Abstract
Aim
Biodiversity losses under the species level may have been severely underestimated in future global climate change scenarios. Therefore, it is important to characterize the diversity units at this level, as well as to understand their ecological responses to climatic forcings. We have chosen an endemic rodent from a highly endangered ecogeographic area as a model to look for distributional responses below the species level: Phyllotis darwini.
Location
The central Chile biodiversity hotspot: This area harbours a high number of endemic species, and it is known to have experienced vegetational displacements between two mountain systems during and after the Last Glacial Maximum.
Methods
We have characterized cryptic lineages inside P. darwini in a classic phylogeographic approach; those intraspecific lineages were considered as relevant units to construct distribution models at Last Glacial Maximum and at present, as border climatic conditions. Differences in distribution between border conditions for each lineage were interpreted as distributional responses to post-glacial climate change.
Results
The species is composed of two major phylogroups: one of them has a broad distribution mainly across the valley but also in mountain ranges, whereas the other displays a disjunct distribution across both mountain ranges and always above 1500 m. The lineage distribution model under LGM climatic conditions suggests that both lineages were co-distributed in the southern portion of P. darwini’s current geographic range, mainly at the valley and at the coast.
Main conclusions
Present distribution of lineages in P. darwini is the consequence of a cryptic distributional response to climate change after LGM: postglacial northward colonization, with strict altitudinal segregation of both phylogroups.
Keywords: Biodiversity hotspots, central Chile, Phyllotis darwini, distribution models, phylogeography, cryptic diversity, Last Glacial Maximum, past climate change
INTRODUCTION
Climate regimes may affect species distribution and therefore future climate change could potentially induce geographical range dynamics such as contraction, expansion or geographical range shifts (Walther et al., 2002; Pearson & Dawson, 2003; Summers et al., 2012). It is also agreed that the climatic impact on species distribution can be extrapolated in community and ecosystem shifts (Walther et al., 2002; Hamann & Wang, 2006). For these reasons, the species level might be critical for conservation issues in a global climate change scenario (GCC).
At the species level, climate change is expected to affect the geographical range mainly through physiological restrictions as temperature and precipitation tolerances in conjunction with species dispersal abilities (Walther et al., 2002). Altogether, those factors may determine species ability to keep up with climate change. From an ecological perspective, expected species responses to climate change are i) to tolerate or spread in the new climatic set up (either by physiological tolerance or phenotypic plasticity) ii) to change its distribution in order to catch up the new climate regime iii) to become extinct (Pettorelli, 2012). Range shifts may change the distribution of genetic diversity and range contractions will most likely reduce the genetic diversity (Alsos et al., 2012; Pauls et al., 2012). Nevertheless, the emphasis on the role of evolution in species responses to climate change has been usually focused on evolutionary adaptations and the relationship between species adaptation speed and climatic change rate (Hoffmann & Sgrò, 2011).
It has been recently proposed about the need to consider GCC effects on biodiversity below the species level, other than the usually advocated adaption potential and phenotypical plasticity issues. There exists a severe lack of studies on GCC effects on biodiversity at the intraspecific genetic level. This is highly evident if we compare it with the vast amount of publications at the ecosystem, community and species levels (Fraser & Bernatchez, 2001), despite the general agreement on the importance of Evolutionary Significant Units (ESUs) for conservation planning. Bálint et al. (2011) demonstrated that species with strong population genetic structure will face massive losses of diversity at the intraspecific level in a future GCC scenario; these authors refer to this as “cryptic diversity”. This term has been coined to appoint for biodiversity units below the species level, such as haplotypes, ESUSs and Molecular Operational Taxonomic Units (MOTUs). The term should not be confused with the classical concept of cryptic species. Therefore, conservation strategies based only on species, community, or ecosystem organization levels could be an oversimplified approach.
For the above reasons, it might be critical for future conservation planning not just to quantify the amount of cryptic diversity at risk, but to understand what could be the ecological and evolutionary responses of ESUs below the species level to GCC. There exists an important number of publications trying to understand the relationship between evolutionary and ecological species responses to climate change, mainly through phylogeographic information and species distribution models (SDMs; Carstens & Richards, 2007; Waltari et al., 2007; Kozak et al., 2008; Provan & Bennett, 2008; Cordellier & Pfenninger, 2009; Marske et al., 2009; Waltari & Guralnick, 2009; Buckley et al., 2010; Lim et al., 2010; Allal et al., 2011; Eckert, 2011; Gugger et al., 2011; Marske et al., 2011; Svenning et al., 2011; Marske et al., 2012; Qi et al., 2012). However, approaches using intraspecific phylogenetic information to build lineage distribution models (lineages below the species level) are less frequent, even though this approach has been used with success in systematics for species delimitation (Raxworthy et al., 2007; Rissler & Apodaca, 2007; Engelbrecht et al., 2011; du Toit et al., 2012; Florio et al., 2012). Studies evaluating the relationship between lineage formation and variation of the ecological niche in the Peromyscus maniculatus species group (Kalkvik et al., 2011) demonstrated that the majority of genetic lineages within species occupy distinct environmental niches. In another integrated approach, Fontanella et al. (2012) concluded that two main haploclades in the lizard Liolaemus petrophilus shared different distributional responses to climate change during the Pleistocene. Therefore, realistic predictions of range shifts for future climate change scenarios should consider phylogenetic information in order to perform lineage-specific distribution models, because species might not necessarily respond as an ecological unit to future GCC.
In this work, we evaluated the geographic range responses of an endemic rodent species from a biodiversity hotspot to past climate change. We specifically evaluated the responses to climate change after Last Glacial Maximum (LGM), integrating intraspecific diversity information and intraspecific lineage distribution models for present and past climatic conditions. This information is valuable to understand what should be the relevant implications of considering cryptic diversity in conservation strategies planning, compared to approaches based only in species or ecosystem levels. This information should be relevant for conservation strategies, by setting more realistic assumptions on species responses to GCC. We chose a sigmodontine rodent species as study model, endemic to a biodiversity hotspot area in central Chile because: i) We wanted to know how vertebrate species have responded to climate change in a currently endangered area; ii) There is abundant evidence showing that general genetic structure of species in a hotspot shows features that are characteristic of the region and its particular evolutionary and geo-climatic history (Calsbeek et al. 2003, Tolley et al., 2006, Carnaval et al. 2009, Tolley et al. 2009, Verboom et al. 2009a, Verboom et al., 2009b). Former evidence suggest that biodiversity hotspots may be appropriate systems to study the relationship between climate dynamics and genetic differentiation in endangered areas.
Sixty one species of endemic vertebrates have been described in the hotspot of central Chile (Simonetti, 1999). The latter area is one of the 25 areas proposed as hotspots based on endemism and threat of habitats (Myers et al., 2000). Of the 150 mammal species described for Chile, 56 are distributed in central Chile with nine endemics, with rodents representing the greatest diversity (12 species; Palma, 2007). One of these endemics is the sigmodontine Phyllotis darwini (Waterhouse, 1837), the Darwin’s leaf-eared mouse distributed from Atacama to Bío Bío regions (27 to 36° S), and from coastal areas up to 2000 m (Redford & Eisenberg, 1992). The species is part of the tribe Phyllotini (46 species, 19 in Chile; Spotorno et al., 2001) with an hypothesized origin in the southern Altiplano (Reig, 1986; Spotorno et al., 2001) and none recognized subspecies (Walker at al. 1984, Steppan et al. 2007) .
We expected that potential genetic splits in Phyllotis darwini might be associated to the altitudinal gradient given by the Andes, the valleys, and the Coastal Cordillera. We also expected that the history of the species’ geographic range shows signatures of the LGM in central Chile. This event was chosen to test the hypothesis because it is known that Quaternary glaciations were one of the most important factors in determining current genetic structure of many populations, species and communities (Hewitt, 2000). Although central Chile was not extensively glaciated at LGM, the glacial advanced northwards through the Andes (Clapperton, 1990; Clapperton, 1994) descending to about 1,100 m with a subsequent drop in the temperature and an increased rainfall (Heusser, 1983; Heusser, 1990; Lamy et al., 1999). The latter scenario may have triggered an Andean vegetational shift downwards to the valley during glacial periods, and a subsequent movement upwards during interglacial. These shifts may have given rise to current biogeographical insulas in the Andes and the Coastal cordilleras of central Chile (Darwin, 1859; Simpson, 1983; Heusser, 1990; Villagrán & Armesto, 1991; Villagrán & Hinojosa, 2005). In addition, it has been suggested that lizards and rodents species diversity in central Chile is explained by different speciation modes, as a result of differential interaction between mountain geography, Quaternary glaciations and ecological features (Fuentes & Jaksic, 1979).
Having the former scenario, the goals of this paper were i) to evaluate the genetic and phylogenetic structure of Phyllotis darwini to look for cryptic intraspecific lineages; ii) to build lineage distribution models at present and at LGM iii) to investigate if species have behaved as a single distributional unit in response to past climate change. To these goals, we sequenced the Hypervariable domain II of the mitochondrial DNA (mtDNA) control region in Phyllotis darwini to recover the morphologically cryptic phylogenetic lineages, and build distribution models for each lineage at present and at LGM. We expected to discern if Phyllotis darwini have shifted its geographical range as a single ecological unit since LGM to present or if there exists cryptic phylogenetic lineages which shared independent distributional responses to past climate change after glacial retreat.
METHODS
(Because of space reasons, fully detailed Methodology is available in AppendixS2).
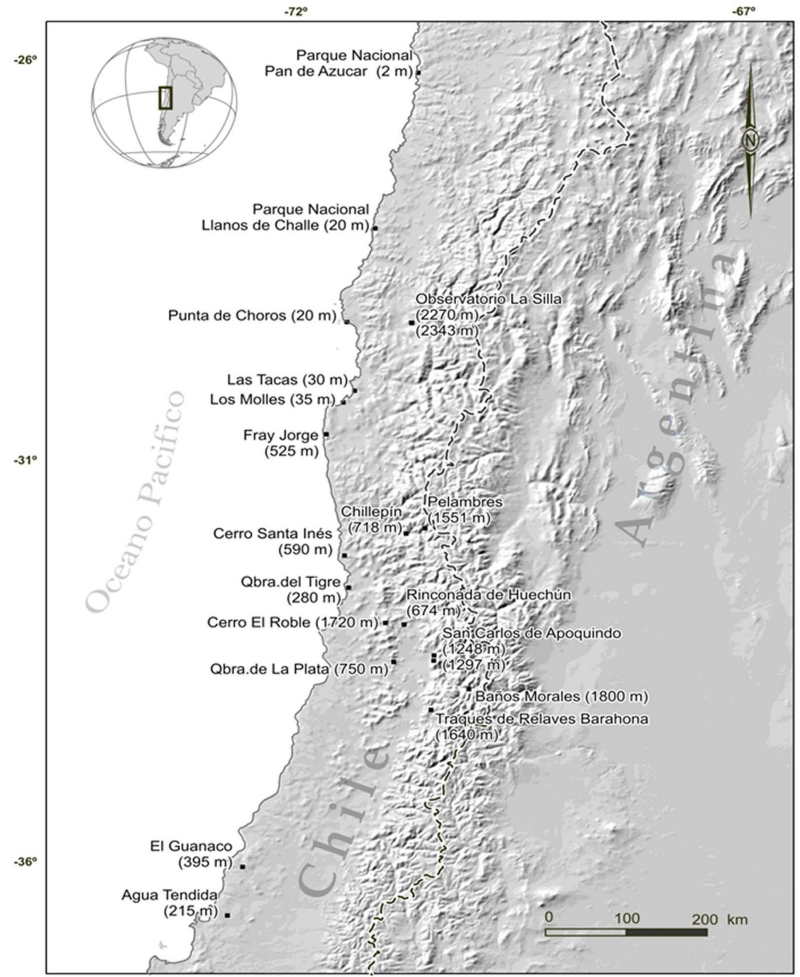
Sixty eight specimens of Phyllotis darwini were analyzed representing 18 localities across central Chile (Fig. 1). The list of specimens, localities and abbreviations is given in Appendix S1. We followed the ASM guidelines during the collection and care of the animals used in this work (Sikes & Gannon, 2011).
Figure 1.
Geographic distribution of localities sampled in Phyllotis darwini distributional range.
We used frozen liver for DNA extraction. The DNA extraction in Phyllotis magister (used as outgroup) was performed from ethanol preserved ear tissue. We sequenced the Hypervariable domain II (HV2) of the mitochondrial DNA (mtDNA) control region in 72 individuals.
Genetic data was analyzed in a phylogeographic context, using the software BAYES PHYLOGENIES ( Pagel & Meade, 2004) for phylogenetic reconstruction, GENELAND (Guilliot et al., 2005) for genetic units delimitation and BEAST v.1.7.4 (Drummond et al., 2006) for time calibrations.
Climatic niche models were built in the software package MAXENT v. 3.2.1 (Phillips et al., 2006) (current and LGM climatic conditions). Trapping coordinates of each individual captured for DNA extraction were used as presence points.
RESULTS
Molecular marker
The Tajima’s D value of 0.54865 was not significative (p > 0.10), thus the neutral mutation hypothesis could not be rejected for the HVII domain. The Index of Substitution Saturation (Iss) obtained with the Xia test (Xia et al., 2003b) was significantly lower than the Critical Index of Substitution Saturation Value (Iss.c), with Iss=0.4211 Iss.c=0.7035 and a p value of 0.0053. Therefore, the molecular marker shows a small saturation, meeting the neutrality and non-saturation assumptions.
Thirty seven haplotypes were recovered representing 68 sequences of P. darwini, with 32 private haplotypes, whereas 27 were represented by a single individual. Among the five most frequent haplotypes, three were shared haplotypes and two had a broad geographic distribution encompassing several localities: DIV1 (11 individuals from Llanos de Challe and Observatorio la Silla, four individuals from Fray Jorge, two from Pelambres, two from Chillepín and one from Quebrada del Tigre); DIV2 (five individuals from Fray Jorge, Chillepín, Cerro Santa Inés and two from San Carlos de Apoquindo).
Intraspecific phylogeny, haplotype network and molecular clock
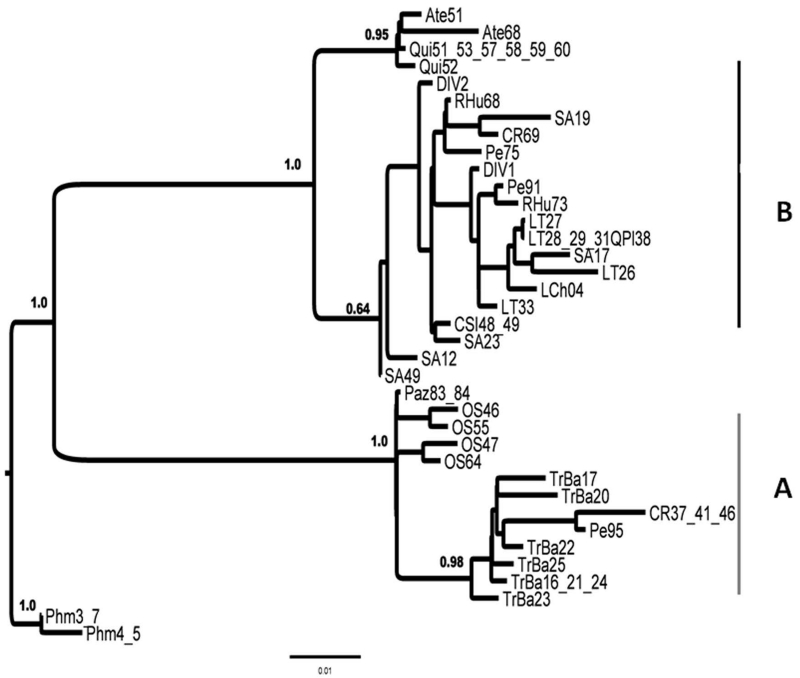
The intraspecific phylogeny confirmed Phyllotis darwini as a monophyletic group, with two major clusters and a posterior probability value of 1.0 (Fig.2). These results disagree with the genetic homogeneity suggested for this species by Steppan (2007). One of the recovered clades included haplotypes from almost all localities sampled in this work (hereafter “Lineage B”; Fig. 2). Lineage B included two widely distributed haplotypes, namely DIV1 and DIV2, distributed from Llanos de Challe to Quebrada del Tigre (28°-32° S), and from Fray Jorge to San Carlos de Apoquindo (30°-33° S), respectively (Figs. 1 and 2). The phylogenetic topology suggests some structuring within lineage B towards the southernmost distributional range (Agua Tendida and Quirihue, 36° S; Fig.1). Despite the low number of localities representing the southernmost range, we recovered the haplotypes from Agua Tendida and Quirihue as a well-supported subclade, reciprocally monophyletic with respect to the northernmost haplotypes belonging to lineage B. Taken together, haplotypes from B are distributed throughout almost the entire latitudinal range of P. darwini. The other supported group (hereafter Lineage A) is restricted to the northernmost locality of Pan de Azúcar National Park (26°S) in the Coastal Desert, and to the highland localities of Observatorio La Silla, Pelambres, and Tranque de Relaves Barahona in the Andean Cordillera, and to Cerro el Roble in the Coastal Cordilllera (Figs. 1 and 2). Lineage A is clearly differentiated from the geographically widespread lineage B. All haplotypes in lineage A were sampled in disjunct localities, with altitudes above 1500 m with the exception of one haplotype sampled in the coast (Pan de Azúcar National Park).
Figure 2.
Phyllotis darwini intraspecific phylogeny based on Bayes Markov Chain Monte Carlo method (BMCMC). The phylogeny was obtained for the Hypervariable Domain II (HV2) from the mitochondrial control region sequence data, whereas for BMCMC represents a consensus tree from the n = 9950 trees from the converged Markov chain. Posterior probability values over 0.5 are represented on each node.
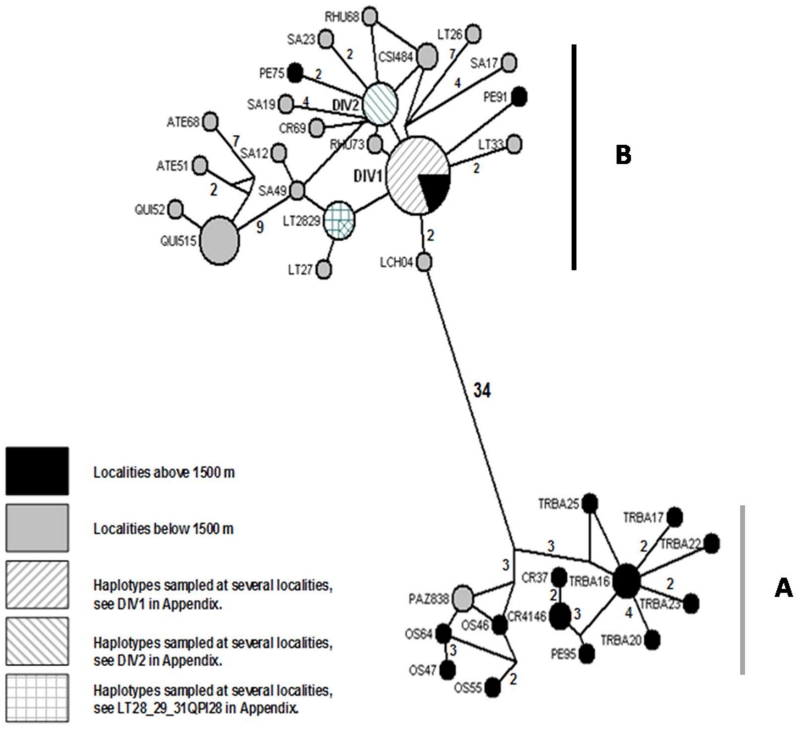
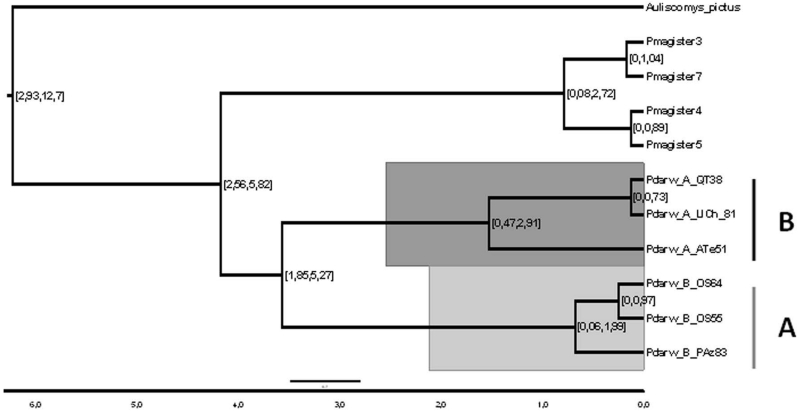
The Median Joining Network was congruent in recovering lineages A and B inferred from MultiBayes (Figs. 2 and 3). Lineage B included the most frequent and widespread haplotypes with a large number of mutational steps (34) separating lineages A and B, despite the geographical proximity between both phylogroups is less than 30 km. This contrasts with the nine mutational steps that separate Agua Tendida and Quirihue haplotypes (Fig. 1), 400 km away from the rest of haplotypes of lineage B. Finally, the 95% highest posterior density (HDP) interval for node ages were 0.47 – 2.9 MYa for lineage B, and 0.06 – 1.99 Mya for lineage A (Fig 5), suggesting that main intraspecific lineages in Phyllotis darwini were established long before LGM (21 Kya).
Figure 3.
Median-Joining haplotype network for the Phyllotis darwini mitochondrial DNA dataset. The size of the Haplotype tip is proportional to its frequency. Numbers on the branches are mutational steps between haplotype tips; when the branch has no number the tips are separated by just one mutational step. Black and grey filled circles are private haplotypes sampled at localities above and below 1500 m altitude respectively. Dashed circles represent shared haplotypes. The DIV 1 is a shared haplotype, but the proportion of individuals sampled above 1500 m altitude is designated by a black filled slice.
Figure 5.
D-loop based Phylogeny for intraspecific lineages in P. darwini and other related species. Diversification times (expressed in millions years) appears below in scale bar. Numbers on the nodes represent 95% highest prior density estimates (95% HPD) for the node age.
Population genetics analyses
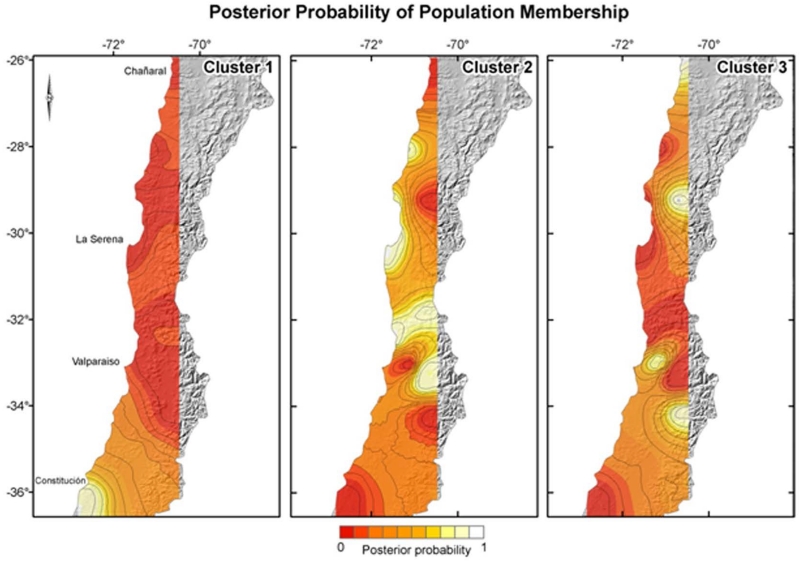
The highest posterior probability density occurred in a value of three for the number of populations parameter. Thus, we set the number of populations at three for the next independent runs; we choose the highest average posterior probability density as the better run to estimate both, the posterior probability cluster membership of each individual, and the geographic position of clusters. The output of the best run was used to draw a map with an estimation of the geographic position of each cluster, as well as the isoclines of population membership (Fig 4). We recovered three populations within the species, one restricted to the southernmost part of the distribution range around 36° S at the coast (Fig. 4, cluster 1). The geographic extension of this population could be broader than the estimated because we failed to capture individuals around Quirihue and Agua Tendida neither northward nor in the Andean cordillera. The second population (Fig. 4, cluster 2) ranges between 28° S and 34° S, mostly in the central valley and the Andean slopes up to1500 m. This population encompasses the major portion of the species distributional range. Individuals from the third population (cluster 3; Fig. 4) appeared as belonging to three disjunct high altitude locations and to the northernmost locality of Pan de Azúcar in the Coastal Desert. Those three populations inferred within the species agreed exactly with the major genealogical clusters inferred from the intraspecific phylogeny and the haplotype network analysis: Clusters 1 and 2 altogether corresponded to the distribution of lineage B, displaying an haplotype diversity (Hd) of 0,812 (sd=0.06) and nucleotide diversity (Pi) of 0.005; cluster 3 corresponded to lineage A with an Hd value of 0.978 (sd=0.05) and Pi = 0.01.
Figure 4.
Map of population membership posterior probability. According to Geneland analysis, the species is composed of three genetic units, designated as cluster 1, 2 and 3. Map depicts posterior probability iso-lines of belong to each cluster.
We evaluated if there was evidence of isolation by distance. We divided the localities in the two major clades because we hypothesized the broad latitudinal extension of each clade as a potential source of genetic differentiation in a stepping stone pattern. Lineage B included localities from the coast and from the valleys, as well as localities from the slopes of the Andes and Coastal cordillera. Lineage A included high altitude localities from the Andes and Coastal cordillera, being Pan de Azúcar the exception to this group since it is a lowland locality. The results indicated that there exists a significant correlation between genetic differentiation and geographic distance into the populations belonging to lineage B with a p-value of 0.049. Meanwhile, in the localities belonging to lineage A the correlation was not significantly distinct from the expected by chance with a p-value of 0.4915. We repeated the test for lineage A now excluding the locality of Pelambres since this shares haplotypes assigned to both major phylogroups. However, the correlation was still not significantly distinct from the expected by chance with a p-value of 0.2073.
Since there is an haplogroup distributed throughout the range of P. darwini (lineage B), whereas the other it is mainly restricted to high altitude locations (lineage A), we considered each lineage as separate demes. We evaluated if each haplogroup had independently experienced an abrupt demographic expansion during its evolutionary history through the “distribution of pairwise genetic differences”. To test the goodness of fit from the observed mismatch distribution to the simulated distribution under the assumption of sudden expansion, we implemented the least square deviation method. The Sum of Square Deviations (SSD) value was 0.025 for lineage B, and 0.031 for lineage A; its associated p-value was 0.85 and 0.29, respectively. Accordingly, the hypothesis of sudden expansion cannot be rejected and we concluded that populations of both clades have experienced at least one important and sudden population size expansion across its evolutionary history
Distribution models
To test the assumption that overlapped lineage distribution models may approximate whole species distribution models, we compared our estimations of Phyllotis darwini’s distribution range from i) all trapping localities as a single distributional unit (Table 1), and ii) models for each intraspecific lineage as an independent distributional unit (Fig 6, Tables 1 and 2). The result shows that the whole species range model is an accurate approximation of the observed distribution range for P. darwini (26°S to 36°S observed, 25°S to 36.5°S predicted; map not shown) and also, with high model performance (AUC, Table 1), whereas overlapped lineage distribution models appeared to slightly over-predict northern distribution (22°S to 36.5°S). Both models are good and consistent approximations of current Phyllotis darwini’s geographic range.
Table 1.
AUC values and standard deviation for whole species’ distribution and individual lineage’s distribution models.
| Model | AUC | AUC stdv. |
|---|---|---|
| P. darwini whole specie’s range |
0.971 | 0.022 |
| P darwini lineage A | 0.968 | 0.025 |
| P. darwini lineage B | 0.83 | 0.15 |
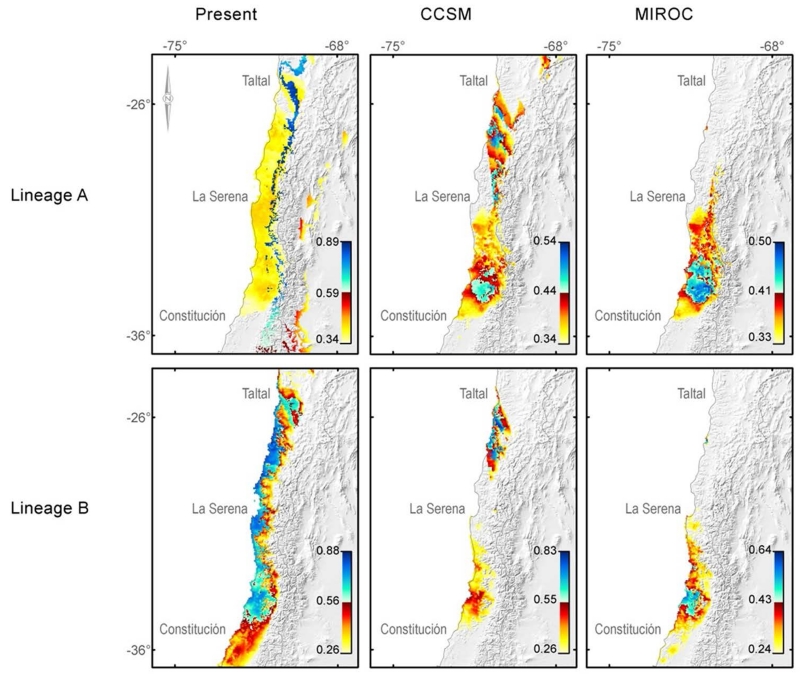
Figure 6.
Lineage distribution models. The figure shows the species distribution models for present and two LGM climatic models (columns). Models were built independently for each P. darwini’s intraspecific lineage (rows). Colour scale represents the full distribution of logistic probability values inside the suitability area determined by the maximum training sensitivity plus specificity logistic threshold; the white-blue portion of the colour scale represents highest 50% of logistic probability values observed inside the suitability area.
Table 2.
Lineage’s distribution model summary, at present and at LGM (current conditions, CCSM and MIROC).
| P. darwini lineage A | P. darwini lineage B | |
|---|---|---|
| Present | 24°S - 36°S. Highest logistic probability values at the Andean mountain range. |
25°S - 35°S .Highest logistic probability values at the valley and at the coast. |
| CCSM | Distributional gap between 28°S - 31°S |
Distributional gap between 28°S-31°S. Not distributed in the Andean mountain range. |
| MIROC | 31°S-35°S. Distributed at andes, valley and coast. |
30°S - 35°S Mainly distributed in the valley and the coast. |
According to the maximum training sensitivity plus specificity logistic threshold, the distribution model for lineage B encompasses the whole distributional range of P. darwini (Fig 6, table 2, full colour scale). Considering that same threshold, suitable areas for lineage A at present are highly overlapped with the predicted distribution for lineage B; this model is in conflict with our observation of strict altitudinal segregation, where lineage A is distributed almost exclusively above 1500 m in both cordilleras. Considering the abrupt orographic changes which characterizes central Chile, and the associated risk of over-prediction in the distribution models for patchy environments, we investigated the spatial distribution of logistic probability values inside the suitability area determined by the threshold (Fig 6, white-blue portion of the colour scale): we observed that high logistic probability values have non-random associations with major orographic features, with all the 50% of highest values distributed in the valley and coastal cordillera for lineage B, and exclusively in elevations above 1500 m in both cordilleras for lineage A (Fig 6). In summary, overlapped lineage distribution models had slightly worst performance and some over-prediction compared to the whole species’ distribution model, but when considering the areas with the highest 50% of logistic probability values inside the suitability area, the lineage distribution models reproduced very accurately the particular distributional pattern observed for both phylogroups at present. This information is missed in the whole species’ distribution range model.
Past lineage distribution estimated by both LGM models is conflicting (CCSM and MIROC, Fig. 6, table 2): the CCSM model predicted a distributional gap during LGM for both phylogroups, but MIROC based distribution models predicted that both phylogroups were restricted to the southern portion of Phyllotis present distribution. Nevertheless, both models consistently predicted the area between 32°S-35°S as suitable for both phylogroups during LGM. Those latitudinal distribution dynamics must be considered with caution because downscaled climatic data may not represent local geographic complexity with accuracy; nevertheless, the main altitudinal displacements between glacial-interglacial transitions for intraspecific lineages were consistently supported by both LGM distribution models. It is important to emphasize that altitudinal particularities reported for both lineages at present were already established during LGM: lineages A and B might have been restricted to approximately the same latitude, but only lineage A displayed suitability areas at Andean mountain range during LGM (Table 2), which also has been sampled mainly at the Andes and at localities above 1500 m. at present.
DISCUSSION
One of the most important features found in the genetic structure of Phyllotis darwini was the major split between the widespread haplogroup (lineage B) and the high altitude haplogroup (lineage A). The phylogroup B has the most frequent and widely distributed haplotypes, whereas lineage A shares only private haplotypes separated by 34 mutational steps from lineage B, and it is mostly restricted to high altitude localities. Lineage ages are at least 47 and 57 Kya for lineages A and B, respectively, (lowest 95% HPD value), and according to GENELAND analysis, the inter-lineage gene flow appeared to be restricted at present. Meanwhile both lineages displayed signals of past population expansions, only lineage B had a significative isolation by distance pattern. Altogether, those phylogenetic and populational features suggested that main lineages in P. darwini had ancient and independent evolutionary trajectories. Fuentes & Jaksic (1979) hypothesized that there exist two asynchronous speciation modes for lizards and rodents in central Chile: i) Mountain speciation occurred during interglacial periods, because of high species replacement with altitude and between mountains isolation, and ii) valley speciation should occur during glacial periods, when species may not reach high altitude elevations and connectivity in the valleys is reduced. Given the ecological attributes of both groups, lizards are expected to display both speciation modes because they might be affected by severe decrease in connectivity in the valley during glacial periods. They are also restricted in high altitude localities during interglacial periods because its low vagility, high habitat specificity, and high species turnover between mountains. Rodents would only exhibit the valley speciation mode because they might have been affected by a decrease in connectivity in the valley during glacial periods, but not by high altitude isolation during interglacial, because their high vagility and lower “between-mountains species turnover” compared to lizards. Those differences in speciation modes would be finally explained by differences in mobility between groups, as a consequence of their different thermoregulation modes and energy requirements (Fuentes & Jaksic, 1979). Our results show that P. darwini displayed differentiation inside the lowland phylogroup (Lineage B) with an isolation by distance pattern and restricted gene flow between subgroups. This could be considered as evidence of the valley speciation mode inside this endemic rodent species. Nevertheless, the fact that postglacial recolonization in mountain ranges has occurred only in the apparently high-altitude adapted lineage A, suggests that mountain speciation mode could be most likely the cause of the origin for this lineage, which does not show latitudinal structure despite being distributed in several disjunct localities along mountain ranges. In conclusion, and contrary to the “lizards and rodents” hypothesis, the lineages in P. darwini appears to have originated by the same mechanism which drives the speciation mode suggested for lizards in the central Chile area (both, valley and mountain speciation modes in the whole species). The specific historic event which could have triggered those intraspecific diversification events remains elusive, because lineage ages in P. darwini could be older than Quaternary times.
The fact that P. darwini’s distributional range at present can be estimated by overlapping lineage distribution models is non-trivial. Even though model performance is slightly worst in lineage distribution models compared to whole species distribution model, it is clear that by using below species level ESUs in SDMs, we are able to recover very important distributional information. In fact, in this approach we have demonstrated that P. darwini is composed of two ancient lineages which, despite their latitudinal overlap, it shares a very strict altitudinal segregation at present. An important methodological consideration is that intraspecific lineages may have more restricted climatic niches than the whole species, and given the low resolution of downscaled climatic models regards local conditions, it is not surprising that the distribution models at intraspecific level suffered more over-prediction than the whole species distribution models (Merow et al., 1992; Laughlin et al., 2012). Therefore, the use of more restrictive probability thresholds for distribution models below species level must be encouraged, especially when environmental conditions changes abruptly, such as altitude across small geographic distances, as occurs in the valley and cordilleras from central Chile.
Once we have established the phylogenetic and population structure, main lineage’s ages and meaningful lineage distribution models at present, the next step was to project our lineage distribution model to climatic conditions at LGM. The rationale behind this procedure is as follows: if we can estimate geographic distribution for intraspecific lineages at present and also at some point in the past, with very different climatic conditions, then we can compare past to present lineage distribution, and interpret the differences between those geographic ranges as distributional responses to climate change. LGM was chosen because is one of the biggest recent climatic events (Heusser, 1990; Clapperton, 1994; Heusser et al., 2006), and it has been hypothesized as a major forcing in vegetation range dynamics in the central Chile biodiversity hotspot (Villagrán & Armesto, 1991; Villagrán & Hinojosa, 2005). In this context, the results shows that at LGM, both lineages were restricted to aproximately the same latitudes (31°S-35°S, probably with a disjunction in lineage B according to the CCSM model) but only lineage A displayed suitability areas at the high altitude Andean mountain ranges. After the LGM event, temperature may have rise and precipitation may have declined at those latitudes, and besides other minor climatic oscilations, present day temperature is higher and precipitation is lower than during LGM. Therefore, this comparison suggests that after post-LGM warming, both lineages expanded their northern distribution to their present geographic range limits around 26°S (or establishing a continuum distribution in the CCSM model for lineage B). However, only lineage A has colonized Andean mountain ranges above 1500 m altitude, being the lineage that retained its Andean distribution during the maximum glacial advance through the Andes during LGM (Clapperton, 1990; Clapperton, 1994). In conclusion, after glaciation, both lineages expanded their distribution reaching the same latitudes (northward expansion for both lineages according to MIROC, and only for lineage A according to CCSM), but clearly not to the same altitudes. This would explain the present segregation of altitudinal limits, where lineage B shows a wide distribution although restricted to the lowlands and the coast, whereas lineage A is mainly distributed through the Andes and the coastal mountain ranges above a threshold of 1500 m approximately. Therefore, the distributional response to an increase of temperature and a decline of precipitation was independent for each lineage: lineage B colonized a broad latitudinal range but restricted to low elevations, whereas lineage A was able to colonize mountain ranges.Thus,we hypothesize that both lineages will display independent distributional responses to future GCC scenarios, as they did in the past. This conclusion would be impossible to achieve if we consider that species behaves as simple ecological units to climate change.
It is important to notice that both lineages seem to have been co-distributed between 31°S-35°S during LGM according to MIROC based estimations, but the distribution model projected in CCSM climatic data predicts a second relict between 25°S-28°S. We can not rule out the latter possibility; in fact it could be a good explanation for the only lowland locality in wich lineage A has been sampled, the current northern P. darwini distribution’s limit. In summary, the area across 31°S-35°S is predicted as suitable by both LGM distribution models, and the oppossite for the high altitude Andean areas. We do not expect to provide precise distribution models because high resolution climatic models wich reproduce the local conditions and the complex geography of the central Chile hotspot are lacking. Nevertheless, the essential altitudinal pattern and independent post-LGM colonization with altitudinal segregation for intraspecific lineages is supported by both, CCSM and MIROC based distribution models, and additionally by our field observations at present. All those distribution dynamics relies on the strong assumption that the climatic niche of these two ancient lineages has remained approximatelly constant since the last 20 Kya. This seems a reasonable assumption given the estimated lineage ages and the dramatic distributional shifts reported, but a note of caution must be given because of the possibility of rapid niche evolution has not been evaluated here. This could be an interesting research topic for further investigations on P. darwini.
Regarding concrete applications of our results for conservation purposes in the central Chile hotspot, the intraspecific distributional dynamics of P. darwini suggests that coastal and Andean cordilleras may behave as a biological corridor for terrestrial vertebrates during interglacial conditions, whereas during glacial conditions the valley and the coastal cordillera could be considered as a refuge for lineages from highlands and lowlands. If this pattern is confirmed for another species, a sensible conservation strategy should be to prioritize a chain of several small protected areas with high connectivity in the Andes, as a biological corridor during current interglacial conditions. In a long term perspective, this strategy should prioritize protected areas with great surface, preferibly located in the coastal cordillera, between 31°S-35°S, which is the area that apparently behave as a refuge during glacial conditions.
In conclusion, our study brings evidence of an endemic species from an endangered area that contains cryptic diversity below the species level, and whose intraspecific lineages have responded independently to climate change in the past. Importantly, the study suggests that lineages of P. darwini may not behave as a single ecological unit to future GCC scenarios. Given that high levels of cryptic diversity below species level is a frequent pattern (Bálint et al., 2011), the integration of genetic and ecological tools must be encouraged (May et al., 2011) as a way to understand the complex distributional responses of species and biotas, in order to prevent massive cryptic biodiversity losses in the future. The methodology proposed in this work could be a way to make more realistic predictions for conservation planning in the future global change scenario.
Supplementary Material
Acknowledgements
We appreciate the technical advice provided by Ricardo Cancino, Iván Barría and Patricio Pliscoff. Some tissue samples were kindly loaned by Dr. Angel Spotorno and Roberto Thomson.
The first author of this work appreciates the uninterested support of Dr. Adam Leland and Dr. Mathew Graham. This work was funded by the following projects: FONDECYT 1100558, 1130467; Hantavirus NIH and Fondecyt CASEB 1501-0001.
Biographies
Biosketches
Pablo A. Gutiérrez-Tapia is primarily interested in evolutionary aspects of biogeography and ecology of vertebrates.
R. Eduardo Palma is primarily interested in evolutionary biology of small mammals, molecular systematics, phylogeny, phylogeography and biogeography.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Appendix S1 List of individuals by catalogue number and GeneBank accession number.
Appendix S2 Detailed methodology.
REFERENCES
- Alsos IG, Ehrich D, Thuiller W, Eidesen PB, Tribsch A, Schönswetter P, Lagaye C, Taberlet P, Brochmann C. Genetic consequences of climate change for northern plants. Proceedings. Biological sciences / The Royal Society. 2012;279:2042–2051. doi: 10.1098/rspb.2011.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allal F, Sanou H, Millet L, Vaillant a., Camus-Kulandaivelu L, Logossa Z.a., Lefèvre F, Bouvet J-M. Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity. 2011;107:174–86. doi: 10.1038/hdy.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacigalupe LD, Nespolo RF, Opazo JC, Bozinovic F. Phenotypic Flexibility in a Novel Thermal Environment: Phylogenetic Inertia in Thermogenic Capacity and Evolutionary Adaptation in Organ Size. Physiological and Biochemical Zoology. 2004;77:805–815. doi: 10.1086/422229. [DOI] [PubMed] [Google Scholar]
- Bálint M, Domisch S, Engelhardt CHM, Haase P, Lehrian S, Sauer J, Theissinger K, Pauls SU, Nowak C. Cryptic biodiversity loss linked to global climate change. Nature Climate Change. 2011;1:313–318. [Google Scholar]
- Bandelt H-J, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Braconnot P, Otto-Bliesner B, Harrison S, Joussaume S, Peterchmitt JY, Abe-Ouchi A, Crucifix M, Driesschaert E, Fichefet T, Hewitt CD, Kageyama M, Kitoh A, Lainé A, Loutre M-F, Marti O, Merkel u., Ramstein G, Valdes P, Weber SL, Yu Y, Zhao Y. Results of PMIP2 coupled simulations of the Mid-Holocene and Last Glacial Maximum – Part 1: experiments and large-scale features. Climate of the Past. 2007;3:261–277. [Google Scholar]
- Brown GG. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. Journal of Molecular Biology. 1986;192:503–511. doi: 10.1016/0022-2836(86)90272-x. [DOI] [PubMed] [Google Scholar]
- Buckley TR, Marske K, Attanayake D. Phylogeography and ecological niche modelling of the New Zealand stick insect Clitarchus hookeri (White) support survival in multiple coastal refugia. Journal of Biogeography. 2010;37:682–695. [Google Scholar]
- Calsbeek R, Thompson JN, Richardson JE. Patterns of molecular evolution and diversification in a biodiversity hotspot: the California Floristic Province. Molecular ecology. 2003;12:1021–9. doi: 10.1046/j.1365-294x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science. 2009;323:785–789. doi: 10.1126/science.1166955. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Richards CL. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution; international journal of organic evolution. 2007;61:1439–54. doi: 10.1111/j.1558-5646.2007.00117.x. [DOI] [PubMed] [Google Scholar]
- Clapperton C. The quaternary glaciation of Chile: a review. Revista Chilena de Historia Natural. 1994;67:369–383. [Google Scholar]
- Clapperton J.R.a.C.M. Quaternary glaciations of the southern Andes. Quaternary Science Reviews. 1990;9:153–174. [Google Scholar]
- Cordellier M, Pfenninger M. Inferring the past to predict the future: climate modelling predictions and phylogeography for the freshwater gastropod Radix balthica (Pulmonata, Basommatophora) Molecular ecology. 2009;18:534–44. doi: 10.1111/j.1365-294X.2008.04042.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. The origin of species. Penguin Books; Oxford, United Kingdom: 1859. [Google Scholar]
- Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidenc. Plos Biology. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Toit N, van Vuuren BJ, Matthee S, Matthee C.a. Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Molecular phylogenetics and evolution. 2012;65:75–86. doi: 10.1016/j.ympev.2012.05.036. [DOI] [PubMed] [Google Scholar]
- Eckert A. Seeing the forest for the trees: statistical phylogeography in a changing world. New Phytologist. 2011;189:894–897. doi: 10.1111/j.1469-8137.2010.03631.x. [DOI] [PubMed] [Google Scholar]
- Elith J, Graham CH, Anderson RP, Dudik M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton JMM, Peterson AT, Phillips SJ, Richardson K, Scachetti-Pereira R, Schapire RE, Soberón J, Williams S, Wisz MS, Zimmermann NE. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- Engelbrecht A, Taylor PJ, Daniels SR, Rambau RV. Cryptic speciation in the southern African vlei rat Otomys irroratus complex: evidence derived from mitochondrial cyt b and niche modelling. Biological Journal of …. 2011;104:192–206. [Google Scholar]
- Fielding a.H., Bell J.f. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environmental Conservation. 1997;24:38–49. [Google Scholar]
- Florio a.M., Ingram CM, Rakotondravony H.a., Louis EE, Raxworthy CJ. Detecting cryptic speciation in the widespread and morphologically conservative carpet chameleon (Furcifer lateralis) of Madagascar. Journal of evolutionary biology. 2012;25:1399–14414. doi: 10.1111/j.1420-9101.2012.02528.x. [DOI] [PubMed] [Google Scholar]
- Fontanella FM, Feltrin N, Avila LJ, Sites JW, Morando M. Early stages of divergence: phylogeography, climate modeling, and morphological differentiation in the South American lizard Liolaemus petrophilus (Squamata: Liolaemidae) Ecology and evolution. 2012;2:792–808. doi: 10.1002/ece3.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DJ, Bernatchez L. Adaptive evolutionary conservation : towards a unified concept for defining conservation units. Molecular Ecology. 2001;10:2741–2752. [PubMed] [Google Scholar]
- Fuentes ER, Jaksic FM. Lizards and rodents: an explanation for their relative species diversity in Chile. Arch. Biol. Med. Exper. 1979;12:179–190. [Google Scholar]
- Gugger PF, González-Rodríguez A, Rodríguez-Correa H, Sugita S, Cavender-Bares J. Southward Pleistocene migration of Douglas-fir into Mexico: phylogeography, ecological niche modeling, and conservation of ‘rear edge’ populations. The New phytologist. 2011;189:1185–99. doi: 10.1111/j.1469-8137.2010.03559.x. [DOI] [PubMed] [Google Scholar]
- Guilliot G, Mortier F, Estoup A. Geneland: a computer package for landscape genetics. Molecular Ecology. 2005;5:712–715. [Google Scholar]
- Guilliot G, Santos F, Estoup A. Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics. 2008;24:1406–1407. doi: 10.1093/bioinformatics/btn136. [DOI] [PubMed] [Google Scholar]
- Hall BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. 1999;41:95–98. [Google Scholar]
- Hamann A, Wang T. Potential effects of climate change on ecosystem and tree species distribution in british Columbia. Ecology. 2006;87:2773–2786. doi: 10.1890/0012-9658(2006)87[2773:peocco]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Harrison S. Palaeoenvironmental data sets and model evaluation in PMIP. In: Braconnot P, editor. Proceedings of the Third PMIP Workshop. La Huardiere; Canada: 2000. pp. 9–25. [Google Scholar]
- Hasumi H, Emori S. K-1 coupled model (MIROC) description. K-1 Tech. Rep. 1. University of Tokyo, Center for climate system research; 2004. p. 34. [Google Scholar]
- Heusser C. Quaternary Pollen Record from Laguna de Tagua Tagua, Chile. Science. 1983;219:1429–1432. doi: 10.1126/science.219.4591.1429. [DOI] [PubMed] [Google Scholar]
- Heusser CJ. Ice age vegetation and climate of subtropical Chile. Palaeogeography,Palaeoclimatology, Palaeoecology. 1990;80:107–127. [Google Scholar]
- Heusser L, Heusser C, Mix A, McManus J. Chilean and Southeast Pacific paleoclimate variations during the last glacial cycle: directly correlated pollen and δ18O records from ODP Site 1234. Quaternary Science Reviews. 2006;25:3404–3415. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–85. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hood GM. PopTools version 3.2.3. 2010. [Google Scholar]
- Iriarte A. Mamíferos de Chile. Lynx editions; Barcelona, España: 2008. [Google Scholar]
- Jezkova T, Jaeger JR, Marshall ZL, Riddle BR. Pleistocene Impacts on the Phylogeography of the Desert Pocket Mouse (Chaetodipus penicillatus) Journal of Mammalogy. 2009;90:306–320. [Google Scholar]
- Kalkvik HM, Stout IJ, Doonan TJ, Parkinson CL. Investigating niche and lineage diversification in widely distributed taxa: phylogeography and ecological niche modeling of the Peromyscus maniculatus species group. Ecography. 2011;35:54–64. [Google Scholar]
- Kozak KH, Graham CH, Wiens JJ. Integrating GIS-based environmental data into evolutionary biology. Trends in ecology & evolution. 2008;23:141–8. doi: 10.1016/j.tree.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Lamy F, Hebbeln D, Wefer G. High-Resolution Marine Record of Climatic Change in Mid-latitude Chile during the Last 28,000 Years Based on Terrigenous Sediment Parameters. Quaternary Research. 1999;51:83–93. [Google Scholar]
- Larizza A, Pesole G, Reyes A, Sbisa E, Saccone C. Lineage Specificity of the Evolutionary Dynamics of the mtDNA D-Loop region in Rodents. Journal of Molecular Evolution. 2002;54:145–155. doi: 10.1007/s00239-001-0063-4. [DOI] [PubMed] [Google Scholar]
- Laughlin DC, Joshi C, van Bodegom PM, Bastow ZA, Z. FP. A predictive model of community assembly that incorporates intraspecific trait variation. Ecology letters. 2012;15:1291–1299. doi: 10.1111/j.1461-0248.2012.01852.x. [DOI] [PubMed] [Google Scholar]
- Lim HC, Rahman M.a., Lim SLH, Moyle RG, Sheldon FH. Revisiting Wallace’s haunt: coalescent simulations and comparative niche modeling reveal historical mechanisms that promoted avian population divergence in the Malay Archipelago. Evolution; international journal of organic evolution. 2010;65:321–334. doi: 10.1111/j.1558-5646.2010.01105.x. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clusstering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Marske K, Leschen R, Buckley T. Concerted versus independent evolution and the search for multiple refugia: Comparative phylogeography of four forest beetles. Evolution. 2012;66:1862–1877. doi: 10.1111/j.1558-5646.2011.01538.x. [DOI] [PubMed] [Google Scholar]
- Marske K.a., Leschen R.a.B., Buckley TR. Reconciling phylogeography and ecological niche models for New Zealand beetles: Looking beyond glacial refugia. Molecular phylogenetics and evolution. 2011;59:89–102. doi: 10.1016/j.ympev.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Marske K.a., Leschen R.a.B., Barker GM, Buckley TR. Phylogeography and ecological niche modelling implicate coastal refugia and trans-alpine dispersal of a New Zealand fungus beetle. Molecular ecology. 2009;18:5126–42. doi: 10.1111/j.1365-294X.2009.04418.x. [DOI] [PubMed] [Google Scholar]
- May SE, Medley K.a., Johnson S.a., Hoffman E.a. Combining genetic structure and ecological niche modeling to establish units of conservation: A case study of an imperiled salamander. Biological Conservation. 2011;144:1441–1450. [Google Scholar]
- Merow C, Latimer AM, Silander JA. Can entropy maximization use functional traits to explain species abundances? A comprehensive evaluation. Ecology. 1992;92:1523–1537. doi: 10.1890/10-1174.1. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Otto-Bliesner B, Brady E, Clauzet G, Tomas R, Levis S, Kothavala Z. Last Glacial Maximum and Holocene Climate in CCSM3. Journal of Climate. 2006;19:2526–2544. [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character state data. Systematic Biology. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- Palma RE. Estado actual de la Mastozoología en Chile. Mastozoología Neotropical. 2007;14:5–9. [Google Scholar]
- Pauls SU, Nowak C, Bálint M, Pfenninger M. The impact of global climate change on genetic diversity within populations and species. Molecular Ecology. 2012 doi: 10.1111/mec.12152. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- Pearson RG, Dawson TP. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecology and Biogeography. 2003;12:361–371. [Google Scholar]
- Pearson RG, Dawson TP, Berry PM, Harrison PA. SPECIES: A Spatial Evaluation of Climate Impact on the Envelope of Species. Ecological Modelling. 2002;154:289–300. [Google Scholar]
- Peterson TA, Nyári AS. Ecological niche conservatism and pleistocene refugia in the trush-like mourner, Schiffornis sp., in the Neotropics. Evolution. 2007;62:173–183. doi: 10.1111/j.1558-5646.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- Pettorelli N. Climate change as a main driver of ecological research. Journal of Applied Ecology. 2012;49:542–545. [Google Scholar]
- Phillips SJ, Dudik M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. [Google Scholar]
- Phillips SJ, Dudik M, Schapire RE. Proceedings of the twenty-first international conference on machine learning. Banff, Canada: 2004. A maximum entropy approach to species distribution modeling; pp. 655–662. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends in ecology & evolution. 2008;23:564–71. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Qi X-S, Chen C, Comes HP, Sakaguchi S, Liu Y-H, Tanaka N, Sakio H, Qiu Y-X. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae) The New phytologist. 2012;196:617–630. doi: 10.1111/j.1469-8137.2012.04242.x. [DOI] [PubMed] [Google Scholar]
- Raes N, ter Steege H. A null-model for significance testing of presence-only species distribution models. Ecography. 2007;30:727–736. [Google Scholar]
- Randin CF, Dirnbock T, Dullinger s., Zimmermann NE, Zappa M, Guisan A. Are niche-based species distribution models transferable in space? Journal of Biogeography. 2006;33:1689–1703. [Google Scholar]
- Raxworthy CJ, Ingram CM, Rabibisoa N, Pearson RG. Applications of ecological niche modeling for species delimitation: a review and empirical evaluation using day geckos (Phelsuma) from Madagascar. Systematic biology. 2007;56:907–923. doi: 10.1080/10635150701775111. [DOI] [PubMed] [Google Scholar]
- Redford KH, Eisenberg JF. Mammals of the Neotropics. The University of Chicago Press; 1992. [Google Scholar]
- Reig OA. Diversitty patterns and differentiation of high Andean rodents. In: Monasterio F.V.a.M., editor. High Altitude tropical Biogeography. Oxford University press; London: 1986. pp. 404–439. [Google Scholar]
- Rissler LJ, Apodaca JJ. Adding more ecology into species delimitation: ecological niche models and phylogeography help define cryptic species in the black salamander (Aneides flavipunctatus) Systematic biology. 2007;56:924–42. doi: 10.1080/10635150701703063. [DOI] [PubMed] [Google Scholar]
- Rogers A, Harpending H. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rohl A, Mihn L. Network: A Program Package for Calculating Phylogenetic Networks. 1997 [Google Scholar]
- Rousset F. Genetic Differentiation and Estimation of Gene Flow from F-Statistics Under Isolation by Distance. Genetics. 1997;145 doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-Del Barrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of Past Demographic Parameters From the Distribution of Pairwise Differences When the Mutation Rates Vary Among Sites: Application to Human Mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: A Software for population genetics data analysis. 2000 [PMC free article] [PubMed] [Google Scholar]
- Sikes RS, Gannon WL. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy. 2011;92:235–253. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti JA. Diversity and Conservation of terrestrial vertebrates in mediterranean Chile. Revista Chilena de Historia Natural. 1999;72:493–500. [Google Scholar]
- Simpson BB. An Historical Phytogeography og the High Andean Flora. Revista Chilena de Historia Natural. 1983;56:109–122. [Google Scholar]
- Spotorno AE, Walker LI, Flores SV, Yevenes M, Marín JC, Zuleta C. Evolución de los filotinos (Rodentia, Muridae) en los Andes del Sur. Evolution of phyllotines (Rodentia, Muridae) in the southern Andes. Revista Chilena de Historia Natural. 2001;74:151–166. [Google Scholar]
- Steppan S, Ramírez O, Banbury J, Huchon D, Pacheco V, Walker LI, Spotorno AE. A Molecular Reappraisal of the Leaf-Eared Mice Phyllotis and their Relatives. In: Dougals JS-B, Kelt A, Patton James L., editors. The Quintessential naturalist: Honoring the Life and Legacy of Oliver. P. Pearson. 2007. pp. 799–826. [Google Scholar]
- Steppan SJ. Phylognetic Relationships and Species Limits within Phyllotis (Rodentia: Sigmodontinae): Concordance between mtDNA Sequence and Morphology. Journal of Mammalogy. 1998;79:573–593. [Google Scholar]
- Summers DM, Bryan B.a., Crossman ND, Meyer WS. Species vulnerability to climate change: impacts on spatial conservation priorities and species representation. Global Change Biology. 2012;18:2335–2348. [Google Scholar]
- Svenning J-C, Fløjgaard C, Marske K.a., Nógues-Bravo D, Normand S. Applications of species distribution modeling to paleobiology. Quaternary Science Reviews. 2011;30:2930–2947. [Google Scholar]
- Tajima F. Statistical Method for testing the Neutral Mutation Hypotesis by DNA Polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller W, Brotons L, Araújo MB, Lavorel S. Effects of restricting environmental range of data to project current and future species distributions. Ecography. 2004;27:165–172. [Google Scholar]
- Tolley K.a., Burguer M, Turner AA, Matthee CA. Biogeographic patterns and phylogeography of dwarf chameleons (Bradypodion) in an African biodiversity hotspot. Molecular Ecology. 2006;15:781–793. doi: 10.1111/j.1365-294X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Tolley K.a., Makokha JS, Houniet DT, Swart BL, Matthee C.a. The potential for predicted climate shifts to impact genetic landscapes of lizards in the South African Cape Floristic Region. Molecular phylogenetics and evolution. 2009;51:120–30. doi: 10.1016/j.ympev.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Verboom G.a., Dreyer LL, Savolainen V. Understanding the origins and evolution of the world’s biodiversity hotspots: The biota of the African ‘Cape Floristic Region’ as a case study. Molecular Phylogenetics and Evolution. 2009a;51:1–4. doi: 10.1016/j.ympev.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Verboom GA, Archibald JK, Bakker FT, Bellstedt DU, Conrad F, Dreyer LL, Forest F, Galley C, Goldblatt P, Henning JF. Origin and diversification of the Greater Cape flora: Ancient species repository, hot-bed of recent radiation, or both? Molecular Phylogenetics and Evolution. 2009b;51:44–53. doi: 10.1016/j.ympev.2008.01.037. [DOI] [PubMed] [Google Scholar]
- Villagrán C, Armesto J. Historical phytogeography of the Chilean Coastal Range. Revista Chilena de Historia Natural. 1991;64:105–123. [Google Scholar]
- Villagrán C, Hinojosa L. Esquema biogeográfico de Chile. In: Bousquets J.J.M. Jorge Llorente., editor. Regionalización Biogeográfica en Iberoámeríca y tópicos afines. Ediciones de la Universidad Nacional Autónoma de México, Jiménez Editores; México: 2005. pp. 551–577. [Google Scholar]
- Walker LI, Spotorno AE, Arrau J. Cytogenetic and Reproductive Studies of Two nominal Subspecies of Phyllotis darwini and Their Experimental Hybrids. Journal of Mammalogy. 1984;65:220–230. [Google Scholar]
- Waltari E, Guralnick RP. Ecological niche modelling of montane mammals in the Great Basin, North America: examining past and present connectivity of species across basins and ranges. Journal of Biogeography. 2009;36:148–161. [Google Scholar]
- Waltari E, Hijmans RJ, Peterson a.T., Nyári ÁS, Perkins SL, Guralnick RP. Locating Pleistocene Refugia: Comparing Phylogeographic and Ecological Niche Model Predictions. PLoS ONE. 2007;2:e563. doi: 10.1371/journal.pone.0000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin J-M, Hoegh-Guldberg O, Bairlein F. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: Data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003a;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003b;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.