Abstract
Inflammatory bowel diseases (IBDs) are complex, multifactorial disorders that comprise Crohn's disease (CD) and ulcerative colitis (UC). Genome-wide association studies have identified approximately 100 loci that are significantly associated with IBD. These loci implicate a diverse array of genes and pathophysiologic mechanisms, including microbe recognition, lymphocyte activation, cytokine signaling, and intestinal epithelial defense. Consistent with epidemio-logic predictions, many IBD-associated loci demonstrate genome-wide significant associations to both CD and UC, notably, genes whose products function in the interleukin-23 pathway, and transcription factors, including NK2 transcription factor related, locus 3 (NKX2-3), SMAD3, STAT3, ZMIZ1, and c-REL. Although CD and UC are both associated with genomic regions that implicate products of genes involved in leukocyte trafficking, there is evidence for association patterns that are distinct between CD and UC. CD-predominant associations include NOD2 and genes that regulate autophagy. In UC, the predominant association signal is on chromosome 6p21, in the major histocompatibility complex region, near HLA class II genes. UC-predominant loci have also implicated genes mediating epithelial defense function. There is a striking overlap of loci between diseases, which could provide comparative insight into mechanisms of disease pathogenesis. Genes that encode factors that function in the interleukin-23 pathway have been associated with a number of chronic inflammatory diseases, notably psoriasis and anky-losing spondylitis. Distinct genetic associations indicate that the colitis associated with primary sclerosing cholangitis is pathophysiologically distinct from UC that is not associated with primary sclerosing cholangitis. As many as 14 susceptibility loci are shared between IBD and celiac disease, indicating significant overlap in pathophysiology. Future genetic studies will be directed toward identifying un common variations with potentially greater statistical effects, defining population differences, and more completely accounting for familial transmission of disease.
Keywords: Crohn's Disease, Ulcerative Colitis, Genetics, Autophagy, Interleukin 23, Genome-Wide Association Studies
Genetic Epidemiology
Population-based studies have provided compelling evidence that genetic factors contribute to the pathogenesis of inflammatory bowel disease (IBD); they demonstrated an 8- to 10-fold greater risk of IBD among relatives of ulcerative colitis (UC) and Crohn's disease (CD) probands1; that Ashkenazi Jews have an increased risk for IBD2; and, most importantly, that there is concordance between twins. Overall, rates of concordance between twins are more modest for CD (30.3% in monozygotic vs 3.6% in dizygotic twins) and are somewhat greater for UC (15.4% in monozygotic vs 3.9% in dizygotic twins) than shown previously.3 The significantly greater concordance for CD vs UC in monozygotic twins indicates that genetic factors make a larger contribution to CD than UC. However, it is also clear that environmental and developmental factors are required for most cases of IBD to develop.
Exceedingly rare, autosomal, recessive mutations in the gene encoding interleukin (IL)-10 receptor4 and the IL-10 cytokine5 have been associated with severe forms of CD in infants born to consanguinous parents. The IL-10 receptor is a tetramer comprising the products of the IL10RA (chromosome 11q21) and IL10RB (chromosome 21q22) genes. Loss-of-function, autosomal, recessive mutations in either receptor subunit are sufficient to cause disease.4 IL-10 inhibits expression of proinflammatory cytokines and increases expression of anti-inflammatory cytokines. These findings demonstrate that loss of IL-10 or IL-10R function can result in a severe CD phenotype without any apparent environmental trigger. Patients with the mutations in the IL-10 receptor4 or cytokine5 have been successfully treated by bone marrow transplantation.
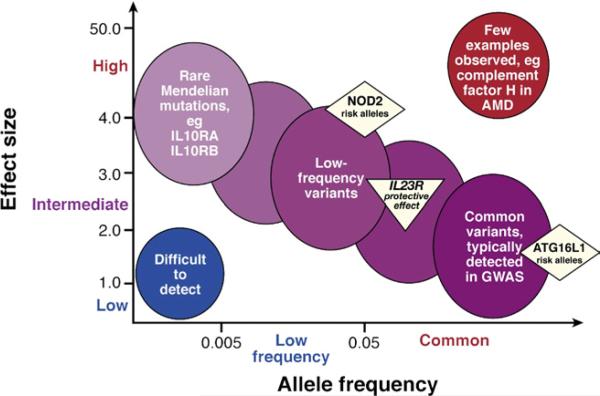
A few genetic disorders, such as those that affect neutrophil function (X-linked chronic granulomatous disease from mutations in cytochrome b-245, β polypeptide) and glycogen storage disease type 1B (from mutations in glucose-6-phosphate transporter SLC37A4), have very high rates of IBD.6,7 The extent to which Mendelian and non-Mendelian forms of IBD share pathogenesis is not known. However, significant associations at chromosome 1q32 near the IL10 gene have been observed in common, non-Mendelian forms of CD8 and UC.9 For the most part, the non-Mendelian forms of IBD have been associated with common (often defined as those polymorphisms where the less common minor allele is present in >5% of chromosomes), relatively low-penetrance genetic variations. Among the >100 IBD genes and loci defined, only nucleotide binding oligomerization domain protein 2 (NOD2) (5% penetrance or ~ 20-fold risk in mutation homozygotes)10 has a meaningful contribution to [CD] risk alone. Nearly all remaining genes and loci contribute modestly to IBD risk, having a relative risk of <1.5-fold and marginal increase in disease penetrance (Figure 1). We have not comprehensively identified genes and variants that mediate familial IBD, found in up to 30% of IBD probands.
Figure 1.
Features of disease-associated genetic variation. Most cases of inflammatory bowel disease (IBD) are multifactorial in etiology, reflecting the effects of multiple genetic risk alleles and developmental and environmental factors. Rare cases of early-onset IBD, with extreme phenotypes, might be single-gene, Mendelian disorders (eg, autosomal recessive mutations in IL10RA or ILRB)—extremely rare mutations with strong effects. In contrast, most of the alleles identified by genome-wide association studies are relatively common (allele frequencies >5%), with modest or low effects and odds ratios <1.5. Greater effects (intermediate to high) include the relatively low-frequency risk alleles in NOD2 and the variant that encodes Arg381Gln in IL23R. Among complex disorders, it is unusual to have common alleles that have strong effects; an exception is that some alleles of the gene that encodes complement factor H confer high-risk for age-related macular degeneration (AMD).
Gene Mapping Approaches
Of the numerous candidate gene studies tested before linkage and genome-wide association studies (GWAS) were performed, only major histocompatibility complex class II alleles were consistently associated with IBD.11 Linkage mapping studies identified segments of human chromosomes shared among affected relatives, greater than expected by chance, and described the IBD1 CD locus on chromosome 16 in 1996, where the NOD2 gene was later identified. GWAS began in 2005, with a modest-sized Japanese case-control cohort of patients with CD and a relatively low-density genome-wide panel of small nucleotide polymorphisms (SNPs).12 This study mapped the region that confers the highest risk for CD among Pacific Asian populations; it contains tumor necrosis factor [TNF] superfamily [ligand] member 15 (TNFSF15) that encodes TNF ligand related molecule 1 (TL1A). Distinct patterns of association have been observed in IBD patients of European ancestry, and studies of Asian and European ancestry-associated polymorphisms indicate that disease-associated haplotypes might affect expression of TNFSF15.13,14 GWAS in North American and European white populations have used arrays of several hundred thousand SNP markers, which comprehensively tag common SNPs throughout the human genome. By combining large datasets, meta-analyses powered to identify loci that have even smaller effects on IBD risk than those identified through individual GWAS have been performed.9,15
With few exceptions, the precise functional allele, or even the causative genes in the GWAS-identified association regions, have not been defined. Many of the loci associated with IBD by GWAS contain multiple genes and some contain no genes. Missense polymorphisms conserved between species and predicted to alter protein structure and/or function are strong candidates for functional, disease-associated polymorphisms. Many of the observed associations are believed to mediate individual differences in messenger RNA expression: the SNPs that have the highest levels of association with IBD may cause significant differences in messenger RNA expression, supporting their role in pathogenesis. Although it is logical to prioritize the potential pathogenic role of expressed transcripts contained within regions of maximal genetic association, it is important to remember that regulatory domains are often distant from the protein-coding regions of genes. Table 1 lists the genetic regions most significantly associated with CD and UC, and select loci that demonstrate genome-wide significance in association with CD and UC.
Table 1.
Genomic Regions Significantly Associated with Inflammatory Bowel Disease in Genome-Wide Association Studies of Patients of European Ancestry
| Definition of association—signal or genes in region | Population differences | Other disease associations | |
|---|---|---|---|
| Predominantly associated with CD | |||
| NOD2 (16q12) | Multiple, uncommon, European-derived, loss-of-function alleles | Similar heterozygote risk in African Americans with CD22; mutations associated with patients of European ancestry not observed in Asians23,24 | Graft-vs-host disease,83 gain-of-function mutations associated with Blau syndrome72a |
| 5q31 | Multiple | No association in Asian populations84,85 | Psoriasis,46 modest association with UCa |
| 9q32 | ZNF365 (encodes zinc finger 365) | Not reported | Not reported |
| 10q21 | Multiple | Not reported | Not reported |
| 18p11 | PTPN2 | Not reported | Type 1 diabetes mellitus,86 celiac disease87 |
| 22q13 | Multiple | Not reported | Not reported |
| Encode factors in the autophagy pathway | |||
| ATG16L1 (2q37) | Common, loss-of-function variant Thr300Ala29 | No association in Asian populations88,89 | Not reported |
| IRGM (5q33.1) | Polymorphism in promoter that affects copy number35 | No association with Japanese cases of CD36 | Different allele found to protect against mycobacterium tuberculosis90 |
| 12q12 | LRRK2 and MUC19 | Not reported | Parkinson's disease,91a leprosy24 |
| Predominantly associated with UC | |||
| Major histocompatibility complex region (6p21) | Multiple, including class II genes | Predominant locus in European and Asian patients with UC | Distinct association patterns in multiple other diseases, including CD |
| FCGR2A (1q23, Fc fragment receptor) | Associated with histidine allele at His131Arg, which increases affinity for Fc fragment | Also associated with a Japanese cohort of patients with UC92 | Arginine allele associated with systemic lupus erythematosus, type 1 diabetes)93-95 |
| 1p36 | Multiple genes | Association with SNPs at 1p36 in Japanese patients with UC92 | No reports in other chronic inflammatory diseases |
| 12q14 | Interferon-γ, IL26, IL22 | Not reported | Not reported |
| Loci that affect epithelial defense | |||
| 7q22 | Multiple, including LAMB1 (encodes laminin β1) | Not reported | Not reported |
| 20q13 | Multiple, including HNF4a (encodes hepatocyte nuclear factor 4α) | Not reported | Not reported |
| Genome-wide significant associations with CD and UC | |||
| Genes that encode factors in the IL-23 pathway | See Figure 2 | Arg381Gln in IL23R (uncommon protective allele), absent in Asian populations89; common alleles associated with IBD in Asians | See Figure 2 |
| 1q32 | Multiple, including IL10 | Not reported | Type 1 diabetes,86a systemic lupus erythematosus,60 Behcet's disease70,71 |
| 5p13 | Gene desert adjacent to PTGER4 (encodes prostaglandin receptor 4) | Not reported | Multiple sclerosis96a |
| 9q32 | TNFSF8, TNFSF15 | Distinct alleles that confer higher risk (>2-fold) in Japanese and Korean subjects with CD12,97 than Europeans with IBD | Leprosy26 |
| 9q34 | Multiple, including CARD9 (encodes caspase recruitment domain family, member 9) | Not reported | Ankylosing spondylitis77 |
| Transcription factors | |||
| 10q22 | ZMIZ1 (encodes zinc finger, MIZ-type containing 1) | Not reported | Celiac disease,87 multiple sclerosis,96 vitiligo98 |
| 10q24 | NKX2-3 (encodes NK2 transcription factor) | Association replicated in Japanese CD99 | No reports in other chronic inflammatory diseases |
| 15q22 | SMAD3 | Not reported | Asthma100 |
NOTE. Loci with P values <10–16 for Crohn's disease (CD) or ulcerative colitis (UC) are presented, excluding loci not containing genes. For a more comprehensive listing of inflammatory bowel disease (IBD) susceptibility loci, see IBD meta-analyses.9,15
Association signals differ between disease and IBD.
Genes That Affect Microbe Recognition by the Innate Immune System—NOD2
The most common mutations in NOD2 that are associated with CD (Arg702Trp [rs2066844], Gly908Arg [rs2066845], and Leu1007fsinsC [rs41450053]) lie either within or near the C-terminal, leucine-rich repeat domain, which is required for microbial sensing.16,17 NOD2 is expressed by many leukocytes, including antigen presenting cells, macrophages, and lymphocytes, as well as ileal Paneth cells, fibroblasts, and epithelial cells. Activation of NOD2 by microbial ligands activates the transcription factor nuclear factor–κB (NF-κB) and mitogen-activated protein kinase signaling, and functions as a positive regulator of immune defense. Studies in primary human cells stimulated with muramyl dipeptide (a ligand of NOD2 and a bioactive component of a peptidoglycan in cell walls of gram-positive and gram-negative bacteria) demonstrated that homozygous or compound heterozygous mutations in NOD2 reduced activation of NF-κB, compared to wild-type NOD2.18
In individuals of European ancestry, heterozygous carriage of one of the major risk alleles confers a 2.4-fold increase in risk for CD; homozygous or compound heterozygous carriage confers a 17.1-fold increase in risk for CD.19 NOD2 mutations are consistently associated with ileal location and stricturing disease.19,20 In African Americans, NOD2 mutations are much less common and only heterozygous, occurring at 20% the frequency observed in Americans of European ancestry—proportionate to the degree of European admixture.21,22 However, risk for CD among carriers of heterozygous mutations is similar (odds ratio = 4.1) to that observed in European ancestry cohorts.22 The NOD2 mutations associated with CD are not observed in Asian or sub-Saharan African populations.22–24 Loss-of-function mutations in NOD2 alone are not sufficient to cause CD. However, NOD2-deficient mice express lower levels of antimicrobial defensins and have increased colonization by commensal bacteria and reduced capacity to clear pathogens. These findings indicate that one major pathogenic mechanism of NOD2 mutations may involve impaired clearance of bacteria that increases their invasion and contributes to the deeper, often transmural inflammation observed with ileal CD.
The NOD2 ligand muramyl dipeptide is ubiquitous, indicating that broad classes of bacteria are capable of activating NOD2. However, the N-glycolyated form of muramyl dipeptide found in mycobacteria and actinomycetes more potently activates NOD2 compared to the N-acetylated form, found more frequently in gram-positive and gram-negative bacteria.25 Particular patterns of bacterial colonization might therefore contribute to CD. A recent Chinese GWAS of leprosy (Mycobacterium leprae) made an interesting association with CD. Whereas major histocompatibility complex genes were most significantly associated with leprosy, genes in the region of NOD2 were also significantly associated, although the SNPs identified were distinct from the variants associated with CD in patients of European ancestry. Receptor interactive protein 2, which is required for NOD2 activation of NF-κB, has not been genetically associated with CD in patients of European ancestry, but SNPs in its gene (RIP kinase 2) were associated with leprosy, as were SNPs in TNFSF15.26
Genes That Regulate Autophagy
Autophagy degrades damaged organelles and proteins, in homeostasis and as a response to starvation, and is important for the clearance of pathogens (xenophagy), which is required for immunity to multiple different types of bacteria. Autophagy 16-like 1 (ATG16L1) has been strongly associated with CD and encodes a protein component of the autophagy complex.27 ATG16L1 is broadly expressed, including in small intestinal Paneth cells28 where it mediates exocytosis of secretory granules that contain antimicrobial peptides. A single Thr300Ala substitution in ATG16L1 has been associated with CD. Thr300Ala (notably, the major allele in European ancestry cohorts) has reduced ability to capture bacteria.29 Mice that are hypomorphic for ATG16L1 and infected with the intestinal pathogen norovirus develop Paneth-cell abnormalities, aberrant responses to injury, and increased susceptibility to ileal injury.30 Despite the demonstration of a functional interaction between NOD2 and ATG16L1,31,32 statistical risks to CD are additive, not synergistic.
Genes that encode immunity-related guanosine triphosphatase M (IRGM) and leucine-rich repeat kinase 2 (LRRK2), which also regulate autophagy, have been associated with CD in GWAS.33,34 Genetic polymorphisms in IRGM that were associated with CD appear to reduce its expression; they include a 20-kb deletion polymorphism 1.6 kb upstream of the IRGM promoter35 and a recently described tetranucleotide insertion.36 Interestingly, although both variants are several-fold more frequent among Japanese people, compared with people of European ancestry, neither variant was associated with CD in Japanese cohorts. Leucine-rich repeat kinase 2 (LRRK2), which has also been associated with Parkinson's disease,37 is located in the CD-associated region on chromosome 12q12,33 along with MUC19 (encodes mucin 19). Mice deficient in LRRK2 have impairments in shuttling of autophagosomes to lysosomes and have increased apoptosis, inflammatory responses, and oxidative damage.38 Alterations in autophagy therefore have important roles in pathogenesis of CD, possibly because of the close apposition of microbial components with high cellular turnover of the intestinal environment.
Lymphocyte Activation, Survival, and Growth
HLA class II genes have been significantly associated with UC. HLA genes are frequently associated with chronic inflammatory genetic disorders, probably because of the enormous genetic and functional diversity contained within this region and its role in regulating interactions between host cells and pathogens. A comprehensive meta-analysis of HLA candidate gene studies reported that DRB1*0103 (odds ratio = 4.6) and DRB1*1502 (odds ratio = 3.3) conferred the greatest risk for UC, whereas DRB1*0410 (odds ratio = 3.9), DQB*0401 (odds ratio = 2.8), and DRB1*0103 (odds ratio = 2.07) conferred the greatest risk for CD among HLA class II variants.39 Among HLA class I genes, B52 conferred the greatest risk for UC (odds ratio = 3.3), whereas Cw8 and B21 (odds ratios = 3.4 and 2.3, respectively) conferred the greatest risk for CD.39 Overall, genes associated with UC confer greater risk with greater consistency than those associated with CD. GWAS studies have highlighted the importance of the HLA region in IBD, with greater association evidence of HLA variations to risk with UC than CD. A recent meta-analysis of nearly 7000 cases of UC and 20,000 controls reported the strongest association with SNP rs9268853, near HLA DR9 (P = 1.4 × 10−55).9 In contrast, a meta-analysis of 6300 cases of CD and 15000 controls found a relatively modest level of association for CD within the HLA region, strongest with SNP rs1799964 (P = 4.0 × 10−11); 21 loci outside the HLA region had more robust significance.15
T-cell activation by HLA class II tetramers presenting peptide antigens involves costimulatory molecules and downstream signaling intermediates. A region on chromo-some 21q22, near ICOSLG (inducible T-cell costimulator ligand), which promotes T-cell proliferation and cytokine secretion, has been associated with CD and UC.33 PTPN22 encodes a lymphocyte-specific protein tyrosine phosphatase that down-regulates lymphocyte signaling; the variant Arg620Trp increases risk for several autoimmune diseases (eg, type I diabetes, rheumatoid arthritis, autoimmune thyroiditis, and systemic lupus erythematosus).33,40,41 Arg620Trp increases the phosphatase activity of PTPN22, compared to Arg620, to inhibit B- and T-cell activation.40 Interestingly, the wildtype arginine rather than threonine at codon 620 was associated with increased risk for CD.33 The chromosome 6q25 region, which includes T-cell activation guanosine triphosphatase-activating protein (TAGAP) was recently associated with CD. This region has been associated with a number of autoimmune diseases; T-cell activation guanosine triphosphatase-activating protein has an expression pattern similar to that of IL-2.15
Cytokines and their receptors regulate T-cell survival and proliferation and have been associated with IBD. A region of chromosome 5p13 that contains IL7R9, which encodes the α subunit of the functional IL-7 receptor, was associated with UC. IL-7 signaling controls survival of naïve and memory T cells, and the IL7R region has been associated with multiple sclerosis.42 Multiple sclerosis42 and IBD15 are also both genetically associated with a region of chromosome 10p15 that contains IL2RA (IL-2 receptor, α subunit). IL-2 controls T-cell proliferation; its effects have complex mechanisms of regulation, based on low- and high-affinity binding to its receptor. Homodimers of IL2RA form a low-affinity receptor for IL-2, whereas the high-affinity receptor comprises α, β, and γ subunits. Finally, multiple sclerosis43 and IBD are both associated with alterations in IL-23 signaling, although in multiple sclerosis (unlike IBD) genetic associations with IL-23 pathway genes have not been established.
IL-23
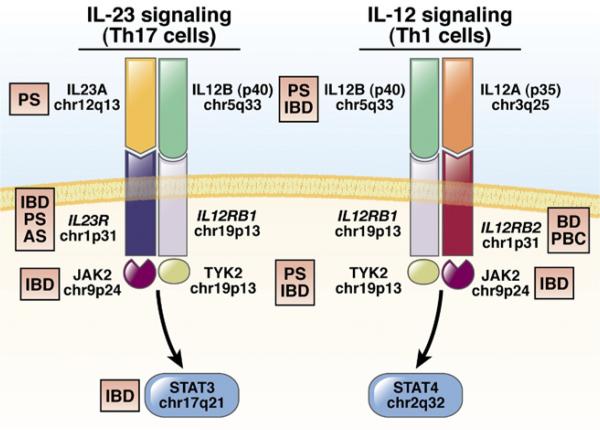
After variants in NOD2, the variant most significantly associated with CD encodes the amino acid change Arg381Gln in the IL-23 receptor (IL23R). Glutamine 381, present in approximately 14% of healthy individuals of European ancestry, reduces risk for CD by nearly 3-fold and for UC by slightly less, compared with Arg381 carriers. Other uncommon alleles that reduce risk have been reported recently.44 Several more common polymorphisms in IL23R have independent contributions to IBD risk. IL23R is a component of the heterodimeric receptor for IL-23 (Figure 2). In addition to their association with IBD, polymorphisms in IL23R have been associated with psoriasis45,46 and ankylosing spondylitis.47 In preliminary studies, antibodies against the p40 subunit that is common to IL-23 and IL-12 were effective in treating patients with CD48; a similar antibody was recently approved for treatment of psoriasis.49 Other genes whose products function in the IL-23 pathway have been associated with IBD (Figure 2). GWAS have therefore established a strong genetic signature for genes that regulate IL-23 signaling in the development of IBDs.50,51
Figure 2.
Interleukin (IL)-23 vs IL-12 signaling. IL-23 and IL-12 signaling are mediated by binding of the cytokine heterodimer to a heterodimeric receptor. These pathways share components of the cytokine (IL12B, p40), receptor (IL12RB1), and downstream signaling components (JAK2 and TYK2). Activation and phosphorylation of downstream components result in the recruitment and activation of signal transducer and activator of transcription (STAT) proteins (STAT3 in IL-23 signaling, STAT4 in IL-12 signaling), which subsequently translocate to the nucleus to activate transcription. Inflammatory bowel disease (IBD) and psoriasis (PS) are each associated with variants in IL23R, IL12B (p40) and TYK2. IBD, PS, and ankylosing spondylitis (AS) are all associated with the chromosome 1p31 region that includes the C-terminal 7 exons of IL23R, and extends into the intergenic region between IL23R and IL12RB2. In contrast, Behçet's disease (BD) and primary biliary cirrhosis (PBC) are associated with the inter-genic region, between IL23R and IL12RB2.
Comparing CD and UC Associations Identified by GWAS
The approximately 100 genomic loci that have been significantly associated with IBD contain candidate genes whose products mediate a variety of cell functions, including microbial recognition, lymphocyte activation, cytokine signaling, metabolism (FUT2, fucosytransferase),52 endoplasmic reticulum stress responses (eg, X box binding protein–1),53 physicochemical defense (eg, MUC1 and MUC19, mucins), and epithelial barrier function.33 Interestingly, a nonsense polymorphism in FUT2 that determines susceptibility to norovirus infection was associated with CD.15,54 Reflecting the clinical and epidemiologic overlap between the subtypes of IBD, it is not surprising that many of these loci demonstrate genome-wide significant associations for both CD and UC55,56 (Table 1).
Many of the loci that have significant associations with CD and UC contain genes that encode transcription factors, especially those that regulate cytokine expression and function (Table 1 and Figure 2). Signal transducer and activator of transcription 3 (STAT3), gene located on chromosome 17q21, is activated in response to a variety of cytokines and growth factors, including IL-6, −10, −21, −22, −23, and −26. Similarly, SMAD family member 3 (SMAD3), gene located on chromosome 15q22, regulates transcription in response to signaling by transforming growth factor–β.57 CD and UC are associated with the region on chromosome 10q21 that encodes zinc finger MIZ type 1 (ZMIZ1). ZMIZ1 increases SMAD signaling and is induced by retinoic acid,58 which broadly mediates immune tolerance in the intestine. IBD is also associated with chromosome 2p16, which contains REL, which encodes c-Rel, a subunit of NF-κB. NF-κB is activated in response to signaling from pattern recognition receptors and tumor necrosis factor–α. The gene that encodes PR domain zinc finger protein 1 (PRDM1), on chromosome 6q21, represses expression of interferon-β and is associated with CD9 and UC15 as well as rheumatoid arthritis59 and systemic lupus erythematosus.60 Finally, both CD and UC demonstrate significant associations with a region on chromosome 10q24 that encodes the transcription factor NK2 transcription factor related, locus 3 (NKX2-3). Mice deficient in NKX2-3 have hyposplenia and defects in intestinal development.61
Although association studies indicate that CD and UC have similar mechanisms of pathogenesis9,15 that involve some of the same transcription factors, comparative analyses of genes that regulate leukocyte mobility provide subtle evidence for different pathogenic mechanisms. For example, CD is significantly associated with the chromo-some 17q12 region that contains chemokine C-C motif ligand (CCL2) and 7 (CCL7), which encode small chemotactic proteins that promote migration of monocytes and macrophage.15 In contrast, UC has a consistent but nominal association with this region.9 Similarly, although CD is associated with a chromosome 6q27 region that contains chemokine C-C motif receptor 6 (CCR6), UC is not.9 CCR6 is specifically expressed by immature dendritic cells and memory T cells; it mediates leukocyte recruitment and migration under inflammatory conditions in which its ligand, chemokine C-C motif ligand 20 (CCL20), is expressed.62 In contrast, the chromosome 11q15 region contains the gene lymphocyte specific protein 1 (LSP1) and is significantly associated with UC, but not CD.9 Although LSP1 was initially thought to be expressed only in leukocytes, it was also found to be expressed in endothelial cells and to be essential for neutrophil extravasation in response to TNF–α and IL-1β.63
Many of the loci that have the strongest associations with UC do not have compelling candidate genes (eg, chr1p36 locus64; Table 1). A number of loci that contain UC-predominant signals include genes that regulate epithelial barrier function, including GNA12 (encodes gua-nine nucleotide binding protein α12, which has a role in tight-junction assembly) on chromosome 7p22, HNF4A (encodes hepatocyte nuclear factor 4, α), CDH (encodes cadherin 1, epithelial, which mediates calcium-dependent glycoprotein binding) on chromosome 16q21, and LAMB1 (laminin β1, which encodes a constituent of basement membranes) on chromosome 7q31.9 Finally, one of the genetic variants that is predominantly associated with UC in patients of European ancestry and in Japanese is a polymorphism that encodes the amino acid change His131Arg in the Fcγ receptor FCGR2A. FCGR2A is found on the surface of a variety of immune cells, including neutrophils and macrophage, and is involved in phagocytosis and clearance of immune complexes.65
Associations Across Chronic Inflammatory Diseases
The overlap between CD- and UC-associated genetic loci reflects the significant epidemiologic and clinical overlap. Extraintestinal manifestations of IBD include primary sclerosing cholangitis (PSC), uveitis, and ankylosing spondylitis, with frequencies of at most 2.4%, 4.5%, and 4.1%, respectively, based on data from a population-based study of IBD.66 Colitis is diagnosed in approximately 50% of patients with PSC, based on clinical evidence, and >80% on histopathology findings; it might be a disorder of overlapping phenotype, but of a distinct etiology, from UC. In a GWAS of PSC, a region that was just toward the centromere from HLA-B had the strongest association.67
Support for the concept that the colitis associated with PSC is pathophysiologically distinct comes from the observation that it is associated with distinct HLA genes, compared with UC (Table 2). Furthermore, PSC is not associated with non-HLA UC regions associated with UC, such as chromosome 1p36 (Table 1) or IL23R; however larger subsequent studies may yet demonstrate associations. In a recent GWAS, PSC was associated with a gene-rich region of 3p21 that was previously associated with IBD (Table 2), indicating some overlap in patho-physiology.68 Primary biliary cirrhosis69 and Behçet's disease70,71 were associated with the intergenic region between IL23R and IL12RB2, although there were slightly different patterns of allelic association compared with IBD (see Figure 2). Gain-of-function mutations in NOD2 are responsible for autosomal-dominant Blau syndrome, which can manifest with iritis and/or uveitis.72 Uveitis occurs in patients with Behçet's disease, ankylosing spondylitis, sarcoidosis, and psoriatic arthritis; a GWAS of all these disorders might be useful in identification of uveitis-associated risk variants.
Table 2.
Comparative Associations Between Inflammatory Bowel Disease and Related Chronic Inflammatory Diseases
| Loci | IBD | Psoriasis | Celiac disease | Primary sclerosing cholangitis | Ankylosing spondylitis |
|---|---|---|---|---|---|
| Gene product in the IL-23 pathway | Yes | Yes | No | No | Yes |
| Major histocompatibility complex | Class II predominant | Class I predominant | Class II predominant | Class I predominant | Class I predominant |
| Loci of genome-wide significance shared with IBD | — | 4 | 14 | 3p21(a gene-rich region that includes MST1)68 | 2: ERAP1a at chr5p15; CARD9 at chr9q34 |
NOTE. The large number of loci shared between inflammatory bowel disease (IBD) and celiac disease might reflect the larger number of loci established for celiac disease, compared with psoriasis, ankylosing spondylitis, and primary sclerosing cholangitis.
IL-23, interleukin-23.
The association with ERAP1 is distinct from the association with Crohn's disease.
Like IBD and psoriasis, the Arg381Gln polymorphism in IL23R reduces risk for ankylosing spondylitis.47 IL23R was 1 of the 2 most significant non-HLA genes in a GWAS of ankylosing spondylitis (the other is the 5q15 region that contains endoplasmic reticulum aminopeptidase 1 (ERAP1), which was also associated with CD and with Psoriasis).15,73,74 A recent study in Han Chinese found nominal evidence for an association of SNPs in janus kinase 2 (JAK2) and STAT3 with ankylosing spondylitis; Arg 381Gln was not found in this population.75 The other 2 loci that have shown significant associations with both ankylosing spondylitis and IBD are caspase recruitment domain family, member 9 (CARD9)76,77 and the 21q22 region.78,79
Overall, there has been a striking overlap in loci associated with different chronic inflammatory diseases. Many genes that encode factors in the IL-23 pathway have been associated with both psoriasis and IBD (IL23R, IL12B, and tyrosine kinase 2 [TYK2]),45,74 indicating the role of the IL-23 in mediating mucosal immunity (in the skin and intestine) (Figure 2). Numerous loci have been associated with both IBD and celiac disease, including those near IL2 and IL21 (chr4q21) and IL18RAP (IL-18 receptor accessory protein, 2q33).9,15,80,81
Future Directions—Uncommon Alleles, Population Differences, and Family Studies
The contributions of genetic factors to IBD are complex. Despite the success of GWAS in identifying significantly associated loci, they are estimated to account for <25% of predicted heritability.15 GWAS find common, human genetic variants associated with disease, but it is possible that uncommon polymorphisms also contribute to IBD; these may be identified through whole-genome sequencing approaches. Thus far, with the exception of the region of IRGM,35 large copy-number polymorphisms have not been associated with IBD.82 Polymorphisms that have a stronger effect on disease development (Figure 1) might be maintained at a lower frequency in the population (uncommon alleles) through negative selection, because they reduce reproductive fitness.
If uncommon variants make significant contributions to complex disorders such as IBD, consideration of population differences will be particularly informative, as less common alleles are more likely to be population specific. African populations have the greatest genetic diversity and will undoubtedly provide important comparative insights. Studies in cohorts of Ashkenazi Jews might be particularly valuable because of a high prevalence of disease and greater allelic homogeneity from endogamy.
A main goal of human genetic studies is to better understand familial transmission of diseases and identify uncommon variants that make large contributions to pathogenesis. Uncommon alleles that are shared among affected individuals within a family are more likely to contribute to familial clustering of disease than common alleles carried within a population. In addition to uncommon, disease-associated alleles, families are more likely to share features of the intestinal microbiome; a functional integration of genetic and family-based microbiome analyses might help us better understand the complex causes of IBD.
Acknowledgments
Funding
We gratefully acknowledge the following: National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) U01 DK062429; U01 DK062422; RC1 DK086800; National Center for Research Resources: KL2RR024138, CCFA; R01 GM059507; David Wermuth; Bohmfalk Medical Foundation; PSC Partners for a Cure; 2P30 DK034989; U19 A1082713 (J.H.C.). NIDDK R01 DK83553; U01 DK062431; The W. Buford and Linda M. Lewis family, the Atran Foundation, and the Harvey M. and Lyn P. Meyerhoff Inflammatory Bowel Disease Center (S.R.B.).
Abbreviations used in this paper
- CD
Crohn's disease
- GWAS
genome-wide association studies
- IBD
inflammatory bowel disease
- IL
interleukin
- IL23R
interleukin-23 receptor
- IRGM
immunity-related guanosine triphosphatase M
- NF-κB
nuclear factor–κB
- PSC
primary sclerosing cholangitis
- SNP
small nucleotide polymorphism
- STAT
signal transducer and activator of transcription
- UC
ulcerative colitis
Biography
 Judy H. Cho
Judy H. Cho
 Steven R. Brant
Steven R. Brant
Footnotes
Supplementary Material
Note: The first 50 references associated with this article are available below in print. The remaining references accompanying this article are available online only with the electronic version of the article. Visit the online version of Gastroenterology at www.gastrojournal.org, and at doi:10.1053/j.gastro.2011.02.046.
Conflicts of interest The authors disclose no conflicts.
References
- 1.Orholm M, Munkholm P, Langholz E, et al. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 2.Lowe AM, Roy PO, B-Poulin M, et al. Epidemiology of Crohn's disease in Quebec, Canada. Inflamm Bowel Dis. 2009;15:429–435. doi: 10.1002/ibd.20756. [DOI] [PubMed] [Google Scholar]
- 3.Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011;17:1–5. doi: 10.1002/ibd.21385. [DOI] [PubMed] [Google Scholar]
- 4.Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glocker EO, Frede N, Perro M, et al. Infant colitis—it's in the genes. Lancet. 376:1272. doi: 10.1016/S0140-6736(10)61008-2. [DOI] [PubMed] [Google Scholar]
- 6.Marciano BE, Rosenzweig SD, Kleiner DE, et al. Gastrointestinal involvement in chronic granulomatous disease. Pediatrics. 2004;114:462–468. doi: 10.1542/peds.114.2.462. [DOI] [PubMed] [Google Scholar]
- 7.Dieckgraefe BK, Korzenik JR, Husain A, et al. Association of glycogen storage disease 1b and Crohn disease: results of a North American survey. Eur J Pediatr. 2002;161(Suppl 1):S88–S92. doi: 10.1007/s00431-002-1011-z. [DOI] [PubMed] [Google Scholar]
- 8.Franke A, Balschun T, Karlsen TH, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 9.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brant SR, Wang MH, Rawsthorne P, et al. A population-based case-control study of CARD15 and other risk factors in Crohn's disease and ulcerative colitis. Am J Gastroenterol. 2007;102:313–323. doi: 10.1111/j.1572-0241.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 11.Stokkers PC, Reitsma PH, Tytgat GN, et al. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395–401. doi: 10.1136/gut.45.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 13.Kakuta Y, Ueki N, Kinouchi Y, et al. TNFSF15 transcripts from risk haplotype for Crohn's disease are overexpressed in stimulated T cells. Hum Mol Genet. 2009;18:1089–1098. doi: 10.1093/hmg/ddp005. [DOI] [PubMed] [Google Scholar]
- 14.Michelsen KS, Thomas LS, Taylor KD, et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One. 2009;4:e4719. doi: 10.1371/journal.pone.0004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 17.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 18.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflamm Bowel Dis. 2006;12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 19.Economou M, Trikalinos TA, Loizou KT, et al. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 20.Lesage S, Zouali H, Cezard JP, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kugathasan S, Loizides A, Babusukumar U, et al. Comparative phenotypic and CARD15 mutational analysis among African American, Hispanic, and White children with Crohn's disease. Inflamm Bowel Dis. 2005;11:631–638. doi: 10.1097/01.mib.0000171279.05471.21. [DOI] [PubMed] [Google Scholar]
- 22.Dassopoulos T, Nguyen GC, Talor MV, et al. NOD2 mutations and anti-Saccharomyces cerevisiae antibodies are risk factors for Crohn's disease in African Americans. Am J Gastroenterol. 2010;105:378–386. doi: 10.1038/ajg.2009.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue N, Tamura K, Kinouchi Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki K, Takazoe M, Tanaka T, et al. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn's disease. J Hum Genet. 2002;47:469–472. doi: 10.1007/s100380200067. [DOI] [PubMed] [Google Scholar]
- 25.Coulombe F, Divangahi M, Veyrier F, et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med. 2009;206:1709–1716. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 27.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuballa P, Huett A, Rioux JD, et al. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 32.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prescott NJ, Dominy KM, Kubo M, et al. Independent and population-specific association of risk variants at the IRGM locus with Crohn's disease. Hum Mol Genet. 2010;19:1828–1839. doi: 10.1093/hmg/ddq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skipper L, Li Y, Bonnard C, et al. Comprehensive evaluation of common genetic variation within LRRK2 reveals evidence for association with sporadic Parkinson's disease. Hum Mol Genet. 2005;14:3549–3556. doi: 10.1093/hmg/ddi376. [DOI] [PubMed] [Google Scholar]
- 38.Tong Y, Yamaguchi H, Giaime E, et al. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci U S A. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 41.Criswell LA, Pfeiffer KA, Lum RF, et al. Analysis of Families in the Multiple Autoimmune Disease Genetics Consortium (MADGC) Collection: the PTPN22 620W Allele Associates with Multiple Autoimmune Phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hafler DA, Compston A, Sawcer S, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 43.Cua DJ, Sherlock J, Chen Y, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 44.Momozawa Y, Mni M, Nakamura K, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2010;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 45.Cargill M, Schrodi SJ, Chang M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burton PR, Clayton DG, Cardon LR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies auto-immunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mannon PJ, Fuss IJ, Mayer L, et al. Anti-interleukin-12 antibody for active Crohn's disease. N Engl J Med. 2004;351:2069–2079. doi: 10.1056/NEJMoa033402. [DOI] [PubMed] [Google Scholar]
- 49.Krueger GG, Langley RG, Leonardi C, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356:580–592. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Zhang H, Kugathasan S, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 51.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 52.McGovern DP, Jones MR, Taylor KD, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlsson B, Kindberg E, Buesa J, et al. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 Norovirus infection. PLoS One. 2009;4:e5593. doi: 10.1371/journal.pone.0005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franke A, Balschun T, Karlsen TH, et al. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 57.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Thyssen G, Beliakoff J, et al. The novel PIAS-like protein hZimp10 enhances Smad transcriptional activity. J Biol Chem. 2006;281:23748–23756. doi: 10.1074/jbc.M508365200. [DOI] [PubMed] [Google Scholar]
- 59.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 62.Nishimura M, Kuboi Y, Muramoto K, et al. Chemokines as novel therapeutic targets for inflammatory bowel disease. Ann N Y Acad Sci. 2009;1173:350–356. doi: 10.1111/j.1749-6632.2009.04738.x. [DOI] [PubMed] [Google Scholar]
- 63.Liu L, Cara DC, Kaur J, et al. LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med. 2005;201:409–418. doi: 10.1084/jem.20040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silverberg MS, Cho JH, Rioux JD, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nimmerjahn FRJ. Fcgamma receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 66.Bernstein CN, Blanchard JF, Rawsthorne P, et al. The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study. Am J Gastroenterol. 2001;96:1116–1122. doi: 10.1111/j.1572-0241.2001.03756.x. [DOI] [PubMed] [Google Scholar]
- 67.Karlsen TH, Franke A, Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138:1102–1111. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 68.Melum E, Franke A, Schramm C, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Remmers EF, Cosan F, Kirino Y, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23RIL12RB2 regions associated with Behcet's disease. Nat Genet. 2010;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mizuki N, Meguro A, Ota M, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet's disease susceptibility loci. Nat Genet. 2010;42:703–706. doi: 10.1038/ng.624. [DOI] [PubMed] [Google Scholar]
- 72.Miceli-Richard C, Lesage S, Rybojad M, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 73.Reveille JD, Sims AM, Danoy P, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Zhang X, Wang Y. Analysis of JAK2 and STAT3 polymorphisms in patients with ankylosing spondylitis in Chinese Han population. Clin Immunol. 2010;136:442–446. doi: 10.1016/j.clim.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 76.Zhernakova A, Festen EM, Franke L, et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pointon JJ, Harvey D, Karaderi T, et al. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes Immun. 2010;11:490–496. doi: 10.1038/gene.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas GP, Brown MA. Genetics and genomics of ankylosing spondylitis. Immunol Rev. 2010;233:162–180. doi: 10.1111/j.0105-2896.2009.00852.x. [DOI] [PubMed] [Google Scholar]
- 80.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holler E, Rogler G, Herfarth H, et al. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood. 2004;104:889–894. doi: 10.1182/blood-2003-10-3543. [DOI] [PubMed] [Google Scholar]
- 84.Yamazaki K, Takazoe M, Tanaka T, et al. Association analysis of SLC22A4, SLC22A5 and DLG5 in Japanese patients with Crohn disease. J Hum Genet. 2004;49:664–668. doi: 10.1007/s10038-004-0204-x. [DOI] [PubMed] [Google Scholar]
- 85.Tosa M, Negoro K, Kinouchi Y, et al. Lack of association between IBD5 and Crohn's disease in Japanese patients demonstrates population-specific differences in inflammatory bowel disease. Scand J Gastroenterol. 2006;41:48–53. doi: 10.1080/00365520510023864. [DOI] [PubMed] [Google Scholar]
- 86.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dubois PCTG, Franke L, Hunt KA, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang SK, Park M, Lim J, et al. Contribution of IL23R but not ATG16L1 to Crohn's disease susceptibility in Koreans. Inflamm Bowel Dis. 2009;15:1385–1390. doi: 10.1002/ibd.20921. [DOI] [PubMed] [Google Scholar]
- 89.Yamazaki K, Onouchi Y, Takazoe M, et al. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn's disease in Japanese patients. J Hum Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 90.Intemann CD, Thye T, Niemann S, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5(9):e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 92.Asano K, Matsushita T, Umeno J, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. 2009;41:1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- 93.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genomewide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salmon JE, Millard S, Schachter LA, et al. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alizadeh BZ, Valdigem G, Coenen MJ, et al. Association analysis of functional variants of the FcgRIIa and FcgRIIIa genes with type 1 diabetes, celiac disease and rheumatoid arthritis. Hum Mol Genet. 2007;16:2552–2559. doi: 10.1093/hmg/ddm194. [DOI] [PubMed] [Google Scholar]
- 96.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang SK, Lim J, Chang HS, et al. Association of TNFSF15 with Crohn's disease in Koreans. Am J Gastroenterol. 2008;103:1437–1442. doi: 10.1111/j.1572-0241.2007.01752.x. [DOI] [PubMed] [Google Scholar]
- 98.Quan C, Ren YQ, Xiang LH, et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet. 2010;42:614–618. doi: 10.1038/ng.603. [DOI] [PubMed] [Google Scholar]
- 99.Yamazaki K, Takahashi A, Takazoe M, et al. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn's disease in Japanese patients. Gut. 2009;58:228–232. doi: 10.1136/gut.2007.140764. [DOI] [PubMed] [Google Scholar]
- 100.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]