Abstract
Alterations in the epigenetic landscape are fundamental drivers of aberrant gene expression that contribute to cancer progression and pathology. Understanding specific modes of epigenetic regulation can be used to identify novel biomarkers or targets for therapeutic intervention to clinically treat solid tumors and leukemias. The bivalent marking of gene promoters by H3K4me3 and H3K27me3 is a primary mechanism to poise genes for expression in pluripotent embryonic stem cells (ESC). In this study we interrogated three well-established mammary cell lines to model epigenetic programming observed among breast cancer subtypes. Evidence is provided for a distinct bivalent signature, activating and repressive histone marks co-residing at the same gene promoter, in the MCF7 (ESR/PGR+) luminal breast cancer cell line. We identified a subset of genes, enriched for developmental pathways that regulate cellular phenotype and signaling, and partially recapitulate the bivalent character observed in embryonic stem cells. We validated the biological relevance of this “oncofetal epigenetic” signature using data from ESR/PGR+ tumor samples from breast cancer patients. This signature of oncofetal epigenetic control is an informative biomarker and may provide novel therapeutic targets, selective for both recurring and treatment-resistant cancers.
Keywords: Bivalency, MCF10A, MCF7, MDA-MB-231, Chromatin, Epigenetics, Histone H3K4, H3K4me3, H3K27me3
Introduction
Breast cancer is a heterogeneous disease influenced by genetic and epigenetic changes that result in aberrant gene function, deregulated gene expression, and genomic instability. Many of the gene expression changes reported in breast cancer have been associated with histone post-translational modifications and/or DNA methylation (Coradini and Oriana, 2014; Dumitrescu, 2012; Maruyama et al., 2011). These modifications exert their function through recruitment of specific transcriptional regulators and chromatin modifying enzymes, resulting in conformational changes to chromatin structure that in turn regulate gene expression. Epigenetic regulation plays a key role in normal cellular development and perturbations in this regulation have emerged as a significant mechanism for cancer development (Baylin and Jones, 2011; Suva et al., 2013; Timp and Feinberg, 2013)
The co-occupancy at promoters of two transcriptionally opposing marks, the active H3K4me3 mark and the repressive H3K27me3 mark, were first observed in embryonic stem cells (ESC) (Azuara et al., 2006; Bernstein et al., 2006). These bivalent domains are believed to provide regulatory flexibility for maintaining phenotype-specifying genes of ESC in a poised state until a signal initiates activation leading to lineage commitment and cellular differentiation. The complexity of bivalency-mediated epigenetic control in pluripotent cells is demonstrated by the finding that H3K4me3 is selectively maintained in mitosis, whereas H3K27me3 remains constitutively marked during the cell cycle (Grandy et al., 2015). Epigenetic bivalency has been predominantly observed in ESC; however there is considerable evidence suggesting that tumors contain subpopulations of cells that display stem cell-like phenotypic properties (Beachy et al., 2004; Ben-Porath et al., 2008; Fillmore and Kuperwasser, 2008). In fact, stem cells are believed to reside in many tissues, including breast epithelium, suggesting that epigenetic reprogramming could lead to cell-identity disruption, triggering cancer initiation and progression (Bapat et al., 2014; Fillmore and Kuperwasser, 2008; Stricker and Pollard, 2014). Several cancer cell lines have been reported to be enriched for stem-like cells, supporting the concept of a sub-population of cancer stem cells (CSC) which can self-renew or differentiate into the heterogeneous phenotypes observed in tumors (Chaffer et al., 2011; Gupta et al., 2011). In fact, cultivation of breast epithelial cells in an anchorage-independent manner can maintain breast CSCs in culture (Ponti et al., 2005). These cells have been shown to support tumor growth, metastasis, and resistance to therapy (Brooks et al., 2015; Sehl et al., 2015)
Cancer stem cells share many features with ESC, including self-renewal and unrestricted proliferation (Ben-Porath et al., 2008; Reya et al., 2001). We hypothesize that the bivalent epigenetic signature present in breast cancer cells is indicative of reprogrammed stem-like cells. Focusing on two well-characterized histone methylation marks, H3K4me3 that is associated with active gene transcription and H3K27me3 with gene repression, we utilized three well-established human mammary cell lines to model breast cancer-associated subtypes to circumvent the difficulty of mirroring the heterogeneity of clinical samples with a single cell line. We selected cell lines that represent a normal-like subtype (MCF10A; fibrocystic disease) and two epithelial cancer subtypes, luminal (MCF7; ESR/PGR+) and basal-like metastatic (MDA-MB-231; ESR/PGR/HER2−). We provide evidence of a distinct bivalent signature in the MCF7 (ESR/PGR+) luminal cell line that partially recapitulates the epigenetic status of ESC at promoters for genes involved in regulation of cellular phenotype and signaling. We validated the biological relevance of this “oncofetal epigenetic” signature using data from ESR/PGR+ tumor samples from breast cancer patients (Jansen et al., 2013). This signature of oncofetal epigenetic control is an informative biomarker and may provide novel therapeutic targets, selective for both recurring and treatment-resistant cancers.
Materials and Methods
Cell culture and reagents
The established cell lines, MCF10A, MCF7, and MDA-MB-231 were utilized in this study. MCF10A cells were grown in DMEM: F12 (Hyclone-SH30271with phenol red), 5% (v/v) horse serum (Gibco #16050 lot #1075876) + 10ug/ml human insulin (Sigma I-1882)+ 20ng/ml recombinant hEGF (Peprotech AF-100-15) + 100ng/ml Cholera toxin (Sigma C-8052) + 0.5 ug/ml Hydrocortisone (Sigma H-0888) Pen/Strep (Life Technologies #15140163) and Glutamine (Life Technologies #25030081, Carlsbad, CA). MCF7 and MDA-MB-231 cells were grown in DMEM: F12 (Hyclone-SH30271 with phenol red) + 10% (v/v) FBS (Atlanta Biologicals #S11550 lot #H1030, Lawrenceville, GA). Each cell line was authenticated using short tandem repeat (STR) cell identification analysis and q-PCR characterized by expression of key cellular markers attributed to each cell type. STR - DNA fingerprinting was performed by the University of Vermont Cancer Center DNA Analysis Facility using the Promega GenePrint® 10 System according to manufacturer’s instructions (Promega #B9510). The STR profiles were compared to known ATCC fingerprints (ATCC.org) and/or the Cell Line Integrated Molecular Authentication database (CLIMA) database version 0.1.200808 (Romano et al., 2009) (http://bioinformatics.istge.it/clima/).
Chromatin Immunoprecipitation
Cells were seeded at 1.5×106 cells per 100mm dish and grown to 80% confluence prior to cross linking of the protein-DNA complex by fixation with a final concentration of 0.8% formaldehyde (Thermo-Scientific Prod # 28908 lot N16114410, Rockford, IL) for 10 min at room temperature. Crosslinking was neutralized by the addition of glycine (final concentration 0.125M) for an additional 5 min. Cells were washed with ice-cold PBS containing protease inhibitors (Roche-Protease Inhibitor cocktail) prior to final harvest and then flash frozen and stored at −80°C. Nuclei extraction was performed using a protocol modified from Dignam et al (1983). Isolated nuclei were sonicated using a Covaris S-220 Ultrasonic Processor to obtain sheared chromatin ranging from 200–800 bp with an average peak size of 500 bp. A total of 30 μg of sheared chromatin was used for ChIP with either 10 ug of anti-H3K4me3 (Abcam, ab1012 lot#GR80367-1) or 12 ug of anti-H3K27me3 (Millipore -07-449 lot#1764447 or 2475696 ) used at a ratio of 3:1 or 2.5:1 chromatin to antibody, respectively. Immunoprecipitated complexes were purified using Protein-G Dynabeads (Life Technologies 10004D lot#123085320) The ChIP-DNA-bead complexes were washed 4 times with 1ml cold RIPA buffer (150mM salt), followed by 1–2 washes with 1 ml cold high-salt RIPA buffer (500mM salt), and a final wash with TE + NaCl (10mM Tris, 1mM EDTA, and 50mM NaCl). DNA fragments were isolated by reversing the crosslinks of the protein-DNA complex overnight at 65°C. The eluted ChIP-DNA were treated with RNase A (0.2mg/ml final concentration-Life Technologies Cat # AM2269), proteinase K treatment (Life Technologies Cat # AM2548) and then phenol extracted, precipitated with ethanol, and resuspended in 10mM Tris. DNA was quantified using a Qubit fluorimeter (Life Technologies) prior to the library preparation.
Library and sequencing preparation
Sequencing libraries were prepared using the TruSeq ChIP sample preparation kit (Illumina Cat #9235121 lot #15027084, San Diego, CA) following instructions provided by the manufacturer. Libraries were then size selected, purified and quantified prior to sequencing by automated DNA sequencing performed in the University of Vermont Cancer Center Advanced Genome Technologies Core (Messier et al., 2016).
Bioinformatics Analysis
Sequencing base calls were generated on the HiSeq 1500 instrument in the Advanced Genome Technologies Core Massively Parallel Sequencing Facility. Fastq conversion and demultiplexing were done using bcl2fastq (Illumina, v1.8.4), evaluated (Fastqc), processed to remove low quality reads, and trimmed (FastX toolkit). Reads were mapped to the human genome (hg38) using STAR aligner (version 2.4) with splicing disabled (--alignIntronMax 1) (Dobin et al., 2013). Enriched regions (narrowPeak calls) for each replicate were generated using MACS2 (Feng et al., 2012) and replicates were then evaluated using bamCorrelate and IDR (where applicable, see Supplemental Table SI). After pooling replicates, MACS2 was used on H3K4me3 to call narrowPeak at high stringency (p-value < 10e-5) and SICER V1.1 (Xu et al., 2014) was used to identify broad H3K27me3 enrichment domains. SICER results were filtered by consensus using a range of window-gap combinations (windows 200, 300, 375, and 400; gaps 600, 1200, 1125, and 2400 respectively). LogFE wiggle tracks were generated using bdgcmp and pileups from MACS2. Data from H7-hESC (GSE35583) and data from clinical samples of fresh frozen ESR/PGR+ tumors from 12 patients with good or poor outcomes after aromatase therapy (GSE40867) were processed in the same manner (Jansen et al., 2013). Our data sets were deposited in the NCBI Gene Expression Omnibus (GEO) (GSE77772).
A comprehensive promoter annotation was derived from GENCODE V21 transcripts. To be considered, a transcript’s TSS had to be within 500 bp of an H3K4me3 narrowPeak summit. In cases of multiple transcripts overlapping, transcript support level and finally distance to peak summit of TSS were used to select a single transcript per site. Each TSS was then extended by +/− 2kb. A matching (mutually exclusive) gene body annotation was created by trimming established promoter regions. Fold-enrichment at each feature was calculated using read counts calculated by a customized HTSEQ script. Gene promoters were called as being marked by H3K4me3 if a narrowPeak overlapped with a promoter (bedtools intersect −f 0.5). Gene bodies were called as being marked by H3K27me3 if a SICER domain in at least 2 of the 4 window-gap combinations overlapped with the gene body. Gene promoters were called as being promoter enriched for H3K27me3 if the promoter met the same set of SICER requirements and had a fold enrichment (FE) at the promoter greater than the FE across the gene body.
Profile data for heatmaps was calculated using the ngsplot R package (version 2.47) (Shen et al., 2014). To prevent distortion of enrichment profiles around promoters, gene annotations were shortened by 2kb on the 5′ end and ngsplot was run with a 12kb flanking region, the result being the region from −10kb to +2kb from TSS having a constant bp scale. Heat maps were generated using a combination of k-means clustering and sorting. R’s built-in k-means algorithm was initialized with random centers and allowed to run for 10 iterations or until convergence.
Gene ontology (GO) enrichment analysis was done using the PANTHER Overrepresentation Test in the complete biological process GO (release 20150430) with the 2015-08-06 release of the GO database (Mi et al., 2016). GO terms and their Bonferroni corrected p-values were then input into REViGO (Supek et al., 2011) to determine a representative set of GO terms. GO terms were selected as representative if they had an overrepresentation p-value less than 10e-3 and a REViGO dispensability metric less than 0.3.
Results
Breast epithelial cell types share bivalent characteristics with embryonic stem cells
To investigate the genome-wide epigenetic landscape in cell lines representing basal and luminal stages of breast cancer, we examined the distribution of the activating H3K4me3 histone modification at gene promoters, the repressive H3K27me3 histone modification across the gene body, or the bivalent co-occupancy of both activating and repressive marks at gene promoters, using chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq). The two breast cancer subtypes -- MCF7, an ESR/PGR+ luminal cell line, and MDA-MB-231, a triple-negative basal metastatic cell line, were compared to MCF10A mammary epithelial normal-like cell line and to each other.
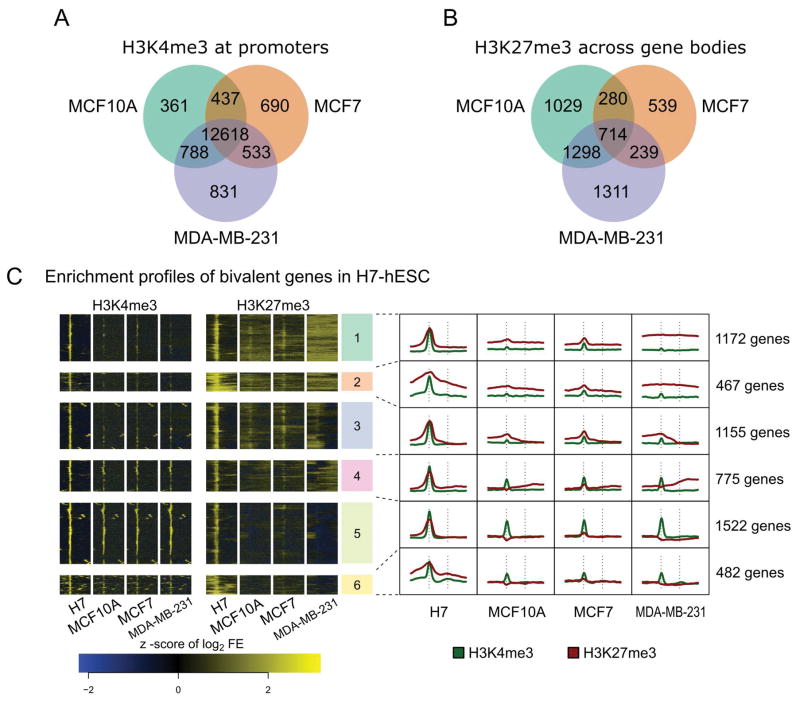
Comparison between the three breast cell types revealed a large number of genes enriched for H3K4me3 (Fig. 1A; Supplemental Table SIIA), with fewer enriched for H3K27me3 (Fig. 1B; Supplemental Table SIIB). Most H3K4me3-enriched gene promoters are marked in all three cell lines (85–89% or 12,618 gene promoters) suggesting that the majority of genes have a common activation status. In contrast, only 20–40% (714) of H3K27me3 enriched genes are marked in all three cell lines, suggesting that repression status is cell type specific. The ESR/PGR+ MCF7 cell type exhibits the fewest H3K27me3 marked genes, with only 1772 compared to 3321 and 3562 in the normal-like MCF10A and triple-negative MDA-MB-231, respectively. As a majority of H3K27me3-marked genes are shared between MCF10A and MDA-MB-231, this finding suggests that these cells have a more repressive chromatin state in contrast to MCF7 cells that exhibit phenotypic plasticity (Ponti et al., 2005; Visvader and Stingl, 2014)
Figure 1. H3K4me3 and H3K27me3 histone modifications in cancer cell lines and compared to H7-hESC bivalently marked genes.
A: The activation associated H3K4me3 mark is largely shared between the MCF10A, MCF7, and MDA-MB-231 breast cell lines. 12,618 genes are marked by H3K4me3 in all three cell lines studied. Fewer than 1000 genes are marked uniquely within any one cell subtype. B: The repression associated H3K27me3 mark on gene bodies shows more variability between the three breast cell subtypes. Only 714 genes are marked in all 3 cell lines. MCF7 cells exhibit the least number of genes uniquely marked, only 539 compared to 1029 in MCF10A and 1311 in MDA-MB-231 cells. C: Clustering of H3K4me3 and H3K27me3 enrichment profiles in hESC and 3 breast cell lines. Six clusters were calculated by k-means clustering. The heatmap on the left presents enrichment profiles for all 5573 genes found to be bivalent in H7-hESC. The line plots on the right summarize each cluster. The TSS and TES of each gene are indicated by vertical dashed lines in the summarization plots. Note the similarity of H3K27me3 near promoters between MCF7 and H7-hESC, and the lack of the same in MDA-MB-231.
Due to the differences in H3K27me3 observed between the three cell lines and evidence of an ESC-like state in cancer, we compared bivalency (H3K4me3 and H3K27me3 co-localized at gene promoters) in our cell lines to that observed in H7-hESC. We calculated enrichment profiles for H7-hESC and the three breast epithelial cell subtypes. A total of 5573 bivalent genes were identified in H7-hESC line and unsupervised k-means clustering was applied to this set, yielding 6 clusters with distinct patterns for the two histone marks across all four cell lines (Fig. 1C; Supplemental Table SIIC). Based on the overall patterns of H3K4me3 and H3K27me3 occupancy, we identified gene promoters bivalently marked in both H7-hESC and each of the breast epithelial cell types. MCF7 profiles were the most similar to H7-hESC, as demonstrated by clusters 1–5, which appear bivalent in both H7 and MCF7 (Fig. 1C). We also observed some genes with similar enrichment profiles between H7-hESC and the MCF10A cell type; however, this was limited primarily to clusters 1–3. MDA-MB-231 was mostly characterized by broad H3K27me3 marking across the gene body, most prominently demonstrated by clusters 1, 2 and 4. This observation suggests that repression by H3K27me3 plays a role in establishing the MDA-MB-231 aggressive phenotype, as has been reported for other cancers (Agarwal et al., 2016; Takeshima et al., 2015)
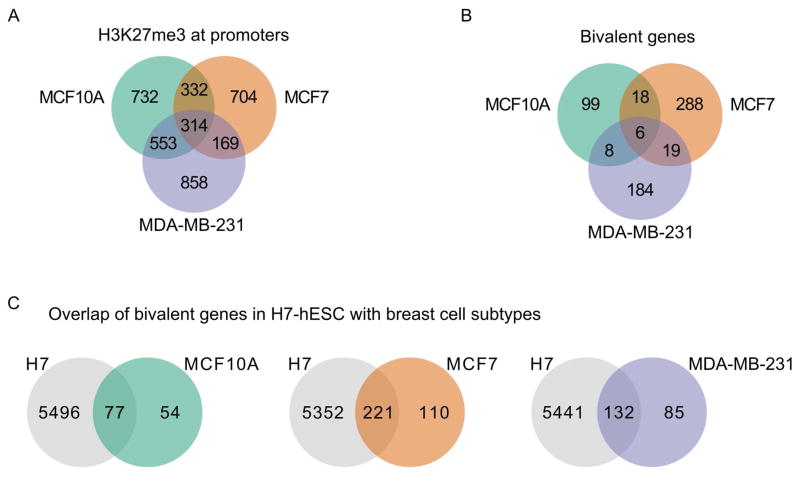
Within the bivalency patterns of breast epithelial cell types (Fig. 1C), we observed H3K27me3 enrichment in both promoters and gene bodies. To identify genes with a hESC-like pattern of promoter-associated bivalency, genes that had a greater enrichment of H3K27me3 at promoters compared to gene body (assessed by relative fold-enrichment (FE)) were determined and compared across the three cell types (Fig. 2A; Supplemental Table SIIIA). The pattern of H3K27me3 enrichment was localized at promoters in the MCF7 cell line (86% of genes), with a more equal distribution of H3K27me3 between promoter and gene body in MCF-10A and MDA-MB-231 cells (58% and 53% of genes, respectively).
Figure 2. Promoter enrichment for H3K27me3 defined bivalency in breast cell subtypes.
A: Genes with H3K27me3 enriched at promoters are present in all three breast cell lines. Unlike H3K4me3 at promoters (Fig. 1A), relatively few gene promoters are enriched with H3K27me3 in all 3 cell lines. Note that the higher levels of H3K27me3 seen in MCF10A and MDA-MB-231 compared to MCF7 cells is seen both across gene bodies and at gene promoters. B: Genes marked by both H3K4me3 and H3K27me3 at promoters are bivalent. Very few genes were bivalently marked in more than one cell line, indicating that the majority of bivalent genes are unique to each breast cell subtype. MCF7 had the most, with 331 total, MDA-MB-231 was next with 217, and MCF10A had 131 genes identified as bivalent. C: Bivalent genes present in the 3 breast cell subtypes are found as bivalently marked genes in the H7 hESC cells. The fraction of bivalent genes shared between H7 hESC and each of the 3 breast epithelial cells was similar, ranging from 0.59 to 0.67.
We identified bivalent marked genes in each of the breast epithelial cell types based on co-occupancy of histone H3K4me3 and H3K27me3 at gene promoters (Fig. 2B; Supplemental Table SIIIB). The normal-like breast epithelial cell line MCF10A had 131 promoter-bivalent genes, while the cancer cell lines had increased bivalency (MCF7, 331 genes; MDA-MB-231, 217 genes). Among the three breast cell lines, there was minimal overlap of promoter-bivalent genes, with most being unique to each breast cell type (76% for MCF10A, 87% for MCF7 and 85% for MDA-MB-231). These unique bivalent genes may be poised to respond to particular environmental cues or may relate to the different biological processes associated with the phenotypic characteristics of each – benign hyperplastic proliferation, steroid-responsive tumorigenesis, or metastasis. Strikingly, of the bivalent marked genes in each cell type many are bivalent in the H7 human embryonic stem cells, ranging from 59%–67% (Fig. 2C; Supplemental Tables SIIIC–E). The substantially higher number of promoter-bivalent genes in the MCF7 cell type (331 vs 131 (MCF10A) and 217 (MDA-MB-231)) may relate to epigenetically mediated loss of mammary epithelial cell properties and acquisition of breast cancer cell characteristics.
Biological Implications of Bivalent Marked Genes
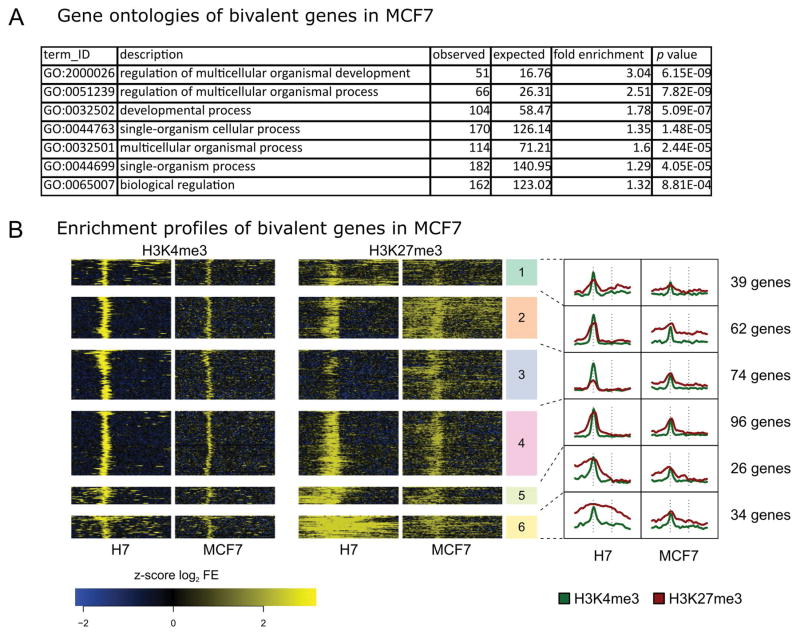
Using Gene Ontology (GO) analysis, we examined the biological pathways or processes associated with the unique subset of bivalent gene promoters among each of the three cell types. For the subsets of bivalent gene promoters in the MCF10A and MDA-MB-231 cell lines, there were no specific pathways or processes identified by GO analysis. For the MCF7 cell type, which represents an early stage tumor, the bivalent marked gene promoters yielded GO terms that included regulation of multicellular organismal development, organismal process, single and multi-cellular process, and biological regulation. These GO terms are indicative of biological processes during early development (Fig. 3A; Supplemental Table SIVA). Unsupervised k-means clustering was applied to enrichment profiles of the bivalent genes in MCF7 and H7-hESC for the 331 bivalent gene promoters ascribed to the MCF7 luminal cell line (Fig. 3B; Supplemental Table SIVB). Overall, we observed a strong relationship between MCF7 and H7-hES cells for both H3K4me3 and H3K27me3 bivalency patterns. These results suggest the possibility of epigenetic programming to a more embryonic-like state within the MCF7 cell type that is consistent with loss of mammary epithelial cell regulation and acquisition of breast cancer stem cell-like properties.
Figure 3. Genes classified as bivalent in MCF7 have comparable enrichment profiles to those of H7-hESC.
A: The 331 genes identified as bivalent in MCF7 cells exhibit statistically significant gene ontology enrichment for many terms associated with regulation of development as well as with cell-cell signaling (p values are Bonferroni corrected). Only a representative set of GO terms is presented, see Supplemental Table SIVA for complete results. B: Clustered histone profiles for bivalent genes in MCF7 cells. Clustering and figure composition are the same as in Figure 1C.
In vivo relevance of a bivalent epigenetic signature in ESR/PGR+ breast cancer patient tumors
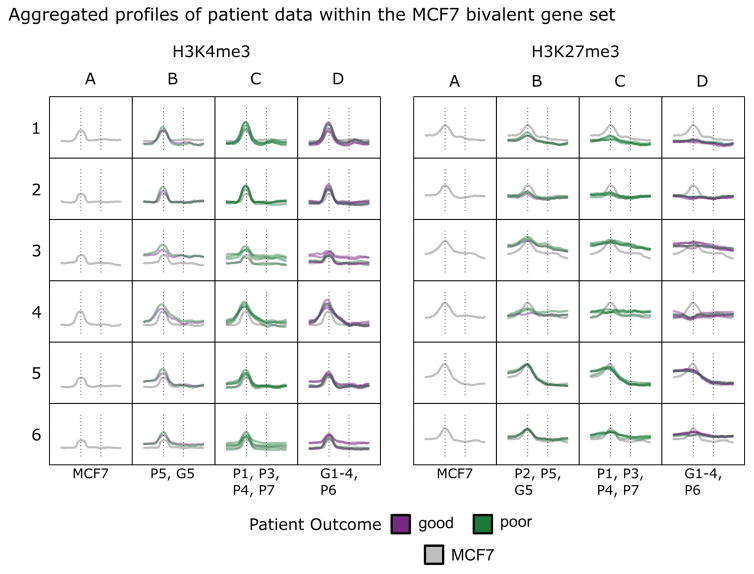
To establish the clinical relevance of our findings, we performed integrated analysis using the distinct bivalent signature identified in the MCF7 luminal cell line with genome-wide H3K4me3 and H3K27me3 enrichment profiles from patient tumor samples. Using clinical samples of ESR/PGR+ tumors from patients with good or poor outcomes after aromatase therapy (Jansen et al., 2013), we compared the 331 bivalent gene promoters in MCF7 to the patient samples by k-means clustering. The tumor samples consisted of 5 individual patients with good outcome (>24 months to disease progression; G1–G5) and 7 with poor outcome (<12 months to disease progression; P1–P7). Patient sample P2 was excluded in their analysis due to the lack of data for the H3K4me3 ChIP; however we included this patient in our analysis as data were available for H3K27me3 ChIP.
As reported by Jansen et al, our analysis revealed similar profiles for H3K4me3 among all patient samples, consistent with our observations for the breast epithelial cell lines in our study (Fig. 4; Supplemental Table SV and Supplemental Figure SI). However, patterns of H3K27me3 enrichment at promoters and gene bodies were variable amongst patients. A pattern of H3K27me3 promoter bivalency consistent with the pattern observed in the MCF7 cell types is most evident in clusters 5 and 6. In these clusters, we observed three patterns of bivalency in patients: the first group is highly similar to MCF7 with well-defined enrichment at promoters (Fig. 4, column B), the second group is less defined with an intermediate signal (column C), and the third group lacks enrichment at promoters (column D). The segregation of patient outcomes of these three groups is indicated in Figure 4 with bivalency being more prominent in patients with a shorter time to disease progression (columns B and C).
Figure 4. ES/PGR+ breast cancer patients display histone modification patterns similar to those observed in MCF7 cells.
The 331 bivalent gene profiles identified in MCF7 cells were used to interrogate patient breast cancer data sets (Jansen, Knijnenburg et al. 2013: GSE40867). Patients with poor (P) outcomes had disease progression within 1 year and those with good (G) outcomes had disease progression after more than 2 years. Patients P2, P5, and G5 had levels of H3K27me3 enrichment at promoters similar to those seen in MCF7 cells within clusters 5 and 6. Patients G1 thru G4 and P6 had essentially no enrichment at bivalent promoters.
DISCUSSION
Our observation of a bivalent profile in the MCF7 luminal cell line that overlaps with the H7-hESC line led us to hypothesize that this epigenetic signature may reflect a subset of breast CSC-like cells that are in dynamic equilibrium with the non-CSC cells (summarized in Fig. 5). Although our profile of 331 bivalent genes in MCF7 cells was selected with high stringency, there are more genes that appear bivalent (see Fig. 1C). This weak but consistent enrichment could originate from a CSC-like subpopulation. We established clinical relevance for key components of our observations using data derived from epigenetic analysis of patient samples from breast cancer tumors (Jansen et al., 2013). We demonstrated an embryonic stem cell-like profile shared by MCF7 and by patient tumors. Examples of genes shared by embryonic stems cells, MCF7, and the patient tumor samples are the stem cell transcriptional regulatory factor, LHX4 (Wang et al., 2014), orphan nuclear receptor estrogen related β (ESRRB) (Lu et al., 2015) and a tumor suppressor, SOX9, which is involved in the APC pathway and in cell survival (Guo et al., 2012).
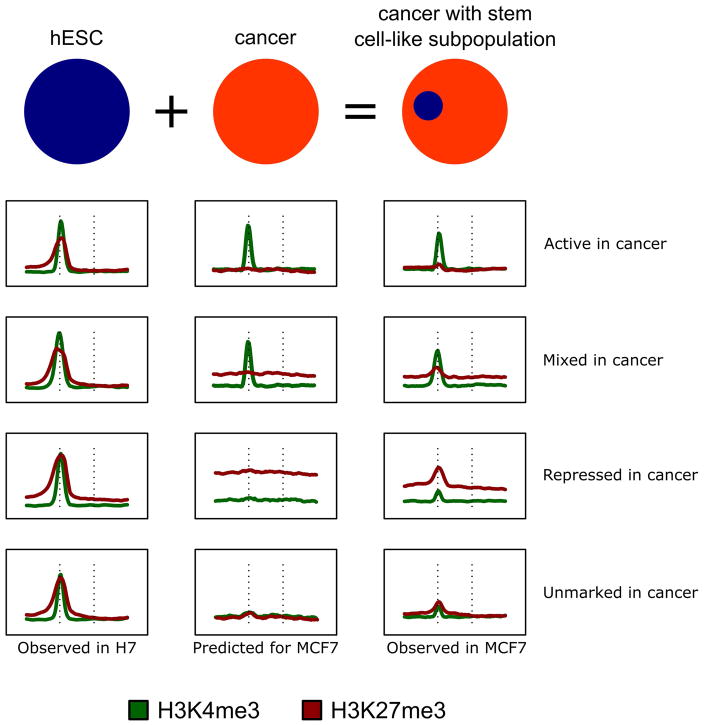
Figure 5. Bivalency in the cancer-stem cell subpopulation is obscured by the histone modification profile of the bulk cell population.
Bivalent mark profiles seen in H7-hESC are visible in MCF7 cells for the majority of genes (Fig. 1C). In contrast to the robust signal seen in H7, these marks in MCF7 cells often had enrichment only marginally greater than that in the surrounding region; only a small subset could be confidently called using MAC2 (H3K4me3) and SICER (H3K27me3). The schematic presents observed histone profiles for H7-hESC (left) and MCF7 (right), with a predictive histone profile pattern for cancer cells with no stem-cell character (middle).
The observation of tumor heterogeneity and differences in relapse time post therapy are the basis for the cancer stem cell or cancer initiating cell models (Ben-Porath et al., 2008; Brooks et al., 2015). Properties of cancer stem cells include dormancy that renders them refractory to initial therapy, and resistance to therapy upon resumption of proliferation. Cancer stem cells share many phenotypic features and gene expression profiles with embryonic stem cells (Beachy et al., 2004; Reya et al., 2001; Stingl and Caldas, 2007). Breast tumors often over-express genes known to mediate physiological control in ESC. ESC-like expression signatures have been observed in both high grade ESR-positive and poorly differentiated ESR-negative breast tumors (Ben-Porath et al., 2008). Expression of ESC genes in breast cancer is consistent with the well-established concept of oncofetal protein expression. These proteins that are stringently regulated during the earliest stages of development recapitulate expression in cancer cells, but with compromised control. While oncofetal proteins have provided effective biomarkers for cancer detection and therapeutic responsiveness they have been largely ineffective as therapeutic targets.
Epigenetic control is emerging as a key regulatory mechanism in cancer biology and pathology. Discoveries in epigenetic control are providing insights leading to novel biomarkers, and inhibitors of epigenetic regulation are proving to be clinically effective in treating solid tumors. Our demonstration of bivalent histone modifications in breast epithelial cancer cells may reflect partial recapitulation of an epigenetic mechanism that is operative in pluripotent embryonic stem cells. Further characterization of oncofetal epigenetic bivalency has the potential to reveal informative biomarkers and therapeutic targets selective for both recurring and treatment-resistant tumors with minimal off-target consequences.
Supplementary Material
Acknowledgments
Funding: Contract grant sponsor: Pfizer Investigator-Initiated Research Award; Contract grant number: IIR Grant WS2049100 (GSS); Contract grant sponsor: NIH; Contract grant number: R37DE012528 (JBL), P01 CA082834 (GSS; JLS).
Next-generation sequencing was performed in the Advanced Genome Technologies Core Massively Parallel Sequencing Facility at the University of Vermont and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, UVM College of Agriculture and Life Sciences, and the UVM College of Medicine. Computing support was provided by the Molecular Bioinformatics Shared Resources Core (MBSR) and secure storage resources were provided by UVM College of Medicine Information Systems (COMIS) and UVM Advanced Computing Core (VACC). Special thanks to S. Tighe, T. Hunter, J. Dragon, B. Devins and Dr. J. Bond for technical and analytical support associated with Massive Parallel Sequencing. The authors also thank Drs. S. K. Zaidi and P. N. Ghule for insightful comments and L. Martin-Buley for editorial assistance.
References
- Agarwal P, Alzrigat M, Parraga AA, Enroth S, Singh U, Ungerstedt J, Osterborg A, Brown PJ, Ma A, Jin J, Nilsson K, Oberg F, Kalushkova A, Jernberg-Wiklund H. Genome-wide profiling of histone H3 lysine 27 and lysine 4 trimethylation in multiple myeloma reveals the importance of Polycomb gene targeting and highlights EZH2 as a potential therapeutic target. Oncotarget. 2016;7(6):6809–6823. doi: 10.18632/oncotarget.6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bapat SA, Jin V, Berry N, Balch C, Sharma N, Kurrey N, Zhang S, Fang F, Lan X, Li M, Kennedy B, Bigsby RM, Huang THM, Nephew KP. Multivalent epigenetic marks confer microenvironment-responsive epigenetic plasticity to ovarian cancer cells. Epigenetics. 2014;5(8):716–729. doi: 10.4161/epi.5.8.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brooks MD, Burness ML, Wicha MS. Therapeutic Implications of Cellular Heterogeneity and Plasticity in Breast Cancer. Cell Stem Cell. 2015;17(3):260–271. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradini D, Oriana S. The role of maintenance proteins in the preservation of epithelial cell identity during mammary gland remodeling and breast cancer initiation. Chin J Cancer. 2014;33(2):51–67. doi: 10.5732/cjc.013.10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu RG. DNA methylation and histone modifications in breast cancer. Methods Mol Biol. 2012;863:35–45. doi: 10.1007/978-1-61779-612-8_3. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature Protoc. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandy RA, Whitfield TW, Wu H, Fitzgerald MP, VanOudenhove JJ, Zaidi SK, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Genome-Wide Studies Reveal that H3K4me3 Modification in Bivalent Genes Is Dynamically Regulated during the Pluripotent Cell Cycle and Stabilized upon Differentiation. Mol Cell Biol. 2015;36(4):615–627. doi: 10.1128/MCB.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Jansen MP, Knijnenburg T, Reijm EA, Simon I, Kerkhoven R, Droog M, Velds A, van Laere S, Dirix L, Alexi X, Foekens JA, Wessels L, Linn SC, Berns EM, Zwart W. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. 2013;73(22):6632–6641. doi: 10.1158/0008-5472.CAN-13-0704. [DOI] [PubMed] [Google Scholar]
- Lu Y, Li J, Cheng J, Lubahn DB. Messenger RNA profile analysis deciphers new Esrrb responsive genes in prostate cancer cells. BMC Mol Biol. 2015;16(1):21. doi: 10.1186/s12867-015-0049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Choudhury S, Kowalczyk A, Bessarabova M, Beresford-Smith B, Conway T, Kaspi A, Wu Z, Nikolskaya T, Merino VF, Lo PK, Liu XS, Nikolsky Y, Sukumar S, Haviv I, Polyak K. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet. 2011;7(4):e1001369. doi: 10.1371/journal.pgen.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier TL, Gordon JA, Boyd JR, Tye CE, Browne G, Stein JL, Lian JB, Stein GS. Histone H3 lysine 4 acetylation and methylation dynamics define breast cancer subtypes. Oncotarget. 2016;7(5):5094–5109. doi: 10.18632/oncotarget.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic acids research. 2016;44(D1):D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37(Database issue):D925–932. doi: 10.1093/nar/gkn730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS. Modeling of Cancer Stem Cell State Transitions Predicts Therapeutic Response. PLoS One. 2015;10(9):e0135797. doi: 10.1371/journal.pone.0135797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7(10):791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Stricker S, Pollard S. Reprogramming cancer cells to pluripotency: an experimental tool for exploring cancer epigenetics. Epigenetics. 2014;9(6):798–802. doi: 10.4161/epi.28600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339(6127):1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Identification of coexistence of DNA methylation and H3K27me3 specifically in cancer cells as a promising target for epigenetic therapy. Carcinogenesis. 2015;36(2):192–201. doi: 10.1093/carcin/bgu238. [DOI] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13(7):497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He C, Hu X. LIM homeobox transcription factors, a novel subfamily which plays an important role in cancer (review) Oncol Rep. 2014;31(5):1975–1985. doi: 10.3892/or.2014.3112. [DOI] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, Peng W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol. 2014;1150:97–111. doi: 10.1007/978-1-4939-0512-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.