Tang et al. show that in fission yeast Myo51 is a single-headed class V myosin with two actin-binding sites: A typical ATP-dependent binding site and a novel ATP-independent site in its tail that requires binding partners Rng8/9. This molecular architecture allows Myo51 to slide actin-tropomyosin filaments relative to each other during contractile ring assembly.
Abstract
Myo51, a class V myosin in fission yeast, localizes to and assists in the assembly of the contractile ring, a conserved eukaryotic actomyosin structure that facilitates cytokinesis. Rng8 and Rng9 are binding partners that dictate the cellular localization and function of Myo51. Myo51 was expressed in insect cells in the presence or absence of Rng8/9. Surprisingly, electron microscopy of negatively stained images and hydrodynamic measurements showed that Myo51 is single headed, unlike most class V myosins. When Myo51–Rng8/9 was bound to actin-tropomyosin, two attachment sites were observed: the typical ATP-dependent motor domain attachment and a novel ATP-independent binding of the tail mediated by Rng8/9. A modified motility assay showed that this additional binding site anchors Myo51–Rng8/9 so that it can cross-link and slide actin-tropomyosin filaments relative to one another, functions that may explain the role of this motor in contractile ring assembly.
Introduction
The separation of daughter cells at the end of mitosis depends on the function of the contractile ring, a structure that is conserved from yeast to animals. The fission yeast Schizosaccharomyces pombe is an excellent model system to study cytokinesis because it is genetically tractable and amenable to quantitative light microscopy. These features have allowed quantification of the amounts, and timeline of appearance, of many key proteins involved in this process (Pollard and Wu, 2010). Assembly and constriction of the contractile ring involves an actomyosin-based system. In fission yeast, this process requires two class II myosins (Myo2 and Myp2) and one class V myosin (Myo51), which is the focus of this paper. Myo2 appears at the contractile ring first, followed by Myo51, and lastly Myp2 (Laplante et al., 2015). The three myosins act synergistically to accomplish cytokinesis, with Myo2 and Myo51 being more important in ring assembly and Myp2 the largest contributor to ring constriction (Laplante et al., 2015). The nascent contractile ring first appears as a broad band of medially accumulated precursor nodes, which contain Myo2 and the formin Cdc12. A leading model of ring assembly called search, capture, pull, and release (SCPR) postulates that formin-polymerized actin filaments from one node are captured by Myo2 from adjacent nodes. The nodes coalesce as Myo2 moves toward the barbed end of actin and exerts force (Vavylonis et al., 2008).
Although Myo2 is recruited to the nodes independent of actin, Myp2 and Myo51 rely on actin to localize to the contractile ring at different stages (Win et al., 2001; Lo Presti et al., 2012; Takaine et al., 2015). Myo51 localizes between the nodes during ring assembly in a motor domain–independent fashion (Wang et al., 2014). Fission yeast cells lacking Myo51 take significantly longer to assemble the contractile ring (Wang et al., 2014; Laplante et al., 2015). The mechanism for the delay is unknown, because Myo51 has not yet been characterized in detail at the molecular level. It has been proposed that Myo51 may enhance node condensation by stabilizing actin filaments (Wang et al., 2014). Alternatively, the assembly delay may be caused by defective cortical flow, an additional process that Myo51 is implicated in (Huang et al., 2012). Cortical flow of actin filaments nucleated outside the division site is threaded into the early contractile ring during the assembly phase (Arai and Mabuchi, 2002; Huang et al., 2012; Coffman et al., 2013).
It was recently shown that Myo51 is associated with two additional proteins, Rng8 and Rng9 (Rng8/9), which dictate its localization and function (Wang et al., 2014). Deletion of either Rng8 or Rng9 abolishes Myo51 localization to the ring. This study also showed that Rng8/9 binds to a region in the tail between the light chain–binding lever arm and the presumptive cargo-binding motif. Based on the observation that purified Rng8/9 tends to oligomerize in the absence of Myo51, it was proposed that Rng8/9 clusters multiple Myo51 dimers to regulate Myo51 localization and function (Wang et al., 2014).
To provide further insight into how Myo51 contributes to cytokinesis, we expressed and purified the Myo51–Rng8/9 complex using the baculovirus/insect cell expression system. Surprisingly, EM showed that Myo51 is a single-headed motor both in the absence or presence of Rng8/9. Hydrodynamic data provided independent support for a single-headed motor. In addition to the typical MgATP-sensitive binding of the motor domain to actin, the Myo51–Rng8/9 complex also shows a novel second ATP-insensitive binding site to actin decorated with Cdc8, the sole tropomyosin (Tpm) in fission yeast and a critical component of the contractile ring. This second binding site provides an ATP-insensitive anchorage that enables Myo51 to translocate actin-Tpm filaments relative to each other. This unusual structure and mode of action offers mechanistic insight into the function of Myo51 in the contractile ring during cytokinesis and in other Tpm-bound actin structures in fission yeast.
Results
Rng8 and Rng9 copurify with expressed Myo51
The full-length Myo51 heavy chain and its associated light chains (essential light chain Cdc4 and calmodulin Cam1) were coexpressed using the baculovirus/Sf9 insect cell expression system. The Myo51 heavy chain contained a C-terminal biotin tag for attachment to streptavidin coated surfaces, followed by a FLAG tag to facilitate purification by affinity chromatography (Fig. 1 A). On SDS-PAGE, the purified protein consisted of heavy and light chains (Fig. 1 B, lane 3). A small proportion of Myo51 contained bacterially expressed Cam1 that was added during purification to facilitate full occupancy of the light chain–binding domain of the heavy chain (Fig. 1 B, dashed line).
Figure 1.
Domain structure and SDS gels of expressed Myo51. (A) Schematic of the domain structure of the Myo51 heavy chain. Residue numbers for individual domains of the heavy chain are given. The green curves represent Rng8/9 binding to the tail of Myo51 based on a previous study (Wang et al., 2014). (B) 4–20% SDS-PAGE gel showing molecular mass standards (lane 1), purified Myo51 coexpressed only with light chains Cam1 and Cdc4 (lane 2), and purified Myo51 coexpressed with light chains Cam1 and Cdc4, and Rng8 and Rng9 (lane 3). The band with an asterisk indicates a mixture of small degradation products (lane 3) identified by mass spectrometry. The dashed line (lane 3) is a small amount of bacterially expressed Cam1 that was supplemented throughout purification to facilitate light chain association. The bacterially expressed light chains migrate slower than those expressed in Sf9 cells, with no differences in primary protein sequences. (C) Predicted α-helical coiled-coil probability for the tail regions of Myo51 and vertebrate MyoVa (Paircoil2 analysis). Amino acids 893–1,471 of Myo51 and amino acids 908–1,853 of mouse MyoVa are shown. The arrow indicates the starting residue of the globular tail domain (amino acid 1,461) of MyoVa and dashed lines indicate the 0.5 probability cutoff. Myo51 is predicted to have only four heptad repeats.
When the Myo51-binding partners Rng8 and Rng9 were coexpressed in addition to the Myo51 heavy and light chains, Rng8/9 remained bound throughout the purification procedure, indicating that these two proteins have a strong and specific interaction with the Myo51 heavy chain (Fig. 1 B, lane 2). Mass spectrometry analysis validated the identity of these copurifying proteins as Rng8/9.
Myo51–Rng8/9 is a single-headed motor
Dimerization of most myosins occurs through a region of the tail called the rod, a domain that typically forms an α-helical coiled coil. The propensity to form an α-helical coiled coil is based on the strength of the heptad repeat, a seven–amino acid motif in which the first and fourth positions, which form the internal seam of the coiled coil, are small hydrophobic amino acids. Analysis of the Myo51 heavy chain sequence for its propensity to form an α-helical coiled coil revealed that the tail region has only one short predicted coiled-coil patch consisting of a total of four heptad repeats (Fig. 1 C), raising the question of whether it is a single- or double-headed motor. The much higher extent of predicted coiled coil in a known mouse dimeric class Va myosin from vertebrates is shown for comparison.
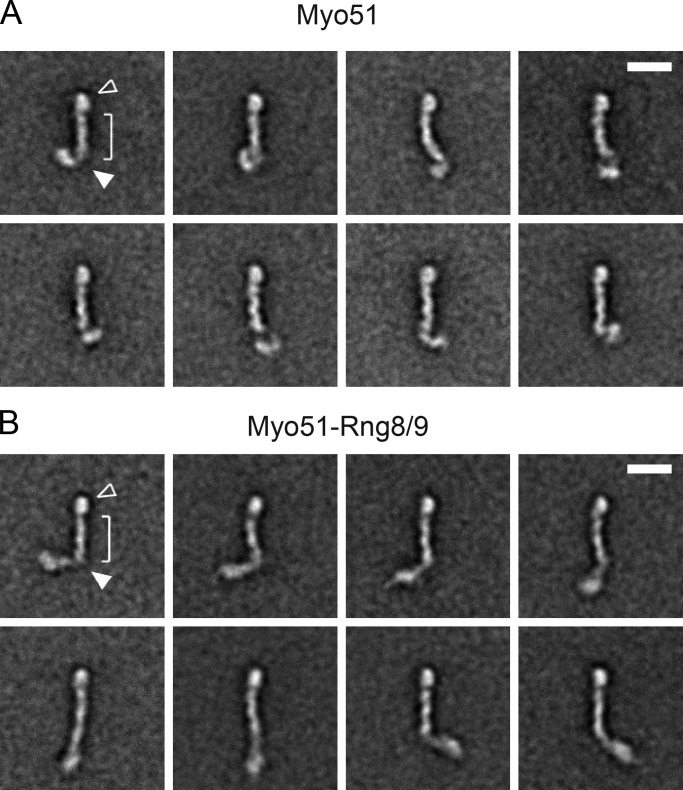
To directly show the state of oligomerization of Myo51, we visualized the motor by EM of negatively stained samples (Fig. S1). The images showed a single-headed motor, consistent with the low predicted coiled-coil propensity of the tail. Representative class average images of Myo51 are shown in Fig. 2 A. The class averages are characterized by a globular motor domain (Fig. 2 A, open arrowhead) connected to an ∼22-nm-long light chain–bound lever arm (bracket). The motor domain undergoes a conformational change in the presence of MgATP, bending relative to the lever arm (Fig. S2 and Video 1). The remainder of the molecule is globular and appears to be flexibly attached to the end of the lever arm (Fig. 2 A, closed arrowhead).
Figure 2.
Representative single-molecule class averages of Myo51 and Myo51–Rng8/9 derived from alignment and classification of EM images. (A) Eight class averages of Myo51 (n = 2510). (B) Eight class averages of Myo51–Rng8/9 (n = 1489). Open arrowhead, globular motor domain; single bracket, lever arm. Closed arrowhead indicates a hinge region between the lever arm and tail. Bars, 20 nm.
In the presence of Rng8/9, the appearance of the Myo51 motor domain and lever arm is unaffected, but the region following the lever arm adopts a more extended structure, consistent with binding of Rng8/9 to the tail region (Fig. 2 B; Wang et al., 2014). The Myo51–Rng8/9 complex shows a prominent bend after the lever arm, approximately two thirds of the total length of the myosin molecule (Fig. 2, closed arrowhead). There is no evidence for a more symmetrical “V” shaped dimer typical of vertebrate myosin Va. The end of the tail appendage is somewhat bulky and highly variable in position, different from the motor domain that is consistently globular. These images show that Myo51 is a single-headed motor in vitro, irrespective of the presence of Rng8/9. Occasionally, we observed two adjacent Myo51 or Myo51–Rng8/9 complexes in electron micrographs that could be interpreted as double-headed molecules (Fig. S1, arrows). Molecules that could be interpreted as dimers appeared at a very low frequency in EM fields of both Myo51 (7.1%, n = 1092) and Myo51–Rng8/9 (7.5%, n = 1010).
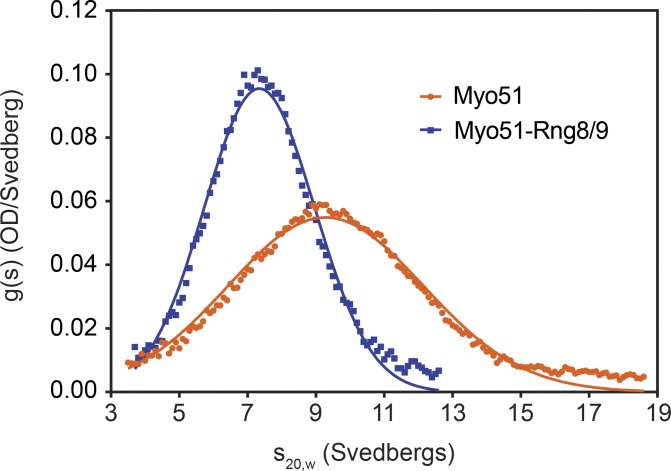
To provide independent validation that the myosin structures observed by EM are prevalent in solution, we performed sedimentation velocity and sedimentation equilibrium experiments with Myo51 and Myo51–Rng8/9 in the analytical ultracentrifuge. Myo51 and Myo51–Rng8/9 each sedimented as a single boundary (Fig. 3), with a mean sedimentation coefficient of 9.6S or 7.5S, respectively (Table 1). Given that Myo51–Rng8/9 has a higher molecular mass than Myo51 alone, the lower sedimentation coefficient indicates formation of a more asymmetric structure, consistent with the more extended tail structure observed by EM. The molecular masses obtained by sedimentation equilibrium for Myo51–Rng8/9 are close to that calculated for one heavy chain, six light chains, and one copy each of Rng8 and Rng9 (Table 1). The observed molecular mass of Myo51 alone (390 kD) is higher than that calculated for one heavy chain and six light chains (Table 1). Combined with the broader boundary seen by sedimentation velocity for Myo51 alone (Fig. 3), it is likely that Myo51 exists as a reversible equilibrium between predominantly monomers and some dimers at the higher concentrations used for these experiments. Overall, the hydrodynamic analysis suggests that binding of Rng8/9 to Myo51 stabilizes the motor complex in a homogenous, single-headed conformation.
Figure 3.
Sedimentation velocity of Myo51 and Myo51–Rng8/9. A representative analytical centrifugation run of Myo51 (orange circles) and Myo51–Rng8/9 (blue squares). The sedimentation coefficient was 9.6S for Myo51 and 7.5S for Myo51–Rng8/9. OD, optical density.
Table 1. Values obtained from sedimentation velocity and sedimentation equilibrium .
| Construct | Calculated molecular massa | Observed molecular massb | s20,w | |
|---|---|---|---|---|
| Monomer | Dimer | |||
| kD | kD | kD | ||
| Myo51 | 276 | 552 | 390 ± 12 (n = 6) | 9.6 (n = 2)c |
| Myo51–Rng8/9 | 346 | 692 | 301 ±10 (n = 6) | 7.5 (n = 2) |
The calculated molecular masses include the tags, light chains, and one copy each of Rng8 and Rng9 when present.
The molecular mass values (mean ± SD) were obtained from two independent protein preparations.
Mean s20,w from two independent preparations. The values obtained from the two experiments were the same.
Furthermore, we tested if Myo51–Rng8/9 can generate multiple 36-nm steps on actin, a hallmark of the processive dimeric motor, vertebrate myoVa. Myo51–Rng8/9, or a control constitutively active dimeric form of myoVa (myoVa–HMM), was attached to streptavidin-coated quantum dots (Qdots) via its C-terminal biotin tag at a ratio of 1 motor to ≥6 Qdots to remain primarily in the single molecule regime. The stepping pattern was followed by total internal reflection fluorescence (TIRF) microscopy. Approximately 80% of all control myoVa–HMM Qdots moved with its characteristic 36 nm stepping pattern (Mehta et al., 1999; Rief et al., 2000; Warshaw et al., 2005; Fig. S3). In contrast, most Myo51–Rng8/9 Qdots transiently associated with actin without any net motion. The few Myo51–Rng8/9 Qdots that did move showed a highly variable stepping pattern, including back steps and forward leaps (unpublished data). These functional data are consistent with the observations from EM and analytical ultracentrifugation that Myo51–Rng8/9 molecules are single headed in vitro and thus unable to support continuous motion on actin as a single molecule.
Motion of motor ensembles
We next tested if multiple Myo51 or Myo51–Rng8/9 motors bound to a Qdot would support continuous motion on actin. Motors were mixed with streptavidin Qdots in a 24:1 molar ratio to ensure saturation of the Qdot. Based on geometric considerations, however, each Qdot can only accommodate approximately three to five motors, which we used to simulate multiple motor transport (Hodges et al., 2009). Small ensembles of either Myo51 or Myo51–Rng8/9 moved continuously on bare actin with the same run length, speed, and run frequency (Fig. 4). Because actin cables and the contractile ring in fission yeast are extensively decorated with Tpm, we also tested motion on actin-Tpm. Small ensembles of Myo51 showed a 38% longer run length on actin-Tpm compared with bare actin (P < 0.01, Mann–Whitney test; Fig. 4 B), suggesting a possible increase in duty ratio and/or cooperative recruitment of heads in the presence of Tpm. Speed of Myo51 was 19% slower on actin-Tpm than on bare actin (P < 0.001, Mann–Whitney test), but had the same run frequency (Fig. 4 C).
Figure 4.
Movement of small ensembles of motors bound to a Qdot, visualized by TIRF microscopy. (A) Kymographs of movement of multiple motors (either Myo51 or Myo51–Rng8/9) bound to a Qdot on either actin or actin-Tpm tracks. Note that Myo51–Rng8/9 shows no motion on actin-Tpm. (B) Histograms of run lengths of Myo51 on actin (n = 231) or actin-Tpm (n = 221), and Myo51–Rng8/9 on actin (n = 288). The red lines are exponential fits (y = Ae−x/λ) to the data. The run length constants λ are indicated. The run length of Myo51 is higher on actin-Tpm compared with bare actin (P < 0.01, Mann–Whitney test). (C) Normalized run frequencies and run speed distributions. The mean run frequency of Myo51 on bare actin (number of events per µm actin per µM Myo51 per second) was normalized to one. Mean speed is indicated by the red lines. Mean ± SD values are indicated. Myo51 run speed is slower on actin-Tpm compared with bare actin (P < 0.001, Mann–Whitney test). On bare actin, the presence of Rng8/9 bound to Myo51 caused no statistical difference in run length, run frequency, or speed.
Unexpectedly, small ensembles of Myo51–Rng8/9 did not move on actin-Tpm. The myosin-coated Qdots bound to the actin-Tpm filaments were stationary, with only occasional, short runs (Fig. 4 A and Video 2). Static decoration of the actin track is typically seen in the absence of MgATP or when myosin is inactive, but we showed that the Myo51–Rng8/9 complex was fully active on bare actin and that the Myo51 motor domain interacts favorably with actin-Tpm.
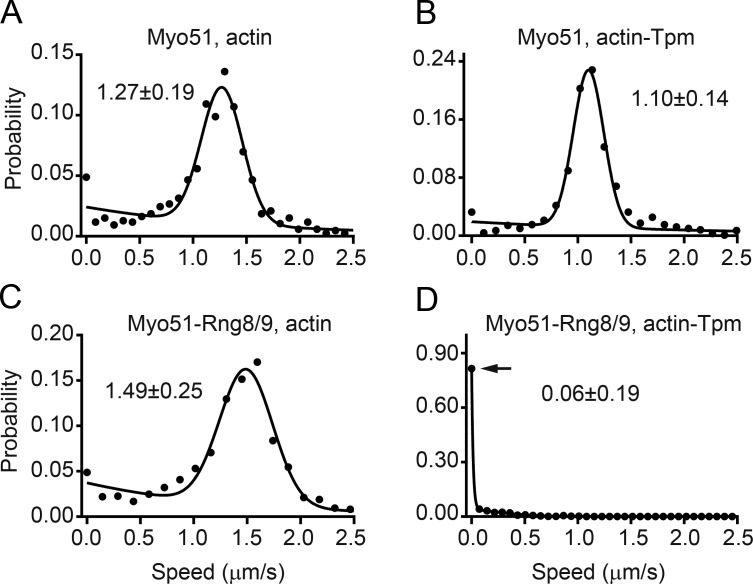
To further investigate this unexpected lack of motion, we used a standard ensemble in vitro motility assay. Myosin was specifically adhered to a neutravidin–coated coverslip via its C-terminal biotin tag to ensure that all motor domains are free to interact with actin. Movement of fluorescently labeled actin over this bed of motors was quantified. Myo51–Rng8/9 moved bare actin filaments ∼15% faster (P < 0.001) than Myo51. Addition of Tpm slowed the movement of Myo51 by 15% (P < 0.05), showing a similar trend to that observed with Qdot ensemble motility (Fig. 5 and Table 2). As seen with the inverted Qdot assay, motion was inhibited only when Myo51–Rng8/9 and actin-Tpm were paired. Approximately 80% (Fig. 5 D, arrow) of the actin-Tpm filaments remained static, whereas a small proportion (∼20%) of the filaments showed gliding at a speed five or six times lower than all the other conditions (Fig. 5 and Video 3). The slowed or stopped motility is reminiscent of myosin sliding actin filaments under a load exerted by an actin-binding protein such as α-actinin (Bing et al., 2000). This observation supports the idea that static filaments are not caused by loss of motor activity but mechanistically resembles slowing by tethering. These assays suggest the existence of an additional strong actin-Tpm–binding motif on Myo51–Rng8/9 that is distinct from the motor domain and absent in Myo51 alone.
Figure 5.
Speed distribution of actin or actin-Tpm movement by ensembles of surface-immobilized motors. (A) Actin moved by Myo51 (n = 860), (B) actin-Tpm moved by Myo51 (n = 981), (C) actin moved by Myo51–Rng8/9 (n = 1,374), (D) actin-Tpm and Myo51–Rng8/9 (n = 1,515). The arrow indicates that ∼80% of all actin-Tpm filaments are nonmotile with Myo51–Rng8/9. The lines indicate the nonlinear fit of the data to the sum of an exponential decay and a Gaussian distribution (see supplemental Materials and methods). The mean ± SD of the Gaussian distribution is shown. Tpm slows motion powered by Myo51 (P < 0.05, t test comparing the Gaussian distributions). The presence of Rng8/9 on Myo51 increased the actin gliding speed of bare actin (P < 0.001, t test comparing Gaussian distributions).
Table 2. Speeds from motility assays with different geometric arrangementsa.
| Motor | Track | Actin glidingb | Qdot ensemble motilityc | Dual-color actin-gliding motilityd |
|---|---|---|---|---|
| µm/s | µm/s | µm/s | ||
| Myo51 | Actin | 1.27 ± 0.19 (860) | 1.74 ± 0.50 (231) | Brownian motione |
| Myo51 | Actin-Tpm | 1.10 ± 0.14 (981) | 1.46 ± 0.46 (222) | Brownian motion |
| Myo51–Rng8/9 | Actin | 1.49 ± 0.25 (1,374) | 1.66 ± 0.49 (288) | Brownian motion |
| Myo51–Rng8/9 | Actin-Tpm | 0.06 ± 0.19 (1,515) | 0 | 1.28 ± 0.39 (114) |
Data are mean ± SD; the number of tracked filaments or Qdots are given in parentheses.
Standard ensemble motility assay with motors immobilized on the surface.
Multiple motors attached to a Qdot moving on an immobilized track.
Green fluorescent–labeled actin or actin-Tpm immobilized on the surface as the substratum for ATP-insensitive motor attachment. Red fluorescent–labeled actin or actin-Tpm was flowed in to monitor motility.
Red filaments showed longitudinal Brownian motion but no directed motor-driven motion.
In fission yeast, acetylated Tpm is the predominant species that decorates actin in the contractile ring, whereas unacetylated Tpm binds actin cables (Skoumpla et al., 2007; Coulton et al., 2010). The unacetylated Tpm acted indistinguishably in our gliding motility experiments compared with Tpm containing an Ala-Ser N-terminal extension, which has been used as an acetylation mimic (unpublished data).
Myo51–Rng8/9 has an ATP-independent binding site on actin-Tpm
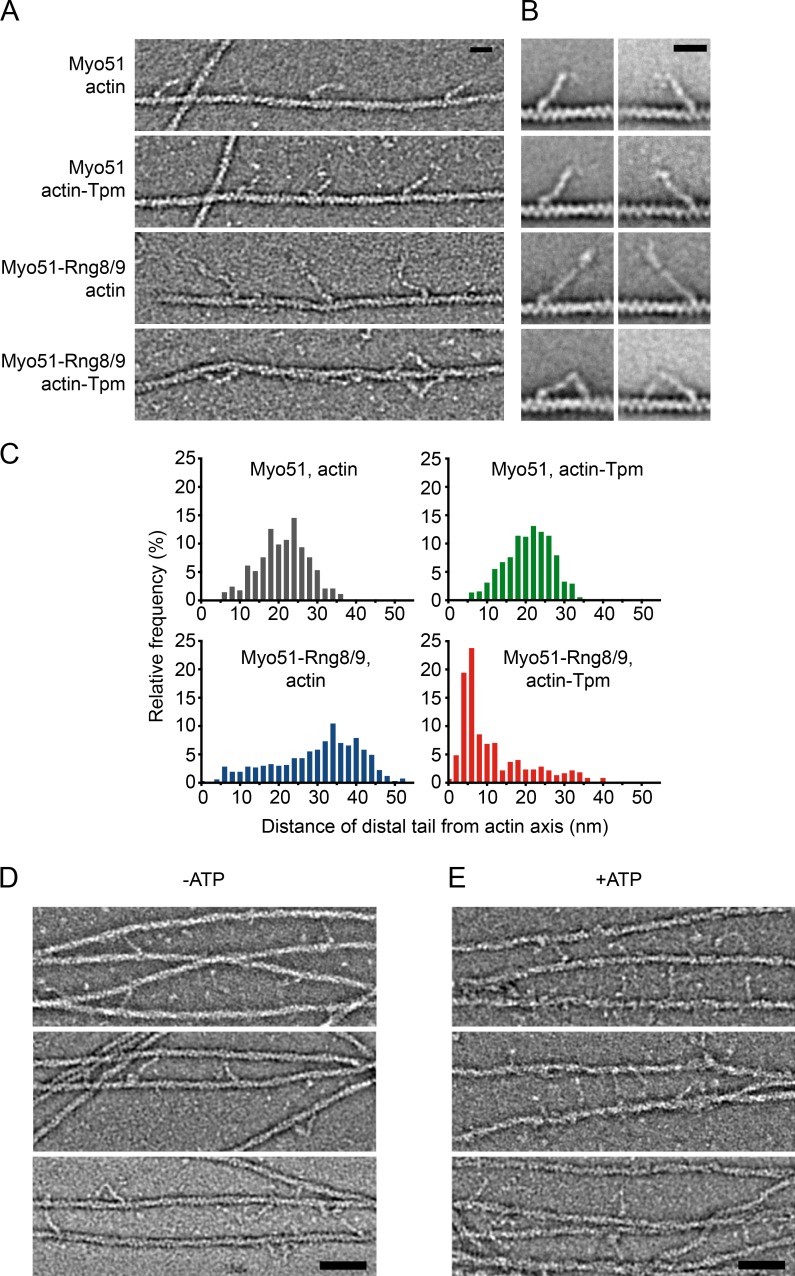
EM was used to visualize how Myo51 or Myo51–Rng8/9 interacts with actin and actin-Tpm. In the absence of MgATP, the myosin motor domains bind strongly to actin. As expected, Myo51 showed a single point of attachment with actin or actin-Tpm, as did Myo51–Rng8/9 bound to bare actin (Fig. 6, A and B). In contrast, a single molecule of Myo51–Rng8/9 showed two binding sites on actin-Tpm, made possible by a sharp kink in the molecule (Fig. 6, A and B), a structural characteristic also seen with Myo51–Rng8/9 that was not bound to actin (Fig. 2 B). The longer segment corresponds to the motor domain and the lever arm, and the shorter segment corresponds to the Rng8/9-bound tail (Fig. 6 B). The consistently asymmetric appearance and overall size of the bound Myo51–Rng8/9 complex suggest that it binds actin-Tpm through its motor domain and a secondary site, rather than as a dimer that binds with two heads. Measurements of the distance between the end of the tail and the axis of the actin/actin-Tpm track show that this two-site contact was unique, as the tail is only in close proximity with the track when the tail is bound to Rng8/9 and the track is decorated with Tpm (Fig. 6 C). It is not seen with Myo51–Rng8/9 on bare actin or with Myo51 on either actin or actin-Tpm (Fig. 6 C). Given that the myosin motor domain attaches to the actin with a characteristic angle that indicates the actin polarity, we found that Myo51–Rng8/9 tail binds to actin-Tpm ahead of the motor domain, further toward the plus end. This orientation suggests that it is unfavorable for Myo51–Rng8/9 to move in an “inchworm” fashion (Fig. 6 B). In the presence of MgATP, Myo51 and Myo51–Rng8/9 did not bind to bare actin, nor did Myo51 bind to actin-Tpm. In contrast, the distal region of Myo51–Rng8/9 remained bound to actin-Tpm in the presence of MgATP (Fig. S4). These EM images support the hypothesis that the Rng8/9-specific anchorage on actin-Tpm is nucleotide insensitive.
Figure 6.
Negatively stained images of various motor-track combinations in nucleotide-free conditions. (A) Fields of view showing the typical attachment appearance for Myo51 with or without Rng8/9 bound to bare actin or actin-Tpm. (B) Representative class averages showing bound molecules in each condition. Bars, 20 nm. (C) Histograms of the distance distribution between distal end of the tails of Myo51 or Myo51–Rng8/9 and the axis of the bound actin or actin-Tpm filament. Myo51 with actin, n = 619; Myo51 with actin-Tpm, n = 580; Myo51–Rng8/9 with actin, n = 670; Myo51–Rng8/9 with actin-Tpm, n = 597. (D and E) Myo51–Rng8/9 cross-links and bundles actin-Tpm filaments. Examples showing actin-Tpm cross-linked by Myo51–Rng8/9 in the absence (D) or presence (E) of MgATP. Bars, 50 nm.
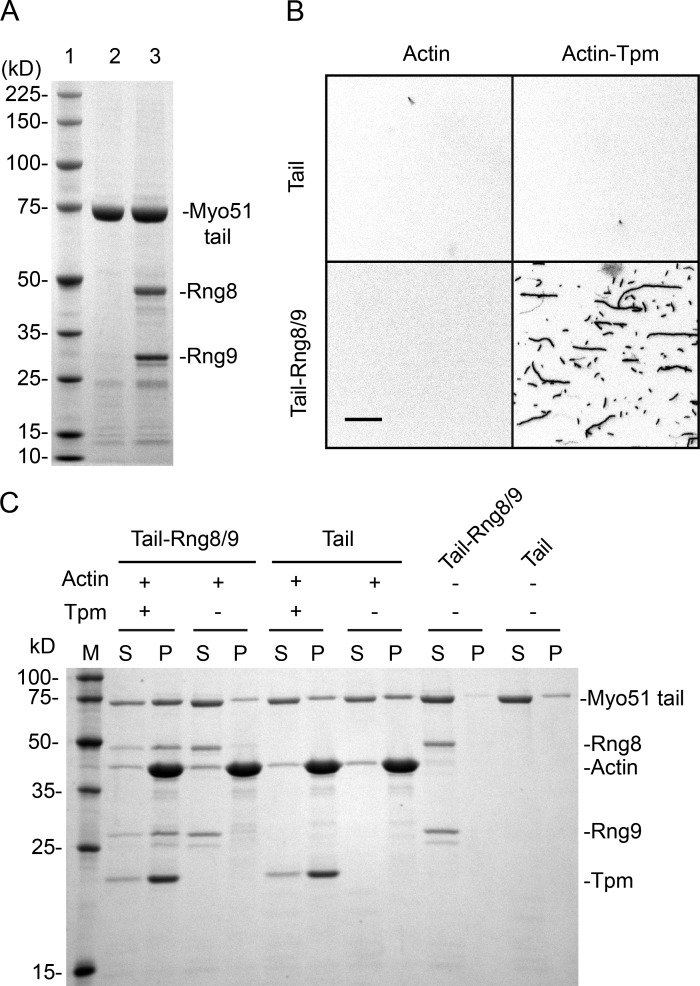
To confirm that a second ATP-independent binding site exists in the tail, we expressed the Myo51 tail domain (with N-terminal FLAG and biotin tags) with or without Rng8/9. Rng8/9 copurified with the Myo51 tail domain when the three proteins were coexpressed (Fig. 7 A), showing that the tail region of Myo51 is sufficient for Rng8/9 association, consistent with our EM analysis and a previous study (Wang et al., 2014). The Myo51 tail (± Rng8/9) was bound to a neutravidin-coated surface, and the ability of actin (±Tpm) filaments to be captured was assessed. Only actin filaments decorated with Tpm bound to the Myo51 tail–Rng8/9 complex; assays with all other combinations showed no binding (Fig. 7 B). In complementary actin-pelleting assays, the majority of Myo51 tail–Rng8/9 cosedimented with actin-Tpm, whereas very little pelleted with bare actin. Most of the Myo51 tail alone did not bind to either actin or actin-Tpm (Fig. 7 C). Similar results were observed in the presence of MgATP (unpublished data).
Figure 7.
Binding of Myo51 tail constructs to actin or actin-Tpm. (A) 4–20% SDS gel showing FLAG affinity purified N-FLAG-Biotin Myo51 tail (amino acids 893–1,471 of full-length Myo51 heavy chain) constructs. Lane 1, molecular mass standards. Lane 2, Myo51 tail. Lane 3, Myo51 tail coexpressed with Rng8 and Rng9. (B) Actin or actin-Tpm filaments bound to Myo51 tail or Myo51 tail-Rng8/9 coated surfaces. Bar, 15 µm. (C) 12% SDS gel showing the supernatant (S) and the pellet (P) fractions after high-speed centrifugation of Myo51 tail or Myo51 tail–Rng8/9 incubated with or without actin/actin-Tpm. M, molecular mass standards. The majority of Myo51 tail-Rng8/9 cosedimented with actin-Tpm, but not with bare actin. Myo51 tail mostly stayed unbound to actin or actin-Tpm.
Myo51–Rng8/9 strongly promotes actin-Tpm bundle formation, both in the absence or presence of MgATP. This tendency to bundle was reduced by lowering the amount of Myo51–Rng8/9 relative to actin-Tpm. In relatively thin areas of these bundles, individual Myo51–Rng8/9 molecules could be visualized extending and cross-linking two actin-Tpm filaments by the head and the second binding site, respectively (Fig. 6, D and E). The cross-linked actin-Tpm filaments can be either parallel or antiparallel in polarity (n = 25). Orientation of each actin-Tpm filament is based on the angular orientation at which the motor domain is attached. The observation that bundles also form in the presence of MgATP may suggest that the duty ratio of Myo51 is relatively high, which would allow the motor domain to stay strongly bound to actin-Tpm for a significant fraction of its ATPase cycle. In summary, the data show that Rng8/9 forms an appendage structure that allows Myo51 to attach to actin-Tpm at a site independent of the motor domain.
Myo51–Rng8/9 slides actin-Tpm filaments relative to one another
The dual binding sites of Myo51–Rng8/9 on actin-Tpm prompted us to consider whether Myo51–Rng8/9 could power the sliding of two actin-Tpm filaments relative to each other provided that the geometry was correct. The motility assays described earlier did not allow the Myo51–Rng8/9 complex to anchor to one actin-Tpm filament via its ATP-independent site and move another filament via its ATP-sensitive motor domain.
To test this possibility, we employed a modified dual-colored actin gliding assay (Fig. 8 A). A dense layer of BODIPY-phallacidin–labeled actin-Tpm (green actin) was adhered to a nitrocellulose-coated coverslip. Myo51–Rng8/9 was incubated with the green actin-coated surface in the presence of MgATP to enrich for binding via the nucleotide-insensitive tail. Rhodamine-phalloidin–labeled actin-Tpm (red actin) and excess Tpm was then flowed into the chamber to initiate motility. The bottom actin-Tpm layer becomes tightly bundled immediately after Myo51–Rng8/9 was infused into the chamber (Fig. 8, B and C; and Videos 4, 5, 6, 7, and 8), consistent with the EM observations showing a propensity for Myo51–Rng8/9 to cross-link actin-Tpm (Fig. 6, D and E). The strong bundle formation redistributed the green actin-Tpm heterogeneously on the surface, such that green actin-Tpm was highly enriched in some areas and almost absent in others (Fig. 8, B and C). Importantly, Myo51–Rng8/9 was now able to power the motion of the actin-Tpm in solution via its motor domain, relative to the fixed layer of actin-Tpm bundles on the coverslip that engaged the ATP-insensitive actin-Tpm–binding site. Red actin-Tpm gliding activity was observed where the green actin bundle was present and was most robust on the thickest bundled areas, where Myo51–Rng8/9 is presumably the most highly concentrated. Interestingly, in some cases, the green actin bundle spontaneously formed a ring-shaped structure upon which numerous short red actin filaments moved in a circle (Video 8). Often, the motile red actin-Tpm filaments tend to densely swarm near or at the center of the green actin-Tpm (Videos 6, 7, and 8). We manually tracked filaments that showed continuous gliding for at least 5 s before merging paths with other filaments or before it stopped moving. The speed distribution showed a mean speed of 1.28 µm/s, similar to that of Myo51 with actin or actin-Tpm observed in a standard in vitro motility assay (Fig. 8 D, Fig. 5, and Table 2), suggesting that the motor domain is fully active and can reach a robust actin gliding speed when the tail is anchored to a separate actin-Tpm layer.
Figure 8.
Modified ensemble motility assay showing that Myo51–Rng8/9 slides actin-Tpm filaments relative to each other. (A) Schematic of Myo51 attached to surface-immobilized green actin-Tpm (labeled with BODIPY-phallacidin) via the Rng8/9 appendage. The green actin-Tpm is bundled as Myo51–Rng8/9 cross-links filaments. Some Myo51 motor domains remain available to move red actin-Tpm filaments (labeled with rhodamine-phalloidin), whereas others are involved in bundle formation. (B and C) Top: kymographs of a red actin-Tpm filament that traveled along the green actin bundles formed by Myo51–Rng8/9. The white arrowheads indicate the moving red actin-Tpm filament at various time points. Note that in B, the filament paused midway in the trajectory and resumed motility. Bottom: maximum projection of the 60 s motility shown in the top panel. Bars, 5 µm. The red filament trajectory overlaps with the ridge of green actin-Tpm bundles. (D) Motile red filament speed distribution (n = 114). The mean ± SD is indicated.
Control experiments verified that this experimental setup allows actin-Tpm in solution to be moved only when the myosin can bind to the surface-adhered actin-Tpm layer in an ATP-insensitive manner (Table 2). Relative sliding was not supported by Myo51 on either bare actin or actin-Tpm (Video 9, A and B) or by Myo51–Rng8/9 on bare actin (Video 9 C).
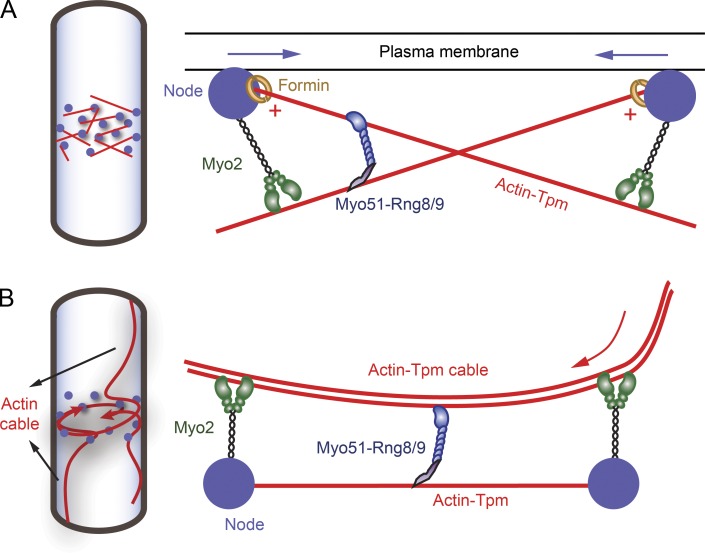
The dual-actin layer motility assay demonstrated that Myo51–Rng8/9 is capable of sliding actin-Tpm filaments relative to one another and that this behavior is highly selective for Tpm-decorated actin filaments. The sliding and bundling activities of Myo51–Rng8/9 suggest a mechanism by which Myo51 can recruit actin-Tpm cables into the contractile ring or facilitate the compaction of dispersed actin-Tpm filaments into a tight ring structure (Fig. 9).
Figure 9.
Models illustrating how Myo51–Rng8/9 may function during fission yeast contractile ring assembly. (A) In the search, capture, pull and release (SCPR) model, actin filaments (red lines) were polymerized by formin (open yellow circles) in the precursor nodes (filled blue circles) that were accumulated equatorially at the early stage of cytokinesis and captured predominantly by Myo2 (green, a class II myosin) from adjacent nodes. Actin filaments are bound to acetylated fission yeast tropomyosin. Myo2 exerts force on actin-Tpm and moves toward the plus end (red plus symbols) of the filament (where the formin is bound), thus moving the nodes closer to each other (blue arrows indicate the movement of the nodes). Myo51–Rng8/9 anchors its tail on one actin-Tpm filament while sliding a nearby actin-Tpm filament, providing additional force and movement, presumably toward the plus end of actin-Tpm as well, resulting in efficient ring compaction. (B) In the cortical flow model, actin cables polymerized nonmedially are recruited into the contractile ring during ring assembly, in addition to the actin polymerized at the nodes (SCPR model). The actin cables are bound to and stabilized by fission yeast Tpm. The cable movement (indicated by the red arrow) into the ring and the subsequent compaction is driven by Myo2 and Myo51–Rng8/9. Myo51–Rng8/9 tail binds to an actin-Tpm filament or cable captured between the nodes and its motor domain assists the continuous transport of the actin cable into the contractile ring.
Discussion
In fission yeast, Myo51 plays a role in contractile ring assembly and actin cable maintenance during vegetative growth (Lo Presti et al., 2012; Wang et al., 2014; Laplante et al., 2015). Recently identified binding partners Rng8/9 dictate the cellular localization and function of Myo51, and it is likely that Myo51 is associated with Rng8/9 for most or all of its cellular activities (Wang et al., 2014). Here, we show by EM that expressed Myo51 is a single-headed motor, in the absence or presence of Rng8/9, consistent with the relatively short stretch of predicted α-helical coiled coil in the tail. Hydrodynamic analysis confirmed that Myo51–Rng8/9 forms a homogeneous population of single-headed motors. Moreover, binding of Rng8/9 abolishes the slight tendency of Myo51 alone to reversibly dimerize at higher protein concentrations. Rng8/9 binds to the tail region (Fig. 7; Wang et al., 2014), and EM showed that association with these binding partners extends and appears to stabilize the Myo51 tail but does not cause dimerization. In contrast, all vertebrate class V myosins studied to date are double-headed, which enables them to move in a hand-over-hand manner on actin for long distances before dissociation, a feature known as processivity. Although there is precedent in budding yeast for a single-headed class V motor called Myo4-She3 (Krementsova et al., 2011; Sladewski et al., 2013), it is dimerized by the mRNA binding protein She2. Once dimerized by its binding partner, Myo4 moves processively on actin much like a class V myosin that is dimerized via its α-helical coiled coil.
A novel feature of the Myo51–Rng8/9 complex is a strong ATP-insensitive binding site to actin-Tpm, in addition to the typical ATP-sensitive binding of the motor domain. This anchorage only occurs when Rng8/9 is complexed to Myo51 and when fission yeast Tpm is bound to actin. In certain in vitro motility geometries, the Rng8/9-dependent tethering is strong enough to prevent movement. No motion of actin-Tpm is seen when Myo51–Rng8/9 is immobilized on a coverslip, nor do small ensembles of Myo51–Rng8/9 attached to a Qdot move on surface immobilized actin-Tpm. This result implies that when a motor engages both Rng8/9 and the motor domain on the same actin-Tpm filament, motion ceases.
This strong tethering is distinct from the diffusive tail tether that may allow an “inchworm”-like motility for myosin III on actin (Les Erickson et al., 2003; Merritt et al., 2012) or for promoting processivity of kinesin-8s (Stumpff et al., 2011; Su et al., 2011) and the minus-end–directed kinesin-14A (Ncd) on microtubules (Furuta and Toyoshima, 2008). Earlier studies showed that some class I myosins from Dictyostelium and Acanthamoeba have a nucleotide-insensitive actin-binding site in their tails (Lynch et al., 1986; Jung and Hammer, 1994). The tail homology domain 2 of these myosins contains large amounts of Gly and Pro, a feature not shared with Rng8/9, which are predicted to form α-helical coiled coils. With myosin I, it was proposed that the function of this second actin-binding site was to generate tension in the cortical actin meshwork.
A modified motility assay provided insight into how Myo51–Rng8/9 may function in the contractile ring. When Myo51–Rng8/9 is bound to surface-immobilized actin-Tpm via its ATP-insensitive site, which strongly engages Rng8/9 with the track, the motor domain of the Myo51–Rng8/9 complex is free to power ATP-dependent sliding of adjacent actin-Tpm filaments. This observation suggests that Myo51–Rng8/9 may rearrange and slide actin-Tpm filaments in the contractile ring (Fig. 9 A). A model based on the steady-state kinetics of a dual-actin–binding Acanthamoeba myosin I proposed that the ratio of actin to myosin affects the outcome: at a very low actin to myosin ratio, cross-linking of the actin filaments by one myosin cooperatively facilitates the cross-linking activity of neighboring myosin molecules, whereas very high ratios of actin to myosin favor noncooperative activation of myosin ATPase activity (Albanesi et al., 1985). The contractile ring is composed of ∼1,000–2,000 actin filaments (Kamasaki et al., 2007) that result in a local concentration of actin >500 µM (Wu and Pollard, 2005), whereas there are an estimated 300–400 molecules of Myo51 at the ring (Wang et al., 2014). This ratio between actin and Myo51 is more than sufficient to allow the motor domain and tail of Myo51–Rng8/9 to bind to different filaments, thus favoring relative sliding of actin-Tpm. It has been shown that during the early stages of cytokinesis, the ring consists of parallel actin filaments of opposite directionality, which then rearrange to a mixed directionality before contraction begins (Kamasaki et al., 2007). Myo51–Rng8/9 localizes between nodes in the contractile ring (Wang et al., 2014) and thus is in a position to locally slide and compact actin-Tpm filaments. At the same time, cables decorated by Myo51–Rng8/9 are pulled toward the nascent ring structure by both myosin II (Huang et al., 2012) at the nodes and perhaps by the Myo51–Rng8/9 complex as well (Fig. 9 B).
Relative sliding of tracks was observed with the microtubule-based motor kinesin-1 (Urrutia et al., 1991; Navone et al., 1992; Jolly et al., 2010), kinesin-5 (Eg5; Kapitein et al., 2005), kinesin-8 (Su et al., 2011, 2013; Roostalu and Surrey, 2013), and kinesin-14A (Ncd; Furuta and Toyoshima, 2008). These kinesins also have microtubule cross-linking/bundling activity and are involved in regulating microtubule dynamics of the mitotic/meiotic spindle. Their tails contain an ATP-independent microtubule-binding motif. The comparable function of ATP-independent tethers in both kinesins and Myo51–Rng8/9 suggests a common mechanism for molecular motors to slide tracks relative to one another.
Another cellular role for Myo51 is in actin cable organization, in conjunction with Myo52, the only other class V myosin in fission yeast (Motegi et al., 2001; Win et al., 2001). Interphase actin cables are dynamic bundles originating at the cell poles and assembled by formin. They provide tracks for Myo52 to transport material toward the growing tips of the cell, a process critical for the maintenance of cell polarity. The role of Myo52 in cable organization is thought to be through force generation to extend the bundles and through transport of regulators of actin polymerization to the cell tip (Lo Presti et al., 2012).
The strong tendency of Myo51–Rng8/9 to bundle actin-Tpm suggests that Myo51 may contribute to cable tension by cross-linking the cable filaments. The role of Myo51 in maintaining cable morphology was shown to be independent of the C-terminal cargo-binding motif, in contrast to Myo52 whose role required the globular tail (Lo Presti et al., 2012; Wang et al., 2014). Knowing that Rng8/9 binds to a region of the tail that precedes the cargo-binding motif, and that Rng8/9 is necessary for ATP-insensitive binding to the actin-Tpm track, explains why the C terminus of Myo51 is not required for these functions. Interestingly, the role of Myo51 in mating fusion focus, which requires Tpm, was also shown to be independent of the cargo-binding motif (Kurahashi et al., 2002; Dudin et al., 2015).
We cannot rule out that a yet unidentified protein can bind to the globular tail of Myo51 and cause it to dimerize and move processively under certain conditions. Recently, Tor1 kinase was proposed to interact with the Myo51 globular tail and require Myo51 to localize to the contractile ring (Baker et al., 2016). During meiosis, Myo51 colocalizes with the microtubule organizing center in fission yeast in a globular tail–dependent fashion and plays a role in spore formation (Doyle et al., 2009).
In summary, we show that Myo51–Rng8/9 is a single-headed motor for which actin-Tpm is both a track and a cargo. This newly discovered feature explains how Myo51–Rng8/9 can play a role in assembly and compaction of the cytokinetic ring by enabling the relative sliding of actin-Tpm filaments. The ability of Myo51–Rng8/9 to cross-link filaments could contribute to maintenance of actin cables/asters by straightening and rigidifying these structures. In addition to passive linkers, fission yeast has evolved to use energetically expensive motor molecules to promote actomyosin self-organization. A key question that remains is how Myo51 collaborates with the two myosin IIs at the contractile ring, given its biochemical characteristics. Future studies combining structural information and defined geometric arrangement/distribution of these myosins will help to understand this conserved and fundamental biological process.
Materials and methods
Expression constructs
The Sf9 cell codon–optimized coding sequence of full-length Myo51 was synthesized (Integrated DNA Technologies) to eliminate an alternative internal promoter sequence that resulted in expression of a truncated protein. The Myo51 coding sequence followed by a C-terminal biotin tag and FLAG tag was then ligated into baculovirus vector pAcSG2 (BD). The coding sequence of Myo51 tail domain (amino acids 893–1,471) preceded by N-terminal FLAG tag and biotin tag was ligated to pFastBac-1 vector (Thermo Fisher Scientific). Mouse myosin V-HMM (truncated at residue 1,098) followed by C-terminal biotin and FLAG tags was cloned into pAcSG2 vector (Krementsov et al., 2004; Ali et al., 2013). The biotin tag provides a specific attachment site for binding to streptavidin-coated Qdots or neutravidin-coated glass surfaces. The FLAG tag facilitates purification by affinity chromatography. The Rng8 and Rng9 cDNA sequences were amplified from fission yeast genomic DNA, and each was cloned into baculovirus vector pAcUW51 (BD). Open reading frames of these constructs were confirmed by sequencing. The cDNAs of Myo51 light chains Cam1 and Cdc4 were cloned into pET3a bacterial expression vector (EMD Millipore) and pAcUW51 (BD) Sf9 cell expression vector. The cDNA of fission yeast tropomyosin Cdc8 (nonacetylated) and its acetylation mimic containing an N-terminal Met-Ala-Ser peptide extension were cloned into pET3a vector (EMD Millipore) for bacterial expression (Clayton et al., 2014).
Protein purification
For Myo51 constructs, Sf9 cells were coinfected with recombinant baculovirus for the Myo51 heavy-chain tail domain (amino acids 893–1,471) or full-length heavy chain and light chains of Myo51, with or without Rng8 and Rng9. For the MyoVa construct, Sf9 cells were coinfected with baculovirus encoding MyoVa HMM and calmodulin. The cells were harvested after 72 h, spun down at 5,000 g for 10 min at 4°C, and resuspended in ice-cold lysis buffer (300 mM NaCl, 10 mM imidazole, pH 7.4, 5 mM MgCl2, and 1 mM EGTA) supplemented with 2 mM DTT, protease inhibitors (0.5 mM 4–[2-aminoethyl] benzenesulfonyl fluoride hydrochloride [Thermo Fisher Scientific]; 5 µg/ml leupeptin [Acros Organics]; 0.5 mM PMSF [MP Biomedical]; and 0.4 mg/ml benzamidine [Sigma-Aldrich]), and 20 µg/ml of bacterially expressed Cam1 and Cdc4 (for Myo51) or 25 µg/ml Cam (for MyoVa). Cells were lysed by sonication on ice and supplemented with 2 mM MgATP before the cell debris was spun down by ultracentrifugation at 250,000 g for 20 min at 4°C. The supernatant was batch-incubated with FLAG-affinity resin (Sigma-Aldrich) for 1 h at 4°C, washed with lysis buffer, and eluted with 100 µg/ml FLAG peptide (Sigma-Aldrich). The eluate was concentrated by Amicon-Ultra filtration (EMD Millipore) and dialyzed at 4°C against buffer (300 mM NaCl, 10 mM imidazole, pH 7.4, 1 mM EGTA, 1 mM NaN3, and 50% glycerol [vol/vol] supplemented with 2 mM DTT and 1 µg/ml leupeptin), and stored at −20°C. Proteins purified for EM were isolated similarly but dialyzed against buffer containing 300 mM KCl, 10 mM imidazole, pH 7.4, 0.1 mM EGTA, 1 mM MgCl2, and 1 mM Tris (2-carboxyethyl) phosphine hydrochloride (Sigma-Aldrich).
Actin was extracted from chicken muscle acetone powder at 0°C with G buffer (2 mM Tris-Cl, pH 8.0, at 25°C, 0.2 mM ATP, 0.5 mM β-mercaptoethanol, and 0.2 mM CaCl2) and filtered. The flow through actin was polymerized, pelleted, and depolymerized by dialyzing into G buffer for 3 days (Spudich and Watt, 1971). The G-actin were flash-frozen and stored at −80°C.
Bacterial expression constructs containing fission yeast cDNA Cam1, Cdc4, Cdc8, and Cdc8 with N-terminal Ala-Ser (acetylation mimic) were transformed into BL21-DE3 competent Escherichia coli cells (EMD Millipore). After growth to OD600 0.8–1, protein expression was induced with 0.4 mM IPTG in Luria–Bertani broth for 5 h at 25°C. The bacteria were spun down and resuspended in ice-cold buffer A (20 mM imidazole, pH 7.5, 10 mM NaCl, and 2 mM EDTA) supplemented with 1 mM DTT and 1 µg/ml leupeptin, 0.5 mM PMSF, and 0.4 mg/ml benzamidine. The cells were lysed by sonication on ice, and the cell debris was spun down at 30,000 g for 15 min at 4°C. The supernatant was boiled for 5 min with constant stirring and cooled to 25°C. Denatured proteins were spun down at 30,000 g for 15 min at 4°C. Cam1, Cdc4, and Cdc8 were precipitated by adjusting the pH of the supernatant to their respective isoelectric focusing points, pelleted down at 30,000 g for 15 min, and dissolved in buffer A plus 1 mM DTT and 1 µg/ml leupeptin by gentle agitation for 4 h or overnight at 4°C. Cam1 and Cdc4 were dialyzed against buffer B (20 mM imidazole, pH 7.5, 50 mM NaCl, 2 mM EDTA, and 50% glycerol [vol/vol]) supplemented with 1 mM DTT and 1 µg/ml leupeptin. Cdc8 was dialyzed against buffer C (50 mM NaCl, 10 mM imidazole, pH 7.5, and 1 mM DTT).
In vitro motility assays
Myo51 or Myo51–Rng8/9 was mixed with a twofold molar excess of actin and 5 mM MgATP and clarified at 400,000 g for 20 min at 4°C to remove ATP-insensitive motor heads. A nitrocellulose-coated chamber was infused with 0.5 mg/ml biotinylated BSA (Thermo Fisher Scientific) in Motility buffer A (50 mM KCl, 4 mM MgCl2, 10 mM imidazole, pH 7.4, 1 mM EGTA, and 2 mM DTT) for 1 min, followed by three washes with motility buffer A containing 0.5 mg/ml BSA, and then incubation with 50 µg/ml neutravidin (Thermo Fisher Scientific) in Motility buffer A for 1 min. The neutravidin-coated surface was then washed three times with Motility buffer A. Myosins were diluted to 170 nM, infused into the chamber, and allowed to bind for 1 min. The chamber was then washed three times with Motility buffer A supplemented with 0.5 mg/ml BSA, followed by two washes (30 s each) with 1 µM vortexed, unlabeled actin filaments in Motility buffer A to block MgATP-insensitive motors. The flow cell was then washed three times with Motility buffer A containing 2 mM MgATP, followed by three washes with Motility buffer A. 25 nM rhodamine-phalloidin–labeled actin or actin-Tpm filaments were infused twice into the chamber, followed by two washes with Motility buffer A. The chamber was then washed once with Motility buffer B (150 mM KCl, 10 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 20 mM DTT, oxygen scavengers [50 µg/ml catalase, 130 µg/ml glucose oxidase, and 3 mg/ml glucose], 0.5% [wt/vol] methylcellulose, 70 µg/ml Cam1, and 70 µg/ml Cdc4) and twice with Motility buffer B containing 1.5 mM MgATP. Assays were performed at 25°C.
In the dual-color motility assay, 10 µM BODIPY-phallacidin (Thermo Fisher Scientific)–labeled actin or actin-Tpm filaments in Motility buffer A were infused into the nitrocellulose-coated chamber and allowed to adhere for 10 min at 25°C. The chamber was then washed three times with Motility buffer C (50 mM KCl, 10 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 1 mg/ml BSA, 0.2 mg/ml κ-casein [Sigma-Aldrich], 0.5% [wt/vol] Pluronic F68 [Sigma-Aldrich], and 2 mM DTT). Myosins were centrifuged with a twofold molar excess of actin and 5 mM MgATP at 400,000 g for 20 min at 4°C to remove MgATP-insensitive motor heads and diluted to 40 nM in Motility buffer C with 5 mM MgATP, infused into the chamber, and allowed to attach for 45 s to 1 min, followed by three washes with Motility buffer C with 5 mM MgATP and three washes of Motility buffer C without MgATP. 30 nM rhodamine-phalloidin–labeled actin or actin-Tpm filaments in Motility buffer A were flowed in twice (30 s each time), followed by two washes with Motility buffer C. Finally the chamber was washed once with Motility buffer D (50 mM KCl, 10 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 20 mM DTT, oxygen scavengers [50 µg/ml catalase, 130 µg/ml glucose oxidase, and 3 mg/ml glucose], 0.5% [wt/vol] methylcellulose, 70 µg/ml Cam1, and 70 µg/ml Cdc4) and twice with Motility buffer D containing 1.5 mM MgATP. 1 µM Tpm dimer was included in Motility buffer A, B, C, and D in conditions when actin-Tpm filaments were tested. Assays were performed at 25°C.
Filament-tail–binding assay
A nitrocellulose-coated chamber was infused with 0.5 mg/ml biotinylated BSA in Motility buffer A for 1 min, followed by three washes with motility buffer A containing 1 mg/ml BSA, and then incubation with 50 µg/ml neutravidin (Thermo Fisher Scientific) in Motility buffer A for 1 min. The neutravidin-coated surface was then washed three times with binding buffer (150 mM KCl, 10 mM imidazole, pH7.5, 4 mM MgCl2, 1 mM EGTA, and 2 mM DTT). Myo51 tail proteins were diluted to 170 nM, infused into the chamber, and allowed to bind for 1 min. The chamber was then washed three times with binding buffer supplemented with 0.5 mg/ml BSA. 25 nM rhodamine-phalloidin–labeled actin or actin-Tpm filaments was infused twice into the chamber, followed by two washes with binding buffer (1 µM Tpm dimer was also included in the final washes when actin-Tpm was tested) and imaged. The image of the surface was captured as one still image in the tetramethylrhodamine (TRITC) channel and was inverted using ImageJ. The assay was performed at 25°C.
Actin pelleting assay
Preclarified 1 µM Myo51 tail and Myo51 tail–Rng8/9 were incubated with or without 5 µM actin or actin-Tpm in at 25°C in the buffer containing 150 mM NaCl, 10 mM imidazole, pH 7.5, 5 mM MgCl2, 1 mM EGTA, and 2 mM DTT for 10 min. The mixtures were pelleted at 400,000 g for 20 min at 4°C. The supernatant was removed and the pellet was washed twice with G buffer. 1.5 mM MgATP was included in the buffer when ATP sensitivity was tested. The supernatant and the pellet fractions were analyzed by SDS-PAGE.
Fluorescent imaging
Filament gliding images and filament tai–binding images were collected by a CoolSNAP HQ2 14-bit camera (Photometrics) driven by NIS elements software (Nikon) using an inverted microscope (TE2000-E2; Nikon) with a Plan-Apo 60× (1.45 NA) objective lens. For the filament tail–binding assay, the image of the surface was captured as one still image in the TRITC channel. For gliding filaments, images were collected in the TRITC channel for 60 to 120s at 1-s intervals at 25°C in Motility buffer B containing 1.5 mM MgATP with or without Tpm. Actin-gliding speeds were calculated from the coordinates of ≥860 filaments per condition tracked by ImageJ with Multitracker plugin (http://imagej.nih.gov/ij). The histograms of speed distribution were fit to an equation:
where A is amplitude 1, B is amplitude 2, λ is the decay coefficient, and SD is the standard deviation. In the dual-color motility assay, the BODIPY-phallacidin–labeled (green) layer was captured as one still image in the FITC channel before the recording of the gliding rhodamine-phalloidin–labeled (red) filaments in TRITC channel. The relative gliding motility movies were assembled as merged channels using ImageJ. The speeds of 114 red actin-Tpm filaments were manually tracked using ImageJ with MtrackJ plugin.
EM and image processing
For Myo51 or Myo51–Rng8/9 examined in the absence of actin, samples were diluted to 50 nM in buffer containing 10 mM MOPS, pH 7.0, 100 mM KCl, 2 mM MgCl2, and 0.1 mM EGTA (supplemented with 100 µM MgATP where indicated). For Myo51 or Myo51–Rng8/9 examined in the presence of actin or actin-Tpm, all samples were diluted in buffer containing 10 mM MOPS, pH 7.0, 50 mM KCl, 2 mM MgCl2, and 0.1 mM EGTA. F-actin was initially diluted to 20 µM (with or without 10 µM Tpm, acetylation mimic) in buffer supplemented with 40 µM phalloidin and incubated for a minimum of 1 h. Immediately before making EM grids, Myo51 or Myo51–Rng8/9 samples were diluted to 200 nM (supplemented with 100 µM MgATP where indicated). Actin or actin-Tpm samples were diluted to 2 µM (supplemented with 100 µM MgATP where indicated). Equal volumes of myosin and actin solutions were then mixed and applied to the grid after ∼30 s. All EM samples were applied to UV-treated, carbon-coated copper grids as 3-µl drops and stained immediately with 1% uranyl acetate. Micrographs were recorded on a JEOL 1200EX II microscope operating at room temperature at a nominal magnification of 40,000. Data were recorded on an AMT XR-60 CCD camera. Catalase crystals were used as a size calibration standard. Image processing was performed using SPIDER software as described previously (Burgess et al., 2004). For analysis of myosins bound to actin, particles with levers pointing in opposite directions were selected and analyzed separately (with respect to an actin filament rotated to be horizontal with the myosin bound on the upper side of the actin filament). The following size datasets were used for image processing, where M is number of micrographs, P is the number of particles, A refers to myosins bound to actin with the lever emerging from the right-hand side of the bound motor, and B refers to myosins with the lever emerging from the left-hand side of the bound motor: Myo51 apo, M = 50, P = 2,510. Myo51–Rng8/9 apo, M = 60, P = 1,489. Myo51 + ATP, M = 50, P = 854. Myo51 + actin M = 150, PA = 402, PB = 241. Myo51–Rng8/9 + actin M = 100, PA = 341, PB = 339. Myo51 + actin-Tpm M = 100, PA = 351, PB = 245. Myo51–Rng8/9 + actin-Tpm M = 184, PA = 359, PB = 282. For histograms of the distance between the myosin tail and the track, the axis of the track and the most distal part of the molecule was marked and the distance between them was measured.
TIRF microscopy
Myo51 or Myo51–Rng8/9 was centrifuged with a twofold molar excess of actin and 5 mM MgATP at 400,000 g for 20 min at 4°C to remove MgATP-insensitive motor heads. Myosins were then incubated with Qdot 655 Streptavidin conjugate (Invitrogen) at a motor/Qdot molar ratio of 24:1 (Fig. 3). Each Qdot can accommodate three to five motors for multiple motor conditions, given geometric considerations (Hodges et al., 2009). A glass chamber was infused with 0.1 mg/ml N-ethylmaleimide–modified myosin for 10 min at 23°C and washed five times with Motility buffer C. 60 nM rhodamine-phalloidin–labeled actin or actin-Tpm filaments was flowed in and allowed to bind for 4 min, followed by two washes with Motility buffer A and one wash with Motility buffer C. Myosin/Qdot mixtures were then diluted to 20 nM myosin (Fig. 4) in assay buffer containing 50 mM KCl, 10 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 1 mg/ml BSA, 2 mM MgATP, 0.2 mg/ml κ-casein, 70 µg/ml Cam1, 70 µg/ml Cdc4, 0.5% (wt/vol) Pluronic F68, 50 mM DTT, and oxygen scavengers and infused into the glass chamber for visualization. For conditions testing actin-Tpm tracks, 1 µM Tpm was included in the assay buffer.
For step-size measurements, Myo51–Rng8/9 or vertebrate MyoVa-HMM was mixed with a six- or tenfold molar excess of Qdots, respectively, in Motility buffer A, and the mixture was diluted to a final concentration of 0.04 nM myosin (dimer concentration for MyoVa-HMM) in assay buffer containing 50 mM KCl, 10 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 1 mg/ml BSA, 1 µM MgATP, and 100 µg/ml CamΔall (a calmodulin mutant lacking all calcium binding sites [Krementsov et al., 2004]) for MyoVa-HMM or 70 µg/ml Cam1 and 70 µg/ml Cdc4 for Myo51–Rng8/9, ATP-regenerating system (0.5 mM phosphoenolpyruvate and 100 U/ml pyruvate kinase), 50 mM DTT, and oxygen scavengers.
TIRF microscopy was performed at 25°C using an Eclipse Ti-U microscope (Nikon) with a 100× Plan-Apo objective lens (1.49 NA) with additional 1.5× magnification. Qdot 655 was excited at 473 nm and rhodamine-phalloidin–labeled actin or actin-Tpm was excited at 532 nm. Images were collected at 20–30 frames/s with 93 nm/pixel resolution using XR/Turbo-Z camera driven by Piper Control software. For assays with 2 mM ATP, Qdot movements were tracked manually using ImageJ with MtrackJ plugin. For step size measurement, a custom 2D-Gaussian fitting MATLAB (MathWorks) program was used to track Qdot movements, and a MATLAB Kerssemakers step-finding algorithm was used to calculate step size based on Qdot movement tracks (Kerssemakers et al., 2006).
Analytical ultracentrifugation
Sedimentation coefficients of Myo51 and Myo51–Rng8/9 complexes were determined using the Optima XL-1 analytical ultracentrifuge (Beckman Coulter) with the An-60 Ti rotor. Sedimentation velocity runs were performed at 40,000 rpm and 20°C in buffer containing 10 mM imidazole, pH 7.4, 300 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 1 mM DTT. Equilibrium runs were performed in the same buffer at 8,000 rpm and 4°C. The sedimentation coefficients were calculated using DCDT+ (v.2.4.3) software by John Philo, and molecular mass was calculated using Optima XL-A/XL-I data analysis software (v.6.03). Values were corrected for buffer composition and temperature using Sednterp software.
Online supplemental material
Fig. S1 shows the representative EM fields of Myo51 and Myo51–Rng8/9. Fig. S2 shows representative class average images of Myo51 in the presence of MgATP. Fig. S3 shows the step size of vertebrate MyoVa-HMM attached to a Qdot. Fig. S4 shows the EM images of Myo51or Myo51–Rng8/9 with actin or actin-Tpm in the presence of ATP. Video 1 shows Myo51 motor domain bends relative to lever arm with ATP (related to Fig. 2 and Fig. S2). Video 2 (related to Fig. 4) shows Qdots bound by multiple Myo51–Rng8/9 motors do not move on actin-Tpm tracks. Video 3 (related to Fig. 5) shows standard actin or actin-Tpm gliding motility by Myo51 or Myo51–Rng8/9. Videos 4 and 5 (related to Fig. 8) each show a single red actin-Tpm filament traveling along a green actin-Tpm bundle propelled by Myo51–Rng8/9. Videos 6–8 (related to Fig. 8) show multiple red actin-Tpm filaments gliding along the thickest areas of green actin-Tpm bundle by Myo51–Rng8/9 in dual-color motility assays. Video 9 shows a dual-color motility assay by Myo51 with actin or actin-Tpm and by Myo51–Rng8/9 with actin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201511102/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201511102.dv.
Supplementary Material
Acknowledgments
We thank Jian-Qiu Wu for sharing information on Rng8/9 during early stages of this project, David Warshaw and Guy Kennedy for the use of the TIRF microscope, Thomas Sladewski for help with single-molecule step size measurement, Luther Pollard for quantitative analysis of in vitro motility, and Susan Lowey and Enrique De La Cruz for helpful discussion. We thank the National Heart, Lung, and Blood Institute Electron Microscopy Core Facility for the use of facilities.
This work was supported by National Institutes of Health grant GM097193 to M. Lord and GM078097 to K.M. Trybus. Mass spectrometry protein identification of expressed protein bands was performed by the Vermont Genetics Network Core Facility, which is supported by National Institutes of Health Institutional Development Award P20GM103449.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Cdc8
- fission yeast tropomyosin
- myoVa-HMM
- a truncated constitutively active heavy meromyosin domain of vertebrate myosin Va
- Qdot
- quantum dot
- SCPR
- search, capture, pull, and release
- TIRF
- total internal reflection fluorescence
- Tpm
- tropomyosin
References
- Albanesi J.P., Coué M., Fujisaki H., and Korn E.D.. 1985. Effect of actin filament length and filament number concentration on the actin-activated ATPase activity of Acanthamoeba myosin I. J. Biol. Chem. 260:13276–13280. [PubMed] [Google Scholar]
- Ali M.Y., Previs S.B., Trybus K.M., Sweeney H.L., and Warshaw D.M.. 2013. Myosin VI has a one track mind versus myosin Va when moving on actin bundles or at an intersection. Traffic. 14:70–81. 10.1111/tra.12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R., and Mabuchi I.. 2002. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 115:887–898. [DOI] [PubMed] [Google Scholar]
- Baker K., Kirkham S., Halova L., Atkin J., Franz-Wachtel M., Cobley D., Krug K., Maček B., Mulvihill D.P., and Petersen J.. 2016. TOR complex 2 localises to the cytokinetic actomyosin ring and controls the fidelity of cytokinesis. J. Cell Sci.:jcs.190124 10.1242/jcs.190124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing W., Knott A., and Marston S.B.. 2000. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem. J. 350:693–699. 10.1042/bj3500693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.A., Walker M.L., Thirumurugan K., Trinick J., and Knight P.J.. 2004. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 147:247–258. 10.1016/j.jsb.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Clayton J.E., Pollard L.W., Sckolnick M., Bookwalter C.S., Hodges A.R., Trybus K.M., and Lord M.. 2014. Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol. Biol. Cell. 25:66–75. 10.1091/mbc.E13-04-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Sees J.A., Kovar D.R., and Wu J.Q.. 2013. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J. Cell Biol. 203:101–114. 10.1083/jcb.201305022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulton A.T., East D.A., Galinska-Rakoczy A., Lehman W., and Mulvihill D.P.. 2010. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J. Cell Sci. 123:3235–3243. 10.1242/jcs.069971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A., Martín-García R., Coulton A.T., Bagley S., and Mulvihill D.P.. 2009. Fission yeast Myo51 is a meiotic spindle pole body component with discrete roles during cell fusion and spore formation. J. Cell Sci. 122:4330–4340. 10.1242/jcs.055202 [DOI] [PubMed] [Google Scholar]
- Dudin O., Bendezú F.O., Groux R., Laroche T., Seitz A., and Martin S.G.. 2015. A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J. Cell Biol. 208:897–911. 10.1083/jcb.201411124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., and Toyoshima Y.Y.. 2008. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18:152–157. 10.1016/j.cub.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Hodges A.R., Bookwalter C.S., Krementsova E.B., and Trybus K.M.. 2009. A nonprocessive class V myosin drives cargo processively when a kinesin- related protein is a passenger. Curr. Biol. 19:2121–2125. 10.1016/j.cub.2009.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huang Y., Yu H., Subramanian D., Padmanabhan A., Thadani R., Tao Y., Tang X., Wedlich-Soldner R., and Balasubramanian M.K.. 2012. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J. Cell Biol. 199:831–847. 10.1083/jcb.201209044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A.L., Kim H., Srinivasan D., Lakonishok M., Larson A.G., and Gelfand V.I.. 2010. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc. Natl. Acad. Sci. USA. 107:12151–12156. 10.1073/pnas.1004736107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., and Hammer J.A. III. 1994. The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2). FEBS Lett. 342:197–202. 10.1016/0014-5793(94)80500-8 [DOI] [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., and Mabuchi I.. 2007. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 178:765–771. 10.1083/jcb.200612018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kerssemakers J.W.J., Munteanu E.L., Laan L., Noetzel T.L., Janson M.E., and Dogterom M.. 2006. Assembly dynamics of microtubules at molecular resolution. Nature. 442:709–712. 10.1038/nature04928 [DOI] [PubMed] [Google Scholar]
- Krementsov D.N., Krementsova E.B., and Trybus K.M.. 2004. Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 164:877–886. 10.1083/jcb.200310065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsova E.B., Hodges A.R., Bookwalter C.S., Sladewski T.E., Travaglia M., Sweeney H.L., and Trybus K.M.. 2011. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J. Cell Biol. 195:631–641. 10.1083/jcb.201106146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Imai Y., and Yamamoto M.. 2002. Tropomyosin is required for the cell fusion process during conjugation in fission yeast. Genes Cells. 7:375–384. 10.1046/j.1365-2443.2002.00526.x [DOI] [PubMed] [Google Scholar]
- Laplante C., Berro J., Karatekin E., Hernandez-Leyva A., Lee R., and Pollard T.D.. 2015. Three myosins contribute uniquely to the assembly and constriction of the fission yeast cytokinetic contractile ring. Curr. Biol. 25:1955–1965. 10.1016/j.cub.2015.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Les Erickson F., Corsa A.C., Dosé A.C., and Burnside B.. 2003. Localization of a class III myosin to filopodia tips in transfected HeLa cells requires an actin-binding site in its tail domain. Mol. Biol. Cell. 14:4173–4180. 10.1091/mbc.E02-10-0656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L., Chang F., and Martin S.G.. 2012. Myosin Vs organize actin cables in fission yeast. Mol. Biol. Cell. 23:4579–4591. 10.1091/mbc.E12-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T.J., Albanesi J.P., Korn E.D., Robinson E.A., Bowers B., and Fujisaki H.. 1986. ATPase activities and actin-binding properties of subfragments of Acanthamoeba myosin IA. J. Biol. Chem. 261:17156–17162. [PubMed] [Google Scholar]
- Mehta A.D., Rock R.S., Rief M., Spudich J.A., Mooseker M.S., and Cheney R.E.. 1999. Myosin-V is a processive actin-based motor. Nature. 400:590–593. 10.1038/23072 [DOI] [PubMed] [Google Scholar]
- Merritt R.C., Manor U., Salles F.T., Grati M., Dose A.C., Unrath W.C., Quintero O.A., Yengo C.M., and Kachar B.. 2012. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Curr. Biol. 22:320–325. 10.1016/j.cub.2011.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F., Arai R., and Mabuchi I.. 2001. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol. Biol. Cell. 12:1367–1380. 10.1091/mbc.12.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navone F., Niclas J., Hom-Booher N., Sparks L., Bernstein H.D., McCaffrey G., and Vale R.D.. 1992. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J. Cell Biol. 117:1263–1275. 10.1083/jcb.117.6.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., and Wu J.Q.. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155. 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M., Rock R.S., Mehta A.D., Mooseker M.S., Cheney R.E., and Spudich J.A.. 2000. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 97:9482–9486. 10.1073/pnas.97.17.9482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J., and Surrey T.. 2013. The multiple talents of kinesin-8. Nat. Cell Biol. 15:889–891. 10.1038/ncb2820 [DOI] [PubMed] [Google Scholar]
- Skoumpla K., Coulton A.T., Lehman W., Geeves M.A., and Mulvihill D.P.. 2007. Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 120:1635–1645. 10.1242/jcs.001115 [DOI] [PubMed] [Google Scholar]
- Sladewski T.E., Bookwalter C.S., Hong M.S., and Trybus K.M.. 2013. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat. Struct. Mol. Biol. 20:952–957. 10.1038/nsmb.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., and Watt S.. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- Stumpff J., Du Y., English C.A., Maliga Z., Wagenbach M., Asbury C.L., Wordeman L., and Ohi R.. 2011. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell. 43:764–775. 10.1016/j.molcel.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Qiu W., Gupta M.L. Jr., Pereira-Leal J.B., Reck-Peterson S.L., and Pellman D.. 2011. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell. 43:751–763. 10.1016/j.molcel.2011.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Arellano-Santoyo H., Portran D., Gaillard J., Vantard M., Thery M., and Pellman D.. 2013. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 15:948–957. 10.1038/ncb2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M., Numata O., and Nakano K.. 2015. An actin-myosin-II interaction is involved in maintaining the contractile ring in fission yeast. J. Cell Sci. 128:2903–2918. 10.1242/jcs.171264 [DOI] [PubMed] [Google Scholar]
- Urrutia R., McNiven M.A., Albanesi J.P., Murphy D.B., and Kachar B.. 1991. Purified kinesin promotes vesicle motility and induces active sliding between microtubules in vitro. Proc. Natl. Acad. Sci. USA. 88:6701–6705. 10.1073/pnas.88.15.6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.Q., Hao S., O’Shaughnessy B., and Pollard T.D.. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100. 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wang N., Lo Presti L., Zhu Y.-H., Kang M., Wu Z., Martin S.G., and Wu J.-Q.. 2014. The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J. Cell Biol. 205:357–375. 10.1083/jcb.201308146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw D.M., Kennedy G.G., Work S.S., Krementsova E.B., Beck S., and Trybus K.M.. 2005. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys. J. 88:L30–L32. 10.1529/biophysj.105.061903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T.Z., Gachet Y., Mulvihill D.P., May K.M., and Hyams J.S.. 2001. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114:69–79. [DOI] [PubMed] [Google Scholar]
- Wu J.Q., and Pollard T.D.. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314. 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.