNET subtypes. P values correspond to the Fisher exact test. ns, not significant. B, visualization of different histologic subtypes of RT2 tumors with hematoxylin and eosin (H&E) staining, and immunostaining for the driving oncoprotein (SV40 T-antigen) and insulin. Images are representative of the analysis from 60 tumor-bearing RT2 B6, B6AF1, and B6D2F1 mice (see Methods section for different strains of the RT2 mouse model). Scale bar, 200 μm. C, heatmap showing median expression of MLP-specific genes (biomarkers) in distinct stages of the RT2 mouse model of PanNET. N, normal islet; H, hyperplastic/dysplastic islet; and A, angiogenic islet. D and E, expression of PanNET subtype genes in five different laser capture microdissection–derived (LCMD) samples from tissue sections of tumors representing IT, Ins-hi MLP, and Ins-lo MLP subtypes using (D) RT-PCR (Ins1 and Enpp2) and (E) microarray (multiple PanNET subtype genes) methodologies. In D, * represents statistical P value < 0.05 as measured using the Student t test comparing relative Ins1 or Enpp2 expression in IT versus Ins-hi and Ins-lo MLP tumors. F, visualization of different histologic subtypes of RT2 tumors within a single pancreas by immunostaining for ENPP2 and insulin protein expression. Images are representative of the analysis from 8 tumor-bearing RT2 mice. Scale bar, 50 μm. G, ENPP2 and insulin protein expression in human PanNETs as assessed using IHC. DAPI (4′,6-diamidino-2-phenylindole) staining represents cellular nucleus. Scale bar, 50 μm.
Also of note, 42% (n = 24) and 28% (n = 25) of the human MEN1-like/intermediate and MLP subtype tumors, respectively, had recently identified PanNET-specific mutations in DAXX/ATRX (24), whereas none of the IT tumors exhibited this mutation (P < 0.05; Fisher exact test; Fig. 3A), confirming recent reports that DAXX/ATRX mutations are enriched in more aggressive nonfunctional PanNETs (24, 38). We observed that 21% (n = 24) of the intermediate/MEN1-like tumors had both MEN1 and DAXX/ATRX mutations [29% (n = 24) of the intermediate/MEN1-like tumors did not have either MEN1 or DAXX/ATRX mutations]. In RT2 mice, we did not detect Daxx/Atrx mutations, as assessed by DNA sequencing of 24 RT2 mouse tumor samples, including 14 MLPs and 10 ITs.
In addition, we evaluated the association of the mTOR pathway genes TSC2 and PTEN, which are mutated in Pan-NETs, with the human transcriptome subtypes (24). We observed that both MLP (24%; n = 25) and IT (19%; n = 16) subtypes are associated with TSC2 or PTEN mutations. In contrast, only 4% (n = 24) of the intermediate/MEN1-like tumors have TSC2 or PTEN mutations (Fig. 3A). Similar to the mTOR pathway genes, the DNA repair pathway gene ATM is also mutated in PanNETs (24). Again, MLP (16%; n = 25) and IT (25%; n = 16) subtypes are identifiable (not statistically significant) with ATM mutations, whereas only 4% (n = 24) of the intermediate/MEN1-like tumors have this mutation (Fig. 3A). However, these associations of mTOR pathway genes (TSC2 and PTEN) or ATM mutations with subtypes are not statistically significant (Fig. 3A), which may be attributed to the limited sample size of tumors.
Overall, this integrated analysis revealed that MEN1 mutations are significantly enriched in the intermediate subtype (which is renamed accordingly as the MEN1-like subtype); DAXX/ATRX mutations significantly associated with the MLP and MEN1-like/intermediate subtype; and TSC2/PTEN/ATM mutations associated (not statistically significant) with the MLP and IT subtypes. These findings warrant further validation using increased sample size.
Subtype-Specific Associations in Tumor Functionality, Biomarker Expression, Proliferative Index, and Vascularity
Next, we sought to histologically characterize the transcriptome subtypes of PanNET based on tumor differentiation state and insulin protein expression. We identified three histopathologic subtypes (histotypes) of PanNET in the mouse model, with the following distinctive characteristics: (a) moderately- to well-differentiated tumors that stained highly positive for insulin, ostensibly the IT subtype (Fig. 3B, top), (b) less-differentiated tumors that stained variably positive for insulin expression, ascribed as insulin-high MLP (Ins-hi MLP; Fig. 3B, middle), and (c) a group of poorly differentiated RT2 tumors that did not express insulin and were therefore designated as insulin-low MLP (Ins-lo MLP; Fig. 3B, bottom). Notably, tumors with Ins-lo MLP characteristics have recently been identified in the RT2 model of PanNET and referred to as poorly differentiated invasive carcinoma (PDIC; ref. 39). Given the many characteristics this classification shares with the one we define herein as Ins-lo MLP, it is considered and renamed as such in the present study.
These three mouse PanNET histotypes cannot be linked directly per se with the transcriptome subtypes. In order to investigate a possible association, we identified a small number of genes that readily distinguish mouse MLP from IT tumors based on our transcriptome profiling analyses (Fig. 2E). These included Enpp2 (autotaxin, ATX), Sema3e (semaphorin 3e), Epha3 (ephrin A3), Gpm6a (glycoprotein m6a), and the genes encoding for the metabolic enzymes HK1 (hexokinase) and MCT1 (SLC16A1; monocarboxylate transporter; Fig. 3C); all are expressed at higher levels in MLP than IT. Seeking to associate the PanNET histotypes with the molecular subtypes, we performed laser capture microdissection (LCM) from tissue sections of mouse RT2 tumors (n = 5) that were either positive or negative for insulin protein expression by IHC. After microdissection, expression of the Ins1 and Enpp2 genes in LCM tumors was assessed using qRT-PCR to associate the different histotypes with the delineated transcriptome subtypes. Four of the five tumors expressed Ins1, of which two had relatively low Enpp2 expression, identifying them as IT; the two others had high Enpp2 expression and were identified as Ins-hi MLP. In addition, one tumor had low Ins1 expression and high Enpp2 expression, consistent with the Ins-lo MLP subtype (Fig. 3D). Next, we performed microarray analysis of these LCM tumor samples followed by hierarchical clustering using a selected set of genes representing IT and MLP transcriptome subtypes. Concordant with the RT-PCR results, the two Ins-hi MLP samples clustered with Ins-lo MLP, whereas the two IT tumors clustered separately. Figure 3E shows relatively higher expression of MLP markers in Ins-hi and Ins-lo MLP tumors compared with IT tumors; in contrast, IT markers were more highly expressed in IT compared with Ins-hi and Ins-lo MLP tumors. Furthermore, in order to identify and establish IHC markers for these subtypes, we evaluated Enpp2 as a marker by IHC for ENPP2 protein expression. Consistent with the gene expression data from Fig. 3D and E, we observed significantly increased ENPP2 protein expression in Ins-hi and Ins-lo MLP, whereas there was decreased protein expression in the IT subtype (Fig. 3F).
In addition to insulin, we observed that both the IT and Ins-hi MLP tumors expressed higher levels of chromogranin A (Chga) and secretogogin (Scgn; Fig. 3E) genes compared with the Ins-lo MLP tumors, suggesting that the insulin-secretory pathway is still active in the IT and Ins-hi MLP tumors, consistent with their insulin IHC staining in Fig. 3F. Hence, by gene expression profile and histology, the mouse Ins-lo MLP and IT subtypes are evidently similar to the human MLP (incorporating both human MLP-1 and MLP-2 sub-subtypes) and IT subtypes, respectively. Overall, these results clearly link the transcriptome and histotypes of human and mouse PanNET and demonstrate the existence of two robust subtypes in common: IT and MLP.
We further corroborated ENPP2 as a marker in human PanNETs by analyzing ENPP2 expression in our core clinical gene expression dataset. We found that tumors of the MLP-2 subtype have relatively high ENPP2 gene expression compared with MLP-1 and IT. In addition, MLP-2 subtype tumors have relatively low gene expression of insulin (Supplementary Fig. S4). Next, we tested ENPP2 protein expression in human PanNETs using a tissue microarray (TMA; n = 20) and IHC. Interestingly, 11 human PanNETs were negative for insulin, but positive for ENPP2 (Fig. 3G; Supplementary Table S3A). These tumors are similar to the mouse Ins-lo MLPs and probably represent human Ins-lo MLPs. However, some of these human ENPP2 -high and insulin-negative tumors could represent intermediate/MEN1-like subtype tumors (Supplementary Fig. S4). In addition, we observed two human PanNETs in our TMA were positive for insulin, but negative for ENPP2, probably representing the IT subtype. There were also three human PanNETs that were positive for both insulin and ENPP2, which likely represent the Ins-hi MLP subtype (Supplementary Table S3A). However, an increased sample size is required for definitive evaluation of the human PanNET subtype protein markers by IHC. Collectively, these results suggest that ENPP2 may serve as a biomarker that—along with insulin—can be used to distinguish human and mouse PanNET subtypes (and even sub-subtypes of MLP).
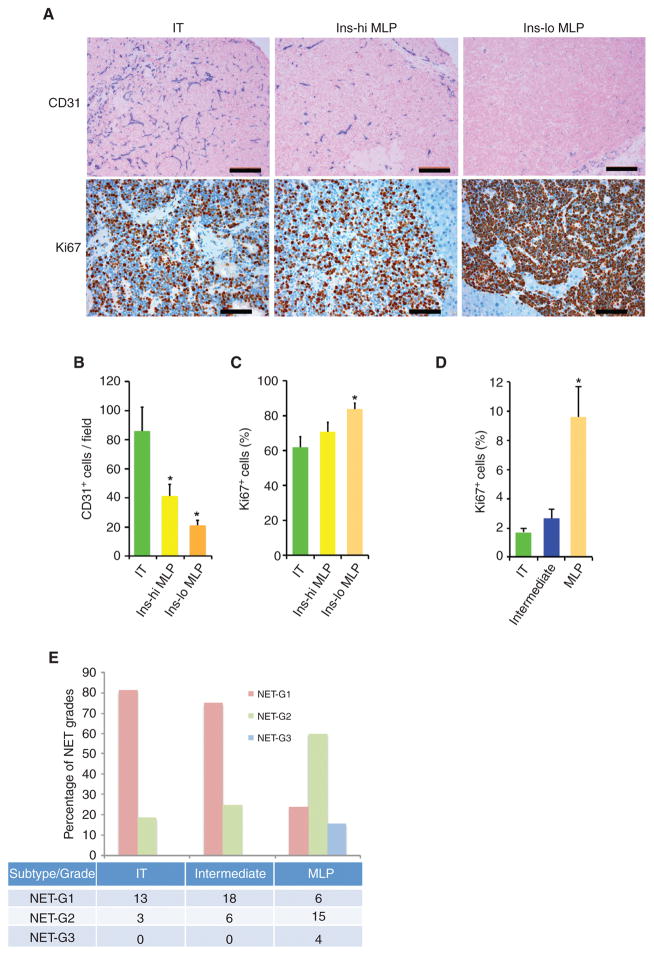
To further characterize the phenotypes of distinct PanNET subtypes in RT2 mice, we evaluated the tumor vasculature by immunostaining for CD31. As presented in Fig. 4A, top, and consistent with the clinical presentation of human IT and our previous studies in the RT2 model, mouse IT samples were highly vascularized. Interestingly, the MLP tumors were comparatively less vascularized. IT samples contained about 40% to 60% more CD31-positive cells than Ins-hi and Ins-lo MLP tumors (Fig. 4B).
Figure 4.
Vascular and proliferative characteristics of human and mouse PanNETs. A, CD31 (top) and Ki67 (bottom) staining in RT2 tumor subtypes. Images are representative of the analysis of 60 tumor-bearing RT2 B6, B6AF1, and B6D2F1 mice (see Methods section for different strains of the RT2 mouse model). In the top plot, blue represents CD31, pink represents nucleus, and scale bar represents 200 μm; in the bottom plot, red represents Ki67, blue represents nucleus, and scale bar represents 100 μm. B and C, quantification of CD31-positive blood vessels (B) and Ki67-positive tumor cells (C) in different subtypes of RT2 tumor subtypes. The quantitation is representative of the analysis of 10 random fields from 10 different tumor-bearing RT2 B6, B6AF1, and B6D2F1 mice from the analysis illustrated in A. D, percentage of Ki67-positive cells in human PanNET subtypes (from the samples of the core clinical gene expression dataset; see Methods section for detailed description about quantitation). * represents statistical P value < 0.05 as measured using the Student t test comparing IT versus Ins-hi MLP and IT versus Ins-lo MLP. Error bars in the graphs represent SD. E, identifiable percentage and summary (number of samples in the table) of NET grades in human Pan-NET subtypes. NET grades are significantly (P < 0.05; Fisher exact test) associated with PanNET subtypes.
We additionally assessed cancer cell proliferation in human and mouse tumors by Ki67 staining, which is clinically used for assessing NET grades (17–21). We observed a statistically significant increase in Ki67 staining in the mouse Ins-lo MLP tumors compared with the IT samples, whereas the differences with Ins-hi MLP tumors were not significant (Fig. 4A, bottom and C). This characteristic of increased Ki67 staining in the RT2 Ins-lo MLP sub-subtype is also evident in the human tumors, where MLP tumors (equivalent to mouse Inslo tumors) exhibit a substantially higher Ki67 staining compared with MEN1-like/intermediate and insulinoma subtype PanNETs (less than 3%), albeit lower overall (Fig. 4D). The difference in frequencies of Ki67-positive cells between mouse and human PanNETs may be attributable to the effects of the driving oncogene—SV40 large T-antigen—in the mouse PanNET cancer cells.
Association of NET Grades with PanNET Transcriptome Subtypes
In order to ascertain whether the transcriptome subtypes are associated with the WHO classification of NET grades, we sought to associate the core clinical mRNA dataset with NET grade information. A majority of the IT (81%) and intermediate (75%) subtypes are identifiable as NET G1, whereas the remainder are NET G2. In contrast, 60% of the MLP subtype PanNETs are NET G2, and 16% are NET G3; the remaining 24% are NET G1 (Fig. 4E; Supplementary Table S2). Thus lower-grade (including NET G2) human PanNETs are heterogeneous, variably associating with all three transcriptome subtypes, whereas high-grade NET G3 tumors are exclusively associated with the MLP subtype.
Genes Regulating Early Pancreas Development Distinguish MLP from IT
Given the highly distinctive mRNA and miRNA profiles of the PanNET subtypes, we hypothesized that these subtypes may be the result of tumors arising from different cellular origins. To identify the putative cell of origin for the IT and MLP subtypes, we examined the expression of various pancreatic stem/progenitor- and differentiation-specific genes. In both human and mouse PanNETs, the MLP samples were enriched for expression of pancreatic progenitor-specific genes, such as HNF1B (40) and GATA6 (though clustered with mature β-cell genes in human PanNETs; ref. 41), and the stem cell factor LIN28B (ref. 42; Lin28b significant only in mouse PanNETs), relative to the IT samples (Fig. 5A and B). Collectively, this pattern of gene expression suggests that the MLP subtypes share characteristics with β-cell precursors or immature β cells. Conversely, the IT subtype differentially expresses a number of mature β-cell–specific genes, including PDX1, INS1, GCK (GK), and SLC2A2 (GLUT2), and the insulinoma-specific gene INSM1 (Fig. 5A and B). These results provide evidence that the IT subtype resembles mature β cells.
Figure 5.
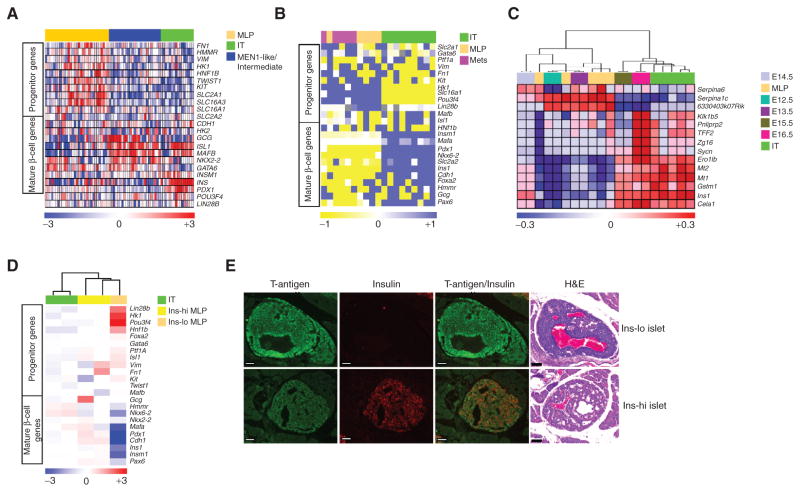
Stem cell and differentiated phenotypes of PanNET subtypes. A and B, heatmaps showing differential expression of genes that distinguish islet progenitor cells from differentiated β cells, as well as metabolic pathway genes (including metabolic genes that are disallowed in mature β cells; ref. 46) showing differential expression, in PanNET samples from (A) human and (B) mouse. Human PanNET data are from gene expression microarrays, whereas mouse PanNET data are from RT-PCR analysis. The relative (to housekeeping gene PPIA) gene expression from mouse PanNET RT-PCR data was multiplied by 10,000 units before performing the heatmap analysis. Gray color in the heatmap of B represents not available value. C, heatmap comparing the ComBat-merged gene expression profile datasets of IT and MLP subtypes from the RT2 mouse model versus embryonic pancreas (from stages e12.5 to e16.5; data from GSE8070). D, heatmap showing differential expression of genes (measured using microarrays) distinguishing islet progenitor cells from differentiated β cells, as well as variably expressed metabolic pathway genes, in mouse LCM-derived tissue samples from IT, Ins-hi MLP, and Ins-lo MLP PanNET tumor sections, from Fig. 3D and E. E, hyperplastic/dysplastic islets in pancreas tissue sections from RT2 mice showing regions of Ins-lo (top) and Ins-hi (bottom) islets, as visualized with hematoxylin and eosin (H&E) and insulin staining. Images are representative of the analysis from 12 tumor-bearing RT2 mice (8–10 weeks of age). Scale bar, 50 μm.
To further investigate a possible stem/progenitor pancreatic cell origin of MLPs, we compared the gene expression profiles of the PanNET subtypes with those of stem/progenitor islet cells from early pancreatic developmental stages. Islet β cells originate during two sequential regulatory phases of pancreas development, termed the primary and secondary transitions. The primary transition occurs between e9.5 and e12.5, whereas the secondary transition begins at e13.5 and ends at e15.5. During the primary transition, the developing pancreas primarily contains multilineage precursor cells and a small number of immature β cells. These precursor and immature cells undergo a differentiation process during the secondary transition to commit to separate pancreatic cell lineages, including one that produces the islet β cells (43). We used the ComBat algorithm to merge the gene expression profiles from RT2 tumors (IT and MLP) with a published dataset (GSE8070) consisting of serial samples collected from the dorsal pancreas of the mouse embryo at each day from e12.5 to e16.5. We observed that the samples from e12.5 to e14.5, which are part of the late primary transition and early secondary transition, clustered tightly with the MLP samples, whereas the majority of the IT samples clustered with the e15.5 and e16.5 samples that are representative of the late secondary transition period (Fig. 5C). It is notable that the RIP-TAG2 transgene is activated at e9.5 (44) and is demonstrably expressed in the multipotential progenitors; its expression in immature β cells can be surmised by its activation during the primary transition. These results indicate that MLPs share characteristics with immature β cells of the pancreas and presumably with the recently described rare adult pancreatic stem/progenitor cells (45). In contrast, the IT cancer cells are more closely related to mature islet β cells of the pancreas than to these progenitors. Moreover, a set of epithelial– mesenchymal transition (EMT)-specific genes—including Vim —was highly expressed in MLPs compared with ITs (Fig. 5B). The expression of EMT genes in MLP in Fig. 5B along with our previous report of EMT in RT2 mouse cell lines (28) suggest that a dedifferentiation process probably also takes place in a subset of tumors that progressed from IT into MLP tumors.
To gain further insights into the canonical IT multistage tumorigenesis pathway, we used the ComBat-based merging of data to compare the gene expression profiles of samples from the different stages of tumor progression with profiles (from Chun and Hanahan; ref. 25) of microdissected noninvasive IT and the broadly invasive IC2 that had been microdissected from tissue sections of RT2 tumors. Interestingly, we observed that MLP tumors have similarities to those of the microdissected invasive regions of IC2 tumors (Supplementary Fig. S5A). Similar to MLP, the microdissected invasive IC2s showed increased expression of the EMT gene— Vim. On the other hand, the noninvasive IT tumors showed increased expression of mature β-cell genes, such as Ins1, Pdx1, and Insm1 (Supplementary Fig. S5B). These data suggest that IC2s arising via progression from ITs switch on an invasive program similar to that of MLP tumors.
In order to clarify whether both Ins-hi and Ins-lo MLP tumors have increased expression of β-cell precursor genes, we further examined the LCM-derived tumor samples described in Fig. 3D and E. Interestingly, we observed that only the Ins-lo MLP tumors have increased expression of a majority of the β-cell precursor genes compared with the Ins-hi MLP and IT tumors (Fig. 5D). The above results motivated us to further examine the histologic differences in hyperplastic/dysplastic islets in the RT2 mouse model, a premalignant stage we had historically considered to be homogeneous. We analyzed 8- to 10-week-old mice and observed heterogeneity in this premalignant stage: Some hyperplastic/dysplastic islets had characteristics of Ins-lo MLP, whereas the majority had the expected characteristics of Ins-hi MLP or IT (Fig. 5E). Although dysplastic islets can be detected with poorly differentiated features and reduced insulin secretion, similar to Ins-lo MLP tumors, they are infrequent (2.8% of 145 premalignant hyperplastic islet lesions evaluated from 12 mice of 8 and 10 weeks old had this phenotype; Fig. 5E). Overall, these results, along with the above-mentioned analysis comparing PanNET tumors with the stem/progenitor cells of the pancreas, suggest the existence of an early and separate cell- and/or pathway-of-origin for prometastatic Ins-lo MLP tumors, whereas Ins-hi MLP tumors arise either directly from mature β cells or via progression of β-cell–derived IT tumors.
Differences in Central Carbon Metabolism Illustrate Subtype-Specific Similarities to Distinct Stages of β-Cell Development
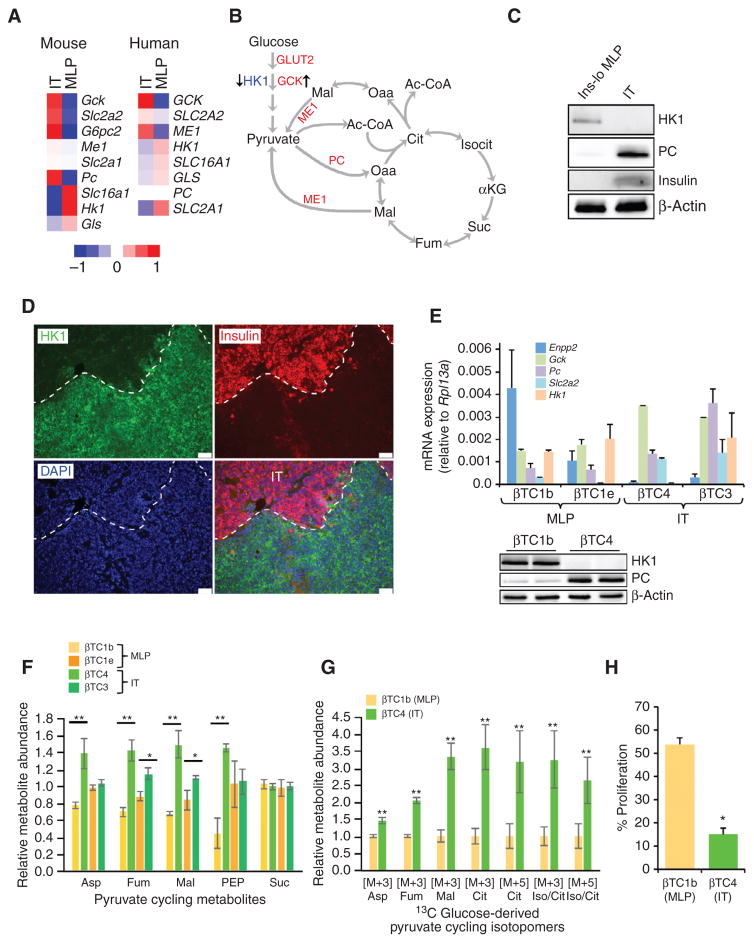
Among the panel of genes differentially expressed between the MLP and IT subtypes was a set involved in central carbon metabolism. Tumors from the IT subtype demonstrably expressed metabolic enzymes of the β-cell lineage, including the glucose transporter GLUT2 (SLC2A2), glucose 6-phosphatase (G6PC2), and glucokinase (GCK) genes. In contrast, the lactate transporter MCT1 (SLC16A1) and hexokinase (HK1) genes, which are not expressed in mature β cells (46), were considerably enriched in both mouse and human samples from the MLP subtype (Fig. 6A). In addition to differences in expression of enzymes involved in glucose uptake and glycolysis, human and murine IT tumors exhibited increased gene expression of pyruvate carboxylase (Pc) and cytoplasmic malic enzyme 1 (Me1), relative to MLP subtype tumors (Fig. 6A). Expression of these enzymes is consistent with pyruvate cycling, a phenomenon observed in mature β cells and insulinomas that facilitates the generation of energy in the mitochondria to sustain glucose-stimulated insulin secretion (refs. 47–49; Fig. 6B). The differential expression of metabolic genes in the IT and MLP subtypes was further validated at the protein expression level. Global, mass spectrometry– based proteomic analyses of mouse IT and Ins-lo MLP tumor samples confirmed that GLUT2 and PC were highly expressed in IT but not in Ins-lo MLP, whereas MCT1 and HK1 protein levels were increased in Ins-lo MLP compared with IT (Supplementary Table S3B). In addition, protein expression of HK1 and PC protein in the Ins-lo MLP subtype was confirmed by immunoblot and IHC (Fig. 6C and D).
Figure 6.
Metabolic characteristics of PanNET subtypes. A, heatmap showing median expression of metabolic genes that are differentially expressed in human and mouse IT versus MLP subtypes. The comparisons involved 5 IT and 5 MLP tumors from the mouse model, and 16 IT and 31 MLP from humans. B, a simplified model of glycolysis and the tricarboxylic acid cycle, overlaid with the pyruvate cycle (arrows in green), highlighting genes upregulated (in red) and downregulated (in blue) in the IT subtype compared with the MLP subtype. C and D, protein expression in IT and Ins-lo MLP determined by immunoblot for HK1, PC, and Insulin (C) and IHC for HK1 and Insulin (D). Scale bars, 50 μm. DAPI staining represents cellular nucleus. E, expression of MLP-specific genes in mouse PanNET cancer cell lines (βTC) representing different subtypes of PanNET. The gene expression is relative to the expression of a housekeeping gene, Rpl13a (top). Immunoblot analysis of βTC cells using antibodies against HK1, PC, and β-actin (bottom). F and G, relative abundance of pyruvate cycle metabolites in mouse PanNET subtype-specific cell lines cultured under normal conditions (F) or in the presence of 13 C-labeled glucose (G). Metabolites in F are median-centered across samples for each metabolite, and this was done separately for two different (batches of) metabolic profiling experiments either using βTC1b (MLP)/βTC4 (IT) or βTC1e (MLP)/βTC3 (IT); and labeled metabolites in G are normalized to their characteristically lower level in the MLP subtype. The M+3 isotopomers of aspartate (Asp), malate (Mal), fumarate (Fum), citrate (Cit), and isocitrate (Iso/Cit) are biosynthesized from glucose by way of PC. The M+5 species of Cit and Iso/Cit are M+3 labeled by PC. H, proliferation of IT versus MLP cancer cell lines in the absence of glucose in the media. * (comparison between βTC1e and βTC3) or ** (comparison between βTC1b and βTC4) represents statistical P value < 0.05 as measured using the Student t test. Error bars in the graphs represent SD or SEM.
Given these observations, we predicted that the metabolic phenotypes of these two subtypes would be similarly distinct. To study PanNET metabolism in vitro, we sought to identify mouse PanNET cell lines that represent each subtype. To this end, we classified cell lines derived from RT2 mouse tumors using two genes that are among those most highly expressed in mouse MLP samples relative to IT samples (Enpp2 and Hk1) and three other genes that conversely are highly expressed in IT cell lines compared with MLP [ Gck, Pc, and Glut2 (Slc2a2); Fig. 6E, top]. qRT-PCR analyses revealed that two RT2 Pan- NET-derived cell lines, βTC1b and βTC1e (50), have MLP subtype characteristics, displaying high expression of Enpp2 and Hk1 in both the cell lines and low expression of Gck, Pc, and Glut2 in both the cell lines. In contrast, two other lines, βTC4 and βTC3, had low expression of the MLP genes and high expression of the IT genes (except for Hk1, which is heterogeneous in these two cell lines), and are thus representative of the IT subtype (Fig. 6E, top). The MLP subtype of βTC1b and the IT characteristics of βTC4 were further validated by immunoblot analyses, confirming that the MLP cell line exhibited low expression of PC and high expression of HK1 compared with the IT cell line (Fig. 6E, bottom).
With IT and MLP cell lines in hand, we performed metabolomic profiling using a LC/MS-MS–based metabolomics platform (51). Consistent with our expectation, we found that the IT (βTC4 and βTC3) and MLP (βTC1b and βTC1e) cells had distinctive global metabolic profiles (Supplementary Fig. S6A and S6B). Among these differences, we observed an increase in the relative abundance of metabolites involved in pyruvate cycling, including aspartate, fumarate, and malate (Fig. 6F).
To further explore differences in metabolites as well as features of central carbon metabolism between these subtype-representative cell lines, we traced glucose metabolism using isotopically carbon-13 (13 C)–labeled glucose in cell lines representing the IT (βTC4) and MLP (βTC1b) subtypes. Cells were grown overnight in media containing uniformly 13 C-labeled glucose to achieve steady-state saturation of label, and then subjected to targeted LC/MS-MS, as above. Here, incorporation of 13 C was monitored at each carbon position for metabolites in pyruvate cycling and the tricarboxylic acid (TCA) cycle. Indeed, we observed an elevation in abundance of the glucose-derived M+3 isotopomer in several TCA intermediates in the IT cells (Fig. 6G), indicative of pyruvate entry into the TCA cycle via PC. In addition, we observed that the IT cell line had a 5-fold increased dependence on glucose for proliferation (Fig. 6H). Glucose-derived carbon may be particularly important to these cells for energy generation via pyruvate cycling, consistent with the metabolic characteristics of mature β cells, which harness such energy to produce and release insulin (52). The increased pyruvate cycling observed in IT cells provides evidence for the retention of mature β-cell metabolism in ITs, in contrast with the distinctive metabolic characteristics of the MLP cell line. Thus, the distinctive metabolic phenotypes of these subtypes of PanNET further delineate them as such.
DISCUSSION
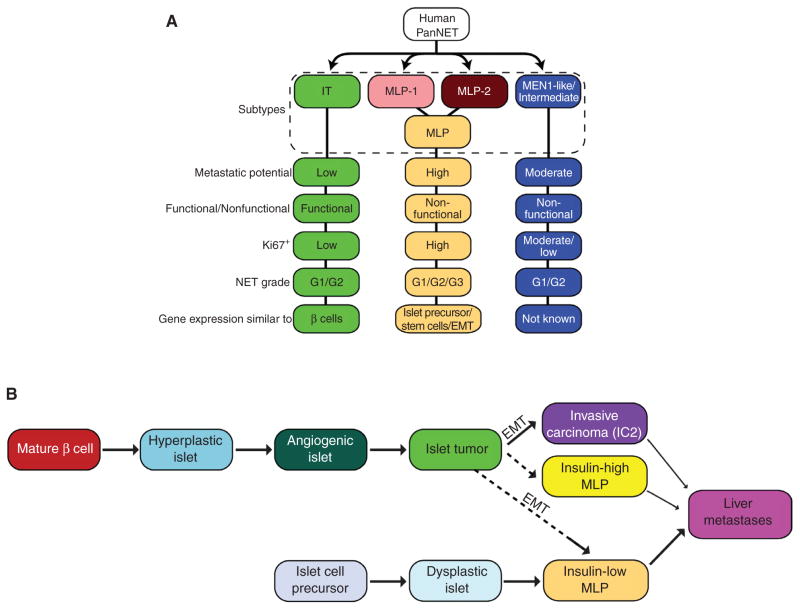
The low incidence of PanNET has made it difficult to correlate apparent heterogeneity in clinical presentation with responses to therapies (23). By using dual miRNA and mRNA transcriptome profiling analysis, we have revealed the existence of three distinctive molecular subtypes and associated biomarkers in human PanNET. Remarkably, despite its rapid and synchronous ontogeny, a PanNET genetically engineered mouse model (GEMM; RT2) is demonstrably recapitulating two of these subtypes based on analysis of the transcriptome, the metabolome, selected protein expression, and histologic and phenotypic presentation, including distant metastasis. Hence, the distinct pathophysiology and molecular mechanisms behind two of the three human PanNET subtypes, MLP and IT, can be further studied using the RT2 model as a surrogate for human (both insulinoma and nonfunctional) tumors. In addition, the RT2 model reveals distinctive angiogenic phenotypes between PanNET subtypes, with decreased blood vessels in MLP compared with IT, predicting analogous differences in human PanNETs that are worthy of future investigation. Although the RT2 model does not represent the MEN1-like/intermediate subtype, due to the apparent necessity of MEN1 mutations to elicit this tumor type, its selective presence in a discrete subtype of human PanNET tumors provides clarity in regard to heretofore unrecognized heterogeneity among nonfunctional PanNETs. The characteristics of the distinct subtypes of human and mouse PanNETs are summarized in Fig. 7A and B.
Figure 7.
A new view of PanNET tumor heterogeneity. A, summary of human PanNET subtypes and their characteristics. B, a revised schematic of the parallel pathways of PanNET tumorigenesis, evidently with distinctive cells of origin, leading to the IT and MLP subtypes, as revealed in the RT2 mouse model, and inferred in the human from the concordance of transcriptomic and phenotypic data presented for mouse and human PanNET.
Although treatment decisions for PanNET patients are based on NET grade, which is assessed in part using Ki67 labeling, there is heterogeneity in patient responses to standard-of-care therapies (23, 53). This heterogeneity in NET G2, for example, may be explained by the observation that G2 tumors are distributed across all three transcriptome subtypes with highest prevalence in the metastasis-associated MLP subtype. In addition, the observation that there is a higher density of CD31-positive microvessels in the less-aggressive mouse IT subtype compared with the MLP subtypes corroborates data indicating that increased microvessel density is associated with better histoprognostic factor (and negatively correlated with tumor progression) and vice versa in PanNET patients (54–57). This variable association of transcriptome subtypes with histologic grades and microvessel density in regard to therapeutic responses should be further assessed in additional patient samples.
Previous efforts to molecularly subclassify other human tumor types have implicated distinctive cells of origin/phenotypes in several instances (32, 58, 59). In the current study, expression of progenitor-specific genes in the MLP subtype implicates a stem cell/progenitor origin. In addition, however, the variable expression of certain pancreatic progenitor genes among the mouse MLP samples (Fig. 5B) implies that tumors of this subtype may originate from more than one progenitor cell, a possibility consistent with the observation that the SV40 oncogene is expressed in multiple β-cell precursors of the embryonic mouse pancreas (44). Further support for the proposition that MLP tumors are not derived from differentiated β cells comes from the observation that the mature β-cell–specific transcription factor MAFA is poorly expressed in MLP tumors (Fig. 5B; refs. 41, 43).
The data from this and a previous report (28) implicate two pathways to invasive carcinomas, which either have the complete MLP signature or a subset of it. Tumors with a partial MLP signature are envisaged to arise out of the canonical islet tumorigenesis pathway that leads to IT, which then progress to IC2/MLP tumors. The second pathway, already invasive in small islet-like lesions (Fig. 5E), has the full MLP signature, including expression of pancreatic progenitor genes, and we hypothesize that it originates from islet progenitor cells. The alternative pathways to MLP in RT2 mouse are presented in Fig. 7B.
Beyond molecular profiles of stem cell– and lineage-specific gene expression, we also observed subtype-correlated variation in metabolic enzymes, and in metabolic activities, providing additional evidence in support of different cells of origin/phenotypes for IT and MLP tumors. During terminal β-cell differentiation, glucokinase (GK)—with its lower affinity for glucose—replaces HK1; the Km of GK is in the physiologic range for blood glucose, and thus GK can act as a sensor, relating glucose concentration in the circulation to insulin production and secretion. Similarly, expression of the MCT family of cation transporters is turned off in terminally differentiated β cells, because lactate/pyruvate uptake would feed directly into the TCA cycle, bypassing the glucose-sensing feature of GK (46). Thus, MLP express HK1 and MCT1, much like progenitors, whereas IT express GK but not HK1 or MCT1, much like normal islet β cells. Moreover, our metabolomic analysis (both metabolite profiles in Fig. 6F and Supplementary Fig. S6 and 13 C-labeling experiments in Fig. 6G) of PanNET cancer cells revealed distinct metabolic profiles for IT and MLP cell lines, which provide further support for distinctive cells of origin. Namely, we observed enriched activity in pyruvate cycling, a β-cell phenomenon (47–49), in the IT subtype both in terms of metabolic enzyme expression and metabolite profiles. Taken together, our data suggest that islet tumors have mature β-cell characteristics, whereas MLP tumors have islet cell precursor characteristics. A revised summary of tumor progression in the RT2 model (3), reflective of what our results suggest for the human IT and MLP subtypes, and now including information regarding the cell of origin/phenotype, is presented in Fig. 7B.
Human PanNETs show considerable genetic heterogeneity and malignant potential. Notably, Jiao and colleagues recently sequenced the exomes of 10 nonfamilial PanNETs, seeking to identify the most frequent mutations in this tumor type (24); their analysis revealed a variety of low-frequency mutations with potential functional roles, but no prevalent mutations in commonly known driving oncogenes or tumor suppressor genes (TSG). Now, we have stratified human and mouse Pan-NETs into subtypes, utilizing a wealth of data from miRNA, mRNA, mutation, and metabolome profiling. Furthermore, our current study has revealed that MEN1 -mutant tumors are significantly enriched in the intermediate human subtype we defined; additional studies will be required to determine if those MEN-like/intermediate subtype tumors that have not suffered MEN1 loss-of-function mutations have found alternative means to abrogate the MEN1 TSG pathway. Notably, Jiao and colleagues reported new inactivating mutations in DAXX/ATRX in nonfunctional PanNETs (24). Now, we demonstrate that tumors carrying these mutations can be alternatively segregated into MEN1-like/intermediate and MLP subtypes with distinct transcriptomic profiles. Our finding that 21% of MEN1-like/intermediate subtype tumors also contained DAXX/ATRX alterations suggests that these mutations are probably late events in PanNET pathogenesis driven by MEN1 loss, consistent with the detection of DAXX/ATRX mutations in PanNET patients with MEN1 syndrome by de Wilde and colleagues (60).
In order to apply these transcriptomic subtypes clinically, it will be important to identify robust biomarkers and assays that can discriminate each of these subtypes similar to that in our previous colorectal cancer study (32). Based on the analysis of both human and mouse tissue, we suggest ENPP2 and INS as possible biomarkers that can distinguish IT and MLP subtypes via IHC and RT-PCR assays. These biomarkers warrant prospective and retrospective validation. Additional biomarkers along with these two will be required to unambiguously identify the MEN1-like/intermediate subtype.
In summary, we have used an integrated multiomics data analysis approach that includes transcriptomics (mRNA and miRNA), mutatomics (selected mutations), and metabolomics along with histology and clinical information to evaluate PanNET from patient samples and a GEMM as well as derivative cancer cell lines. This cross-species analysis has revealed and characterized previously unrecognized subtypes of PanNET in both mouse and humans, and assigned to these tumor subtypes different mutations and phenotypic, clinical, and pathologic properties that underlie the heterogeneous biology of this disease, knowledge that may prove applicable to the development of subtype-selective (personalized) therapies.
METHODS
PanNET Patient Samples and RNA Isolation for Validation Studies
The miRNA expression validation (n = 50) and matched gene expression validation (n = 29; except one unmatched sample) datasets (Supplementary Figs. S1B–S1C and S2B–S2C) involved a total of 51 primary and metastatic PanNET samples. For gene expression analysis, total RNA was isolated from tumor tissues using the Ambion RiboPure Kit (Life Technologies) as per the manufacturer’s instructions. For miRNA analysis, tumor tissues were sectioned using cryotome, every sixth section was evaluated using hematoxylin and eosin (H&E) staining, and total RNA was isolated using the miRNeasy Kit (Qiagen) as per the manufacturer’s instructions. This study was conducted in accordance with the Declaration of Helsinki. It was approved by the ethics committee of Charité - Universitätsmedizin Berlin, and informed consent was obtained from all patients.
RT2 Transgenic Mouse Model of PanNET
The breeding and generation of RT2 mice have been described previously (2). Although the majority of the mice in this study were on a C57BL/6 background, which is typically used for this model, we also crossed C57BL/6J (B6) RT2 mice with A/J (B6AF1) or DBA/2J (B6D2F1) mice to produce F1 mice. All animal research was performed under the auspices of animal protocols approved by Experience sur animaux (EXPANIM)—Service de la consommation et des Affaires veterinaries (SCAV) in Switzerland.
Isolation of RT2 Tumor and Premalignant Lesions
RT2 mouse PanNET tumors, liver metastases, normal, hyperplastic, and angiogenic islets were dissected out or isolated as described previously (25, 28, 61).
RT2 Mouse Tumor Cell Lines
βTC cell lines were previously derived by us from the RT2 mouse model (50). These cell lines (passages between 20 and 30) were authenticated by growth, morphologic characteristics, and insulin gene and protein expression at certain intervals between the years 2010 and 2015. These cell lines were also tested for Mycoplasma.
Processing of Microarray Data and Subtype Identification
The core clinical gene and miRNA expression datasets were from Missiaglia and colleagues (GEO Omnibus ID—GSE73338; ref. 34) and Roldo and colleagues (GSE73350; ref. 29). Gene expression was assessed using Affymetrix GeneChip Human and Mouse Gene 1.0 ST arrays (Affymetrix; GSE73339—patient gene expression validation samples and GSE73514—mouse RT2 samples). Human microarrays were analyzed using a statistical framework R and Bioconductor (62) as described earlier (31, 32) and mouse microarrays using aroma. affymetrix (63). miRNA expression was assessed using Agilent Human miRNA Microarrays (based on miRBase release 16; Agilent Technologies; for patient miRNA validation samples; GSE73367). Normalization of the miRNA profile data was performed as described (64).
For subtype identification, the variable genes (from core clinical gene expression dataset) with SD > 0.8 or miRNAs (from core clinical miRNA expression dataset) with SD > 0.5 from the normalized data were median-centered across patient samples for each gene and used for subtype identification by applying the NMF algorithm (30) followed by SAM analysis (33) to identify significantly differentially expressed features (mRNA or miRNA), as described previously (31, 32). For (both gene and miRNA) validation datasets, SAM genes from the core clinical datasets were mapped followed by NMF analysis for subtype identification.
Cross-Species Analysis
ComBat (“sva” Biocoductor package; ref. 35) or DWD (36, 65) was used to correct the batch effects and merge human and mouse gene expression data after selecting variable genes with SD > 0.8 across samples from each dataset. For merging the human and mouse data, we used official gene names from the HUGO Gene Nomenclature Committee (HGNC) as references.
Cross-Comparison of Gene and miRNA Subtypes
The hypergeometric test was performed to reconcile gene and miRNA subtypes, as described (66).
Laser Capture Microdissection
Snap-frozen pancreatic sections (10 μm) from 15-week-old RT2 mice were mounted onto PET-membrane slides (MMI). Sections were incubated with ice-cold 70% ethanol and stained with cresyl violet. Individual tumors were microdissected using a PALM laser dissecting microscope (Zeiss) followed by RNA isolation and hybridization to Affymetrix GeneChip Mouse Gene 1.0 ST arrays.
IHC
IHC was performed as described in our previous studies (32, 67, 68) using the primary antibodies listed in the Supplementary Information. Ki67 scoring was performed as previously described (34). For mouse samples, ImageJ software was used to count the total number of Ki67-positive cells. The number of CD31-positive blood vessels in random 20× magnification fields was determined independently by two investigators, and averaged.
Metabolic Profiling
To characterize central carbon metabolism, targeted LC/MS-MS was performed (51). Briefly, βTC cells were grown in DMEM (25 mmol/L glucose, 2 mmol/L glutamine, without pyruvate) to 80% confluency, and metabolites were extracted using 80% methanol at dry ice temperatures. Metabolite fractions were normalized to protein concentration from a biologic replicate processed in parallel. For 13 C-labeling experiments, cells were grown in DMEM (without pyruvate, glucose, and glutamine) in the presence of 10% dialyzed FBS, supplemented with 13 C-glucose (25 mmol/L; Cambridge Isotope Laboratories) for 20 hours.
Next-Generation Sequencing
Human DNA samples were analyzed with a panel targeting all coding sequences of MEN1, ATRX, DAXX, PTEN, TSC2, and ATM. Next-generation targeted sequencing was performed as previously described (69).
Gene-specific primers used for qRT-PCR using mouse samples are provided in Supplementary Table S4.
R scripts associated with data analysis are available from github (https://github.com/syspremed/PanNETassigner).
A detailed description of Materials and Methods is available in Supplementary Information.
Supplementary Material
Acknowledgments
The authors thank I. Michael for insightful comments on the manuscript; P. Olson (Pfizer) for preparing RNA samples from the mouse tumors for gene expression profiling while he was working in Hanahan lab; K. Homicsko [Swiss Federal Institute of Lausanne (EPFL)] and O. Michielin (Swiss Institute of Bioinformatics) for helpful scientific discussions; E. Drori, L. Ostos, M. Schnetz, M. Peng, R. Prakash, B. Torchia, and other Hanahan lab members (EPFL) for help with mouse colonies and other experiments; and P. Poudel and G. Nyamundanda [Institute of Cancer Research (ICR)] for help with validating the R scripts, data, and certain results and making an R package. The authors also thank the following core facilities for their services: the Histology Core Facility, EPFL, Switzerland; Lausanne Genomic Technologies Facility, Center for Integrative genomics (CIG), University of Lausanne, Lausanne, Switzerland; the Mass Spectrometry Facility, EPFL(including R. Hamelin for processing samples and help with the data analysis); and the Genomics Core Laboratory (University of California, San Francisco, San Francisco, CA).
Grant Support
A. Sadanandam was partially supported by a U.S. Department of Defense Postdoctoral Fellowship (BC087768). C.A. Lyssiotis was partially supported by a PanCAN-AACR Pathway to Leadership award and a Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation (DFS-09-14). The work on mutational analysis of human PanNET was supported by the Italian Cancer Genome Project (FIRB RBAP10AHJB), Associazione Italiana Ricerca Cancro (grant no. 12182), and Fondazione Italiana Malattie Pancreas – Ministero Salute (CUP_J33G13000210001) to A. Scarpa. L.C. Cantley was supported by P01 grant CA117969. While A. Sadanandam performed a majority of the research at the EPFL, a portion of the work was also performed at the ICR, London. A. Sadanandam acknowledges National Health Service funding to the National Institute for Health Research Biomedical Research Centre at The Royal Marsden and the ICR. This study was primarily supported by a Swiss National Science Foundation research grant (to D. Hanahan).
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
L.C. Cantley serves on the Board of Directors of Agios Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
One of the Editors-in-Chief of Cancer Discovery is an author of this article. In keeping with the AACR’s editorial policy, the paper was peer reviewed and an AACR journal editor not affiliated with Cancer Discovery rendered the decision concerning acceptability.
Authors’ Contributions
Conception and design: A. Sadanandam, B. Wiedenmann, A. Scarpa, D. Hanahan
Development of methodology: A. Sadanandam, J. Körner, A. Scarpa
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A. Sadanandam, S. Wullschleger, C.A. Lyssiotis, C. Grötzinger, S. Barbi, I. Wafy, R.T. Lawlor, J.M. Asara, B. Wiedenmann, A. Scarpa
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Sadanandam, S. Wullschleger, C.A. Lyssiotis, S. Barbi, S. Bersani, I. Wafy, A. Mafficini, M. Simbolo, J.M. Asara, H. Bläker, B. Wiedenmann, A. Scarpa, D. Hanahan
Writing, review, and/or revision of the manuscript: A. Sadanandam, S. Wullschleger, C.A. Lyssiotis, C. Grötzinger, J.M. Asara, L.C. Cantley, B. Wiedenmann, A. Scarpa, D. Hanahan
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A. Sadanandam, J. Körner, A. Scarpa
Study supervision: A. Sadanandam, L.C. Cantley, A. Scarpa, D. Hanahan
Other (provided samples): R. Lawlor
Other (mouse genome target sequencing): M. Simbolo
References
- 1.Hanahan D, Wagner EF, Palmiter RD. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 2007;21:2258–70. doi: 10.1101/gad.1583307. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–22. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D. Dissecting multistep tumorigenesis in transgenic mice. Annu Rev Genet. 1988;22:479–519. doi: 10.1146/annurev.ge.22.120188.002403. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 5.Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–8. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- 6.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–3. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 7.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443– 53. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 8.Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2002;1:339–53. doi: 10.1016/s1535-6108(02)00055-7. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Hanahan D. Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell. 2013;153:86–100. doi: 10.1016/j.cell.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 10.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietras K, Pahler J, Bergers G, Hanahan D. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med. 2008;5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casanovas O, Hicklin DJ, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell. 2005;8:299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Bergers G, Hanahan D. Combining antiangiogenic agents with metronomic chemotherapy enhances efficacy against late-stage pancreatic islet carcinomas in mice. Cold Spring Harb Symp Quant Biol. 2002;67:293– 300. doi: 10.1101/sqb.2002.67.293. [DOI] [PubMed] [Google Scholar]
- 14.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111:1287–95. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu CW, Nozawa H, Hanahan D. Survival benefit with proapoptotic molecular and pathologic responses from dual targeting of mammalian target of rapamycin and epidermal growth factor receptor in a preclinical model of pancreatic neuroendocrine carcinogenesis. J Clin Oncol. 2010;28:4425–33. doi: 10.1200/JCO.2010.28.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymond E, Dahan L, Raoul JL, Bang YJ, Borbath I, Lombard-Bohas C, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:501–13. doi: 10.1056/NEJMoa1003825. [DOI] [PubMed] [Google Scholar]
- 17.Rindi G, Wiedenmann B. Neuroendocrine neoplasms of the gut and pancreas: new insights. Nat Rev Endocrinol. 2011;8:54–64. doi: 10.1038/nrendo.2011.120. [DOI] [PubMed] [Google Scholar]
- 18.Bosman FT. Classification of tumours of the digestive system. In: Bosman FT, editor. WHO classification of tumours of the digestive system. Lyon, France: World Health Organization; 2010. [Google Scholar]
- 19.Sobin LH, Gospodarowicz MK, Wittekind CC, editors. TNM classification of malignant tumors. Oxford, UK: Wiley-Blackwell; 2009. [Google Scholar]
- 20.Modlin IM, Kidd M, Latich I, Zikusoka MN, Shapiro MD. Current status of gastrointestinal carcinoids. Gastroenterology. 2005;128:1717– 51. doi: 10.1053/j.gastro.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Scarpa A, Mantovani W, Capelli P, Beghelli S, Boninsegna L, Bettini R, et al. Pancreatic endocrine tumors: improved TNM staging and histopathological grading permit a clinically efficient prognostic stratification of patients. Mod Pathol. 2010;23:824–33. doi: 10.1038/modpathol.2010.58. [DOI] [PubMed] [Google Scholar]
- 22.Velayoudom-Cephise FL, Duvillard P, Foucan L, Hadoux J, Chougnet CN, Leboulleux S, et al. Are G3 ENETS neuroendocrine neoplasms heterogeneous ? Endocr Relat Cancer. 2013;20:649–57. doi: 10.1530/ERC-13-0027. [DOI] [PubMed] [Google Scholar]
- 23.de Wilde RF, Edil BH, Hruban RH, Maitra A. Well-differentiated pancreatic neuroendocrine tumors: from genetics to therapy. Nat Rev Gastroenterol Hepatol. 2012;9:199–208. doi: 10.1038/nrgastro.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun MG, Hanahan D. Genetic deletion of the desmosomal component desmoplakin promotes tumor microinvasion in a mouse model of pancreatic neuroendocrine carcinogenesis. PLoS Genet. 2010;6:e1001120. doi: 10.1371/journal.pgen.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–64. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 27.Hager JH, Hodgson JG, Fridlyand J, Hariono S, Gray JW, Hanahan D. Oncogene expression and genetic background influence the frequency of DNA copy number abnormalities in mouse pancreatic islet cell carcinomas. Cancer Res. 2004;64:2406–10. doi: 10.1158/0008-5472.can-03-3522. [DOI] [PubMed] [Google Scholar]
- 28.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 30.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164– 9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619– 25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, della Peruta M, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J Clin Oncol. 2010;28:245–55. doi: 10.1200/JCO.2008.21.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 36.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–14. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 37.Pieterman CR, Conemans EB, Dreijerink KM, de Laat JM, Timmers HT, Vriens MR, et al. Thoracic and duodenopancreatic neuroendocrine tumors in multiple endocrine neoplasia type 1: natural history and function of menin in tumorigenesis. Endocr Relat Cancer. 2014;21:R121–42. doi: 10.1530/ERC-13-0482. [DOI] [PubMed] [Google Scholar]
- 38.Cao Y, Gao Z, Li L, Jiang X, Shan A, Cai J, et al. Whole exome sequencing of insulinoma reveals recurrent T372R mutations in YY1. Nat Commun. 2013;4:2810. doi: 10.1038/ncomms3810. [DOI] [PubMed] [Google Scholar]
- 39.Hunter KE, Quick ML, Sadanandam A, Hanahan D, Joyce JA. Identification and characterization of poorly differentiated invasive carcinomas in a mouse model of pancreatic neuroendocrine tumorigenesis. PloS One. 2013;8:e64472. doi: 10.1371/journal.pone.0064472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 41.Benitez CM, Goodyer WR, Kim SK. Deconstructing pancreas developmental biology. Cold Spring Harbor Perspect Biol. 2012;4:a012401. doi: 10.1101/cshperspect.a012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395– 406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hang Y, Stein R. MafA and MafB activity in pancreatic beta cells. Trends Endocrinol Metab. 2011;22:364–73. doi: 10.1016/j.tem.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alpert S, Hanahan D, Teitelman G. Hybrid insulin genes reveal a developmental lineage for pancreatic endocrine cells and imply a relationship with neurons. Cell. 1988;53:295–308. doi: 10.1016/0092-8674(88)90391-1. [DOI] [PubMed] [Google Scholar]
- 45.Smukler SR, Arntfield ME, Razavi R, Bikopoulos G, Karpowicz P, Seaberg R, et al. The adult mouse and human pancreas contain rare multipotent stem cells that express insulin. Cell Stem Cell. 2011;8:281–93. doi: 10.1016/j.stem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Quintens R, Hendrickx N, Lemaire K, Schuit F. Why expression of some genes is disallowed in beta-cells. Biochem Soc Trans. 2008;36:300–5. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- 47.Jensen MV, Joseph JW, Ronnebaum SM, Burgess SC, Sherry AD, Newgard CB. Metabolic cycling in control of glucose-stimulated insulin secretion. Am J Physiol Cell Physiol. 2008;295:E1287–97. doi: 10.1152/ajpendo.90604.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS) Proc Natl Acad Sci U S A. 2002;99:2708–13. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pongratz RL, Kibbey RG, Shulman GI, Cline GW. Cytosolic and mitochondrial malic enzyme isoforms differentially control insulin secretion. J Biol Chem. 2007;282:200–7. doi: 10.1074/jbc.M602954200. [DOI] [PubMed] [Google Scholar]
- 50.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, et al. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc Natl Acad Sci U S A. 1988;85:9037–41. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden MC, Holness MJ. The pyruvate carboxylase-pyruvate dehydrogenase axis in islet pyruvate metabolism: Going round in circles ? Islets. 2011;3:302–19. doi: 10.4161/isl.3.6.17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oberg K, Akerstrom G, Rindi G, Jelic S, Group EGW. Neuroendocrine gastroenteropancreatic tumours: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21:v223–7. doi: 10.1093/annonc/mdq192. [DOI] [PubMed] [Google Scholar]
- 54.Marion-Audibert AM, Barel C, Gouysse G, Dumortier J, Pilleul F, Pourreyron C, et al. Low microvessel density is an unfavorable histoprognostic factor in pancreatic endocrine tumors. Gastroenterology. 2003;125:1094–104. doi: 10.1016/s0016-5085(03)01198-3. [DOI] [PubMed] [Google Scholar]
- 55.Couvelard A, O’Toole D, Turley H, Leek R, Sauvanet A, Degott C, et al. Microvascular density and hypoxia-inducible factor pathway in pancreatic endocrine tumours: negative correlation of microvascular density and VEGF expression with tumour progression. Br J Cancer. 2005;92:94–101. doi: 10.1038/sj.bjc.6602245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi Y, Akishima-Fukasawa Y, Kobayashi N, Sano T, Kosuge T, Nimura Y, et al. Prognostic value of tumor architecture, tumor-associated vascular characteristics, and expression of angiogenic molecules in pancreatic endocrine tumors. Clin Cancer Res. 2007;13:187–96. doi: 10.1158/1078-0432.CCR-06-1408. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Jia Z, Li Q, Wang L, Rashid A, Zhu Z, et al. Elevated expression of vascular endothelial growth factor correlates with increased angiogenesis and decreased progression-free survival among patients with low-grade neuroendocrine tumors. Cancer. 2007;109:1478–86. doi: 10.1002/cncr.22554. [DOI] [PubMed] [Google Scholar]
- 58.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 59.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2009;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Wilde RF, Heaphy CM, Maitra A, Meeker AK, Edil BH, Wolfgang CL, et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod Pathol. 2012;25:1033–9. doi: 10.1038/modpathol.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parangi S, Dietrich W, Christofori G, Lander ES, Hanahan D. Tumor suppressor loci on mouse chromosomes 9 and 16 are lost at distinct stages of tumorigenesis in a transgenic model of islet cell carcinoma. Cancer Res. 1995;55:6071–6. [PubMed] [Google Scholar]
- 62.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bengtsson H, Simpson K, Bullard J, Hansen K. Tech Report #745. Berkeley, CA: Department of Statistics, University of California; 2008. aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. [Google Scholar]
- 64.Pradervand S, Weber J, Thomas J, Bueno M, Wirapati P, Lefort K, et al. Impact of normalization on miRNA microarray expression profiling. RNA (New York, NY) 2009;15:493– 501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadanandam A, Wang X, de Sousa EMF, Gray JW, Vermeulen L, Hanahan D, et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle. 2014;13:353–7. doi: 10.4161/cc.27769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sadanandam A, Varney ML, Singh S, Ashour AE, Moniaux N, Deb S, et al. High gene expression of semaphorin 5A in pancreatic cancer is associated with tumor growth, invasion and metastasis. Int J Cancer. 2010;127:1373–83. doi: 10.1002/ijc.25166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sadanandam A, Sidhu SS, Wullschleger S, Singh S, Varney ML, Yang CS, et al. Secreted semaphorin 5A suppressed pancreatic tumour burden but increased metastasis and endothelial cell proliferation. Br J Cancer. 2012;107:501–7. doi: 10.1038/bjc.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amato E, Molin MD, Mafficini A, Yu J, Malleo G, Rusev B, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. 2014;233:217–27. doi: 10.1002/path.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.