Abstract
Cu, Zn-superoxide dismutase (SOD1), an enzyme implicated in the progression of familial amyotrophic lateral sclerosis (fALS), forms amyloid fibrils under certain experimental conditions. As part of our efforts to understand ALS pathogenesis, in this study we found that reduction of the intramolecular disulfide bond destabilized the tertiary structure of metal free wild-type SOD1 and greatly enhanced fibril formation in vitro. We also identified fibril core peptides that are resistant to protease digestion by using mass spectroscopy and Edman degradation analyses. Three regions dispersed throughout the sequence were detected as fibril core sequences of SOD1. Interestingly, by using three synthetic peptides that correspond to these identified regions, we determined that each region was capable of fibril formation, either alone or in a mixture containing multiple peptides. It was also revealed that by reducing the disulfide bond and causing a decrease in the structural stability, the amyloid fibril formation of a familial mutant SOD1 G93A was accelerated even under physiological conditions. These results demonstrate that by destabilizing the structure of SOD1 by removing metal ions and breaking the intramolecular disulfide bridge, multiple fibril-forming core regions are exposed, which then interact with each another and form amyloid fibrils under physiological conditions.
Key words: amyloid fibril core, ALS, mechanism, SOD1G93A mutant, structural stability
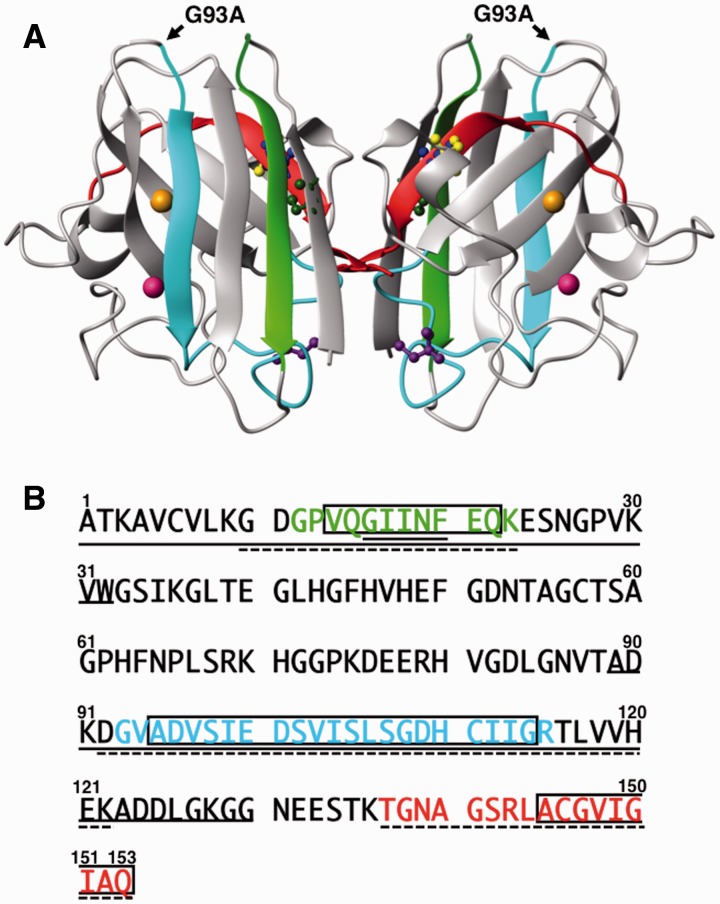
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that specifically targets motor neurons and leads to a progressive loss of function that is ultimately fatal, usually within 3–5 years after onset (1). Mutations in Cu, Zn-superoxide dismutase (SOD1) are known to cause a familial form of ALS (fALS) (2–4). SOD1 is a highly conserved enzyme (5) that is the primary scavenger of superoxide radicals in the cytosol. SOD1 is a dimer composed of two identical 153-amino acid residue subunits (Fig. 1) (6). The overall structure of the subunit assumes a β-sandwich structure containing several β-strands (7). Each subunit binds one copper and one zinc ion. Four cysteine residues are located at Positions 6, 57, 111 and 146 in the amino acid sequence. Interestingly, Cys57 and Cys146 form a disulfide bond even in the highly reductive environment of the eukaryotic cytosol, whereas Cys6 and Cys111 are found in the reduced thiol (SH) form (8). Previous studies have established that SOD1 is implicated in ALS pathogenesis not through loss of superoxide dismutase activity but rather by acquisition of some unknown toxic function(s), probably due to misfolding of the SOD1 protein (9–11). Previous research has demonstrated that inclusion bodies may be observed in spinal cords of human patients afflicted by both sporadic ALS (12–14) and fALS (13–15), and in spinal cord samples from transgenic mice that express human fALS SOD1 variants (16–23). These results demonstrate the strong relationship between SOD1 misfolding, SOD1 aggregation and ALS pathogenesis.
Fig. 1.
Structural characteristics of SOD1. (A) SOD1 has a total of four cysteine residues: Cys57 (yellow) and Cys146 (blue) form an intramolecular disulfide bond, whereas Cys6 (green) and Cys111 (purple) remain as free cysteine forms. The copper and zinc ions are shown as orange color and pink spheres, respectively. Synthetic core regions P1 (Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys), P2 (Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Leu-Ser-Gly-Asp-His-Cys-Ile-Ile-Gly-Arg) and P3 (Thr-Gly-Asn-Ala-Gly-Ser-Arg-Leu-Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln) are shown as greenish-yellow, blue-green and red colors, respectively. The G93A mutation site is indicated by a black arrow. (B) Amino acid sequence alignment of SOD1. P1–P3 are colored by greenish-yellow, blue-green and red, respectively. Open squares indicate the possible amyloidogenic regions, which were deduced by the Waltz program (http://waltz.switchlab.org/) developed by the Switch Laboratory. The continuous line and the dashed line indicate regions correspond to the PK- and API-derived peptide sequences determined in this study, respectively (see main text).
SOD1 aggregates also share some common characteristics with amyloid fibrils (17, 24) that are isolated from tissues of patients afflicted by neurodegenerative diseases such as Alzheimer disease and Parkinson disease (25, 26). Previously, it was reported that SOD1 that is chemically denatured by guanidine hydrochloride (Gdn-HCl) or organic solvents (27–29) is capable of forming amyloid fibrils. However, the experimental conditions under which these fibrils were formed differed greatly from the conditions in an actual cell. As SOD1 normally exists in the cytosol where a reducing environment is predominant (30), it was suggested that reduction of the intramolecular disulfide bond in native SOD1 may act as a trigger for fibril formation in vivo, and this idea was subsequently demonstrated in transgenic mice (19, 21). Thus, to comprehend the pathogenesis of ALS, it is important to understand the structural characteristics and structural stability of SOD1 under physiological conditions.
In this study, we first focused on the consequences of forming or reducing the Cys57-Cys146 intramolecular disulfide bond of wild-type SOD1 with regard to the formation of SOD1 amyloid fibrils. Our results indicated that removal of metal ions and reduction of the Cys57-Cys146 disulfide bond destabilized the SOD1 tertiary structure and greatly enhanced fibril formation in vitro. Additionally, we identified three distinct amino acid sequence regions that were resistant to protease digestion in the final fibril state. Interestingly, three synthetic peptides that corresponded to each of these regions could form typical amyloid fibrils, either solely or in a mixture of one or more peptide species. To our knowledge, this finding is the first to demonstrate fibril formation by multiple specific short peptides derived from the SOD1 fibril core. Additionally, for a G93A mutant implicated in familial ALS, the reduction of this disulfide bond was in fact a prerequisite to trigger amyloid fibril formation. These results suggested that the structure of SOD1 was largely destabilized by removal of the metal ions and reduction of the intramolecular disulfide bond, which led to the exposure of core amyloid-forming regions. The exposed core regions subsequently interact with one another and ultimately this interaction leads to the formation of amyloid fibrils. This understanding of the underlying molecular mechanism of SOD1 fibril formation may eventually lead to a specific medical treatment for preventing and curing ALS.
Materials and Methods
Expression and purification of SOD1
Wild-type SOD1
The human SOD1 gene was amplified using PCR from cDNA preparations of human brain (Cap site cDNA dT: Nippon gene) and subcloned into the NdeI and XhoI multi-cloning site of the expression vector pET23a(+) (Novagen) to produce pEThSOD1, and used to transform Escherichia coli BLR(DE3). Cultivated BLR(DE3)/pEThSOD1 cells were suspended in lysis buffer (50 mM Tris-HCl, pH 8.0, containing 150 mM KCl, 0.1 mM EDTA and 1 mM dithiothreitol (DTT)), disrupted using sonication and centrifuged to remove debris (14,000 rpm, 30 min). Streptomycin sulfate (final 2.5%) was added to the supernatant to remove nucleic acids. After removal of nucleic acids by centrifugation, ammonium sulfate was added to the supernatant to 65% saturation. After this treatment, SOD1 remained in the supernatant fraction and was recovered by centrifugation. The supernatant was dialyzed thoroughly at 4°C against 20 mM potassium phosphate buffer, pH 6.5, containing 0.01 mM EDTA, and applied onto a Q-Sepharose ion exchange column (GE healthcare) at 25°C (elution was achieved by a 0–0.5 M KCl linear gradient). Fractions containing SOD1 were pooled and solid ammonium sulfate was added to 2 M. Then, the SOD1 sample was subjected to a Phenyl-Toyopearl 650 hydrophobic chromatography column (Tosoh) at 25°C, using 50 mM sodium phosphate buffer, pH 6.5, containing 2 M ammonium sulfate, 150 mM NaCl, 0.1 mM EDTA, 0.25 mM DTT. Samples were eluted with a linear gradient of 2–0 M ammonium sulfate. The wild-type SOD1 protein purified according to the method above lost a significant fraction of its bound Cu, Zn-ligands and required a reconstitution step to recover holo SOD1 protein (see below). Concentrations of wild-type SOD1 were determined by either using a protein assay kit (Bio-Rad Laboratories) using bovine serum albumin (Sigma-Aldrich) as a standard protein or by direct spectrophotometric measurement using a molar absorption coefficient of ε280 nm = 10,800 M−1 cm−1 (31).
SOD1 with mutation of G93A (G93ASOD1)
The familial human SOD1 mutant (G93A) gene was derived from the wild-type gene through point mutation using the QuickChange site-directed mutagenesis kit (Agilent Technologies). The mutation in pEThSOD1G93A was confirmed by DNA sequence analysis of the entire SOD1 coding region. Cell pastes of E.coli BLR(DE3)/pEThSOD1G93A cells were suspended in lysis buffer (50 mM Tris-HCl, pH 8.0, containing 150 mM KCl, 0.1 mM EDTA, 1 mM DTT), disrupted using sonication and centrifuged (14,500 rpm, 30 min). Under our conditions, G93A SOD1 protein was recovered in inclusion bodies. The precipitate was suspended in lysis buffer containing 4 M Gdn-HCl and dialyzed thoroughly at 4°C against 20 mM potassium phosphate buffer, pH 6.5, containing 0.01 mM EDTA. Through this step, soluble Cu, Zn-free G93ASOD1 (Apo-G93ASOD1) was obtained. After dialysis, samples were centrifuged at 14,500 rpm for 30 min and the supernatant was applied onto a Q-Sepharose ion exchange column (GE Healthcare) at 25°C (elution: 0–0.7 M KCl linear gradient). Concentrations of Apo-G93ASOD1 were determined using a protein assay kit (Bio-Rad Laboratories) using bovine serum albumin (Sigma-Aldrich) as a standard protein.
Preparation of SOD1 derivatives
Since the purified wild-type SOD1 was not recovered in the fully Cu, Zn-bound, holo form, Cu, Zn-liganded SOD1 (Holo-SOD1) was reconstituted by incubating purified wild-type SOD1 protein in 50 mM HEPES-KOH, pH 7.0, containing 0.1 mM CuCl2, 6 M urea and 0.1 mM Zn(CH3COO)2 overnight at 25°C. After incubation, the sample was desalted with a PD-10 column (GE Healthcare) to obtain Holo-SOD1. Holo-G93ASOD1 was prepared through a similar process that used a lower concentration of urea, i.e. Apo-G93ASOD1 was incubated in 50 mM HEPES-KOH, pH 7.0, containing 0.1 mM CuCl2, 1 M urea and 0.1 mM Zn(CH3COO)2 overnight at 25°C, followed by desalting. Wild-type apo-SOD1 (Apo-SOD1) was prepared by incubation of the purified wild-type SOD1 in 50 mM HEPES-KOH, pH 7.0, containing 10 mM EDTA and 3 M Gdn-HCl for 1 h at 25°C. The Cu and Zn metal content of Holo- and Apo-SOD1 proteins were quantified using inductively coupled plasma atomic emission spectroscopy, SPECTRO CIROS CCD (SPECTRO), after desalting the samples with PD-10 column equilibrated with Milli-Q water. Holo-SOD1 typically contained 0.76-mol Cu and 1.08-mol Zn per 1-mol SOD1 subunit, while Apo-SOD1 typically contained 0.01-mol Cu and 0.04-mol Zn per 1-mol SOD1 subunit. The detected Cu and Zn content for Holo-G93ASOD1 samples were 0.84 mol and 1.28 mol per mol subunit, respectively. The corresponding values for Apo-G93ASOD1 were 0.06 mol (Cu) and 0.1 mol (Zn) per subunit.
SOD1 derivative where the free thiols of Cys6 and Cys111 are carboxyamidated (2SCAM-SOD1) was prepared as follows: Apo-SOD1 dissolved in 50 mM Tris-HCl, pH 7.0, containing 150 mM NaCl was treated by the addition of 2 M Gdn-HCl and 10 mM ICH2CONH2 for 30 min at room temperature to modify the free SH groups of Cys6 and Cys111. Samples were then desalted using PD-10 columns. SOD1 derivative in which the free thiols of Cys6 and Cys111 have been carboxyamidated and the intra-disulfide bond of Cys57-Cys146 has been reduced (2SCAMRed-SOD1) was prepared by adding 5 mM DTT to 2SCAM-SOD1 preparations and incubating for 30 min at room temperature, followed by desalting using a PD-10 column equilibrated with 50 mM Tris-HCl, pH 7.0, containing 150 mM NaCl and 1 mM tris-(2-carboxyethyl) phosphine (TCEP, Molecular Probes). TCEP was used to maintain the reduced thiol groups of Cys57 and Cys146. Red-SOD1 and Red-G93ASOD1 were prepared by the addition of 2 M Gdn-HCl and 5 mM DTT to protein solutions, followed by incubation for 30 min at room temperature, then the samples were desalted using PD-10 columns equilibrated with 50 mM Tris-HCl (pH 7.0) containing 150 mM NaCl and 1 mM TCEP. SOD1 derivative where all of the thiols in Cys6, Cys 57, Cys 111 and Cys146 have been carboxyamidated (4SCAM-SOD1) was prepared by extensive alkylation of all SH groups in the protein, by adding 10 mM ICH2CONH2 to Red-SOD1 sample and incubation for 30 min in the dark at room temperature, followed by desalting using PD-10 columns equilibrated with 50 mM Tris-HCl, pH 7.0, containing 150 mM NaCl.
Structural unfolding measurements
Unfolding experiments of SOD1 in Gdn-HCl monitored using circular dichroism (CD) and fluorescence were carried out in 50 mM Tris-HCl, pH 7.0, containing 150 mM NaCl at 25°C. Prior to measurement, prepared samples were allowed to achieve equilibrium overnight at 25°C. Unfolding transition curves were measured by monitoring either the fluorescence at 350 nm with excitation at 295 nm or the CD signal at 216 nm. Fluorescence was measured on a Jasco FP-6300 fluorescence spectrophotometer equipped with a constant temperature cell holder. CD spectra were measured using a Jasco J-720 or J-820 spectrophotometer equipped with a constant temperature cell holder at 25°C. Far-UV CD spectra were recorded using 1 mm light path-length cells. The protein concentrations of the samples were fixed at 0.15 mg/ml. To convert the raw CD and fluorescence values to the fraction of unfolded protein (fD), straight lines were fit through the upper and lower plateau regions of the sigmoidal plot. We used the following equation for normalizing the data: fD = (N − Nl)/(Nu − Nl), where Nl and Nu were the fits of the lines through the lower and upper plateau regions, respectively, and N was the observed raw data at a given concentration of Gdn-HCl. Finally, transition curves using these fD values were plotted as a function of the Gdn-HCl concentration. Midpoint denaturant concentration (Cm) for unfolding was determined by averaging the values at fD = 0.5 from CD and fluorescence measurements.
Amyloid fibril formation and ThioT binding assay
Fibril formation of SOD1 proteins (1 mg/ml: 62.5 µM) was achieved by linear (back and forth) shaking at a rate of 170 repetitions/min of samples prepared in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl with or without 1 mM TCEP at 37°C. For experiments using synthetic core peptides (0.2 mg/ml: 85–150 µM), a modified buffer (50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl, 1 mM TCEP and 0.4 M Gdn-HCl) was used; 0.4 M Gdn-HCl was added to this buffer to solubilize the peptides completely. Samples were withdrawn at appropriate times and mixed with 25 µM Thioflavin T (ThioT) (Wako) in phosphate-buffered saline. Fibril formation was monitored by ThioT binding fluorescence assays; the fluorescence intensity at 480 nm was monitored at an excitation wavelength of 440 nm using a Hitachi F-4500 fluorescence spectrophotometer.
To monitor the fibril formation of G93ASOD1 proteins, protein (1 mg/ml; 62.5 µM) was prepared in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl, 2 mM DTT and 20 µM ThioT and 150 µl aliquots were deposited into 96-well plates. Fibril formation was monitored by measuring fluorescence intensities of ThioT at 486 nm at an excitation wavelength of 450 nm using a microplate reader (Perkin Elmer ARVOx) at 37°C with agitation.
Transmission electron microscopy measurements
Transmission electron microscopy (TEM) measurements were performed on a JEOL-100CX or JEOL-1400 plus transmission electron microscope operating at 80 kV. Samples were diluted 5- to 10-fold with water and negatively stained with 2% (w/v) uranyl acetate solution on copper grids (400-mesh) covered by carbon-coated collodion film (Nisshin EM). Samples were observed at magnifications of 27,000–50,000.
Determination of amyloid fibril core region
Fibril core regions in SOD1 were determined according to the method described previously (32, 33). Briefly, isolated precipitates (fibril core region) representing fractions of amyloid fibrils that had avoided digestion by A.lyticus protease I (API, Wako) or proteinase K (PK, Roche) were dissolved in 7.5 M Gdn-HCl, and individual peptides were isolated using reversed-phase high performance liquid chromatography (HPLC) on a CAPCELL PAK C-18 reverse phase column (Shiseido) and a Gilson HPLC system. The identities of the isolated peptides were determined using mass spectrometry measurements (BRUKER Autoflex matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) Mass spectrometer) and Edman degradation (Shimadzu PPSQ-10 protein sequencer).
Synthetic core peptides of SOD1
Custom syntheses of peptide samples were performed by SCRUM Inc. (Tokyo) or Peptide 2.0 Inc. (Chantilly, USA). Peptide purity was ∼>95% as determined by HPLC and electrospray mass spectrometry. The peptide concentrations were estimated by weighing the dried peptide samples on an electron micro balance (Excellence XS205Du, Mettler Toledo).
Visualization of amyloid fibrils formed by SOD1 peptides
Synthetic core region peptides P1 (Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys), P2 (Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Lue-Ser-Gly-Asp-His-Cys-Ile-Ile-Gly-Arg) and P3 (Thr-Gly-Asn-Ala-Gly-Ser-Arg-Lue-Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln) (Fig. 1B) were labelled with fluorescein 5-isothiocyanate (FITC) (Sigma), N-(iodoacetyl)-N'-(5-sulfo-l- naphthyl)-ethylenediamine (1,5-IAEDANS) (Molecular Probes) or Cy3 maleimide (Cy3) (GE Healthcare), respectively. P2 and P3 peptides each contain one native cysteine residue in the sequence. Peptides (0.2 mg/ml, i.e. 150 µM P1, 85 µM P2 and 126 µM P3) were dissolved in basal labelling buffer (50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl, 1 mM TCEP and 0.4 M Gdn-HCl) containing 0.7 molar equivalents of the respective fluorescence label (FITC for P1 or 1,5-IAEDANS for P2) and the samples were incubated at 25°C for 30 min. Labelling of peptide P3 with Cy3 was performed according to the manufacturer’s protocols using the same buffer with added label. After the reaction, labelled peptides were allowed to form fibrils by linear shaking (170 min−1) in sealed siliconized test tubes at 37°C. For fluorescence microscopy (FM) experiments, after mixing either two or three labelled peptides in an equimolar ratio, aliquots (10 µl) of this sample solution was deposited immediately on a non-fluorescent glass slide (Matsunami Glass Industry Ltd., Japan), tightly sealed with a cover slip, and incubated at 37°C for 24 h in the dark. After incubation, samples were observed on a ZEISS Axiovert 200 system equipped with the appropriate fluorescence filters.
Results
Structural stability and amyloid fibril formation
To study the structural stability of SOD1, we prepared metal-free SOD1 proteins where the Cu and Zn ions had been removed (Apo-SOD1), as well as reduced SOD1 proteins where the intramolecular disulfide bond between Cys57 and Cys146 in the apo-SOD1 preparations had been severed by reduction (Red-SOD1). We also prepared alkylated versions of each SOD1 preparation, in which the free thiols were carboxyamidated (2SCAM-SOD1, 2SCAMRed-SOD1 and 4SCAM-SOD1). These protein preparations are hereafter referred to by their respective designations, in line with the designation Holo-SOD1 for the intact native enzyme containing Cu and Zn metals.
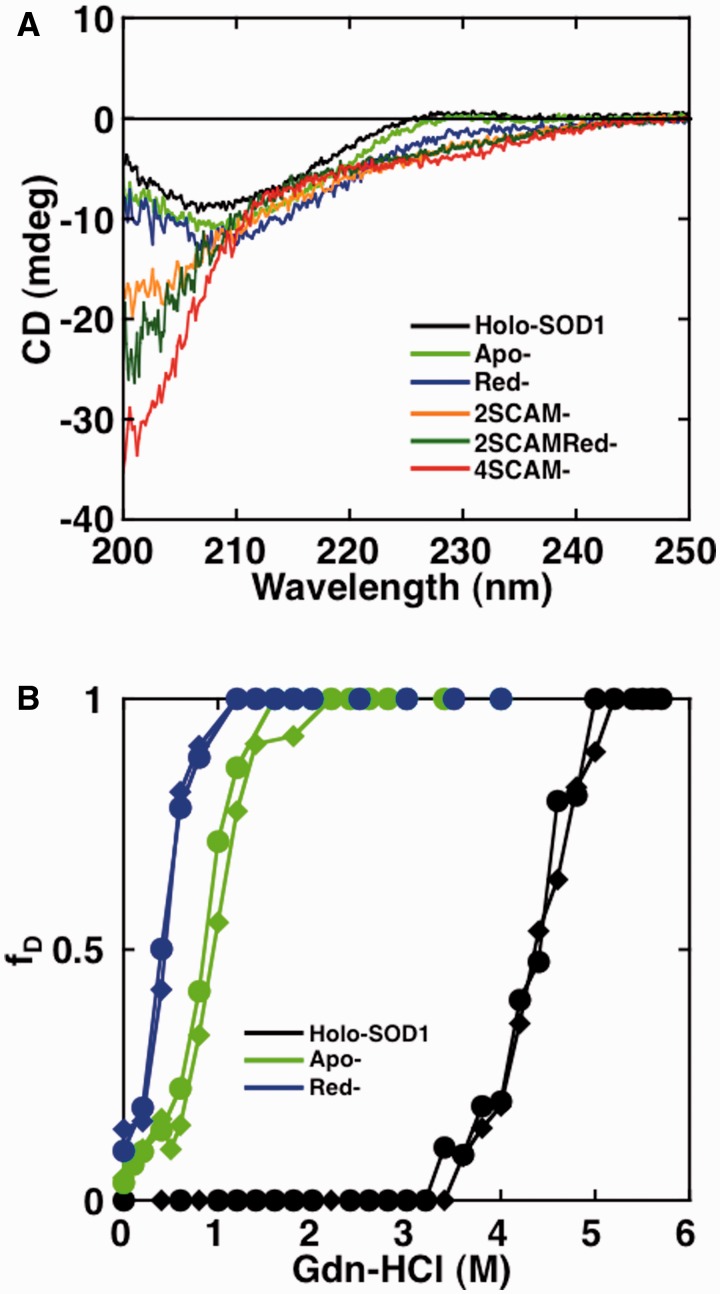
First, we measured the CD spectra of the protein samples to evaluate their respective secondary structures. As shown in Fig. 2A, Holo-SOD1 displayed a CD spectrum with a negative maximum peak at around 208 nm, which is characteristic of β-sheets (34). Apo-SOD1 and Red-SOD1 both displayed CD spectra that were similar to Holo-SOD1, demonstrating that the secondary structures of these samples remained largely intact, even after removal of metal ions and intramolecular disulfide bond. However, when all of the Cys residues in each protein were alkylated, the structure was completely unfolded, as shown in the CD spectrum of 4SCAM-SOD1. The CD spectra of 2SCAM-SOD1 and 2SCAMRed-SOD1, in contrast, suggested that these proteins were only partially unfolded. This result suggested that alkylation on Cys residues, especially Cys57 and Cys146, greatly perturbed the overall structure of SOD1 and induced unfolding.
Fig. 2.
The secondary structure and structural unfolding of wild-type SOD1 by Gdn-HCl. (A) CD spectra of Holo-SOD1, Apo-SOD1, Red-SOD1, 2SCAM-SOD1, 2SCAMRed-SOD1 and 4SCAM-SOD1. (B) Structural unfolding transition curves of Holo-SOD1, Apo-SOD1 and Red-SOD1. Measurements of CD (closed circle) and fluorescence (closed diamond) were performed in various concentrations of Gdn-HCl. The CD intensities at 216 nm and the fluorescence intensities at 350 nm (excitation at 295 nm) were converted to the fraction of unfolded protein (fD) and plotted against the Gdn-HCl concentration. See online version for details regarding color in this and subsequent Figures.
Next, we examined the structural stabilities of Holo-SOD1, Apo-SOD1 and Red-SOD1 by monitoring the unfolding transitions in Gdn-HCl using CD and fluorescence (Fig. 2B). Since native Holo-SOD1 and Apo-SOD1 retained their dimer structure as confirmed in gel-filtration assays, the experiments for structural unfolding, especially of Holo-SOD1, were not monitored under strict equilibrium conditions. Even under these conditions of quasi-equilibrium, however, we were able to obtain information regarding the structural stability of these proteins; the Cm values of Holo-SOD1, Apo-SOD1 and Red-SOD1 were 4.37, 0.90 and 0.42 M Gdn-HCl, respectively (Table I). Holo-SOD1 was the most stable derivative. Removal of metal ions (Apo-SOD1) resulted in a remarkable decrease in stability, and the breakage of the intramolecular disulfide bond further destabilized the structure (Red-SOD1). From the transition curves, it was found that both Red-SOD1 and Apo-SOD1 were only marginally stable at 0 M Gdn-HCl.
Table I.
Denaturation midpoint (Cm) of SOD1 samples
| Holo- (M) | Apo- (M) | Red- (M) | |
|---|---|---|---|
| Wild-type SOD1 | 4.37 | 0.90 | 0.42 |
| G93ASOD1 | 2.85 | 0.27 | N.D. a |
aN.D.: not determined.
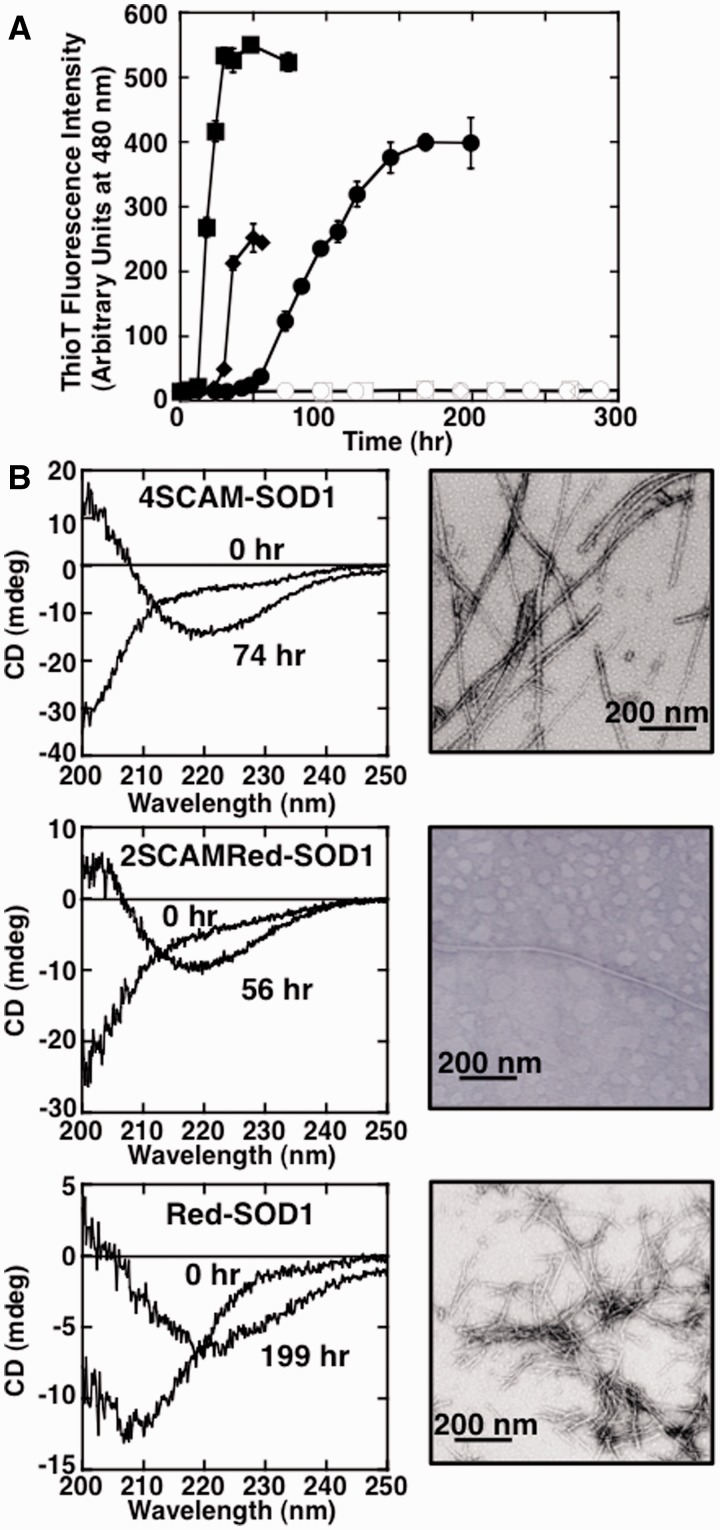
To examine whether Holo-SOD1, Apo-SOD1, Red-SOD1, 2SCAM-SOD1, 2SCAMRed-SOD1 and 4SCAM-SOD1 could form amyloid fibrils, each sample was incubated at 37°C with linear shaking. As shown in Fig. 3, 4SCAM-SOD1, 2SCAMRed-SOD1 and Red-SOD1, which all lack the intramolecular disulfide bond, showed a significant increase in ThioT fluorescence, β-structural characteristics in CD spectral analyses and a fibrous shape in TEM measurements after prolonged shaking. The fibril widths of 4SCAM-SOD1, 2SCAMRed-SOD1 and Red-SOD1 were 13.2 ± 0.6 nm, 13.6 ± 2.4 nm and 10.9 ± 0.6 nm, respectively. In an interesting contrast, Holo-SOD1, Apo-SOD1 and 2SCAM-SOD1 all failed to show any increases in ThioT fluorescence and CD spectra even after a 300 h incubation. Although Red-SOD1 and Apo-SOD1 displayed similar structural stabilities in unfolding transition assays (Cm = 0.42 and 0.90 M Gdn-HCl, respectively, Fig. 2), only Red-SOD1 formed amyloid fibrils. These findings demonstrated that reduction of the intramolecular disulfide bond was the crucial element in forming SOD1 amyloid fibrils; once the intramolecular disulfide bond was reduced, fibril formation was accelerated by further structural destabilization (4SCAM-SOD1 > 2SCAMRed-SOD1 > Red-SOD1).
Fig. 3.
Fibril formation of wild-type SOD1 samples. (A) Amyloid fibril formation monitored by ThioT binding assay. Conditions were 62.5 µM Holo-SOD1 (opened square), Apo-SOD1 (opened circle), 2SCAM-SOD1 (opened diamond) and 4SCAM-SOD1 (closed square) in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl at 37°C, and 62.5 µM 2SCAMRed-SOD1 (closed diamond) and Red-SOD1 (closed circle) in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl and 1 mM TCEP at 37°C. Standard error bars from at least three independent measurements are also shown. (B) CD (left panel) and TEM (right panel) measurements of 4SCAM-SOD1 fibrils after 74 h incubation (upper), 2SCAMRed-SOD1 fibrils after 56 h incubation (middle) and Red-SOD1 fibrils after 199 h incubation (bottom), respectively. The scale bar in each panel of TEM measurement represents 200 nm.
Determination of fibril core region
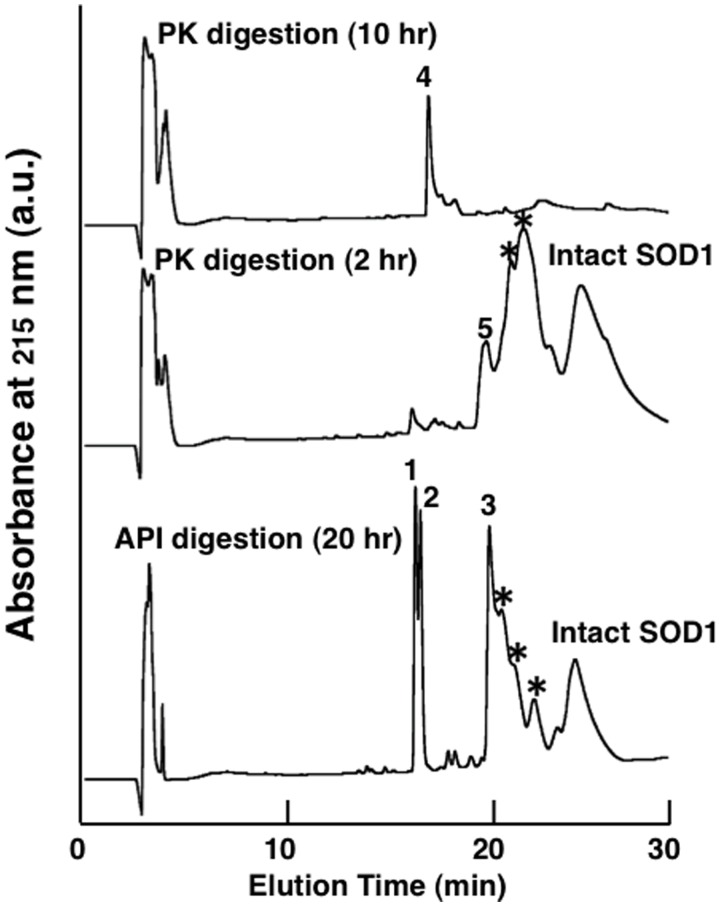
To further understand the SOD1 fibril formation mechanism, we identified the fibril core regions, which are presumably composed of β-strands and possesses protease-resistance (35). We used a lysine specific protease, API (36) and a broadly specific PK to digest Red-SOD1 amyloid fibrils. After digestion with API for 20 h, it was observed in TEM experiments that the diameter of the remaining fibrils had been reduced to 5.2 ± 1.0 nm, indicating that the fibrils had been partially digested (not shown). The precipitates that remained after centrifugation were dissolved in 7.5 M Gdn-HCl and isolated using reversed phase HPLC. A typical HPLC elution pattern of API digested Red-SOD1 fibril samples is shown in Fig. 4. Several peaks were observed after API proteolysis of Red-SOD1 fibrils and analysed by MALDI-TOF-MS and Edman degradation. Although intact SOD1 protein and intermediate large peptides that evaded digestion were also detected, three peaks (designated as Peaks 1–3), eluted at around 16–20 min, were identified as core regions. The corresponding sequences of these three peptides were determined to be 137Thr-Gly-Asn-Ala-Gly-Ser-Arg-Leu-Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln153 for Peak 1, 10Gly-Asp-Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys23 for Peak 2 and 92Asp-Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Leu-Ser-Gly-His-Asp-Cys-Ile-Ile-Gly-Arg-Thr-Leu-Val-Val-His-Glu-Lys122 for Peak 3.
Fig. 4.
Typical reverse phase HPLC patterns of protease resistant core peptides in Red-SOD1. Reverse phase HPLC patterns of fibril core peptides that were resistant to API and PK digestions. Bottom trace shows Red-SOD1 fibril core peptides remained after 20 h digestion by API at 37°C. The protease resistant precipitates were dissolved in 7.5 M Gdn-HCl and subjected to HPLC analysis. Three peaks (numbered as 1–3) at around 16–20 min as shown in Table II, and the broad peak at around 25 min was determined to be intact Red-SOD1. Middle and upper traces show Red-SOD1 fibril core peptides (numbered as 4, 5) remained after 10 and 2 h digestions by PK at 37°C, respectively. Asterisks (*) indicate partially digested intermediate peptides.
We also digested and analysed Red-SOD1 fibrils using a non-specific protease PK, anticipating that we could obtain shorter, more specific fibril core peptides. After a 10-h digestion, only one peak (Peak 4 in Fig. 4) was detected, which suggested that the digestion of fibrils proceeded at a rate that was faster than that for API digestion under the conditions that we used, probably due to the broad specificity of PK. When Red-SOD1 fibrils were digested for a shorter interval of 2 h, a broad Peak 5 was observed and isolated as seen in Fig. 4. This peak also contained intact and intermediate large peptides. Analyses of Peaks 4 and 5 revealed that the sequence of Peak 4 was 16Gly-Ile-Ile-Asn-Phe20 and Peak 5 was found to be a mixture of two peptides, 1Ala-Thr-Lys-Ala-Val-Cys-Val-Leu-Lys-Gly-Asp-Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys-Glu-Ser-Asn-Gly-Pro-Val-Lys-Val-Trp32 and 89Ala-Asp-Lys-Asp-Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Leu-Ser-Gly-Asp-His-Cys-Ile-Ile-Gly-Arg-Thr-Leu-Val-Val-His-Glu-Lys-Ala-Asp-Asp-Leu-Gly-Lys-Gly-Gly130. Of note was the identification of peptide (Gly-Ile-Ile-Asn-Phe) that was relatively short in length as a participant in fibril core formation. Intuitively, such a short peptide fragment seems to be an unlikely candidate to form any specific interactions that may lead to amyloid fibrils. However, it has been reported that a synthetic 5 residue peptide fragment derived from human islet amyloid polypeptide (37), as well as a seven amino acid peptide derived from Sup35 (38), are capable of forming typical amyloid fibrils. The identification of peptides are summarized in Table II and also visualized in Fig. 1B. We also determined the fibril core regions of 4SCAM-SOD1 fibrils and determined that the core peptides isolated were identical to that for Red-SOD1. Allowing for differences derived from the substrate specificities of API and PK, the core peptide regions identified from both protease digestions were consistent. We concluded that SOD1 amyloid fibril formation involved three specific core peptide regions located in different segments of the sequence; N-terminal (10Gly-Asp-Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys23), internal (92Asp-Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Leu-Ser-Gly-His-Asp-Cys-Ile-Ile-Gly-Arg-Thr-Leu-Val-Val-His-Glu-Lys122) and C-terminal (137Thr-Gly-Asn-Ala-Gly-Ser-Arg-Leu-Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln153) as shown in Fig. 1B. A measurable seeding effect for Red-SOD1 fibril formation was observed when samples retrieved after digestion of Red-SOD1 fibrils by API or PK were added to fresh Red-SOD1 solutions (data not shown).
Table II.
Identification of core peptides of Red-SOD1 fibrils
| Peak No.a | Detected mass number |
Assignment (Theoretical molecular weight) | |
|---|---|---|---|
| API digestion | PK digestion | ||
| 1 | 1587.6 | 137TGNAGSRLACGVIGIAQ153 (1587.7) | |
| 2 | 1501.6 | 10GDGPVQGIINFEQK23 (1501.5) | |
| 3 | 3264.4 | 92DGVADVSIEDSVISLSGDHCIIGRTLVVHEK122 (3264.6) | |
| 4 | 563.7 | 16GIINF20 (562.7) | |
| 5 | 3412.2 | 1ATKAVCVLKGDGPVQGIINFEQKESNGPVKVW32 (3412.8) | |
| 4292.4 | 89ADKDGVADVSIEDSVISLSGDHCIIGRTLVVHEKADDLGKGG130 (4293.7) | ||
aPeak No. denotes peaks that were identified in the reverse phase HPLC traces in Fig. 4.
Amyloid fibril formation of the fibril core peptides
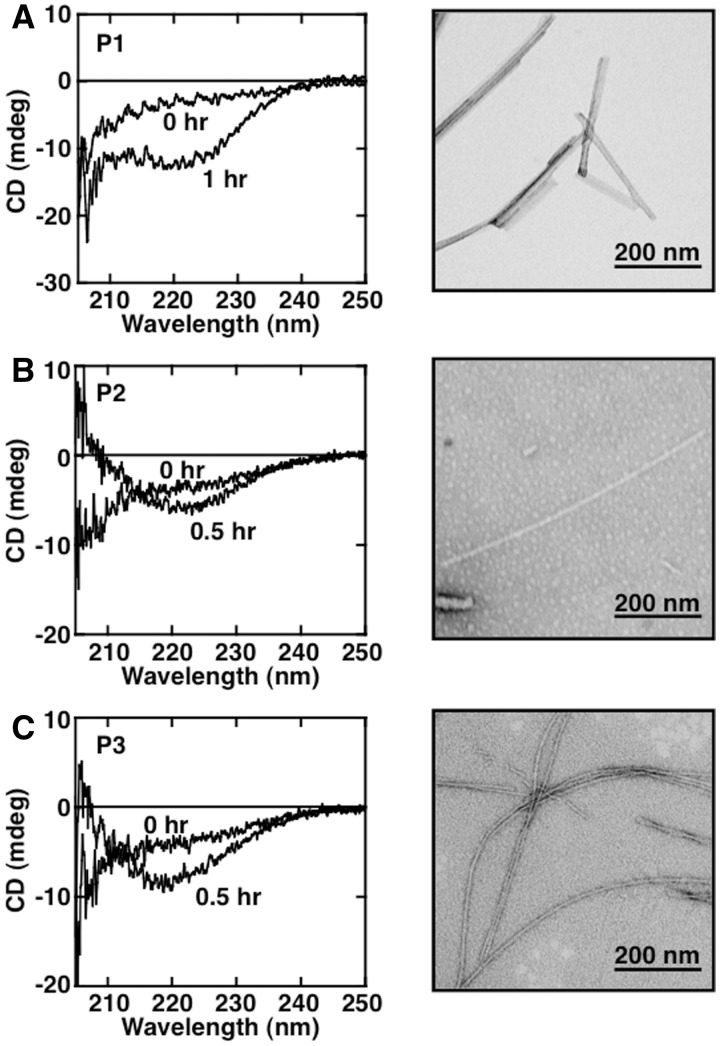
Next, to examine the tendency of these three core regions to form amyloid fibrils, we synthesized three peptides corresponding to the sequence of these regions. P1 (Gly-Pro-Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln-Lys), P2 (Gly-Val-Ala-Asp-Val-Ser-Ile-Glu-Ap-Ser-Val-Ile-Ser-Leu-Ser-Gly-Asp-His-Cys-Ile-Ile-Gly-Arg) and P3 (Thr-Gly-Asn-Ala-Gly-Ser-Arg-Leu-Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln) were designed on the basis of the results from protease-digestion experiments coupled with Waltz prediction algorithm estimates by the Switch Laboratory (http://waltz.switchlab.org/). First, we examined whether each peptide could form amyloid fibrils independently. Each peptide displayed an increase in ThioT fluorescence after incubation for several hour with linear shaking at 37°C. CD spectra and TEM images of the samples are shown in Fig. 5. Characteristics consistent with β-structure were detected in CD spectra, and a fibrous image could be identified in TEM observations with samples derived from each peptide (P1–P3). The widths of the fibrils derived from P1–P3 samples were 15.0 ± 1.1 nm, 10.9 ± 0.6 nm and 13.2 ± 0.6 nm, respectively.
Fig. 5.
Fibril formation of various SOD1 core peptides. (A–C) CD (left panel) and TEM (right panel) measurements of synthesized core peptides; (A) P1 (GPVQGIINFEQK, 150 µM), (B) P2 (GVADVSIEDSVISLSGDHCIIGR, 85 µM), (C) P3 (TGNAGSRLACGVIGIAQ, 126 µM) incubated in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl, 1 mM TCEP and 0.4 M Gdn-HCl at 37°C. For TEM measurements, data were collected after 3 h incubation. The scale bar in each panel of TEM measurement represents 200 nm.
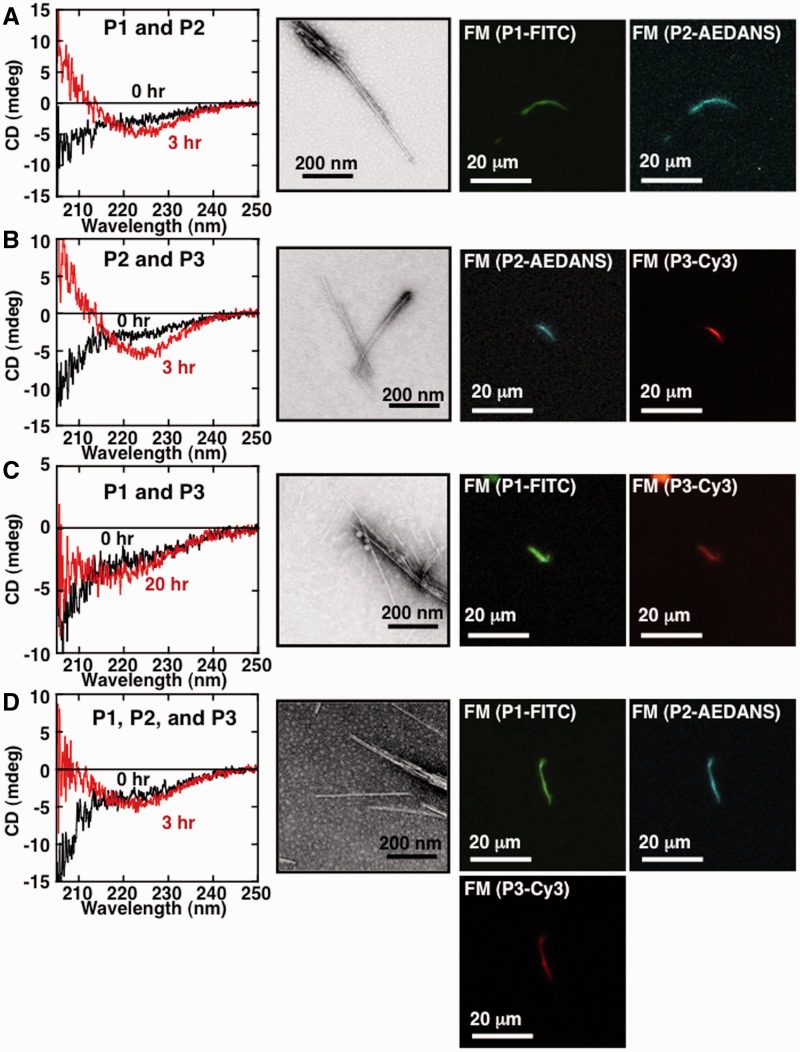
Next, to further test whether a mixture of two or three core peptides could form mixed amyloid fibrils, we incubated peptide mixtures under agitation and monitored fibril formation using CD and TEM, and additionally, visualization experiments using FM with fluorescent dyes. P1–P3 were, respectively, labelled before the experiments with FITC, 1,5-IAEDANS and Cy3. Preliminary experiments showed that fluorescent dye labelling had no effect on the fibril formation tendencies of each individual peptide. As shown in Fig. 6, CD measurements showed that all combinations of peptides tested underwent a conformational change to β-rich structure, and amyloid fibrils were observed in TEM measurements, although the morphologies were differed slightly depending on the combination of peptides used. We examined whether all of the labelled peptides used were in fact incorporated into the resultant fibrils by using FM. As shown in Fig. 6A, for fibrils formed from a mixture of P1 and P2 labelled peptides, both greenish-yellow fluorescence corresponding to FITC and blue-green fluorescence corresponding to AEDANS could be observed in the same fibril. We concluded from this that fibrils formed from a mixture of P1 and P2 peptides incorporated both peptides into the fibril. Similarly, FM of Fig. 6B–D showed that in each case, resultant fibril contained each peptide that was included in the initial reaction mixture. These results suggested that two and even three peptides could be readily incorporated into mixed amyloid fibrils.
Fig. 6.
Fibril formation of mixed core peptides. (A–D) CD (left panel), TEM (middle panel) and FM (right two panels) measurements of fibrils formed from an equimolar mixture of two or three core peptide fibrils. Peptide mixture combinations are (A) P1 and P2, (B) P2 and P3, (C) P1 and P3 and (D) P1–P3. CD and TEM analysis of samples were performed after a 3 or 20 h incubation in a siliconized test tube at 37°C with shaking (170 min−1). The scale bar in each panel of TEM measurement represents 200 nm. FM was measured using samples prepared on a non-fluorescent glass slide, and tightly sealed with a cover slip, and incubated at 37°C for 24 h in darkness. Fluorescence intensities specific for P1-FITC, P2-AEDANS and P3-Cy3 were observed using the appropriate emission filters. The scale bar in each panel of FM measurement represents 20 µm.
G93ASOD1 amyloid fibril formation
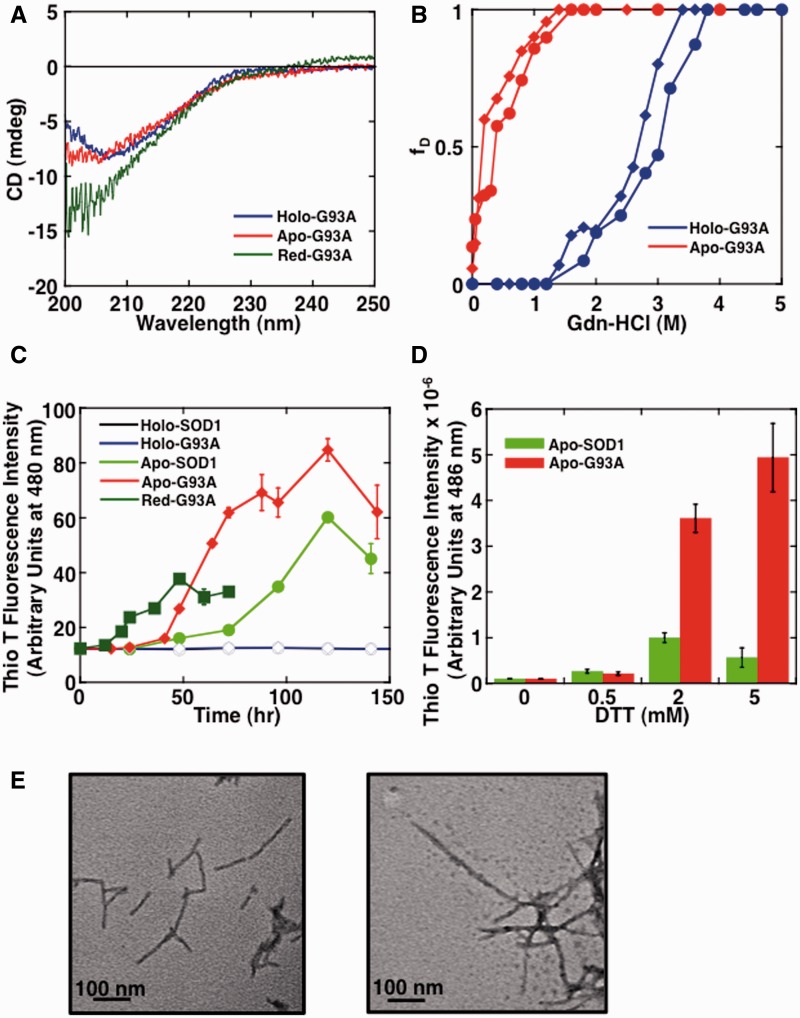
To confirm that the reduction of the Cys57-Cys146 intradisulfide bond triggers structural perturbations that are closely related to fibril formation in familial SOD1 mutants as well, we next performed analogous experiments on the familial ALS mutant G93ASOD1. In a manner similar to wild-type SOD1, we first observed the secondary structures and structural stabilities of G93ASOD1 and derivatives. As shown in Fig. 7A, the secondary structure of Holo-G93ASOD1 was similar to that of wild-type holo-SOD1 (Holo-SOD1). On the other hand, we observed that Apo-G93ASOD1 was slightly unfolded relative to that of wild-type apo-SOD1 (Apo-SOD1) as measured by CD. As shown in Fig. 7B, the Cm values of Holo-G93ASOD1 and Apo-G93ASOD1 unfolding were 2.85 M and 0.27 M Gdn-HCl, respectively. The results suggested that G93ASOD1 was significantly destabilized compared with wild-type SOD1, and this destabilization was irrespective of the presence or absence of metal ions (Table I). Next, to observe the formation of amyloid fibrils, Holo-SOD1, Apo-SOD1, Holo-G93ASOD1, Apo-G93ASOD1 and Red-G93ASOD1 were agitated by linear shaking under neutral pH in the presence of 2 mM DTT at 37°C. As shown in Fig. 7C, a dramatic increase in ThioT fluorescence was observed for all apo-samples in the presence of reducing reagent. On the other hand, no measurable increase was detected in the holo-samples. Interestingly, Apo-G93ASOD1 seemed to form fibrils at a faster rate than Apo-SOD1. However, the reduced mutant Red-G93ASOD1 formed fibrils the fastest among the samples tested. TEM measurements confirmed the presence of fibrils in each of the samples, and as shown in Fig. 7E, the widths of the Apo-SOD1 and Apo-G93ASOD1 fibrils were 12.0 ± 1.6 nm and 14.8 ± 1.6 nm, respectively. It should be noted that Apo-G93ASOD1 could not form any amyloid fibrils in the absence of reducing reagent, despite the fact that the stability of Apo-G93ASOD1 was lower than that of Red-SOD1 (based on comparison of the Cm value: Apo-G93ASOD1 (0.27 M) versus Red-SOD1 (0.42 M)). This result indicates that the reduction of intradisulfide bond is a prerequisite for triggering fibril formation.
Fig. 7.
Structural unfolding and amyloid fibrils formation of G93ASOD1. (A) CD spectra of Holo-G93ASOD1, Apo-G93ASOD1 and Red-G93ASOD1. (B) Structural unfolding transition curves of Holo-G93ASOD1 and Apo-G93ASOD1. The intensities of CD at 216 nm (closed circle) and fluorescence at 350 nm upon extension at 295 nm (closed diamond) at various concentrations of Gdn-HCl were normalized and plotted in terms of the fraction of unfolded protein (fD). (C, D) Amyloid fibril formation monitored by ThioT binding assay. Standard error bars from at least three independent measurements are also shown. In panel (C), conditions were 62.5 µM Holo-SOD1 (opened circle), Apo-SOD1 (closed circle), Holo-G93ASOD1 (opened diamond), Apo-G93ASOD1 (closed diamond) and Red-G93ASOD1 (closed square) in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl and 2 mM DTT at 37°C with shaking (160 min−1). In the panel (D), conditions were 62.5 µM Apo-SOD1 and Apo-G93ASOD1 in 50 mM Tris-HCl buffer, pH 7.0, containing 150 mM NaCl and 0–5 mM DTT at 37°C with shaking using Perkin Elmer ARVOx after 4 h incubations. (E) After testing in (C), TEM images were indicated for Apo-SOD1 fibrils (left panel) and Apo-G93ASOD1 fibrils (right panel) after 144 h incubation. The scale bar in each panel represents 100 nm.
To confirm that the added reducing reagent DTT does in fact reduce the Cys57-Cys146 intradisulfide bond, we next determined the amount of free SH groups in the amyloid fibril samples. The amount of free thiols in soluble Apo-SOD1 and Apo-G93ASOD1 samples were determined to be 1.46 mole SH/mole and 1.47 mole SH/mole, respectively, for a rough estimate of two free cysteine residues per SOD1 subunit. In contrast, after fibril formation, the amounts of free thiols in samples prepared by solubilizing fibril samples obtained from these two forms of SOD1 were estimated to be 2.97 mole SH/mole subunit. Since the amount of free thiols in extensively reduced SOD1 samples was determined to be 3.0 mol SH/mole using the same protocol, it was demonstrated that the intradisulfide bond had indeed been reduced almost completely in the fibrils. This clearly indicates that reduction of the intradisulfide bond between Cys57 and Cys146 is conductive to amyloid fibril formation. Additionally, we also investigated the effects of changing the concentration of DTT. As shown in Fig. 7D, after shaking for 4 h in a plate reader, the ThioT fluorescence intensity of Apo-G93ASOD1 increased in accordance with an increase in DTT concentration. This tendency was less pronounced in Apo-SOD1. This result suggested that Apo-G93ASOD1 is more susceptible to the reduction of the disulfide bond rather than Apo-SOD1 and consequently forms amyloid fibrils more readily. From these results, it is suggested strongly that the structural integrity of G93A is compromised by the removal of the two metal ions, resulting in an exposure of the intradisulfide bond to agents such as DTT. The consequence of this exposure would be reduction of this disulfide bond and the initiation of the fibrillation process.
Discussion
Structural stability and amyloid fibril formation
The focus of our research was to understand the structural basis for amyloid fibril formation of SOD1 in light of its close association to the neurological disease ALS (12, 13, 15). We compared the characteristics of two SOD1 types: wild-type and the familial ALS-affiliated G93A mutant, which is often used as a model protein for fALS studies (18–20, 27, 31, 39). We found that the removal of metal ions destabilized the structures of both wild-type and mutant SOD1 considerably (Figs 2 and 7, Table I). In particular for Apo-G93ASOD1, we deduced that the structure had been partially unfolded, judging from CD spectra comparison (Fig. 7A). This destabilization could be explained by the idea that the metal ions stabilize the Phe50-Gly82 loop of SOD1 and contribute thereby to structural stability (40, 41).
In addition, we found that the intramolecular disulfide bond that bridges Cys57 and Cys146 significantly contributed to the structural stability of SOD1. In the case of wild-type SOD1, although the reduction of the disulfide bond had almost no effects on the native secondary structure in CD measurements, the structural stability was remarkably decreased (Fig. 2). In the case of G93ASOD1, however, it was shown that the secondary structure was completely unfolded upon reduction of the intramolecular disulfide bond (Fig. 7A). The disulfide-reduced proteins (Red-SOD1, 2SCAMRed-SOD1 and Red-G93ASOD1) formed typical fibrils in vitro (Figs 3 and 7B). In contrast, although 2SCAM-SOD1 was seen to be partially unfolded (Fig. 2A) and Apo-G93ASOD1 was determined to be less stable than Red-SOD1 in denaturation experiments (Table I), these two SOD1 derivatives did not form fibrils. This finding clearly indicated that the reduction of the intramolecular disulfide bond, rather than differences in overall structural stability, is the critical deciding factor for triggering the formation of fibrils. Several groups have reported that removal of metal ions and reduction of the disulfide bond of SOD1 lead to increased flexibility of loops Phe50-Gly82 and Glu121-Ser142 (40), exposes Cys57 (42) and promotes dissociation of native dimer (40, 43, 44), underscoring the importance of this disulfide bond in molecular flexibility. Although Red-SOD1 initially formed fibrils at a slower rate compared with 4SCAM-SOD1, the rate of this fibril formation could be significantly accelerated by the addition of modest concentrations of Gdn-HCl (0.4 M; corresponding to the Cm of the Red-SOD1 unfolding transition (Fig. 2B)). This enhanced rate (data not shown) was similar to the rate of 4SCAM-SOD1 fibrillation as shown in Fig. 3A. It has been demonstrated that the addition of denaturants such as Gdn-HCl or urea promotes fibril formation of proteins such as insulin (45, 46) and GroES protein (47), suggesting that a globally unfolded protein conformation favours fibril formation. This idea is also supported by previous observations that hydrophobic regions, which are normally sequestered inside the folded structure, may be exposed by partial unfolding, and that this exposure sometimes leads to fibrils (48). These results indicated that once the reduction of the disulfide bond occurs, fluctuations and global unfolding may follow, and then fibril formation of SOD1 will be promoted. Thus, the reduction of disulfide bond acts as a trigger of fibril formation.
Fibril formation in the presence of DTT that mimics the physiological conditions
As shown in the Results section, apo-proteins formed fibrils only when they were incubated with the reducing agent DTT. However, as shown in Fig. 7C and D, Apo-G93ASOD1 formed fibrils faster than Apo-SOD1 in the presence of DTT, and the fibril formation rate of Apo-G93ASOD1 was accelerated in proportion to increased concentrations of DTT. This result could be explained by considering dynamic as well as static aspects of structural stability; Apo-G93ASOD1 is partially unfolded with a high degree of dynamic structural fluctuation and so DTT is able to interact more easily with the buried disulfide bond (42, 46). Similar findings were reported for β2-microglobulin, in which a disulfide bond buried in the interior of the molecule (49) was reduced rapidly in the partially unfolded state (50–52). In contrast, holo-proteins did not form fibrils even in the presence of DTT. This was thought to be due to the idea that holo-SOD1 was quite stable, with a low degree of structural fluctuation due to stabilization through binding of metal ions (40, 41, 43, 53). The structural rigidity brought about by metal binding precludes access of DTT to the buried disulfide bond. Such relationships between molecular dynamics and reduction of disulfide bond located in the interior of molecule have also been reported for immunoglobulin light chain (54–57).
From our results that found that the reduction of the buried disulfide bond of apo-SOD1 proteins acted as a trigger for amyloid fibrils formation in the presence of DTT, it is intriguing to consider a plausible mechanism of SOD1 fibril formation that may be relevant in the cell. SOD1 is highly conserved and located in the cytosol (5), where conditions favour a reducing environment maintained by reduced glutathione (58). If the presence of DTT may be interpreted to mimic this reductive cytosolic environment, the appearance of significant fractions of apo-SOD1 may be a contributing factor to the onset of ALS. This notion is attractive to us because we discovered that Apo-G93ASOD1 formed fibrils more readily than Apo-SOD1 in the presence of DTT (Fig. 7C), which suggests a more easily triggered fibrillation mechanism in the mutant enzyme. Also, aggregates composed of the reduced form of SOD1 protein are indeed found in the spinal cord of G85RSOD1 transgenic mice (19, 21), suggesting that reduction of the disulfide bond does occur in vivo (30, 58, 59) and does form deposits in afflicted mice. Since Holo-G93ASOD1 did not form any amyloid fibrils under physiological conditions (Fig. 7C), maintaining the holo-form may be important in preventing the onset of ALS. In this context, a recent report that G93ASOD1 transgenic mice that are also deficient in metallothioneins that can function as zinc chaperones for apo-SOD1 display significantly higher mortality rates compared with G93ASOD1 mice without this additional deficiency (60). Perhaps the enhanced supply of Zn ions through chaperoning act as an additional preventive measure for amyloidosis in wild-type mice.
Identification of core peptides and its fibrillation
Our experiments identified three distinct peptide sequence regions in Red-SOD1 that formed the protease-resistant core of SOD1 fibrils. Interestingly, these three regions were dispersed throughout the amino acid sequence (N-terminal, internal and C-terminal regions; Table II and Fig. 1). The Waltz amyloidogenic sequence identification program (Switch Laboratory), predicted that the amyloidogenic regions in the SOD1 amino acid sequence were 14Val-Gln-Gly-Ile-Ile-Asn-Phe-Glu-Gln22, 89Ala-Asp-Val-Ser-Ile-Glu-Asp-Ser-Val-Ile-Ser-Leu-Ser-Gly-Asp-His-Cys-Ile-Ile-Gly114 and 145Ala-Cys-Gly-Val-Ile-Gly-Ile-Ala-Gln153. These identified regions were very consistent with the regions experimentally determined by us for Red-SOD1 (Fig. 1). Notably, since the C-terminal core region includes Cys146, that forms the stabilizing intramolecular disulfide bond with Cys57, this region is of particular interest when we consider the possible sequence of events that lead up to fibril formation.
As shown in Figs 5 and 6, the three core peptides were all capable of forming amyloid fibrils, either individually or in mixtures of peptides in all possible combinations; no example of a peptide interfering with the fibrils formation of another was detected. Additionally, we determined that all three peptides were incorporated into the resultant fibrils, and no biases among the three peptides were seen in the finished fibrils (as determined by FM). Our results were the first to demonstrate fibril formation by using these specific short peptides, although a similar study using core regions was reported for several SOD1 mutants (61). It had been an open question as to how exactly do these three core regions participate in fibril formation. An example where a single core peptide region among several potential core regions was able to form fibrils was also observed for His-tagged halophilic protein (62). Various details, such as the precise ratio of the core peptides incorporated into the finished fibrils, as well as the order of interaction of each peptide within the fibrils, remain to be determined. Our results are in line with other reports that show fibril formation of heterologous proteins, such as a short peptide derived from transthyretin and insulin (63), α-synuclein and GroES (64) and lysozyme and ovalbumin (65).
Fibril formation mechanism of SOD1
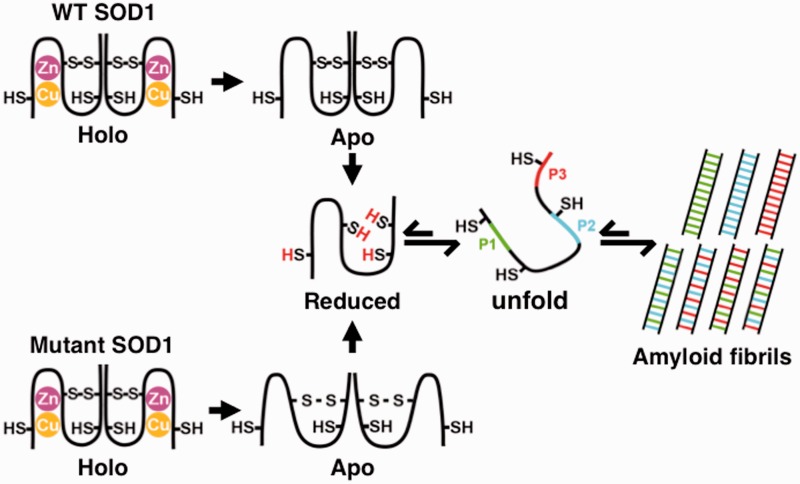
Based on the results in our study, a possible mechanism of SOD1 amyloid fibril formation may be summarized as shown in Fig. 8. The holo-form of SOD1, which has one intramolecular disulfide bond between Cys57 and Cys146 and Cu and Zn metals bound forms a stable dimer. When the Cu and Zn metals are removed to yield apo-form, the structural stability decreases significantly. If the disulfide bond of wild-type Apo-SOD1 is then reduced, Red-SOD1 dissociates to monomer with a large fluctuation. This monomer species unfold readily. Amyloid fibrils are begun via this reduced unfolded species. This unfolded species in the equilibrium state are able to be enhanced by alkylation of the reduced cysteine residues. Three specific peptide regions in the SOD1 sequence, P1–P3, interact to form the amyloid fibril core. Regarding the familial ALS mutant G93ASOD1, the apo-form of the mutant is more unstable than the apo-form of wild-type SOD1 and is therefore more sensitive to reduction of its important intradisulfide bond by reducing reagents, and as a consequence, is more susceptible to amyloid fibril formation. When taken together with the normal reductive environment of the cytosol, the mutant SOD1 would be more likely to form amyloid fibrils, which may hasten the onset of ALS.
Fig. 8.
Plausible mechanism of amyloid fibril formation of SOD1. A schematic representation of structural states and amyloid fibril formation mechanism of wild-type SOD1 and G93ASOD1. When Cu and Zn ions in holo-state are removed, apo-SOD1 is formed. The structural stability of the apo-form is significantly lowered and overall molecular fluctuation is increased. When reduction of the intramolecular disulfide bond occurs, a more unstable monomeric SOD1 with high fluctuation is formed. For the familial mutant G93A, the mutation destabilizes the subunit stability, which increases dynamic fluctuation and facilitates disulfide reduction, ultimately leading to fibril formation. Three specific peptide regions, dispersed through the SOD1 sequence (N-terminal, internal and C-terminal regions) that are responsible for the proteolysis-resistant fibril core formation, interact one another to form mature amyloid fibrils. See text for detail.
Although the characteristics of fibril formation and onset of ALS were somewhat different depending on the mutants (28, 66), it was shown that most mutations resulted in destabilization of SOD1 structure and dynamic conformational alteration (67, 68). From our findings in this study, it would be reasonable to consider that those mutants are susceptible to reduction of the intramolecular disulfide bond not only in vitro (31) but also in vivo. This understanding on the fundamental mechanism of fibril formation may shed a light to a medical treatment and/or prevention of the onset of ALS.
Acknowledgements
The authors thank E. Kawahara of Tottori University for technical assistance in measurements of electron microscopy.
Funding
This work was supported in part by Grant-in-Aid for Scientific Research (C) (No.25330027 to Y.K.) from the Japan Society for the Promotion of Science (JSPS), Grant-in-Aid for Scientific Research on Innovative Areas (No.24113716 to Y.K.) and the X-Ray Free Electron Laser Priority Strategy Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). This study has also been partially supported by financial aid from Wakasa Seikatsu Co. Japan.
Conflict of Interest
None declared.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- API
Achromobacter lyticus protease I
- Apo-
Cu and Zn metal free form
- CD
circular dichroism
- Cy3
Cy3 maleimide
- DTT
dithiothreitol
- fALS
familial form of amyotrophic lateral sclerosis
- FITC
fluorescein 5-isothiocyanate
- FM
fluorescence microscopy
- Gdn-HCl
guanidine hydrochloride
- G93ASOD1
SOD1 with mutation of G93A
- Holo-
Cu and Zn metal bound form
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- PK
proteinase K
- Red-
reduced form of intramolecular disulfide bond between Cys57 and Cys146
- SOD1
Cu, Zn-superoxide dismutase
- TCEP
tris(2-carboxyethyl)phosphine
- TEM
transmission electron microscopy
- ThioT
Thioflavin T
- 1
5-IAEDANS, N-(iodoacetyl)-N'-(5-sulfo-l-naphthyl)-ethylenediamine
- 2SCAM-SOD1
SOD1 derivative where the free thiols of Cys6 and Cys111 are carboxyamidated
- 2SCAMRed-SOD1
SOD1 derivative in which the free thiols of Cys6 and Cys111 have been carboxyamidated and the intra-disulfide bond of Cys57-Cys146 has been reduced
- 4SCAM-SOD1
SOD1 derivative where all of the thiols in Cys6, Cys 57, Cys 111 and Cys146 have been carboxyamidated
References
- 1.Bruijn L.I., Miller T.M., Cleveland D.W. (2004) Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 27, 723–749 [DOI] [PubMed] [Google Scholar]
- 2.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.X., Rahmani Z., Krizus A., McKenna-Yasek D., Cayabyab A., Gaston S. M., Berger R., Tanzi R. E., Halperin J. J., Herzfeldt B., Van den Bergh R., Hung W., Bird T., Deng G., Mulder D. W., Smyth C., Laing N. G., Soriano E., Pericak–Vance M. A., Haines J., Rouleau G. A., Gusella J. S., Horvitz H. R., Brown R. H., Jr (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 3.Orrell R.W. (2000) Amyotrophic lateral sclerosis: copper/zinc superoxide dismutase (SOD1) gene mutations. Neuromuscul. Disord. 10, 63–68 [DOI] [PubMed] [Google Scholar]
- 4.Valentine J.S., Doucette P.A., Zittin Potter S. (2005) Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 74, 563–593 [DOI] [PubMed] [Google Scholar]
- 5.Crapo J.D., Oury T., Rabouille C., Slot J.W., Chang L.Y. (1992) Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. USA 89, 10405–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barra D., Martini F., Bannister J.V., Schinina M.E., Rotilio G., Bannister W.H., Bossa F. (1980) The complete amino acid sequence of human Cu/Zn superoxide dismutase. FEBS Lett. 120, 53–56 [DOI] [PubMed] [Google Scholar]
- 7.Strange R.W., Antonyuk S., Hough M.A., Doucette P.A., Rodriguez J.A., Hart P.J., Hayward L.J., Valentine J.S., Hasnain S.S. (2003) The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 328, 877–891 [DOI] [PubMed] [Google Scholar]
- 8.Briggs R.G., Fee J.A. (1978) Further characterization of human erythrocyte superoxide dismutase. Biochim. Biophys. Acta 537, 86–99 [DOI] [PubMed] [Google Scholar]
- 9.Reaume A.G., Elliott J.L., Hoffman E.K., Kowall N.W., Ferrante R.J., Siwek D.F., Wilcox H.M., Flood D.G., Beal M.F., Brown R.H., Jr., Scott R.W., Snider W.D. (1996) Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 13, 43–47 [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Xu G., Li H., Gonzales V., Fromholt D., Karch C., Copeland N.G., Jenkins N.A., Borchelt D.R. (2005) Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: alphaB-crystallin modulates aggregation. Hum. Mol. Genet. 14, 2335–2347 [DOI] [PubMed] [Google Scholar]
- 11.Rakhit R., Chakrabartty A. (2006) Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1762, 1025–1037 [DOI] [PubMed] [Google Scholar]
- 12.Shibata N., Hirano A., Kobayashi M., Sasaki S., Kato T., Matsumoto S., Shiozawa Z., Komori T., Ikemoto A., Umahara T., Asayama K. (1994) Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci. Lett. 179, 149–152 [DOI] [PubMed] [Google Scholar]
- 13.Forsberg K., Jonsson P.A., Andersen P.M., Bergemalm D., Graffmo K.S., Hultdin M., Jacobsson J., Rosquist R., Marklund S.L., Brannstrom T. (2010) Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS One 5, e11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad L.I. (2014) Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 111, 3620–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata N., Hirano A., Kobayashi M., Asayama K., Umahara T., Komori T., Ikemoto A. (1993) lmmunohistochemical demonstration of Cu/Zn superoxide dismutase in the spinal cord of patients with familial amyotrophic lateral sclerosis. Acta Histochem. Cytochem. 26, 619–622 [Google Scholar]
- 16.Watanabe M., Dykes-Hoberg M., Culotta V.C., Price D.L., Wong P.C., Rothstein J.D. (2001) Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 8, 933–941 [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. (2003) Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 12, 2753–2764 [DOI] [PubMed] [Google Scholar]
- 18.Basso M., Massignan T., Samengo G., Cheroni C., De Biasi S., Salmona M., Bendotti C., Bonetto V. (2006) Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J. Biol. Chem. 281, 33325–33335 [DOI] [PubMed] [Google Scholar]
- 19.Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Lindberg M., Oliveberg M., Marklund S.L. (2006) Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain 129, 451–464 [DOI] [PubMed] [Google Scholar]
- 20.Rakhit R., Robertson J., Vande Velde C., Horne P., Ruth D.M., Griffin J., Cleveland D.W., Cashman N.R., Chakrabartty A. (2007) An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat. Med. 13, 754–759 [DOI] [PubMed] [Google Scholar]
- 21.Zetterstrom P., Stewart H.G., Bergemalm D., Jonsson P.A., Graffmo K. S., Andersen P.M., Brannstrom T., Oliveberg M., Marklund S.L. (2007) Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl. Acad. Sci. USA 104, 14157–14162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaarsma D., Teuling E., Haasdijk E.D., De Zeeuw C.I., Hoogenraad C.C. (2008) Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J. Neurosci. 28, 2075–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karch C.M., Prudencio M., Winkler D.D., Hart P.J., Borchelt D.R. (2009) Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl. Acad. Sci. USA 106, 7774–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa Y., Kaneko K., Yamanaka K., O’Halloran T.V., Nukina N. (2008) Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 283, 24167–24176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selkoe D.J. (2003) Folding proteins in fatal ways. Nature 426, 900–904 [DOI] [PubMed] [Google Scholar]
- 26.Shastry B.S. (2003) Neurodegenerative disorders of protein aggregation. Neurochem. Int. 43, 1–7 [DOI] [PubMed] [Google Scholar]
- 27.Chattopadhyay M., Durazo A., Sohn S.H., Strong C.D., Gralla E.B., Whitelegge J.P., Valentine J.S. (2008) Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. USA 105, 18663–18668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oztug Durer Z.A., Cohlberg J.A., Dinh P., Padua S., Ehrenclou K., Downes S., Tan J.K., Nakano Y., Bowman C.J., Hoskins J.L., Kwon C., Mason A.Z., Rodriguez J.A., Doucette P.A., Shaw B.F., Valentine J.S. (2009) Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One 4, e5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toichi K., Yamanaka K., Furukawa Y. (2013) Disulfide scrambling describes the oligomer formation of superoxide dismutase (SOD1) proteins in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 288, 4970–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer F.Q., Buettner G.R. (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212 [DOI] [PubMed] [Google Scholar]
- 31.Tiwari A., Hayward L.J. (2003) Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J. Biol. Chem. 278, 5984–5992 [DOI] [PubMed] [Google Scholar]
- 32.Yagi H., Sato A., Yoshida A., Hattori Y., Hara M., Shimamura J., Sakane I., Hongo K., Mizobata T., Kawata Y. (2008) Fibril formation of hsp10 homologue proteins and determination of fibril core regions: differences in fibril core regions dependent on subtle differences in amino acid sequence. J. Mol. Biol. 377, 1593–1606 [DOI] [PubMed] [Google Scholar]
- 33.Yagi H., Takeuchi H., Ogawa S., Ito N., Sakane I., Hongo K., Mizobata T., Goto Y., Kawata Y. (2010) Isolation of short peptide fragments from alpha-synuclein fibril core identifies a residue important for fibril nucleation: a possible implication for diagnostic applications. Biochim. Biophys. Acta 1804, 2077–2087 [DOI] [PubMed] [Google Scholar]
- 34.Higurashi T., Nosaka K., Mizobata T., Nagai J., Kawata Y. (1999) Unfolding and refolding of Escherichia coli chaperonin GroES is expressed by a three-state model. J. Mol. Biol. 291, 703–713 [DOI] [PubMed] [Google Scholar]
- 35.Serpell L.C. (2000) Alzheimer’s amyloid fibrils: structure and assembly. Biochim. Biophys. Acta 1502, 16–30 [DOI] [PubMed] [Google Scholar]
- 36.Sakiyama F., Masaki T. (1994) Lysyl endopeptidase of Achromobacter lyticus. Methods Enzymol. 244, 126–137 [DOI] [PubMed] [Google Scholar]
- 37.Tenidis K., Waldner M., Bernhagen J., Fischle W., Bergmann M., Weber M., Merkle M.L., Voelter W., Brunner H., Kapurniotu A. (2000) Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 295, 1055–1071 [DOI] [PubMed] [Google Scholar]
- 38.Nelson R., Sawaya M.R., Balbirnie M., Madsen A.O., Riekel C., Grothe R., Eisenberg D. (2005) Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindberg M.J., Tibell L., Oliveberg M. (2002) Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc. Natl. Acad. Sci. USA 99, 16607–16612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding F., Dokholyan N.V. (2008) Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc. Natl. Acad. Sci. USA 105, 19696–19701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teilum K., Smith M.H., Schulz E., Christensen L.C., Solomentsev G., Oliveberg M., Akke M. (2009) Transient structural distortion of metal-free Cu/Zn superoxide dismutase triggers aberrant oligomerization. Proc. Natl. Acad. Sci. USA 106, 18273–18278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banci L., Bertini I., Cantini F., D’Amelio N., Gaggelli E. (2006) Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form. J. Biol. Chem. 281, 2333–2337 [DOI] [PubMed] [Google Scholar]
- 43.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O’Halloran T.V. (2004) The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 279, 47998–48003 [DOI] [PubMed] [Google Scholar]
- 44.Doucette P.A., Whitson L.J., Cao X., Schirf V., Demeler B., Valentine J.S., Hansen J.C., Hart P.J. (2004) Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J. Biol. Chem. 279, 54558–54566 [DOI] [PubMed] [Google Scholar]
- 45.Ahmad A., Millett I.S., Doniach S., Uversky V.N., Fink A.L. (2003) Partially folded intermediates in insulin fibrillation. Biochemistry 42, 11404–11416 [DOI] [PubMed] [Google Scholar]
- 46.Ahmad S., Gromiha M., Fawareh H., Sarai A. (2004) ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higurashi T., Yagi H., Mizobata T., Kawata Y. (2005) Amyloid-like fibril formation of co-chaperonin GroES: nucleation and extension prefer different degrees of molecular compactness. J. Mol. Biol. 351, 1057–1069 [DOI] [PubMed] [Google Scholar]
- 48.Dobson C.M. (2003) Protein folding and misfolding. Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 49.Hong D.P., Gozu M., Hasegawa K., Naiki H., Goto Y. (2002) Conformation of beta 2-microglobulin amyloid fibrils analyzed by reduction of the disulfide bond. J. Biol. Chem. 277, 21554–21560 [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto S., Hasegawa K., Yamaguchi I., Tsutsumi S., Kardos J., Goto Y., Gejyo F., Naiki H. (2004) Low concentrations of sodium dodecyl sulfate induce the extension of beta 2-microglobulin-related amyloid fibrils at a neutral pH. Biochemistry 43, 11075–11082 [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto S., Yamaguchi I., Hasegawa K., Tsutsumi S., Goto Y., Gejyo F., Naiki H. (2004) Glycosaminoglycans enhance the trifluoroethanol-induced extension of beta 2-microglobulin-related amyloid fibrils at a neutral pH. J. Am. Soc. Nephrol. 15, 126–133 [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto K., Yagi H., Ozawa D., Sasahara K., Naiki H., Goto Y. (2008) Thiol compounds inhibit the formation of amyloid fibrils by beta 2-microglobulin at neutral pH. J. Mol. Biol. 376, 258–268 [DOI] [PubMed] [Google Scholar]
- 53.Hornberg A., Logan D.T., Marklund S.L., Oliveberg M. (2007) The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J. Mol. Biol. 365, 333–342 [DOI] [PubMed] [Google Scholar]
- 54.Goto Y., Hamaguchi K. (1981) Formation of the intrachain disulfide bond in the constant fragment of the immunoglobulin light chain. J. Mol. Biol. 146, 321–340 [DOI] [PubMed] [Google Scholar]
- 55.Goto Y., Hamaguchi K. (1982) Unfolding and refolding of the reduced constant fragment of the immunoglobulin light chain. Kinetic role of the intrachain disulfide bond. J. Mol. Biol. 156, 911–926 [DOI] [PubMed] [Google Scholar]
- 56.Kawata Y., Goto Y., Hamaguchi K., Hayashi F., Kobayashi Y., Kyogoku Y. (1988) Hydrogen-exchange kinetics of the indole NH proton of the buried tryptophan in the constant fragment of the immunoglobulin light chain. Biochemistry 27, 346–350 [DOI] [PubMed] [Google Scholar]
- 57.Kawata Y., Hamaguchi K. (1990) Global fluctuations of the immunoglobulin domains under physiological conditions. Biopolymers 30, 389–394 [DOI] [PubMed] [Google Scholar]
- 58.Hwang C., Sinskey A.J., Lodish H.F. (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 [DOI] [PubMed] [Google Scholar]
- 59.Sevier C.S., Kaiser C.A. (2002) Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 3, 836–847 [DOI] [PubMed] [Google Scholar]
- 60.Puttaparthi K., Gitomer W.L., Krishnan U., Son M., Rajendran B., Elliott J.L. (2002) Disease progression in a transgenic model of familial amyotrophic lateral sclerosis is dependent on both neuronal and non-neuronal zinc binding proteins. J. Neurosci. 22, 8790–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furukawa Y., Kaneko K., Yamanaka K., Nukina N. (2010) Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 285, 22221–22231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokunaga Y., Matsumoto M., Tokunaga M., Arakawa T., Sugimoto Y. (2013) Amyloid fibril formation in vitro from halophilic metal binding protein: its high solubility and reversibility minimized formation of amorphous protein aggregations. Protein Sci. 22, 1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacPhee C.E., Dobson C.M. (2000) Formation of mixed fibrils demonstrates the generic nature and potential utility of amyloid nanostructures. J. Am. Chem. Soc. 122, 12707–12713 [Google Scholar]
- 64.Yagi H., Kusaka E., Hongo K., Mizobata T., Kawata Y. (2005) Amyloid fibril formation of alpha-synuclein is accelerated by preformed amyloid seeds of other proteins: implications for the mechanism of transmissible conformational diseases. J. Biol. Chem. 280, 38609–38616 [DOI] [PubMed] [Google Scholar]
- 65.Sugimoto Y., Kamada Y., Tokunaga Y., Shinohara H., Matsumoto M., Kusakabe T., Ohkuri T., Ueda T. (2011) Aggregates with lysozyme and ovalbumin show features of amyloid-like fibrils. Biochem. Cell Biol. 89, 533–544 [DOI] [PubMed] [Google Scholar]
- 66.Wang Q., Johnson J.L., Agar N.Y., Agar J.N. (2008) Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 6, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uversky V.N., Fink A.L. (2004) Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta 1698, 131–153 [DOI] [PubMed] [Google Scholar]
- 68.Molnar K.S., Karabacak N.M., Johnson J.L., Wang Q., Tiwari A., Hayward L.J., Coales S.J., Hamuro Y., Agar J.N. (2009) A common property of amyotrophic lateral sclerosis-associated variants: destabilization of the copper/zinc superoxide dismutase electrostatic loop. J. Biol. Chem. 284, 30965–30973 [DOI] [PMC free article] [PubMed] [Google Scholar]