Abstract
Fatty acyl reductases (FARs) constitute an evolutionarily conserved gene family found in all kingdoms of life. Members of the FAR gene family play diverse roles, including seed oil synthesis, insect pheromone biosynthesis, and mammalian wax biosynthesis. In insects, FAR genes dedicated to sex pheromone biosynthesis (pheromone-gland-specific fatty acyl reductase, pgFAR) form a unique clade that exhibits substantial modifications in gene structure and possesses unique specificity and selectivity for fatty acyl substrates. Highly selective and semi-selective ‘single pgFARs’ produce single and multicomponent pheromone signals in bombycid, pyralid, yponomeutid and noctuid moths. An intriguing question is how a ‘single reductase’ can direct the synthesis of several fatty alcohols of various chain lengths and isomeric forms. Here, we report two active pgFARs in the pheromone gland of Spodoptera, namely a semi-selective, C14:acyl-specific pgFAR and a highly selective, C16:acyl-specific pgFAR, and demonstrate that these pgFARs play a pivotal role in the formation of species-specific signals, a finding that is strongly supported by functional gene expression data. The study envisages a new area of research for disclosing evolutionary changes associated with C14- and C16-specific FARs in moth pheromone biosynthesis.
Insects use sophisticated chemical communication systems, including behavior-modifying pheromones, and these systems reinforce intra- and interspecific reproductive isolation and thus play pivotal roles in the evolutionary process. The insect order Lepidoptera (moth and butterflies) contains the second largest number of species (approximately 180,000 species described to date), and their richness and diversity are linked to pheromone-based communication and sophisticated chemosensory systems1,2,3. In moths, chemical communication is the major factor in the premating isolation mechanism, and sex pheromone differences inhibit successful mating and disfavor reproductive success4. The female signal and male response are highly species specific, but the mechanism regulating concomitant changes in signals and responses has not been established. These changes must occur at the gene level, and it is important to identify and characterize the genes responsible for pheromone production and to thus provide conclusive proof regarding pheromone-driven evolution in Lepidoptera5,6. The pheromone race is generally described in moths, and the variability in chemical mating signals and responses is genetically determined and under stabilizing selection7,8,9. Studies over the last two decades have pinpointed that the epigenetic effect of pheromone-driven adaptive evolution is one of the major factors driving the successful diversification of Lepidopteran insects10. In moths, a few substitutions in critical amino acids in the key pheromone biosynthetic enzymes are sufficient to create a novel pheromone component11,12. Few studies have reported the genetic basis of pheromone divergence in insects due to a lack of information on the key pheromone biosynthetic enzymes and the absence of a model insect species.
Moth pheromone compounds are derived from saturated C10-C18 fatty acyl moieties that are modified in the pheromone gland (PG) through the addition of functional groups, such as alcohol, acetate ester, or aldehyde. The biosynthesis of pheromone bouquets involves specialized enzymes, including fatty acyl-CoA desaturases, fatty acyl-CoA reductases (FARs), alcohol oxidases and fatty alcohol acetyltransferases13. One of the key enzymes involved in the production of oxygenated functional groups is FAR, which catalyzes the reduction of fatty acyl-CoA precursors to the corresponding alcohols13. Many of the pgFARs reported to date are selective regarding the carbon chain length of the substrate and show reduced activity to substrate chains that are shorter or longer than those of the original precursors7,11,14,15,16,17,18. The current information reveals a single pgFAR that acts on several C14 or C16 acyls11,14,16,17,19. However, several moths, including Spodoptera species, utilize multicomponent C12, C14 and C16 alcohol and acetate ester pheromone blends. Of the thirty Spodoptera species, the pheromone mixtures identified from 18 species (see Table S1) are composed of a multicomponent blend of isomerically related mono- and di-unsaturated C12, C14, and C16 fatty alcohols and acetates20,21,22. In the PG, an intriguing question is how a single reductase enzyme system can act on several fatty acyls of various chain lengths and isomeric forms. The present study describes an extensive search of the Spodoptera exigua PG that revealed two active pgFAR candidates, and a heterologous gene expression study in yeast demonstrated that each FAR plays a specific role in the reduction step. The two pgFARs belong to a unique pgFAR subfamily of Lepidoptera and are expressed solely in the PG of S. exigua. One enzyme exhibits broad selectivity toward C14 fatty acyl compounds, and the other has high specificity toward C16 fatty acyl compounds. Our study provides the first description of the involvement of multiple pgFARs with different selectivities and preferences for C14 and C16 fatty acyl moieties in pheromone biosynthesis in moths.
Results
Cloning of FAR candidates
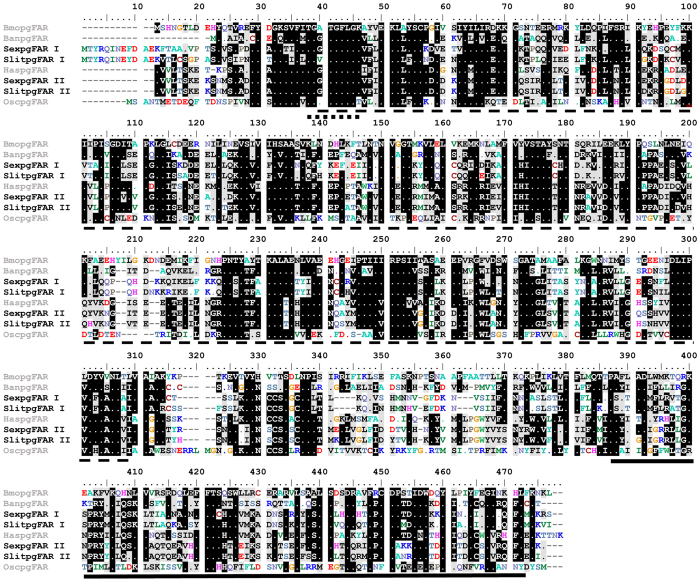
In the search for pgFARs, we identified eight FAR-like genes (FAR-I to FAR-VIII, GenBank Accession Nos. KF805977-KF805983 and KR781119-KR781121) based on their deduced amino acid sequences, which contain a conserved NAD(P)H-binding motif 23,24,25. BLASTx searches of the non-redundant (nr) protein database identified two candidate FARs from S. exigua that show significant identity with those of the nymphalid butterfly Bicyclus anynana (AGD98718) [50% amino acid (aa) identity and 67% similarity]26 and the noctuid moth Helicoverpa assulta (45% identity and 65% similarity)17. We named the candidates SexpgFAR I and SexpgFAR II. The full-length SexpgFAR I and SexpgFAR II cDNA transcripts contain 1,616 bp and 1,526 bp, respectively, and encompass open reading frames (ORFs) of 1,377 base pair (bp) and 1,362 bp, corresponding to proteins of 459 aa (SexpgFAR I) and 454 aa (SexpgFAR II) with predicted molecular weights of 51.7 and 51.4 kDa, respectively. SexpgFAR I and SexpgFAR II share 39% and 42% identity with the B. mori pgFAR16, 33% and 34% identity with the human FAR27, and 27% and 25% identity with the Jojoba FAR28, respectively (Table 1). The nucleotide and aa comparison of SexpgFAR I and SexpgFAR II with other moth pgFARs is provided in Table 1. SexpgFAR I and SexpgFAR II share 44% aa identity (<65% similarity) and more than 50% nucleotide identity (62% similarity) and possess highly conserved NAD(P)H-binding Rossmann fold domains (Fig. 1). The pgFARs contain a GXXGXX(G/A) motif at their N terminus that resembles the canonical ADP-binding domain, and this motif is likely involved in binding to NAD(P)H; these proteins also contain a C-terminal sterile domain and an epimerase domain (Fig. 1)23,24,25. Both pgFARs contain the classic YXXXK active-site motif of the short-chain dehydrogenase/reductase superfamily and are members of the short-chain dehydrogenase/reductase (SDR) family29. The results of a sequence analysis using the GenBank database identified both pgFARs as alcohol-forming long-chain FARs (E.C.1.2.1.50). The endoplasmic reticulum localization of SexpgFAR I and SexpgFAR II, which is a typical characteristic of pgFAR proteins, was identified using the Euk-mPloc 2.0 server30. We also cloned the pgFARs from S. littoralis (hereafter named SlitpgFAR I and SlitpgFAR II; GenBank Accession Nos. KR781119 and KR781120) by RACE-PCR cDNA amplification and subsequent Sanger sequencing. These pgFARs encompass ORFs of 1,438 and 1,362 bp, corresponding to proteins of 476 and 454 aa, respectively. SlitpgFAR I and SexpgFAR I share 84% aa identity, SlitpgFAR II and SexpgFAR II share 93% aa identity, and SlitpgFAR I and SlitpgFAR II share 44% aa identity. A pair-wise sequence comparison of SexpgFAR I and SlitpgFAR I with other noctuid pgFARs (Heliothis and Helicoverpa spp.) showed more than 44% aa identity, whereas a comparison of SexpgFAR II and SlitpgFAR II with the same FARs (Heliothis and Helicoverpa spp.) showed 73% aa identity (Table 1).
Table 1. Percentages of fatty acyl reductase (FAR) nucleotide and amino acid identity and similarity.
| FAR (GenBank Acc. Nos.) |
SexpgFAR I |
SlitpgFAR I |
SexpgFAR II |
SlitpgFAR II |
||||
|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | nt | aa | |
| BmorpgFAR (NM_001043502) | 51.30 (60.27) | 38.99 (61.87) | 50.46 (59.72) | 40.21 (62.37) | 53.04 (63.11) | 41.62 (66.38) | 52.23 (62.5) | 41.18 (66.05) |
| OscpgFAR E (EU817405) | 46.88 (55.93) | 32.46 (54.68) | 46.0 (55.48) | 35.06 (55.01) | 44.68 (55.32) | 31.71 (54.51) | 44.39 (55.09) | 31.71 (54.34) |
| OscpgFAR Z (AB506111) | 46.73 (55.82) | 30.93 (53.84) | 46.0 (55.48) | 33.98 (54.18) | 44.54 (55.20) | 30.17 (54.18) | 44.39 (55.09) | 30.39 (54.01) |
| YevpgFAR (ADD62439) | 51.70 (60.38) | 44.98 (64.04) | 50.88 (59.77) | 45.65 (64.71) | 50.81 (61.78) | 43.42 (66.55) | 49.85 (61.05) | 42.76 (65.71) |
| HaspgFAR (AFD04727) | 54.26 (62.45) | 44.73 (64.88) | 55.28 (63.23) | 45.61 (64.54) | 71.35 (77.66) | 74.22 (86.45) | 70.91 (77.32) | 73.34 (85.28) |
| HarpgFAR (AFD04728) | 54.45 (62.72) | 46.37 (65.05) | 55.4 (63.45) | 47.03 (64.71) | 70.91 (77.54) | 72.68 (86.12) | 71.06 (77.66) | 72.46 (84.94) |
| AsepgFAR (AGP26039) | 53.74 (61.94) | 44.10 (64.21) | 53.74 (61.94) | 44.54 (63.54) | 71.5 (77.66) | 68.72 (84.44) | 71.86 (77.93) | 69.16 (84.28) |
| Ban-wFAR 1 (AGD98718.1) | 55.63 (62.84) | 50.33 (66.55) | 54.57 (62.06) | 48.98 (65.21) | 56.98 (66.12) | 49.66 (72.9) | 55.48 (65.01) | 49.43 (72.07) |
| Ban-wFAR 2 (AGD98719) | 53.40 (60.27) | 45.96 (62.87) | 53.05 (60.27) | 45.45 (61.87) | 55.82 (61.94) | 48.89 (67.05) | 55.09 (61.39) | 48.23 (66.55) |
| HsaFAR (AY600449) | 45.72 (44.84) | 33.11 (46.48) | 45.21 (44.67) | 34.41 (46.32) | 46.15 (46.62) | 33.92 (50.33) | 46.0 (46.51) | 34.14 (49.66) |
| SchiFAR (AF149917) | 41.95 (45.68) | 27.45 (43.31) | 41.75 (45.73) | 27.92 (43.47) | 42.85 (48.18) | 24.88 (45.15) | 41.75 (47.35) | 24.22 (45.15) |
The pairwise sequence identity and similarity from the Clustal W multiple sequence alignments49 were calculated using the SIAS program (http://imed.med.ucm.es/Tools/sias.html).
Numbers in parenthesis are the similarity values. nt: nucleotide; aa: amino acid. SexpgFAR: Spodoptera exigua pgFAR; SlitpgFAR: S. littoralis pgFAR; BmorpgFAR: Bombyx mori pgFAR; OscpgFAR E: Ostrinia scapulalis (E-strain) pgFAR; OscpgFAR Z: Ostrinia scapulalis (Z-strain ) pgFAR; YevpgFAR: Yponomeuta evonymellus pgFAR; HaspgFAR: Helicoverpa assulta pgFAR; HarpgFAR: H. armigera pgFAR; AsepgFAR: Agrotis segetum pgFAR; Ban-wFAR 1: Bicyclus anynana FAR 1; Ban-wFAR 2: B. anynana FAR 2; HsaFAR: Homo sapiens FAR; SchiFAR: Simmondsia chinensis FAR.
Figure 1. Protein sequence alignment of SexpgFAR I and SexpgFAR II with B. mori (BmopgFAR) (GenBank accession no. BAC79426), O. scapularis (OscpgFAR) (ACJ06520) B. anynana (BanpgFAR) (AGD98719) and H. assulta (HaspgFAR) (JF709977).
The sequence alignments were computed in Clustal W and edited in BioEdit (v.7.2.5). The amino acids shaded in black or grey indicate identical residues or conserved substitutions. The FAR protein structure includes an N-terminal Rossmann fold (NAD(P)(+)-binding domain) (dashed line), the NADPH-binding motif (dotted line), and the sterile domain (thick dark line).
Tissue specificity studies and quantitative PCR identified a pheromone gland-specific FAR
Our RT-PCR results of the pgFAR gene expression pattern in different tissues from male and female S. exigua moths revealed that both SexpgFAR I and SexpgFAR II were expressed only in females and were exclusively expressed in the PG (Fig. 2). The PG-specific expression of both FARs suggests that these proteins may play a selective role in pheromone biosynthesis31. In contrast, all other FARs (FARI-FARVI) were found to be broadly distributed in various tissues, and their expression was not female-specific (data not shown). We performed quantitative PCR (qPCR) using cDNA synthesized from 2- to 3-day-old female moths, and our data showed that transcript abundances of both SexpgFAR I and SexpgFAR II were the same in female moths, indicating that both pgFARs exhibit a similar gene expression pattern in the PG (Fig. S1).
Figure 2. Tissue-specific expression of SexpgFAR I and SexpgFAR II by RT-PCR.
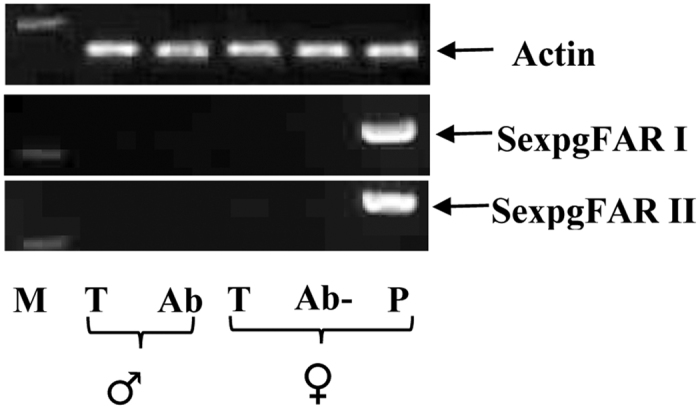
Amplicon sizes: SexpgFAR I, 262 bp; SexpgFAR II, 286 bp and actin, 217 bp. The 250 bp DNA marker (M) is shown. T: thorax; Ab: abdomen; P: pheromone gland; Ab-: abdomen without PG.
Phylogenetic analysis revealed that SexpgFAR I and SexpgFAR II are orthologous to Lepidopteran pgFARs
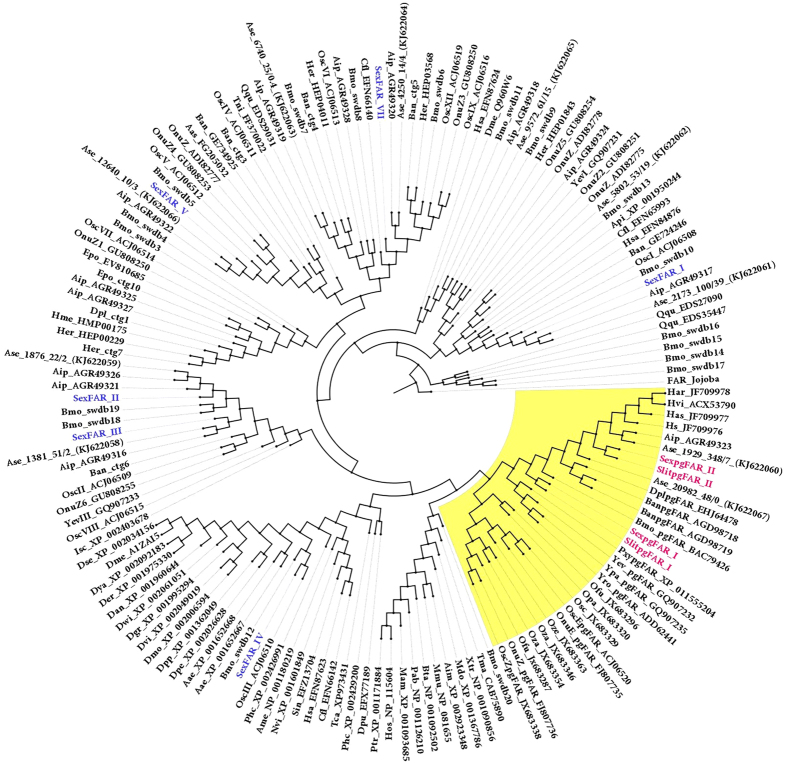
We constructed a phylogeny of Lepidopteran FARs using the FAR genes collected from the GenBank, genomic, transcriptomic and EST databases. Both pgFARs from S. exigua fall under the Lepidopteran pgFAR clade and were grouped with the pgFARs from B. mori16, Ostrinia spp.7,11,14, Yponomeuta spp.19, Agrotis spp.15,32 and Heliothis and Helicoverpa spp.17 (Fig. 3). Interestingly, SexpgFAR I formed a cluster with the B. anynana (BanpgFAR) and B. mori (BmopgFAR) pgFARs, which are reported to be C16-acyl-specific FARs16,26, and with the Danaus plexippus (DplpgFAR) and Plutella xylostella (PxypgFAR) pgFARs (Fig. 3), which are uncharacterized but predicted to be C16-acyl-specific FARs because the pheromone compounds are C16-acyl derivatives. SexpgFAR II was grouped with the noctuid pgFARs, which are known to be semi-selective C14 acyl-specific FARs (Fig. 3). In the gene tree, the Spodoptera pgFAR II was clustered in proximity to those pgFARs with broad specificity, whereas the Ostrinia pgFARs remain a separate group that is highly selective11. The orthologous SlitpgFAR I and SlitpgFAR II from S. littoralis were also grouped into the Lepidopteran pgFAR clade (Fig. 3).
Figure 3. Maximum likelihood (ML) tree of the fatty acyl reductase proteins from Lepidoptera.
The ML analysis was computed using MEGA (v.6.0)49. [The Jones-Taylor-Thornton (JTT) model for the ML heuristic search methods was the Nearest-Neighbor Interchange]. The Lepidopteran FAR-like sequences were retrieved from the GenBank, EST and TSA databases using BLASTx searches, and SexpgFAR I and SexpgFAR II were used as the query. The protein sequences were aligned using MUSCLE48. The branch containing Jojoba FAR28 was used as outgroup to root the tree. Other FAR-like proteins characterized from S. exigua are shown in blue. The pgFAR clade is highlighted in yellow. The GenBank accession numbers are indicated.
A yeast functional assay revealed that SexpgFAR I and SexpgFAR II prefer C16 and C14 fatty acyl substrates, respectively
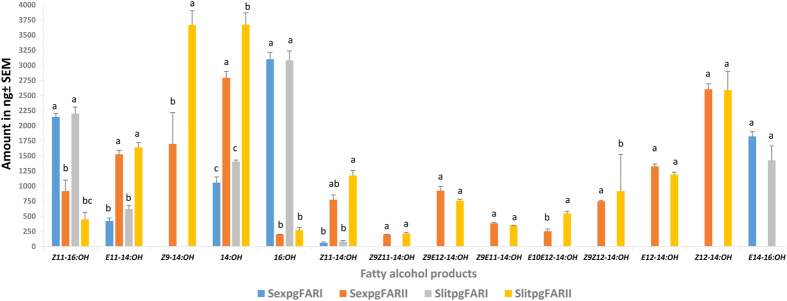
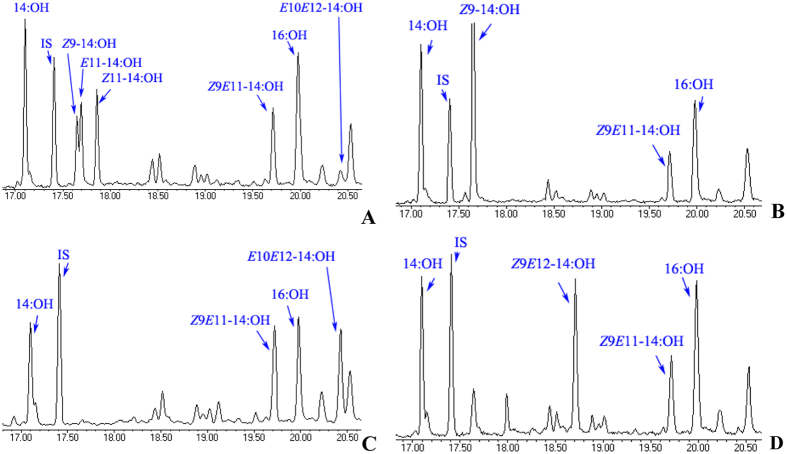
To address the functional role of the S. exigua pgFARs, we cloned the SexpgFAR I and SexpgFAR II ORFs into the pYES2.1 shuttle vector and transformed them into InvSc1 yeast cells. We also established InvSc1 yeast cells that were transformed with an empty vector (negative control) and B. mori pgFAR (positive control)16 to ensure that the production of alcohol in the yeast cells was due to expression of the recombinant pgFAR genes. We initially performed the functional assay with the SexpgFAR I- and SexpgFAR II-transformed yeast cells supplemented with 0.5 mM hexadecanoic acid methyl ester (C16:COOMe) and tetradecanoic acid methyl ester (C14:COOMe). Both pgFARs were found to be capable of reducing both the exogenous and endogenous C14 acyl and C16 acyl compounds that naturally occur in yeast (Fig. S2A). However, the SexpgFAR I-transformed yeast cells produced a significantly higher quantity of C16 fatty alcohol, whereas the SexpgFAR II-transformed yeast cells selectively reduced the amount of C14 fatty acids and produced significantly less C16 alcohol (Figs S2A and 4; Table 2). The SexpgFAR I-transformed yeast cells produced more than five fold greater amounts of 16:OH compared with the SexpgFAR II-transformed yeast cells, which is an even higher concentration of 16:OH than that produced by BmopgFAR (Fig. S2; Table 2). SexpgFAR II produced a significantly higher concentration of C14:OH (Figs S2A and 4). None of the corresponding C14 and C16 fatty alcohol compounds were observed in the negative control experiment (Fig. S2A–C). To determine whether both FARs reduce the (Z)-11-hexdecenoic acid (Z11-16:acyl), one of the minor pheromone precursors, we performed a functional assay with Z11-16:acyl, and both pgFAR-transformed yeast constructs produced Z11-16:OH. However, SexpgFAR I produced significantly more Z11-16:OH than SexpgFAR II (Figs S2B and 4). The SexpgFAR II-transformed yeast cells produced minor amounts of Z11-16:OH (6% of the corresponding production of saturated C16 alcohol), indicating reduced activity (Table 2). The majority of the unused Z11-16:COOMe from the yeast cells is retained in the hexane extract (Fig. S2B). We then tested a rare C16 pheromone precursor (E14-16:acyl, (E)-14-hexdecenoic acid), an intermediate pheromone precursor of the Asian corn borer (ACB) O. furnacalis that is not found in the glands of Spodoptera species (Table S1), to test the substrate selectivity of SexpgFAR I and determine whether it can reduce unsaturated C16-acyls other than Z11-16:acid. The SexpgFAR I-transformed yeast cells produced high levels of E14-16:OH, and these levels were even higher than those found with the positive control B. mori pgFAR (Figs S2C and 4). SexpgFAR II was unable to convert E14-16:COOMe to the corresponding alcohol (Fig. S2C). A summary of pgFAR activities based on the corresponding fatty alcohol derivatives produced from different fatty acyl substrates is provided in Table 2. These data suggest that Spodoptera pgFAR I has evolved the ability to reduce a broad set of C16 fatty acyls. We repeated the functional assay experiment with the SlitpgFAR I and SlitpgFAR II yeast constructs and obtained similar results (Fig. 4).
Table 2. Summary of the pgFAR activities toward different substrates.
| Substrate tested | SexpgFAR I | SlitpgFAR I | SexpgFAR II | SlitpgFAR II |
|---|---|---|---|---|
| Saturated fatty acids | ||||
| C14:Acyl/C16:Acyl | C16:Acyl | C16:Acyl | C14:Acyl | C14:Acyl |
| C14:Acyl | C14:Acyl | C16:Acyl | C16:Acyl | |
| Monounsaturated fatty acids | ||||
| Z11-16:Acyl | Z11-16:Acyl | Z11-16:Acyl | Z12-14:Acyl | Z9-14: Acyl |
| E11-14:Acyl | E14-16:Acyl | E14-16:Acyl | Z9-14: Acyl | Z12-14:Acyl |
| Z9-14:Acyl | E11-14: Acyl | E11-14: Acyl | E11-14: Acyl | E11-14:Acyl |
| E12-14:Acyl | Z11-14: Acyl | Z11-14: Acyl | E12-14:Acyl | Z11-14:Acyl |
| Z12-14:Acyl | Z11-16:Acyl | E12-14:Acyl | ||
| Z7-12:Acyl* | Z11-14: Acyl | Z11-16:Acyl | ||
| E14-16:Acyl | ||||
| Z5-10:Acyl* | ||||
| Z11-14:Acyl | ||||
| Diunsaturated fatty acids | ||||
| Z9Z11-14:Acyl | — | — | Z9E12-14:Acyl | Z9Z12-14:Acyl |
| Z9E12-14:Acyl | Z9Z12-14:Acyl | Z9E12-14:Acyl | ||
| Z9E11-14:Acyl | Z9E11-14:Acyl | Z9E11-14:Acyl | ||
| E10E12-14:Acyl | E10E12-14:Acyl | E10E12-14:Acyl | ||
| Z9Z12-14:Acyl | Z9Z11-14:Acyl | Z9Z11-14:Acyl | ||
The total fatty alcohol production from three biological replicates was quantified11, and based on the quantity of the fatty alcohols produced in yeast assays, the preferred substrates were arranged from highest to lowest in each row.
Not reducing to alcohol; —indicate no enzyme activity.
Figure 4. Analysis of the yeast construct functional assay showing the fatty alcohol production of SexpgFAR I and SexpgFAR II from S. exigua and SlitpgFAR I and SlitpgFAR II from S. littoralis.
The recombinant yeast strain S. cerevisiae was supplemented with 0.5 mM concentrations of various fatty-acyl methyl ester (FAME) substrates for 48 h at 30 °C, and the n-hexane extracts of the yeast pellets were subjected to GC-MS analysis. The fatty alcohol production was calculated using a previously described method11. The bars represent the SEMs of fatty alcohol production (in nanograms) of 3 biological replicates. The bars with different letters within each series are significantly different (P < 0.05).
SexpgFAR I is selective for Z11-16:acyl, and SexpgFAR II is involved in saturated and unsaturated C14 fatty acyl reduction
A series of yeast functional assay experiments was performed with different saturated and monounsaturated Spodoptera pheromone precursors. We supplemented the SexpgFAR I-transformed yeast cells with Z11-16:COOMe, (E)-11-tetradecenoic acid Me (E11-14:COOMe), (Z)-9-tetradecenoic acid Me (Z9-14:COOMe) and (Z)-11-tetradecenoic acid Me (Z11-14:COOMe) in separate experiments (single-substrate assay). Hexane extraction of the yeast pellets revealed large amounts of Z11-16:OH (Figs S2B and 4) and traces of E11-14:OH. None of the other substrates could be reduced to the corresponding alcohols (Fig. S3A–C). The supplementation of the SexpgFAR II-transformed yeast cells with the aforementioned compounds in single-substrate assay experiments converted all of the 14C fatty acid methyl ester (FAME) precursors into the corresponding alcohols (Fig. S3A–C). These studies demonstrated that SexpgFAR I is actively involved in the reduction of Z11-16:acyl (Fig. S2B) and that SexpgFAR II actively reduces all of the C14 mono fatty acyl moieties (Figs S2, S3A–C and 4). Similar functional assay experiments were performed with SlitpgFAR I and SlitpgFAR II, and these yielded similar results (Fig. 4).
To further test the selectivity of pgFAR II, we performed an additional functional assay with E- and Z12-14:acid, a rare pheromone precursor of ACB O. furnacalis33 that is not found in the PG of Spodoptera (Table S1). Interestingly, the SexpgFAR II-transformed yeast cells reduced (E)-12-tetradecenoic acid Me (E12-14:COOMe) and (Z)-12-tetradecenoic acid Me (Z12-14:COOMe) to the corresponding alcohols, and we observed significant Z12-14:OH production compared with all the other monounsaturated compounds (Figs 4 and S4). The results indicate the broad selectivity of pgFAR II toward C14 acyls. SexpgFAR I was unable to reduce the E- and Z12-14:acid compounds (Fig. S4). We repeated the same experiment with B. mori pgFAR, and, as expected, BmopgFAR was unable to reduce the E- and Z12-14:acids (data not shown), indicating the functional similarity of S. exigua pgFAR I and B. mori pgFAR. Both have evolved to reduce C16 acyl compounds.
We performed another set of experiments with di-unsaturated pheromone precursors. None of the corresponding fatty alcohol compounds were produced in the yeast SexpgFAR I functional assay (Fig. S3D–H). The SexpgFAR II-transformed yeast cells produced the corresponding alcohols from the di-unsaturated C14 fatty acid pheromone precursors (Fig. S3D–H). Similar results were obtained with SlitpgFAR I and SlitpgFAR II (Fig. 4).
Spodoptera pgFAR II is selective for Z9-14:acid and Z9E12-14:acid precursors
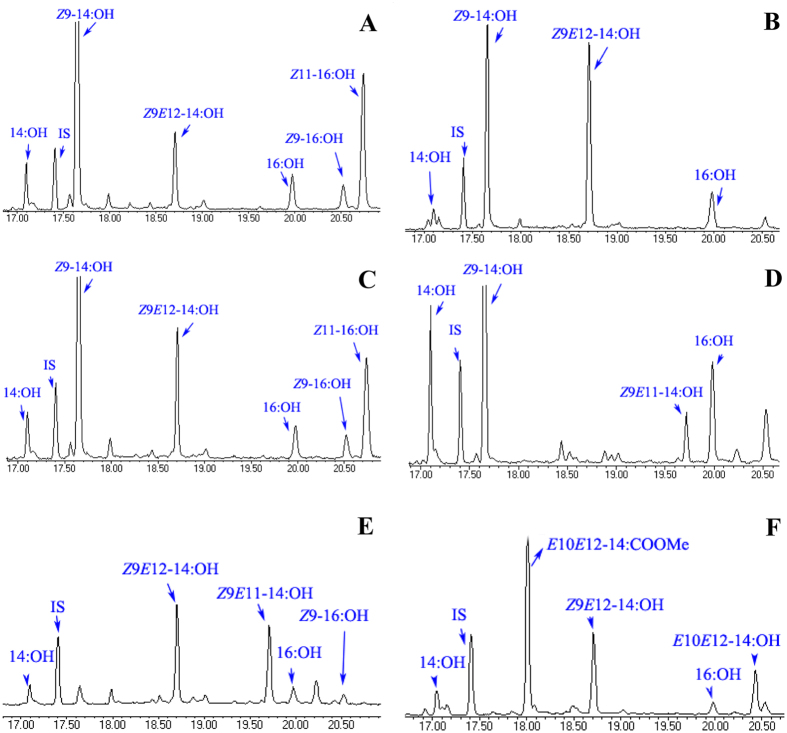
The hexane extract of the female S. exigua pheromone gland contained five major pheromone compounds22. However, the solid-phase microextraction (SPME) analysis revealed a mixture of (Z,E)-9,12-tetradecadienyl acetate (Z9E12-14:OAc), Z9-14:OAc, Z11-16:OAc and Z9-14:OH at a 34:40:4:22 ratio22. Thus, in the functional assay, we supplemented the SexpgFAR II-transformed yeast cells with the above-described mixture of three FAME compounds. Interestingly, we obtained 62% Z9-14:OH and 22% Z11-16:OH, but the yeast cells produced significantly less Z9E12-14:OH (16%) (Fig. 5A). We compared the production of Z11-16:OH with that observed in our previous single-substrate assay (see Fig. 4) and found that the same level of Z11-16:OH production by SexpgFAR II. Because we supplemented the yeast cells with higher quantities of Z11-16:COOMe (0.5 mM) in the single-substrate assays, SexpgFAR II, despite its low reductase activity to the Z11-16:acid, produced nearly half of the Z11-16:OH amount produced by SexpgFAR I (Fig. 4). The reason for this finding might be the availability of a high quantity of substrate [produced by elongation of the Z9-14:acid to the Z11-16:acid through endogenous yeast elongase activity34] to produce higher levels of Z11-16:OH in the S. exigua functional assay. However, in a yeast functional assay with an S. exigua blend, SexpgFAR II showed higher reductase activity toward the Z9-14:acid and produced the same amount of Z9-14:OH. These results indicate that SexpgFAR II is highly selective for the Z9-14:acid precursors and that SexpgFAR II activity might be highly restricted to C14 acyls in vivo. Alternatively, the introduction of the C16:acyl substrates (as in the case of the Z11-16:acid in our experiment) would reduce enzyme activity and decrease production of the di-unsaturated fatty alcohol (Fig. 5A). This experiment also supports the notion that SexpgFAR I is actively involved in Z11-16:OH synthesis in the PG. Additional functional assays were independently performed using ratios of Z9-14:COOMe to Z9E12-14:COOMe equal to 62:34 and 1:1. Both cases showed increased Z9-14:OH production (Fig. 5B), and we obtained fatty alcohol ratios of 52:48 and 74:26, respectively, which are slightly higher than the Z9-14:OH production level that was previously reported22. These results reiterate the selective nature of SexpgFAR II in systems supplemented with only C14 mono and di-unsaturated precursors. However, significantly higher amounts of Z9-14:OH were produced in the 1:1 functional assay (Fig. 5C). To further confirm the selectivity of SexpgFAR II toward Z9-14:acyls, we conducted another functional assay with a ratio of Z9-14:acyl to Z9E11-14:acyl (major pheromone precursor of S. littoralis)21 equal to 1:1 and obtained significantly higher Z9-14:OH production with a blend ratio of 73:27 (Fig. 5D). These experiments reiterate the fact that SexpgFAR II prefers the Z9-14:acid substrate.
Figure 5.
(A–F) Functional assay and GC-MS analysis of yeast (InvSc1) transformed with the SexpgFAR II construct and supplemented with different blends of 0.5 mM (total concentration) FAMEs: (A) Z9E12-14:COOMe, Z9-14:COOMe and Z11-16:COOMe (34:62:4), (B) Z9E12-14:COOMe and Z9-14:COOMe (34:62), and (C) Z9E12-14:COOMe, Z11-16:COOMe and Z9-14:COOMe (1:1), (D) Z9E11-14:COOMe and Z9-14:COOMe (1:1), (E) Z9E12-14:COOMe and Z9E11-14:COOMe (1:1) and (F) Z9E12-14:COOMe and E10E12-14:COOMe (1:1). Spodoptera pgFAR reduces the 14:acid, 16:acid and Z9-16:acid compounds that are naturally present in the yeast.
To identify the second most preferred substrate of S. exigua pgFAR, we conducted two separate multi-substrate assay experiments, one with concentration ratios of Z9E12-14:COOMe to Z9E11-14:COOMe equal to 1:1 and another with concentration ratios of Z9E12-14:COOMe and E10E12-14:COOMe (pheromone component of S. littoralis21) equal to 1.1. In both experiments, we obtained higher Z9E12-14:OH ratios, indicating that the Z9E12-14:acid is the second most preferred precursor for SexpgFAR II (Fig. 5E,F). Taken together, these results demonstrate that SexpgFAR II exhibits specificity for Z9-14:acid and Z9E12-14:acid precursors.
S. littoralis uses the pheromone blend of (Z,E)-9,11-tetradecadienyl acetate (Z9E11-14:OAc), Z9-14:OAc, E11-14:OAc, 14:OAc, Z11-14:OAc, and (E,E)-10,12-tetradecadienyl acetate (E10E12-14:OAc) at a ratio of 57:11:11:1:7:1421. SlitpgFAR II from S. littoralis was recently characterized, but the reductase specificity for shaping the pheromone blend has not been demonstrated18. Hence, we attempted to perform a functional assay in which SlitpgFAR II-transformed yeast cells were supplemented with the above-mentioned precursor FAME compounds at the indicated ratios. We obtained similar quantities of Z9E11-14:OH (28%), E11-14:OH (25%), Z11-14:OH (27%) and Z9-14:OH (20%) and a lower concentration of E10E12-14OH (Fig. 6A). The results indicate the different enzyme activities of the S. exigua and S. littoralis reductase systems toward di-unsaturated pheromone precursors. The former is selective for the Z9-14:acid, whereas the latter is selective for the Z9E11-14:acid.
Figure 6.
(A–D) Functional assay and GC-MS analysis of yeast (InvSc1) transformed with the SlitpgFAR II construct and supplemented with different ratios of 0.5 mM FAMEs: (A) Z9E11-14:COOMe, Z9-14:COOMe, E11-14:COOMe, 14:COOMe, Z11-14:COOMe, and E10E11-14:COOMe (57:11:11:1:7:14)21 (B) Z9E12-14:COOMe and Z9-14:COOMe (1:1), (C) Z9E11-14:COOMe and E10E12-14:COOMe (1:1), and (D) Z9E12-14:COOMe and Z9E11-14:COOMe (1:1). Spodoptera pgFAR essentially reduces the 14:acid, 16:acid and Z9-16:acid compounds that are naturally present in the yeast to alcohol.
Because we obtained selectivity in the S. littoralis pgFAR system for C14 mono acyls and Z9E11-14:acids, which contradicts the results reported by Carot-Sans et al.18, we performed additional substrate specificity assays with S. littoralis pgFAR II. In a functional assay with SlitpgFAR II-transformed yeast cells supplemented with methyl esters of the Z9-14:acid and Z9E11-14:acid at a ratio of 1:1, we obtained a significantly higher concentration of Z9-14:OH (Fig. 6B). We performed another functional assay with a ratio of Z9E11-14:COOMe to E10E12-14:COOMe equal to 1:1 in yeast selective media, and both compounds were reduced to the corresponding alcohols. The concentration of Z9E11-14:OH (S. littoralis major component) was similar to that of E10E12-14:OH (Fig. 6C). To confirm the similarity of the S. exigua and S. littoralis pgFAR activities, we performed a functional assay with SlitpgFAR II-transformed yeast cells supplemented with an equal concentration of Z9E12-14:COOMe (S. exigua major precursor) and Z9E11-14:COOMe (S. littoralis major precursor). S. littoralis pgFARII preferentially converted the Z9E12-14:acid to the corresponding alcohol (Fig. 6D).
SexpgFAR II is selective and semi-selective for C14 mono- and di-unsaturated precursors, respectively, and SexpgFAR I is highly selective for C16 acyls
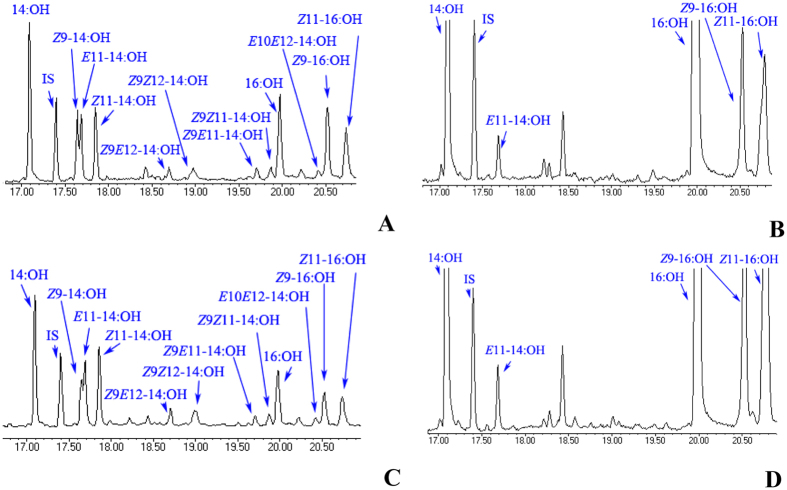
Among the 18 Spodoptera spp. pheromones identified, most species use Z9E12-14:OAc as a major pheromone compound, followed by Z9E11-14:OAc and E10E12-14:OAc (Table S1). Only four Spodoptera spp. (S. frugiperda, S. pectinicornis, S. praefica and S. triturate) exclusively use the C12 and C14 mono-unsaturated fatty acetate compounds as a pheromone (specifically, Z7-12:OAc and Z9-14:OAc). We performed a functional assay with SexpgFAR II-transformed yeast cells supplemented with equal concentrations of five mono-unsaturated and di-unsaturated compounds (Fig. 7) in 5 mL of selective media. Z9-14:OH, 14:OH, Z11-14:OH and E11-14:OH production was significantly increased compared with di-unsaturated alcohol production (Fig. 7A). We also performed a functional assay with SlitpgFAR II by supplementing yeast cells with equal concentrations of 10 precursor compounds and obtained the results similar to those obtained with SexpgFAR II (Fig. 7C). In another set of functional assays, SexpgFAR I-transformed yeast cells were supplemented with equal concentrations of all 10 compounds, and as expected, we obtained only 14:OH, Z11-16:OH and a trace amount of E11-14:OH (Fig. 7B). The same functional assay experiments were repeated with SlitpgFAR I-transformed yeast cells and yielded the same results (Fig. 7D). These findings indicate that the substrate specificities of pgFAR I in S. exigua and S. littoralis are similar.
Figure 7.
(A–D) Total ion chromatogram (TIC) showing the fatty alcohol products extracted from the yeast cells. The yeast (InvSc1) were transformed with the pgFAR construct and supplemented with a blend of equal concentrations (0.5 mM total) of Z11-16:COOMe, C14:COOMe, E11-14:COOMe, Z9-14:COOMe, Z11-14:COOMe, Z9Z11-14:COOMe, Z9E12-14:COOMe, Z9E11-14:COOMe, E10E12-14:COOMe and Z9Z12-14:COOMe in 5 mL of selective media. Spodoptera pgFAR reduces the 14:acid, 16:acid and Z9-16:acid compounds that are naturally present in the yeast. (A) SexpgFAR II, (B) SexpgFAR I, (C) SlitpgFAR II and (D) SlitpgFAR I.
The yeast functional assay (multi-substrate) results for SexpgFAR II also showed that all of the monounsaturated methyl esters were converted to alcohols at approximately the same level; however Z9-14:OH presented the highest production, followed by Z11-14:OH and E11-14:OH (Fig. 7A). We obtained lower quantities of the fatty alcohols derived from the di-unsaturated compounds (Fig. 7A). This study indicates that SexpgFAR II is selective for C14 and C16 mono-unsaturated fatty acyls. Thus, SexpgFAR II is semi-selective for reduction of the Z9E12-14:acid and highly selective for reduction of mono-unsaturated C14-fatty acids to alcohols. Nevertheless, Spodoptera pgFAR I is unable to reduce the C14 mono- and di-unsaturated pheromone precursors, with the exception of E11-14:COOMe, and is selective for C16 fatty acid compounds.
A yeast functional assay revealed that both SexpgFAR I and SexpgFAR II cannot reduce C10 and C12 fatty acid precursors
Among the 18 Spodoptera spp. reported, S. frugiperda, S. pectinicornis and S. praefica use saturated and mono-unsaturated C12 acetate esters as pheromones9. Through a single-substrate assay, we tested the substrate chain length preference of SexpgFAR I and SexpgFAR II by supplementing the yeast cells with 0.5 mM (Z)-5-decenyl acid Me (Z5-10:COOMe) and (Z)-7-dodecenyl acid Me (Z7-12:COOMe). None of these compounds were converted into the corresponding alcohols (results not shown). The results indicate that SexpgFAR I and SexpgFAR II are unable to reduce any fatty acyl substrates shorter than C14.
Discussion
We identified two active FARs that are exclusively expressed in the PG and exhibit specificity for C16 and C14 acyls, respectively, and demonstrate that both FARs are involved in the Spodoptera pheromone biosynthesis pathway to produce different precursor alcohol compounds. Our yeast functional assay data confirm that pgFAR I can primarily reduce C16 fatty acyl precursors, whereas pgFAR II was found to be involved in the reduction of a broad range of saturated and unsaturated C14 acyls. FAR could reduce neither C10 nor C12 fatty acyl precursors. The discovery of a C16-specific function of FAR I may not be consistent with the function of the previously identified FARs from corn borer moths7, small ermine moths19 and heliothine moths17 that produce a broad range of alcohols. However, the degree of substrate specialization and the close relationship between SexpgFAR I and B. anynana FAR26 led us to assume that the pheromone biosynthesis pathway is shared between moths and butterflies. Furthermore, SexpgFAR II, which is similar to all of the broad-range FARs mentioned above, is capable of reducing C14, adding to the degree of specialization, as observed with the production of Z9E12-14:OH in S. exigua and Z9E11-14:OH in S. littoralis. The enzymatic activity and substrate specificity identified in this study clearly suggest that the Spodoptera female sex pheromone components are produced via de novo common biosynthetic routes13 that involve both FARs (Fig. 8). We identified eight duplicate FAR genes from the pheromone gland of S. exigua, found that SexpgFAR I and SexpgFAR II belong to the Lepidoptera-specific pgFAR lineage and demonstrated that these two genes encode specialized pheromone biosynthetic reductases. A recent study of the evolution of FAR genes in plants and animals clearly indicated that insects have undergone large expansions relative to the other taxa examined, which is consistent with a hypothesis that FAR is a multigene family that evolved through the birth-death evolution35. Extensive studies on the evolutionary pattern of FAR genes in plants and animals show considerable losses and gains of novel functions as a result of gene duplication events, and insects gained a substantial number of FAR genes when they diverged from other animals35. There are two reductase genes present in the vertebrate genomes, whereas more than a dozen are present in Lepidoptera14,36. Among these genes, a sub-group of FARs gained the ability to convert palmitic acid (C16: acid) into hexadecanol (C16:OH). These FARs are involved in pheromone biosynthesis in moths16 and butterflies26. In moths, FAR expansions have facilitated the adaptive evolution of a variety of specialized functions involving the precursors that serve as substrates for reductase genes. In S. exigua and S. littoralis, the presence of mono- and di-unsaturated C14 and C16 fatty acyl precursors might prime the expansion of two pgFARs in these species.
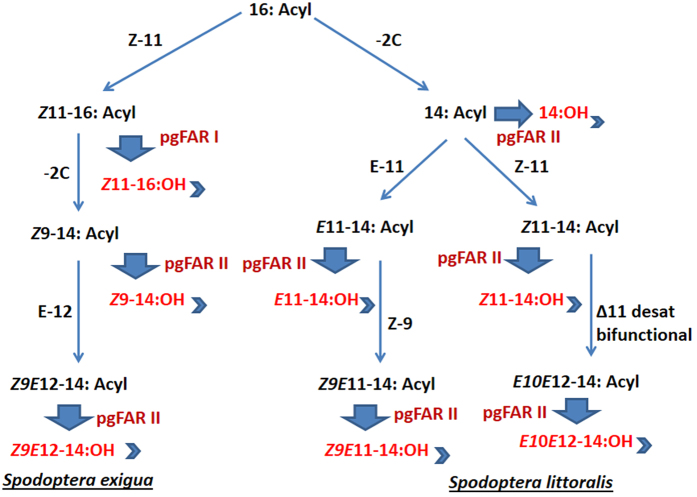
Figure 8. Spodoptera exigua and S. littoralis pheromone biosynthesis pathway for the production of pheromone precursors.
The de novo biosynthesis of all precursors starts from palmitoyl-CoA (16:acyl). In S. exigua, a Δ11 desaturase catalyzes the production of (Z)-11-hexadecenyl [(Z)-11–16:acyl], which undergoes one cycle of β-oxidation to produce (Z)-9-tetradecenyl [(Z)-9–14:acyl]22. A Δ12 desaturase catalyses (Z)-9–14:acyl to produce (Z,E)-9,12- tetradecadienyl acyl (Z9E12-14:acyl). In S. littoralis, 16:acyl undergoes one cycle of β-oxidation to produce myristoyl-CoA (14:acyl). A Δ11 desaturase acts on 14:Acyl to produce (E)- and (Z)-11-tetradecenoyl [(E) + (Z)-11–14:acyl]21. A Δ9 desaturase acts on (E)-11–14:acyl to produce (Z,E)-9,11- tetradecadienyl acyl (Z9E11-14:acyl), and a bifunctional Δ11 desaturase acts on (Z)-11–14:acyl to produce (E,E)-10,12- tetradecadienyl acyl (E10E12-14:acyl)50. Each compound undergoes a reduction step catalysed by pgFAR I and pgFAR II to produce the corresponding fatty alcohol derivatives. Finally, an acetyltransferase catalyses the conversion of the fatty alcohol precursors into the corresponding acetate esters (shown with an arrowhead).
Several pgFAR orthologs that are specific to C16 acyl precursors are reported to produce traces of C14 saturated and unsaturated fatty alcohols, as reported in B. mori16 and B. anynana26. Hence, we hypothesize that the production of trace amounts of C14:OH and E11-14:OH by SexpgFAR I may result from an ancestral feature that is capable of reducing C14 and C16 acyl precursors. However, the ancestral activity was less specialized and reduced C14:acyl and E11-14:acyl. To confirm this hypothesis, we performed multi-substrate functional assays with BmopgFAR-transformed yeast cells supplemented with 10 different precursors as a positive control (Fig. S5). Similarly to SexpgFAR I, BmorpgFAR was unable to reduce the di-unsaturated C14 fatty acid precursors. However, trace amounts of E11-14:OH, Z11-14:OH and a large amount of Z11-16:OH were observed in the yeast cell extract (Fig. S5). Another functional assay with a rare C16 precursor, E14-16:acid, revealed that SexpgFAR I and BmopgFAR but not SexpgFAR II can reduce the E14-16:acid. These results indicate the functional similarity of B. mori, S. exigua and S. littoralis FAR I, which are specialized to reduce C16 acid derivatives. The single-substrate functional assay confirmed that SexpgFAR II is involved in the reduction of a broad range of saturated and unsaturated C14 acyls and that SexpgFAR I could reduce only Z11-16:acyls. Many Spodoptera species use Z11-16:OAc as a minor pheromone compound (Table S1). Several orthologous pgFARs that are specific for C14 acyl precursors are reported to produce traces of C16 saturated and unsaturated fatty alcohols11,19. Therefore, the production of trace amounts of Z11-16:OH derivatives by SexpgFAR II may reflect an ancestral feature. These preadaptations of the reductase system have also been reported in O. furnacalis, which uniquely possesses a ∆12-tetradecenyl precursor as a consequence of the activity of ∆14 desaturases33,37; however, pgFAR can reduce E/Z11-16 acyls11.
The yeast functional assay results show that SexpgFAR II presents activity toward a wide range of fatty acyl precursors. Although we did not achieve the same blend ratio as was previously reported21, a preference of SlitpgFAR II toward the Z9E11-14:acid was observed, and the results contradict those published previously18. One reason for this discrepancy may be that the pheromone blend ratio reported by Munoz et al.21 involved a PG extract lipid analysis, and an SPME analysis of S. littoralis has not been performed. In contrast, an SPME analysis of S. exigua PG has revealed a ratio that differs from that obtained through a gland extract analysis22. Several insect pheromone blend ratios were recently revised through an SPME analysis (for example, Batrachedra amydraula)38,39. Hence, we reiterate that the selectivity of SlitpgFAR II favors the Z9E11-14:acid. In contrast to the reported PG extract ratio of E10E12-14:OAc (14%), we found significantly lower quantities of E10E12-14:OH (>4%) (Fig. 6A). The explanation for the SPME analysis also applies in this case. However, we cannot rule out the selectivity of desaturases and acetyltransferases in shaping the final pheromone blend in Spodoptera. Similarly to yponomeutid and heliothine pgFARs17,19, Spodoptera pgFAR II accepts a broad range of substrates, suggesting that their function in the PG evolved in the genus prior to species diversification. Hence, evolutionary divergence probably occurred upstream of the desaturases or downstream of the acetyltransferases. In this case, the production of a specific blend of pheromone compounds depends on the available fatty acyl precursors or the selectivity of the acetyltransferase. The acetyltransferase has not been characterized at the molecular level in any insect13,40,41,42. Additional comparative functional analyses of the desaturase, reductase and acetyltransferase activities must be conducted to prove this hypothesis.
The multi-substrate functional assay of SexpgFAR II with different blend ratios of Z9-14:COOMe and Z9E12-14:COOMe resulted in higher Z9-14:OH production, indicating the selectivity of the S. exigua pgFAR system. We obtained pheromone blend ratios of the major pheromone compounds that were similar to those previously reported22. Our multi-substrate functional assays proved that both Spodoptera pgFARs are selective for the Z9-14:acid, and S. exigua and S. littoralis exhibit a preference for its major precursors: Z9-14:acid, Z9E12-14:acid and Z9E11-14:acid, respectively. Of the 18 identified Spodoptera spp. pheromones, Z9-14:OAc is a major pheromone component, and 14 Spodoptera spp. use Z9E12-14:OAc in their pheromone blends (Table S1). Our functional assays highlight the fact that all of the present-day Spodoptera pgFARs have the ability to reduce substantial quantities of these two compounds, as demonstrated for S. exigua and S. littoralis in our study (Figs 5 and 6). Although S. littoralis does not use Z9E12-14:OAc as a pheromone compound, several studies have demonstrated the presence of this compound in the PG extract43,44. The results indicate that both S. exigua and S. littoralis are selective for Z9E12-14:acid among all di-unsaturated C14 fatty acids. We hypothesized the existence of a functional site or domains in the FARs that define this property, which is highly conserved among all Spodoptera pgFARs. Previous studies of heliothine pgFARs demonstrated a preference for Z9-14:acid compounds17. The functional ability of S. exigua pgFAR II to reduce Z9-14:acid might be an ancestral feature that is retained in noctuid moths. However, the ability of the Spodoptera pgFAR to reduce the Z9E12-14:acid (or Z9E11-14:acid) is a characteristic of this genus. Until now, no other Lepidoptera pgFARs have been reported to exhibit multifunctional activity toward mono- and di-unsaturated pheromone precursors7,11,14,15,16,17,19.
A single pgFAR with broad selectivity for C16 fatty acyl precursors with a distinct substrate specificity for E10Z12-16:acid has been reported in B. mori16. Highly selective pgFARs that are unable to reduce any precursors other than the natural pheromone precursors, E11-14:acid and Z11-14:acid, have been reported in O. nubilalis7. In contrast, the orthologous reductases from Yponomeuta spp., Agrotis spp. Heliothis and Helicoverpa spp. reduce a broad range of C12, C14 and C16 acyl precursors but present clear specificity for the natural C14 pheromone precursors15,17,19. In the present study, we identified two pgFARs in S. exigua and S. littoralis. pgFAR I in both species possesses similar functional activity, and its function might have evolved before radiation within the Spodoptera genus. We reiterate that the Spodoptera pgFAR I selectively recognizes the chain length of the substrates for pheromone biosynthesis, whereas pgFAR II accepts a broad range of substrates. pgFAR II is selective for the major pheromone precursors (Z9-14:acyl and Z9E12-14:acyl for S. exigua; Z9E11-14:acyl for S. littoralis), which unequivocally supports the hypothesis that the S. exigua and S. littoralis pgFARs play important roles in shaping the final pheromone blend ratio. Overall, our findings demonstrate that two biosynthetic pgFARs are involved in Spodoptera. pgFAR II exhibits broad selectivity and acts on C14 mono- and di-unsaturated precursors, and pgFAR I exhibits narrow selectivity and acts on Z11-16:acyl to produce the moth pheromone signals. The mechanism through which Spodoptera pgFARs coordinate the reduction of the mono- and di-unsaturated C14 and C16 precursors remains to be elucidated. In most Spodoptera species, the precursor and pheromone component ratios exhibit significant disparities (Table S1), and the pgFAR substrate preference is necessary to explain the production of species-specific pheromone blends. Our yeast functional assay revealed that both FARs cannot reduce C10 and C12 acyl precursors, which indicates that another potentially active FAR candidate may be involved in the reduction pathways in S. frugiperda. S. praefica and S. pectinicornis (see Table S1). Our study of the two pgFARs in Spodoptera will form a basis for the identification of FARs in other Spodoptera moths. Our major findings are that a subgroup of FARs (pgFAR I) dedicated to the reduction of C16:acid is functionally conserved in moth and butterfly lepidopteran lineages and that a multifunctional pgFAR II acts on the mono and di-unsaturated C14:acid precursors that are collectively involved in Spodoptera pheromone biosynthesis. Our study opens the door for a new area of research aiming to disclose the evolutionary changes associated with C14-specific and C16-specific FARs in moth pheromone biosynthesis.
Materials and Methods
Chemicals
The chemicals and authentic standard compounds mentioned in the supporting information (SI) were purchased from Pest Control of India Private Limited (Mumbai, India) and Pherobank (Netherlands). The compounds used for the functional assays were diluted in 95% ethanol (0.5 mM), and the standard compounds were diluted in n-hexane (250 ng/μL).
Insects
Spodoptera exigua and S. littoralis pupae were originally collected from a vegetable garden [Al-Kharj, (24.1500°N, 47.3000°E) Saudi Arabia], and laboratory cultures were maintained on an artificial diet45. Adult moths were collected from private properties for which permits were not required. S. exigua and S. littoralis are not endangered or protected species. The emerged females were separated daily prior to reaching scotophase, and their pheromone glands were dissected14.
PCR amplification and cloning of the pgFAR transcripts
We extracted the total RNA from the pooled pheromone glands of 0- to 3-day-old virgin S. exigua females using an RNeasy isolation kit (Qiagen). First-strand cDNAs were prepared using ArrayScript reverse transcriptase (Life Technologies). We created a dataset of FARs using EST, as previously reported14, and the degenerate primers are listed in Table S2. Briefly, 500 ng of the PG cDNAs was used to amplify the FARs, and the PCR product was gel purified (Promega), cloned into a pGEMt vector and transformed into chemically competent Escherichia coli JM109 cells (Promega). A total of 400 clones (100 for each degenerate PCR, see Table S2) was selected. The plasmid DNA was purified, and sequencing reactions were performed using a BigDye terminator kit v3.1 (Applied Biosystems) and subsequently analyzed on an ABI PRISM 3500 genetic analyzer (Life Technologies). The sequences were identified through BLASTx searches using the S. exigua FAR sequences as the queries. The 5′ and 3′ cDNA specimens were prepared with the SMART RACE amplification kit (Clontech) according to the manufacturer’s instructions. Gene-specific primers were designed (Table S2) and used in the PCR reactions to amplify the PCR products, which were gel-purified and ligated into the pGEM-T easy vector (Promega). The ligation products were transformed into Escherichia coli JM109 chemically competent cells (Promega). Plasmid DNA specimens were purified, sequenced, and analyzed using an ABI 3500 genetic analyzer. Separate sequencing reactions were established for primer walking to obtain the full-length pgFAR sequences, and the data were compiled using the BioEdit program46.
Tissue-specific expression analysis
The total RNA from the entire thorax and abdomen (without PG) of 3-day-old female S. exigua moths (n = 3) and from the entire thorax and abdomen of male moths (n = 3) was prepared using an RNeasy mini kit (Qiagen). The first-strand cDNAs were synthesized from 1 μg of the total RNA using an ArrayScript RT kit (Life Technologies). Touchdown PCR was performed using 200 ng of the cDNA template and primer sets designed for the specific amplification of each FAR-like sequence (Table 2). The conditions were 94 °C for 1 min, 10 cycles of 94 °C for 30 s, 60 °C for 1 min (reduced by 1 °C/cycle), and 72 °C for 1 min, 20 cycles of 94 °C for 30 s, 50 °C for 1 min and 72 °C for 1 min, and a final extension at 72 °C for 7 min. The PCR products were electrophoretically separated on a 2% agarose gel.
Analysis of the pgFAR transcript abundance through a quantitative PCR analysis
Three PGs were dissected from 2- to 3-day-old female S. exigua moths at mid-scotophase. Total RNA was extracted (Qiagen), and cDNAs were synthesized using MultiScribe™ Reverse Transcriptase (Life Technologies). Three biological cDNA samples were prepared, and quadruplicate 25-μL quantitative PCR reactions for each transcript (SexpgFAR I, SexpgFAR II and β-actin) were performed on an ABI 7500 instrument (Life Technologies). The template was 100 ng of PG cDNA, and SexpgFAR I, SexpgFAR II, and β-actin primers at a concentration of 10 μM were used (Table S2) (Tm = 60 °C). Premier Primer software (Biosoft International) and the Power SYBR Green qPCR MasterMix were used (Life Technologies). The amplification efficiency (E) of each primer pair was initially assessed from serial dilutions of the cDNAs, and the raw cycle threshold values (Ct) were averaged from the quadruplicate reactions. The β-actin RNA was used for normalization, and all relative expression levels were calculated using the ABI 7500 program (Life Technologies). The fluorescence background baselines and amplification thresholds were calculated automatically. The fold changes in expression levels were calculated against a normalized (β-actin) sample and subjected to one-way analysis of variance (ANOVA). Means were compared using Tukey’s HSD (honestly significant difference) test in the SPSS program47.
Phylogenetic analysis
The S. exigua and S. littoralis FAR nucleotide sequences were used as queries for BLASTx searches in the GenBank database, and the nucleotide and amino acid sequences from different insect species were retrieved to construct a phylogenetic tree. Similarity analyses of the DNA and protein sequences were conducted, and multiple-sequence alignment was performed using the MUSCLE program48. A maximum likelihood analysis was performed, and a dendrogram was constructed using MEGA v6.049.
Functional gene expression and assay
A yeast expression system (Invitrogen) was used to functionally express the S. exigua and S. littoralis pgFAR transcripts according to a previously described protocol7,19. We used pYES2.1/V5-His TOPO (Invitrogen) as a shuttle vector for the pgFAR transcripts, and the plasmids were transformed into Saccharomyces cerevisiae (INVSc1 strain, Invitrogen) according to the protocol listed in the SI Materials and Methods. For each test sample in the functional assay, we included a negative control (vector only) and a positive control (B. mori pgFAR)16. All oligonucleotide primers and a synthetic gene representing the ORF of B. mori pgFAR were purchased from Integrated DNA Technologies (Belgium).
pgFAR substrate preference analysis
The pgFAR yeast transformants were supplemented with different ratios of the precursors that were added at a total concentration of 0.5 mM. Single- and multi-substrate assays with different blend ratios (mentioned in the results) were performed using three replicates for each experiment as well as a negative control (vector only) and a positive control (B. mori pgFAR)16. The incubation and extraction protocols and the gas chromatography coupled to mass spectrometry (GC-MS) analysis were performed as described in the SI Materials and Methods7,19. The final fatty alcohol production was calculated as previously described7,11. The standard deviations (SDs) and standard errors of the mean (SEMs) were calculated using MS Excel. The significant differences in alcohol production in each functional assay were calculated via ANOVA tests followed by Tukey’s test, and homogeneity-of-variance tests were performed using the SPSS program.
Additional Information
Accession codes: The sequences reported in this paper have been deposited in the GenBank database (Accession Nos KF805977-KF805983 and KR781119-KR781121).
How to cite this article: Antony, B. et al. Two fatty acyl reductases involved in moth pheromone biosynthesis. Sci. Rep. 6, 29927; doi: 10.1038/srep29927 (2016).
Supplementary Material
Acknowledgments
We thank Prof. Christer Löfstedt for the valuable discussion and comments. This work was supported by Grants-in-aid for Scientific Research No. 12-AGR2554-02 from King Abdul Aziz City for Science and Technology—National Plan for Science, Technology and Innovation (KACST-NSTIP), Saudi Arabia to BA. We thank the KSU-Deanship of Scientific Research, Research Chair Program, Saudi Arabia, and Prof. Christer Löfstedt for supporting BD’s involvement in this work (Vetenskapsrådet 621-2010-5430). We thank our doctoral students Alan and Khalid for raising the insects and providing technical assistance. We are grateful to the reviewers for their numerous perceptive and constructive comments, which helped us improve our manuscript.
Footnotes
Author Contributions B.A., K.I.M. and B.-J.D. conceived the study. B.A. and B.-J.D. participated in its design and coordination. S.A.A. and A.S.A. provided the Spodoptera exigua and S. littoralis specimens. B.A. and B.-J.D. conducted the experiments and compiled the data. B.A. wrote the paper with contributions from K.I.M. and B.-J.D. All authors have read and approved the final manuscript.
References
- Grimaldi D. & Engel M. S. Evolution of the Insects . (Cambridge University Press, 2005). [Google Scholar]
- Leal W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J., Walker W. B. & Wang G. Pheromone reception in moths: from molecules to behaviors. Prog. Mol. Biol. Transl. Sci. 130, 109–128 (2015). [DOI] [PubMed] [Google Scholar]
- Smadja C. & Butlin R. K. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity 102, 77–97, 10.1038/hdy.2008.55 (2009). [DOI] [PubMed] [Google Scholar]
- Lofstedt C. Moth pheromone genetics and evolution. Philos. Trans. R. Soc. B 340, 167–177 (1993). [Google Scholar]
- Groot A. T., Dekker T. & Heckel D. G. The Genetic Basis of Pheromone Evolution in Moths. Annu. Rev. Entomol. 61 (2016). [DOI] [PubMed] [Google Scholar]
- Lassance J. M. et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature 466, 486–489 (2010). [DOI] [PubMed] [Google Scholar]
- Groot A. T. et al. Within-population variability in a moth sex pheromone blend: genetic basis and behavioural consequences. Proc. R. Soc. London, Ser. B 281, 20133054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehend M. et al. Geographic variation in sexual attraction of Spodoptera frugiperda corn-and rice-strain males to pheromone lures. PLOS ONE 9, e89255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J. V. Nutrient-dependent/pheromone-controlled adaptive evolution: a model. Socioaff. Neurosci. Psyc. 3, doi: http://dx.doi.org/10.3402/snp.v3i0.20553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassance J. M. et al. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc. Natl. Acad. Sci.USA 110, 3967–3972 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buček A. et al. Evolution of moth sex pheromone composition by a single amino acid substitution in a fatty acid desaturase. Proc. Natl. Acad. Sci.USA 112, 12586–12591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurenka R. In The Chemistry of Pheromones and Other Semiochemicals 97–132 (Springer, 2004). [Google Scholar]
- Antony B. et al. Pheromone-gland-specific fatty-acyl reductase in the adzuki bean borer, Ostrinia scapulalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 39, 90–95 (2009). [DOI] [PubMed] [Google Scholar]
- Ding B.-J. & Löfstedt C. Analysis of the agrotis segetum pheromone gland transcriptome in the light of Sex pheromone biosynthesis. BMC genomics 16, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moto K. et al. Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc. Natl. Acad. Sci.USA 100, 9156–9161 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagström Å. K., Liénard M. A., Groot A. T., Hedenström E. & Löfstedt C. Semi–selective fatty acyl reductases from four heliothine moths influence the specific pheromone composition. PLOS ONE 7, e37230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carot‐Sans G., Muñoz L., Piulachs M., Guerrero A. & Rosell G. Identification and characterization of a fatty acyl reductase from a Spodoptera littoralis female gland involved in pheromone biosynthesis. Insect Mol. Biol. 24, 82–92 (2015). [DOI] [PubMed] [Google Scholar]
- Liénard M. A., Hagström A. K., Lassance J. M. & Löfstedt C. Evolution of multicomponent pheromone signals in small ermine moths involves a single fatty-acyl reductase gene. Proc. Natl. Acad. Sci.USA 107, 10955–10960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas P. et al. Phylogenetic molecular species delimitations unravel potential new species in the pest genus Spodoptera Guenée, 1852 (Lepidoptera, Noctuidae). PLOS ONE 10, e0122407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz L., Rosell G., Quero C. & Guerrero A. Biosynthetic pathways of the pheromone of the Egyptian armyworm Spodoptera littoralis. Physiol. Entomol. 33, 275–290 (2008). [Google Scholar]
- Acín P., Rosell G., Guerrero A. & Quero C. Sex pheromone of the Spanish population of the beet armyworm Spodoptera exigua. J. Chem. Ecol. 36, 778–786 (2010). [DOI] [PubMed] [Google Scholar]
- Eggink G., Engel H., Vriend G., Terpstra P. & Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J. Mol. Biol. 212, 135–142 (1990). [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P. & Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187, 101–107 (1986). [DOI] [PubMed] [Google Scholar]
- Aarts M. G. et al. The Arabidopsis male sterility 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12, 615–623 (1997). [DOI] [PubMed] [Google Scholar]
- Liénard M. A., Wang H.-L., Lassance J.-M. & Löfstedt C. Sex pheromone biosynthetic pathways are conserved between moths and the butterfly Bicyclus anynana. Nat. Commun. 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. B. & Russell D. W. Mammalian wax biosynthesis I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 279, 37789–37797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J. G., Pollard M. R., Anderson L., Hayes T. R. & Lassner M. W. Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 122, 635–644 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron M.-B. et al. A Conserved Domain Database for the functional annotation of proteins. Nucl. Acids Res. 39, D225–D229, 10.1093/nar/gkq1189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.-C. & Shen H.-B. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLOS ONE 5, e9931 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony B. & Ishikawa Y. Tissue‐specific expression of the pheromone gland‐specific fatty acyl reductase gene in Ostrinia scapulalis. Entomol. Exp. Appl. 149, 94–98 (2013). [Google Scholar]
- Gu S. H. et al. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC genomics 14, 636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Löfstedt C. & Wang X. Sex pheromone biosynthesis in the Asian corn borer Ostrinia furnacalis (II): Biosynthesis of (E)‐and (Z)‐12‐tetradecenyl acetate involves Δ14 desaturation. Arch. Insect Biochem. Physiol. 15, 57–65 (1990). [Google Scholar]
- Meyer A. et al. Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J. Lipid Res. 45, 1899–1909 (2004). [DOI] [PubMed] [Google Scholar]
- Eirín-López J. M., Rebordinos L., Rooney A. P. & Rozas J. The birth-and-death evolution of multigene families revisited. Genome Dyn. 7, 170–196 (2012). [DOI] [PubMed] [Google Scholar]
- Antony B. et al. Genes involved in sex pheromone biosynthesis of Ephestia cautella, an important food storage pest, are determined by transcriptome sequencing. BMC genomics 16, 532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R., Fukuzawa M., Nakano R., Tatsuki S. & Ishikawa Y. Alternative suppression of transcription from two desaturase genes is the key for species-specific sex pheromone biosynthesis in two Ostrinia moths. Insect Biochem. Mol. Biol. 39, 62–67 (2009). [DOI] [PubMed] [Google Scholar]
- Levi-Zada A. et al. Identification of the sex pheromone of the lesser date moth, Batrachedra amydraula, using sequential SPME auto-sampling. Tetrahedron Lett. 52, 4550–4553 (2011). [Google Scholar]
- Levi-Zada A. et al. Reevaluation of the sex pheromone of the lesser date moth, Batrachedra amydraula, using autosampling SPME-GC/MS and field bioassays. Chemoecology 23, 13–20 (2013). [Google Scholar]
- Blomquist G. J., Jurenka R., Schal C. & Tittiger C. Pheromone Production: Biochemistry and Molecular Biology in Insect Endocrinology (eds Gilbert L. I. ) 523–553 (Academic press, London 2011). [Google Scholar]
- Ding B. J. et al. A plant factory for moth pheromone production. Nat. Commun. 5, 3353, 10.1038/ncomms4353 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurenka R. A. & Roelofs W. L. Characterization of the acetyltransferase used in pheromone biosynthesis in moths: specificity for the Z isomer in Tortricidae. Insect Biochem. 19, 639–644 (1989). [Google Scholar]
- Tamaki Y. & Yushima T. Sex pheromone of the cotton leafworm, Spodoptera littoralis. J. Insect Physiol. 20, 1005–1014 (1974). [DOI] [PubMed] [Google Scholar]
- Dunkelblum E. & Kehat M. Sex pheromone precursors in Spodoptera littoralis (Lepidoptera: Noctuidae). Insect Biochem. 17, 877–881 (1987). [Google Scholar]
- Elvira S., Gorría N., Muñoz D., Williams T. & Caballero P. A simplified low-cost diet for rearing Spodoptera exigua (Lepidoptera: Noctuidae) and its effect on S. exigua nucleopolyhedrovirus production. J. Econ. Entomol. 103, 17–24 (2010). [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2, 60–61 (2011). [Google Scholar]
- Norušis M. J. SPSS professional statistics 6.1 . (Prentice Hall, 1994). [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra M., Piña B., Bujons J., Camps F. & Fabriàs G. Biosynthesis of 10, 12-dienoic fatty acids by a bifunctional Δ11 desaturase in Spodoptera littoralis. Insect Biochem. Mol. Biol. 36, 634–641 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.