Abstract
Direct comparison of human diseases with model phenotypes allows exploration of key areas of human biology which are often inaccessible for practical or ethical reasons. We review recent developments in comparative evolutionary approaches for finding models for genetic disease, including high-throughput generation of gene/phenotype relationship data, the linking of orthologous genes and phenotypes across species, and statistical methods for linking human diseases to model phenotypes.
INTRODUCTION
In a natural extension of the traditional model organism approach, new data sources and techniques are allowing connections to be drawn between human and model systems, even when phenotypes don’t obviously match. As organisms diverge over evolutionary time, the relationship between genes and the phenotypes they encode often also diverge. Many novel phenotypes arise from repurposed gene networks, rather than novel genes [1 and 2], while, conversely, molecular networks can lose their associations with conserved phenotypes [3]. Such complexity gives rise to a wealth of potential model systems, each capable of providing useful insights into human disease.
Such comparative evolutionary approaches to study human disease are rooted in the traditional use of experimental and genetic data from diverse organisms to explore mechanisms of human genetics. However, new methods for discovering relevant organismal models for human disease are being developed, most notably methods drawing on computer science and evolutionary analyses to incorporate the growing wealth of genetic and phenotypic data in increasingly diverse species.
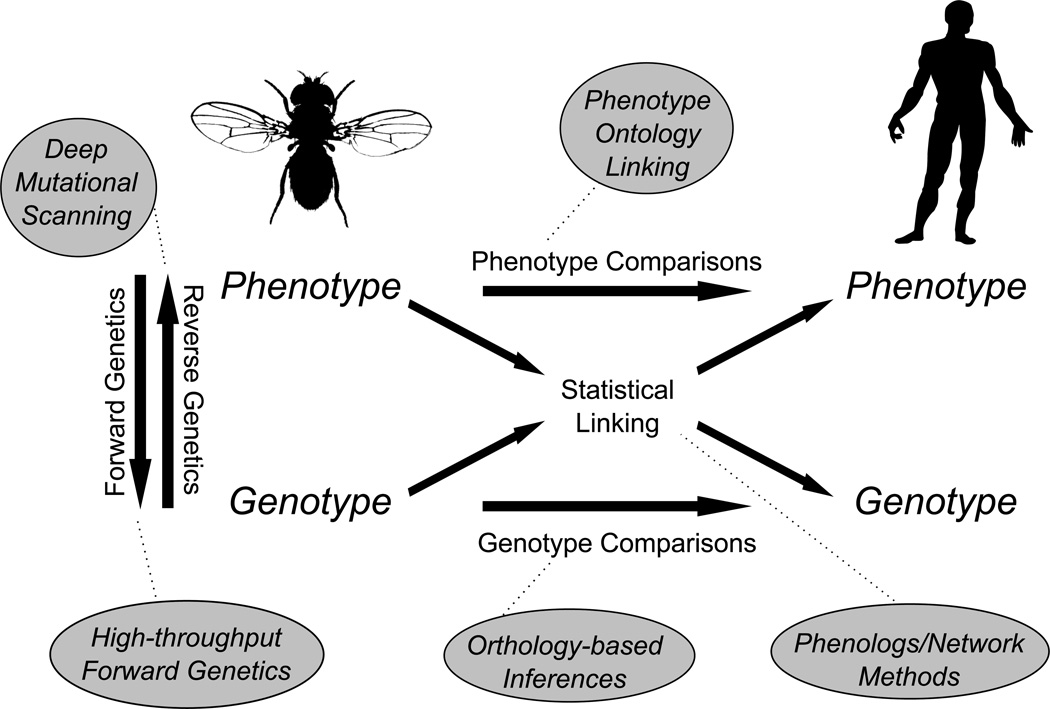
Here, we review recent advances in using semantic, genetic, and evolutionary information in both model and non-model organisms to rationally identify the genetic underpinnings of human disease. Figure 1 introduces a general framework that categorizes elements of comparative approaches. Typical approaches include identifying relationships between genes and phenotypes in a model organism via forward and/or reverse genetics, followed by comparison to human disease via gene orthology, phenotype similarity, or a combination of both. The different components have all been touched by an ever-increasing emphasis on high-throughput methods. Using the framework in Figure 1, we categorize the major approaches taken by researchers, and illustrate these approaches with several examples of how the new methods are applied to the study of human diseases.
Figure 1.
Components of comparative methods for rationally identifying human disease genes. Silhouettes are from PhyloPic (URL: http://phylopic.org/).
GENERATING DATA WITH FORWARD AND REVERSE GENETICS
High-throughput identification of mutant phenotypes and their underlying genetics often provide the raw material for human disease model identification. The comparative approach often begins with a genetic screen in a model system to identify relevant pathways and genes. Genetic screens are traditionally divided into reverse genetics, which perturbs specific genes and looks for phenotypic effects, and forward genetics, which identifies phenotypes of interest and then uncovers their genetic basis. Generation of mutants by both approaches, and the corresponding large-scale identification of phenotypes, has been revolutionized by use of high-throughput sequencing, synthetic biology techniques, and image analysis.
In particular, over the last decade, reverse genetics has been scaled up by high-throughput knockdowns, especially RNAi screens and comprehensive gene knockout collections in model species that span the tree of life [4, 5, 6, 7 and 8]. More recently, CRISPR screens, which are potentially amenable to any organism of interest, have been used to create libraries of human cell line knockouts [9 and 10], allowing reverse genetics to be applied to human systems; phenotypic analysis of the resulting CRISPR libraries now seems to be the bottleneck. At least on the single gene level, a group of methods termed “Deep Mutational Scanning” allows the systematic switching of, for instance, every codon in an open reading frame to every other codon in search of phenotype-causing substitutions [11*, 12 and 13].
Similar gains in throughput are now being seen in forward genetics screens as well. Often, forward genetic screens introduce untargeted mutations into a genome, and then identify lines with phenotypes of interest before screening for mutations. Recent efforts have utilized high-throughput sequencing to make this approach feasible in mammals [14] and on much larger scales than in previous generations [15]. Other methods utilize the diversity of natural populations as a basis for phenotypic screening [16* and 17]. Platforms in yeast, plants, worms, and fruit flies now exist to record quantitative data on multiple levels, including morphology, metabolism, transcription, and translation [16*, 18, 19, 20 and 21]. Combining these measurements make it possible not only to uncover the biology of model systems, but also to screen human genes and candidate disease alleles in a model system background, a method that Jasper Rine and colleagues termed “surrogate genetics” [22 and 23*]. Although model organisms will remain a mainstay of human disease genetics, the advent of these novel molecular tools has raised the possibility of high-throughput screens of genotype/phenotype relations in any organism of interest, blurring the lines between model and non-model organism. Importantly, the advances in both forward and reverse genetics have produced hundreds of thousands of gene-phenotype associations across multiple organisms [24, 25, 26 and 27], providing deep datasets that now make computational analyses of new disease genes increasingly possible.
FINDING MODELS THROUGH PHENOTYPE COMPARISON
Traditionally, non-human models of disease are often identified by the direct comparison of a model organism phenotype with traits of a human disease. However, such comparisons have historically relied only on the expertise of researchers, and tended to make use of organism-specific language to describe phenotypes. The development of ontologies, formal hierarchies of descriptive annotations [28, 29 and 30], now allows researchers to find new human disease models by directly searching for homologous phenotypes using phenotype ontologies, an approach easily scalable to large phenotypic datasets. Multiple ontologies [31, 32 and 33] have been developed, enabling systematic analyses of phenotypes in a way that is descriptive, robust, programmatically accessible, and extensible across species. Notably, major organismal databases now use ontologies to describe phenotypes, e.g. as for the Worm [34], Human [35], and Mammalian [36] Phenotype Ontologies.
Formal ontologies allow phenotype databases to be cross referenced, much as researchers might search for homologous sequences across organisms. This functionality can significantly improve the throughput and sensitivity of the comparative approach, and can be used to identify disease models based on phenotypic qualities alone. As one such example, PhenomeNET [37], for instance, employs the Phenotype and Trait Ontology (PATO) developed by Gkoutos and colleagues, and was used to suggest novel genes involved in the Tetralogy of Fallot, a congenital heart defect. Other algorithms have also made use of phenotype annotations to facilitate the discovery of candidate disease genes [38**, 39, 40, 41 and 42].
FINDING MODELS THROUGH GENETIC COMPARISON
Just as phenotype ontologies can be used to identify disease models based on phenotypes alone, appropriate models can also be selected using orthologs [43] of human disease genes identified in model organisms (Figure 1). Although this approach is limited by its reliance on a priori knowledge of the genetic basis of a disease, the basic step of determining gene orthology between organisms of interest still forms the core of most comparative studies. This is because orthologs, which are separated historically only by speciation events, tend to be more closely related in function than paralogs [44–46], which result from shared ancestral gene duplications and can often partition an ancestral function, or take on whole new functions [47, 48 and 49].
New methods for inferring orthologs, and new databases for storing this information are proliferating. As of 2015, there are at least 37 different orthology databases using a variety of algorithms, reviewed in detail by Sonnhammer et al. 2014 [50**]. In spite of this tremendous focus of community effort, benchmarking suggests that no one method outperforms all others, with methods differing in their precise definition of “orthology” as well as in their tendency to favor either precision or recall for discovery of correct orthologs [51 and 52]. Meta-analyses that compare and compile information from different algorithms and databases are therefore expected to significantly improve performance [53 and 54]. Another promising direction is to use information about the species phylogeny to probabilistically inform gene tree inference [55, 56 and 57]. When the gene of interest for a disease has not yet been identified, computational strategies now exist for prioritizing potential candidate genes, as we discuss next, but even these methods usually require knowledge of orthologs as a starting point.
STATISTICALLY ASSOCIATING GENES AND DISEASES
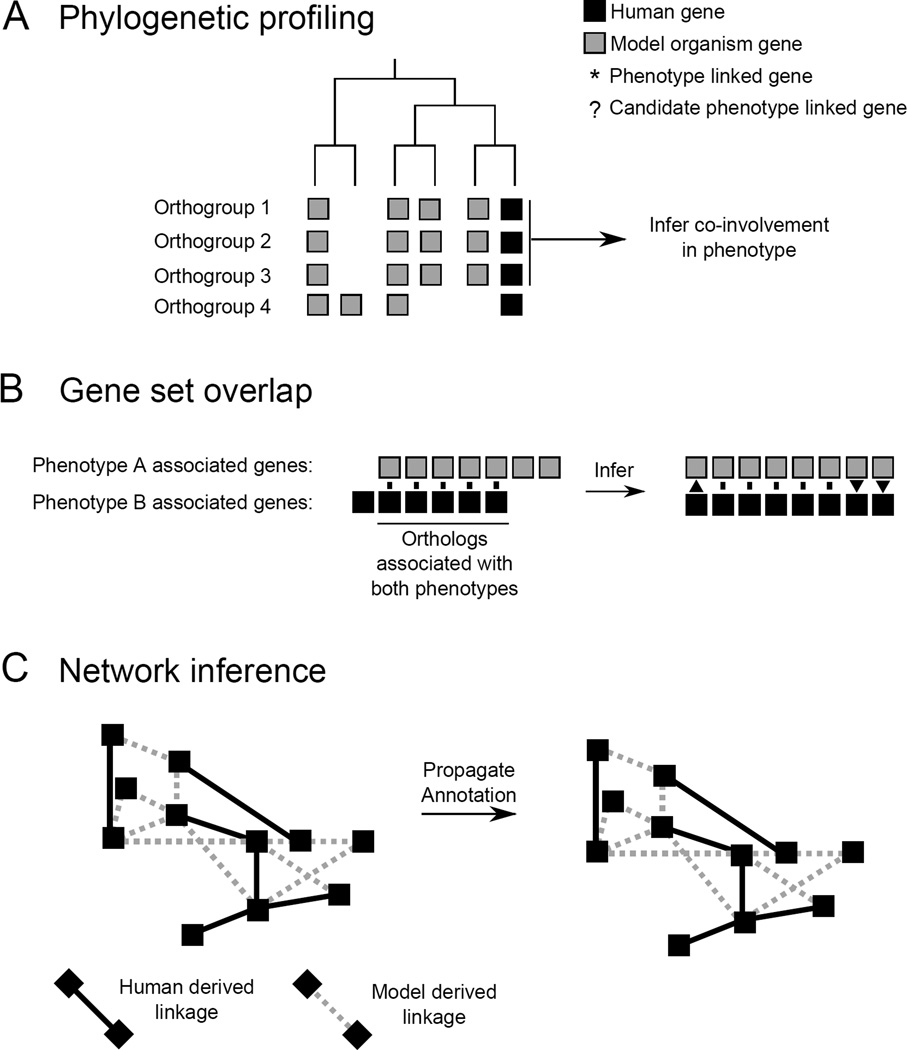
The methods described above either rely on prior information about the genetic nature of a human disease, or on a clear phenotypic similarity between organisms. However, phenotypes arising from conserved genetic pathways may have diverged so far that their homology is unrecognizable. A number of methods have been developed to derive useful information from such cases, as well as to facilitate the synthesis of data from multiple species (Figure 2). These methods have the added benefits of identifying both novel human disease genes and appropriate model systems for studying those genes. A common principle of these methods is to group genes together by some criterion that reports on function (“statistical association”). These groupings then enable the statistical inference of novel disease-associated genes. Methods vary in way that they group genes and associate them with phenotypes (Figure 2). Below, we highlight some of the most common methods, and some recent innovations in this area.
Figure 2.
Components of statistical linking methods for rationally identifying human disease genes.
Phylogenetic profiles
Genes with linked function tend to have similar patterns of presence/absence across species, and this presence/absence vector is termed a “phylogenetic profile” (Figure 2A). Phylogenetic profiling allows the search for candidate genes which may have co-evolved with the disease-linked gene, and be involved in the same disease-causing process. Inferring gene functions by their phylogenetic profile, as proposed by Pellegrini et al. 1999, is not new [58, 59 and 60], however, these methods are increasingly being applied to the genetics of human disease [61**]. Recent improvements to this technique, such as using orthologous groups of genes, weighting by species divergence, and ancestral state inference, have increased the power of phylogenetic profiling methods, and by extension their application to candidate gene discovery [62**, 63**, 64 and 65]. As one recent example, Dey and colleagues applied phylogenetic profiling to discover genes involved in ciliary and centrosomal defects [62**].
Gene set overlap approaches
Gene set overlap methods determine if two groups of phenotype-associated genes in different species significantly share (orthologous) members (Figure 2B). These methods employ a statistical model to test the significance (commonly, the hypergeometric probability) of two phenotype-associated groupings from two species sharing a set of orthologous genes. This has proven to be an effective approach for identifying extremely divergent model phenotypes which employ the same genetic pathways involved in human diseases [27 and 66], such as, for example, the identification of a plant model of human Waardenburg syndrome [66]. Because these phenotypes employ orthologous genetic mechanisms, they have been termed “phenologs” [66]. The original approach inferred pairwise phenologs [66], but has been subsequently improved upon by extension to multiple species [27]. The success of this extension indicates an important fact: more comparative data means more inferential power for discovering novel genes associated with human disease.
Involvement in a particular phenotype is not the only way to classify genes in overlap-based studies. Korcsmáros et al. 2011 used pathway annotations in model species to predict more complete, integrated human signaling pathways [67]. Increasingly, multiple sources of data are being combined in databases [68 and 69*] that allow comprehensive comparison of overlap between gene sets.
Network approaches
Gene network-based approaches use networks to provide statistical frameworks for inferring new gene functions or disease associations. A network is first built from interactions between genes (or their encoded proteins); in a comparative gene-discovery framework, these interactions may be experimentally derived from multiple species. The resulting network can then be used to propagate information from genes (network nodes) whose function is known, to genes of unknown function (Figure 2C, [70]), using the interactions (network edges) to functionally annotate new genes [71, 72]. In principle, edges in the network can incorporate interaction data gleaned from any data source or species, and are therefore often a preferred method of generating consensus annotations. Many kinds of functional annotations can be propagated and therefore predicted, including new gene annotations [73] and disease gene associations, such as might arise from genome-wide association scans, e.g. as shown for Crohn’s disease using the human gene network HumanNet [71]. There are many methods used for information propagation (reviewed in depth in Wang and Marcotte 2010 [70]). Networks have also been constructed from genetic interactions, as might be gleaned from, for instance, double deletion screens (74), and used to find new genetic modifiers [75**]. Many gene networks are now available online that combine evidence from different interaction types, enabling the use of network-based inferences in most major model organisms [76, 77, 78*, 79 and 80**].
In one particularly interesting recent application of gene networks, the networks are not just used for candidate gene prioritization or annotation. Vidal and colleagues have suggested that disease phenotypes can be viewed as disruptions between interacting genes within the network structure (“edgetics”) [81, 82 and 83**], suggesting a wider use for gene networks in human disease research in guiding the disruption of only some, but not all, of a given gene’s interactions to affect a specific biological outcome.
RECENT APPLICATIONS
In reviewing the methods above, we have focused on the discovery of novel genes associated with human disease. One ultimate goal of these studies is to identify novel therapeutic agents that ameliorate the disease, which can be a challenge even when the target is known. We briefly highlight three recent studies that use the methods outlined above to identify novel drugs that target cancers, neurodegenerative diseases, and parasites (Figure 3).
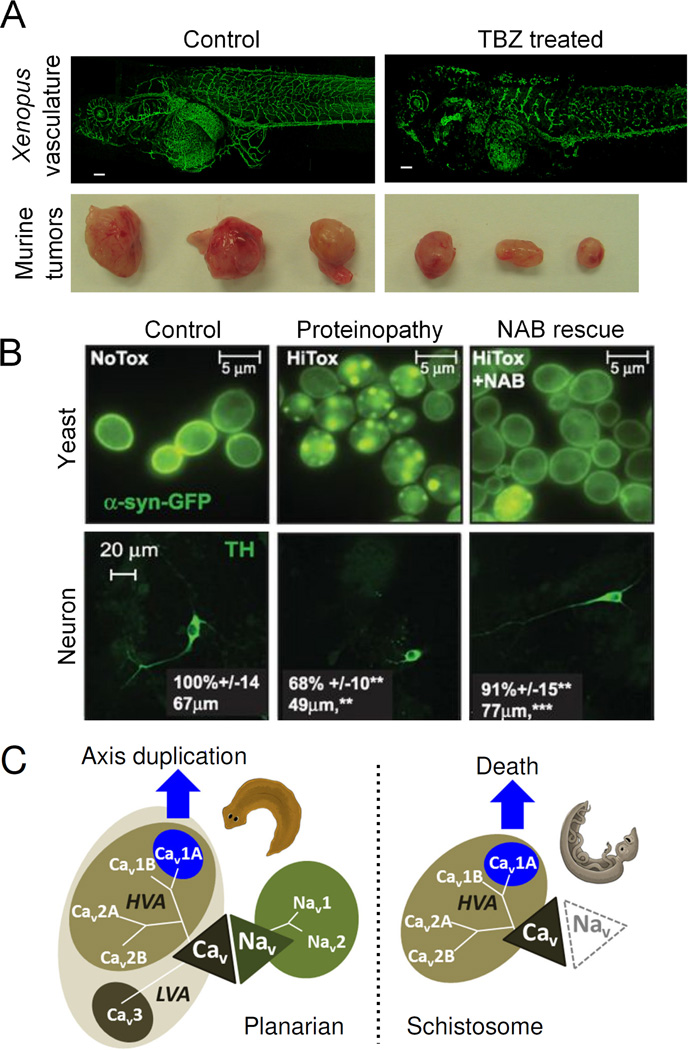
Figure 3.
Recent applications of statistical linking methods to identify and treat human disease. A. Cha et al. 2012 used overlap between vasculature genes and genes linked to antifungal sensitivity in yeast to identify TBZ as a novel vascular disrupting agent. TBZ disassembled vasculature in Xenopus embryos (top panels) and slowed human fibrosarcoma tumor growth in mice (bottom panels). Adapted from Cha et al. 2012 [84]. B. Tardiff et al. 2013 overexpressed α-synuclein and screened for phenotype rescuing compounds. One such compound, NAB, in turn ameliorated neuronal proteinopathies in worm neurons. Adapted from Tardiff et al. 2013 [88]. C. Chan et al. 2014 identified divergent phenotypes in planarians and schistosomes in response to the small molecule PZQ. Cav, voltage-gated calcium channel; Nav, voltage-gated sodium channel; HVA, high-voltage activated; LVA, low-voltage activated. Adapted from Chan et al. 2014 [90].
One of the predictions of the original phenolog study [66] was a yeast gene set that models vertebrate angiogenesis, a key dependency of tumor growth. Cha et al. 2012 [84**] used prior information about gene-drug genetic interactions between the yeast pathway and a variety of small compounds [85] to prioritize drugs that might block blood vessel growth. They identified thiabendazole (TBZ) as a candidate angiogenesis inhibitor, and found that not only did it indeed prevent vascularization in Xenopus embryos, but it disrupted pre-existing immature vasculature and slowed fibrosarcoma tumor growth in a mouse model, making it the first such vascular disrupting agent with FDA approval for human use (here, for its antifungal activity). TBZ illustrates that guilt-by-association approaches can be predictive, even between yeast and vertebrates.
Yeast are especially useful for high-throughput drug and genetic screens, as shown by recent work on the protein α-synuclein. This protein is associated with Parkinson's disease and related neurodegenerative diseases, termed synucleinopathies [86]. Susan Lindquist and colleagues used drug screens to identify compounds that inhibited aggregation of the protein α-synuclein exogenously expressed in yeast [87]. They identified a class of compounds called N-Aryl Benzimidazoles (NABs) that inhibit α-synuclein aggregation in yeast cells and animal neurons [88]. They further utilized the genetic tractability of yeast to identify pathways affected by α-synuclein [89**], suggesting potential new therapeutic avenues for synucleinopathies.
Whereas these two comparative approaches were applied to understand human genetic disease, the same approaches can also inform on infectious disease. Chan et al. 2014 [90**] used planarians, a model platyhelminth worm, to identify the target of the drug praziquantel (PZQ), which kills schistosomes, the pathogenic platyhelminths that cause schistosomiasis, but whose molecular target is unknown. Remarkably, while PZQ kills schistosomes, it causes an axis-duplication phenotype in planarians. This axis duplication phenotype, unlike death, can be explored using RNAi screens, readily applied in planarians. Using this method, Chan et al. identified the cellular basis of the axis duplication phenotype and also identified novel gene targets, as well as new compounds that phenocopy the effect PZQ and are therefore candidate anti-schistosomal agents.
OUTLOOK FOR THE FUTURE
We have touched on recent work using comparative approaches to relate human phenotypes to those in model organisms. We expect opportunities for connecting human genetics with the genetics of non-model organisms to increase considerably over the near time. In particular, data are increasingly available on human genetic variation, including familial inheritance, most recently across the Icelandic population 91], and increasingly provide a reference of human genetic variation for comparative approaches. It also seems a safe bet that the capacity for high-throughput CRISPR screens will dramatically increase known gene-phenotype associations from ever more diverse organisms, including non-traditional models. These developments will only increase in power and accuracy of comparative genomic approaches, and going forward, such methods will serve as a foundation to discover trends across life that point to the cause and treatment of human disease.
Acknowledgments
This work was supported by a fellowship from the N.I.H. to B. J. L. and grants from the N.I.H., N.S.F., C.P.R.I.T., and the Welch Foundation (F-1515) to E.M.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gould SJ, Vrba ES. Exaptation-A Missing Term in the Science of Form. Paleobiology. 1982;8:4–15. [Google Scholar]
- 2.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 3.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 4.Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giaever G, Nislow C. The yeast deletion collection: a decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spirek M, Benko Z, Carnecka M, Rumpf C, Cipak L, Batova M, Marova I, Nam M, Kim D-U, Park H-O, et al. S. pombe genome deletion project: an update. Cell Cycle. 2010;9:2399–2402. doi: 10.4161/cc.9.12.11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. The knockout mouse project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. The authors review the impact of large-scale mutagenesis of proteins on the study of protein properties, and the simulation of human disease states.
- 12.Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold CN, Xia Y, Lin P, Ross C, Schwander M, Smart NG, Müller U, Beutler B. Rapid identification of a disease allele in mouse through whole genome sequencing and bulk segregation analysis. Genetics. 2011;187:633–641. doi: 10.1534/genetics.110.124586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014;15:662–676. doi: 10.1038/nrg3745. [DOI] [PubMed] [Google Scholar]
- 16. Skelly DA, Merrihew GE, Riffle M, Connelly CF, Kerr EO, Johansson M, Jaschob D, Graczyk B, Shulman NJ, Wakefield J, et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res. 2013;23:1496–1504. doi: 10.1101/gr.155762.113. The authors profile 14,000 phenotypic traits of diverse strains of S. cerevisiae, opening the door to prediction of yeast phenotypes.
- 17.Freddolino PL, Goodarzi H, Tavazoie S. Revealing the genetic basis of natural bacterial phenotypic divergence. J. Bacteriol. 2014;196:825–839. doi: 10.1128/JB.01039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat. Rev. Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 19.Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc. Natl. Acad. Sci. U. S. A. 2005;102:19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HYK, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sozzani R, Benfey PN. High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biol. 2011;12:219. doi: 10.1186/gb-2011-12-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayfield JA, Davies MW, Dimster-Denk D, Pleskac N, McCarthy S, Boydston EA, Fink L, Lin XX, Narain AS, Meighan M, et al. Surrogate genetics and metabolic profiling for characterization of human disease alleles. Genetics. 2012;190:1309–1323. doi: 10.1534/genetics.111.137471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dunham MJ, Fowler DM. Contemporary, yeast-based approaches to understanding human genetic variation. Curr. Opin. Genet. Dev. 2013;23:658–664. doi: 10.1016/j.gde.2013.10.001. The authors review yeast as a platform for the systematic study of human variation and disease.
- 24.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT Mouse Genome Database Group. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic acids res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids res. 2011;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods JO, Singh-Blom UM, Laurent JM, McGary KL, Marcotte EM. Prediction of gene-phenotype associations in humans, mice, and plants using phenologs. BMC Bioinformatics. 2013;14:203. doi: 10.1186/1471-2105-14-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gkoutos GV, Schofield PN, Hoehndorf R. Computational tools for comparative phenomics: The role and promise of ontologies. Mamm. Genome. 2012;23:669–679. doi: 10.1007/s00335-012-9404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aberer K, Choi K-S, Noy N, Allemang D, Lee K-I, Nixon L, Golbeck J, Mika P, Maynard D, Mizoguchi R, et al., editors. The Semantic Web. Berlin Heidelberg: Springer; 2007. [Google Scholar]
- 31.Gkoutos GV, Green ECJ, Mallon A-M, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel Ma. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelman G, Fernandes JS, Bastiani Ca, Yook K, Sternberg PW. Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community. BMC Bioinformatics. 2011;12:32. doi: 10.1186/1471-2105-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GCM, Brown DL, Brudno M, Campbell J, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CL, Eppig JT. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: A whole-phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smedley D, Oellrich A, Köhler S, Ruef B, Westerfield M, Robinson P, Lewis S, Mungall C. PhenoDigm: Analyzing curated annotations to associate animal models with human diseases. Database. 2013;2013:1–11. doi: 10.1093/database/bat025. Phenodigm is a platform for semantic linking of model organism phenotypes to human disease phenotypes, making it particularly useful for finding models of diseases where the genetic basis is unknown.
- 39.Tassy O, Pourquié O. Manteia, a predictive data mining system for vertebrate genes and its applications to human genetic diseases. Nucleic Acids Res. 2014;42:D882–D891. doi: 10.1093/nar/gkt807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodenreider O, Burgun A. A framework for comparing phenotype annotations of orthologous genes. Stud. Health Technol. Inform. 2010;160:1309–1313. [PMC free article] [PubMed] [Google Scholar]
- 41.Köhler S, Doelken SC, Ruef BJ, Bauer S, Washington N, Westerfield M, Gkoutos G, Schofield P, Smedley D, Lewis SE, et al. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research. 2013;2:30. doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson PN, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 44.Koonin EV. Paralogs and mutational robustness linked through transcriptional reprogramming. Bioessays. 2005;27:865–868. doi: 10.1002/bies.20296. [DOI] [PubMed] [Google Scholar]
- 45.Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Zhang J. The Ortholog Conjecture Is Untestable by the Current Gene Ontology but Is Supported by RNA Sequencing Data. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohno S. Evolution by Gene Duplication. Berlin Heidelberg: Springer; 1970. [Google Scholar]
- 48.Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- 49.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 50. Sonnhammer E, Gabaldón T, Wilter Sousa da Silva A, Martin M, Robinson-Rechavi M, Boeckmann B, Thomas P, Dessimoz C. Big Data and Other Challenges in the Quest for Orthologs. Bioinformatics. 2014;30:2993–2998. doi: 10.1093/bioinformatics/btu492. Written by the Quest for Orthologs Consortium, this review highlight the state of the art for the accurate characterization of orthologs across species, which forms the foundation for many comparative analyses. They also introduce new community standards, such as benchmarking datasets and formats.
- 51.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dessimoz C, Gabaldón T, Roos DS, Sonnhammer ELL, Herrero J. Toward community standards in the quest for orthologs. Bioinformatics. 2012;28:900–904. doi: 10.1093/bioinformatics/bts050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pryszcz LP, Huerta-Cepas J, Gabaldón T. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2011;39:e32. doi: 10.1093/nar/gkq953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maher MC, Hernandez RD. Rock, Paper, Scissors: Harnessing Complementarity in Ortholog Detection Methods Improves Comparative Genomic Inference. G3: Genes|Genomes|Genetics. 2015;5:629–638. doi: 10.1534/g3.115.017095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boussau B, Szöllosi GJ, Duret L, Gouy M, Tannier E, Daubin V. Genome-scale coestimation of species and gene trees. Genome Res. 2013;23:323–330. doi: 10.1101/gr.141978.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y-C, Rasmussen MD, Bansal MS, Kellis M. TreeFix: statistically informed gene tree error correction using species trees. Syst. Biol. 2013;62:110–120. doi: 10.1093/sysbio/sys076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szöllősi GJ, Tannier E, Daubin V, Boussau B. The inference of gene trees with species trees. Syst. Biol. 2015;64:e42–e62. doi: 10.1093/sysbio/syu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 60.Marcotte EM. Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol. 2000;10:359–365. doi: 10.1016/s0959-440x(00)00097-x. [DOI] [PubMed] [Google Scholar]
- 61. Maxwell EK, Schnitzler CE, Havlak P, Putnam NH, Nguyen A-D, Moreland R, Baxevanis AD. Evolutionary profiling reveals the heterogeneous origins of classes of human disease genes: implications for modeling disease genetics in animals. BMC Evol. Biol. 2014;14:212. doi: 10.1186/s12862-014-0212-1. The authors examine phylogenetic profiles of human disease genes to characterize the evolutionary trait of genes involved in disease. They suggest that model organism selection should be done on a disease-by-disease basis, and informed by evolutionary history of the target disease genes.
- 62. Dey G, Jaimovich A, Collins SR, Seki A, Meyer T. Systematic Discovery of Human Gene Function and Principles of Modular Organization through Phylogenetic Profiling. Cell Rep. 2015;10:993–1006. doi: 10.1016/j.celrep.2015.01.025. The authors apply phylogenetic profiling to groups of orthologous genes in order to predict involvement of uncharacterized genes in functional modules. They predict and experimentally validate novel WASH complex interactors and cilia/basal components.
- 63. Tabach Y, Golan T, Hernandez-Hernandez a, Messer aR, Fukuda T, Kouznetsova a, Liu JG, Lilienthal I, Levy C, Ruvkun G. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol Syst Biol. 2013;9:692. doi: 10.1038/msb.2013.50. The authors determine that genes associated with a disease will have similar phylogenetic profiles. They used this approach to identify co-factors of disease associated transcription factors.
- 64.Liebeskind BJ, Hillis DM, Zakon HH. Convergence of ion channel genome content in early animal evolution. Proc. Natl. Acad. Sci. U. S. A. 2015;112(8):E846–E851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, Omilian AR, Oakley TH. Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol Biol. 2010;10:123. doi: 10.1186/1471-2148-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korcsmáros T, Szalay MS, Rovó P, Palotai R, Fazekas D, Lenti K, FarkasIllé IJ, Csermely P, Vellai T. Signalogs: Orthology-based identification of novel signaling pathway components in three metazoans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ. GeneWeaver: A web-based system for integrative functional genomics. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hwang S, Kim E, Yang S, Marcotte EM, Lee I. MORPHIN: A web tool for human disease research by projecting model organism biology onto a human integrated gene network. Nucleic Acids Res. 2014;42:1–7. doi: 10.1093/nar/gku434. Integrating gene set overlap and network analysis, MORPHIN takes a set of model organism genes as input, and uses an orthology-based projection of those genes onto a human genome network to suggest relevant human diseases.
- 70.Wang PI, Marcotte EM. It’s the machine that matters: Predicting gene function and phenotype from protein networks. J. Proteomics. 2010;73:2277–2289. doi: 10.1016/j.jprot.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hwang S, Rhee SY, Marcotte EM, Lee I. Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network. Nat. Protoc. 2011;6:1429–1442. doi: 10.1038/nprot.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang PI, Hwang S, Kincaid RP, Sullivan CS, Lee I, Marcotte EM. RIDDLE: reflective diffusion and local extension reveal functional associations for unannotated gene sets via proximity in a gene network. Genome Biol. 2012;13:R125. doi: 10.1186/gb-2012-13-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wiley DJ, Juan I, Le H, Cai X, Baumbach L, Beattie C, D’Urso G. Yeast Augmented Network Analysis (YANA): a new systems approach to identify therapeutic targets for human genetic diseases. F1000Research. 2014;3:121. doi: 10.12688/f1000research.4188.1. The authors describe an approach which uses yeast genetics to identify gene networks relevant to a particular human disease. They demonstrate their approach by identifying and validating potential therapeutic targets for the treatment of muscular atrophy disorder.
- 76.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schmitt T, Ogris C, Sonnhammer ELL. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic Acids Res. 2014;42:D380–D388. doi: 10.1093/nar/gkt984. FunCoup 3.0 is a database of functional associations, distinguished by type of coupling interaction. A web interface allows the exploration of networks, and access to experimental data supporting network edges.
- 79.Lee I. Probabilistic functional gene societies. Prog. Biophys. Mol. Biol. 2011;106:435–442. doi: 10.1016/j.pbiomolbio.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 80. Wong AK, Krishnan A, Yao V, Tadych A, Troyanskaya OG. IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv486. Advance Access. The 2.0 update to IMP (Integrative Multi-species Prediction) incorporates functional prediction of human disease with updated integrated networks covering seven organisms, and allowing analysis of candidate gene sets in a network context.
- 81.Sahni N, Yi S, Zhong Q, Jailkhani N, Charloteaux B, Cusick ME, Vidal M. Edgotype: A fundamental link between genotype and phenotype. Curr. Opin. Genet. Dev. 2013;23:649–657. doi: 10.1016/j.gde.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong Q, Simonis N, Li Q-R, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. Widespread Macromolecular Interaction Perturbations in Human Genetic Disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. Using a variety of interaction assays, the authors profile thousands of human missense mutations. They find that disease-linked alleles commonly perturb protein-protein interactions.
- 84. Cha HJ, Byrom M, Mead PE, Ellington AD, Wallingford JB, Marcotte EM. Evolutionarily Repurposed Networks Reveal the Well-Known Antifungal Drug Thiabendazole to Be a Novel Vascular Disrupting Agent. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001379. The authors show that a conserved module used in yeast for cell wall maintenance is repurposed in vertebrates to regulated angiogenesis. An antifungal agent which targets this module causes disassembly of new blood vessels, suggesting an anti-tumor application
- 85.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St.Onge RP, Tyers M, Koller D, et al. The Chemical Genomic Portrait of Yeast: Uncovering a Phenotype for All Genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lashuel HA, Overk CR, Oueslati A, Masliah E. The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013;14:38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tardiff DF, Lindquist S. Phenotypic screens for compounds that target the cellular pathologies underlying Parkinson’s disease. Drug Discov. Today Technol. 2013;10:e121–e128. doi: 10.1016/j.ddtec.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, et al. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates α-synuclein toxicity in neurons. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tardiff DF, Khurana V, Chung CY, Lindquist S. From yeast to patient neurons and back again: powerful new discovery platform. Mov. Disord. 2014;29:1231–1240. doi: 10.1002/mds.25989. The authors present yeast as a model of synucleinopathies and a platform to study the cell biology of neurological disorders.
- 90. Chan JD, Agbedanu PN, Zamanian M, Gruba SM, Haynes CL, Day Ta, Marchant JS. “Death and Axes”: Unexpected Ca2+ Entry Phenologs Predict New Anti-schistosomal Agents. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003942. The authors use divergent outcomes from modulation of orthologous pathways in schistosomes and planarians to identify targets of the anti-parasite drug, PZQ.
- 91.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]