ABSTRACT
In assisted reproduction, about 30% of embryo implantation failures are related to inadequate endometrial receptivity. To identify molecules involved in endometrial receptivity acquisition, we investigated, using a SELDI-TOF approach, the protein expression profile of early-secretory and mid-secretory endometrium samples. Among the proteins upregulated in mid-secretory endometrium, we investigated the function of S100A10 in endometrial receptivity and implantation process. S100A10 was expressed in epithelial and stromal cells of the endometrium of fertile patients during the implantation windows. Conversely, it was downregulated in the mid-secretory endometrium of infertile patients diagnosed as non-receptive. Transcriptome analysis of human endometrial epithelial and stromal cells where S100A10 was silenced by shRNA revealed the deregulation of 37 and 256 genes, respectively, related to components of the extracellular matrix and intercellular connections. Functional annotations of these deregulated genes highlighted alterations of the leukocyte extravasation signaling and angiogenesis pathways that play a crucial role during implantation. S100A10 silencing also affected the migration of primary endometrial epithelial and stromal cells, decidualization and secretory transformation of primary endometrial stromal cells and epithelial cells respectively, and promoted apoptosis in serum-starved endometrial epithelial cells. Our findings identify S100A10 as a player in endometrial receptivity acquisition.
KEYWORDS: early implantation, endometrial receptivity, human, proteome, S100A10
Introduction
Understanding the molecular mechanisms involved in the acquisition of the endometrial receptivity phenotype is important in assisted reproductive technology (ART). Indeed, around 30% of implantation failures are related to inadequate endometrial receptivity. Different gene array studies have determined the mRNA changes during the menstrual cycle.1,2 However, biological processes, including the acquisition of the endometrial receptivity phenotype, are ultimately controlled by proteins and therefore analysis of the changes in the protein profile is required to acquire a complete overview. Indeed, the overall lack of correlation between gene and protein expression data on endometrial receptivity2 underlines the importance of post-transcriptional or translational regulations for the acquisition of the receptive phenotype during the implantation window. Nevertheless, studies on endometrium receptivity using high-throughput mass spectrometry-based proteomic approaches are still scarce (Table 1). So far, only 4 studies compared the endometrial protein expression profiles between the mid- or late-proliferative and mid-secretory stages.3-6 However, the reported protein profiles were completely different, although similar endometrial samples were compared. Similarly, 2 other studies that compared the endometrium proteome during the transition from the early-secretory to the mid-secretory stage did not highlight any shared protein.7,8 Many factors could justify these disparities (different proteomic approaches, recruited patients, number of samples, statistical methodologies).2,9 Particularly, the endometrial sample size was generally low (Table 1) and the clinical populations were mostly not well defined. Moreover, few studies analyzed the protein profile changes between early-secretory and mid-secretory endometrium in the same patients and in a large cohort of patients, to minimize the inter-patient variability.1,2,9 In addition to sufficiently powered studies, validation of selected proteins in an independent cohort of patients remains a key step, which is not always performed.9 Moreover, the determination of the function(s) of selected proteins remains indispensable for understanding the signaling pathways controlling human endometrial receptivity.
Table 1.
Comparison of the study design of the current and previous mass spectrometry-based proteomic analyses of endometrial samples at different phases of the menstrual cycle.
| Study | Number of patients | Number of samples used for proteomic analysis | Patient age (mean ± sem or range) | Patient population | Compared samples (number of samples) | Proteomic approach | |
|---|---|---|---|---|---|---|---|
| 3 | 6 | 6 | ? | Hysterectomy specimens | Proliferative vs. mid-secretory | Isotope-coded affinity tags + Multidimensional | |
| (n = 3) | (n = 3) | liquid chromatography + Nanobore LC-MS/MS | |||||
| 4 | 20 | 6 | ? | Normal fertile women | Mid-proliferative (days 8-10) vs. mid-secretory (days 19-23) | DIGE + MALDI-TOF/TOF MS | |
| (n = 3) | (n = 3) | ||||||
| 5 | 11 | 11 | 25–35 | Healthy women | Late-proliferative vs. mid-secretory (LH+6) | DIGE + MALDI-TOF-TOF | |
| (n = 5) | (n = 6) | ||||||
| 6 | ? | 12 | ? | Normal menstrual cycle and free of | Mid-proliferative (days 7-10) vs. mid-secretory (days 20-24) | DIGE + MALDI-MS-MS | |
| uterine abnormalities | (n = 6) | (n = 6) | |||||
| 7 | 8 | 16 | 23–39 | Ovum donors | Early-secretory (LH+2) vs. mid-secretory (LH+7) | DIGE + MALDI-MS | |
| (n = 8) | (n = 8) | ||||||
| 8,15* | 4 | 8 | 31.3 ± 1.5 | Fertile women | Early-secretory (LH+2) vs. mid-secretory (LH+7) | DIGE + MALDI-TOF/MS | |
| (n = 4) | (n = 4) | ||||||
| Current study | 9 | 18 | 31.3 ± 1.8 | Patients referred for ICSI for male infertility | Early-secretory (LH+2) vs. mid-secretory (LH+7) | DIGE + SELDI-TOF/MS-MS | |
| (n = 9) | (n = 9) | ||||||
*Article in Chinese.
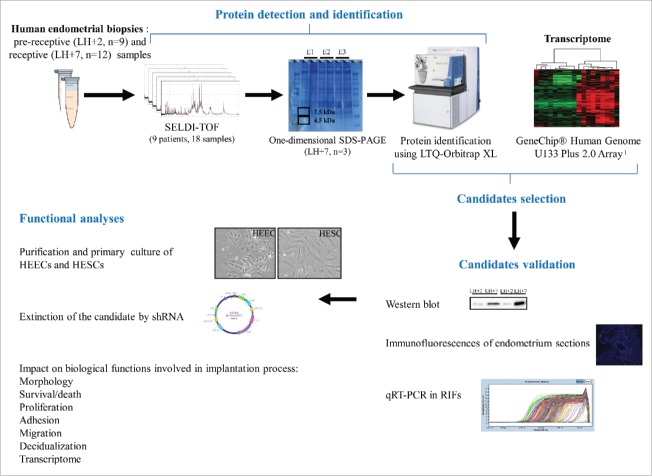
The aim of the present study was to perform a comparative proteomic analysis of endometrium samples collected at the early-secretory and mid-secretory stages from the same normal responder patients undergoing natural cycle. The differential expression of 2 selected candidates (S100A10 and S100A11) in independent endometrium samples was then validated by western blotting, RT-qPCR and/or immunofluorescence. Then, S100A10 was selected for functional analyses in primary human endometrial cells (Fig. 1).
Figure 1.
Study workflow: from proteomic pre-screening up to investigation of the function(s) of candidate proteins. E1-3, endometrial sample 1-3; HEECs, human endometrial epithelial cells; HESCs, human endometrial stromal cells; LH, luteinizing hormone; RIF, repeated implantation failure.
Results
SELDI-TOF profiling of pre-receptive and receptive endometrial samples
To identify proteins that could be involved in the acquisition of the endometrium receptivity phenotype, endometrial biopsies were performed during the pre-receptive [2 days after the LH (luteinizing hormone) surge, LH+2; n = 9 samples] and receptive phases (7 days after the LH surge, LH+7; n = 9 samples) of the same natural cycle in 9 patients (mean ± SEM, age: 31.3 ± 1.8 years) referred for intra-cytoplasmic sperm injection (ICSI) for male infertility. The protein expression profile shift between LH+2 and LH+7 samples was then investigated using the SELDI-TOF technology (Fig. 1 for a summary of the study design). A total of 80 peaks were detected after noise filtering and normalization of SELDI-TOF profiling analyses. Using the Biomarker Wizard Software 3.1 (Mann-Whitney non-parametric statistical test), 4 peaks with a significant difference (P < 0.05) between pre-receptive and receptive endometrial samples were identified [4,470 m/z, P = 0.047; 4,553 m/z, P = 0.019; 4,634 m/z, P = 0.047; 7,465 m/z, P = 0.038, respectively]. The results of the statistical analysis were compared using the Wilcoxon signed-rank test (non-parametric statistical test) and 3 peaks with significant changes were identified [4,470 m/z, P < 0.0001; 6,838 m/z, P < 0.0001; 7,465 m/z, P < 0.0001 respectively]. The two peaks (4,470 m/z and 7,465 m/z) that were found to be significantly differentially expressed by both statistical tests were selected for protein identification. Their peak intensity volume was 7.8 and 2.2 fold higher in LH+7 samples than in LH+2 samples, respectively.
Protein characterization and candidate selection
The proteins included in the 4,470 m/z and 7,465 m/z peaks were then identified using 3 additional endometrial (LH+7) biopsies from 3 patients (age: 29 ± 2.7 years) referred for ICSI (Fig. 1). Protein extracts from these biopsies were separated by one-dimensional SDS (sodium dodecyl sulfate)-PAGE electrophoresis (12% polyacrylamide gel) and 2 gel bands corresponding to 4.5 kDa and 7.5 kDa were excised and analyzed by LC-MS/MS after trypsin digestion (Fig. 1). A total of 157 and 95 proteins in the 4,470 m/z and 7,465 m/z peaks, respectively, were detected and 36 proteins were in common between the peaks. In total, 216 proteins were identified (Table S1). To select candidate proteins involved in endometrial receptivity, this list of proteins was compared with our previous transcriptomic data obtained using LH+7 and LH+2 endometrial samples to identify molecules that were over-expressed (both mRNA and protein) during the implantation window (Fig. 1).1 The identified candidates (n = 16) were then compared with previously published transcriptomic and proteomic data on similar endometrial samples to determine whether they had been previously reported (Table 2).7,10-15 Nine of the 16 identified proteins have been previously described at least once in these studies. As among these candidates there were several members of the S100 family (S100A4, S100A6, S100A10 and S100A11), we decided to focus our investigations on S100A10 and S100A11.
Table 2.
Proteins identified by SELDI-TOF-based proteomic analysis in this study and comparison with the results obtained by previous transcriptomic and proteomic studies comparing pre-receptive and receptive endometrium samples. The fold change relative to pre-receptive endometrium is indicated.
| Present study |
Transcriptomic studies |
Proteomic studies |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein name | Identification peak | Carson et al., 2002 | Riesewijk et al., 2003 | Mirkin et al., 2005 | Talbi et al., 2006 | Haouzi et al., 2009a | Diaz et al., 2011 | Li et al., 2006 | Domínguez et al., 2009 |
| APOC1 | 4,470 m/z | – | – | – | 1.6 | 2.9 | – | – | – |
| FGB | 4,470 m/z, 7,465 m/z | – | – | – | 2 | 34.6 | – | – | – |
| KRT7 | 4,470 m/z | – | 6 | – | – | 8.3 | 5.4 | – | – |
| LCP1 | 7,465 m/z | – | – | 2.6 | 1.6 | – | – | – | 1.6 |
| MYL9 | 4,470 m/z | – | 6 | – | – | 4.7 | – | – | – |
| S100A4 | 7,465 m/z | – | 7 | – | 2.5 | 3.9 | 4.6 | – | – |
| S100A6 | 7,465 m/z | – | – | – | 1.8 | 4.5 | – | – | – |
| TAGLN | 4,470 m/z | – | 6 | – | – | 5.9 | – | – | 1.7 |
| S100A10 | 7,465 m/z | – | – | – | – | 3.5 | – | – | 4.8 |
| ENO2 | 4,470 m/z, 7,465 m/z | – | – | – | – | 2.3 | – | – | – |
| GLRX | 7,465 m/z | – | – | – | – | 3.4 | – | – | – |
| LMOD1 | 4,470 m/z | – | – | – | – | 10.2 | – | – | – |
| PTRF | 4,470 m/z, 7,465 m/z | – | – | – | – | 2.0 | – | – | – |
| PYGB | 4,470 m/z | – | – | – | – | 4.8 | – | – | – |
| S100A11 | 7,465 m/z | – | – | – | – | 2.1 | – | – | – |
| SYNPO2 | 4,470 m/z | – | – | – | – | 2.1 | – | – | – |
S100A10 and S100A11 are overexpressed during the implantation window in the endometrium of fertile women in natural cycle
The upregulation of these 2 candidates in receptive LH+7 endometrium was first validated by western blot analysis of LH+2 and LH+7 endometrium biopsies from 2 volunteer fertile women (age: 45.5 ± 1.5 years) during the same natural cycle. Densitometric analysis indicated that S100A10 and S100A11 protein levels (relative to GAPDH) were 7.5 and 1.85 times higher during the mid-secretory stage (LH+7) compared to the early-secretory phase (LH+2) (Fig. 2A).
Figure 2.
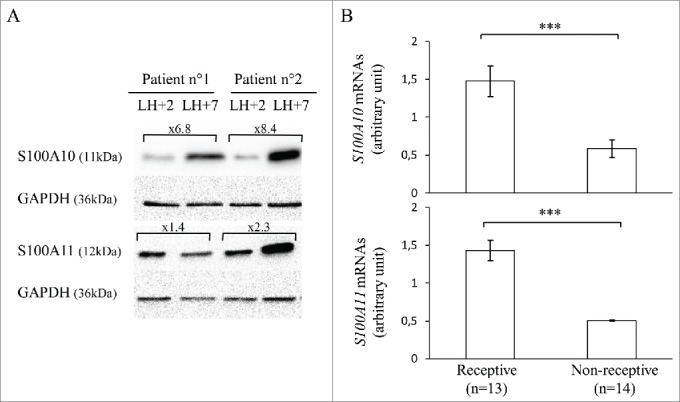
S100A10 and S100A11 expression in endometrium samples from fertile women and patients with RIF. (A) Western blot analysis of S100A10, S100A11 and GAPDH expression in pre-receptive (LH+2) and receptive (LH+7) endometrium samples obtained from 2 fertile women during the same menstrual cycle. (B) RT-qPCR analysis of S100A10 (upper panel) and S100A11 (lower panel) mRNA expression in LH+7 endometrium samples from patients with RIF classified as receptive or non-receptive using the Win-Test. S100A10 and S100A11 mRNA expression were calculated relative to HPRT1 expression. The error bars represent the SEM; n, number of samples; *** P ≤ 0.001.
S100A10 and S100A11 are downregulated in patients with repeated implantation failure and a non-receptive endometrium
S100A10 and S100A11 expression was then evaluated by RT-qPCR in the endometrium (LH+7 samples) of infertile women (n = 27; age: 34.6 ± 4.6 years) with repeated implantation failure (RIF) during previous in vitro fertilization (IVF) cycles (≥ 3). Samples were classified as receptive (n = 13) or non-receptive (n = 14) using the Win-Test.38 S100A10 and S100A11 mRNA expression level were significantly downregulated (by 2.5- and 2.8-fold, respectively) in non-receptive endometrium samples compared with receptive endometrium samples (P = 0.0012 and 0.0001 respectively) (Fig. 2B).
For the functional analysis, we then focused only on S100A10 because of its stronger protein upregulation in mid-secretory endometrium compared to early-secretory samples.
S100A10 is expressed in both HESCs and HEECs and it is maintained during culture
Immunofluorescence analysis of paraffin-embedded receptive endometrium tissue sections from the 2 fertile women showed that S100A10 was expressed in both luminal and glandular endometrial epithelial cells and in endometrial stromal cells (Fig. 3A). To investigate the functional roles of S100A10 in endometrium, human endometrial stromal cells (HESCs) and human endometrial epithelial cells (HEECs) were isolated from the endometrial biopsies (LH+7) of the 2 fertile women. After one passage, the purity of HESC and HEEC cultures was > 95%, based on their morphological features, and reached 98% at passage 5 (Fig. 3B). Primary HESCs and HEECs could be maintained in culture up to passage 10 before the onset of senescence. Analysis of S100A10 protein expression at different passages confirmed that it was expressed in both HESCs and HEECs (Fig. 3C).
Figure 3.
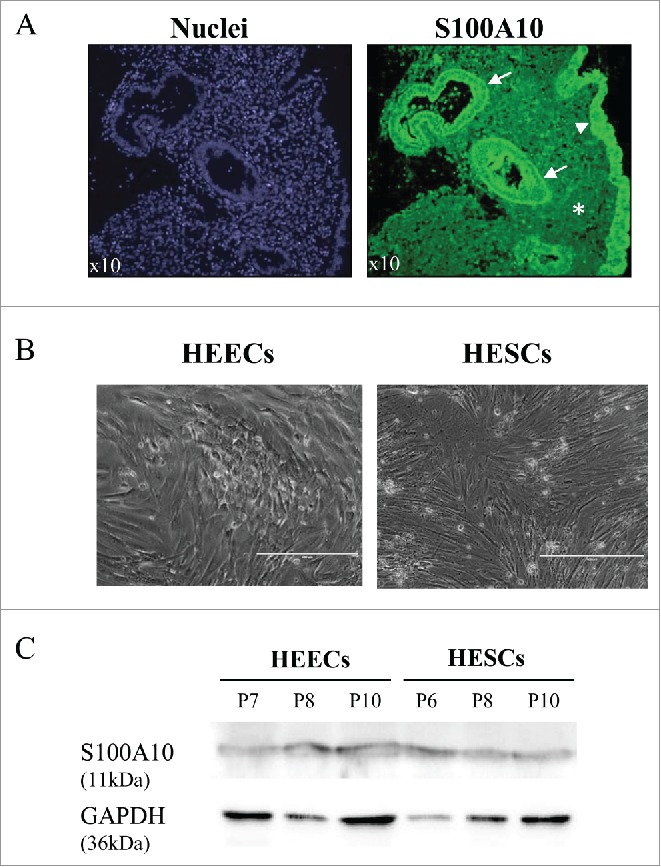
S100A10 protein expression in endometrial samples from fertile women and in primary endometrial cells. (A) Immunofluorescence analysis of paraffin-embedded endometrium tissue sections using an anti-S100A10 antibody (green) shows S100A10 expression in the endometrial glandular epithelium (arrow), luminal epithelium (arrowhead) and in stromal cells (asterisk). Nuclei (blue) were stained with DAPI. (B) Phase-contrast image of purified primary HEECs and HESCs from endometrium biopsies of 2 fertile women. Bars, 400 µm. (C) Western blot analysis of S100A10 and GAPDH protein expression in purified primary HESCs and HEECs at different passages (P).
S100A10 silencing does not affect primary HESC and HEEC morphological features and proliferation
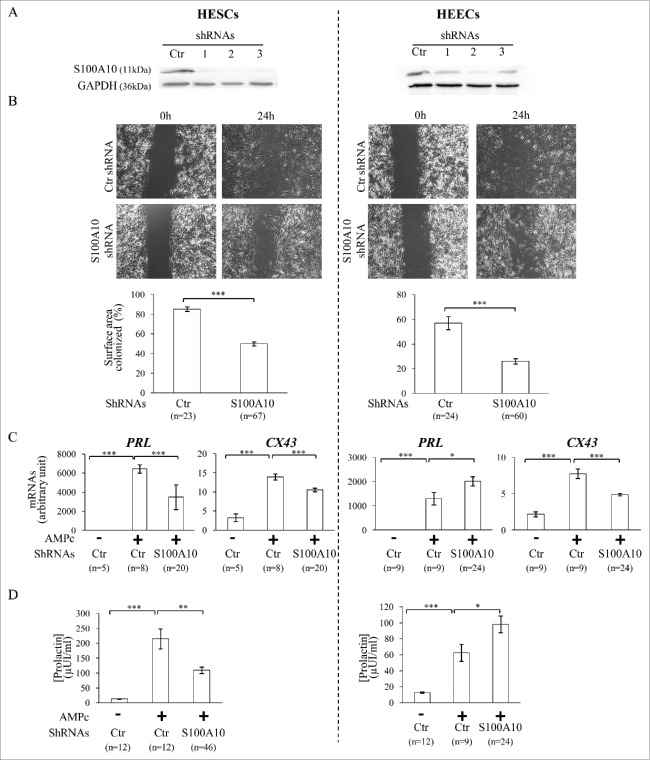
Seventy-two hours after transduction of HESCs and HEECs at passage 6 with the anti-S100A10 or control GFP (green fluorescence protein)-tagged shRNAs, 98% of cells were GFP-positive. Moreover, S100A10 protein levels were significantly reduced in HESCs and HEECs transduced with the specific S100A10 shRNAs compared to control shRNA. S100A10 protein was similarly downregulated by the different S100A10 shRNAs (1 to 3) with a mean reduction of 86 ± 7% (P = 0.0007) and 63 ± 9% (P = 0.045) in HESCs and HEECs, respectively, compared to control shRNA (Fig. 4A).
Figure 4.
(A) Western blot analysis of S100A10 and GAPDH protein levels in HESCs and HEECs transduced with control shRNA (Ctr) and 3 different shRNAs (1-3) that target specifically S100A10. These results validate the efficiency of the S100A10 shRNAs. (B) Phase contrast images of wound healing assays to test the effect of S100A10 silencing on cell migration. Images show the surface colonized by HESCs (left panels) and HEECs (right panels) transduced with control shRNA or S100A10 shRNAs at 0 and 24 hours after removal of the inserts. The lower histograms show the quantification of the results (percentage of colonized surface after 24 hours). (C) PRL and CX43 mRNA levels (markers of decidualization) in HESCs and HEECs transduced with control shRNA (Ctr) or S100A10 shRNAs after incubation (+) or not (−) with 8-Br-cAMP (cAMP). (D) Prolactin secretion in culture medium of HESCs and HEECs transduced with control shRNA (Ctr) and S100A10 shRNAs after incubation (+) or not (−) with 8-Br-cAMP (cAMP). The error bars represent the SEM. n, number of samples. *P ≤ 0.05, **P ≤ 0.01 and *** P ≤ 0.001 (Student's t test).
Compared to control shRNA, S100A10 silencing did not affect the morphological features of HESCs and HEECs. Similarly, proliferation indexes over 120 hours were not significantly different between S100A10 shRNAs and control shRNA in HESCs (2.89 ± 0.06 vs. 2.99 ± 0.1, P = 0.36) and HEECs (4.05 ± 0.21 vs. 3.48 ± 0.15, P = 0.1).
S100A10 silencing increases apoptosis in serum-starved HEECs
Following culture in medium with 1% foetal bovine serum (FBS) for 3 days, apoptosis was assessed by calculating the percentage of caspase-3-positive cells. Caspase-3 activation was observed in 35.2 ± 3.4% of S100A10-silenced HEECs (n = 10,062 cells) and in 0.8 ± 0.3% of control shRNA cells (n = 6,357 cells, P < 0.0001) and in 0.03 ± 0.01% of S100A10-silenced HESCs (n = 8,907 cells) and in 0.06 ± 0.04% of control shRNA cells (n = 14,183 cells, P = 0.34). This indicates that upon serum starvation, S100A10 silencing promotes apoptosis in HEECs, but not in HESCs.
S100A10 silencing decreases primary HESC and HEEC migration
Wound healing assay was used to assess whether S100A10 knockdown had any effect on the migration of HESCs and HEECs. After 24 hours, cell migration was significantly reduced in HESCs cells transduced with S100A10 shRNA compared to cell transduced with control shRNA (26 ± 2% of colonized surface compared with 57 ± 5%, P < 0.0001) (Fig. 4B, left panels). Similar results were obtained in HEECs (50 ± 2% of colonized surface in S100A10-silenced cells compared with 85 ± 2% in control shRNA cells, P < 0.0001) (Fig. 4B, right panels).
S100A10 silencing affects decidualization and secretory transformation of primary HESCs and HEECs, respectively
To determine whether S100A10 had a role in the decidualization of endometrial stromal cells and the secretory transformation of epithelial cells, HESCs and HEECs transduced with S100A10 shRNAs or control shRNA were incubated with 8-bromoadenosine 3′:5′-cyclic monophosphate (8-Br-cAMP) for 9 days. This treatment induced a morphological change, characterized by a typical epithelial-like morphology, in HESCs and HEECs transduced with control shRNA. This effect was confirmed by the increased mRNA expression of connexin 43 (CX43) and prolactin (PRL), 2 decidualization markers, in 8-Br-cAMP-treated compared with untreated control cells (Fig. 4C; + and – Ctr, respectively). CX43 mRNA upregulation was reduced by 45% (P = 0.0005) in S100A10-silenced HESCs and by 37% (P < 0.0001) in S100A10-silenced HEECs compared with cells transduced with control shRNA (Fig. 4C). PRL expression was also significantly reduced by 54% (P < 0.0001) in S100A10-silenced HESCs, whereas it was strongly increased by 64% (P = 0.047) in S100A10-silenced HEECs compared with cells transduced with control shRNA (Fig. 4C). These findings were validated by quantification of prolactin in the culture medium. Prolactin secretion was increased by 16.3-fold in the culture medium of 8-Br-cAMP-treated HESCs transduced with control shRNA compared with untreated cells (215 ± 33 vs. 13 ± 1 µUI/ml, P < 0.0001) and only by 8.3-fold in S100A10-silenced HESCs (110 ± 10 µUI/ml, P = 0.009, compared to control shRNA), indicating an inhibitory effect of S100A10 silencing on prolactin secretion by HESCs. Conversely, 8-Br-cAMP treatment induced prolactin secretion in both HEECs transduced with control shRNA (62 ± 11 vs. 13 ± 1 µUI/ml, P < 0.0001 compared to untreated cells) and with the S100A10 shRNAs (98 ± 10 µUI/ml, P = 0.025, compared to control shRNA), indicating that in HEECs, S100A10 silencing promotes prolactin secretion (Fig. 4D).
S100A10 is not involved in trophoblast cell adhesion
To mimic the trophoblast-decidual interface in early pregnancy, spheroids formed by JAR trophoblast cells were placed on a monolayer of 8-Br-cAMP-treated or undifferentiated HESCs or HEECs transduced with S100A10 shRNAs or control shRNA. After 30 min, the percentage of JAR spheroids attached to undifferentiated HESCs and HEECs transduced with S100A10 shRNAs or control shRNA was not significantly different (40% and 56% vs. 54% and 58%; P = 0.33 and 0.78, respectively). Similar results were obtained when JAR spheroids were placed on 8-Br-cAMP-treated HESCs and HEECs transduced with S100A10 shRNAs or control shRNA (66% and 58% vs. 62% and 63%, P = 0.57 and 0.64, respectively).
Effect of S100A10 knockdown on the transcriptome profiles of primary HEECs and HESCs
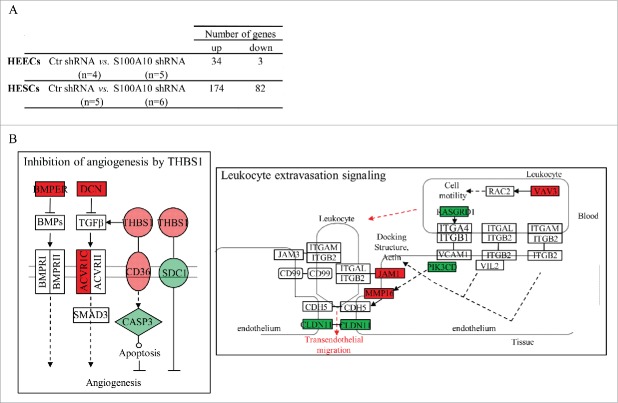
Finally, to investigate the effect of S100A10 knockdown on the transcriptome profile of primary endometrial epithelial and stromal cells, cRNA samples from undifferentiated HESCs and HEECs transduced with anti-S100A10 (n = 6 and 5, respectively) or control shRNAs (n = 5 and 4, respectively) were hybridized to HG-U133 plus 2.0 arrays. Concerning the HEEC samples, after the first selection based on the ‘present’ detection call (in at least 4 samples), a list delimiting 2,908 genes was submitted to SAM. 37 genes were differentially expressed (34 upregulated and 3 downregulated, including S100A10: x-4.4, FDR < 0.0001) in S100A10-silenced HEECs compared with cells transduced with control shRNA (Fig. 5A and complete list in Table S2). Functional annotation using Ingenuity indicated that the 37 differential expressed genes did not affect any canonical pathway.
Figure 5.
Transcriptome analysis by DNA microarrays of HEECs and HESCs transduced with control shRNA or S100A10 shRNAs. (A) Number of genes that are differentially expressed in S100A10-silenced HEECs and HESCs compared to control cells (control shRNA). (B) Deregulation of genes involved in the angiogenesis inhibition by THBS1 and leucocyte extravasation signaling pathways in S100A10-silenced HESCs compared to control cells. Green, genes downregulated by S100A10 knockdown; red, genes upregulated by S100A10 knockdown. n, number of samples.
For HESCs, SAM analysis performed on the pre-selected list that included 2,061 genes (CV ≥ 40% and a ‘present’ detection call in at least 5 samples) found 256 genes that were differentially expressed in S100A10-silenced HESCs (174 upregulated and 82 downregulated genes, including S100A10: x-9.1, FDR < 0.0001) compared with cells transduced with control shRNA (Fig. 5A and complete list in Table S3). Functional annotation of this list of genes identified 2 canonical pathways, ‘the inhibition of angiogenesis by THBS1’ [ACVR1C (x2.8), BMPER (x2.3), CASP3 (x-2.1), CD36 (x3.9), DCN (x3.7), SDC1 (x-2.1) and THBS1 (x3.9), P = 1.34E-03] and ‘the leukocyte extravasation signaling’ [CLDN11 (x-8.3), JAM1 (x2.3), MMP16 (x2.9), PIK3CD (x-2.1), RASGRD1 (x-2.1) and VAV3 (x2.5), P = 1.49E-02], that were significantly affected by S100A10 silencing in HESCs (Fig. 5B).
Discussion
Improving our knowledge on the factors that regulate the acquisition of the endometrial receptivity phenotype is crucial to better understand the mechanisms that control the embryo-endometrium dialog during human implantation. Here, we performed a proteomic study to identify proteins that are differentially expressed in pre-receptive and receptive secretory stages of human endometrium and then assessed the biological functions of one of these molecules (S100A10) in primary HEECs and HESCs. S100A10 silencing affected the migration of primary HEECs and HESCs, decidualization of HESCs as well as secretory transformation of HEECs, and promoted apoptosis in serum-starved HEECs. Moreover, transcriptome analysis showed that S100A10 silencing in human endometrial epithelial and stromal cells led to the deregulation of 37 and 256 genes, respectively, related to components of the extracellular matrix and to intercellular connections.
Here, we used the SELDI-TOF technique to determine the proteomic profiles of pre-receptive (LH+2) and receptive (LH+7) endometrial samples. Differently from other mass spectrometry approaches, this technique allows retaining proteins of a sample in a selective way on various chemical surfaces. This selective fixation, combined with the modulation of the stringency of the washes, gives the possibility to detect and analyze minority proteins or proteins with low molecular weight (< 20 kDa), such as S100A10, that would otherwise have been masked by the presence of the most abundant proteins in the sample. Due to its high sensibility, the SELDI-TOF technology allows the analysis of proteins for which the classic techniques do not give conclusive results. Indeed, due to its resolution and detection limits, the classical 2D-electrophoresis-based proteomic approach could not identify differentially expressed peaks between pre-receptive and receptive samples. For this reason, we performed proteins identification by LC-MS/MS after excision of the 2 peaks of interest (4.5 and 7.5 kDa). The potential of these 2 peaks as biomarkers of endometrial receptivity has not been evaluated. We then compared the present proteomic data with our previous transcriptomic data1 to select candidates the mRNA and protein of which are consistently over-expressed during the implantation window. Using this strategy, we identified 16 potential candidates.
Among the 16 candidate proteins there were several S100 family members (S100A4, S100A6, S100A10 and S100A11). The S100 family includes a group of low molecular weight, acidic proteins that contain 2 EF-hand calcium-binding motifs. Binding to Ca2+ results in a conformational change of S100 proteins that exposes the hydrophobic residues of the C-terminal extension, thus enabling their interaction with various target proteins. The only exception is S100A10 that is permanently locked in an active conformation.16 They are expressed in many cell types and have a broad range of extracellular and intracellular functions, including cell growth, proliferation, differentiation, apoptosis, inflammation, motility, migration and invasion.17 Elevated expression of S100 proteins is associated with several cancers. They are involved in tumor progression because of their roles in cell proliferation, metastasis, angiogenesis and immune evasion.18 S100A10 and S100A11 also have roles in fertility. Several studies have reported S100A10 upregulation during the implantation windows in humans7 and rhesus monkey19 and specifically at the implantation site in rhesus monkey. Moreover, downregulation of S100A11 during the implantation window has been related to pregnancy failure in humans.20 Here, we confirmed the upregulation of S100A10 and S100A11 mRNA and protein expression during the implantation window in receptive human endometrial samples. In addition, we found that S100A10 and S100A11 mRNA level were significantly reduced in non-receptive endometrium compared to receptive endometrium in patients with RIF. These findings suggest a key role of these 2 proteins in the acquisition of the endometrial receptivity phenotype.
To investigate S100A10 role in endometrial receptivity, we first assessed its expression. Immunofluorescence analysis of endometrial tissue sections showed that S100A10 was expressed in epithelial luminal and glandular endometrial epithelial cells and in endometrial stromal cells. This is in agreement with a recent study showing S100A10 expression in both epithelial and stromal cells, mainly localized in the cytoplasm and cytomembrane of luminal and glandular epithelia.21 Then, S100A10 silencing in primary HESCs and HEECs resulted in migration inhibition. A similar effect was previously reported in human macrophages and S100A10 role in cell migration seems to be plasmin-dependent.22,23 Indeed, S100A10 is typically found in a heterotetrameric complex with annexin A2. This complex binds to plasminogen, tissue plasminogen activator or urokinase-type plasminogen activator, leading to the conversion of plasminogen into plasmin.24 Plasmin contribution to cell migration is dependent on its ability to degrade extracellular matrix (ECM) proteins and activate other proteinases with matrix-degrading activity called metalloproteinases.23,25 In agreement, here we found that S100A10 silencing in primary endometrial cells induced the transcriptional activation of several ECM components: collagens, proteoglycans and COL4A6 in both HEECs and HESCs, COL4A5 and FN1 in HEECs, COL11A1, LAMA1, MMP16, SERPINI1, VCAN, DCN, FMOD, MCAM and NCAM2 in HESCs. Cell polarity remodelling, especially at the apico-basolateral of luminal epithelial cells, is a prerequisite for successful implantation, although we did not find any significant difference in JAR spheroid attachment rate to HESC and HEEC monolayers, and this, independently of the endometrial cell status (differentiated or undifferentiated). Moreover, functional annotation of S100A10-silencend HESCs revealed deregulation of the angiogenesis and the leukocyte extravasation signaling cascades, 2 pathways involved in the implantation process.26,27
Serum withdrawal in S100A10-silenced primary HEECs induced apoptosis. As apoptosis plays a crucial role in the endometrium during the implantation window, deregulation of this signaling pathway due to S100A10 downregulation could explain implantation failures in RIF. Indeed, embryo attachment and invasion through the endometrial epithelial layer and finally implantation in the decidualized stroma are crucial step for a successful pregnancy. Although disruption of the endometrial epithelial layer was already correlated with apoptosis,28 our data provide evidence that S100A10 plays a crucial role in apoptosis of endometrial epithelial cells. S100A10 role in apoptotic signaling could also be associated with the formation of the heterotetrameric complex with annexin A2 because this complex has been implicated also in apoptosis and tissue integrity.29 The mechanism whereby S100A10 downregulation affects apoptosis, and consequently implantation, needs to be precisely investigated.
S100A10 silencing affected the decidualization and the secretory transformation of primary HESCs and HEECs, respectively. Decidualization is a complex maturation process that involves morphological and functional changes in the endometrial stromal cell structure and physiology during the secretory phase of the menstrual cycle. More precisely, decidualization characterizes the differentiation of endometrial stromal fibroblasts (undifferentiated cells) into specialized secretory decidual cells (differentiated cells). It could be considered as the postovulatory endometrial remodelling in preparation for pregnancy. One of the hallmarks of decidualization induction is the expression of specific markers such as prolactin. Decidualization of endometrial stromal cells is mainly induced by ovarian steroids and progesterone-dependent decidualization is mediated in part by the second messenger cAMP.30 On the other hand, decidualization is endowed with the secretory transformation of the uterine glands, influx of specialized uterine natural killer cells and vascular remodelling.31 Uterine endometrial glands and their secretory products are critical for the establishment of uterine receptivity, and therefore, for successful implantation. As for decidualisation, uterine gland maturation is mediated through cAMP allowing expression of implantation-related factors including decidual markers.32,33 Decidualization has been widely investigated using in vitro culture models of stromal endometrial cells.34 However, the behavior of epithelial endometrial cells during the decidualization process of stromal cells has rarely been studied.35 Yet, they are many evidences that both luminal and glandular uterine epithelial cells play a key role in the regulation of stromal cell decidualization.36,37 Our data indicate that following incubation with 8-Br-cAMP, HESCs and HEECs secrete prolactin, a hormone playing a key role in the establishment and maintenance of pregnancy. S100A10 silencing inhibited prolactin secretion in HESCs, but significantly enhanced it in HEECs. The molecular mechanisms by which S100A10 deficiency affects differently the prolactin secretion must be investigated. Nevertheless, as decidualization of stromal cells as well as the secretory transformation of glandular epithelial cells are essential for the acquisition of the endometrial receptivity phenotype, both up- or downregulation of molecules involved in this process, such as S100A10, could contribute to embryo implantation failure.
Embryo implantation is a tremendously complicated process that involves the specific remodelling of the endometrium to become receptive, a competent embryo and the synchronization between endometrium and embryo age. Therefore, by affecting migration, decidualization and apoptosis, some major biological functions involved in the implantation process, S100A10 might play a key role during implantation. These findings suggest that S100A10 is a relevant candidate biomarker for predicting implantation failure, particularly due to inadequate endometrial receptivity.
Material and methods
Endometrial samples
Endometrial sample were from women recruited after written informed consent. For protein profiling using the SELDI-TOF technology, human endometrial biopsies were performed during the pre-receptive (LH+2) and receptive (LH+7) phases of the same natural cycle in 9 patients (mean ± SEM, age: 31.3 ± 1.8 years) referred for ICSI due to male infertility. For protein identification by the LC/MS/MS, 3 additional endometrial biopsies performed during the mid-secretory phase (LH+7) from 3 patients (age: 29 ± 2.7 years) referred for ICSI were included (Fig. 1). To validate the selected protein candidates by western blotting, immunofluorescence staining of paraffin-embedded endometrium tissue sections and quantitative RT-PCR (Fig. 1), paired endometrial biopsies (LH+2 and LH+7 during the same natural cycle) from 2 volunteer fertile women (age: 45.5 ± 1.5 years) were used. The LH surge was estimated with LH urinary kits. These women had natural pregnancies, had not taken any treatment in the previous 3 months and had no uterine pathologies. In addition, single endometrial biopsy during the implantation window (at LH+7-8, or progesterone +6-8 for patients under natural and hormonal replacement therapy, respectively) were obtained from 27 patients (age: 34.6 ± 4.6 years) with RIFs during previous IVF cycles (≥ 3).
Preparation of endometrial samples for SELDI-TOF MS/MS analysis
Total proteins were concurrently extracted from endometrial biopsies with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Pellets were resuspended in 50 µl of 50mM Tris pH 7.5 with 0.5% Triton X-100. 5 µl of each sample were used to determine the protein concentration with the bicinchoninic acid (BCA) assay (Sigma-Aldrich, Saint-Quentin Fallavier, France). Protein samples were stored at −80°C until analysis.
Profiling of endometrium proteins using SELDI-TOF MS
For SELDI-TOF analysis, 40µg of each protein sample was diluted 10 times with binding buffer (100 mM Tris pH 9 + 0.1% Triton X-100) for application on Q10 (anion exchange) ProteinChip arrays (BioRad, Hercules, CA, USA). Q10 ProteinChip arrays were pre-equilibrated with 150μL of binding buffer in a 96-well bioprocessor and incubated with gentle agitation for 5min. After removing the binding buffer from the wells, samples were added and incubated on a plate shaker at room temperature (RT) for 1 hour. Wells were washed twice with binding buffer and once with 100 mM Tris pH 9 for 5 min, followed by a final brief rinse with water. Q10 ProteinChip arrays were removed from the bioprocessor and air-dried. Finally, 0.8 μL of saturated sinapinic acid solution was applied twice to each spot and chips were allowed to air-dry. Mass spectrometric analysis was performed using SELDI-TOF in a PCS4000 ProteinChip reader (BioRad) and the same settings for all samples and for data collection (calibration, focusing mass, laser intensity and detector sensitivity). Each spectrum was an average of 530 laser shots. External calibration was done with the All-in-1 Protein Standard II (BioRad). Spectra analysis was carried out using the ProteinChip Data Manager 3.5 (BioRad). The background was subtracted using the default software settings. Peaks with a ratio signal/noise above 3 were identified by the ProteinChip software. Differentially expressed mass peaks (P < 0.05) were selected for protein identification using the Biomarker wizard software 3.1 (Ciphergen Biosystems, Fremont, CA, USA).
One-dimensional SDS-PAGE
Proteins from 3 additional endometrial biopsies (LH+7) were extracted with the RNeasy Mini Kit, as described above. The NuPAGE Electrophoresis system (Invitrogen, Saint Aubin, France) was used for protein separation. Briefly, following the manufacturer's protocol, 40 µg of each sample in 30 µl of 4X LDS (lithium dodecyl sulfate) buffer were loaded on 12% Bis-Tris gels. Protein separation was performed in MOPS running buffer at 100 V for 1 hour. Gels were stained with colloidal Coomassie following the manufacturer's protocol (GelCode, Thermo Fisher Scientific, Illkirch, France) and then scanned using an Epson 1680 scanner. Two gel bands were manually excised at around 4.5 and 7.5 kDa and stored in PCR tubes (Eppendorf, Montesson, France).
Protein identification using high resolution mass spectrometry
The excised gel bands were spun to remove the excess water. 150 µl of 50% ethanol and 50 mM ammonium bicarbonate (pH 8.4) were then added at RT for 20 min. The gel bands were dried on a vacuum concentrator (Labconco, Kansas City, MO, USA) and processed using a Bravo AssayMAP platform (Agilent, Les Ulis, France). Briefly, 30µL of denaturing solution (20 mM DTT, 100 mM Tris pH 8.5) was added at 37°C under agitation for 1 hour. Alkylation was then performed by adding 6µL of alkylation solution (400 mM iodoacetamide, 1M Tris pH 11) at 37°C for 30 min. Before the digestion step, samples were diluted with 210 µL of 20 mM Tris pH 8.5/2 mM DTT. Protein digestion with 0.5 µg trypsin was carried out at 37°C overnight and then stopped by adding 15 μL formic acid (pH < 4). The obtained peptides were desalted using C18 AssayMAP tips according to the manufacturer's instructions. Samples were then transferred to LoBind tubes (Eppendorf), dried in a vacuum concentrator (Labconco) and resuspended in 20 µL of 2% acetonitrile/0.1% formic acid/97.9% water for 10 min under agitation.
Peptides were concentrated on a pre-column (Dionex, C18 PepMap100, 300 µm × 5 mm, 5 µm, 100 A) and separated through a reversed-phase capillary column (Dionex, C18 PepMap100, 75 µm × 150 mm, 2 µm, 100 A) over 45 min. Peptide fragmentation was carried out with an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nano-ESI source and performed in the positive ion mode. Xcalibur 2.0.7 (Thermo Fisher Scientific) was used for data processing and result delivery.
Mass spectrometry (MS) data were interrogated with Mascot v2.3 against CPS_human (CPS_human_20120125). The selected enzyme was trypsin. The interrogation parameters accepted one putative missed cleavage, a 15 ppm and 0.05 Da mass range for the parent peptide and for the MS/MS fragment, respectively. As proteins were reduced and alkylated, carbamidomethylation was selected as fixed modification and methionine oxidation as variable modification. Management and validation of MS data were performed using the Proteome Discoverer software (Thermo Fisher Scientific) and Mascot V2.3 (Matrix Sciences, London, UK) with significance threshold P < 0.01, with a minimum of one peptide per protein (Table S1).
Isolation and culture of primary human endometrial cells
A portion of the endometrial biopsies (LH+7) from the 2 fertile patients was rinsed with phosphate buffered saline (PBS), minced into small fragments (0.5 – 1 mm) and digested by incubation with 300 µg/ml collagenase Type III (Sigma-Aldrich) and 40 µg/ml deoxyribonuclease I (StemCell Technologies, Inc. Inc.., Vancouver, Canada) in PBS at 37°C for 30 min. For HEEC isolation, Dynabeads Epithelial Enrich beads coated with the monoclonal antibody BerEP4 against the human epithelial antigen EpCAM (Invitrogen) were used. Then, the supernatant was incubated with Dynabeads bound to the antibody against CD45 (Invitrogen) that targets leukocytes, thus leaving purified HESCs. Primary endometrial cells were culture in Dulbecco's modified Eagle medium (DMEM)/F12 supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin solution (Invitrogen). HEEC cultures were enriched with 10 ng/ml of recombinant human long epidermal growth factor (GroPep Bioreagents, Thebarton, Australia). HESCs and HEECs were grown in 5% CO2 at 37°C. Culture medium was routinely changed and sub-confluent monolayers were passaged with 0.25% trypsin-EDTA (Invitrogen). The purity of HESC and HEEC cultures was evaluated based on their morphological features.
Western blotting
Total proteins from endometrial biopsies (n=2 LH+2 and n = 2 LH+7 samples) from the 2 fertile women and from primary endometrial cells were directly extracted in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100 supplemented with 1X protease inhibitors cocktail (Roche, Mannheim, Germany)). After tissue or cell dissociation by pipetting, suspensions were incubated at 4°C for 30 min and centrifuged at 16100g for 10 min. Supernatants were recovered for protein quantification using the BCA assay kit. Proteins from endometrial biopsies (20 µg) or primary endometrial cells (15 µg) were separated by SDS-polyacrylamide gel electrophoresis (12% polyacrylamide), transferred to nitrocellulose membranes and incubated with 2 µg/ml mouse anti-human S100A11 monoclonal antibody (R&D systems, Lille, France) or 0.4 µg/ml rabbit anti-human S100A10 polyclonal antibody (Sigma-Aldrich), and 0.05 µg/ml mouse anti-human GAPDH monoclonal antibody (R&D systems). Blots were then incubated with the relevant peroxidase-conjugated anti-immunoglobulin and revealed using an ECL detection system (SuperSignal West Pico chemiluminescent substrate, Thermo Fisher Scientific). Bands were visualized using the molecular imager ChemiDoc XRS System (BioRad). For quantification, the expression of the target protein was normalized to GAPDH and expressed as arbitrary unit.
Immunofluorescence staining
Endometrial biopsies from the 2 fertile women were fixed in 4 % neutrally buffered formaldehyde (DiaPath S.p.A, Italy) and paraffin-embedded. Tissue sections (5µm thickness) were deparaffinized and rehydrated and rinsed 3 times with PBS for 10 min. Then, they were permeabilized in 0.1% Triton X-100 for 10 min, rinsed in PBS and blocked with 1% bovine serum albumin (BSA). Sections were incubated with 1 µg/ml rabbit polyclonal anti-S100A10 antibody (Sigma-Aldrich) at 4°C overnight, followed by the appropriate fluorochrome-labeled secondary antibody for 1 hour at room temperature. Control sections were stained with the secondary antibody only. Nuclei were counterstained with 10 µg/ml DAPI. DAKO fluorescent mounting medium was used (DAKO, Les Ulis, France). Images were acquired with an Axiovert 200 inverted fluorescence microscope (Carl Zeiss, France).
ShRNA-mediated silencing
S100A10 MISSION Lentiviral transduction particles (S100A10 shRNA) and non-target scrambled DNA (control shRNA) were from Sigma-Aldrich. The shRNA vector was pLKO.1-puro-CMV-TurboGFP and the DNA sequences are reported in Table S4. Primary endometrial cells were seeded in T-175 flasks to reach 50-80% confluence in 24 hours. Cells were transduced with 8 µg/ml hexadimethrine bromide (Sigma-Aldrich) and the lentiviral particles at a multiplicity of infection of 20. After 24 hours, the medium containing the viral particles was replaced with fresh medium. The day after, infected cells were selected with 1 µg/ml puromycin dihydrochloride (Sigma-Aldrich) for 48 hours and then selection was maintained by adding 0.1 µg/ml puromycin dihydrochloride to the medium that was changed every 3 days. GFP expression in the infected cells was observed using an EVOS digital inverted microscope (Advanced Microscopy Group, Thermo Fisher Scientific). Gene silencing was validated by western blotting.
Migration assay and time-lapse video microscopy
For the wound healing experiments, 25,000 cells in 70 µl of culture medium were plated in each IBIDI culture insert in a 12-well plate to reach sub-confluence in 24 hours (Biovalley, Marne la Vallée, France). Then, the inserts were removed to form a 500 µm cell-free gap. Cells were rinsed carefully with PBS and fresh medium was added. Cell migration was monitored by time-lapse video-microscopy using an Axiovert 200 motorized inverted microscope at 37°C in 5% CO2 (5x magnification). Images were acquired at 5 min intervals for 24 hours. Three independent experiments were performed corresponding to 90 IBIDI culture inserts for HESCs (23 for cells transduced with control shRNA and 67 for S100A10 shRNAs) and 84 inserts HEECs, (24 for cells transduced with control shRNA and 60 for S100A10 shRNAs). Images were analyzed with the ImageJ macro MRI Wound healing tool (Montpellier RIO Imaging, Montpellier, France). The percentage of colonized surface was measured 24 hours after insert removal.
Decidualization assay
Cells were seeded in 12-well plates at a density of 120,000 cells/well (HESCs) and 135,000 cells/well (HEECs) to reach 80% confluence after 24 hours. Cells were then incubated with 0.5 mM 8-Br-cAMP (Sigma-Aldrich). Cells were photographed with an EVOS digital inverted microscope (AMG). After 9 days of treatment (medium with 8-Br-cAMP was changed every 3 days), cells were harvested in RLT buffer (RNeasy Mini Kit, Qiagen) to prepare RNA for RT-qPCR analysis of biomarkers of cells decidualization (CX43 and PRL). Spent culture medium was also collected and stored at −80°C for prolactin quantification.
Prolactin quantification
Prolactin secretion in the spent medium of primary endometrial cell cultures incubated or not with 8-Br-cAMP was measured with the automated immunoassay system BRAHMS KRYPTOR (BRAHMS, Clichy, France). The inter-assay and intra-assay coefficients of variation were 3.7% and 3.5%, respectively, and the limit of sensitivity was 5 µUI/ml.
JAR spheroid attachment assay
To study the interactions of human trophoblast cells with endometrium we employed an in vitro model where spheroids made using the human choriocarcinoma cell line JAR (ATCC, Molsheim, France) were co-cultured on a monolayer of primary endometrial cells. Multicellular JAR spheroids were prepared by culturing a cell suspension of 850,000 JAR cells per ml in RPMI 1640 medium supplemented with L-glutamine (Thermo Fisher scientific), 10% heat-inactivated FBS and 1% penicillin/streptomycin solution with shaking at 40 rpm at 37°C for 24 hours. Control and S100A10 shRNA-expressing HEECs and HESCs were cultured to reach confluence in Lab-Tek Chamber Slide System wells (8 wells, Dominique Dutscher, Brumath, France). Medium was removed from confluent monolayers of HEECs and HESCs and replaced by fresh RPMI medium supplemented, as described above. JAR spheroids (100-150 µm) were deposited on confluent HEEC and HESC monolayers (5-10 spheroids per well). After 30 min, non-adherent spheroids were removed by gentle pipetting. The number of attached JAR spheroids was counted under a binocular microscope. Spheroid attachment to endometrial cell monolayers was expressed as the percentage of adherent spheroids divided by the total number of deposited spheroids. The same experiment was performed also with control and S100A10 shRNA-expressing HEECs and HESCs after treatment with 8-Br-cAMP for 9 days.
Apoptosis quantification
Control and S100A10 shRNA-expressing cells were plated on coverslips in 24-well plates at a density of 65,000 (HESCs) and 80,000 (HEECs) cells/well and cultured in complete DMEM/F12 medium with 1% heat-inactivated FBS. After 3 days, cells were fixed in 4% paraformaldehyde for 10 min, rinsed in PBS and blocked with 1% BSA. Cells were incubated with the polyclonal anti-caspase-3 antibody (Cell Signaling) followed by the appropriate fluorochrome-labeled secondary antibody (both at RT for 1 hour). Nuclei were counterstained with 10 µg/ml DAPI and cells mounted with DAKO fluorescent mounting medium. Images were acquired with an Axiovert 200 inverted fluorescence microscope. The apoptosis rate was calculated as the percentage of caspase-3-positive cells relative to the total number of cells visualized by DAPI.
Quantitative RT-PCR analyses
500 µg of RNA from endometrial biopsies and 200 µg of RNA from primary endometrial cells were used for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis according to the manufacturer's recommendations (Applied Biosystems, Villebon sur Yvette, France). Endometrium samples were obtained from patients with RIF diagnosed as receptive (n = 13) and non-receptive (n = 14) using the Win-Test (Window Implantation Test).38 For qPCR, 2µl (of a 1:5 dilution) first strand DNA were added to a 10 µl reaction mixture containing 0.25 µM of each primer and 5 µl of 2X LightCycler 480 SYBR Green I Master mix (Roche). DNA was amplified for 45 cycles with annealing temperature at 63°C using the Light Cycler 480 detection system (Roche). Gene expression was normalized to HPRT1 expression using the following formula: Etested geneΔCt / EHPRT1ΔCt (E =10−1/slope), ΔCt = Ct control – Ct unknown, where E corresponds to the efficiency of the PCR reaction. The E value was obtained by a standard curve that varies in function of the used primers. A receptive endometrium sample was used as control. Each sample was analyzed in duplicate and multiple water blanks were included. The primer sequences are in Table S5.
Microarray hybridization and data analysis
Affymetrix microarrays were processed at the Microarray Core Facility of the Institute for Regenerative Medicine and Biotherapy, CHRU-INSERM-UM Montpellier (http://irmb.chu-montpellier.fr). Total RNA (100 ng) was used to prepare twice-amplified and labeled cRNA samples for hybridization with HG-U133 plus 2.0 arrays (Affymetrix™, United Kingdom, UK), as described in Haouzi.38 Each endometrial sample was processed individually on a separate DNA microarray chip. Scanned GeneChip images were processed using the Affymetrix GCOS 1.4 software to obtain the intensity value signal and the absent/present detection call for each probe set using the default analysis settings and global scaling as first normalization method. Probe intensities were derived using the MAS5.0 algorithm.
To determine the impact of shRNA-mediated silencing of S100A10 in HEECs and HESCs, we compared gene expression profiles between cells transduced with control shRNA (n = 4 for HEECs and n = 5 for HESCs) and with S100A10 shRNAs (n = 5 for HEECs and n = 6 for HESCs). A first selection was carried out using the detection call (present in a least 4 HEEC samples and 5 HESC samples) and a coefficient of variation (CV) ≥ 40 % between HEECs and HESCs. Then, the significant analysis of microarrays39 (SAM, Stanford University, USA) was used to identify genes that were significantly differentially expressed in S100A10 shRNA-expressing HEECs or HESCs compared with control shRNA-expressing HEECs or HESCs. The list of differentially expressed genes (Fold change, FC >2; False discovery rate, FDR < 5%) was submitted to Ingenuity (http://www.ingenuity.com) to identify the signaling pathways altered in S100A10-silenced endometrial cells.
Statistical analyses
Excepted for proteomics and transcriptomic data, statistical analyses were performed with the GraphPad InStat 3 software. Data are expressed as the mean ± SEM and differences between groups were considered significant when the Student's t-test gave a P < 0.05.
Supplementary Material
Abbreviations
- 8-Br-cAMP
8-bromoadenosine 3′:5′-cyclic monopho-sphate
- COL
collagen
- CX43
connexin 43
- DCN
decorin
- ECM
extracellular matrix
- FBS
foetal bovine serum
- FDR
false discovery rate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HEECs
human endometrial epithelial cells
- HESCs
human endometrial stromal cells
- IVF
in vitro fertilization
- ICSI
intra-cytoplasmic sperm injection
- LC
liquid chromatography
- LH
luteinizing hormone
- MMP
matrix metallopeptidase
- MS/MS
tandem mass spectrometry
- PRL
prolactin
- RIF
repeated implantation failure
- S100A10
S100 calcium binding protein A10
- S100A11
S100 calcium binding protein A11
- SDS
sodium dodecyl sulfate
- SELDI-TOF
Surface-enhanced laser desorption/ionization time-of-fligh
- shRNA
short hairpin RNA
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the ART teams from Montpellier Hospital for their assistance during this study. We thank Dr. Paris F. for her help and implication in prolactin quantification in culture medium, as well as Dr Jacques Faidherbe for his help in patients' recruitment.
Author contributions
LB performed most of the experiments and analyses and participated in the paper redaction; LD performed the apoptosis experiments and their quantification; YA performed RT-qPCR in RIF patients; LT, CH, SL performed proteomic experiments and analyses, contributed to proteomic data interpretation and participated in the paper redaction; IJK, FB participated to the patients' recruitment; HP performed endometrial tissue sections; DH, SH participated in the study conception/design, data interpretation, paper redaction and final approval.
Funding
This work was partially supported by a grant from the Ferring Pharmaceutical Company and FEDER (Fonds Européen de Développement Régional).
References
- [1].Haouzi D, Mahmoud K, Fourar M, Bendhaou K, Dechaud H, De Vos J, Rème T, Dewailly D, Hamamah S. Identification of new biomarkers of human endometrial receptivity in the natural cycle. Hum Reprod 2009; 24:198-205; PMID:18835874; http://dx.doi.org/doi/ 10.1093/humrep/den360 [DOI] [PubMed] [Google Scholar]
- [2].Haouzi D, Dechaud H, Assou S, De Vos J, Hamamah S. Insights into human endometrial receptivity from transcriptomic and proteomic data. Reprod Biomed Online 2012; 24:23-34; PMID:22119322; http://dx.doi.org/doi/ 10.1016/j.rbmo.2011.09.009 [DOI] [PubMed] [Google Scholar]
- [3].DeSouza L, Diehl G, Yang EC, Guo J, Rodrigues MJ, Romaschin AD, Colgan TJ, Siu KW. Proteomic analysis of the proliferative and secretory phases of the human endometrium: protein identification and differential protein expression. Proteomics 2005; 5:270-81; PMID:15602768 [DOI] [PubMed] [Google Scholar]
- [4].Chen JI, Hannan NJ, Mak Y, Nicholls PK, Zhang J, Rainczuk A, Stanton PG, Robertson DM, Salamonsen LA, Stephens AN. Proteomic characterization of midproliferative and midsecretory human endometrium. J Proteome Res 2009; 8:2032-44; PMID:19714818 [DOI] [PubMed] [Google Scholar]
- [5].Parmar T, Gadkar-Sable S, Savardekar L, Katkam R, Dharma S, Meherji P, Puri CP, Sachdeva G. Protein profiling of human endometrial tissues in the midsecretory and proliferative phases of the menstrual cycle. Fertil Steril 2009; 92:1091-03; PMID:18793766; http://dx.doi.org/doi/ 10.1016/j.fertnstert.2008.07.1734 [DOI] [PubMed] [Google Scholar]
- [6].Rai P, Kota V, Sundaram CS, Deendayal M, Shivaji S. Proteome of human endometrium: identification of differentially expressed proteins in proliferative and secretory phase endometrium. Proteomics Clin Appl 2010; 4:48-59; PMID:21137015; http://dx.doi.org/doi/ 10.1002/prca.2009-00094 [DOI] [PubMed] [Google Scholar]
- [7].Domínguez F, Garrido-Gómez T, López JA, Camafeita E, Quiñonero A, Pellicer A, Simón C. Proteomic analysis of the human receptive versus non-receptive endometrium using differential in-gel electrophoresis and MALDI-MS unveils stathmin 1 and annexin A2 as differentially regulated. Hum Reprod 2009; 24:2607-17; PMID:19556289; http://dx.doi.org/ 10.1093/humrep/dep230 [DOI] [PubMed] [Google Scholar]
- [8].Li J, Tan Z, Li M, Xia T, Liu P, Yu W. Proteomic analysis of endometrium in fertile women during the prereceptive and receptive phases after luteinizing hormone surge. Fertil Steril 2011; 95:1161-63; PMID:20979995; http://dx.doi.org/doi/ 10.1016/j.fertnstert.2010.09.033 [DOI] [PubMed] [Google Scholar]
- [9].Altmäe S, Esteban FJ, Stavreus-Evers A, Simón C, Giudice L, Lessey BA, Horcajadas JA, Macklon NS, D'Hooghe T, Campoy C, et al.. Guidelines for the design, analysis and interpretation of ‘omics’ data: focus on human endometrium. Hum Reprod Update 2014; 20:12-28; PMID:24082038; http://dx.doi.org/ 10.1093/humupd/dmt048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carson DD, Lagow E, Thathiah A, Al-Shami R, Farach-Carson MC, Vernon M, Yuan L, Fritz MA, Lessey B. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod 2002; 8:871-79; PMID:12200466 [DOI] [PubMed] [Google Scholar]
- [11].Riesewijk A, Martín J, van Os R, Horcajadas JA, Polman J, Pellicer A, Mosselman S, Simón C. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod 2003; 9:253-64; PMID:12728018 [DOI] [PubMed] [Google Scholar]
- [12].Mirkin S, Arslan M, Churikov D, Corica A, Diaz JI, Williams S, Bocca S, Oehninger S. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod 2005; 20:2104-17; PMID:15878921 [DOI] [PubMed] [Google Scholar]
- [13].Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA, et al.. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006; 147:1097-1121; PMID:16306079 [DOI] [PubMed] [Google Scholar]
- [14].Díaz-Gimeno P, Horcajadas JA, Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A, Simón C. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril 2011; 95:50-60, 60.e1-15. PMID:20619403; http://dx.doi.org/ 10.1016/j.fertnstert.2010.04.063 [DOI] [PubMed] [Google Scholar]
- [15].Li J, Tan Z, Li MT, Liu YL, Liu Q, Gu XF, Zhou JZ, Zhuang GL. [Study of altered expression of annexin IV and human endometrial receptivity]. Zhonghua Fu Chan KeZaZhi 2006; 41:803-5; PMID:17327107 [PubMed] [Google Scholar]
- [16].Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J 2006; 396:201-214; PMID:16683912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med 2013; 13:24-57; PMID:22834835 [PMC free article] [PubMed] [Google Scholar]
- [18].Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015; 15:96-109; PMID:25614008; http://dx.doi.org/ 10.1038/nrc3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun XY, Li FX, Li J, Tan YF, Piao YS, Tang S, Wang YL. Determination of genes involved in the early process of embryonic implantation in rhesus monkey (Macaca mulatta) by suppression subtractive hybridization. Biol Reprod 2004; 70:1365-73; PMID:14724130 [DOI] [PubMed] [Google Scholar]
- [20].Liu XM, Ding GL, Jiang Y, Pan HJ, Zhang D, Wang TT, Zhang RJ, Shu J, Sheng JZ, Huang HF. Down-regulation of S100A11, a calcium-binding protein, in human endometrium may cause reproductive failure. J Clin Endocrinol Metab 2012; 97:3672-83; PMID:22869607; http://dx.doi.org/doi/ 10.1210/jc.2012-2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wei X, Tong S, Yan Q. Cyclic changes of S100A10 expression in human endometrium [in Chinese]. Zhonghua Yi Xue Za Zhi 2014; 94:2152-2155; PMID:25327866 [PubMed] [Google Scholar]
- [22].O'Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood 2010; 116:1136-46; PMID:20424186; http://dx.doi.org/doi/ 10.1182/blood-2010-01-264754 [DOI] [PubMed] [Google Scholar]
- [23].Phipps KD, Surette AP, O'Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. 2011; 71:6676-83; PMID:22042827; http://dx.doi.org/doi/ 10.1158/0008-5472 [DOI] [PubMed] [Google Scholar]
- [24].Kwon M, MacLeod TJ, Zhang Y, Waisman DM. S100A10, annexin A2, and annexin a2 heterotetramer as candidate plasminogen receptors. Front Biosci. 2005; 10:300-25; PMID:15574370 [DOI] [PubMed] [Google Scholar]
- [25].Gross SR, Sin CG, Barraclough R, Rudland PS. Joining S100 proteins and migration: for better or for worse, in sickness and in health. Cell Mol Life Sci 2014; 71:1551-79; PMID:23811936; http://dx.doi.org/ 10.1007/s00018-013-1400-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dominguez F, Yáñez-Mó M, Sanchez-Madrid F, Simón C. Embryonic implantation and leukocyte transendothelial migration: different processes with similar players? FASEB J 2005; 19:1056-60; PMID:15985528 [DOI] [PubMed] [Google Scholar]
- [27].Haouzi D, Assou S, Mahmoud K, Tondeur S, Rème T, Hedon B, De Vos J, Hamamah S. Gene expression profile of human endometrial receptivity: comparison between natural and stimulated cycles for the same patients. Hum Reprod 2009; 24:1436-45; PMID:19246470; http://dx.doi.org/doi/ 10.1093/humrep/dep039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Boeddeker SJ, Hess AP. The role of apoptosis in human embryo implantation. J Reprod Immunol 2015; 108:114-22. PMID:25779030; http://dx.doi.org/ 10.1016/j.jri.2015.02.002 [DOI] [PubMed] [Google Scholar]
- [29].Hitchcock JK, Katz AA, Schäfer G. Dynamic reciprocity: the role of annexin A2 in tissue integrity. J Cell Commun Signal 2014; 8:125-33; PMID:24838661; http://dx.doi.org/doi/ 10.1007/s12079-014-0231-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brar AK, Frank GR, Kessler CA, Cedars MI, Handwerger S. Progesterone-dependent decidualization of the human endometrium is mediated by cAMP. Endocrine 1997; 6:301-07; PMID:9368687 [DOI] [PubMed] [Google Scholar]
- [31].Gellersen B, Brosens IA, Brosens JJ. Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med 2007; 25:445-53; PMID:17960529 [DOI] [PubMed] [Google Scholar]
- [32].Kusama K, Yoshie M, Tamura K, Imakawa K, Tachikawa E. EPAC2-mediated calreticulin regulates LIF and COX2 expression in human endometrial glandular cells. J Mol Endocrinol 2015; 54:17-24; PMID:25378661; http://dx.doi.org/ 10.1530/JME-14-0162 [DOI] [PubMed] [Google Scholar]
- [33].Kusama K, Yoshie M, Tamura K, Imakawa K, Isaka K, Tachikawa E. Regulatory Action of Calcium Ion on Cyclic AMP-Enhanced Expression of Implantation-Related Factors in Human Endometrial Cells. PLoS One 2015; 10:e0132017; PMID:26161798; http://dx.doi.org/doi/ 10.1371/journal.pone.0132017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Blois SM, Klapp BF, Barrientos G. Decidualization and angiogenesis in early pregnancy: unravelling the functions of DC and NK cells. J Reprod Immunol 2011; 88:86-92; PMID:21227511; http://dx.doi.org/ 10.1016/j.jri.2010.11.002 [DOI] [PubMed] [Google Scholar]
- [35].Wang H, Pilla F, Anderson S, Martínez-Escribano S, Herrer I, Moreno-Moya JM, Musti S, Bocca S, Oehninger S, Horcajadas JA. A novel model of human implantation: 3D endometrium-like culture system to study attachment of human trophoblast (Jar) cell spheroids. Mol Hum Reprod 2012; 18:33-43; PMID:21989169; http://dx.doi.org/doi/ 10.1093/molehr/gar064 [DOI] [PubMed] [Google Scholar]
- [36].Pawar S, Starosvetsky E, Orvis GD, Behringer RR, Bagchi IC, Bagchi MK. STAT3 regulates uterine epithelial remodeling and epithelial-stromal crosstalk during implantation. Mol Endocrinol 2013; 27:1996-2012; PMID:24100212; http://dx.doi.org/doi/ 10.1210/me.2013-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pawar S, Laws MJ, Bagchi IC, Bagchi MK. Uterine epithelial estrogen receptor alpha controls decidualization via a paracrine mechanism. Mol Endocrinol 2015; 29:1362-74; PMID:26241389; http://dx.doi.org/doi/ 10.1210/me.2015-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Haouzi D, Bissonnette L, Gala A, Assou S, Entezami F, Perrochia H, Dechaud H, Hugues JN, Hamamah S. Endometrial receptivity profile in patients with premature progesterone elevation on the day of HCG administration. Biomed Res Int 2014; 2014:951937; PMID:24877150; http://dx.doi.org/doi/ 10.1155/2014/951937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98:5116-21; PMID:11309499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.