Summary
The tropical Andes of South America, the world's richest biodiversity hotspot, are home to many rapid radiations. While geological, climatic, and ecological processes collectively explain such radiations, their relative contributions are seldom examined within a single clade.
We explore the contribution of these factors by applying a series of diversification models that incorporate mountain building, climate change, and trait evolution to the first dated phylogeny of Andean bellflowers (Campanulaceae: Lobelioideae). Our framework is novel for its direct incorporation of geological data on Andean uplift into a macroevolutionary model.
We show that speciation and extinction are differentially influenced by abiotic factors: speciation rates rose concurrently with Andean elevation, while extinction rates decreased during global cooling. Pollination syndrome and fruit type, both biotic traits known to facilitate mutualisms, played an additional role in driving diversification. These abiotic and biotic factors resulted in one of the fastest radiations reported to date: the centropogonids, whose 550 species arose in the last 5 million yr.
Our study represents a significant advance in our understanding of plant evolution in Andean cloud forests. It further highlights the power of combining phylogenetic and Earth science models to explore the interplay of geology, climate, and ecology in generating the world's biodiversity.
Keywords: Andes, biodiversity hotspot, climate change, diversification, Lobelioideae, Neotropics, pollination syndromes, rapid radiation
Short abstract
See also the Commentary on this article by Hughes, 210: 1152–1154.
Introduction
Species‐rich rapid radiations are a conspicuous ecological and evolutionary phenomenon in the Tree of Life. Clades that have undergone such diversification are often documented in insular environments, including islands (Baldwin & Sanderson, 1998; Givnish et al., 2009; Lapoint et al., 2014), lakes (Wagner et al., 2012), and mountains (McGuire et al., 2007; Hoorn et al., 2013; Hughes & Atchison, 2015; Merckx et al., 2015). Although they represent just one‐eighth of terrestrial land surface, mountains are home to one‐third of all species (Antonelli, 2015) and a large number of species‐rich radiations (Hughes & Atchison, 2015; Schwery et al., 2015), including some of the fastest diversification rates reported to date (Madriñán et al., 2013). Of particular importance are the tropical Andes, which stretch from Venezuela to northern Argentina along the western coast of South America. These incredibly species‐rich mountains (Barthlott et al., 2005; Kreft & Jetz, 2007) are home to c. 15% of all flowering plant species, half of which are endemic to the region (Myers et al., 2000). The extent of this biodiversity is especially striking considering the recency of mountain uplift: despite debate over the precise timing and rates of uplift (Sempere et al., 2006; Ehlers & Poulsen, 2009), an increasing body of evidence suggests that > 60% of the current elevation of the central Andes was attained within the last 10 million yr (Myr) (Gregory‐Wodzicki, 2000; Garzione et al., 2006, 2008, 2014). Although the onset of Andean orogeny dates to the Paleocene and Eocene (66–33.9 Myr), including in regions as far north as the modern Eastern Cordillera of Colombia (Parra et al., 2009), exceptionally rapid surface uplift occurred during the late Miocene (c. 10–6 Myr) and early Pliocene (c. 4.5 Myr) (Garzione et al., 2008; Hoorn et al., 2010; Mulch et al., 2010). Such mountain building is thought to promote diversification in a variety of ways, including by increasing physiographic heterogeneity, affecting local and regional climate, facilitating the immigration of preadapted species, and creating opportunities for extensive, parallel geographic speciation and adaptive radiation in island‐ and archipelago‐like venues (Hoorn et al., 2013; Givnish et al., 2014; Mulch, 2016).
The role of the rise of the Andes in the origination of biodiversity has been implicated in clades as diverse as birds (McGuire et al., 2014), butterflies (Elias et al., 2009), and angiosperms (Hughes & Eastwood, 2006; Madriñán et al., 2013). Within angiosperms, previous work has emphasized that the timing and geotemporal trajectories of diversification have been very different across Andean biomes (Pennington et al., 2010). While high‐elevation grassland clades are exceptionally young, fast, and species‐rich (e.g. Hughes & Eastwood, 2006), clades in lower‐elevation dry inter‐Andean valleys are much older, slower, and species‐poor (Särkinen et al., 2012). Despite their species richness, however, cloud forest biomes have been largely unexplored with respect to angiosperm diversification, although data suggest that they are intermediate in terms of age and tempo (Särkinen et al., 2012).
Identifying the causes of species‐rich rapid radiations, such as those that characterize Andean high‐elevation grasslands and cloud forests, is a major goal in ecology and evolutionary biology. While rapid diversification is undoubtedly the product of both abiotic and biotic factors acting on different spatial and temporal scales (Antonelli & Sanmartín, 2011; Ezard et al., 2011; Bouchenak‐Khelladi et al., 2015), a tendency to explain a given diversity pattern with a single process has persisted until recently. For montane Andean radiations, this was exemplified by invoking molecular divergence time estimates contemporaneous with the final stages of Andean uplift, during which new habitats formed and provided ecological opportunity for diversification (Hughes & Eastwood, 2006; McGuire et al., 2014). However, other factors are probably at play simultaneously. For example, rapid plant diversification in the region has also been attributed to specialized pollination relationships with hummingbirds (Schmidt‐Lebuhn et al., 2007; Abrahamczyk et al., 2014; Givnish et al., 2014), shifts between pollination syndromes (Kay et al., 2005), and the acquisition of fleshy fruits in understory taxa (Smith, 2001; Givnish, 2010). Further, these factors are not likely to be independent of one another, e.g. mountain building affects the local climate (Armijo et al., 2015), which, in turn, alters biotic interactions (Blois et al., 2013). Not surprisingly, diversification studies using molecular phylogenies have become more integrated in their scope as methodology improves, and analyzing multiple factors underlying diversification patterns is becoming tractable (Ezard et al., 2011; Wagner et al., 2012; Givnish et al., 2014; Donoghue & Sanderson, 2015; Hughes et al., 2015).
Here, we seek to untangle the ecological and historical factors that have contributed to the generation of Andean megadiversity, especially in mesic mid‐montane forests, using the Neotropical bellflowers as a model system. The Neotropical bellflowers (Campanulaceae: Lobelioideae) are an Andean‐centered (Gentry, 1982) clade of c. 600 morphologically and ecologically diverse species found from lowland Amazonia to high‐elevation grasslands above 5000 m, although the vast majority of species are found in cloud forests (Lagomarsino et al., 2014). The group was recently resolved into three well‐supported subclades: the Chilean lobelias (Lobelia section Tupa; four species) of the temperate southern Andes; the páramo and puna endemic Lysipomia (50 species); and the centropogonid clade of primarily cloud forest endemics (Centropogon, Burmeistera, and Siphocampylus; c. 550 species) (Lagomarsino et al., 2014). Lysipomia are diminutive herbs, while the other subclades are robust and mostly woody. The centropogonids are particularly diverse in their growth form: most species are vines with woody bases, but herbs, shrubs, and trees are all represented (Fig. 1). Similar phenotypic diversity is also apparent in two traits known to facilitate plant–animal interactions: fruit type and floral morphology. Abiotically dispersed capsules characterize c. 40% of the species, while the remaining species produce fleshy, animal‐dispersed berries, which evolved at least seven times from capsular‐fruited ancestors (Lagomarsino et al., 2014). Floral morphology is also highly variable. Lysipomia produces small, white flowers indicative of invertebrate pollination (Faegri & van der Pijl, 1979), while both the Chilean lobelias and the centropogonids produce robust flowers that are usually vertebrate‐pollinated. Flowers in the centropogonid clade are particularly diverse (Fig. 2) and principally adapted to pollination by hummingbirds and nectar bats (Colwell et al., 1974; Stein, 1992; Muchhala, 2006; Muchhala & Potts, 2007).
Figure 1.

Growth form diversity in Neotropical bellflowers. (a) Siphocampylus tunarensis Zahlbr., a tree c. 7–10 m. (b) Centropogon congestus Gleason, a clonal herbaceous species of wet soil, c. 1.5 m. (c) Centropogon pulcher Zahlbr., a hanging vine. (d) Burmeistera sp., a hemi‐epiphytic herb. (e) Siphocampylus tunicatus Zahlbr., a tall shrub, with individual stems c. 5 m tall, arising from a single point. (f) Lysipomia muscoides Hook f., a minute rosette herb growing among moss in puna habitat. (g) Siphocampylus jelskii Zahlbr., a giant rosette shrub with elongated stems, each c. 1–2 m tall, apparently clonal, growing in high‐altitude grasslands. (h) Siphocamplus smilax Lammers, a xerophyte with a densely woody stem (probably water‐storing) growing in sandstone. (i) Siphocampylus williamsii Rubsy, a plant producing a small number of narrow aerial stems arising from a xylopodium, or underground tuber‐like stem. Photographs by: L. Lagomarsino (b–h), D. Santamaría‐Aguilar (a), and A. Fuentes (i).
Figure 2.
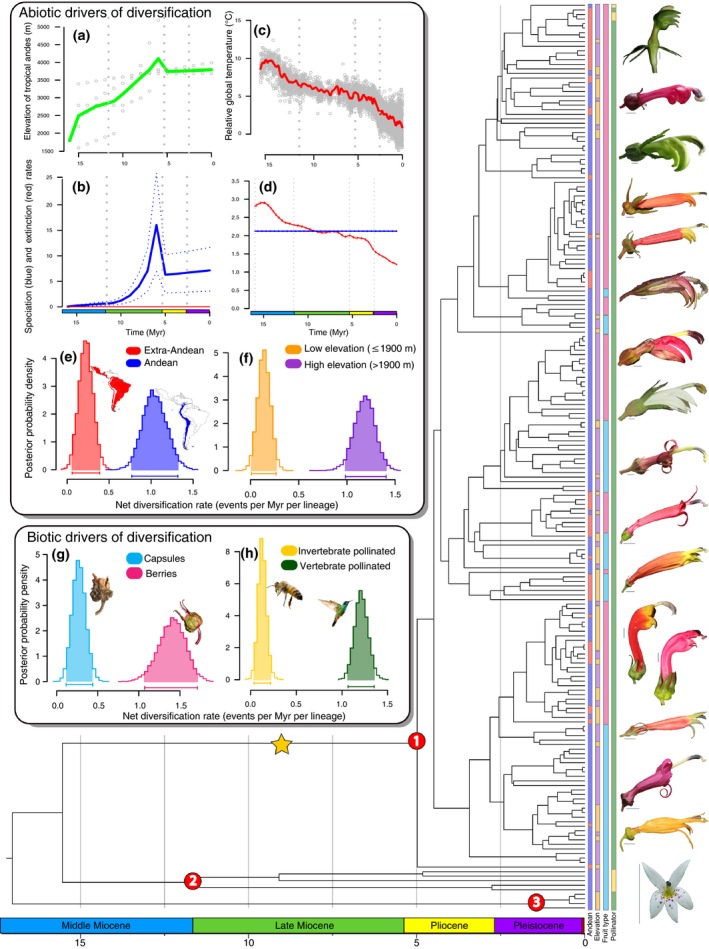
Diversification of Neotropical bellflowers. A time‐calibrated species‐level phylogeny shows the c. 5 Myr age of the largely Andean centropogonid clade (c. 550 species, node 1), whose origin is associated with a significant increase in diversification rate (yellow star) as detected by Bayesian Analysis of Macroevolutionary Mixture (BAMM). Lysipomia (c. 50 species) and Chilean Lobelia (four species) are indicated (nodes 2 and 3, respectively). Representative floral diversity, shown on the right, illustrates the striking phenotypic diversity in the clade (see Fig. 1 for comparable growth form diversity); scale bars, 0.5 cm. Diversification models examining the abiotic correlates of this rapid diversification include average paleoelevation of the tropical Andes through time (a) and global temperatures through time (c). Gray dots in (a) and (c) represent individual data points utilized to create the curves. Results from these models show inferred speciation (blue) and extinction (red) rates through time under models depending on paleoelevation (b) and paleotemperature (d). Additional diversification analyses using binary state‐speciation and extinction (BiSSE) demonstrate the effect of two abiotic and two biotic traits on net diversification rate: Andean occurrence (e; extra‐Andean (red) vs Andean (blue)), elevation (f; low elevation, ≤ 1900 m (orange) vs high elevation, > 1900 m (purple)), fruit type (g; dry capsules (light blue) vs fleshy berries (pink)), and pollinator type (h; invertebrate (yellow) vs vertebrate (green)). Trait scorings are color‐coded to the right of phylogeny. Outgroups were removed, and taxon names omitted because of space constraints. Photographs by L. Lagomarsino. Geological timescale shown at bottom from Walker et al. (2012).
Owing to their broad Andean distribution and remarkable floral and fruit diversity, the Neotropical bellflowers represent a model group to examine the interaction of abiotic and biotic factors that trigger mountain radiations. To accomplish this goal, we develop and apply numerous statistical models to investigate diversification dynamics in the Neotropical bellflowers. Moreover, our analyses are among the first to incorporate geological data into a model of evolutionary diversification (also see Valente et al., 2014 and Mulch, 2016). In this framework, we explicitly investigate the influence of geology, climate, and ecological traits on species diversification using the first well‐sampled, time‐calibrated phylogeny of the group.
Materials and Methods
Taxon sampling and molecular dataset
One‐third of Andean bellflower species were sampled, including all four species of Lobelia section Tupa, five of the c. 40 species of Lysipomia, and 191 species of the c. 550 species of the centropogonid clade (Supporting Information Table S1). Our sampling includes representatives from all taxonomic subdivisions of Centropogon, Burmeistera, and Siphocampylus (Lagomarsino et al., 2014). We assembled a concatenated molecular matrix that includes seven plastid regions totaling 11 990 bp. Taxon sampling, molecular methods for newly generated sequences, and alignment protocols followed Lagomarsino et al. (2014). Outgroup sampling includes broad representation across Lobelioideae. Nine representatives of Campanuloideae were used as outgroups to root the phylogeny and provide appropriate nodes for fossil calibration.
Phylogenetics and dating
Phylogenies were inferred using maximum likelihood (ML) and Bayesian inference as implemented in RAxML 8.0 (Stamatakis, 2014) and Beast 2.1.3 (Bouckaert et al., 2014). Four calibration points were used to estimate divergence times: the fossil seed of †Campanula paleopyramidalis as a minimum age constraint of 16 Myr for the most recent common ancestor of C. pyramidalis and C. carpatica (Cellinese et al., 2009; Crowl et al., 2014); a geological maximum age constraint of 29.8 Myr, which corresponds to the age of the Kure atoll, the oldest emerged island of the Hawaiian Ridge (Clague, 1996), for the crown group of the endemic Hawaiian clade, encompassing c. 125 spp. in six genera represented in our sampling by Brighamia, Cyanea, Clermontia and Delissea (Antonelli, 2009); and two secondary age constraints from Bell et al. (2010): 41–67 Myr ago for crown group Campanulaceae, and 28–56 Myr ago for crown group Campanuloideae. The fossil record for Campanulaceae is very poor, and †C. paleopyramidalis, described from Miocene deposits in the Nowy Sacz Basin in the West Carpathians of Poland (Łancucka‐Srodoniowa, 1977), is the only fossil appropriate for calibration of divergence time estimates in this family (Antonelli, 2009; Cellinese et al., 2009). It is assigned as a close relative of C. pyramidalis on the basis of their shared reticulate seed coats, a feature uncommon in the genus (Łancucka‐Srodoniowa, 1977; Cellinese et al., 2009).
The optimal RAxML tree was dated using penalized likelihood in treePL (Smith & O'Meara, 2012) with hard minimum and maximum age constraints. Confidence intervals were generated using 1000 RAxML bootstrap trees. We then simultaneously inferred phylogenetic relationships and divergence times using both relaxed uncorrelated lognormal and exponential clock models in Beast. A lognormal prior (mean of 1.5 and SD of 1.0) was assigned to the fossil calibration age of 16 Myr, a uniform prior was assigned to the geological age constraint at 29.8 Myr (maximum hard bound), and normal priors were placed on the two secondary age constraints (mean age of 56.0 Myr and SD of 8.3 Myr for Campanulaceae; mean age of 43.0 Myr and SD of 8.0 Myr for Campanuloideae). The dated tree from treePL was specified as the starting tree in each of eight separate Beast runs, which were each conducted for 10 million generations of Markov chain Monte Carlo (MCMC). Convergence was assessed using effective sample size (ESS) values of the runs in Tracer 1.6 (Rambaut et al., 2014), applying a cutoff value of 200. The maximum clade credibility tree, including credibility intervals (CIs) for ages and posterior probabilities (PPs) for node support, was then assembled using TreeAnnotator (Bouckaert et al., 2014).
Robustness of age estimates was assessed by removing one or a series of calibration points. The following sets of calibration points were used for this purpose: the Campanulaceae secondary constraint only; the fossil and the Campanulaceae secondary age constraint; and the fossil and both secondary age constraints. Divergence time estimation can be sensitive to branch length variation, which is potentially influenced by growth form (Smith & Donoghue, 2008). As they are generally herbaceous and have longer internal branches than their woody relatives, we suspected this may be the case for Lysipomia. We thus removed Lysipomia species entirely, and re‐estimated molecular divergence times to determine if these differences had an impact on results. We find no evidence that the results are affected by dating method, calibration strategy, or branch length heterogeneity: the 95% CIs of the re‐estimated ages overlap with the 95% CIs from the Beast analysis that we use for diversification analyses (see also Table S2).
Trait categorization
Species traits were newly obtained from a variety of sources, including: online databases (GBIF, http://www.gbif.org/ and Tropicos, http://www.tropicos.org/), herbarium specimens, taxonomic literature (Wimmer, 1943, 1953), and field collections. Morphological characters and area of occurrence were coded from type specimens available on the JSTOR Global Plants database (http://plants.jstor.org/) for nearly all 594 species, many of which were not represented in our phylogenetic sampling but were used to account for sampling biases. We scored four binary traits related to the biology and geographic occurrence of all Andean bellflower species for our diversification analyses: fruit type (berry or capsule), pollinator type (vertebrate or invertebrate), elevation (median species occurrence ≤ 1900 m vs > 1900 m), and presence/absence in the Andes. Pollinator type was coded according to classical morphological definitions of pollination syndromes (Faegri & van der Pijl, 1979; Muchhala, 2006). Because they are not sensitive to outliers, we used median elevation across all specimen locality data to determine each species value for elevation. We further used the global median of these values across species as our cutoff for high vs low elevation because this resulted in an equal distribution of the character states (high vs low) across the phylogeny, which is ideal for binary state‐speciation and extinction (BiSSE) analyses (Maddison & FitzJohn, 2015). All trait data are provided in Table S3 and in Fig. 2 and are deposited in Dryad (doi: 10.5061/dryad.7h4j).
Diversification analyses
We applied a series of birth–death models to quantify the effects of abiotic and biotic correlates of speciation and extinction in Neotropical bellflowers. We assessed the robustness of our results in a variety of ways. First, to accommodate phylogenetic and dating uncertainties (except in the case of the diversity‐dependent analyses, which are computationally intensive), we conducted our analyses across 500 randomly sampled trees from the Beast posterior distribution (outgroups removed). Second, we implemented a sampling fraction to account for nonsampled species diversity. We also applied a sampling fraction, including trait data for species not sampled in our phylogeny, to account for any sampling bias in trait space for BiSSE analyses (Maddison et al., 2007). Third, for each of our diversification analyses except for those using Bayesian Analysis of Macroevolutionary Mixture (BAMM 2.2.2, Rabosky, 2014), we selected the best‐fitting model by computing the corrected Akaike information criterion (AICc) based on the log‐likelihood and the number of free parameters for each model. The model supported by the lowest AICc was considered best if it was at least two ∆AIC units better than the model with the second lowest score. We also used likelihood ratio tests (significant at P < 0.05) to estimate support for the best‐fitting model when compared with rival models.
The diversification models we apply assume that extinction rates can be estimated from molecular phylogenetic branching patterns, as originally proposed by Nee et al. (1994). It is important to note this is a controversial practice (Rabosky, 2010, 2015), especially without incorporating paleontological data (Quental & Marshall, 2010). Despite this, recent studies have suggested that unbiased estimates of extinction rates can be obtained solely from molecular phylogenies (Morlon et al., 2011; Beaulieu & O'Meara, 2015). In this spirit, we explore the individual components of diversification rate in our discussion of results, particularly with respect to paleoenvironmental change.
Time‐dependent diversification
We used BAMM to estimate speciation and extinction rates through time and to identify shifts in diversification rate. We accounted for incomplete taxon sampling by applying clade‐specific sampling fractions in each of the three principal Andean bellflower subclades: the Chilean lobelias (1.0), Lysipomia (0.12), and the centropogonids (0.35). We were unable to provide finer‐scale sampling fractions within the centropogonids because of nonmonophyly of genera and the difficulty of placing species into their constituent clades, particularly within Siphocampylus (Lagomarsino et al., 2014). While our taxon sampling is fairly low, recent studies exploring the effect of incomplete taxon sampling on the identification of rate shifts suggest that, while results may be incomplete, they should be accurate (Spriggs et al., 2015). We ran BAMM with four reversible jump MCMC chains, each for five million generations. ESS values (> 200) were used to assess convergence. The posterior distribution was used to estimate the configuration of the diversification rate shifts, and alternative diversification models were compared using Bayes factors. Results were analyzed and plotted using the R package BAMMtools 2.0.2 (Rabosky et al., 2014).
Paleoenvironment‐dependent diversification
We quantified the effect of the past environment (i.e. climate change and Andean surface uplift) on speciation and extinction rates in Neotropical bellflowers via an approach that builds on time‐dependent diversification models (Nee et al., 1994; Morlon et al., 2011). The method we utilized allows speciation and extinction rates to correlate not only with time, but also with a quantitative external variable that is time‐dependent (see Condamine et al., 2013 for details). These paleoenvironment‐dependent birth–death models are implemented in the R package rpanda (Morlon et al., 2016). We incorporated two quantitative external paleoenvironmental variables: the global Cenozoic deep‐sea oxygen isotope record as a proxy for global temperature (Zachos et al., 2008), and a generalized model of the paleoelevation history of the tropical Andes. The latter was compiled from several references (Garzione et al., 2006, 2008, 2014; Ehlers & Poulsen, 2009; Leier et al., 2013) and is available on the Dryad repository (doi: 10.5061/dryad.7h4j).
The R package pspline was used to interpolate a smooth line for each environmental variable. This smooth line was sampled during each birth–death modeling process to give the value of the paleoenvironmental variable at each time point. Speciation and/or extinction rates were then estimated as a function of these values along the dated phylogenies according to the parameters of the following models. For both temperature and Andean elevation, we first applied two standard models, i.e. constant speciation without extinction (Yule model) and constant‐rate birth–death, as null references for model comparison. We then applied four models in which the paleoenvironment dependency was exponential (Table S4): speciation rate varies with the environmental variable and no extinction; speciation rate varies with the environmental variable and extinction rate is constant; speciation rate is constant and extinction rate varies with the environmental variable; and both speciation and extinction rates vary with the environmental variable. We repeated these models with linear dependence on the environmental variable (Table S4).
Trait‐dependent diversification
We modeled the impact of traits on the diversification of Neotropical bellflowers by concurrently estimating their impact on speciation, extinction, and transition rates using BiSSE (Maddison et al., 2007). For each of the four traits (Andean occurrence, elevation, pollination syndrome, and fruit type), we evaluated eight BiSSE diversification models of increasing complexity in which speciation, extinction, and transition rates were allowed to either vary or remain equal between traits (Table S5). Analyses were performed using the R package diversitree 0.7‐6 (FitzJohn, 2012). Once the best‐fitting model was selected, CIs for each parameter were estimated for the tree. We used an exponential prior following FitzJohn (2012), and began the MCMC with the ML estimates. We ran MCMC for 20 000 generations and applied a burn‐in of 2000 steps. We then computed the net diversification rates for each trait. Finally, to determine if these traits were correlated, pairwise trait comparisons were performed across a random sampling of 500 trees from the posterior distribution in both a ML and Bayesian framework in BayesTraits 2.0 (Pagel & Meade, 2006). The significance of binary trait dependence was assessed against a rival model where these traits evolved independently using likelihood ratio tests (ML) and Bayes factors of the harmonic mean of likelihood values.
Binary state‐speciation and extinction models have received recent criticism, including high type I error rates (Maddison & FitzJohn, 2015; Rabosky & Goldberg, 2015), especially for trees with fewer than 300 terminals and for traits present in < 10% of taxa (Davis et al., 2013). The distribution of characters states is also important; for example, rapid diversification rates in a region of a phylogeny can be erroneously attributed to a particular character state when the trait is characterized by high transition rates (Rabosky & Goldberg, 2015). However, despite these known issues, SSE models remain a viable framework to test the effect of particular traits on species diversification, particularly when these caveats have been mitigated (Ng & Smith, 2014), as we have attempted to do here. Where possible, we also compare the trait‐dependent results with the inferences made with nontrait‐dependent models (i.e. BAMM and RPANDA) to show that that they are consistent.
Diversity‐dependent diversification
We applied models that allow speciation and extinction to vary according to the number of lineages in the phylogeny. Using the R package DDD 2.7 (Etienne et al., 2012), we built two models: speciation declines with diversity, assuming no extinction, and speciation declines with diversity, allowing for extinction. The initial carrying capacity was set to the current species diversity, and the final carrying capacity was estimated according to the model and corresponding parameters. These models were run on the maximum clade credibility (MCC) tree from our Beast analysis.
Results
Molecular phylogeny and divergence time estimation
The final alignment for phylogenetic analysis comprised seven loci (c. 12 000 bp) sequenced for 275 species (doi: 10.5061/dryad.7h4j) (Table S1). The concatenated Bayesian phylogenetic analysis using the best‐fitting partitioning scheme yielded a highly resolved and strongly supported tree compatible with the ML tree and previously published results (Lagomarsino et al., 2014). Sequence divergence, particularly in the centropogonid clade, was low, as reflected by the shallow branching pattern toward the tips. Despite short internal branches, indicative of a recent radiation, the phylogeny is generally well supported: 66% of nodes are recovered with moderate support (PP > 0.90) and 28% are well supported (PP > 0.95). Our results support the monophyly of the subfamilies Campanuloideae and Lobelioideae and the Neotropical bellflowers (the centropogonids, Lysipomia, and Chilean lobelias) with maximum nodal support (PP = 1).
Results of the dating analyses are presented in Fig. 2 (see Figs S1 and S2 for the 95% CI for each node). Similar estimates were obtained using different dating methods and with alternative calibration scenarios and settings (Table S2). This suggests that potential biases such as long internal branch lengths, possibly attributed to growth form variation, do not have a large effect on our age estimates. Based on the molecular divergence time estimates from Beast (Bouckaert et al., 2014) and treePL (Smith & O'Meara, 2012), we place the origin of the extant Neotropical bellflowers diversity in the Miocene, c. 17.1 Myr ago (95% CI: 14.32–20.32 Myr ago). We further found that the crown groups of their three main subclades – Chilean lobelias, Lysipomia, and the centropogonids – originated 1.45 Myr ago (95% CI: 0.42–2.78 Myr ago), 11.87 Myr ago (95% CI: 9.00–15.24 Myr ago), and 5.02 Myr ago (95% CI 3.95–6.13 Myr ago), respectively (Figs 2, S1). All branching events in the most species‐rich clade, the centropogonids, occur in the Plio‐Pleistocene (5.3–0.01 Myr ago).
The age inferred for Lysipomia is surprisingly old considering that the genus is currently restricted to páramo and puna vegetations, the youngest of Andean biomes (van der Hammen & Cleef, 1986). This older age is potentially an artifact caused by the substantial DNA substitution rate heterogeneity observed between Lysipomia and the rest of the phylogeny, as illustrated in the phylogram in Fig. S3. This elevated substitution rate may be explained by Lysipomia's herbaceous habit or high‐altitude occurrence (Lagomarsino et al., 2014). It is likely that this substantial rate heterogeneity is not adequately accommodated by the rate smoothing in Beast, as documented in other groups that exhibit between‐clade rate variation (Beaulieu et al., 2015). The age estimates for Lysipomia are thus potentially overestimated as a result, which may impact diversification analyses. However, we have shown that this rate heterogeneity does not affect the estimated age of the other two major subclades of Neotropical bellflowers, and as such the impact should be minimal (Table S2).
Diversification analyses
A time‐dependent diversification analysis in BAMM strongly rejects constant diversification. Instead, it indicates that the Neotropical bellflowers underwent a significant shift in net diversification rate coinciding with the recent origin of the centropogonid subclade (c. 5 Myr ago), which includes 550 of the 600 species (Figs 2, S4). This analysis further provides evidence for a 3.7‐fold increase in diversification rate (1.83 vs 0.5 events Myr–1 per lineage); net diversification rate is similarly high using an alternative metric (Magallón & Sanderson, 2001) (0.82–1.15 events Myr–1 per lineage under high (ε = 0.90) and low (ε = 0) extinction fractions, respectively). As illustrated in Fig. S4(b), the diversification rate for the centropogonid clade is much higher than its background rate.
We then adapted a recently developed paleoenvironment‐dependent diversification model that explicitly accommodates the effect of changing environment through time (Condamine et al., 2013) to examine the effect of Andean paleoelevation (Fig. 2a) and past temperature on diversification rate (Fig. 2d). This novel integration of Andean geological data into an analysis of evolutionary diversification reveals that speciation rates in the Neotropical bellflowers are positively correlated with the elevation history of the Andes, while extinction rates appear to be unlinked to mountain uplift (Fig. 2b; Table S4). Here, we reconstruct a continuous increase in speciation rate beginning at c. 12 Myr ago (mid‐Miocene) and peaking at c. 5 Myr ago (early Pliocene), consistent with our result from BAMM. This coincides with the attainment of maximum paleoelevation of the Andes (> 4000 m), at which point we infer an unprecedented ~15 speciation events Myr–1 per lineage for Neotropical bellflowers. Because of the magnitude of this result, we ran additional analyses implementing a trait‐dependent diversification model using BiSSE (Maddison et al., 2007). We investigated two traits that serve as a proxy for the effect of Andean orogeny on diversification in this framework: Andean presence (Andean vs extra‐Andean) and elevational distribution (high elevation (> 1900 m) vs low elevation (≤ 1900 m)). The best‐fitting model for these BiSSE analyses inferred significantly higher speciation rates for Andean species and for species occurring at high elevation (Figs 2e,f, S5; Table S5), respectively. The similar results of these BiSSE analyses are potentially due to the correlated nature of elevational distribution and Andean occurrence that we identified in BayesTraits (Pagel & Meade, 2006) (Table S6); this correlation is not surprising given that the Andes represent the majority of the topographic relief across the Neotropical bellflowers’ range. Together, these results are consistent with an integral role of Andean surface uplift as a trigger of rapid diversification for Neotropical bellflowers.
Using the same paleoenvironmental birth–death model, but instead incorporating relative global temperature proxies that span the last c. 20 Myr of the Cenozoic (Zachos et al., 2008), we demonstrate a significant correlation between climate change and Andean bellflower diversification (Fig. 2d). In contrast to Andean orogeny, which influenced primarily speciation, the best‐fitting paleoclimatic model suggests that climate change had a significant impact only on extinction. Beginning c. 15 Myr ago (mid‐Miocene), extinction rates decreased dramatically towards the present as a function of global temperature (Fig. 2d; Table S4).
Additional BiSSE analyses demonstrate that various clade‐specific biotic traits influence diversification rates in Neotropical bellflowers. BayesTraits analyses showed uncorrelated evolution of these traits with respect to one another, and with both the abiotic factors discussed earlier (Table S6). The best models, as supported by both AICc and likelihood ratio tests (Table S5), infer higher net diversification rates in lineages with two character states: fleshy fruits (vs dry fruits; Figs 2g, S5), and vertebrate pollination syndromes (vs invertebrate pollination syndromes; Figs 2h, S5). These traits acted differently on the two components of diversification: fleshy fruits confer lower extinction rates than capsules, while species pollinated by vertebrates have higher speciation rates than invertebrate‐pollinated species.
Finally, a diversity‐dependent analysis inferred that the Neotropical bellflowers have not reached diversity equilibrium: the estimated carrying capacity (i.e. number of species potentially sustained in the clade) far exceeds the current extant diversity of the clade (Table S7).
Discussion
The Andes are famous for their high species richness (Myers et al., 2000) and the rapid diversification rates that characterize many of their emblematic clades (Hughes & Eastwood, 2006; Madriñán et al., 2013; Givnish et al., 2014). Although numerous studies have documented this pattern, there have been few attempts to ascertain the processes that underlie this megadiversity, especially in the cloud forests where Neotropical bellflower diversity is concentrated. Our study tackles this issue with an interdisciplinary approach that paves the way for future inquiry.
The impact of Andean orogeny and climate change on species diversification
Within the morphologically and ecologically diverse Neotropical bellflower clade, a shift to increased diversification rates on the branch subtending the centropogonid subclade resulted in a cloud forest‐centered radiation with c. 550 extant species. The diversification rate for the centropogonids, estimated to be between 0.65 and 1.42 events Myr–1 per lineage, makes them one of fastest species radiations reported to date (Table 1), with average overall rates of speciation exceeding iconic groups ranging from Espeletia (Madriñán et al., 2013) and hummingbirds (McGuire et al., 2014) in the Andes to the silverswords (Baldwin & Sanderson, 1998) and Drosophilidae of Hawaii (Lapoint et al., 2014).
Table 1.
Net diversification rates in the fastest species radiations in the Tree of Life, including the centropogonids
| Clade | Area | Organism | Species richness | Age (95% CI) | r (ε = 0) | r (ε = 0.5) | r (ε = 0.9) |
|---|---|---|---|---|---|---|---|
| Dianthus (carnations) (Valente et al., 2010) | Eurasia | Plant | 200 | 1.5 (0.9–2.1) | 3.07 (2.12–5.12) | 2.88 (2.06–4.80) | 1.99 (1.42–3.32) |
| African cichlids (Salzburger et al., 2005) | East African Lakes | Fishes | 1800 | 2.4 (1.22–4.02) | 2.83 (1.69–5.76) | 2.71 (1.62–5.34) | 2.14 (1.28–4.22) |
| Lupinus (Andean clade) (Hughes & Eastwood, 2006) | Andes | Plant | 81 | 1.47 (1.18–1.76) | 2.52 (2.10–3.14) | 2.33 (1.95–2.90) | 1.46 (1.22–1.82) |
| Neo‐astragalus (clade F) (Scherson et al., 2008) | Andes | Plant | 90 | 1.89 (1.71–2.07) | 2.04 (1.84–2.23) | 1.87 (1.70–2.06) | 1.19 (1.08–1.31) |
| Zosterops (Moyle et al., 2009) | Cosmopolitan | Birds | 80 | 1.84 (1.40–1.89) | 2.0 (1.95–2.63) | 1.85 (1.80–2.44) | 1.16 (1.13–1.52) |
| Neo‐astragalus (clade G) (Scherson et al., 2008) | Andes | Plant | 15 | 0.98 (0.79–1.17) | 2.06 (1.72–2.55) | 1.82 (1.52–2.26) | 0.84 (0.70–1.04) |
| Gentianella (von Hagen & Kadereit, 2001) | Andes | Plant | 170 | 1.6–3.0 | 1.48–2.78 | 1.39–2.6 | 0.94–1.77 |
| Cistus (Guzmán et al., 2009) | Mediterranean | Plant | 10 | 1.04 (0.66–1.29) | 1.54 (1.25–2.44) | 1.35 (1.09–2.13) | 0.57 (0.46–0.89) |
| Paussus (ant‐nest beetles) (Moore & Robertson, 2014) | Madagascar | Insects | 86 | 2.6 (1.7–3.6) | 1.45 (1.04–2.21) | 1.34 (0.97–2.05) | 0.85 (0.61–1.29) |
| Centropogonids (this study) | Andes | Plant | 556 | 5.02 (3.95–6.13) | 1.12 (0.91–1.42) | 1.06 (0.87–1.35) | 0.79 (0.65–1.00) |
| Espeletiinae (Madriñán et al., 2013) | Andes | Plant | 120 | 4.04 (2.42–5.92) | 1.01 (0.69–1.69) | 0.944 (0.64–1.58) | 0.62 (0.42–1.04) |
| AMC clade of Hawaiian Drosophila (Lapoint et al., 2014) | Hawaii | Insect | 91 | 4.40 (3.45–11.82) | 0.94 (0.35–1.2) | 0.88 (0.33–1.12) | 0.58 (0.22–0.74) |
| Inga (Richardson et al., 2001) | Neotropics | Plant | 300 | 5.9 (2.0–13.4) | 0.85 (0.37–2.5) | 0.8 (0.35–2.36) | 0.57 (0.25–1.69) |
| Ruschioideae (Klak et al., 2003) | South Africa | Plant | 1563 | 3.8–8.7 | 0.7–1.75 | 0.73–1.68 | 0.58–1.31 |
| Bee hummingbirds (McGuire et al., 2014) | Andes | Birds | 36 | 5 | 0.58 | 0.53 | 0.29 |
| Silversword alliance (Baldwin & Sanderson, 1998) | Hawaii | Plants | 28 | 5.2 (4.4–6.0) | 0.51 (0.44–60) | 0.46 (0.40–0.54) | 0.24 (0.21–0.29) |
| Costus subgenus Costus (Kay et al., 2005) | Neotropics | Plant | 51 | 1.5–7.1 | 0.46–2.16 | 0.42–1.98 | 0.24–1.16 |
| Lupinus ‘super radiation’ (Drummond et al., 2012) | Western Americas | Plant | 196 | 5.0–13.2 | 0.35–0.92 | 0.33–0.86 | 0.22–0.59 |
| Hawaiian lobeliads (Givnish et al., 2009) | Hawaii | Plants | 126 | 13.6 (10.49–16.71) | 0.30 (0.25–0.39) | 0.28 (0.23–0.37) | 0.19 (0.15–0.24) |
| Hawaiian Drosophilidae (Lapoint et al., 2014) | Hawaii | Insect | 1000 | 25.15 (23.90–27.46) | 0.25 (0.23–0.26) | 0.24 (0.22–0.25) | 0.18 (0.17–0.19) |
| Plethodon, glutinosus group (Kozak et al., 2006) | North America | Amphibian | 30 | 8–11 | 0.24–0.34 | 0.22–0.31 | 0.12–0.16 |
| Hummingbirds (all) (McGuire et al., 2014) | Neotropics | Bird | 338 | 22.4 (20.3–24.7) | 0.23 (0.21–0.25) | 0.22 (0.20–0.24) | 0.16 (0.14–0.17) |
| Ctenotus + Lerista (Rabosky et al., 2007) | Australia | Reptile | 174 | c. 20 | 0.22 | 0.21 | 0.14 |
Rates of diversification as calculated using the statistic of Magallón & Sanderson (2001) under low, medium, and high estimated extinction (ε) for the centropogonid clade (bold text) rank ordered with other published rapid radiations. All rates in events Myr–1 per lineage. Estimates derived from reported ages and species richness from the relevant literature are cited. When possible, 95% confidence intervals (CIs) for ages are reported, except in cases where sufficient data were unavailable.
In addition, our analyses provide key insights into the abiotic and biotic drivers of rapid diversification in the Andes. Using diversification models that incorporate geological data, we demonstrate that mountain building is correlated with increased diversification rates. Specifically, we demonstrate that the final stages of Andean uplift are strongly associated with the origin and rise of prolific speciation rates in the Neotropical bellflowers (Fig. 2b; Table S4). These rates peaked at a remarkable c. 15 events Myr–1 per lineage, concurrent with the attainment of the maximum past elevations of the Andes. Although not sustained, this diversification rate exceeds the highest reported in plants, including in the Mediterranean carnations (2.2–7.6 events Myr–1 per lineage, Valente et al., 2010) and Andean lupines (2.49–5.21 events Myr–1 per lineage, Hughes & Eastwood, 2006; Drummond et al., 2012). These results are corroborated by the shift to faster speciation rates that was detected by BAMM along the branch that subtends the centropogonid clade, during the period of final Andean uplift. Furthermore, BiSSE analyses bolster our findings that the Andes in general are a major driver of species diversification in this clade: Andean taxa have significantly higher net diversification rates than extra‐Andean taxa (Figs 2e, S5; Table S5). Moreover, species diversification is faster at higher elevations (> 1900 m) (Figs 2f, S5: Table S5). The likely driver of this impressive diversification was not paleoelevation per se (as would be expected under a predominantly vicariant model of speciation), but rather ecological opportunity triggered by the availability of wet montane forests. These habitats emerged and greatly expanded across the eastern flanks of the Amazon in the late Miocene and early Pliocene during periods of active mountain building (van der Hammen & Cleef, 1986).
We further present evidence that past climate change has independently influenced diversification rates in the Neotropical bellflowers. Our paleotemperature‐dependency analysis shows that, while speciation rates remained high but constant, extinction rates are positively correlated with past temperature. Consequently, inferred extinction rates were lowest during the coolest past intervals, and have generally been declining since the origin of the clade, a result that is particularly striking in today's world of anthropogenic warming. Although our proxy of climate change is global, it is almost certain that cooling since the mid‐Miocene has affected the Andes, especially at mid‐to‐high elevations, where Neotropical bellflowers are most diverse. We hypothesize that Neotropical bellflowers evolved their tolerance to cool habitats in southern, perhaps temperate, regions of South America, similar to where Chilean lobelias now occur, before major tropical Andean surface uplift. This climatic preadaptation may explain their northward expansion and decreased extinction rates as this clade exploited cooler niches associated with the rising Andes. These results significantly improve our understanding of the abiotic determinants that have triggered Andean diversification, and collectively indicate that mountain building and climate are key contributors to increases in diversification rate in Neotropical bellflowers, although they have acted on different components of net diversification.
Biotic traits that facilitate mutualisms are associated with elevated rates of net diversification
In addition to orogeny and climate change, our trait‐dependent analyses point to a role of species ecology in influencing species diversification in Neotropical bellflowers (Table S5). Specifically, we identify that mutualistic interactions with seed dispersers and pollinators promote diversification. Here, we demonstrate that bird‐dispersed berries and vertebrate‐pollinated flowers are associated with significantly higher rates of diversification than their counterparts (Figs 2g,h, S4; Table S5). Species with berries have a net diversification rate that is c. 3.5 times higher than capsular species, whose seeds are abiotically dispersed (Figs 2g, S4). As in other groups, fleshy fruits in Neotropical bellflowers are generally associated with densely forested understory habitats where abiotic dispersal is not as effective as in open habitats, where capsular taxa dominate (Lagomarsino et al., 2014). Owing to the limited movement of understory birds, dispersal of fleshy fruits in these habitats is typically across very short distances (Theim et al., 2014). When coupled with rare long‐distance dispersal events, these short dispersal distances can lead to rampant allopatric speciation (Givnish et al., 2005, 2009; Givnish, 2010). Further, the repeated evolution of berries in the centropogonid clade may be indicative of multiple parallel radiations across the Andes, each with rapid speciation rates, consistent with a model of cordilleran diversification, as recently proposed in bromeliads (Givnish et al., 2014).
Pollination by vertebrates appears to have played an even bigger role in species diversification than seed dispersal. Here, we see an approximately sixfold increase in diversification rate relative to invertebrate pollination (Figs 2h, S5; Table S5). Vertebrate pollinators, namely hummingbirds and bats, are hypothesized to be more effective pollinators than invertebrates, in part because they carry higher pollen loads over longer distances (Fleming et al., 2009). The increased speciation rates among vertebrate‐pollinated taxa (Table S5; Fig. S5) are a likely result of floral isolation, which allows prezygotic reproductive isolation to be achieved via an interplay of floral morphology and pollinator behavior (Fulton & Hodges, 1999; Muchhala, 2003; Schiestl & Schlüter, 2009). Supporting this, many Andean bellflower species have sympatric distributions that appear to have been facilitated by either shifts between different classes of vertebrate pollinators (Muchhala, 2006) or character partitioning of floral traits in species that share pollinators (Muchhala & Potts, 2007).
Our analyses of biotic traits indicate that ecological factors, particularly mutualisms with other species, constitute additional triggers of rapid diversification rates in Neotropical bellflowers, as has been demonstrated in extra‐Andean plant groups (Weber & Agrawal, 2014). This effect of species ecology is especially pronounced within the centropogonids, in which more than half of the species are vertebrate‐pollinated and/or produce berries, and in which diversification rates are particularly elevated (Fig. S4). Our results further demonstrate that diversity‐dependent processes did not govern diversification, suggesting that the clade is in its early phases of diversification and is not presently bounded by ecological limits.
A synergy of abiotic and biotic factors drives diversification in Andean cloud forests
Although the interplay between abiotic and biotic drivers has long been recognized as fundamental for regulating diversity, progress toward understanding their interaction has been slow. Our study is among the first to explore this interaction in a clade distributed predominantly in Andean cloud forests. Using the Neotropical bellflowers as a model system, we first provide important evidence that the age and tempo of diversification of cloud forest clades are intermediate between those in the older inter‐Andean valleys and more recent high‐elevation grasslands (Pennington et al., 2010; Särkinen et al., 2012). We then demonstrate that Andean surface uplift and climate are both significantly correlated with the rapid radiation of Neotropical bellflowers. In the context of Andean biodiversity, dramatic geological and environmental transformations, often resulting in habitat barriers and steep environmental gradients (e.g. high ridges and low valleys), characterized the matrix in which plant lineages and their mutualists diversified. Our results indicate that key differences in biotic traits, including fruit type and pollination syndrome, confer enhanced diversification capacity among closely related species co‐occurring in this rapidly changing environment. We hypothesize that preadaptation to cooler climates as the tropical Andes rose allowed this clade to achieve its broad Andean distribution, centered in wet, mid‐elevation cloud forests. This, in turn, precipitated mutualistic interactions with groups showing similarly high Andean diversity (e.g. hummingbirds), further enhancing diversification. Within the context of fruit type, limited dispersibility coupled with rare long‐distance dispersal events in berry‐producing lineages resulted in increased diversification rates, probably via allopatric speciation, as shown in the closely related Hawaiian lobeliads (Givnish et al., 2009). At the same time, an association with vertebrate pollinators resulted in increased rates of speciation via floral isolation. As also demonstrated in bromeliads (Givnish et al., 2014), the repeated evolution of fruit types and pollination syndromes in the complex landscape of the Andes led to multiple parallel radiations in Neotropical bellflowers, which together resulted in a very large, very fast radiation: the centropogonid clade. We argue that while the Andes, and the habitats and habitat heterogeneity created by their orogeny, acted as a montane species pump across many clades, it is an additive interaction of ecological and environmental factors underlies this region's megadiversity.
More generally, rapid plant diversification in the world's montane regions is frequently a product of island‐like ecological opportunity following mountain uplift, further stimulated by evolutionary innovation (Hughes & Eastwood, 2006; Hughes & Atchison, 2015). Our analyses demonstrate that this model applies to Neotropical bellflowers, and is likely relevant to other Andean cloud forest clades. However, why are the Andes more diverse than other montane systems? It is likely that the recency of Andean mountain building partially explains this phenomenon: many clades may still be in the early phases of explosive radiation and thus are not yet greatly impacted by extinction. In addition, the Andes are characterized by deeply dissected topography, including some of the steepest environmental gradients known (Hughes & Atchison, 2015). These characteristics, which almost certainly enhance the island‐like nature of this system and further intensify ecological opportunity, are shared with the Hengduan Mountains (Favre et al., 2014), another of the world's biodiversity hotspots (Myers et al., 2000; Kreft & Jetz, 2007). We are likely to gain insight into why the Andes are so diverse by comparing their clades’ diversification histories with those in other species‐rich montane regions. Our framework, including the integration of climatic, paleoaltimetry, and trait data, can be used towards this end.
Author contributions
L.P.L., F.L.C., A.A. and C.C.D. designed the study; A.M. compiled the paleoaltimetry data; L.P.L. assembled all other data; L.P.L. and F.L.C. analyzed data; and L.P.L. and C.C.D. wrote the paper, with significant contributions from F.L.C. and A.A. and revisions from A.M.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Time‐calibrated phylogeny of Neotropical Campanulaceae, with outgroup representatives of Lobelioideae and Campanuloideae collapsed.
Fig. S2 Time‐calibrated phylogeny of Neotropical Campanulaceae, with the ingroup collapsed and showing outgroup relationships.
Fig. S3 Phylogram showing molecular‐proportional branch lengths, illustrating substitution rate heterogeneity within clade.
Fig. S4 Diversification rate analysis for Neotropical Campanulaceae in BAMM.
Fig. S5 Posterior distributions of trait‐dependent speciation, extinction, transition, and net diversification rates as estimated by BiSSE.
Table S1 Taxon sampling including voucher information, country of origin, and GenBank accession numbers for seven plastid loci
Table S2 Divergence time estimation to assess sensitivity to dating method, calibration selection, and branch length heterogeniety
Table S3 Trait codings for the four traits used in our BiSSE analyses
Table S4 Paleoenvironmental‐dependent diversification analyses using paleoaltimetry and Cenozoic climate data
Table S5 Model comparison for the four BiSSE analyses presented in the main text
Table S6 Correlated evolution of binary traits using BayesTraits
Table S7 Model comparison for the two diversity‐dependence analyses presented, with mean parameter estimates for each model
Acknowledgements
We thank members of the Davis and Antonelli research groups for helpful feedback, and Suzette Flantua, Elisabeth Forrestel, Carina Hoorn, Daniel Santamaría‐Aguilar, and Zhenxiang Xi for insightful conversations. The directors and curators at the following herbaria provided important access to their collections: BOLV, CR, GB, GH, INB, LPB, MO, MOL, NY, PMA, SCZ, SMF and USZ (acronyms follow the Index Herbariorum, Theirs et al., 2013 (continuously updated)). We are also very thankful for the detailed and insightful comments from two anonymous reviewers and the editors. Funding was provided by a National Science Foundation Doctoral Dissertation Improvement Grant DEB‐1210401 to L.P.L. and C.C.D.; by the Swedish Research Council (B05 69601), the European Research Council under the European Union's Seventh Framework Programme (FP/2007‐2013, ERC Grant Agreement no. 331024), and a Wallenberg Academy Fellowship to A.A.; by the Carl Tryggers Stiftelse (CTS 12:14) and a Marie Curie Actions (BIOMME project, IOF‐627684) to F.L.C.; and by the Department of Organismic and Evolutionary Biology, the Botanical Society of America, the American Society of Plant Taxonomists, the Arnold Arboretum's Deland Award, the Explorer's Club, and the Rockefeller Center for Latin American Studies to L.P.L.
See also the Commentary on this article by Hughes, 210: 1152–1154.
References
- Abrahamczyk S, Souto‐Vilarós D, Renner SS. 2014. Escape from extreme specialization: passionflowers, bats and the sword‐billed hummingbird. Proceedings of the Royal Society of London. Series B: Biological Sciences 281: 20140888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A. 2009. Have giant lobelias evolved several times independently? Life form shifts and historical biogeography of the cosmopolitan and highly diverse subfamily Lobelioideae (Campanulaceae). BMC Biology 7: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A. 2015. Biodiversity: multiple origins of mountain life. Nature 524: 300–301. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Sanmartín I. 2011. Why are there so many plant species in the Neotropics? Taxon 60: 403–414. [Google Scholar]
- Armijo R, Lacassin R, Coudurier‐Curveur A, Carrizo D. 2015. Coupled tectonic evolution of Andean orogeny and global climate. Earth‐Science Reviews 143: 1–35. [Google Scholar]
- Baldwin BG, Sanderson MJ. 1998. Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proceedings of the National Academy of Sciences, USA 95: 9402–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthlott W, Mutke J, Rafiqpoor D, Kier G, Kreft H. 2005. Global centers of vascular plant diversity. Nova Acta Leopoldina 92: 61–83. [Google Scholar]
- Beaulieu JM, O'Meara BC. 2015. Extinction can be estimated from moderately sized molecular phylogenies. Evolution 69: 1036–1043. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, O'Meara BC, Crane P, Donoghue MJ. 2015. Heterogenous rates of molecular evolution and diversification could explain the Triassic age estimate for angiosperms. Systematic Biology 64: 869–878. [DOI] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re‐revisited. American Journal of Botany 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Blois JL, Zarnetske PL, Fitzpatrick MC, Finnegan S. 2013. Climate change and the past, present, and future of biotic interactions. Science 341: 499–504. [DOI] [PubMed] [Google Scholar]
- Bouchenak‐Khelladi Y, Onstein RE, Xing Y, Schwery O, Linder HP. 2015. On the complexity of triggering evolutionary radiations. New Phytologist 207: 313–326. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellinese N, Smith SA, Edwards EJ, Kim S‐T, Haberle RC, Avramakis M, Donoghue MJ. 2009. Historical biogeography of the endemic Campanulaceae of Crete. Journal of Biogeography 36: 1253–1269. [Google Scholar]
- Clague D. 1996. Growth and subsidence of the Hawaiian‐Emperor volcanic chain In: Keast A, Mille S, eds. The origin and evolution of Pacific island biotas, New Guinea to Eastern Polynesia: patterns and processes. Amsterdam, the Netherlands: SPB Academic Press, 35–50. [Google Scholar]
- Colwell RK, Betts BJ, Bunnell P, Carpenter FL, Feinsinger P. 1974. Competition for the nectar of Centropogon valerii by the hummingbird Colibri thalassinus and the flower‐piercer Diglossa plumbea, and its evolutionary implications. The Condor 76: 447–452. [Google Scholar]
- Condamine FL, Rolland J, Morlon H. 2013. Macroevolutionary perspectives to environmental change. Ecology Letters 16: 72–85. [DOI] [PubMed] [Google Scholar]
- Crowl AA, Mavrodiev E, Mansion G, Haberle R, Pistarino A, Kamari G, Phitos D, Borsch T, Cellinese N. 2014. Phylogeny of Campanuloideae (Campanulaceae) with emphasis on the utility of nuclear pentatricopeptide repeat (PPR) genes. PLoS ONE 9: e94199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP, Midford PE, Maddison W. 2013. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evolutionary Biology 13: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MJ, Sanderson MJ. 2015. Confluence, synnovation, and depauperons in plant diversification. New Phytologist 207: 260–274. [DOI] [PubMed] [Google Scholar]
- Drummond CS, Eastwood RJ, Miotto STS, Hughes CE. 2012. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Systematic Biology 61: 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers TA, Poulsen CJ. 2009. Influence of Andean uplift on climate and paleoaltimetry estimates. Earth and Planetary Science Letters 281: 238–248. [Google Scholar]
- Elias M, Joron M, Willmott K, Silva‐Brandão KL, Kaiser V, Arias CF, Gomez Piñerez LM, Uribe S, Brower VZ, Freitas VL et al 2009. Out of the Andes: patterns of diversification in clearwing butterflies. Molecular Ecology 18: 1716–1729. [DOI] [PubMed] [Google Scholar]
- Etienne RS, Haegeman B, Stadler T, Aze T, Pearson PN, Purvis A, Phillimore AB. 2012. Diversity‐dependence brings molecular phylogenies closer to agreement with the fossil record. Proceedings of the Royal Society of London. Series B: Biological Sciences 279: 1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezard THG, Aze T, Pearson PN, Purvis A. 2011. Interplay between changing climate and species’ ecology drives macroevolutionary dynamics. Science (New York, N.Y.) 332: 349–351. [DOI] [PubMed] [Google Scholar]
- Faegri K, van der Pijl L. 1979. The principles of pollination ecology. Oxford, UK: Pergamon Press. [Google Scholar]
- Favre A, Päckert M, Pauls SU, Jähnig SC, Uhl D, Michalak I, Muellner‐Riehl AN. 2014. The role of the uplift of the Qinghai‐Tibetan Plateau for the evolution of Tibetan biotas. Biological Reviews 90: 236–253. [DOI] [PubMed] [Google Scholar]
- FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution 3: 1084–1092. [Google Scholar]
- Fleming TH, Geiselman C, Kress WJ. 2009. The evolution of bat pollination: a phylogenetic perspective. Annals of Botany 104: 1017–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton M, Hodges SA. 1999. Floral isolation between Aquilegia formosa and Aquilegia pubescens . Proceedings of the Royal Society of London. Series B: Biological Sciences 266: 2247–2252. [Google Scholar]
- Garzione CN, Auerbach DJ, Jin‐Sook Smith J, Rosario JJ, Passey BH, Jordan TE, Eiler JM. 2014. Clumped isotope evidence for diachronous surface cooling of the Altiplano and pulsed surface uplift of the Central Andes. Earth and Planetary Science Letters 393: 173–181. [Google Scholar]
- Garzione CN, Hoke GD, Libarkin JC, Withers S, MacFadden B, Eiler J, Ghosh P, Mulch A. 2008. Rise of the Andes. Science (New York, N.Y.) 320: 1304–1307. [DOI] [PubMed] [Google Scholar]
- Garzione CN, Molnar P, Libarkin JC, MacFadden BJ. 2006. Rapid late Miocene rise of the Bolivian Altiplano: evidence for removal of mantle lithosphere. Earth and Planetary Science Letters 241: 543–556. [Google Scholar]
- Gentry AH. 1982. Neotropical floristic diversity: phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Annals of the Missouri Botanical Garden 69: 557–593. [Google Scholar]
- Givnish TJ. 2010. Ecology of plant speciation. Taxon 59: 1326–1366. [Google Scholar]
- Givnish TJ, Barfuss MHJ, Van Ee B, Riina R, Schulte K, Horres R, Gonsiska PA, Jabaily RS, Crayn DM, Smith JAC et al 2014. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Molecular Phylogenetics and Evolution 71: 55–78. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, Henss JM, Smith JF, Wood KR, Sytsma KJ. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proceedings of the Royal Society of London. Series B: Biological Sciences 276: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Pires JC, Graham SW, McPherson MA, Prince LM, Patterson TB, Rai HS, Roalson EH, Evans TM, Hahn WJ et al 2005. Repeated evolution of net venation and fleshy fruits among monocots in shaded habitats confirms a priori predictions: evidence from an ndhF phylogeny. Proceedings of the Royal Society of London. Series B: Biological Sciences 272: 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory‐Wodzicki KM. 2000. Uplift history of the Central and Northern Andes: a review. Geological Society of America Bulletin 112: 1091–1105. [Google Scholar]
- Guzmán B, Lledó MD, Vargas P. 2009. Adaptive radiation in Mediterranean Cistus (Cistaceae). PLoS ONE 4: e6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hagen KB, Kadereit JW. 2001. The phylogeny of Gentianella (Gentianaceae) and its colonization of the southern hemisphere as revealed by nuclear and chloroplast DNA sequence variation. Organisms Diversity & Evolution 1: 61–79. [Google Scholar]
- van der Hammen T, Cleef AM. 1986. Development of the high Andean páramo flora and vegetation In: Vuillemier F, Monasterio M, eds. High altitude tropical biogeography. New York, NY, USA: Oxford University Press, 153–201. [Google Scholar]
- Hoorn C, Mosbrugger V, Mulch A, Antonelli A. 2013. Biodiversity from mountain building. Nature Geoscience 6: 154. [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez‐Meseguer A, Anderson CL, Figueiredo JP et al 2010. Amazonia through time: andean uplift, climate change, landscape evolution, and biodiversity. Science (New York, N.Y.) 330: 927–931. [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences, USA 103: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CE, Atchison GW. 2015. The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytologist 207: 275–282. [DOI] [PubMed] [Google Scholar]
- Hughes CE, Nyffeler R, Linder HP. 2015. Evolutionary plant radiations: where, when, why and how? New Phytologist 207: 249–253. [DOI] [PubMed] [Google Scholar]
- Kay KM, Reeves PA, Olmstead RG, Schemske DW. 2005. Rapid speciation and the evolution of hummingbird pollination in Neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. American Journal of Botany 92: 1899–1910. [DOI] [PubMed] [Google Scholar]
- Klak C, Khunou A, Reeves G, Hedderson T. 2003. A phylogenetic hypothesis for the Aizoaceae (Caryophyllales) based on four plastid DNA regions. American Journal of Botany 90: 1433–1445. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Weisrock DW, Larson A. 2006. Rapid lineage accumulation in a non‐adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon). Proceedings of the Royal Society of London. Series B: Biological Sciences 273: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft H, Jetz W. 2007. Global patterns and determinants of vascular plant diversity. Proceedings of the National Academy of Sciences, USA 104: 5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino LP, Antonelli A, Muchhala N, Timmermann A, Mathews S, Davis CC. 2014. Phylogeny, classification, and fruit evolution of the species‐rich Neotropical bellflowers (Campanulaceae: Lobelioideae). American Journal of Botany 101: 2097–2112. [DOI] [PubMed] [Google Scholar]
- Łancucka‐Srodoniowa M. 1977. New herbs described from the Tertiary of Poland. Acta Palaeobotanica 18: 37–44. [Google Scholar]
- Lapoint RT, Magnacca KN, O'Grady PM. 2014. Phylogenetics of the Antopocerus‐Modified Tarsus clade of Hawaiian Drosophila: diversification across the Hawaiian Islands. PLoS ONE 9: e113227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leier A, McQuarrie N, Garzione C, Eiler J. 2013. Stable isotope evidence for multiple pulses of rapid surface uplift in the Central Andes, Bolivia. Earth and Planetary Science Letters 371–372: 49–58. [Google Scholar]
- Maddison WP, FitzJohn RG. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Systematic Biology 64: 127–136. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Systematic Biology 56: 701–710. [DOI] [PubMed] [Google Scholar]
- Madriñán S, Cortés AJ, Richardson JE. 2013. Páramo is the world's fastest evolving and coolest biodiversity hotspot. Frontiers in Genetics 4: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Sanderson MJ. 2001. Absolute diversification rates in angiosperm clades. Evolution 55: 1762–1780. [DOI] [PubMed] [Google Scholar]
- McGuire JA, Witt CC, Altshuler DL, Remsen JV. 2007. Phylogenetic systematics and biogeography of hummingbirds: Bayesian and maximum likelihood analyses of partitioned data and selection of an appropriate partitioning strategy. Systematic Biology 56: 837–856. [DOI] [PubMed] [Google Scholar]
- McGuire JA, Witt CC, Remsen JV, Corl A, Rabosky DL, Altshuler DL, Dudley R. 2014. Molecular phylogenetics and the diversification of hummingbirds. Current Biology 24: 910–916. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT, Hendriks KP, Beentjes KK, Mennes CB, Becking LE, Peijnenburg KTC, Afendy A, Arumugam N, de Boer H, Biun A et al 2015. Evolution of endemism on a young tropical mountain. Nature 524: 347–350. [DOI] [PubMed] [Google Scholar]
- Moore W, Robertson JA. 2014. Explosive adaptive radiation and extreme phenotypic diversity within ant‐nest beetles. Current Biology 24: 1–5. [DOI] [PubMed] [Google Scholar]
- Morlon H, Lewitus E, Condamine FL, Manceau M, Clavel J, Drury J. 2016. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods in Ecology and Evolution, doi: 10.1111/2041‐210X.12526. [Google Scholar]
- Morlon H, Parsons TL, Plotkin JB. 2011. Reconciling molecular phylogenies with the fossil record. Proceedings of the National Academy of Sciences, USA 108: 16327–16332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle RG, Filardi CE, Smith CE, Diamond J. 2009. Explosive Pleistocene diversification and hemispheric expansion of a ‘great speciator’. Proceedings of the National Academy of Sciences, USA 106: 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhala N. 2003. Exploring the boundary between pollination syndromes: bats and hummingbirds as pollinators of Burmeistera cyclostigmata and B. tenuiflora (Campanulaceae). Oecologia 134: 373–380. [DOI] [PubMed] [Google Scholar]
- Muchhala N. 2006. The pollination biology of Burmeistera (Campanulaceae): specialization and syndromes. American Journal of Botany 93: 1081–1089. [DOI] [PubMed] [Google Scholar]
- Muchhala N, Potts MD. 2007. Character displacement among bat‐pollinated flowers of the genus Burmeistera: analysis of mechanism, process and pattern. Proceedings of the Royal Society of London. Series B: Biological Sciences 274: 2731–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulch A. 2016. Stable isotope paleoaltimetry and the evolution of landscapes and life. Earth and Planetary Science Letters 433: 180–191. [Google Scholar]
- Mulch A, Uba CE, Strecker MR, Schoenberg R, Chamberlain CP. 2010. Late Miocene climate variability and surface elevation in the central Andes. Earth and Planetary Science Letters 290: 173–182. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Nee S, May RM, Harvey PH. 1994. The reconstructed evolutionary process. Philosophical transactions of the Royal Society of London Series B: Biological Sciences 344: 305–311. [DOI] [PubMed] [Google Scholar]
- Ng J, Smith SD. 2014. How traits shape trees: new approaches for detecting character state‐dependent lineage diversification. Journal of Evolutionary Biology 27: 2035–2045. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible‐jump Markov chain Monte Carlo. The American Naturalist 167: 808–825. [DOI] [PubMed] [Google Scholar]
- Parra M, Mora A, Jaramillo C, Strecker MR, Sobel ER, Quiroz L, Rueda M, Torres V. 2009. Orogenic wedge advance in the northern Andes: evidence from the Oligocene‐Miocene sedimentary record of the Medina Basin, Eastern Cordillera, Colombia. Geological Society of America Bulletin 121: 780–800. [Google Scholar]
- Pennington RT, Lavin M, Sarkinen T, Lewis GP, Klitgaard BB, Hughes CE. 2010. Contrasting plant diversification histories within the Andean biodiversity hotspot. Proceedings of the National Academy of Sciences, USA 107: 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quental TB, Marshall CR. 2010. Diversity dynamics: molecular phylogenies need the fossil record. Trends in Ecology & Evolution 25: 434–441. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. 2010. Extinction rates should not be estimated from molecular phylogenies. Evolution; International Journal of Organic Evolution 64: 1816–1824. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity‐dependence on phylogenetic trees. PLoS ONE 9: e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. 2015. Challenges in the estimation of extinction from molecular phylogenies: a response to Beaulieu and O'Meara. Evolution 70: 218–228. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Donnellan SC, Talaba AL, Lovette IJ. 2007. Exceptional among‐lineage variation in diversification rates during the radiation of Australia's most diverse vertebrate clade. Proceedings of the Royal Society of London. Series B: Biological Sciences 274: 2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Goldberg EE. 2015. Model inadequacy and mistaken inferences of trait‐dependent speciation. Systematic Biology 64: 340–355. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, Huang H, Larson JG. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees (Kembel S, ed.). Methods in Ecology and Evolution 5: 701–707. [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. [WWW document] URL http://beast.bio.ed.ac.uk/Tracer.
- Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. 2001. Rapid diversification of a species‐rich genus of neotropical rain forest trees. Science (New York, N.Y.) 293: 2242–2245. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Mack T, Verheyen E, Meyer A. 2005. Out of Tanganyika: genesis, explosive speciation, key‐innovations and phylogeography of the haplochromine cichlid fishes. BMC Evolutionary Biology 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Särkinen T, Pennington RT, Lavin M, Simon MF, Hughes CE. 2012. Evolutionary islands in the Andes: persistence and isolation explain high endemism in Andean dry tropical forests. Journal of Biogeography 39: 884–900. [Google Scholar]
- Scherson RA, Vidal R, Sanderson MJ. 2008. Phylogeny, biogeography, and rates of diversification of New World Astragalus (Leguminosae) with an emphasis on South American radiations. American Journal of Botany 95: 1030–1039. [DOI] [PubMed] [Google Scholar]
- Schiestl FP, Schlüter PM. 2009. Floral isolation, specialized pollination, and pollinator behavior in orchids. Annual Review of Entomology 54: 425–446. [DOI] [PubMed] [Google Scholar]
- Schmidt‐Lebuhn AN, Kessler M, Hensen I. 2007. Hummingbirds as drivers of plant speciation? Trends in Plant Science 12: 329–331. [DOI] [PubMed] [Google Scholar]
- Schwery O, Onstein RE, Bouchenak‐Khelladi Y, Xing Y, Carter RJ, Linder HP. 2015. As old as the mountains: the radiations of the Ericaceae. New Phytologist 207: 355–367. [DOI] [PubMed] [Google Scholar]
- Sempere T, Hartley A, Roperch P. 2006. Comment on ‘Rapid uplift of the Altiplano revealed through 13C‐18O bonds in paleosol carbonates’. Science (New York, N.Y.) 314: 760. [DOI] [PubMed] [Google Scholar]
- Smith JF. 2001. High species diversity in fleshy‐fruited tropical understory plants. The American Naturalist 157: 646–653. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science (New York, N.Y.) 322: 86–89. [DOI] [PubMed] [Google Scholar]
- Smith SA, O'Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Spriggs EL, Clement WL, Sweeney PW, Madriñán S, Edwards EJ, Donoghue MJ. 2015. Temperate radiations and dying embers of a tropical past: the diversification of Viburnum. New Phytologist 207: 340–354. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein BA. 1992. Sicklebill hummingbirds, ants, and flowers: plant–animal interactions and evolutionary relationships in Andean Lobeliaceae. BioScience 42: 27–33. [Google Scholar]
- Theim TJ, Shirk RY, Givnish TJ. 2014. Spatial genetic structure in four understory Psychotria species (Rubiaceae) and implications for tropical forest diversity. American Journal of Botany 101: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Thiers B. 2013. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. [WWW document] URL http://sweetgum.nybg.org/ih (continuously updated).
- Valente LM, Etienne RS, Phillimore AB. 2014. The effects of island ontogeny on species diversity and phylogeny. Proceedings of the Royal Society of London. Series B: Biological Sciences 281: 20133227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LM, Savolainen V, Vargas P. 2010. Unparalleled rates of species diversification in Europe. Proceedings of the Royal Society of London. Series B: Biological Sciences 277: 1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CE, Harmon LJ, Seehausen O. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487: 366–369. [DOI] [PubMed] [Google Scholar]
- Walker JD, Geissman JW, Bowring SA, Babcock LE. 2012. Geologic Time Scale v. 4.0. Geological Society of America, doi: 10.1130/2012.CTS004R3C.
- Weber MG, Agrawal AA. 2014. Defense mutualisms enhance plant diversification. Proceedings of the National Academy of Sciences, USA 111: 16442–16447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer FE. 1943. Campanulaceae‐Lobelioideae. I. Teil In: Mansfeld R, ed. Das Pflanzenreich IV.276b. Leipzig, Germany: Wilhem Engelmann, 1–260. [Google Scholar]
- Wimmer FE. 1953. Campanulaceae‐Lobelioideae, II. Teil In: Mansfeld R, ed. Das Pflanzenreich IV.276b. Berlin, Germany: Akademie‐Verlag, 261–814. [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE. 2008. An early Cenozoic perspective on greenhouse warming and carbon‐cycle dynamics. Nature 451: 279–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Time‐calibrated phylogeny of Neotropical Campanulaceae, with outgroup representatives of Lobelioideae and Campanuloideae collapsed.
Fig. S2 Time‐calibrated phylogeny of Neotropical Campanulaceae, with the ingroup collapsed and showing outgroup relationships.
Fig. S3 Phylogram showing molecular‐proportional branch lengths, illustrating substitution rate heterogeneity within clade.
Fig. S4 Diversification rate analysis for Neotropical Campanulaceae in BAMM.
Fig. S5 Posterior distributions of trait‐dependent speciation, extinction, transition, and net diversification rates as estimated by BiSSE.
Table S1 Taxon sampling including voucher information, country of origin, and GenBank accession numbers for seven plastid loci
Table S2 Divergence time estimation to assess sensitivity to dating method, calibration selection, and branch length heterogeniety
Table S3 Trait codings for the four traits used in our BiSSE analyses
Table S4 Paleoenvironmental‐dependent diversification analyses using paleoaltimetry and Cenozoic climate data
Table S5 Model comparison for the four BiSSE analyses presented in the main text
Table S6 Correlated evolution of binary traits using BayesTraits
Table S7 Model comparison for the two diversity‐dependence analyses presented, with mean parameter estimates for each model


