Abstract
Once perceived as merely selfish, transposable elements (TEs) are now recognized as potent agents of adaptation. One way TEs contribute to evolution is through TE exaptation, a process whereby TEs, which persist by replicating in the genome, transform into novel host genes, which persist by conferring phenotypic benefits. Known exapted TEs (ETEs) contribute diverse and vital functions, and may facilitate punctuated equilibrium, yet little is known about this process. To better understand TE exaptation, we designed an approach to resolve the phylogenetic context and timing of exaptation events and subsequent patterns of ETE diversification. Starting with known ETEs, we search in diverse genomes for basal ETEs and closely related TEs, carefully curate the numerous candidate sequences, and infer detailed phylogenies. To distinguish TEs from ETEs, we also weigh several key genomic characteristics including repetitiveness, terminal repeats, pseudogenic features, and conserved domains. Applying this approach to the well-characterized plant ETEs MUG and FHY3, we show that each group is paraphyletic and we argue that this pattern demonstrates that each originated in not one but multiple exaptation events. These exaptations and subsequent ETE diversification occurred throughout angiosperm evolution including the crown group expansion, the angiosperm radiation, and the primitive evolution of angiosperms. In addition, we detect evidence of several putative novel ETE families. Our findings support the hypothesis that TE exaptation generates novel genes more frequently than is currently thought, often coinciding with key periods of evolution.
Keywords: MUSTANG, FAR1, FHY3, FRS, transposable elements, exaptation, molecular domestication, phylogeny, evolution, mutator, MULE, Phox/Bem1p, PB1, Peptidase C48, angiosperm radiation, adaptation, transposon, co-option, selective constraint.
Background
Transposable elements (TEs) are DNA segments that mediate their own duplication and thereby accumulate to high abundance in most eukaryotic genomes. Because of this, TEs were once perceived as selfish (Doolittle and Sapienza 1980; Orgel and Crick 1980); however, it is now understood that they confer many benefits, such as maintaining genome structure, generating variation, and producing evolutionary innovation (Flagel and Wendel 2009; Lisch 2009; Parisod et al. 2010; Rebollo et al. 2010; Levin and Moran 2011; Pardue and DeBaryshe 2011). In plants, TEs are important contributors to evolution and diversity; for example, in more than 60 reported instances, TEs have modified existing genes or given rise to novel phenotypic genes (Oliver et al. 2013).
One mechanism through which TEs contribute to evolution is exaptation. Although the familiar term “adaptation” refers to biological features selected to increase the benefit of existing roles, the term “exaptation” instead refers to features co-opted to perform entirely new roles (Gould and Vrba 1982; Gould and Lloyd 1999). More specifically, TE exaptation (also referred to as co-option or molecular domestication) (Miller et al. 1992) is a process by which a TE, originally conserved through “self-replicative selection,” transitions to increase the fitness of the organism and becomes conserved through “phenotypic selection” (Doolittle and Sapienza 1980; Hoen and Bureau 2015). In eukaryotes, TE exaptation has made possible major evolutionary innovations, including the vertebrate adaptive immune system (Agrawal et al. 1998; Kapitonov and Jurka 2004, 2005), the mammalian placenta (Rawn and Cross 2008), and human cognition (Zhang et al. 2015). Until recently, most exapted TE genes (ETEs) were discovered fortuitously in forward genetic screens (e.g., Bundock and Hooykaas 2005; Lin et al. 2007); however, advances in sequencing technology have enabled us to directly identify putative ETEs using computational analysis of genomic data (Hoen and Bureau 2015), then validate them by reverse genetic characterization of phenotypes (Cowan et al. 2005; Joly-Lopez et al. 2012). For instance, in a large-scale search for ETEs in one family of plants (the Brassicaceae), we recently identified 67 ETEs, more than half of them novel, suggesting that ETEs may be far more abundant than previously thought (Hoen and Bureau 2015).
Discoveries such as this magnify the importance of better understanding the TE exaptation process. However, like many evolutionary mechanisms involving TEs, investigating exaptation can be difficult. One particularly challenging aspect is reconstructing ETE evolutionary histories in order to determine the number and timing of exaptation events, and the identity of the ancestral genomes in which they arose. Such analyses require identifying both ETEs and closely related TEs. But because TE families frequently go extinct within genomes (Donoghue et al. 2011), TEs directly descended from the progenitors of an ETE family may no longer exist, or if they do exist may be present in only a small fraction of genomes. Furthermore, even if TEs closely related to an ETE family do exist in a sequenced genome, identifying and analyzing them may be difficult given the large number of sequenced genomes and vast number of TEs that each genome may contain. Nevertheless, investigating the evolutionary history of ETEs is worthwhile and increasingly feasible as the number of sequenced genomes continues to grow. A better understanding of the origins of ETEs would also help to guide experimental studies, since ETEs that originated in different exaptation events ought also to have different functions.
We therefore undertook to demonstrate the feasibility and value of such investigations by thoroughly characterizing the evolutionary history of the two largest and best-characterized families of ETEs in plants: MUSTANG (MUG) and FAR1-RELATED SEQUENCES (FRS). MUG, which descended from TEs of the Mutator-like element (MULE) superfamily (Cowan et al. 2005; Joly-Lopez et al. 2012), consists in Arabidopsis thaliana of eight genes equally divided between two subfamilies, MUGA and MUGB (Joly-Lopez et al. 2012). Double mutants within each subfamily (mug1 mug2 in MUGA and mug7 mug8 in MUGB) produce stronger phenotypes than the single mutants and, despite similar broad phenotypes (e.g., delayed flowering and reduced yield), each subfamily exhibits different specific phenotypes such as reduced chlorophyll production in mug1 mug2 (Joly-Lopez et al. 2012). In addition to the conserved domains typically associated with MULE transposases, MUGB but not MUGA genes contain a Phox and Bem1p (PB1) domain, which adopts a ubiquitin-like β-grasp fold structure (Sumimoto et al. 2007). Plant genomes have greater numbers of PB1-containing genes than other eukaryotes, and although a few have been characterized and found to be involved in a wide range of biological processes, little is known about the source or biological function of most plant PB1 domains (Borisov et al. 2003; Prasad et al. 2010; Guilfoyle and Hagen 2012; Trehin et al. 2013; Chardin et al. 2014; Korasick, et al. 2014; Zientara-Rytter and Sirko 2014). MUG genes are present in both monocots and eudicots, and likely in basal angiosperms, but have not been identified in other plants, suggesting that exaptation might have occurred during early angiosperm evolution (Cowan et al. 2005; Saccaro et al. 2007; Joly-Lopez et al. 2012). Interestingly, at the time of the monocot-eudicot split, MUGA had already undergone at least two conserved duplications whereas MUGB had undergone none (Joly-Lopez et al. 2012). Overall, such differences in phenotype, gene structure, and phylogeny show that, despite their close similarity in sequence, MUGA and MUGB followed different evolutionary trajectories; however, it is not yet known whether these differences occurred prior or subsequent to exaptation.
The second well-characterized group of plant ETEs, FRS, consist in A. thaliana of the genes FAR-RED IMPAIRED RESPONSE 1 (FAR1) and FAR-RED ELONGATED HYPOCOTYLS 3 (FHY3) (Whitelam et al. 1993; Hudson et al. 1999), as well as 12 additional FAR1-RELATED SEQUENCE (FRS) (Arabidopsis Genome 2000; Hudson et al. 2003; Lin and Wang 2004). FAR1 and FHY3 encode transcription factors essential for far-red light responses controlled by phytochrome A (Lin et al. 2007). They also play roles in diverse developmental and physiological processes (Lin and Wang 2004; Li et al. 2011; Ouyang et al. 2011; Huang et al. 2012; Stirnberg et al. 2012; Gao et al. 2013; Tang et al. 2013). The remaining FRS genes are thus far not well characterized, but have been suggested to play distinct roles in light-controlled development in Arabidopsis (Lin and Wang 2004). Like MUG, the FRS family was derived from MULEs and is found in monocots as well as eudicots, so are thought to have originated in one or more exaptation events during early angiosperm evolution (Lin et al. 2007).
To better understand TE exaptation and the evolutionary histories of MUG and FRS, we explore the following questions: How many TE exaptation events led to the formation of these two groups of ETEs? When did these exaptations occur and did they happen in close succession or at widely separated times? Did they involve similar or divergent MULE families, and might any characteristics of the progenitor TE families shed light on the structural and functional characteristics of the ETEs? After they had been established, how often and when did each ETE family diversify? Finally, what does the timing of exaptation and diversification events suggest about their potential role in evolution? To answer these questions, we take the following approach: 1) identify sequences closely related to MUG or FRS in diverse genomes, 2) generate curated, reliable phylogenies, and 3) measure genomic attributes that differentiate ETEs from TEs. We then analyze the results to determine how many exaptation events occurred and their timing. Our findings reveal that TE exaptation has contributed more than previously understood to angiosperm evolution, and likely provide many functions of potential practical importance that have yet to be discovered.
Results and Discussion
Identifying Genomes of Interest
To maximize our chances of finding extant TEs closely related to MUG and to sample diverse ETE clades, we searched a large number (62) of genomes including representatives from all major angiosperm lineages (supplementary table S1, Supplementary Material online). We expected to find thousands of sequences (mainly TEs) but did not want to preclude any TEs or ETEs of potential interest, even in such distant genomes as Picea abies. We thus devised a strategy to screen for genomes of potential interest (fig. 1). First, we determined which genomes contained any sequences of interest by searching each genome individually. Then we selected the genomes (22) that appeared, according to their individual phylogenetic trees, to contain TEs descended from the last common ancestor of all MUG query sequences (supplementary fig. S1, Supplementary Material online). We also retained genomes with apparent TEs that contained a PB1 domain (see below). For additional analyses, we added three genomes with key positions in the species phylogeny but which were expressed sequence tag (EST) assemblies rather than fully sequenced: the basal eudicot Nelumbo nucifera, the magnoliid Persea americana, and the basal angiosperm Nuphar advena. In total, this amounted to 25 genomes for subsequent study (table 1).
Fig. 1.
Analysis flowchart. Numbered arrows indicate two separate paths. Arrows with no numbers were performed in both iterations. Arrows numbered (1) indicate the first iteration in which 62 genomes were individually searched to identify genomes of interest. Arrows numbered (2) indicate the second iteration, conducted on all selected genomes with additional curation steps. In bold, programs used. In italics, additional descriptions. Parallelogram, input or output; rectangle, process; diamond, action, which includes manual intervention. In the first iteration, TBLASTN, PHI, MAFFT, and FastTreeMP were performed using TARGeT.
Table 1.
Summary of Selected 25 Genomes Included in the MUG Tree.
| Genome Selection Criteria |
|||||
|---|---|---|---|---|---|
| Species Name | Division, Order | AtMUGA Homologsb | AtMUGB Homologsb | TEs Derived from MUG LCAc | TEs with PB1 |
| Aquilegia coerulea | Eudicot, Ranunculales | 2 | 3 | Yes | No |
| Amborella trichopoda | Basal angiosperm | 3 | 0 | Yes | Yes |
| Citrus clementina | Eudicot, Sapindales | 3 | 4 | No | Yes |
| Citrus sinensis | Eudicot, Sapindales | 3 | 4 | Yes | Yes |
| Elaeis oleifera | Monocot, Arecales | 4 | 2 | Yes | Yes |
| Eutrema salsugineum | Eudicot, Brassicales | 4 | 3 | Yes | No |
| Fragaria vesca | Eudicot, Rosales | 3 | 3 | Yes | No |
| Glycine max | Eudicot, Fabales | 6 | 4 | Yes | Yes |
| Gossypium raimondii | Eudicot, Malvales | 5 | 6 | Yes | No |
| Malus domestica | Eudicot, Rosales | 7 | 8 | Yes | No |
| Manihot esculenta | Eudicot, Malpighiales | 1 | 4 | Yes | Yes |
| Mimulus guttatus | Eudicot, Lamiales | 4 | 4 | Yes | Yes |
| Nelumbo nuciferaa | Eudicot, Proteales | 3 | 0 | No | No |
| Nuphar advenaa | Basal angiosperm | 5 | 5 | No | No |
| Panicum virgatum | Monocot, Poales | 5 | 4 | No | Yes |
| Persea americanaa | Magnoliids, Laurales | 2 | 1 | No | No |
| Phoenix dactylifera | Monocot, Arecales | 3 | 4 | Yes | No |
| Physcomitrella patens | Moss, Funariales | 0 | 0 | Yes | No |
| Prunus persica | Eudicot, Rosales | 4 | 3 | Yes | Yes |
| Setaria italica | Monocot, Poales | 2 | 3 | Yes | No |
| Solanum lycopersicum | Eudicot, Solanales | 1 | 5 | Yes | Yes |
| Solanum tuberosum | Eudicot, Solanales | 1 | 4 | Yes | Yes |
| Theobroma cacao | Eudicot, Malvales | 4 | 5 | Yes | No |
| Vitis vinifera | Eudicot, Vitales | 3 | 3 | Yes | Yes |
| Zea mays | Monocot, Poales | 4 | 2 | No | Yes |
aGenomes included in the final list although they did not fulfill the genome selection criteria. They were included because of their strategic position in the tree. These genomes are not shown in the main tree figures but present in the supplementary tree figures, Supplementary Material online.
bNumber of homologous sequences for each given species.
cLCA, Last common ancestor.
Generating and Curating the Phylogenies
We next constructed a phylogenetic tree that included all ETEs and TEs of interest (fig. 1; iteration 2). Alignment curation is critical to the construction of high quality phylogenies, especially for TE genes because they often contain frameshifts, truncations, deletions, and insertions. If not removed, highly degenerate sequences with long gaps or poor alignment within otherwise well-conserved blocks may reduce the accuracy of phylogenetic inference (Castresana 2000). Conversely, we also did not want to remove all pseudogenes because that would include the majority of TE-derived sequences (see below). We thus devised an approach that combined multiple methods to remove extraneous sequences.
To exclude sequences related to MUG only distantly, we increased the search stringency (see Materials and Methods), resulting in 2,077 sequences. We built a preliminary alignment (MAFFT) and used it to remove problematic sequences. We generated a final alignment (MAFFT), curated it further (GBlocks), and inferred a “full” MUG approximately maximum-likelihood phylogenetic tree (FastTreeMP) (supplementary fig. S2, Supplementary Material online). Lastly, for clarity of presentation, we also generated a “simplified” MUG tree containing only the sequences most closely related to MUG by pruning branches more distant than the last common ancestor of all known MUG genes (fig. 2, supplementary fig. S3, Supplementary Material online). For FRS, we used a similar approach, except the final pruning step was not performed because of the large evolutionary distance between certain FRS subtrees (supplementary figs. S4 and S5, Supplementary Material online). To evaluate the robustness of the resulting MUG and FRS trees, we selected a subset of sequences (131) from each tree, representing all major clades, and performed analyses using two additional phylogenetic methods: a Bayesian (MrBayes) (Ronquist and Huelsenbeck 2003) and a Neighbor joining method (BioNJ) (Gascuel 1997) (see Materials and Methods). The resulting trees agreed with the topologies of the original approximately maximum-likelihood trees, including strong support for all key nodes (supplementary figs. S6–S9, Supplementary Material online).
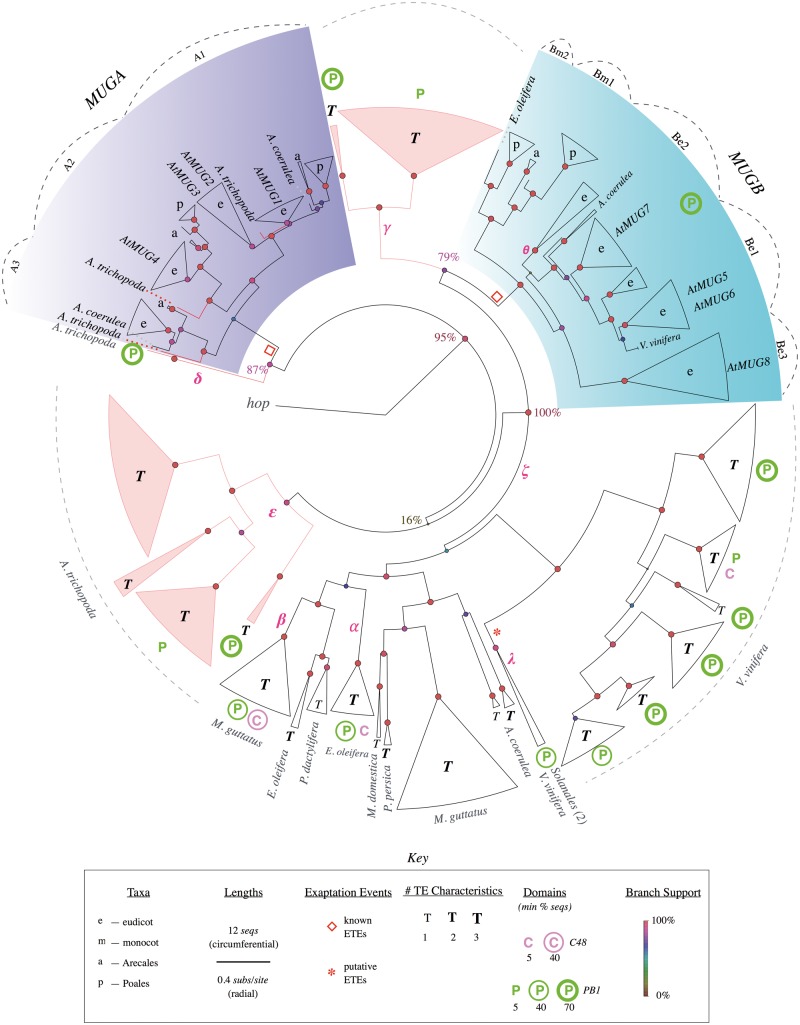
Fig. 2.
Simplified phylogenetic tree of MUG genes and TEs. Curated phylogenetic tree showing sequences descended from the last common ancestor of MUGA and MUGB, rooted at fungal hop. Terminal triangles represent clades, with circumferential width proportional to number of genes (see key). In clades with genes from only one or a few species the species are labeled, otherwise clades are labeled according to taxon. Clades and branches from Amborella trichopoda are colored red. Major clades of MUGA and MUGB are labeled according to Joly-Lopez et al. (2012) and the positions of Arabidopsis thaliana AtMUG1-8 genes are indicated. For simplicity, TE features are categorized by the number of TE characteristics (out of 3) associated with each clade to emphasize differences between clades of known ETEs and clades of putative TEs (see key). For the same tree including detailed TE characteristics, see supplementary figure S3, Supplementary Material online. Radial branch lengths are proportional to the inferred number of substitutions per site (circumferential branch length is arbitrary). Circles at internal nodes have color and size corresponding to “local support values” (Shimodaira–Hasegawa test [Zeh et al. 2009; Oliver et al. 2013]). Empty red diamonds indicate known exaptation events; red asterisks indicate putative novel exaptation events. Greek letters indicate branches referred to in the main text. Dashed lines indicate clades, dotted lines are species labels. Pink branches are used to highlight Am. trichopoda clades or individual sequences. See supplementary figure S2, Supplementary Material online, for a fully expanded phylogenetic tree.
ETEs versus TEs: Two Distinctive Types of Clade
We now had phylogenies that included only the sequences most closely related to MUG or FRS. But how could we determine which clades are ETEs and which are TEs? Certain attributes of individual sequences (and families) can be used to differentiate between TEs and ETEs (Hoen and Bureau 2015). Three such attributes are intrinsic products of phylogenetic analysis (fig. 2): 1) TE genes often have high copy-number (Feschotte and Pritham 2007), which is reflected in the phylogenetic tree by clades with high numbers of paralogs (and low numbers of orthologs). 2) Recently, active TE families may include genes with highly similar sequences, reflected by short terminal branches. 3) Many TEs are lineage-specific, reflected by incongruities between the topologies of the clades and those of the species phylogeny; in other words, whereas phylogenetic sister sequences of ETEs (and regular genes) are usually orthologs from sister species, sister clades of TEs are often from species that are not closely related.
To better distinguish TEs from ETEs, we also evaluated additional sequence characteristics (fig. 2; supplementary fig. S3 and table S2, Supplementary Material online) (Hoen and Bureau 2015): 1) presence of frameshifts and in-frame stop codons; 2) repetitiveness of flanking DNA sequences; 3) presence of potential terminal inverted repeats (TIRs); and 4) presence of a peptidase C48 conserved domain, which is useful in identifying clades of sequences that lack TIRs but nevertheless are TEs (see Materials and Methods). We also searched for the PB1 domain, not because it helps in distinguishing TEs from ETEs, but because of its as-yet unexplained presence in MUGB. 5) In addition, we examine microsynteny, which has previously been used to identify putative ETEs and described for MUG and FRS genes across Brassicaceae genomes (Cowan et al. 2005; Hoen and Bureau 2015). We extend this work by examining microsynteny in the diverse monocot and eudicots genomes examined in this study (supplementary fig. S10, Supplementary Material online).
Combining these phylogenetic and sequence characteristics, two distinctive types of subtree or clade become apparent. The first type is putative ETEs, including all known MUG or FRS genes. Focusing first on MUG (fig. 2), consistent with previous results (Cowan et al. 2005; Joly-Lopez et al. 2012) and our single-species phylogenies (supplementary fig. S1, Supplementary Material online), known MUG genes form two subtrees: MUGA and MUGB. Each subtree has several major clades consisting of orthologs from diverse species (MUGA, 74 sequences; MUGB, 78). Indeed, each subtree includes at least one sequence from every examined angiosperm (except Amborella trichopoda; see below), but does not include any sequence from a nonangiosperm species. The topologies of each subtree and major clade are broadly congruent with the known species phylogeny (Amborella Genome Project 2013); that is, most branches (monocots vs. dicots, Arecales vs. Poales, asterids vs. rosids, Fabidae vs. Malvidae, etc.) agree with the species topology. Finally, in these putative ETE subtrees few sequences have TE characteristics (those that do are presumably false positives).
The second type of subtree or clade is putative TEs. This includes most of the remaining sequences (1,672 of 1,824 sequences in the MUG phylogeny). In contrast to the ETE subtrees, these clades are lineage-specific (they are from single or closely related species) and have sister clades from distantly rather than closely related species. They also often have short terminal branches. Although some small clades are ambiguous, many clades have multiple strong TE characteristics. For example, clade α (fig. 2) has 14 sequences: all are from Elaeis oleifera, all have pseudogenic features (e.g., stop codons: min 2, mean 8), 43% have potential TIRs, and the clade has high DNA repetitiveness (median 51 copies; supplementary table S2, Supplementary Material online). Furthermore, while E. oleifera is a monocot, its nearest sister clade (β) is distant (0.86 subs/site), specific to the eudicot Mimulus guttatus, and itself consists of 29 sequences with strong TE characteristics including several very short terminal branches (e.g., nine are shorter than 0.01 subs/site).
MUGA and MUGB Are Paraphyletic with Respect to TEs
Using our phylogeny labeled with genomic attributes that distinguish ETEs from TEs (fig. 2, supplementary fig. S3, Supplementary Material online), we can now begin to address interesting evolutionary questions. First, did MUGA and MUGB originate together in a single exaptation event, or separately in two (or more) events? As expected, MUGA and MUGB are more closely related to one another (i.e., have a shorter genetic distance between the roots of their respective subtrees) than to the vast majority of apparent TE clades (supplementary fig. S2, Supplementary Material online). However, a few TE clades are more closely related to either MUGA or MUGB; in other words, MUGA and MUGB are paraphyletic with respect to several TE clades. Indeed, in figure 2, all sequences are descended from the last common ancestor of MUGA and MUGB, so this is true of all the apparent TEs included in this figure.
For example, perhaps the most interesting clade of apparent TEs is the sister clade to MUGB (γ), a large family of TEs (53 sequences) in the basal angiosperm Am. trichopoda. Most originated in rapid expansions and all now are apparent pseudogenes (stop codons: min 3, mean 9.5) (supplementary table S2, Supplementary Material online). Consistent with their state of degeneration, only 11 are associated with high-identity TIRs and the median DNA copy-number is 22. Interestingly, many (13) of the sequences are associated with PB1 domains, suggesting that the MUGB PB1 domain originated from its last common ancestor with these TEs (see below). Note that although this phylogeny also suggests that MUGA has a small Am. trichopoda sister clade (two sequences) (δ), the placement of this particular clade is uncertain because these were among the highly degenerate sequences not removed in the final phylogeny, and furthermore the placement of this clade was unstable between multiple independent builds of the full phylogeny (not shown).
In addition to clade γ, there are two additional branches of putative TEs. One is again from Am. trichopoda (ε), but it has low local branch support (16%) so may not truly be a distinct branch. The other includes sequences from eight additional species (ζ), and although not all phylogenetic relationships between these putative subclades are well resolved, there is strong support that this branch as a whole is ancestral to MUGB (100% local branch support), and conversely that the MUGA branch is ancestral to it (95% local branch support). Note that one of the eight species in branch ζ is Vitis vinifera, even though a previous analysis failed to identify any V. vinifera MULEs paraphyletic to MUGA and MUGB (Joly-Lopez et al. 2012), possibly because that analysis included only draft V. vinifera sequences (no other genomes) and a short mudrA subsequence (Benjak et al. 2008). Our topology is also supported by all respective single-genome phylogenies (supplementary fig. S1, Supplementary Material online), by multiple independent builds of the full phylogeny using a range of curation settings (not shown), and by analyses using multiple phylogenetic methods (supplementary figs. S6 and S8, Supplementary Material online).
Paraphyly Implies Separate Exaptation Events Because ETE Reversion Is Unlikely
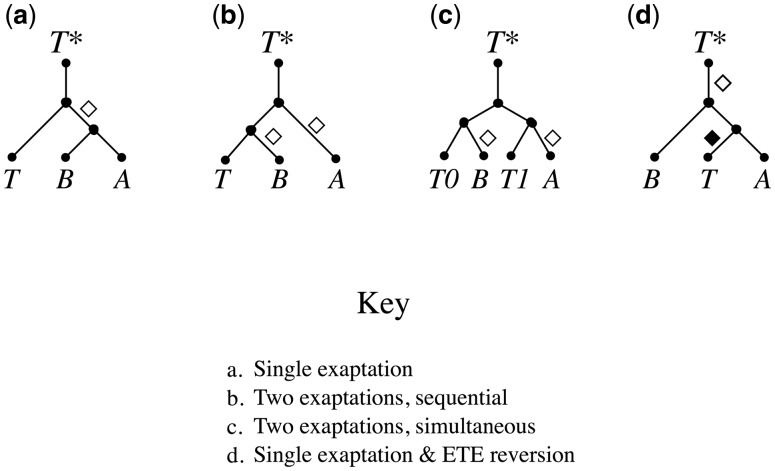
Our results thus provide strong evidence that MUGA and MUGB are paraphyletic with respect to TEs. How might such a topology have arisen? The simplest explanation is straightforward: MUGA and MUGB originated in separate exaptation events (fig. 3b and c) and the TE branches simply reflect the evolutionary history of the progenitor TE families.
Fig. 3.
One exaptation versus two. Differences in phylogenetic relationships that would result if two ETEs originate in a single exaptation versus separate exaptations. T*, common ancestral TE; T, T0, and T1, extant TEs; A and B, ETEs; empty diamonds, exaptation events; filled diamond, hypothetical ETE reversion event. (a) If A and B originate in a single exaptation, then A and B should not be paraphyletic—descended from a common evolutionary ancestor, but not including all the descendant groups—to any TE family (but see case d). (b) If A and B originate in separate exaptations of a single TE family (T), then A and B are paraphyletic with respect to T.(c) If A and B originate in separate exaptations of different (but related) TEs families (T0 and T1), then A and B are paraphyletic with respect to both T0 and T1. (d) Hypothetically, if an ETE were able to revert to being a TE (T) after the ETE family had already differentiated into at least two branches (A and B), it could also result in paraphyly with respect to the TE even if the ETE family had originated in a single exaptation event.
But could there be another explanation? Theoretically, such paraphyly might also arise from a reversal of the TE exaptation process; that is, by ETEs transforming into TEs (fig. 3d). Is ETE reversion plausible? Consider the functional changes and underlying mutations required for TE exaptation versus those that would be required for ETE reversion. Fundamentally, the process of TE exaptation involves a transition from persisting by self-replicative selection to persisting by phenotypic selection. That is, TEs persist by replicating within a genome to escape disabling mutations, whereas ETEs are conserved by conferring phenotypic benefits to the organism. To achieve this transition, TE exaptation entails various functional changes. The most basic of these follows directly from this fundamental change in selection regime: whereas for TEs transposition is essential for the sequence to persist, for ETEs it is not only nonessential, but harmful since transposition may disrupt ETE expression or cause other deleterious mutations. As a consequence, upon exaptation we expect ETE genes to become immobilized and their mobility-related flanking DNA sequences such as TIRs to become degraded or deleted. Indeed, all known (well-supported) ETE genes are immobilized. In addition, the phenotypic functions of ETE-encoded proteins rarely involve mobility-related molecular activities such as DNA cleavage or integration, which again are deleterious to the genome. Thus, with the rare exception of ETEs that have retained such activities in tightly controlled contexts, such as V(D)J recombination (Kapitonov and Jurka 2005), ETEs lose their ability to perform various molecular activities required for transposition. Yet another change is that whereas most TEs are usually silenced, ETEs genes must be expressed at relatively high levels in order to confer phenotypic benefits.
Although TE exaptation involves several functional changes, as evidenced by known ETEs such as MUG and FRS, it does occur at some frequency. We propose two reasons for this. First, each underlying mutation has high probability. For example, any one of many possible point mutations to a transposase could decrease or nullify its ability to catalyze transposition. In addition, TEs frequently sustain deletions, and any sufficiently long deletion in a TIR may lead to immobilization (Feschotte and Pritham 2007; Sinzelle et al. 2009). Furthermore, some mutations can have dual consequences. For example, transcriptional silencing in plants of DNA transposons is largely mediated by RNA-directed DNA methylation focused at the TIRs (Kawashima and Berger 2014); thus, partial deletion of a TIR could result in both immobilization and desilencing. The second reason we propose that TE exaptation can occur despite requiring multiple mutations is that none of these mutations are required for phenotypic selection pressure to begin; thus, they could occur independently and in any order. Fundamentally, this is because the molecular activities that permit an ETE to produce beneficial phenotypes are inherent to the TEs themselves. For example, DNA transposases such as mudrA are often exapted to become transcription factors, utilizing the TEs molecular functions of specific DNA binding and protein-protein interaction. Thus, a transposase could produce a beneficial phenotype and become a nascent ETE even before being immobilized, allowing phenotypic selection to drive the TE exaptation process (Hoen and Bureau 2012).
In contrast, consider the functional changes and mutations that would be required for an ETE to revert to being a TE. Note that for a reversion to be preserved in the phylogenetic record, the ETE family would first need to diversify into at least two well-separated branches; otherwise it would simply appear to be a regular TE family. First, the ETE would need to reacquire mobility-related flanking DNA structures such as TIRs. Unlike deleting them, reacquiring TIRs would seem exceedingly difficult. How might it occur? The following series of four mutations (not necessarily in this order) might for example permit TIR reacquisition: 1) a TE (with TIRs) inserts close to one side of the ETE; 2) a second TE of the same family inserts close to the other side; 3) the interior TIR is deleted from one TE; and 4) the interior TIR is deleted from the second TE. Another possible mechanism of TIR acquisition might be transduplication, which is the direct capture of genomic sequences by certain types of TEs; however, transduplication rarely if ever results in the duplication of entire genes (Juretic et al. 2005). Regardless of how it might occur, TIR acquisition could theoretically allow the ETE to become mobilized in trans by a transposase encoded by the TE family that donated the TIRs. But even this would not be sufficient to reestablish self-mobility and thus selfish selection, because the encoded protein would also need to reacquire any required molecular functions it had lost, such as DNA cleavage and integration, and doing so might require multiple, specific amino acid substitutions. Finally, the revertant transposase would need to be able to specifically bind particular target sequences in the reacquired TIRs, which would require additional specific amino acid substitutions to its DNA-binding domain (except in the seemingly unlikely event that the TIRs were reacquired from same TE family from which the ETE descended, and binding-site specificity had been retained during the intervening evolutionary period). Furthermore, and most crucially, these mutations would all need to occur before selfish selection could even begin to act; that is, they would need to be simultaneous.
So, whereas TE exaptation could occur with relatively few, independent loss-of-function mutations and be driven by phenotypic selection, ETE reversion would require a larger set of gain-of-function mutations that must occur simultaneously. Therefore, while theoretically possible, reversion of a well-established ETE seems extremely improbable. Indeed, while a large and growing number of ETEs have been reported in the literature (Hoen and Bureau 2015), there are no reports of revertant ETEs, nor do we find evidence of it in either the MUGA or MUGB subtrees, nor in any of the FRS subtrees (see below). Finally, suppose that ETE reversion did very occasionally occur. We would still have difficulty explaining the paraphyly of MUGA and MUGB because the tree topology would require not just one, but at least two ETE reversions: one for each of the two (well-supported) TE branches (γ and ζ). Thus, the simplest explanation by far of the observed phylogeny is that MUGA and MUGB formed in separate exaptation events. Indeed, as shown below, differences between the two subtrees suggest that they also originated far apart in time, and thus ought to be considered as separate families, a conclusion further supported by differences in their gene structures (e.g., PB1) and experimental evidence (see below).
MUGB Originated in the Angiosperm Crown Group and Diversified in Monocots and Eudicots
Our approach enabled us to identify TEs closely related to MUGA and MUGB and show that they originated in separate exaptation events. By examining the detailed phylogenies of each ETE subtree, we can also resolve the timing of the original exaptation events, as well as the pattern and timing of diversification within each family subsequent to exaptation.
The MUGB subtree includes clades of single or low copy-number homologs in all examined crown monocots and eudicots (fig. 2, supplementary fig. S2, Supplementary Material online). Consistent with previous results (Joly-Lopez et al. 2012), monocot and eudicot homologs form two monophyletic subtrees. This shows that the progenitor MUGB gene did not undergo duplication prior to the monocot-eudicot split, suggesting it may have originated not long before the split. The basal branches of the MUGB tree confirm this. In the basal-most extant angiosperm genome, Am. trichopoda, even though we detected both a large number of MULEs (see above) and several MUGA homologs (see below), we found no MUGB homolog. We did however find putative MUGB homologs in EST assemblies of the second most basal lineage, Nu. advena, as well as the magnoliids Pe. americana and Liriodendron tulipifera, suggesting that MUGB exaptation likely occurred between the divergence of the Amborellales and the Nymphaeales (supplementary figs. S1 and S2, Supplementary Material online). This places the origin of MUGB near the beginning of the angiosperm radiation (∼145 Ma), a period which produced all five major angiosperm lineages including the magnoliids, the monocots, and the eudicots (Amborella Genome 2013; Zeng et al. 2014).
Not only does the topology near the root of the entire MUGB subtree enable us to resolve the timing of the origin of MUGB, but similarly the topology of its internal clades enables us to resolve the timing of subsequent MUGB duplication events. MUGB has two main monocot-specific clades with long root branches: Bm1 (0.18 subs/site) and Bm2 (0.13 subs/site), suggesting that the single progenitor MUGB gene duplicated once in early monocot evolution. Each of these two clades is composed of diverse species including representatives of both examined monocot orders (Arecales and Poales). In addition to this early duplication, Bm1 and Bm2 each subsequently underwent duplications prior to Poales diversification, as well as further duplications in certain lineages (supplementary fig. S2, Supplementary Material online). Together with losses in certain lineages, these duplications resulted in MUGB having between two and five (median 3) paralogs per monocot genome.
In eudicots, the pattern of MUGB diversification is somewhat different. There are homologs in the basal eudicot Aquilegia coerulea (Ranunculales) but not in N. nucifera (Proteales), and MUGB is divided into three major eudicot clades (Be1, Be2, and Be3) (Joly-Lopez et al. 2012). Clade Be3, which diverged first and has a particularly long root branch (0.27 subs/site), includes homologs in all examined crown eudicot species except Glycine max. The Be3 Brassicales subclade (which includes AtMUG8) has a particularly long root branch (0.42 subs/site), as do the other MUGB Brassicales subclades. The two remaining major eudicot clades, Be1 and Be2, resulted from a far more recent duplication (root branch lengths of 0.08 and 0.07 subs/site, respectively), yet each also includes all examined crown eudicot species, except that Be2 contains no homolog from Eutrema salsugineum and Be1 contains none from Manihot esculenta. These and additional duplications and losses have resulted in MUGB having three to eight (median 4) paralogs per eudicot genome, typically one per subclade. Note that higher copy-numbers in certain genomes resulted from recent duplications and many are pseudogenes; for example, seven of eight Malus domestica MUGB genes have premature stop codons (supplementary table S2, Supplementary Material online). In addition to the three previously assigned clades, this phylogeny suggests that one of the subclades of Be2 (θ) might better be considered as a fourth major eudicot clade.
MUGA Originated and Diversified in the Angiosperm Stem Group
Although MUGB originated during the angiosperm radiation, MUGA originated much earlier. The first clue of this early origin is the absence of any well-supported TE clades closely related to MUGA. Conclusive evidence is provided by the topology and basal branches of the MUGA subtree. MUGA consists of three major clades (A1, A2, and A3) that, unlike the MUGB clades, each include orthologs from all major angiosperm lineages examined, including monocots and eudicots (fig. 2). Thus, MUGA underwent two duplications prior to the divergence of monocots and eudicots. In addition, MUGA includes homologs not only in the magnoliids Pe. americana and L. tulipifera, but importantly also in the basal angiosperm Am. trichopoda. Furthermore, these basal branches do not stem from the base of the MUGA tree, but instead there are Am. trichopoda and other basal angiosperm branches specific to each of the three major MUGA clades. Therefore, MUGA must have originated and diversified at least as early as the angiosperm stem group, prior to the radiation of all extant angiosperms. Indeed, the root branches of the two best-supported major clades (A1 and A2) are long (0.2 subs/site), suggesting that the exaptation and initial diversification of MUGA likely occurred long before this angiosperm radiation.
Subsequent to its initial diversification, MUGA did not undergo any further duplications, except for clade A2 in early core eudicots and certain species-specific duplications. Interestingly, the earliest-diverging clade (A3) is also the least conserved between taxa. For example, although it is present in Carica papaya, a basal Brassicales of the same order as Arabidopsis, it does not include homologs from the Brassicaceae (e.g., A. thaliana) (Joly-Lopez et al. 2012). Also, while it does include monocots of the order Arecales (E. oleifera and P. dactylifera), no homologs were found in the examined Poales (Zea mays, Oryza sativa, Panicum virgatum, and Setaria italica). Conversely, clades A1 and A2 each include homologs from both of these monocot orders. As a consequence of these diversification events, MUGA copy-number ranges from one (Solanum lycopersicum, S. tuberosum, and M. esculenta; all in clade A2) to seven (Ma. domestica), with most angiosperms having three or four MUGA paralogs (median 3).
Could MUGA have originated even earlier, prior to the divergence of angiosperms and gymnosperms? We searched the genomes of nonangiosperm species, including the assembled genome of the gymnosperm P. abies and EST assemblies for Pinus sylvestris, Abies sibirica, Juniperus communis, and Gnetum gnemon, but found no potential MUG homologs. We also found no homologs using supplementary TBLASTN searches (query AtMUG1; default E-value, 1e-3) of the genome assemblies (http://congenie.org, last accessed October 23, 2015) of Picea glauca (white spruce; PG29-v4.0) (Birol et al. 2013) and Pinus taeda (loblolly pine; v1.0) (Zimin et al. 2014). Although negative search results such as these cannot definitively rule out the presence of MUGA, it is revealing that members of both MUG families have been found in virtually every angiosperm that has been searched, including full genome assemblies, EST assemblies, and even in most sufficiently large EST databases (both in this study and in Joly-Lopez et al. [2012]), yet has not been found in any nonangiosperm. Thus, MUGA likely originated early in angiosperm evolution, subsequent to divergence from gymnosperms (estimated at 290–310 Ma), but well before the angiosperm radiation (Zeng et al. 2014). Little is known about this lineage of preangiosperm species (the angiosperm stem group), which recent evidence suggests may have originated around 225–250 Ma in the Late-to-Middle Triassic (Zeng et al. 2014). We are just beginning to address whether MUGA may have played a role in the many crucial adaptations that occurred in the angiosperm stem group.
Experimental Results and dN/dS Analyses Suggest Functional Overlap within Families
As we show above, characterizing the phylogenetic patterns of ETE families and their cognate TEs is useful from an evolutionary standpoint because it elucidates when and how often TE exaptation and ETE diversification occurs. It is also interesting from a practical standpoint, since the evolutionary histories of ETEs may reflect their potential phenotypic functions, molecular interactions, and genetic redundancies.
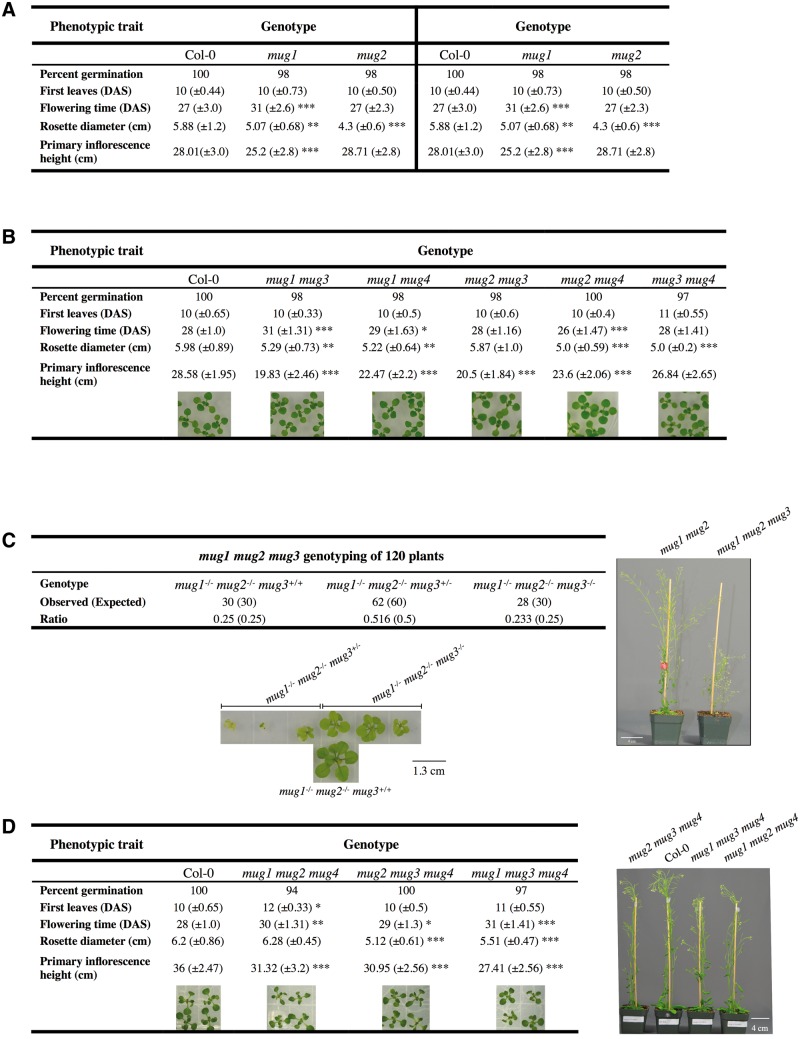
To illustrate, consider the four MUGA paralogs in A. thaliana, which are of particular interest because we previously characterized some of them phenotypically (Joly-Lopez et al. 2012). While single knockout mutants of MUGA genes show only subtle phenotypes under controlled laboratory conditions compared with wild-type Col-0 (fig. 4A), mug1 mug2 double mutants exhibit strong phenotypes for traits usually associated with plant fitness. Similarly, MUGB single mutants do not show strong phenotypes whereas certain double mutants, such as mug7 mug8, do have serious defects (Joly-Lopez, et al. 2012). Here, we show that the other MUGA double mutant combinations, although they do exhibit phenotypes such as delayed flowering time and reduced rosette diameter and inflorescence height, these phenotypes appear under standard laboratory conditions to be weaker than for mug1 mug2 (fig. 4B) (Joly-Lopez et al. 2012).
Fig. 4.
Phenotypes for MUG mutants, including the triple mutant mug1 mug2 mug3 in Arabidopsis thaliana. (A) Phenotypes of wild-type (Col-0), and mug1 to mug4 single mutants, based on traits that have been associated to fitness and cover the lifespan of the plant life cycle. The phenotypic assays for mug1 and mug2 were performed independently in two different growth chambers from mug3 and mug4; hence the two results for Col-0. (B) Results of the phenotypic analysis for the five previously uncharacterized double mutant combinations. n = 60 plants. Images of 2-week-old seedlings grown on one-half MS media and representing double mutant combinations of MUGA. (C) The table shows the results of the segregation ratio for 120 F2 plants following genotyping by PCR. On the bottom, image captures of 3-week-old mug1 mug2 and mug1 mug2 mug3 mutant seedlings heterozygous and homozygous for mug3, all grown on the same one-half MS media supplemented with 2% sucrose. Scale bar = 1.3 cm. On the right, difference in size of mug1 mug2, and mug1 mug2 mug3 mutant plants at 50 days after sterilization. Scale bar = 4 cm. (D) Results of the phenotypic analysis for the other triple mutant combinations. n = 30 plants. On the bottom, images of 2-week-old seedlings grown on one-half MS media. On the right, image of 40-day-old mature plants for the triple mutants compared with Col-0. Scale bar = 4 cm. For the phenotypic analyses, statistical significance is based on a two-sample student t-test ∝= 0.05; *P < 0.05, ** P < 0.01, *** P < 0.001.
The evolutionary history of MUGA is a starting point to explain these differences. MUGA has two major clades that are conserved among all angiosperms: A1 and A2 (A3 is not present in A. thaliana—see above; fig. 2; supplementary figs. S2 and S3, Supplementary Material online). Clade A1 has two subclades that diverged early in eudicot evolution, one of which includes AtMUG1, the other AtMUG2. Although further experiments are warranted to confirm these results, this phylogeny suggests that, whereas AtMUG1 and AtMUG2 may have subfunctionalized to perform eudicot-specific functions that are difficult to detect in single mutants under our growth conditions, they may also have redundancies for more deeply conserved functions that are revealed by the double mutants. Indeed, the topology is similar to that of AtFAR1 and AtFHY3, which are known to have partially redundant functions and direct molecular interactions.
In addition to AtMUG1 and AtMUG2, clade A1 includes a third A. thaliana paralog, AtMUG3. The above phylogeny-based reasoning suggests that mutating all three A1 paralogs should produce even stronger defects. To test this hypothesis, we generated a mug1 mug2 mug3 triple mutant. The progeny of an F2 plant homozygous for mug1 and mug2 and heterozygous for mug3 were screened for triple mutants (n = 120) and homozygotes seedlings were successfully genotyped only when seeds were grown on media supplemented with addition of carbohydrates (2% sucrose vs. 1%). Segregation ratios suggest that mug3 is recessive and segregating independently (fig. 4C). The mug1 mug2 mug3 triple mutant showed an additive phenotype that was more severe than the double mutant: increased pale yellow-green coloration, longer delays in flowering, and smaller overall size (fig. 4C). In addition, whereas wild-type plants produce thousands of seeds, triple mutants yielded two orders of magnitude fewer seeds (average 30), and some plants yielded no seed at all (data not shown). These results support the hypothesis that clade A1 genes together perform critical functions, at least in A. thaliana.
Finally, there is a fourth A. thaliana paralog (AtMUG4), which belongs to a second major clade, A2, that diverged from clade A1 during angiosperm stem group evolution and is itself conserved among all angiosperms. Such a long period of divergence suggests that AtMUG1, AtMUG2, and AtMUG3 may have greater functional overlap or genetic redundancy with one another than with AtMUG4. This phylogeny-based reasoning is supported by our experimental results, which show that double and triple mutant combinations involving AtMUG4 display less severe defects than mug1 mug2 and mug1 mug2 mug3 (fig. 4B and D). Finally, we have not been able to generate quadruple mutant of all A. thaliana MUGA genes, suggesting that these long-diverged lineages may still maintain redundancy for some deeply conserved angiosperm function and that the absence of the MUGA family may be lethal in A. thaliana.
In general, while ETEs from the same family might share functional redundancies or similarities, ETEs derived from separate exaptation events likely do not. This is because, fundamentally, TE exaptation is the acquisition of a novel phenotypic function by a sequence with no prior phenotypic function; thus, the novel functions acquired in different exaptations ought to be independent of one another. Nevertheless, we might expect that certain functional similarities between different ETE families could result from common attributes between the progenitor TEs, such as their molecular activities or expression patterns. Functional similarities might also arise from similar phenotypic selective pressures at their times of exaptation. Applying similar phylogenetic analysis and reasoning to FRS might also aid our understanding of experimental results for that group of ETEs (Lin and Wang 2004).
In addition to experimental analysis, we had previously examined selective pressures in MUGA and MUGB coding regions using dN/dS analysis and found evidence of purifying selection for the entire coding region encompassing three conserved domains found in progenitor TEs (Cowan et al. 2005; Joly-Lopez et al. 2012). To explore this further, we selected 121 representative MUG sequences (fig. 2, supplementary fig. S2, Supplementary Material online), generated a phylogenetic tree (BioNJ; supplementary fig. S11A, Supplementary Material online), and estimated dN/dS ratios using CODEML. Overall, dN/dS for the whole tree suggests that 73% and 26% of sites, respectively, are under negative and positive selection. Results were similar using a branch-site model (test 2): 80% and 20%, respectively (“root branch 1” in supplementary fig. S11B, Supplementary Material online).
To better understand selection on the MUGA subclades, we selected three additional branches labeled “2,” “3,” and “4” to represent clades A3, A2, and A1, respectively (supplementary fig. S11A, Supplementary Material online). The dN/dS ratio for branch 2 (Clade A1), which encompasses MUG1-MUG3 did not show significant positive selection and the branch appears to be mostly fixed under negative selection. In contrast, we detected strong positive selection on branch 3 (P = 0.0057) (Clade A2), which encompasses MUG4 and homologs, where the ω-value for a subset of sites is well above 1, suggesting that certain sites show stronger positive selection than overall identified by a Bayes Empirical Bayes (BEB) analysis for positive sites (“root branch 3” in supplementary fig. S11B, Supplementary Material online). Four positively selected sites were detected in branch 3 (supplementary fig. S11C, Supplementary Material online). Interestingly, these amino acids lie on the border of a conserved domain (supplementary fig. S11D, Supplementary Material online). Although confirmation is needed, the observation that the MUG1-MUG3 subtree branch is more “fixed” than MUG4 suggests that the MUG1-MUG3 ancestral sequence may have acquired a phenotypic function earlier than the MUG4 ancestral sequence, possibly due to being domesticated separately from MUG4, which would be consistent with the tree topology (above). Alternately, it may suggest that the function of MUG1-MUG3 became fixed early while MUG4 continued to undergo subfunctionalization.
FRS Consists of Five Families that Originated and Diversified at Different Times
In addition to MUG, one other group of plant ETEs has been well-characterized: FRS (Lin et al. 2007). Although both MUG and FRS are derived from TEs of the MULE superfamily, their respective TE lineages (mudrA and FAR1) are highly diverged (Lin et al. 2007). Similar to MUG, a previously published phylogeny includes among the descendants of the last common FRS ancestor two branches of TEs (LOM-1 in O. sativa and M. truncatula; and Jittery in Z. mays), suggesting that FRS may have originated in more than one exaptation event (Lin et al. 2007).
To test whether FRS originated in one or in multiple exaptation events, and to resolve the timing of exaptation and subsequent FRS diversification, we followed a similar approach as we did with MUG. We selected a representative query from each of five previously identified FRS lineages (FRS10, FRS6, FHY3, FRS3, and FRS7; supplementary fig. S12, Supplementary Material online) (Lin et al. 2007). We used TARGeT to search for homologs among the 25 final genomes, to which we added O. sativa and M. truncatula in order to include LOM-1. This resulted in 1,117 sequences, to which we added all 14 known A. thaliana FRS sequences, fungal hop as outgroup, and maize Jittery. We generated a multiple sequence alignment (MAFFT), curated it by removing columns with at least 50% gaps and using Gblocks to remove poorly conserved blocks (69% of 602 positions retained), and finally inferred a phylogenetic tree (FastTreeMP). Lastly, we used identical methods as for MUG to identify sequence attributes characteristic of TEs: premature stop codons, frameshifts, DNA repetitiveness, and potential TIRs, as well as the conserved domains PB1 and C48.
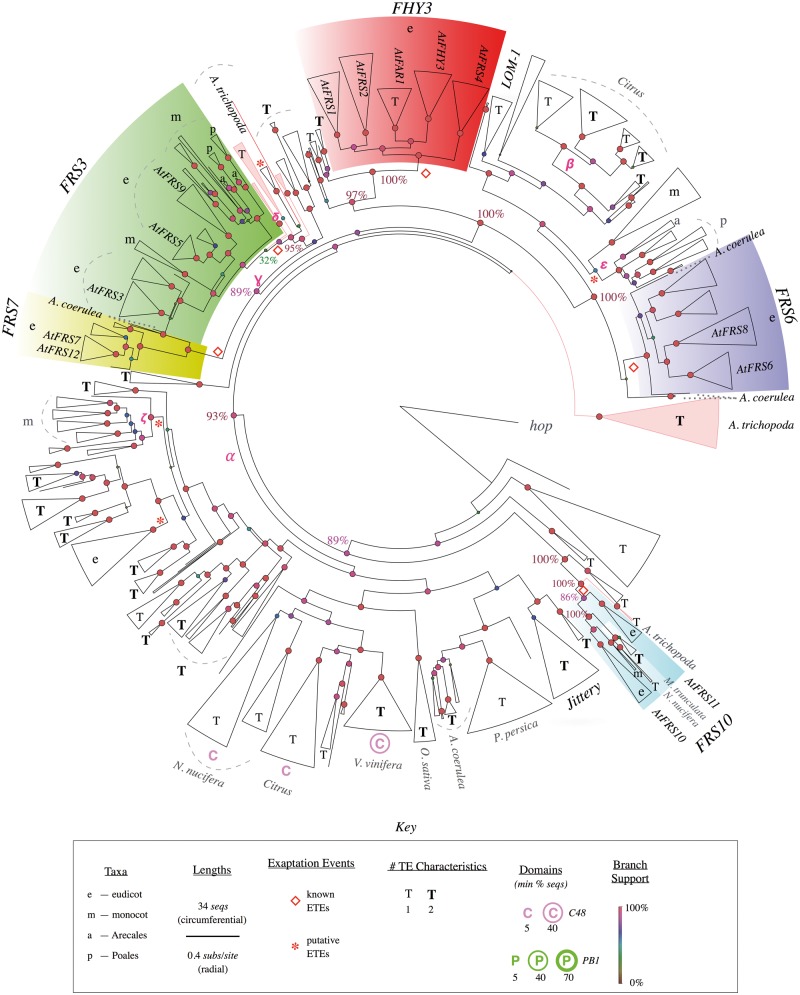
The results, while broadly consistent with the phylogeny of Lin et al. (2007), were nonetheless surprising (fig. 5 tree; supplementary figs. S4 and S5, Supplementary Material online). Not only is FRS paraphyletic with respect to the two TE branches reported by Lin et al., 2007 but moreover all five FRS subtrees are paraphyletic with respect to various apparent TE clades. Thus, each of these five subtrees likely arose in a separate exaptation event, making them separate ETE families (see above). The most obvious case is the FRS10 subtree (18 sequences), which is ancestral to all other FRS subtrees (89% local support), as well as to a large subtree that includes diverse clades of apparent TEs (e.g., α) (supplementary table S2, Supplementary Material online). The four remaining FRS families can be analyzed as two pairs of nearest neighbors (FHY3 and FRS6; FRS7 and FRS3), both of which are also separated by apparent TEs. The FHY3 (98 sequences) and FRS6 (75 sequences) subtrees are paraphyletic with respect to diverse apparent TE clades (e.g., β: 100% local support; 12 sequences; all but one have pseudogenic features; DNA repetitiveness, 56). Similarly, the FRS7 (29 sequences) and FRS3 (123 sequences) subtrees are paraphyletic with respect to apparent TE clades in various eudicots and Am. trichopoda (e.g., γ: 89% local support; five sequences; all five have pseudogenic features; potential TIRs, 60%; DNA repetitiveness, 13).
Fig. 5.
Phylogenetic tree of FRS genes and TEs. Curated phylogenetic tree of all identified FRS sequences, rooted in fungal hop. The five FRS clades are labeled following Lin et al. (2007). Putative TE clades that include Jittery and LOM-1 are indicated. Attributes are labeled only if present in more than one sequence per clade and for clarity only selected putative TE clades are labeled. Terminal triangles represent clades, with circumferential width proportional to number of genes (see key). In clades with genes from only one or a few species the species are labeled, otherwise clades are labeled according to taxon. Clades and branches from Amborella trichopoda are colored red. For simplicity, TE features are categorized by the number of TE characteristics (out of 2) associated with each clade to emphasize differences between clades of known ETEs and clades of putative TEs (see key). For the same tree including detailed TE characteristics, see supplementary figure S5, Supplementary Material online. Radial branch lengths are proportional to the inferred number of substitutions per site (circumferential branch length is arbitrary). Circles at internal nodes have color and size corresponding to their “local support values” (Shimodaira–Hasegawa test [Zeh et al. 2009; Oliver et al. 2013]). Empty red diamonds indicate known exaptation events; red asterisks indicate putative novel exaptation events. Greek letters indicate branches referred to in the main text. Dashed lines indicate clades, dotted lines are species labels. Pink branches are used to highlight Am. trichopoda clades or individual sequences. See supplementary figure S4, Supplementary Material online, for a fully expanded phylogenetic tree.
Furthermore, as for MUGA versus MUGB, multiple exaptation events is also supported by the internal topologies of the five FRS subtrees, which show that the five families originated and diversified at different times (table 2). Each of the FRS subtrees is broadly congruent with the species topology, but each has a different apparent last common ancestor (fig. 5, supplementary fig. S5, Supplementary Material online): two appear to have originated in early angiosperms (FRS3 and FRS10), two in early eudicots (FRS6 and FRS7), and one in early core eudicots (FHY3).
Table 2.
Periods of Origin and Diversification of Known MUG and FRS Familiesa.
| Stem Group Angiospermsb | Basal Angiospermsc | Monocotsd | Eudicotse | Core Eudicotsf | |
|---|---|---|---|---|---|
| MUGA | Origin +2 | — | — | — | 1 |
| MUGB | — | Origin | 1 | 1 | 1 |
| FRS3 | — | Origing | 4 | 2 | 1 |
| FRS10 | — | Origin | — | — | 1 |
| FRS6 | — | — | — | Origin | 3 |
| FRS7 | — | — | — | Origin | — |
| FHY3 | — | — | — | — | Origin +4 |
| Total | 3 | 3 | 5 | 5 | 12 |
aNumerals are the number of postexaptation duplications occurring in a given period (interior nodes).
bIncludes monocots, dicots, and Am. trichopoda.
cIncludes monocots and eudicots but does not include Am. trichopoda.
dIncludes only monocots.
eIncludes only eudicots and includes A. coerulea.
fIncludes only eudicots and does not include A. coerulea.
gMay have originated in stem group (see text).
Specifically, the FRS3 family contains multiple monocot clades that include both Arecales and Poales, and similarly contains multiple clades that include diverse eudicots including the basal eudicot Aq. coerulea. (Note that one monocot clade is not monophyletic with the other monocot clades, but has low local branch support (46%) and thus is likely mislocated.) Interestingly, the sister clade to this well-supported part of the FRS3 subtree is a small Am. trichopoda clade (δ) with no TE characteristics (supplementary table S2, Supplementary Material online) but only low local support (32%), and might also be part of the FRS3 family. This suggests that FRS3 was likely exapted in the basal angiosperms, or perhaps earlier in the angiosperm stem group, and subsequently diversified into various descendant lineages.
The FRS10 family also includes monocot homologs, and although it also has an Am. trichopoda sister clade, both Am. trichopoda sequences contain multiple stop codons and frameshifts. Furthermore, the clade has an ancestral eudicot branch, in violation of the species phylogeny (supplementary fig. S4, Supplementary Material online), which includes a clade of six highly similar sequences in G. max that contain stop codons and frameshifts. These results suggest that AtFRS10 and AtFRS11 might have separate origins, but additional analysis would be required to confirm this. It seems more likely that they do form a single family, which originated in early angiosperms and diversified only once, in early core eudicots.
The remaining three FRS families appear to be eudicot-specific. In the case of the FRS7 family, Lin et al. 2007 reported monocot homologs; however, we found none, even though we included in our search both monocot genomes searched by Lin et al. 2007 (Z. mays and O. sativa). To confirm, we used TARGeT to individually search all 11 monocots in our initial 62 genomes, yet still found no FRS7 homologs (not shown). Instead, the most basal FRS7 homologs we found were in Aq. coerulea, suggesting that FRS7 originated in early eudicots. Interestingly, unlike other FRS and MUG families, FRS7 is single-copy in most genomes, with only one widely conserved duplication, which occurred in the early Brassicaceae.
Lastly, the FHY3 family is of particular interest because it includes AtFHY3 and AtFAR1, currently the best characterized plant ETEs (Wang and Wang 2015). The FHY3 family includes homologs in diverse core eudicots including the asterids (e.g., M. guttatus), but not in Aq. coerulea, suggesting it likely originated in early core eudicots. Thus, interestingly, among the seven MUG and FRS families, the best-characterized family also happens to be the youngest. Furthermore, it has the distinction of being present in more core eudicot-specific clades than any other clade: five of them, each with a single paralogs in most core eudicots, including both rosids and asterids. Consistent with previous results (Lin et al. 2007), AtFHY3 and AtFAR1 are the sole A. thaliana paralogs in neighboring clades.
Potential Novel ETEs
Our results also suggest that the two MUG and five FRS subtrees may not be the only ETEs in these phylogenies. A few additional clades (at least nine clades containing 126 sequences; supplementary table S2, Supplementary Material online) have attributes suggesting that they may also be ETEs rather than TEs, attributes such as low copy-number, high proportions of paralogs to orthologs, and topologies congruent with the known species phylogeny.
In the simplified MUG tree (fig. 2, supplementary fig. S3, Supplementary Material online), there is one such clade (λ): three sequences that are single-copy in V. vinifera, S. lycopersicum, and S. tuberosum, none of which have premature stop codons or frameshifts, TIRs, or repetitive flanking DNA. However, this clade is missing sequences in several sister species, so unlike the MUG and FRS families if this clade is an ETE family it is only weakly conserved.
More convincing cases are found in the FRS tree (fig. 5, supplementary fig. S5, Supplementary Material online). For example, clade ε is closely related to FRS6 but separated by a large family of TEs in citrus (β) and other taxa. The clade includes orthologs from all examined monocots in several species-congruent clades, a topology similar to the monocot clades of the FRS3 family. Only six of the 47 sequences have pseudogenic characteristics (five from E. oleifera alone), and the clade has no other TE characteristics. To further investigate this clade, we examined the six O. sativa paralogs using Genomicus (Louis et al. 2013) to determine whether they have maintained microsynteny with at least two adjacent genes, a characteristic common to most ETEs but not functional TE genes (Hoen and Bureau 2015). Unlike a negative control of 25 known TEs that had no microsynteny beyond Oryza, and similar to a positive control of FRS3 homologs, in clade ϵ five of six O. sativa sequences do have conserved microsynteny among the Poaceae (supplementary table S2, Supplementary Material online). These results suggest that clade ϵ is a family of bona fide monocot-specific ETEs.
An alternative explanation for this topology is that, rather than novel ETEs, clade ε might be part of the FRS6 family, even though the intervening apparent TE clades have high local branch support (100%). However, this is not the only such subtree in the FRS phylogeny. Clade ζ, which is not closely related to any known FRS ETEs, has similar characteristics. Finally, perhaps the strongest example is clade θ, which consists of 25 species-congruent sequences that are conserved in diverse core eudicots and, except a single pseudogene, have no TE characteristics. A caveat to this interpretation is that putative TE sequences in the FRS phylogeny generally have fewer pseudogenic and other TE characteristics than sequences in the MUG phylogeny, making the distinction between ETEs and TEs less apparent (supplementary table S2, Supplementary Material online). Thus, although these clades have intriguing characteristics, further analysis is needed to determine whether they are indeed novel ETEs, unusual TE families, or artifacts.
If these clades do represent novel ETE families, it would be consistent with our recent finding that ETEs may be far more abundant than is currently understood (Hoen and Bureau 2015). Note that none of the novel ETEs we reported in that study are present in these phylogenies because none are related closely enough to MUG or FRS. Conversely, none of the potential novel ETEs reported here could have been found in that previous study because none include an A. thaliana ortholog.
If some of these subtrees do represent novel ETEs, it increases even further the contribution of TE exaptation to angiosperm evolution, especially in monocots. Indeed, this would not be surprising, given that three of five known FRS families are eudicot-specific while none are monocot specific. This is likely due to a selection bias: the initial search for FRS genes was restricted to the A. thaliana genome (Lin and Wang 2004) and subsequent phenotypic characterization was apparently limited to close orthologs of the initial twelve FRS genes found in A. thaliana (Lin et al. 2007).
C48-MULEs Form Widely Diverged Clades, Some with TIRs
In addition to characterizing ETEs, our results also uncovered TE clades with noteworthy characteristics. Along with three conserved domains normally present in the mudrA transposase, certain MULE families include a second gene, Kaonashi (KI), that contains a peptidase C48 domain normally found in ubiquitin-like protein-specific proteases (ULPs) (Hoen et al. 2006; van Leeuwen et al. 2007; Benjak et al. 2008). Although the function of KI is unknown, at least some KI-MULEs do not have easily identifiable TIRs, yet remain capable of transposition (Hoen et al. 2006).
In the full MUG tree (supplementary fig. S2, Supplementary Material online), although three C48-MULE clades are closely related to MUG (in V. vinifera, M. guttatus, and E. oleifera), most are concentrated in the branches furthest from MUG. Consistent with previous results (Le et al. 2000; Hoen et al. 2006; Benjak et al. 2008), most C48-MULE clades have high proportions of associated sequences with potential TIRs. However, some do not; for example, a large clade that appears to have been recently active in the basal eudicot Aq. coerulea. Furthermore, the branches of the tree containing most C48-MULE clades also includes clades that lack C48—some associated with TIRs, some not—a sporadic phylogenetic distribution similar to that of PB1 (see below), suggesting that C48 was lost from various lineages. Interestingly, several MULE clades are associated with both C48 and PB1 domains. Finally, in addition to those in the MUG tree, we found C48-MULEs in the FRS tree, in the genomes of V. vinifera, Citrus clementina, Citrus sinensis, M. truncatula, N. nucifera, Theobroma cacao, and M. guttatus. Consistent with previous results in melon (van Leeuwen et al. 2007), we also identified C48-MULEs in FRS tree (supplementary fig. S5, Supplementary Material online), even though it is widely diverged from MUG.
The PB1 Conserved Domain Is Present in Diverse MULEs
Finally, in addition to peptidase C48, we surprisingly found certain TEs that contain another unusual domain. As discussed above, MUGA and MUGB have a key difference in their gene structures: every known MUGB gene but no MUGA gene contains a PB1 domain (fig. 2). The origin of this domain in MUGB has been a mystery because, unlike other domains present in MUG, no MULE or indeed any TE has been reported to contain PB1.
Surprisingly, here we detected PB1 domains associated with apparent TEs in nine genomes in the MUG tree (fig. 2; supplementary fig. S3, Supplementary Material online), including several potentially active TE clades such as the sister clade of MUGB. PB1-MULEs might have previously remained undetected for several reasons. First, in the literature we could find no previous report of a specific search for the PB1 domain in TEs. Second, PB1-MULEs are present in only a small fraction of genomes (9 of 62 examined). Third, none of the genomes containing PB1-MULEs happen to be model genomes (e.g., A. thaliana and O. sativa contain none). Finally, even among these nine genomes, although PB1 is abundant in MULEs that are closely related to MUG (found in 178 of 397 non-MUG sequences that are paraphyletic to MUGA and MUGB; fig. 2), it is rare among other MULEs (found in only 71 of 658 remaining sequences in the full MUG tree; supplementary figs. S2 and S3, Supplementary Material online) and in none of the sequences of the FRS tree. Discovery of these PB1-MULEs solves the mystery of the origin of the MUGB PB1 domain. Furthermore, these high copy-number PB1-MULE families may explain previous observations that PB1 domains are far more abundant in plants than in other kingdoms.
The detailed phylogenetic pattern of PB1-MULEs is lineage-specific and sporadic. Clades associated with PB1 are tightly interspersed with clades not associated with it, and even clades associated with PB1 have highly variable proportions of PB1-MULEs (fig. 2). This sporadic distribution pattern may have arisen either because PB1 has been acquired multiple times in separate TE branches, or because it has been repeatedly lost. To determine which of these is more likely, we aligned the PB1 amino acid subsequence, including both PB1-MULEs and MUGB members, and inferred a separate phylogenetic tree. Except for minor differences that can be explained by the low information content of this domain, which is short (84 aa in AtMUG7) and has many variable positions (Sumimoto et al. 2007), the topology of the PB1 tree is broadly congruent with the MUG tree (supplementary fig. S13, Supplementary Material online). Although far from conclusive, these results are consistent with a single origin of PB1 in a common MULE ancestor followed by lineage-specific losses.
The origin of PB1 in MULEs is unknown, but given that PB1 has not been reported in other TEs, it may have been acquired through transduplication (Juretic et al. 2005), similar to how peptidase C48 domains may have been acquired (Hoen et al. 2006). This possibility is supported by the MUGB gene structure: compared with the other MULE transposase genes, PB1 occurs in an additional short 5′-exon, consistent with the general pattern of MULE transduplication. Transduplication is also supported by the recurrent deletion of PB1 from various MULE clades, suggesting that although it may somehow improve the transpositional success of PB1-MULE families, it is not essential. Interestingly, transduplication—the co-option of genes by TEs for a “selfish” function—is in a sense the evolutionary inverse of TE exaptation—the co-option of selfish TE genes for a phenotypic function. Thus if a MULE ancestor of MUGB did originally acquire PB1 by transduplication, there is an interesting corollary: the MUGB PB1 domains have likely undergone a complete co-evolutionary cycle, from phenotypic function to selfish function and back again.
How frequently are non-TE conserved domains transduplicated by TEs, then exapted from the TEs, and eventually have all evidence of their origin erased by extinction of the TE family? We can recognize MUGB as an ETE family because of its TE-specific MULE domain; however, many TEs have no TE-specific domain (Hoen and Bureau 2015), or may have lost any TE-specific domains during or following exaptation. Thus perhaps cycles of exaptation have enabled the amplification and diversification of not just PB1, but other sequences as well. If such cycles took place during primordial evolution, few traces of the origins of these sequences would remain in extant genomes (Roussigne et al. 2003; Quesneville et al. 2005; Babu et al. 2006).
Summary and Conclusions
We have shown that through careful phylogenetic analysis of ETE families, we may obtain a better understanding of the evolutionary role of TE exaptation. By analyzing ETEs in angiosperms previously thought to constitute only two families, MUG and FRS, we have shown that they instead likely originated in a total of at least seven separate exaptation events, triple the number of TE exaptation events previously understood for these ETEs, for a total among the 22 final genomes of 281 ETEs out of 2,934 sequences. Furthermore, we report preliminary evidence suggesting that additional ETE families have yet to be characterized. These results confirm and expand upon another recent study in which we showed that the number of ETEs in A. thaliana is more than double that previously reported (Hoen and Bureau 2015).
In addition to improving our theoretical understanding of how TEs have contributed to genome evolution, there is another motivation to better resolving the phylogenetic history of ETEs. As we have shown for MUGA and MUGB, ETEs of different families may have similar broad phenotypes, such as delays in development or decreases in plant size. This may be explained by the fact that many different families often act in concert to generate complex traits. However, it would be surprising if common functions were shared by ETEs from different families that originated in separate exaptation events, especially from widely diverged TE families (e.g., FRS10 vs. the four other FRS families) or greatly separated in time (e.g., MUG1 and MUG7). For instance, while AtFHY3 and AtFAR1 have not only been shown to rescue each other but to heterodimerize (Lin et al. 2008), they have not been shown to complement any FRS protein outside the FRS family. Furthermore, AtFHY3 and AtFAR1 have been well characterized and shown to act as transcription factors, to bind to thousands of sites in the genome, to differentially regulate hundreds of genes under light or dark conditions, and to regulate far-red induced hypocotyl de-etiolation (Wang and Deng 2002). Thus, it is important to emphasize that each of the four additional FRS families, which are thus far largely uncharacterized, each have as much potential as AtFHY3 and AtFAR1 to impact plant function. For example, we have recently shown that ETEs are often involved in abiotic stress responses, including genes in both the MUGA and MUGB families, in at least four FRS families, and in a large set of novel ETEs (unpublished results; Lin and Wang 2004; Lin et al. 2007; Ouyang et al. 2011; Gao et al. 2013).
In conclusion, it has not gone unnoticed that the self-perpetuating nature of TEs, sometimes denigrated as selfish, endows in them the capacity to act as agents of periodic rapid evolution (Hoen and Bureau 2015). This study uses phylogenetic analysis to investigate TE exaptation and highlights the importance of resolving the origin and evolution of ETE families. Such analyses can contribute greatly to our understanding of the potential functions and interactions of ETEs. We have shown that both the MUG and FRS groups of ETEs are not single families, but instead are derived from multiple exaptation events. These TE exaptations and subsequent ETE diversification contributed to all key stages of angiosperm evolution, from the early stem group, to the angiosperm radiation, to recent crown group radiations. In the future, such evolutionary histories will help improve the design and interpretation of experimental studies of ETEs and, we hope, will encourage additional investigations of as-yet uncharacterized ETE families.
Materials and Methods
To maximize our ability to find TEs closely related to MUG or FRS, as well as to identify basal and diverse ETEs in these groups, we searched a large number of genomes (62 species), including representatives from all major angiosperm lineages (49 species: 3 basals, 2 magnoliids, 11 monocots, 33 eudicots), six gymnosperms, five algae, one vascular plant, and one moss species (supplementary table S1, Supplementary Material online). As queries, to maximize search sensitivity we selected seven amino acid sequences representing diverse MUG sequences, including monocots and eudicots from each previously identified MUGA and MUGB clade (Joly-Lopez et al. 2012), or FRS clade (Lin et al. 2007; supplementary fig. S1, Supplementary Material online).
Genomes Selection
Because we expected to find thousands of sequences not closely related to MUG or FRS and therefore not of interest, to reduce the size of final analysis we devised a strategy to identify only the genomes of potential interest (fig. 1). First, we determined which genomes contained any sequence of interest by searching each genome individually. We used a customized command-line version of the TARGeT pipeline (Tree Analysis of Related Genes and Transposons) (v2.00; [Han et al. 2009]; Cavinder B, personal communication) along with TBLASTN (v2.2.26) to align queries and genomes (see below), PHI (v2.4; [Han et al. 2009]; http://target.iplantcollaborative.org/, last accessed February 3, 2016) to join local alignments and count stop codons and frameshifts, MAFFT (v7.158b; option –max_iterate 100; [Han et al. 2009]) to generate multiple alignments, and FastTreeMP (v2.1.7 SSE3 OpenMP; option –gamma; [Katoh and Standley 2013]) to infer phylogenetic trees. For the initial search, we selected a permissive similarity threshold (TBLASTN E-value, 1e-30) in order to identify even distantly related putative MUG homologs.
We selected for further analyses the genomes that fulfilled one or both of the following criteria: they contained apparent TEs that were descended from the last common ancestor of all MUG query sequences, or they had homologs associated with PB1 domains. In addition, we included for further analyses three genomes with key positions in the species phylogeny: the basal eudicot N. nucifera, which has the unique biological feature of being an aquatic herbaceous species; the magnoliid Pe. americana, which is basal to monocots and eudicots; and the basal angiosperm Nu. advena.
Alignment, Curation, and Tree Building
We then searched the genomes of interest, again using a command-line version of TARGeT that we customized to search multiple genomes, using a similarity threshold selected to maximize stringency while still retaining all sequences of interest (TBLASTN E-value, 1e–55). We included the following sequences for the phylogenetic analysis: fungal hop, maize mudrA, maize Jittery, and all previously identified MUG genes (Larsson 2014), which were from the following six genomes: C. papaya, Sorghum bicolor, Oryza sativa, Brachypodium distachyon, Medicago truncatula, and A. thaliana (fig. 2; supplementary figs. S2 and S3, Supplementary Material online). We built a preliminary alignment (MAFFT), then removed 188 problematic sequences that contained long truncations, insertions, deletions, or frameshifts, resulting in poor alignment within highly conserved blocks. We then generated a final multiple alignment (MAFFT), which we curated by first removing columns with gaps in 50% of sequences or more, then using Gblocks (Castresana 2000) to retain only highly conserved alignment blocks (63% of 555 columns). We inferred a final MUG phylogenetic tree (FastTreeMP) (supplementary fig. S3, Supplementary Material online). Lastly, for clarity of presentation, we made a simplified MUG tree containing only the sequences most closely related to MUG by pruning branches more diverged than the last common ancestor of all known MUG genes (fig. 2). FigTree (v1.4.2; http://tree.bio.ed.ac.uk/software/figtree/, last accessed February 3, 2016) was used to visualize the phylogenetic trees.
To validate the phylogenetic analysis, we used two additional methods: 1) neighbor joining using BioNJ/Neighbor (PHYLYP; v3.66; default parameters; 300 bootstraps; http://www.Phylogeny.fr, last accessed February 3, 2016 [Dereeper et al. 2008, 2010]); 2) Bayesian MCMC using MrBayes v3.2.6 (default parameters, except “heating temperature” 0.01 for FRS [Huelsenbeck et al. 2001; Ronquist and Huelsenbeck 2003; Altekar et al. 2004]), obtaining standard deviation of split frequencies of 0.008 after 2,000,000 generations for MUG and 0.052 after 2,000,000 generations for FRS.
Discriminate TEs from ETEs
To discriminate TEs from ETEs, we evaluated four sequence characteristics. 1) To evaluate pseudogenic features, we counted the number of stop codons and frameshifts as identified by PHI (Castresana 2000). 2) To identify flanking repetitive sequences, we aligned the DNA sequence flanking either side (3 kb) of each putative homolog to its respective genome (NCBI BLASTN v.2.2.29+; E-value, 1e-100; [Han et al. 2009]). To reduce artifacts caused by the presence of any repetitive sequences unrelated to the putative homologs (e.g., insertions of other TEs), we calculated for each putative homolog the minimum number of nonself-hits flanking either side, thus eliminating cases where a TE had inserted on only one side of the homolog. We then used the median of these repetitiveness values per clade, so that even if unrelated TEs had inserted on both sides of some elements, the repetitiveness measure would not be unduly affected, especially for large clades. 3) To identify potential TIRs, we again using the DNA sequences flanking each putative homolog, but this time aligned the two sides together (BLASTN; E-value, 0.01; reverse strand). Because the lengths of MULEs vary widely, we analyzed a large range of flanking lengths (1–30 kb), then chose a biologically reasonable representative length (10 kb) that had low (presumably false) positives among known MUGs and low (presumably false) negatives among other sequences. Finally, to detect Peptidase C48 domains we used NCBI RPS-TBLASTN (E-value, 0.01; v.2.2.29+; [Camacho et al. 2009]) and the NCBI Conserved Domain Database (Camacho et al. 2009) to search the genomic DNA sequence corresponding to each putative homolog plus 5 kb flanking each side. The same method was also used to identify PB1 domains.
Plant Material
To characterize the MUG mutant phenotypes, we used the approach and methods as described in Joly-Lopez et al. (2012) (Marchler-Bauer et al. 2011). The mutants mug1-1 (GK_514B01), mug2-3 (SALK_090878), mug3-1 (SALK_053113), and mug4-2 (SALK_036408) were obtained from GABI-Kat (http://www.gabi-kat.de, last accessed February 3, 2016) (Joly-Lopez et al. 2012) and SALK (http://www.arabidopsis.org/abrc, last accessed February 3, 2016) (Rosso et al. 2003) T-DNA insertion populations. Positions of insertion sites in double mutants used in phenotypic analyses were confirmed by sequencing the allele-specific PCR products. Wild-type ecotype Col-0 seeds were originally obtained from Lehle Seeds (www.arabidopsis.com). For the triple mutant genotyping and phenotypic analyses, seeds were plated on one-half MS media supplemented with 2% sucrose instead of 1% as described previously (Joly-Lopez et al. 2012).
dN/dS Analysis
The selective pressure for the MUGA family within the MUG tree was examined using dN/dS analysis. The same amino acid MAFFT (121 sequences) was used as in the MUG BioNJ/Neighbor analysis. Amino acids were replaced with corresponding genomic DNA sequences. Tree adjustments and branch calling were made using the Tree viewer T-Rex (Boc et al. 2012). dN/dS was estimated using CODEML (Phylogenetic Analysis by Maximum Likelihood package (PAML); version 4.8a release August 2014; default parameters except clean data = 0, with fix_omega = 1 for null and fix_omega = 0 for the alternative [model 2]). Sites under positive selection were analyzed using BEB (Yang et al. 2005) and the position of the residues visualized using the alignment viewer Aliview (Larsson 2014). The aligned amino acid sequence of MUG4 was used as query to search the NCBI Conserved Domain database to detect the position of the conserved domains (Marchler-Bauer et al. 2011).
Supplementary Material
Supplementary materials are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
The authors thank Adrian E. Platts for his help with the dN/dS analyses using CODEML and for his very useful advice. The authors also thank Brad Cavinder and James Burnette for supplying a command-line version of the TARGeT pipeline. This research was funded by grants from Genome Québec and Genome Canada to T.E.B. and M.B., and the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery) to T.E.B. The authors declare that they have no conflicts of interest. D.R.H. and Z.J.L. contributed equally. They conceived and designed the study, collected and analyzed the data, and drafted the manuscript. T.E.B. and M.B. participated in conceiving the study and revising the manuscript. All authors read and approved the final manuscript.
References
- Agrawal A, Eastman QM, Schatz DG. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744–751. [DOI] [PubMed] [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. 2004. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342:1241089. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L. 2006. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34:6505–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjak A, Forneck A, Casacuberta JM. 2008. Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PLoS One 3:e3107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MM, Keeling CI, Brand D, Vandervalk BP, et al. 2013. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29:1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. 2003. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 131:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P. 2005. An Arabidopsis hAT-like transposase is essential for plant development. Nature 436:282–284. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chardin C, Girin T, Roudier F, Meyer C, Krapp A. 2014. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J Exp Bot. 65:5577–5587. [DOI] [PubMed] [Google Scholar]
- Cowan R, Hoen D, Schoen D, Bureau T. 2005. MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol. 22:2084–2089. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C. 2011. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 11:47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phnotype paradigm and genome evolution. Nature 284:601–603. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA Transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. New Phytol. 183:557–564. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu H, An C, Shi Y, Liu X, Yuan W, Zhang B, Yang J, Yu C, Gao H. 2013. Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 75:795–807. [DOI] [PubMed] [Google Scholar]
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 14:685–695. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lloyd EA. 1999. Individuality and adaptation across levels of selection: how shall we name and generalize the unit of Darwinism? Proc Natl Acad Sci U S A. 96:11904–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Vrba ES. 1982. Exaptation-a missing term in the science of form. Paleobiology 8:4–15. [Google Scholar]
- Guilfoyle TJ, Hagen G. 2012. Getting a grasp on domain III/IV responsible for auxin response factor-IAA protein interactions. Plant Sci. 190:82–88. [DOI] [PubMed] [Google Scholar]
- Han Y, Burnette JM, 3rd, Wessler SR. 2009. TARGeT: a web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Res. 37:e78.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, Bureau TE. 2012. Transposable element exaptation in plants In: Grandbastien MA, Casacuberta JM, editors. Plant transposable elements. Heidelberg: Springer Berlin; p. 219–251. [Google Scholar]
- Hoen DR, Bureau TE. 2015. Discovery of novel genes derived from transposable elements using integrative genomic analysis. Mol Biol Evol. 32:1487–1506. [DOI] [PubMed] [Google Scholar]
- Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, Cowan RK, Bureau TE. 2006. Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol. 23:1254–1268. [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW. 2012. Arabidopsis FHY3 and HY5 positively mediate induction of cop1 transcription in response to photomorphogenic UV-B light. The Plant Cell 24:4590–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. 1999. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13:2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH. 2003. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34:453–471. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314. [DOI] [PubMed] [Google Scholar]
- Joly-Lopez Z, Forczek E, Hoen DR, Juretic N, Bureau TE. 2012. A Gene family derived from transposable elements during early angiosperm evolution has reproductive fitness benefits in Arabidopsis thaliana. PLoS Genet. 8:e1002931.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic N, Hoen D, Huynh M, Harrison P, Bureau T. 2005. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15:1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. 2004. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 23:311–324. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2005. RAG1 Core and V(D)J recombination signal sequences were derived from transib transposons. PLoS Biol. 3:e181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Berger F. 2014. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet. 15:613–624. [DOI] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. 2014. Molecular basis for auxin response factor protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A. 111:5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le QH, Wright S, Yu Z, Bureau T. 2000. Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 97:7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV. 2011. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 12:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan XY, Li J, Lau OS, Ouyang X, Dai M, Wan J, et al. 2011. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 13:616–622. [DOI] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. 2007. Transposase-derived transcription factors regulate light signaling in arabidopsis. Science 318:1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Teng Y, Park HJ, Ding L, Black C, Fang P, Wang H. 2008. Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol. 148:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H. 2004. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol. 136:4010–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2009. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 60:43–66. [DOI] [PubMed] [Google Scholar]
- Louis A, Muffato M, Roest Crollius H. 2013. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Hagemann S, Reiter E, Pinsker W. 1992. P-element homologous sequences are tandemly repeated in the genome of Drosophila guanche. Proc Natl Acad Sci U S A. 89:4018–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KR, McComb JA, Greene WK. 2013. Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol. 5:1886–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607. [DOI] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. 2011. Genome-Wide binding site analysis of far-red elongated hypocotyl3 reveals its novel function in Arabidopsis development. Plant Cell 23:2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. 2011. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 108:20317–20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA. 2010. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 186:37–45. [DOI] [PubMed] [Google Scholar]
- Prasad BD, Goel S, Krishna P. 2010. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One 5:e12761.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H, Nouaud D, Anxolabehere D. 2005. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol. 22:741–746. [DOI] [PubMed] [Google Scholar]
- Rawn SM, Cross JC. 2008. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 24:159–181. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Horard B, Hubert B, Vieira C. 2010. Jumping genes and epigenetics: towards new species. Gene 454:1–7. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 53:247–259. [DOI] [PubMed] [Google Scholar]
- Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. 2003. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 28:66–69. [DOI] [PubMed] [Google Scholar]
- Saccaro NL, Van Sluys M-A, de Mello Varani A, Rossi M. 2007. MudrA-like sequences from rice and sugarcane cluster as two bona fide transposon clades and two domesticated transposases. Gene 392:117–125. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Izsvak Z, Ivics Z. 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci. 66:1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O. 2012. FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71:907–920. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Kamakura S, Ito T. 2007. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007:re6.. [DOI] [PubMed] [Google Scholar]
- Tang W, Ji Q, Huang Y, Jiang Z, Bao M, Wang H, Lin R. 2013. FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 163:857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trehin C, Schrempp S, Chauvet A, Berne-Dedieu A, Thierry AM, Faure JE, Negrutiu I, Morel P. 2013. QUIRKY interacts with STRUBBELIG and PAL OF QUIRKY to regulate cell growth anisotropy during Arabidopsis gynoecium development. Development 140:4807–4817. [DOI] [PubMed] [Google Scholar]
- van Leeuwen H, Monfort A, Puigdomenech P. 2007. Mutator-like elements identified in melon, Arabidopsis and rice contain ULP1 protease domains. Mol Genet Genomics. 277:357–364. [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW. 2002. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21:1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H. 2015. Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20:453–461. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. 1993. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22:1107–1118. [DOI] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA, Ishida Y. 2009. Transposable elements and an epigenetic basis for punctuated equilibria. BioEssays 31:715–726. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 5:4956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF. 2015. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron 86:490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K, Sirko A. 2014. Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front Plant Sci. 5:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A, Stevens KA, Crepeau MW, Holtz-Morris A, Koriabine M, Marcais G, Puiu D, Roberts M, Wegrzyn JL, de Jong PJ, et al. 2014. Sequencing and assembly of the 22-gb loblolly pine genome. Genetics 196:875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.