Abstract
Mature leaves of plants transferred from low to high light typically increase their photosynthetic capacity. In A rabidopsis thaliana, this dynamic acclimation requires expression of GPT2, a glucose 6‐phosphate/phosphate translocator. Here, we examine the impact of GPT2 on leaf metabolism and photosynthesis. Plants of wild type and of a GPT2 knockout (gpt2.2) grown under low light achieved the same photosynthetic rate despite having different metabolic and transcriptomic strategies. Immediately upon transfer to high light, gpt2.2 plants showed a higher rate of photosynthesis than wild‐type plants (35%); however, over subsequent days, wild‐type plants acclimated photosynthetic capacity, increasing the photosynthesis rate by 100% after 7 d. Wild‐type plants accumulated more starch than gpt2.2 plants throughout acclimation. We suggest that GPT2 activity results in the net import of glucose 6‐phosphate from cytosol to chloroplast, increasing starch synthesis. There was clear acclimation of metabolism, with short‐term changes typically being reversed as plants acclimated. Distinct responses to light were observed in wild‐type and gpt2.2 leaves. Significantly higher levels of sugar phosphates were observed in gpt2.2. We suggest that GPT2 alters the distribution of metabolites between compartments and that this plays an essential role in allowing the cell to interpret environmental signals.
Keywords: high light, sugar signalling, photosynthesis
Short abstract
Acclimation to changes in the environment are crucial to optimising plant growth and productivity. In Arabidopsis thaliana, the glucose‐6‐phosphate/phosphate translocator GPT2 plays an important role in the acclimation to increased irradiance. Here we show that expression of GPT2 influences the partitioning of carbon between different storage pools, enhancing starch synthesis. Acclimation of metabolism occurs but is distinct in plants expressing or lacking GPT2.y
Introduction
Plants in nature experience fluctuations in environmental conditions occurring on timescales ranging from seconds to months. In the very short term, changes in light intensity can give rise to large fluctuations in the redox poise of photosynthetic electron transport and in the concentrations of primary metabolites, which are buffered by rapidly induced regulatory processes (Bailey et al. 2001; Wulff‐Zottele et al. 2010; Dietz & Pfannschmidt 2011). In the longer term, changes in weather patterns can give rise to sustained changes in ambient light (e.g. transition from cloudy to sunny weather). As sessile organisms, plants must respond biochemically to long‐term changes in conditions in order to maintain efficient levels of photosynthesis and avoid stress (Takahashi & Badger 2011).
Variations in light intensity directly affect photosynthesis by changing the amount of energy available for electron transport and carbon fixation. Photosynthetic acclimation allows plants to respond to changes in light intensity, optimizing the balance of light absorption and use, reducing susceptibility to stress (Walters 2005). When plants are subjected to increased light, acclimation can result in sustained increases in photosynthetic capacity (e.g. Murchie et al. 2002; Oguchi et al. 2003; Athanasiou et al. 2010).
During development, photosynthetic acclimation can be achieved by altering plant's morphology; for example, by altering leaf area and thickness and altering root/shoot ratios (Murchie & Horton 1997; Ballaré 1999; Weston et al. 2000; Walters 2005). In mature tissues by contrast, the responses of photosynthesis to changes in light intensity must be achieved dynamically, via changes in protein concentrations in pre‐existing cells (Leister 2003; Li et al. 2009). We previously demonstrated that developmental and dynamic acclimation are, to some extent, distinct processes in Arabidopsis thaliana (Athanasiou et al. 2010).
When a mature plant is exposed to increased light, a multitude of potential signals is generated, which must be sensed and interpreted in a way that gives rise to an increase in the capacity for photosynthesis (for a review, see Hausler et al., 2014). Factors known to change gene expression include signals from light receptors, the redox poise of cellular components and the concentrations of metabolites, in particular various sugars. The cell is capable of detecting this complex range of signals and distinguishing these from other perturbations that have comparable effects on some or all of the signals.
Under conditions where mature, fully expanded leaves are subjected to a sustained increase in light intensity, Arabidopsis plants are typically capable of substantially increasing their photosynthetic capacity (P max; Athanasiou et al. 2010). Arabidopsis accession Wassilewskija‐4 (Ws‐4) shows a strong dynamic acclimation of photosynthetic capacity, increasing P max by up to 40% when moved from a low to a higher irradiance and this ability to acclimate dynamically plays a major role in determining plant's fitness in naturally fluctuating environments (Athanasiou et al. 2010). Regulation of acclimation to high light (HL) has been suggested to involve redox signals from electron transport, changes in carbohydrate/nutrient status and hormone levels, and requires communication between the chloroplast, cytosol and nucleus (retrograde and anterograde signalling) (Hausler et al. 2014). The exact signalling mechanisms giving rise to these increases in P max remain unknown (for a review, see Walters 2005). We previously showed that a gene (At1g61800) encoding a glucose 6‐phosphate/phosphate translocator GPT2 is required for acclimation of mature leaves to an increase in light; in a T‐DNA insertion knockout of GPT2 (gpt2.2), increases in light intensity do not lead to substantial increases in P max following a fourfold increase in light from 100 to 400 μmol m−2 s−1 (Athanasiou et al. 2010). The non‐acclimating gpt2.2 line is therefore a useful tool to study the dynamic acclimation response in Arabidopsis. In addition, the commonly used accession Columbia (Col‐0) has a similar phenotype, in terms of acclimation to increased light, to the gpt2.2 mutant and to the corresponding mutant, gpt2.1, in a Col‐0 background.
There are 16 plastidic phosphate translocator genes found in Arabidopsis, six of which encode functional genes (Knappe et al. 2003; Fischer 2011). These function as antiporters, swapping inorganic phosphate and phosphorylated C3, C5 and C6 compounds across the plastid envelope in a strictly 1:1 exchange (Flügge 1999). Among the characterized translocators, the two GPT proteins are the least substrate specific, although glucose 6‐phosphate (G6P) is the favoured substrate (Kammerer et al. 1998; Eicks et al. 2002). In heterotrophic tissues, it has been proposed that GPT proteins (primarily GPT1) take G6P into plastids for incorporation into starch or to feed the oxidative pentose phosphate pathway (Niewiadomski et al. 2005; Kunz et al. 2010). GPT1 is essential for the growth and development of Arabidopsis; homozygous T‐DNA insertion knockouts of GPT1 are gametophyte lethal (Niewiadomski et al. 2005). GPT2 knockout plants are viable and show phenotypically normal growth under standard laboratory growth conditions. Only under variable growth conditions has a clear growth phenotype been demonstrated (Athanasiou et al. 2010).
Expression of GPT2 can be induced under a range of treatments and at various developmental stages, as well as in a number of mutants impaired in carbon metabolism or its regulation. In wild‐type plants, GPT2 expression is induced in imbibed seeds (Finch‐Savage et al. 2007), in response to exogenous and endogenous increases in sucrose (Lloyd & Zakhleniuk 2004; Gonzali et al. 2006), in response to an increase in light intensity (Athanasiou et al. 2010) and during senescence (Pourtau et al. 2006). Taken together, these results are consistent with GPT2 expression being induced in response to an accumulation of sugars in the cell.
To better elucidate processes occurring during acclimation to increased light and how GPT2 affects those processes, we have examined changes in metabolite levels in wild‐type and gpt2 plants, concentrating on the regulatory events occurring at an early stage in acclimation to increased light, when GPT2 expression is maximal. We show that metabolite pools in both wild‐type and gpt2 plants are altered in response to HL, even though these lines differ in their ability to increase photosynthetic capacity. The metabolic signatures of this acclimation differ between plants, implying that the responses observed are mechanistically different. We suggest that the partitioning of G6P between the chloroplast and the cytosol may provide a signal that allows the plant to sense and respond to environmental changes.
Materials and Methods
Plant growth and tissue preparation
Seeds of Arabidopsis thaliana accession Wassilewskija‐4 (wild‐type) and a homozygote T‐DNA insertion knockout of the GPT2 gene (gpt2.2, FLAG_326E03; INRA, Versailles, France) were used in all experiments. Selected measurements are also shown for Col‐0 and the gpt2.1 mutant. Plants for acclimation experiments were grown with an 8 h day/16 h night cycle for 8 weeks at an irradiance of 100 μmol m−2 s−1 white fluorescent light until mature. Plants were then transferred to an irradiance of 400 μmol m−2 s−1 [high light (HL)] or left at 100 μmol m−2 s−1 [low light (LL)] as a control. Both wild‐type and gpt2 seeds were generated at the same time from plants grown under our LL conditions throughout their life cycle. For photosynthetic measurements, the in situ rate of net CO2 fixation was measured on fully expanded leaves using a CIRAS 1 infrared gas analyser (PP Systems, Amesbury, MA, USA) under ambient light and CO2 conditions at the end of the photoperiod. A two‐way anova was performed on the data to determine significant differences (P < 0.05).
Microarray analysis
Microarray analyses were carried out as in Athanasiou et al. (2010). At each time point, a mature leaf was detached and flash‐frozen in liquid N2. RNA extraction was carried out using an RNeasy mini kit (Qiagen, Venlo, the Netherlands). Total RNA was used to produce cDNA, which was then hybridized to an Arabidopsis (Arabidopsis thaliana) ATH1‐121501 oligonucleotide array (Affymetrix, St Clara, CA, USA) according to the manufacturer's instructions. The arrays were read by an Agilent GeneArray scanner 3000 7G using Affymetrix GCOS (GeneChipOperating Software) V1.4. Quality control of the arrays was carried out to check for outliers using dChip software (Li & Wong 2001). The normalization and expression analysis was performed using robust multichip averaging (Bolstad et al. 2003). Differential expression was assessed with a modified t‐test on logarithmically scaled data using Cyber‐T (Baldi & Long 2001). Differentially expressed genes were identified using the criteria of Cyber‐T P‐values less than 0.01, mean expression level greater than 100 in at least one condition and mean fold change greater than 2. Gene annotation was derived from The Arabidopsis Information Resource (release 10.0; http://www.arabidopsis.org/). The Affymetrix chip analysis was performed at The Bioinformatics Core Facility (BCF) of the University of Manchester (Manchester, UK).
Fourier Transform Infrared (FTIR) spectroscopy
All chemicals, reagents and solvents used were purchased from Sigma‐Aldrich (St Louis, MO, USA). For FTIR spectroscopy analysis, fully expanded leaves were excised under growth light conditions using a razor blade, flash‐frozen and lyophilized overnight, after which the leaves were ground and transferred to −80 °C storage prior to extraction. Leaf tissue [30 mg dry weight (DW)] was homogenized in 1.5 mL of HPLC‐grade water, 15 μL of homogenate was directly loaded on to the wells of a silicon 96‐well IR target plate (Bruker, Coventry, UK) and dried.
Sample homogenates were loaded onto the IR target plate in duplicate and two readings were taken from each sample well to serve as analytical replicates. The plate was loaded onto a motorized high‐throughput stage (HTR‐XT; Bruker, Billerica, MA, USA) attached to a Bruker Equinox 55 FTIR and run in transmission mode according to Winder et al. (2004).
Gas chromatography electron impact time‐of‐flight mass spectrometry (GC‐EI‐TOF/MS) analysis
All chemicals, reagents and solvents used were purchased from Sigma‐Aldrich. Fully expanded leaves were excised using a razor blade, flash‐frozen and lyophilized overnight, ground and transferred to a −80 °C storage prior to extraction. Tissue aliquots (30 mg DW) were taken for the GC‐EI‐TOF/MS extraction procedure, which precisely followed that of Allwood et al. (2009) and Lisec et al. (2006). Polar phase extracts were dried using a centrifugal vacuum concentrator and spiked with 100 μL of an internal standard solution containing 200 μg mL−1 succinic‐d4 acid, glycine‐d5 and malonic‐d2 acid before analysis. Quality control samples were prepared by mixing equal volumes of the upper polar phase of all of the sample extracts, and these were analysed throughout the gas chromatography–mass spectrometry (GC‐MS) analyses so that analytical reproducibility could be assessed, as detailed in Dunn et al. (2011).
Because many of the metabolites within central metabolism are non‐volatile, a two‐step chemical derivatization was applied following the method detailed in Allwood et al. (2009). Samples were analysed within a single randomized analytical block. Quality control samples were interspersed throughout the block, permitting the assessment of instrument stability and removal of irreproducible metabolite features. Analyses were carried out with a Leco Pegasus III (4D) GC × GC/MS in GC/MS mode (Leco Corp., St. Joseph, MO, USA), with a Gerstel MPS‐2 autosampler (Gerstel, Baltimore, MD, USA) and an Agilent 6890N gas chromatograph (Agilent Technologies, Santa Clara, CA, USA), according to the method of Begley et al. (2009).
Raw GC‐EI‐TOF/MS data were baseline‐corrected, aligned and subjected to deconvolution to produce an XY matrix suitable for statistical analyses, following the method of Begley et al. (2009). Peak identities were putatively assigned on the basis of mass spectral similarity to NIST library and Golm Metabolome Database entries (Kopka et al. 2005) or were unambiguously assigned by mass spectral and retention index matching with an in‐house library (Begley et al. 2009) from the Manchester Metabolomics Database (Brown et al. 2009). Metabolite identifications were considered unambiguous when they had a match score of >800 and retention index within 20 units.
Statistical analysis of FTIR and GC‐MS data
The calculated FTIR absorbance spectra were exported directly into Matlab R2008 (The MathWorks Inc., Natwick, MA, USA). Pre‐processing of the FTIR data involved five‐point smoothing by the Savitzky–Golay algorithm (Savitzky & Golay 1964). For GC‐MS, after deconvolution, the metabolite and metabolite level pairs were imported into Matlab. Post pre‐processing, the FTIR or GC‐MS‐derived XY outputs were independently examined using principal component analysis (PCA) as described in Allwood et al. (2006). Further, based upon the PCs generated by PCA, a supervised multivariate technique known as PC‐discriminant function analysis (PC‐DFA) was used to discriminate between the experimental groups based on an a priori knowledge of experimental class structure (Goodacre et al. 1998, 2004). PC‐DFA were generated and validated following the methods of Kaderbhai et al. (2003) and Allwood et al. (2006). N‐way anova was also performed to identify variables that were significant at a 99.9% confidence limit.
Anion‐exchange liquid chromatography–tandem mass spectrometry (LC‐MS/MS)
Mature leaf tissue was excised 8, 56 or 104 h after first exposure to HL. Control (LL) samples were excised after 8 h. Tissue was flash‐frozen in liquid N2 and stored on dry ice until used. Frozen tissue powder was extracted using chloroform–methanol as described in Lunn et al. (2006). Phosphorylated intermediates and organic acids were quantified by high‐performance anion‐exchange chromatography coupled to tandem mass spectrometry, operating in negative ion mode, as described in Lunn et al. (2006). Metabolites were quantified by comparison against authentic standards. Measurements of T6P were corrected for ion suppression using a deuterated‐T6P internal standard. Technical replicates were averaged and a two‐way anova was performed on the data (P < 0.05) using SPSS (IBM, Armonk, NY, USA).
Sucrose and starch analyses
Enzymatic assays were carried out on extracts from three fully expanded leaves from three separate plants. Tissue was excised and flash‐frozen in liquid nitrogen every 2–4 h throughout the photoperiod and stored at −80 °C before analysis. Starch was estimated using a total starch assay (Megazyme, Wicklow, Eire, Method E). Sucrose measurements were collected using a sucrose assay kit (Sigma, Poole, UK). Three biological replicates were assayed at each time point and each measurement was replicated three times. Technical replicates were averaged and a two‐way anova was performed on the data (P < 0.05) using SPSS (IBM).
Results
Loss of acclimation results in a loss of photosynthesis under growth conditions
Previously, we showed that the photosynthetic capacity of wild‐type Ws‐4 plants increases significantly (∼40%) over a 7‐day‐acclimation period following a transfer from 100 (LL) to 400 (HL) μmol m−2 s−1 irradiance, and that this response requires GPT2 (Athanasiou et al. 2010). In order to determine the impact of this on plant's performance under actual growth conditions, measurements of photosynthesis and respiration were made on intact plants in growth chambers at ambient light and CO2 (Fig. 1). At LL, there was no significant difference in the rate of CO2 assimilation between wild‐type and gpt2.2 plants. Higher irradiance resulted in an immediate increase in the rate of photosynthesis (Fig. 1a) in both wild‐type and gpt2.2 plants; however, the proportional increase was smaller than the proportional increase in irradiance, indicating a degree of saturation of photosynthesis. This initial increase was greater in gpt2.2 than it was in wild type, reflecting the fact that there is a higher P max in these plants when grown in LL (Athanasiou et al. 2010). Over subsequent days, however, photosynthetic rates increased significantly in wild‐type plants, whereas no further increase was observed in gpt2.2 plants. This change reflected the differences in P max observed over a 7‐day‐acclimation period in the different plants (Athanasiou et al. 2010). Respiration also increased (Fig. 1b), with significantly higher rates observed in both wild‐type and gpt2.2 plants by the end of the first day. This increase was sustained over subsequent days, with no significant difference between the two lines. Measurements of in situ gas exchange were also performed on plants of Col‐0 and of gpt2.1, a GPT2 insertion mutant in the Col‐0 background (Supporting Information Fig. S1). As expected from previous measurements (Athanasiou et al. 2010), neither of these lines were able to acclimate their photosynthetic capacity in response to an increase in irradiance in mature leaves. Consistent with this, the actual rate of photosynthesis under growth conditions also did not increase following an increase in growth irradiance. No significant differences were observed in gas exchange parameters between Col‐0 and gpt2.1. Based upon the above results, we conclude that net carbon fixation is greater at HL, compared to LL, in both wild‐type and gpt2 plants but that the lines differ from one another as acclimation proceeds.
Figure 1.
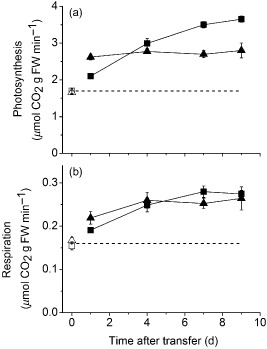
Photosynthetic rates and respiration during acclimation. (a) Photosynthetic rate at the end of the photoperiod measured on attached leaves of plants within growth cabinets. (b) Dark respiration measured at the end of the photoperiod within growth cabinets. Plants of Ws‐4 wild‐type (squares) and gpt2.2 (triangles) lines were grown at low light (LL, 100 μmol m−2 s−1 white light, 8 h light, 20 °C/16 h dark, 18 °C) for 8 weeks. They were then transferred to high light (HL, 400 μmol m−2 s−1 white light, 8 h light, 20 °C/16 h dark, 18 °C; closed symbols) or maintained at LL (open symbols). LL measurements were made at the beginning of the acclimation period for comparison. Points represent the mean of at least three replicates from three separate plants. Error bars represent ±1 SE. Significant differences between samples were determined using a two‐way anova (P < 0.05, see the Results section for details).
Gene expression patterns in response to HL differ in wild‐type and gpt2.2 plants
To examine the impact of loss of GPT2 on overall gene expression, global transcriptome analysis was performed on leaf tissue at LL and after 4 h of HL from gpt2.2 and Ws‐4 wild‐type plants. A comparison of the two lines at LL showed significant differences in the transcript levels for a number of genes, indicating that the low level of GPT2 expression observed under LL (i.e. non‐stressed) conditions still has an impact on transcription.
Using MapMan transcriptomic software, we compared transcription in the two lines under LL conditions. A number of metabolic bins (Thimm et al. 2004) were highlighted as showing large transcript level differences (Supporting Information Fig. S2) including trehalose metabolism (with TPS‐8, TPS‐10 and TPS‐11 overexpressed in LL gpt2.2 plants compared with LL wild type) and lipid metabolism (with genes overexpressed in gpt2.2 plants). Bins containing genes for raffinose, starch and sucrose metabolism also showed differences, with genes both up‐ and down‐regulated in these categories.
Notably, there were striking differences in genes associated with photosynthetic light reactions; after applying stringent criteria to gene expression levels (P < 0.01, fold change above 1.5 and expression levels over 100 in at least one condition, Table 1) 26 out of 111 genes (23%) were significantly different between the two lines, the vast majority (24) of these being greater in gpt2.2 plants. Of these overexpressed light‐reaction genes, 17 are chloroplast‐encoded, which is representative of a general trend in gpt2.2 plants; 36 of 77 chloroplast‐encoded genes identified by MapMan software (47%) are significantly higher in gpt2.2 plants under LL control conditions, with only one being significantly lower (Supporting Information Table S1).
Table 1.
Fold changes in gene expression
| ID | Gene ID | Description | gpt2.2 LL versus Ws‐4 LL | gpt2.2 HL versus gpt2.2 LL | Ws‐4 HL versus Ws‐4 LL |
|---|---|---|---|---|---|
| 244977_at | atcg00730 | PETD, subunit of cytochrome b6/f | 9.147 | 1.226 | 1.448 |
| 245007_at | atcg00350 | PSAA, PSI reaction centre | 8.773 | −1.05 | 2.282 |
| 245047_at | atcg00020 | PSBA, chlorophyll binding protein D1 | 6.765 | 1.125 | 1.045 |
| 244932_at | atcg01060 | PSAC, subunit of PS I | 4.241 | 1.035 | 1.480 |
| 245002_at | atcg00270 | PSBD, PSII D2 protein | 3.77 | 1.294 | 1.495 |
| 244973_at | atcg00690 | PSBT, PS II 5 kD protein subunit PSII‐T. | 3.002 | 1.212 | 1.194 |
| 244972_at | atcg00680 | PSBB, CP47, subunit of PSII | 2.712 | 1.46 | 1.127 |
| 259970_at | at1g76570 | chlorophyll A‐B binding family protein | 2.404 | 1.178 | 1.949 |
| 245021_at | atcg00550 | PSBJ, PSII component | 2.317 | 1.05 | −1.039 |
| 260481_at | at1g10960 | ATFD1, ferredoxin 1 | 2.308 | −1.08 | 1.723 |
| 244964_at | atcg00580 | PSBE, PSII cytochrome b559. | 2.195 | 1.221 | −1.05 |
| 244995_at | atcg00150 | ATPI, subunit of ATPase complex CF0 | 2.028 | −1.484 | 1.191 |
| 245025_at | atcg00130 | ATPF, ATPase F subunit. | 1.843 | −1.15 | −1.045 |
| 253790_at | at4g28660 | PSB28, PS II reaction center | 1.791 | −1.077 | 1.875 |
| 265722_at | at2g40100 | LHCB4.3, light harvesting complex PSII | 1.781 | −1.357 | 1.736 |
| 245026_at | atcg00140 | ATPH, ATPase III subunit | 1.682 | −1.019 | 1.129 |
| 244975_at | atcg00710 | PSBH, 8kD component of PSII | 1.649 | 1.138 | 1.258 |
| 256015_at | at1g19150 | LHCA6 | 1.611 | −1.801 | −1.451 |
| 244963_at | atcg00570 | PSBF, PSII cytochrome b559 | 1.565 | 1.137 | −1.033 |
| 251701_at | at3g56650 | thylakoid lumenal 20 kDa protein | 1.559 | −1.592 | −1.317 |
| 254398_at | at4g21280 | PSBQ, oxygen‐evolving enhancer protein | 1.538 | 1.368 | −1.297 |
| 264959_at | at1g77090 | thylakoid lumenal 29.8 kDa protein | 1.531 | −1.081 | 1.204 |
| 245003_at | atcg00280 | PSBC, CP43 subunit of PSII | 1.521 | −1.036 | −1.011 |
| 258993_at | at3g08940 | LHCB4.2, light harvesting complex PSII | 1.52 | −2.479 | −3.386 |
| 245368_at | at4g15510 | PSII reaction center PsbP family protein | 1.496 | −1.908 | −1.547 |
| 258239_at | at3g27690 | LHCB2.4, light harvesting complex PSII | 1.403 | −2.607 | −2.886 |
| 259491_at | at1g15820 | LHCB6, CP24 | 1.36 | −1.329 | −1.64 |
| 251784_at | at3g55330 | PPL1, PsbP‐like protein | 1.312 | −1.706 | −1.234 |
| 267569_at | at2g30790 | PSBP‐2, PS II subunit P‐2 | 1.131 | −1.565 | −1.371 |
| 261388_at | at1g05385 | PS II 11 kDa protein‐related | 1.082 | −1.64 | −1.363 |
| 263114_at | at1g03130 | PSAD‐2 , PS I subunit D‐2 | 1.067 | −2.29 | −2.837 |
| 244966_at | atcg00600 | PETG, Cytochrome b6‐f subunit V | 1.054 | −4.754 | 1.266 |
| 261769_at | at1g76100 | PETE1, plastocyanin 1 | −1.022 | −1.805 | −2.485 |
| 244974_at | atcg00700 | PSBN, PSII low MW protein | −1.1 | −1.607 | 1.503 |
| 252130_at | at3g50820 | PSBO2, PSII subunit O‐2 | −1.232 | −1.572 | −1.66 |
| 265149_at | at1g51400 | PS II 5 kD protein | −1.272 | −3.062 | −2.758 |
| 245000_at | atcg00210 | YCF6 | −1.556 | 1.856 | 1.059 |
| 245024_at | atcg00120 | ATPA, ATPase alpha subunit | −1.584 | −1.563 | −1.062 |
Genes associated with the light reactions were mined using MapMan software. Of 111 genes, those which changed significantly between Ws‐4 and gpt2.2 at LL, or in either line during HL acclimation, are shown here. Significantly changed genes must show a fold change of more than 1.5‐fold.
HL, high light; LL, low light.
In contrast, when comparing LL and HL plants using MapMan software, changes in genes linked to the photosynthetic light reactions were conspicuously absent (Supporting Information Figs S3 & S4). Despite there being a significant increase in both P max (Athanasiou et al. 2010) and photosynthetic rate under growth conditions (Fig. 1), only 14 of 111 genes (12%; Table 1; Supporting Information Table S1) associated with the light reactions were significantly altered in wild‐type plants in response to HL. In gpt2.2 plants, only 16 of these genes are significantly changed. During developmental acclimation to HL, gene expression is regulated in response to changes in the photosynthetic pathway (Pfannschmidt 2003). However, previous work has shown a lack of correlation between photosynthesis and gene expression at different intensities of steady‐state light (Walters 2005; Piippo et al. 2006). In this experiment, photosynthetic gene expression does not change substantially after transfer to HL in either wild‐type or gpt2.2 plants. This suggests that changes in transcription levels of genes encoding photosynthetic proteins are not necessary to increase photosynthetic capacity in response to increased light in the range examined here.
In wild‐type plants, changes are evident in MapMan bins for cell wall synthesis, modification and degradation, starch metabolism and trehalose metabolism upon transfer to HL (Supporting Information Fig. S3). There were fewer changes in gpt2.2 plants, with fewer genes in the cell wall synthesis, cell wall modification and trehalose metabolism bins showing significant changes (Supporting Information Fig. S4). We observe fewer significant changes in gene expression in gpt2.2 plants versus wild‐type plants (421 versus 666). There are 816 genes that show a significant change in either or both lines. Of these, 33% change similarly in both wild‐type and gpt2.2 plants, highlighting a conserved response to increased light intensity (see Supporting Information Table S1 for genes differentially expressed between lines). However, 395 (48%) of these 816 significantly altered genes are changed in wild‐type plants only, showing that there is a clear response in the acclimating wild‐type line that is not observed in gpt2.2 plants.
Sucrose accumulation is independent of GPT2 expression during acclimation to HL, but starch accumulation is significantly lower in gpt2.2 plants
Given the increase in the net rate of photosynthesis observed in response to HL (Fig. 1), changes in the rate of flux of carbon compounds into different storage pools were to be expected. There are a number of temporary leaf carbon stores, the most prominent being sucrose and starch, both of which are synthesized from triose phosphates produced during photosynthesis. Levels of starch and sucrose were measured in Ws‐4 wild‐type and gpt2.2 leaves through the first and third photoperiods after transfer to HL. Both starch and sucrose levels within leaf tissue exhibit a diurnal cycle of accumulation during the day and degradation at night under both LL and HL conditions (Fig. 2 and data not shown).
Figure 2.
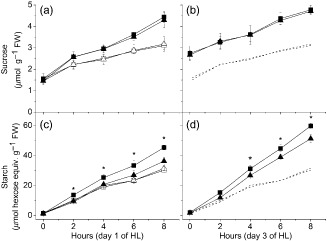
Sucrose and starch accumulation during acclimation. Plants of Ws‐4 wild type (squares) and gpt2‐2 (triangles) were grown at low light (LL, 100 μmol m−2 s−1 white light, 8 h light, 20 °C/16 h dark, 18 °C) for 8 weeks. They were then moved from low to high light (HL, 400 μmol m−2 s−1 white light, 8 h light, 20 °C/16 h dark, 18 °C; closed symbols) at day 0 or maintained at LL (open symbols). (a) Sucrose content measured in Ws‐4 wild‐type and gpt2.2 plants on the first day after transfer to HL. (b) Sucrose content measured in Ws‐4 wild‐type and gpt2.2 plants on the third day after transfer to HL. (c) Levels of starch measured in Ws‐4 wild‐type and gpt2.2 plants on the first day after transfer to HL. (d) Levels of starch measured during acclimation in Ws‐4 wild‐type and gpt2.2 plants on the third day after transfer to HL. Points represent the mean of at least five biological replicates. Error bars represent ±1 SE. Significant differences between samples were determined using a two‐way anova. Asterisks denote where levels in Ws‐4 wild‐type and gpt2.2 plants under HL treatment differ significantly (P < 0.05).
Accelerated net accumulation of sucrose occurred in both lines on the first day of HL treatment, with leaf sucrose content being significantly increased by 2 h of exposure to HL compared with LL, and this higher rate of sucrose accumulation was maintained throughout the photoperiod (Fig. 2a). Overnight, this sucrose pool was not completely lost from the leaf, so that at dawn on day 3, there was significantly more sucrose in HL‐grown plants than in those maintained at LL (Fig. 2b). Unlike on day 1 of HL, the rate of accumulation of sucrose through the photoperiod was similar in both HL and LL conditions on day 3. Notably, no differences were observed between sucrose levels in wild‐type and gpt2.2 plants, either in the rate of accumulation in the light or of the nighttime loss of sucrose from the leaf, in spite of the differing rates of photosynthesis in these plants. This suggests that rates of synthesis and export of sucrose are carefully regulated such that the leaf pools are not affected in response to increased irradiance, except on the first day, when there is a net accumulation of an additional sucrose pool, which is then maintained on subsequent days.
In contrast to sucrose, there were significant differences in the pattern of starch accumulation between Ws‐4 wild‐type and gpt2.2 plants (Fig. 2c,d). In wild‐type plants, there was a rapidly induced increase in the rate of starch accumulation after transfer to HL, with a significant rise in starch already being apparent after 2 h of HL and a faster rate of starch accumulation being maintained throughout the photoperiod (Fig. 2c). In contrast, in gpt2.2 plants, there was no detectable increase in starch accumulation in response to HL in the early part of the photoperiod (0–4 h) with significant differences only occurring towards the end of the day. This was in spite of the higher rate of photosynthesis in gpt2.2 plants in the first photoperiod of HL (Fig. 1).
On day 3, the rate of starch accumulation was higher in HL than on day 1, in both wild‐type and gpt2.2 leaves, implying tha an acclimation of starch synthesis had occurred. However, wild‐type plants accumulated significantly higher amounts of starch than gpt2.2 throughout the HL photoperiod.
Global metabolite analyses show distinct acclimation of metabolism in wild‐type and gpt2 plants
By the end of the first photoperiod of HL treatment, gpt2.2 plants accumulated similar levels of sucrose as that of Ws‐4 wild type but significantly lower levels of starch (Fig. 2), despite showing a higher rate of photosynthesis on that day (Fig. 1). This suggests that plants without GPT2 might be diverting carbon into other pools, possibly including lipids, proteins and cell walls. In order to elucidate processes occurring during HL acclimation and to examine the consequences of these differences, global changes in metabolites were examined in both wild‐type and non‐acclimating gpt2.2 lines. FTIR spectroscopy was used for generating metabolic fingerprints and GC‐MS for non‐targeted metabolite profiling.
Fully expanded leaves were collected at the end of the first and third photoperiods from HL‐treated plants and from controls maintained at LL. PCA was performed on the FTIR data, and PC‐DFA, a supervised multivariate technique, was applied to the principal components to identify the responses of each plant type to HL (Fig. 3a). Data from wild‐type and gpt2.2 LL samples were weakly discriminated by PCA but diverge when subjected to HL treatment (data not shown). In wild‐type plants, the metabolic changes are substantial, and the variance between the days of the HL treatment can be mostly accounted for by PC‐DF loading 1. The changes in gpt2.2 plants are smaller, with the variance accounted for by both PC‐DF loadings 1 and 2. The most substantial changes in metabolites occur on the first day of acclimation in both Ws‐4 wild‐type and gpt2.2 plants, presumably reflecting the direct effects of increased carbon fixation. On day 3 of the HL treatment, Ws‐4 wild‐type and gpt2.2 samples tended to converge on the PC‐DFA plot, suggesting that the main metabolic differences between the lines occur in the early stages of acclimation (Fig. 3a). In terms of the variance between LL controls and day 1 samples, changes in wild‐type metabolism are mainly accounted for by PC‐DF1, and changes in gpt2.2 plants mainly by PC‐DF2, which suggests that there were larger metabolic changes in Ws‐4 wild‐type than gpt2.2. PC‐DF loading plots allow us to examine which types of phenotypic changes underlie the variance between PC‐DFAs and therefore broadly between the two lines on the first day of acclimation (Fig. 3b–d). Loadings for both PC‐DFAs show changes in the fatty acid and polysaccharide regions of the IR spectrum, with substantial changes in the amide region. This suggests significant changes in whole leaf metabolism occurring in both wild‐type and non‐acclimating gpt2.2 plants, with the responses being different in the two lines.
Figure 3.
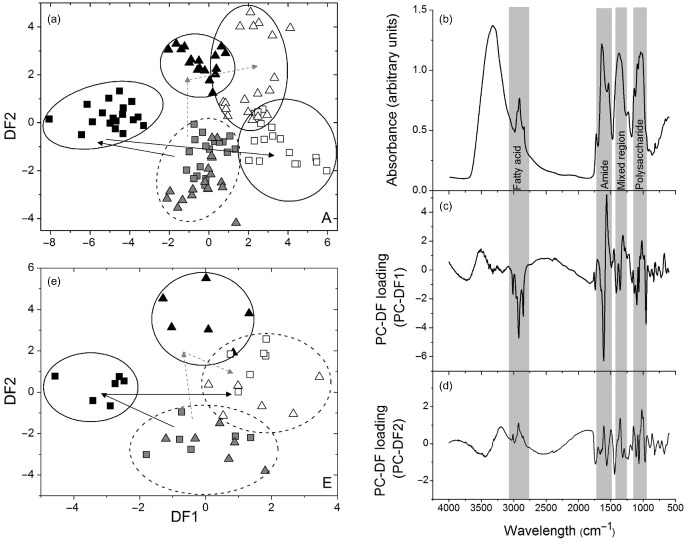
Fourier transform infrared (FTIR) and gas chromatography–mass spectrometry (GC‐MS) analysis of changes in metabolism during acclimation to high light (HL). (a) Principal component‐discriminant function analysis (PC‐DFA) model of FTIR spectra of extracts from Ws‐4 wild‐type and gpt2.2 lines in the first 3 d after transfer to HL conditions. Circles and arrows are indicative of clustering only and have no statistical significance. Plants were grown for 8 weeks at low light (LL, 100 μmol m −2 s−1) and then transferred to HL (400 μmol m−2 s−1) for up to 3 d. Ten PCs were used in the calculation, accounting for 99.1% of the total variance. PC1 explained 56.2% of the total variance, with PC2 explaining a further 24% of the total variance. Grey symbols represent plants at the end of day in LL, black symbols represent plants at the end of 1 photoperiod of HL. Open symbols represent plants at the end of 3 d of HL treatment. (b) A representative FTIR spectrum of an extract from Ws‐4 wild‐type plants grown under LL conditions. (c–d) PC‐DF loadings highlighting parts of the FTIR spectrum where a chemical functional group exhibits a particular absorbance important for the PC‐DFA model. PC‐DFA loading vectors are plotted against wavenumbers (cm−1) for (c) PC‐DF1 and (d) PC‐DF2. (e) PC‐DFA model constructed from the GC‐MS data from extracts from Ws‐4 wild‐type and gpt2.2 lines in the first 3 d after transfer to HL conditions. PC1 accounted for 65% of the total variance, with PC2 accounting for a further 28%. Samples were collected at the end of the photoperiod in plants maintained at LL (grey symbols) and transferred to HL for 1 d (black symbols) and 3 d (open symbols).
PC‐DFA was also carried out on data acquired by GC‐MS. GC‐MS was used to identify 85 polar compounds, with 39 of these being of unknown chemical structure (for full data set, see Supporting Information Table S2). All of the GC‐MS data were used in the PC‐DFA (Fig. 3e). As for the FTIR, there was poor separation of data for LL‐grown plants. When plants were exposed to HL for 1 photoperiod, a clear separation of wild‐type and gpt2.2 plants could be observed, with PC‐DF1 explaining most of the change in wild‐type and PC‐DF2 being responsible for the differences in the mutant. By the third day of acclimation, a convergence of the data was observed, with the wild‐type and gpt2.2 plants no longer being well discriminated. Both the FTIR and the GC‐MS data sets are highly reproducible between technical, analytical and biological replicates within the same experimental classes.
FTIR and GC‐MS analysis were also performed on plants of Col‐0 wild‐type and gpt2.1 (Supporting Information Fig. S5). In contrast to the results comparing Ws‐4 and gpt2.1, no clear discrimination was observed between Col‐0 and the corresponding mutant. This is consistent with the data from other groups on the gpt2.1 mutant and indicates that the major changes in metabolism separating Ws‐4 from gpt2.2 are likely a consequence of different acclimation process occurring in those plants rather than a direct consequence of GPT2 expression.
Targeted analysis of metabolites involved in starch and sucrose synthesis
Given that GPT2 is a G6P translocator, it is likely that expression of this protein will have a direct impact on the pools of metabolites involved in the pathways of sucrose and starch biosynthesis. To quantify these metabolites, targeted LC‐MS/MS was carried out on leaf samples collected at the end of the first, third and fifth photoperiods from plants exposed to HL and from controls maintained at LL. Twenty metabolites were identified by LC‐MS/MS on the first day of acclimation (Table 1; for the full LC‐MS/MS data set, see Supporting Information Table S3).
In plants maintained at LL, the majority of measured metabolites did not differ significantly between Ws‐4 wild‐type and gpt2.2 plants. A notable exception to this was trehalose 6‐phosphate (T6P), with gpt2.2 plants having over twice as much as wild type. T6P has previously been suggested to play a role in regulating photosynthetic capacity (Delatte et al. 2011). The concentration of shikimate was also found to be substantially higher in gpt2.2 leaves than in Ws‐4 wild type under all treatments, while the concentration of succinate was lower.
When plants were transferred to HL, two major responses could be observed among the identified metabolites. Metabolites involved in flux from photosynthesis to either starch or sucrose typically increased upon first transfer to HL, but then decreased again as acclimation proceeded. This pattern of behaviour was notably observed for phosphoenolpyruvate (PEP). Meanwhile, pyruvate and various intermediates of the TCA cycle exhibited a drop in concentration, which was then reversed as plants acclimated (for an overview, see Fig. 5). These two broad responses were observed in both Ws‐4 wild‐type and gpt2.2 plants. Although the responses were qualitatively similar, there were quantitative differences between the lines, with individual metabolites showing exaggerated responses in the mutant (tending to increase further and then decrease more).
Figure 5.

Summary of carbon metabolism changes occurring during acclimation to high light (HL). Increases in light result in rapid changes (within the first photoperiod) in concentrations of primary metabolites within the cell. Starch and sucrose levels increase rapidly (Fig. 3), as do a number of sugar phosphates and nucleotide sugars involved in their synthesis (Fig. 5). Triose phosphate is exported from the chloroplast primarily by the triose phosphate translocator (TPT) and can be channelled into sucrose synthesis via a number of intermediates, including G6P. GPT2 facilitates the movement of glucose 6‐phosphate (G6P) across the chloroplast envelope. It is proposed that the net direction of flux is into the chloroplast. G6P is a substrate for starch synthesis, accounting for the increased starch accumulation observed in wild‐type plants. Metabolites in red increase on day 1 of HL treatment, metabolites in blue decrease, relative to the LL control. Metabolites in black do not change. Metabolites in grey were not measured. In general, metabolites in red increase on day 1 and return to near control levels by day 7 in both Ws‐4 wild‐type and gpt2.2 plants. Metabolites in italics show a different pattern in wild‐type and gpt2.2 plants, and are further discussed in the text.
There were however a number of notable differences between the plants. Seven metabolites directly involved in starch and/or sucrose metabolism were identified [glucose 1‐phosphate (G1P), glucose 6‐phosphate (G6P), fructose 1, 6‐bisphosphate (F16BP), fructose 6‐phosphate (F6P), sucrose 6′‐phosphate (S6P), ADP‐glucose and UDP glucose], in addition to T6P, which is an important sugar signalling metabolite (Lunn et al. 2006). In general, concentrations of these significantly and substantially increased in both Ws‐4 wild‐type and gpt2.2 plants by the end of the first day of HL treatment; only levels of glucose 1‐phosphate (G1P) and UDP‐glucose in wild type did not change significantly. In addition, on the first day of HL treatment, levels of all eight of these metabolites were higher in the gpt2.2 plants than in Ws‐4 wild type, significantly so for all except G1P and S6P (Fig. 4; Table 1). Over longer time periods, the majority of identified sugar phosphates/nucleotide sugars showed a similar pattern in their response to increased light, with increases followed by a decline towards the end of the acclimation period. G6P levels remained significantly elevated relative to LL levels after 5 d HL, with levels in both Ws‐4 wild‐type and gpt2.2 plants being double as those observed in control plants after 5 d at HL (Fig. 4c).
Figure 4.
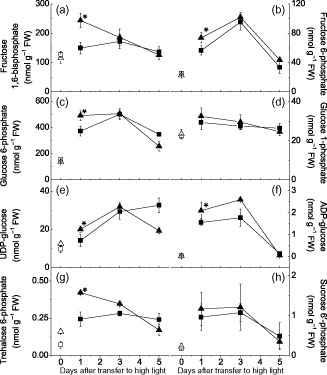
Sugar phosphate and sugar nucleotide levels during acclimation. Sugar phosphate and sugar nucleotide levels were measured using anion‐exchange liquid chromatography–tandem mass spectrometry (LC‐MS/MS) on tissue from plants at low light (LL) or transferred to high light (HL) or up to 5 d. Sugar phosphate/nucleotide levels were determined in plants grown for 8 weeks at LL (100 μmol m−2 s−1, 8 h light, 20 °C/16 h dark, 18 °C) or grown at LL and then transferred to HL (400 μmol m−2 s−1, 8 h light, 20 °C/16 h dark, 18 °C). (a) fructose 1,6‐bisphosphate, (b) fructose 6‐phosphate, (c) glucose 6‐phosphate, (d) glucose 1‐phosphate, (e) UDP‐glucose, (f) ADP‐glucose, (g) trehalose 6‐phosphate and (h) sucrose 6′‐phosphate. Points represent the mean of three to five biological replicates. Error bars represent ±1 SE. Asterisks indicate data points where wild‐type and gpt2.2 plants differ significantly (based on a two‐way anova, P < 0.05).
T6P levels are already significantly different between the two lines under LL control conditions; gpt2.2 leaves had a significantly higher T6P content than wild type (Fig. 4g). T6P levels rose, relative to the LL controls, on the first day of HL in both plants; however, this effect was much more marked in gpt2.2. In the mutant line, T6P levels fell sharply through the acclimation period, falling to LL levels by day 5. In contrast, concentrations in wild type were substantially higher throughout the experiment in HL leaves and remained higher than LL controls at the end of the experiment. In contrast to most sugar phosphates, UDP‐glucose levels rose throughout the HL treatment. In gpt2.2, however, UDP‐glucose concentration fell upon longer exposure of HL, although it remained above the LL level at the end of the experiment. Qualitatively, the responses of PEP were similar to those of UDP‐glucose, with an elevated concentration being observed in the wild type but not in gpt2.2 at the end of the acclimation period.
Discussion
Previously, we demonstrated that the ability of plants to acclimate their photosynthetic apparatus to sustained changes in light has a substantial role in determining yield in naturally fluctuating conditions (Athanasiou et al. 2010). Here, we have shown that plants able to acclimate to higher light are also able to achieve a significantly higher rate of photosynthesis following acclimation (Fig. 1, Supporting Information Fig. S1). In addition, our results show that acclimation to light involves major changes in metabolism, which we suggest allow plants to buffer changes in their environment; increased irradiance results in a transiently altered metabolome; however, the tendency is for metabolites to tend towards starting levels after acclimation.
In plants grown at LL, there is little difference observed between Ws‐4 wild type and gpt2.2 in terms of their physiology, with LL‐grown plants having similar growth, seed yield, photosynthetic/respiration rates and morphology (Athanasiou et al. 2010; Fig. 1). However, there are clear differences between the lines in terms of their transcriptomic profiles accompanied by weaker differences in FTIR analyses (Fig. 3), and GC‐MS and LC‐MS (Figs 3, 4). This discrimination is perhaps not surprising given that GPT2 expression is observed throughout plant development. This suggests that a lack of GPT2 expression has effects on transcription under control conditions despite gpt2.2 plants having no obvious phenotype. Notably, there are differences in photosynthetic gene expression in gpt2.2 plants, with higher transcript levels for genes linked to the light reactions and for most chloroplast‐encoded genes. gpt2.2 plants have a slightly higher photosynthetic capacity at LL (Athanasiou et al. 2010) but nevertheless achieve the same photosynthetic rates at growth irradiance (Fig. 1). gpt2.2 plants therefore have a different transcriptional strategy to achieve similar photosynthetic results. This points to a striking degree of homeostasis being achieved by these plants.
When plants are transferred from LL to HL, there is an immediate increase in the rate of CO2 fixation (Fig. 1), which is significantly greater in gpt2.2 plants than in wild type. This is consistent with the observation that gpt2.2 plants have a greater photosynthetic capacity when grown at LL (Athanasiou et al. 2010). When plants are exposed to HL, leaf concentrations of sucrose and starch increase in wild‐type plants. However, in spite of the higher rate of photosynthesis, net accumulation of sucrose in gpt2.2 is identical and starch synthesis is lower than in wild type. Sucrose content of leaves is a product of rates of synthesis and export or breakdown. The lack of change of the diurnal accumulation of sucrose in response to increased photosynthesis suggests that these rates are carefully balanced. Nevertheless, several primary metabolites, including G6P, FBP and T6P, increase significantly and substantially more in gpt2.2 than in Ws‐4 wild type. This indicates that plants without GPT2 are less able to buffer short‐term changes in carbohydrate metabolism.
There is evidence that the export of triose phosphates, via the triose phosphate translocator, is an important regulator of gene expression, especially at the early stages following a change in irradiance (Vogel et al. 2014). Mutants lacking TPT have previously been shown to be unable to acclimate to high irradiances (Walters et al. 2004. Rapid transient expression of transcription factors observed following an increase in irradiance is suppressed in the tpt2 mutant of Col‐0 (Vogel et al., 2014). This suggests that cytosolic concentrations of triose phosphates and/or other primary metabolites are important in regulating gene expression in a highly dynamic way. Our observation that GPT2 expression is important for the longer term responses to light is consistent with a role for GPT2 in modulating such effects. On the contrary, GPT2 has been shown to not be required for the sugar‐dependent rescue of the high chlorophyll fluorescence (HCF) phenotype observed in mutants deficient in both triose phosphate export and starch synthesis (Schmitz et al. 2012). This HCF phenotype seems to arise due to a mis‐assembly of the thylakoid membranes, with accumulation of disconnected light‐harvesting proteins (Schmitz et al. 2012), probably indicating an effect that is importantly already present under developmental conditions. Our previous data (Athanasiou et al. 2010) indicated that GPT2 was not important during developmental acclimation, consistent with the results from Schmitz et al. (2012). Nevertheless, the results here suggest that under both sets of conditions, the distribution of primary metabolites between chloroplast and cytosol is likely to be important.
The expression of GPT2 within the leaf will allow the exchange of G6P and inorganic phosphate (Pi) across the chloroplast envelope, with the direction determined by the relative concentrations of G6P and Pi. The first alternative is that G6P is exported from the chloroplast, with a concurrent import of Pi. Accumulation of phosphorylated intermediates following the increase in CO2 fixation will lower the levels of free Pi in both the cytosol and the chloroplasts. Low stromal levels of Pi lead to an inhibition of photosynthesis (Sharkey & Vanderveer 1989; Rao & Terry 1995). Export of G6P would supplement TP export, increasing cytosolic substrates for sucrose synthesis. At the same time, given the relatively low substrate specificity of GPT2, it could be acting to supplement TPT activity, exporting TPs. Phosphate released in sucrose synthesis would be reimported into the chloroplast. This scenario would lead to increases in sucrose synthesis due to increased cytosolic G6P and potentially to increases in photosynthetic rate due to an increased availability of Pi in the chloroplast. At the same time, carbon would be diverted away from starch synthesis. During the first 3 d of acclimation, plants without GPT2 maintain similar levels of net sucrose accumulation as that of wild type and have lower starch synthesis, while initially having a higher photosynthetic rate than wild‐type plants. Net export of sugar phosphates via GPT2 seems therefore unlikely, although without measurements of flux through the sucrose pool, this idea cannot be excluded.
The observation that starch accumulation is higher at HL in the wild type indicates that the net direction of G6P flux through GPT2 is more likely to be into the chloroplast. Fixed carbon exported as triose phosphate would be converted to hexose phosphate and then reimported into the plastid for incorporation into starch. By removing G6P from the cytosol, GPT2 may help buffer changes in carbon metabolism and divert more fixed carbon into starch. This model is consistent with the higher starch synthesis in wild type and also with observations from fractionation studies that G6P concentrations are higher in the cytosol than the chloroplast (Gerhardt et al. 1987; Vosloh 2011). The increase in starch synthesis in the wild type is rapid, with a significant increase in starch being detectable within 2 h of HL (Fig. 2), consistent with the rapid induction of GPT2 (20 fold increase within 2 h; Athanasiou et al. 2010).
Expression of GPT2 will allow equilibration of G6P and phosphate across the chloroplast envelope and will, we suggest, lower the cytosolic concentration of G6P. G6P has many direct and indirect effects on metabolism that have already been elucidated. G6P levels are known to positively regulate sucrose synthesis (Doelhert and Huber, 1983). In addition, while G6P is a direct allosteric activator of sucrose phosphate synthase (SPS), Pi is an allosteric inhibitor, so sucrose synthesis is tightly linked to the G6P/Pi ratio. G6P also increases the activity of SPS via regulation of the PKIII protein kinase (Huber & Huber 1996; Toroser et al. 2000). SnRK1, which activates sucrose synthase, is inhibited by G6P (Smeekens et al. 2010). SnRK1 is also necessary for sucrose‐dependent redox activation of ADP‐glucose pyrophosphorylase and is therefore a significant regulator of starch accumulation (Kanegae et al. 2005; Jain et al. 2008). G6P inhibition of SnRK1 could therefore have a significant effect on metabolism as it provides a convergence point for regulatory networks (Cho et al. 2010). By altering the relative concentrations of G6P (and hence other metabolites), GPT2 may thus affect a number of metabolic signals controlling gene expression and/or protein synthesis.
There are significant differences between the two lines in terms of other metabolites that respond to HL treatment. T6P has previously been implicated in the regulation of photosynthesis via a feedback loop involving sucrose and SnRK1, which acts to activate expression of photosynthetic genes. T6P is increased by high sucrose levels (Lunn et al. 2006) and has been shown to inhibit SnRK1 in vitro (Zhang et al. 2009; Delatte et al. 2011; Nunes et al. 2013); however, our data show that there is no simple causal relationship between T6P and photosynthetic capacity. HL induces a much larger transient increase in T6P in gpt2.2 than in wild‐type plants, yet no acclimation of P max occurs in the former case. On the contrary, final steady‐state concentrations of T6P do correlate with photosynthetic capacity. In the mutant, T6P levels return to control levels (Fig. 4) and P max is largely unaltered (Athanasiou et al. 2010). The wild‐type plant maintains a higher T6P concentration at the end of acclimation and has an increased photosynthetic rate and capacity. There is also evidence that T6P is involved in the regulation of starch turnover in the leaf, with increased T6P during the day leading to small increases in starch accumulation and substantial inhibition of starch degradation at night (Martins et al. 2013). However, other factors play important roles in the regulation of starch content and turnover (Pyl et al. 2012; Martins et al. 2013), so it is difficult to interpret these changes in T6P and starch content in isolation. It has recently been proposed that the ratio of T6P:sucrose provides a homeostatic signal allowing plants to maintain appropriate cellular sucrose (Yadav et al. 2014). There is a clear correlation over the first 3 d of HL between sucrose and T6P levels, with both increasing in parallel (Figs 2, 4); however, this correlation breaks down on day 5, when sucrose continues to increase while T6P decreases. This might indicate adjustment of the set point for the sucrose:T6P ratio as the plant acclimates to HL. Although GPT2 expression is important for acclimation of photosynthetic capacity to increased light, it is clearly not required for all components of HL acclimation. There is a clear acclimation of metabolism in both the Ws‐4 and Col‐0 wild‐type and the gpt2 mutants, although global analysis by transcriptomics, FTIR spectroscopy and GC‐MS shows that the nature of this acclimation is different in different cases. The identified intermediates in carbon metabolism show a qualitatively similar response, with steady‐state concentrations of these typically, although not universally, rising initially and then falling to low levels as the plants acclimate to HL. Starch synthesis shows a significant increase in both wild‐type and gpt2.2 plants, although the wild type retains a greater capacity for starch accumulation even after 7‐day‐acclimation period (see Supporting Information Fig. S4). In contrast, the diurnal accumulation of sucrose remained constant throughout HL treatment.
Within primary metabolism, there is a striking dichotomy of response to HL, with metabolites involved in photosynthesis and glycolysis increasing on the first day at HL, while those involved in the TCA cycle decrease (Fig. 5). These data suggest that the conversion of PEP to pyruvate, catalysed by pyruvate kinase, is a key regulatory step that directly responds to HL (Paul & Pellny 2003). Conversion of PEP to shikimate in the chloroplast may also be important in controlling fluxes through the glycolysis pathway. Shikimate concentrations increased in response to HL, while it is constitutively high in the gpt2.2 mutant. The return of most metabolite pools to near LL levels following acclimation is consistent with metabolic acclimation operating to achieve an overall homeostasis of metabolism. This occurs in both wild‐type and gpt2.2 plants, although the rates of change and absolute levels of metabolites are different.
In conclusion, GPT2 allows plants to control the diversion of carbon into transient starch reserves and to regulate the concentrations of various metabolites, notably G6P. These metabolites may not act directly as signals for acclimation; their concentrations change less in acclimating than non‐acclimating plants. Rather, we suggest that the control of metabolite concentrations may provide permissive conditions, allowing photosynthetic acclimation. Expression of GPT2 acts to buffer metabolic changes during acclimation to HL and therefore provides an essential route for antero‐ and retrograde signalling between chloroplast and cytosol.
Supporting information
Figure S1. Photosynthetic rates of plants of Col‐0 (circles) and gpt2.1 (triangles) measured under growth conditions.
Figure S2. MapMan representation of genes showing significant changes in expression in gpt2.2 leaves, compared to Ws‐4 wild type in low light.
Figure S3. MapMan representation of genes showing significant changes in expression in Ws‐4 wild type following transfer from low to high light.
Figure S4. MapMan representation of genes showing significant changes in expression in gpt2.2 following transfer from low to high light.
Figure S5. FT‐IR and GC‐MS analysis of changes in metabolism during acclimation to high light.
Table S1. Microarray analyses were carried out as in Athanasiou et al. (2010).
Table S2. Metabolite levels were obtained using GC‐EI‐TOF/MS on tissue from plants under acclimation conditions.
Table S3. Metabolite levels were obtained using LC‐MS/MS on tissue from plants under acclimation conditions.
Acknowledgments
We would like to thank Dr David Ellis (University of Manchester) and Dr Elon Correa (University of Manchester) for their assistance with the FTIR and statistical analyses. We would also like to thank Dr Leo Zeef and Dr Andy Hayes of the The Bioinformatics Core Facility (BCF) of the University of Manchester for their assistance with the Microarray analysis. This work was supported by a studentship from the United Kingdom Natural Environment Research Council (to B.C.D.), by UK Biotechnology and Biological Sciences Research Council (Grant No. BB/J004103/1) and by the Max Planck Society (R.F. & J.E.L.).
References
- Allwood J.W., Ellis D.I., Heald J.K., Goodacre R. & Mur L.A.J. (2006) Metabolomic approaches reveal that phosphatidic and phosphatidyl glycerol phospholipids are major discriminatory non‐polar metabolites in responses by Brachypodium distachyon to challenge by Magnaporthe grisea . The Plant Journal 46, 351–368. [DOI] [PubMed] [Google Scholar]
- Allwood J.W., Erban A., de Koning S., Dunn W.B., Luedemann A., Lommen A. & Goodacre R. (2009) Inter‐laboratory reproducibility of fast gas chromatography–electron impact–time of flight mass spectrometry (GC‐EI‐TOF/MS) based plant metabolomics. Metabolomics 5, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou K., Dyson B.C., Webster R.E. & Johnson G.N. (2010) Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiology 152, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S., Walters R.G., Jansson S. & Horton P. (2001) Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta 213, 794–801. [DOI] [PubMed] [Google Scholar]
- Baldi P. & Long A.D. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t‐test and statistical inferences of gene changes. Bioinformatics (Oxford, England) 17, 509–519. [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends in Plant Science 4, 97–102. [DOI] [PubMed] [Google Scholar]
- Begley P., Francis‐McIntyre S., Dunn W.B., Broadhurst D.I., Halsall A., Tseng A., … Kell D.B. (2009) Development and performance of a gas chromatography time‐of‐flight mass spectrometry analysis for large‐scale nontargeted metabolomic studies of human serum. Analytical Chemistry 81, 7038–7046. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M. & Speed T.P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics (Oxford, England) 17, 185–193. [DOI] [PubMed] [Google Scholar]
- Brown M., Dunn W.B., Dobson P., Patel Y., Winder C.L., Francis‐McIntyre S., … Kell D.B. (2009) Mass spectrometry tools and metabolite‐specific databases for molecular identification in metabolomics. The Analyst 134, 1322–1332. [DOI] [PubMed] [Google Scholar]
- Cho Y.‐H., Hong J.‐W., Kim E.‐C. & Yoo S.‐D. (2010) Regulatory functions of SnRK1 in stress‐responsive gene expression and in plant growth and development. Plant Physiology 158, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T.L., Sedijani P., Kondou Y., Matsui M., de Jong G.J., Somsen G.W., … Schluepmann H. (2011) Growth arrest by trehalose‐6‐phosphate: an astonishing case of primary metabolite control over growth by way of the Snrk1 signaling pathway. Plant Physiology 157, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.J. & Pfannschmidt T. (2011) Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiology 155, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlert D.C. & Huber S.C. (1983) Phosphate inhibition of spinach leaf sucrose phosphate synthase as affected by glucose‐6‐phosphate and phosphoglucoisomerase. Plant Physiol. 76, 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.B., Broadhurst D., Begley P., Zelena E., Francis‐McIntyre S., Anderson N., … Goodacre R. (2011) Procedures for large‐scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols 6, 1060–1083. [DOI] [PubMed] [Google Scholar]
- Eicks M., Maurino V.N., Knappe S., Flügge U.‐I. & Fischer K. (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiology 128, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch‐Savage W.E., Cadman C.S.C., Toorop P.E., Lynn J.R. & Hilhorst H.W.M. (2007) Seed dormancy release in Arabidopsis Cvi by dry after‐ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. The Plant Journal 51, 60–78. [DOI] [PubMed] [Google Scholar]
- Fischer K. (2011) The import and export business in plastids: transport processes across the inner envelope membrane. Plant Physiology 155, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U.‐I. (1999) Phosphate translocators in plastids. Annual Review of Plant Physiology and Plant Molecular Biology 50, 27–45. [DOI] [PubMed] [Google Scholar]
- Gerhardt R., Stitt M. & Heldt H.W. (1987) Subcellular metabolite levels in spinach leaves: regulation of sucrose synthesis during diurnal alterations in photosynthetic partitioning. Plant Physiology 83, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Solfanelli C., Novi G., Alpi A. & Perata P. (2006) Identification of sugar‐modulated genes and evidence for in vivo sugar sensing in Arabidopsis . Journal of Plant Research 119, 115–123. [DOI] [PubMed] [Google Scholar]
- Goodacre R., Timmins E.M., Burton R., Kaderbhai N., Woodward A.M., Kell D.B. & Rooney P.J. (1998) Rapid identification of urinary tract infection bacteria using hyperspectral whole‐organism fingerprinting and artificial neural networks. Microbiology (Reading, England) 144, 1157–1170. [DOI] [PubMed] [Google Scholar]
- Goodacre R., Vaidyanathan S., Dunn W.B., Harrigan G.G. & Kell D.B. (2004) Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in Biotechnology 22, 245–252. [DOI] [PubMed] [Google Scholar]
- Hausler R.E., Heinrichs L., Schmitz J. & Flugge U.‐I. (2014) How sugars might coordinate chloroplast and nuclear gene expression during acclimation to high light intensities. Molecular Cell 7, 1121–1137. [DOI] [PubMed] [Google Scholar]
- Huber S.C. & Huber J.L. (1996) Role and regulation of sucrose‐phosphate synthase in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 431–444. [DOI] [PubMed] [Google Scholar]
- Jain M., Li Q.‐B. & Chourey P.S. (2008) Cloning and expression analyses of sucrose non‐fermenting‐1‐related kinase 1 (SnRK1b) gene during development of sorghum and maize endosperm and its implicated role in sugar‐to‐starch metabolic transition. Physiologia Plantarum 134, 161–173. [DOI] [PubMed] [Google Scholar]
- Kaderbhai N.N., Broadhurst D.I., Ellis D.I., Goodacre R. & Kell D.B. (2003) Functional genomics via metabolic footprinting: monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT‐IR and direct injection electrospray mass spectrometry. Computational and Functional Genomics 4, 376–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A. & Flügge U.‐I. (1998) Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6‐phosphate/phosphate antiporter. The Plant Cell 10, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae H., Miyoshi K., Hirose T., Tsuchimoto S., Mori M., Nagato Y. & Takano M. (2005) Expressions of rice sucrose non‐fermenting‐1 related protein kinase 1 genes are differently regulated during the caryopsis development. Plant Physiology and Biochemistry 43, 669–679. [DOI] [PubMed] [Google Scholar]
- Knappe S., Flügge U.‐I. & Fischer K. (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator‐homologous transporters, classified by their putative substrate‐binding site. Plant Physiology 131, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B.R., Bergmüller E., … Steinhauser D. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics (Oxford, England) 21, 1635–1638. [DOI] [PubMed] [Google Scholar]
- Kunz H.H., Häusler R.E., Fettke J., Herbst K., Niewiadomski P., Gierth M., … Schneider A. (2010) The role of plastidial glucose‐6‐phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biology 12, 115–128. [DOI] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends in Genetics 19, 47–56. [DOI] [PubMed] [Google Scholar]
- Li C. & Wong W.H. (2001) Model‐based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Science United States of America 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B. & Niyogi K.K. (2009) Sensing and responding to excess light. Annual Review of Plant Biology 60, 239–260. [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L. & Fernie A.R. (2006) Gas chromatography mass spectrometry‐based metabolite profiling in plants. Nature Protocols 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Lloyd J.C. & Zakhleniuk O.V. (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3 . Journal of Experimental Botany 55, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Lunn J.E., Feil R., Hendriks J.H.M., Gibon Y., Morcuende R., Osuna D., … Stitt M. (2006) Sugar‐induced increases in trehalose 6‐phosphate are correlated with redox activation of ADP glucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana . Biochemical Journal 397, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M.C.M., Hejazi M., Fettke J., Steup M., Feil R., Krause U., … Lunn J.E. (2013) Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6‐phosphate. Plant Physiology 163, 1142–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H. & Horton P. (1997) Acclimation of photosynthesis to irradiance and spectral quality in British plant species: chlorophyll content, photosynthetic capacity and habitat preference. Plant, Cell & Environment 20, 438–448. [Google Scholar]
- Murchie E.H., Hubbart S., Chen Y., Peng S. & Horton P. (2002) Acclimation of rice photosynthesis to irradiance under field conditions. Plant Physiology 130, 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U.S., … Schneider A. (2005) The Arabidopsis plastidic glucose 6‐phosphate/phosphate translocator gpt1 is essential for pollen maturation and embryo sac development. The Plant Cell 17, 760–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., Primavesi L.F., Patel M.K., Martinez‐Barajas E., Powers S.J., Sagar R., … Paul M.J. (2013) Inhibition of SnRK1 by metabolites: tissue‐dependent effects and cooperative inhibition by glucose 1‐phosphate in combination with trehalose 6‐phosphate. Plant Physiology and Biochemistry 63, 89–98. [DOI] [PubMed] [Google Scholar]
- Oguchi R., Hikosaka K. & Hirose T. (2003) Does the photosynthetic light‐acclimation need change in leaf anatomy? Plant, Cell & Environment 26, 505–512. [Google Scholar]
- Paul M.J. & Pellny T.K. (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. Journal of Experimental Botany 54, 539–547. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2003) Chloroplast redox signals: how photosynthesis controls its own genes. Trends in Plant Science 8, 33–41. [DOI] [PubMed] [Google Scholar]
- Piippo M., Allahverdiyeva Y., Paakkarinen V., Suoranta U.M., Battchikova N. & Aro E.M. (2006) Chloroplast‐mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiological Genomics 25, 142–152. [DOI] [PubMed] [Google Scholar]
- Pourtau N., Jennings R., Pelzer E., Pallas J. & Wingler A. (2006) Effect of sugar‐induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis . Planta 224, 556–568. [DOI] [PubMed] [Google Scholar]
- Pyl E.T., Piques M., Ivakov A., Schulze W., Ishihara H., Stitt M. & Sulpice R. (2012) Metabolism and growth in Arabidopsis depend on the daytime temperature but are temperature‐compensated against cool nights. The Plant Cell 24, 2443–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao I.M. & Terry N. (1995) Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet (IV. Changes with time following increased supply of phosphate to low‐phosphate plants). Plant Physiology 107, 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitzky A. & Golay M.J.E. (1964) Smoothing and differentiation of data by simplified least squares procedures. Analytical Chemistry 36, 1627–1639. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Schöttler M.A., Krueger S., Geimer S., Schneider A., Kleine T., … Häusler R.E. (2012) Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana . BMC Plant Biology 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T.D. & Vanderveer P.J. (1989) Stromal phosphate concentration is low during feedback limited photosynthesis. Plant Physiology 91, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S., Ma J., Hanson J. & Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology 13, 273–278. [DOI] [PubMed] [Google Scholar]
- Takahashi S. & Badger M.R. (2011) Photoprotection in plants: a new light on photosystem II damage. Trends in Plant Science 16, 53–60. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., … Stitt M. (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal 37, 14–39. [DOI] [PubMed] [Google Scholar]
- Toroser D., Plaut Z. & Huber S.C. (2000) Regulation of a plant SNF1‐related protein kinase by glucose‐6‐phosphate. Plant Physiology 123, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M.O., Moore M., König K., Pecher P., Alsharafa K., Lee J. & Dietz K.‐J. (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN‐ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis . The Plant Cell 26, 1151–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosloh D. (2011) Subcellular compartmentation of primary carbon metabolism in mesophyll cells of Arabidopsis thaliana . PhD thesis, University of Potsdam, Potsdam, Germany. [Google Scholar]
- Walters R.G. (2005) Towards an understanding of photosynthetic acclimation. Journal of Experimental Botany 56, 435–447. [DOI] [PubMed] [Google Scholar]
- Walters R.G., Ibrahim D.G., Horton P. & Kruger N.J. (2004) A mutant of Arabidopsis lacking the triose‐phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiology 135, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston E., Thorogood K., Vinti G. & Lopez‐Juez E. (2000) Light quantity controls leaf‐cell and chloroplast development in Arabidopsis thaliana wild‐type and blue‐light‐perception mutants. Planta 211, 807–815. [DOI] [PubMed] [Google Scholar]
- Winder C.L., Carr E., Goodacre R. & Seviour R. (2004) The rapid identification of Acinetobacter species using Fourier transform infrared spectroscopy. Journal of Applied Microbiology 96, 328–339. [DOI] [PubMed] [Google Scholar]
- Wulff‐Zottele C., Gatzke N., Kopka J., Orellana A., Hoefgen R., Fisahn J. & Hesse H. (2010) Photosynthesis and metabolism interact during acclimation of Arabidopsis thaliana to high irradiance and sulphur depletion. Plant, Cell & Environment 33, 1974–1988. [DOI] [PubMed] [Google Scholar]
- Yadav U.P., Ivakov A., Feil R., Duan G.Y., Walther D., Giavalisco P., … Lunn J.E. (2014) The sucrose‐trehalose 6‐phosphate (Tre6p) nexus: specificity and mechanisms of sucrose signalling by Tre6p. Journal of Experimental Botany 65, 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A.C., Powers S.J., … Paul M.J. (2009) Inhibition of SNF1 related protein kinase1 activity and regulation of metabolic pathways by trehalose‐6‐phosphate. Plant Physiology 149, 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Photosynthetic rates of plants of Col‐0 (circles) and gpt2.1 (triangles) measured under growth conditions.
Figure S2. MapMan representation of genes showing significant changes in expression in gpt2.2 leaves, compared to Ws‐4 wild type in low light.
Figure S3. MapMan representation of genes showing significant changes in expression in Ws‐4 wild type following transfer from low to high light.
Figure S4. MapMan representation of genes showing significant changes in expression in gpt2.2 following transfer from low to high light.
Figure S5. FT‐IR and GC‐MS analysis of changes in metabolism during acclimation to high light.
Table S1. Microarray analyses were carried out as in Athanasiou et al. (2010).
Table S2. Metabolite levels were obtained using GC‐EI‐TOF/MS on tissue from plants under acclimation conditions.
Table S3. Metabolite levels were obtained using LC‐MS/MS on tissue from plants under acclimation conditions.


