Abstract
Gibberellic acid (GA) signaling regulates diverse aspects of plant growth and developmental processes. The DELLA repressors of GA signaling are named for an N-terminal conserved DELLA domain. In this study, four genes encoding DELLA proteins, PeRGA1, PeRGA2, PeGAI1 and PeGAI2, were isolated and characterized in poplar. A gene structural analysis revealed that the DELLA genes were all intron-free. Multiple protein sequence alignments revealed that these proteins contained seven highly conserved domains: the DELLA domain, the TVHYNP domain, leucine heptad repeat I (LHR I), the VHIID domain, leucine heptad repeat II (LHR II), the PFYRE domain, and the SAM domain. Temporal expression patterns of these genes were profiled during the adventitious root development of poplar. The four DELLA genes were expressed in root, stem and leaf in a dynamic manner. The subcellular localization demonstrated that these DELLA genes were mainly localized to the nucleus. These results suggest that the four DELLA genes may play diverse regulatory roles in the adventitious root, stem and leaf development of poplar, and contribute to improving our understanding of conserved and divergent aspects of DELLA proteins that restrain GA signaling in various species.
Electronic supplementary material
The online version of this article (doi:10.1186/s40064-016-2728-x) contains supplementary material, which is available to authorized users.
Keywords: DELLA proteins, GA signaling, Expression profiles, Protoplast transfection
Background
Gibberellic acid (GA) signaling plays a pivotal role in plant developmental processes and adaptive responses. The repression by the DELLA repressors can be relieved in response to GA by their rapid GA-induced degradation via the ubiquitin–proteasome system. DELLA proteins, representing a subset of the plant-specific GRAS family of transcription factors, exhibit considerable sequence homology to each other throughout their carboxyl termini, including two leucine heptad repeats(LHRI and LHRII) and three conserved motifs, VHIID, PFYRE and SAW (Davière and Achard 2016). Additionally, DELLA proteins contain an N-terminal conserved DELLA domain, which is involved in recognizing GA signaling and interacts with the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) (Sun and Gubler 2004; Murase et al. 2008).
Studies have indicated that DELLA proteins negatively regulate the GA signaling pathway and restrain plant growth (Harberd 2003; Achard and Genschik 2009). GA binding to GID1 stimulates the formation of the GA-GID1-DELLA complex, which can induce the degradation of the DELLAs (Davière and Achard 2013). The GA-GID1-DELLA complex can interact with an F-box protein. F-box proteins are components of the SCF (SKP1, CULLIN, F-BOX) E3 ubiquitin-ligase complexes, which catalyze DELLA protein degradation through the 26S proteasome, and then relieve the inhibitory effect of DELLA proteins on plant growth (Lechner et al. 2006). The Arabidopsis thaliana genome codes for at least five DELLA proteins that are assigned to three classes (GAI, RGA and RGA-LIKE proteins). RGA and GAI play important roles in cell division and cell expansion in root, hypocotyl, shoot, and floral induction (Dill and Sun 2001; King et al. 2001; Feng et al. 2008; Lucas et al. 2008; Davière et al. 2014). RGL1 modulates floral development (Tyler et al. 2004). RGL2 is a major inhibitor of seed germination (Lee et al. 2002; Cao et al. 2005), and RGL3 contributes to plant fitness during environmental stress (Achard et al. 2008; Wild et al. 2012).
Despite the detailed analysis of GA signaling in several herbaceous model organisms, there are no reports on the isolation and functional characterization of DELLA genes in poplar. In this study, four genes encoding DELLA proteins involved in adventitious rooting were isolated from poplar, and their genomic structures, sequence similarities, expression patterns and subcellular localizations were revealed.
Methods
Plant materials
All of the sampled plantlets of the elite clone “Nanlin895” poplar (Populus deltoids × Populus euramericana) were cultivated on Murashige and Skoog (MS) medium under 16/8 h of light/dark at corresponding temperatures of 25/18 °C. Newly expanded young leaves from 6-week-old plants were used for protoplast isolation and DNA extraction. Various tissues were harvested at multiple developmental stages (1-, 2-, 3-, and 4-week-old roots: 1WR, 2WR, 3WR, and 4WR, respectively, and 4-week-old leaves and stems: 4WL and 4WS, respectively) during adventitious rooting on 4-week-old stem cuttings Additional file 1: Figure S1, then quickly frozen in liquid nitrogen, and then stored at –80 °C until RNA extraction.
Cloning and sequencing of full-length DELLA genes
Total RNA was extracted from samples using the RNeasy Plant Mini Kit (QIAGEN), and was treated with RNase-free DNase I (TaKaRa). The concentration and integrity of RNA was quantified using the ND-2000 spectrophotometer (Nanodrop) and electrophoresis on 1 % agarose gel respectively. The DNase-treated RNA was used for rapid amplification of cDNA ends (RACE) and then reverse transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa) for RT-PCR. Based on probe sequences provided by the GeneChip Poplar Genome Array, nested primers were designed to amplify the full-length sequences with the 3′-Full RACE Core Set Kit and 5′-Full RACE Kit (TaKaRa) according to the manufacturer’s instructions. The PCR products were purified by the QIAquick Gel Extraction Kit (QIAGEN), ligated into pMD19-T vectors (TaKaRa), and then transformed into competent cells of Escherichia coli strain TOP10. White colonies were checked by PCR, and the positive colonies were sequenced. By comparing and aligning the sequences of 3′-RACE and 5′-RACE and the middle region products, the full-length cDNA sequences were obtained. The predicted open reading frames (ORFs) were subsequently amplified by PCR, and were verified by sequencing. Genomic DNA was extracted from the newly expanded young leaves using a DNeasy Plant Mini Kit (QIAGEN). Genomic DNA sequences of the above genes were amplified with the RNase-treated DNA, and were verified by sequencing. The sequences of the primers are listed in Table 1.
Table 1.
Primer sequences of the four poplar DELLA genes
| Primer_ID | Forward PCR primer (5′–3′) | Reverse PCR primer (5′–3′) |
|---|---|---|
| PeRGA1_3OUTER | CATCAAGAAACCATTGGTGGTGCT | TACCGTCGTTCCACTAGTGATTT |
| PeRGA1_3INNER | CAAAGCTGAATCTTCTTCTTCGTCAAT | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| PeRGA1_5OUTER | CATGGCTACATGCTGACAGCCTA | CCCAACTAGGGTGAGCTTCATTCG |
| PeRGA1_5INNER | CGCGGATCCACAGCCTACTGATGATCAGTCGATG | ATCGAATTCTTGTTGAGGATAGGCAGC |
| PeRGA1_ORF | ATGAAGAGAGATCATCAAGA | TCATTGACTCGGTAGCTCGA |
| PeRGA1_sRT-PCR | AAGCTTGTTTGACTCACTC | ACGTTTCATGTCGCT |
| PeRGA1_qRT-PCR | GTTGGGTTCAAAGCATG | GGGAATTGCTCTGAGAT |
| PeRGA2_3OUTER | CACTATAACCCTTCAGATCTCT | TACCGTCGTTCCACTAGTGATTT |
| PeRGA2_3INNER | TCAACAATCTACCTTCTACTGATCTTGA | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| PeRGA2_5OUTER | CATGGCTACATGCTGACAGCCTA | ATTGCATTCCCTGTTTTAAACC |
| PeRGA2_5INNER | CGCGGATCCACAGCCTACTGATGATCAGTCGATG | CTCGACTTGCATTCGCGAAAGCTTCAAG |
| PeRGA2_ORF | ATGAAGAGAGATCATCAAGAA | TCATTGTTGTGAATCACCAG |
| PeRGA2_sRT-PCR | ACTGATCTTGATTCATCTA | TGCCGGCGCAATATTAGACC |
| PeRGA2_qRT-PCR | CTTAGATTTCCCCAGTAA | CTAGAAAAACCGGACCGT |
| PeGAI1_3OUTER | CTTACTAGCACCGGTACTATGAC | TACCGTCGTTCCACTAGTGATTT |
| PeGAI1_3INNER | TGGATGAACTTTTAGCTGTTTTGGGTTA | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| PeGAI1_5OUTER | CATGGCTACATGCTGACAGCCTA | ACTCACTGATCCCTCCAACGAG |
| PeGAI1_5INNER | CGCGGATCCACAGCCTACTGATGATCAGTCGATG | CTCCGGTTTCATTTGTTTCACAACTGAT |
| PeGAI1_ORF | ATGAAAAGAGAACACTCAA | TTAAGCAGCACCGCCTACTGG |
| PeGAI1_sRT-PCR | GAGATTGTTACTGTCGTTG | CACTGAGTCAGGGTC |
| PeGAI1_qRT-PCR | GTCAAACAAATCGGCTT | AATATCTGAGAGAGAGT |
| PeGAI2_3OUTER | TAAGATCGTCAGACATGGCTGA | TACCGTCGTTCCACTAGTGATTT |
| PeGAI2_3INNER | CACGCGCAAGAAGATGGTCTTTCCCAC | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| PeGAI2_5OUTER | CATGGCTACATGCTGACAGCCTA | CTTCCCCAAGTACACCTCTGAC |
| PeGAI2_5INNER | CGCGGATCCACAGCCTACTGATGATCAGTCGATG | ATTATGGTTCGCTTCTTGCTCAACAACA |
| PeGAI2_ORF | ATGAAAAGAGAACACCCA | CTAAGCAGCACCAACTACCG |
| PeGAI2_sRT-PCR | GCATCTGATTCTGTCCATT | TGGTCGGAGAAATCGATAG |
| PeGAI2_qRT-PCR | GACCCCTCTGCTGATTCTT | TATAGGTCTGTTTTTAA |
| EF1α_qRT-PCR | GGCAAGGAGAAGGTACACAT | CAATCACACGCTTGTCAATA |
| 18S_sRT-PCR | TCAACTTTCGATGGTAGGATAGTG | CCGTGTCAGGATTGGGTAATTT |
Bioinformatics analyses
The BioEdit software was used to analyze the DNA and protein sequences. ORFs were predicted by FGENESH program (http://www.mendel.cs.rhul.ac.uk/mendel.php?topic=fgen). The theoretical isoelectric point (pI), molecular weight (MW) and amino acid composition of the proteins were predicted and calculated using Expasy Protparam (http://www.web.expasy.org/protparam/). Protein transmembrane structures, protein domain and signal peptide cleavage site analyses were preformed using the TMHMM, PROSITE and SignalP online tools, respectively. Secondary structures of amino acid sequences were predicted by the SOPMA program (https://www.npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). The phylogenetic tree was constructed using MEGA6 software with the Neighbor-Joining (NJ) method and 1000 bootstraps (Tamura et al. 2013).
RT-PCR
Total RNA extraction from various tissues and organs, and cDNA reverse transcription were performed as described above. For semi-quantitative RT-PCR, specific primers were designed by Oligo 7 software (Table 1), and they generated a PCR product of 200–400 bp, with the following reaction program: 94 °C for 5 min, followed by 25–28 cycles of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C. For real-time RT-PCR, specific primers were designed to generate an 80–150 bp PCR product (Table 1). Real-time RT-PCR was performed on an ABI ViiA™ 7 Real-time PCR system (Applied Biosystems) using FastStart Universal SYBR Green Master (Rox) for RT-PCR Kit (Roche), according to the manufacturer’s protocol. All reactions were performed in triplicate. The reactions final volume was 20 µL, containing 10 µL of FastStart Universal SYBR Green Master (Rox), 1 µL of each primer, 2 µL of cDNA, and 6 µL dH2O. The real-time PCR program was as follows: initial denaturation at 95 °C for 1 min, followed by 40 cycles of 15 s at 95 and 60 °C annealing extension for 1 min. All of the reactions were performed in triplicate. The calculations of relative expression levels between the target and the internal control EF1α (elongation factor 1-alpha) were performed using the delta-Ct method (Xu et al. 2011).
GFP fusion construct and protoplast transfection
In this study, plasmids were constructed using Gateway technology (Invitrogen), according to the manufacturer’s protocol. The PeRGA (or PeGAI) coding region (without a stop codon) was cloned into the entry vector, pCR8/GW/TOPO (Invitrogen), by a simple TOPO cloning reaction. For the subcellular localization of tagged proteins, the inset from the entry vector was transferred to its destination vector, p2GWF7, with a C-terminal GFP fusion, using an LR clonase enzyme mix (Invitrogen). The generated GFP fusion vectors (35S::PeRGA1-GFP, 35S::PeRGA2-GFP, 35S::PeGAI1-GFP and 35S::PeGAI2-GFP) were high-copy vectors, driven by the promoter of double 35S cauliflower mosaic virus (CaMV), with ampicillin as the bacterial selection marker. Protoplast isolation and polyethylene glycol-mediated transfection were performed using the method of Tan et al. (2013).
Results
Isolation and characterization of DELLA genes
Poplar DELLA genes were successfully isolated and identified by 3′-RACE and 5′-RACE procedures, and termed PeRGA1, PeRGA2, PeGAI1 and PeGAI2. Comparisons of genomic and cDNA sequences showed that these four DELLA genes were all intron-free. The full-length sequences of PeRGA1 cDNA was 2321 bp, containing an ORF of 1770 bp, flanked by 324 bp of 5′-untranslated region (UTR) and a 227 bp 3′-UTR; PeRGA2 was 2504 bp with an ORF of 1824 bp, flanked by 226 and 454 bp 5′- and 3′-UTRs, respectively; PeGAI1 was 2106 bp with an ORF of 1809 bp, flanked by 144 and 153 bp 5′- and 3′-UTRs; and PeGAI2 was 2187 bp with an ORF of 1803 bp, flanked by 61 and 323 bp 5′- and 3′-UTRs (Table 2).
Table 2.
Characteristics of DELLA genes of poplar
| Gene_ID | Full-length | 5′UTR | 3′UTR | ORF | Predicted peptide | Secondary structure prediction | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA (bp) | (bp) | (bp) | (bp) | MW (kDa) | pI | GRAVY | Hh (%) | Ee (%) | Tt (%) | Cc (%) | |
| PeRGA1 | 2321 | 324 | 227 | 1770 | 64.15 | 4.94 | −0.240 | 43.80 | 16.13 | 9.00 | 31.07 |
| PeRGA2 | 2504 | 226 | 454 | 1824 | 65.88 | 4.94 | −0.193 | 42.17 | 13.67 | 9.72 | 34.43 |
| PeGAI1 | 2106 | 144 | 153 | 1809 | 66.31 | 5.68 | −0.279 | 47.18 | 15.78 | 8.64 | 28.41 |
| PeGAI2 | 2187 | 61 | 323 | 1803 | 65.96 | 5.43 | −0.242 | 46.83 | 15.67 | 8.17 | 29.33 |
UTR untranslated region, MW molecular weight (kDa), pI isoelectric point, GRAVY grand average of hydropathicity, Hh alpha helix, Ee extended strand, Tt beta turn, Cc random coil
These cDNAs encode polypeptides of 589, 607, 602 and 600 amino acid residues, respectively. The corresponding MWs, pIs and grand averages of hydropathicity (GRAVY) for these polypeptides were 64.15 kDa, 4.94 and −0.240, respectively; 65.88 kDa, 4.94 and −0.193, respectively; 66.31 kDa, 5.68 and −0.279, respectively; and 65.96 kDa and 5.43, −0.242, respectively (Table 2). The SOPMA program was used to predict the secondary structures of these four proteins. The PeRGA1, PeRGA2, PeGAI1 and PeGAI2 proteins contained 43.80, 42.17, 47.18 and 46.83 % of alpha helices, respectively; and correspondingly 16.13, 13.67, 15.78 and 15.67 % of extended strands, respectively; 9.00, 9.72, 8.64 and 8.17 % of beta turns, respectively; and 31.07, 34.43, 28.41 and 29.33 % of random coils, respectively (Table 2).
The multiple alignment of PeRGA1, PeRGA2, PeGAI1 and PeGAI2 with other plant DELLA proteins (PtoGAI, Paulownia tomentosa, AFP58844.1; AtGAI, Arabidopsis thaliana, Y15193; AtRGL1, A. thaliana, AY048749; AtRGL2, A. thaliana, NP_186995), AtRGL3, A. thaliana, AL391150; PtRGA, Populus trichocarpa, XP_002302975.1; VviGAI1, Vitis vinifera, XP_002266267.1; CusGAI, Cucumis sativus, XP_004155733.1; RicGAI, Ricinus communis, XP_002534030.1; GlyGAI1, Glycine max, NP_001240948.1; BrRGA2, Brassica rapavar. Perviridis, AAX33298.1; OsGAI, Oryza sativa Japonica, NP_001051032.1), revealed that the four poplar DELLAs contained two DELLA-specific domains in the N-terminal, DELLA and TVHYNP, and also included five GRAS-specific motifs, LHRI, VHIID, LHRII, PFYRE and SAW, in the C-termini (Fig. 1).
Fig. 1.
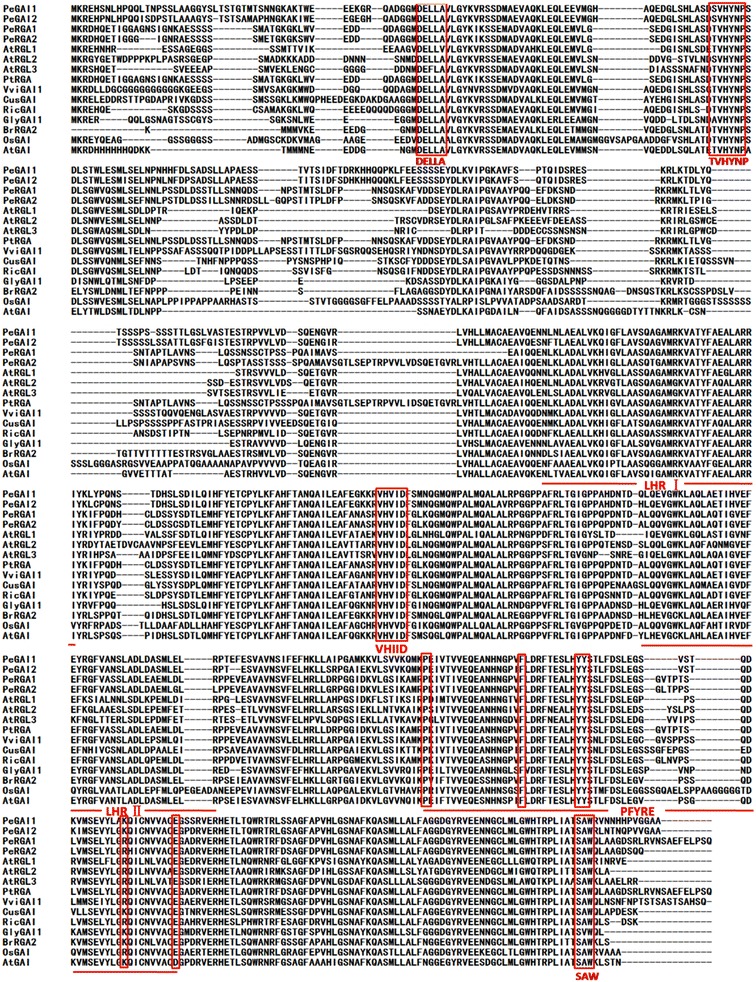
Alignment of the amino acid sequences of various plants DELLA proteins
To understand the evolutionary relationship of DELLA proteins from different species, the amino acid sequences of 18 DELLA proteins were aligned, and an un-rooted NJ phylogenetic tree was constructed using MEGA 6 software with 1000 bootstrap replications. The un-rooted NJ tree based on multiple sequence alignments showed that the 18 proteins were clustered into three distinct groups (Fig. 2). PeGAI2 and PeGAI2, most closely related to PtoGAI, were positioned in the first clade; PeRGA1 and PeRGA2 most closely related to PtRGA, were positioned in the second clade; and the three RGL proteins of A. thaliana were positioned in the third clade.
Fig. 2.
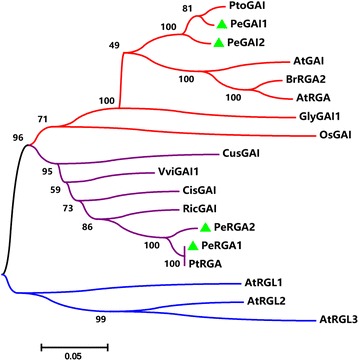
Phylogenetic analysis of the DELLA proteins
Expression patterns of DELLA genes
To analyze the expression patterns of the PeRGA1, PeRGA2, PeGAI1 and PeGAI2 genes in poplar, we measured their transcript levels by semi-quantitative and real-time RT-PCR at various developmental time points and in different tissues, including 1WR, 2WR, 3WR, 4WR, 4WS and 4WL Additional file 1: Figure S1. These four DELLA genes could be expressed at all developmental time points and in various tissues at different expression levels (Fig. 3). In addition to PeRGA1, the mRNA expression patterns of PeRGA2, PeGAI1 and PeGAI2 were similar at different developmental stages of poplar roots, with an obvious increase–decrease trend during the 1–4 weeks. The minimum of all of them occurred in 1WR, and the maximum in 2WR (PeRGA2) and 3WR (PeGAI1 and PeGAI2). However, the expression pattern of PeRGA1 in contrast with the three genes above, which obviously had the decrease-increase pattern in their 1- to 4-week-old roots, was lower in 2WR and 3WR than in 1WR and 4WR, with a minimum in the 2WR and a maximum in the 4WR. In the root and stem, PeGAI2 had the highest expression abundance. PeGAI1 had the lowest expression level in the 4-week-old leaves (Fig. 3).
Fig. 3.
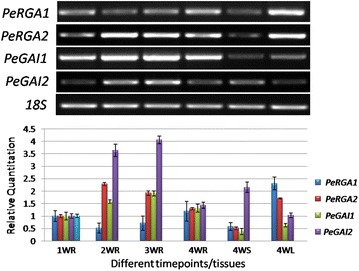
Temporal and spatial expression patterns of poplar DELLA genes by semi-quantitative and real-time RT-PCR. 1WR, 2WR, 3WR and 4WR are 1-, 2-, 3- and 4-week-old roots, respectively; and 4WL and 4WS are 4-week-old leaves and stems, respectively. For semi-quantitative RT-PCR, 18S was used an internal control. For real-time RT-PCR, EF1a was used as an internal control, and the relative transcript levels were calculated using the comparative Ct method. Error bars represent standard deviations for three biological replicates
Subcellular localization of DELLA proteins
To probe the subcellular localizations of the PeRGA1, PeRGA2, PeGAI1 and PeGAI2 proteins, the GFP-fusion vectors (35S::PeRGA1-GFP, 35S::PeRGA2-GFP, 35S::PeGAI1-GFP and 35S:: PeGAI2-GFP) were transformed into Populus protoplasts under control of the double 35S CaMV promoter. Confocal microscopy was used to observe the cellular localization of the fusion proteins. The PeRGA1-GFP and PeGAI1-GFP fusion proteins were located in the nucleus and cytoplasm, while the PeRGA2-GFP and PeGAI2-GFP fusion proteins were located only in the nucleus (Fig. 4). As a positive control, the 35::GFP fusion protein was detected in the nucleus and cytoplasm of Populus protoplasts.
Fig. 4.
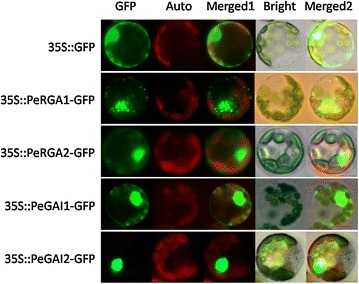
Subcelluar localization of the Populus DELLA proteins. Green fluorescence protein (GFP), chlorophyll autofluorescence (Auto), merged 1, bright and merged 2 images are shown. Scale bar 5 µm. The 35::GFP fusion was used as a positive protein control, and was detected in the nucleus and cytoplasm in protoplasts
Discussion
DELLA proteins are a subgroup of the plant-specific GRAS family (Bolle 2004). They are highly conserved repressors of GA signaling in angiosperms and their rapid GA-induced degradation can activate the GA signaling pathway (Vandenbussche 2007; Locascio et al. 2013; Zhang et al. 2011). The GA-GID1-DELLA complex plays a pivotal role in controlling plant growth and root elongation (Aleman 2008; Dai 2010; Harberd et al. 2009). The DELLA proteins participation in the growth regulation of roots has also been elucidated, such as, GAI and RGA are the major DELLAs controlling cell expansion in hypocotyl, shoot and root (Fu and Harberd 2003; Ubeda-Tomás et al. 2008). Several lines suggest that active GAs may inhibit lateral root formation (Berova and Zlatev, 2000; Chaney 2003; Watson 2004; Grossi et al. 2005; Gou et al. 2010). By heterologous expression of DELLA-less versions of GAI in Populus, the GA signaling was blocked, and the root biomass was increased via lateral root proliferation (Busov et al. 2006). By contrast, GA-overproducing mutations and exogenous GA application in aspen led to suppression of lateral and adventitious root formation (Eriksson et al. 2000).
Our previous study, a whole-genome transcriptional analysis of adventitious rooting in poplar hardwood cuttings was conducted using the GeneChip Poplar Genome Array, revealed the involvement of some putative DELLA genes in the adventitious rooting process. Here, four poplar DELLA genes involved in adventitious root development were isolated and characterized. A gene structure analysis revealed that these four DELLA genes were all intron-free. Previous studies revealed that SCR and SHR genes (also belonging to the GRAS family) of Arabidopsis, maize, rice and soybean were all intron- free (Xuan et al. 2014), but the homologous genes in Pinus contain introns (Laajanen et al. 2007).
Multiple sequence alignments suggest that PeRGA1, PeRGA2, PeGAI1 and PeGAI2 all have DELLA motifs that contain the specific DELLA protein’s domain, and it is necessary for DELLA proteins to interact with GID1 (Sun et al. 2010). The phylogenetic analysis of plant DELLA proteins indicates that these four DELLA proteins of poplar are highly homologous with Arabidopsis DELLA proteins. They may have similar functions, being involved in the GA signaling pathway and then regulating poplar root growth (Heo et al. 2011).
Expression patterns of DELLA genes in poplar showed that PeGAI1 and PeGAI2 sustained increases in expression levels during adventitious early root formation until root maturity, The PeRGA2 expression level was highest during middle root development, then began to decline. The expression level of PeRGA1 was highest in early root development and maturation, but lowest during middle root development. Only PeRGA1 and PeRGA2 had high levels of expression in leaves. The three DELLA genes (PeRGA2, PeGAI1, and PeGAI2) have a peak expression in 2 to 3 week-old adventitious root (Additional file 1: Figure S1). This result indicated that the PeRGA2, PeGAI1 and PeGAI2 might be involved in cell expansion in poplar adventitious root (Fu and Harberd 2003; Ubeda-Tomas et al. 2008). During the growth and development of A. thaliana, RGA and RGL2 are the main negative regulators of flower formation and reproductive growth, and RGA and GAI are the main negative regulators of stem elongation. However, in A. thaliana, four DELLA genes are involved in the regulation of seed germination (Peng et al. 1999; Dill et al. 2001; Dill and Sun 2001).
Conclusions
In this study, four DELLA genes involved in adventitious root development were isolated from poplar, and detailed information about the gene structures, sequence similarities, transcript profile and subcellular localization of the four poplar DELLA genes were revealed. Studies showed that the greater the DELLA protein content in plants, the greater ability to adverse environments (Achard et al. 2006). Therefore, the study of poplar DELLA genes not only increases our understanding of the specific functions of DELLA genes and the DELLA protein pathway in poplar, but also increases our understanding of environmental stress responses in poplar.
Authors’ contributions
MX conceived and designed the experiments. LX performed the experiments and participated in the writing of the manuscript. SL and MX analyzed the data and wrote the manuscript. LAX and MH helped to design and draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This project was supported by grants from the National Natural Science Foundation of China (31570671 and 31100484), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (13KJA220002), the Natural Science Foundation of Jiangsu Province (BK20150551), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- RACE
rapid amplification of cDNA ends
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- ORF
open reading frame
- GA
gibberellic acid
Additional file
10.1186/s40064-016-2728-x Distinct developmental stages of adventitious rooting on poplar stem cuttings.
Footnotes
Sian Liu and Lei Xuan contributed equally to this work
Contributor Information
Sian Liu, Email: sianliu@foxmail.com.
Lei Xuan, Email: 13851991791@163.com.
Li-An Xu, Email: laxu@njfu.edu.cn.
Minren Huang, Email: njmrhuang@njfu.edu.cn.
Meng Xu, Email: xum@njfu.edu.cn.
References
- Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot. 2009;60(4):1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van D, Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthome R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18:656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- Aleman L, Kitamura J, Abdel-mageed H, Lee J, Sun Y, Nakajima M, Ueguchi-Tanaka M, Matsuoka M, Allen RD. Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol Biol. 2008;68:1–16. doi: 10.1007/s11103-008-9347-z. [DOI] [PubMed] [Google Scholar]
- Berova M, Zlatev Z. Physiological response and yield of paclobutrazol treated tomato plants (Lycopersicon esculentum Mill.) Plant Growth Regul. 2000;30:117–123. doi: 10.1023/A:1006300326975. [DOI] [Google Scholar]
- Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- Busov V, Meilan R, Pearce DW, Rood SB, Ma C, Tschaplinski TJ, Strauss SH. Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta. 2006;224:288–299. doi: 10.1007/s00425-005-0213-9. [DOI] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta. 2005;223:105–113. doi: 10.1007/s00425-005-0057-3. [DOI] [PubMed] [Google Scholar]
- Chaney WR. Tree growth retardants: arborists discovering new uses for an old tool. Tree Care Ind. 2003;14:54–59. [Google Scholar]
- Dai C, Xue HW. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29:1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Achard P. Gibberellin signaling in plants. Development. 2013;140(6):1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant. 2016;9(1):10–20. doi: 10.1016/j.molp.2015.09.011. [DOI] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol. 2014;24(16):1923–1928. doi: 10.1016/j.cub.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T. Synergistic de-repression of gibberellins signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci. 2001;98(24):14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol. 2000;18:784–788. doi: 10.1038/77355. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schäfer E, Fu XD, Fan LM, Deng XW. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J, Steven H, Strauss Tsai CJ, Fang K, Chen YR, Jiang XN, Busova VB. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell. 2010;22(3):623–639. doi: 10.1105/tpc.109.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi JA, Moraes PJ, Tinoco SA, Barbosa JG, Finger FL, Cecon PR. Effects of paclobutrazol on growth and fruiting characteristics of Pitanga ornamental pepper. Acta Hortic. 2005;683:333–336. doi: 10.17660/ActaHortic.2005.683.41. [DOI] [Google Scholar]
- Harberd NP. Botany. Relieving DELLA restraint. Science. 2003;299:1853–1854. doi: 10.1126/science.1083217. [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, Lim MM, Lim J. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci. 2011;108(5):2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laajanen K, Vuorinen I, Salo V, Juuti J, Raudaskoski M. Cloning of Pinus sylvestris SCARECROW gene and its expression pattern in the pine root system, mycorrhiza and NPA-treated short roots. New Phytol. 2007;175(2):230–243. doi: 10.1111/j.1469-8137.2007.02102.x. [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D. Genomic analysis of DELLA protein activity. Plant Cell Physiol. 2013;54(8):1229–1237. doi: 10.1093/pcp/pct082. [DOI] [PubMed] [Google Scholar]
- Lucas M, Davie` re JM, Rodrı´guez-Falco´n M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Bla´ zquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456(7221):459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J. Green revolution genes encode mutant gibberellins response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Sun XL, Jones WT, Harvey D, Edwards PJB, Pascal SM, Kirk C, Considine T, Sheerin DJ, Rakonjac J, Oldfield CJ, Xue B, Dunker AK, Uversky CN. N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J Biol Chem. 2010;285:11557–11571. doi: 10.1074/jbc.M109.027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Xu M, Chen Y, Huang M. Transient expression for functional gene analysis using Populus protoplasts Plant Cell. Tissue Organ C (PCTOC) 2013;114(1):11–18. doi: 10.1007/s11240-013-0299-x. [DOI] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, Hedden P, Bhalerao R, Bennett MJ. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Straeten DVD. Evolutionary conservation of plant gibberellin signaling pathway components. BMC Plant Biol. 2007;7:1–17. doi: 10.1186/1471-2229-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson G. Effect of transplanting and paclobutrazol on root growth of ‘Green Column’ black maple and ‘Summit’ green ash. J Environ Hortic. 2004;22:209–212. [Google Scholar]
- Wild M, Davie` re JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 2012;24:3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhang B, Su X, Zhang S, Huang M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem. 2011;408(2):337–339. doi: 10.1016/j.ab.2010.08.044. [DOI] [PubMed] [Google Scholar]
- Xuan L, Xu M, Chen C, Yang C, Xu LA, Huang M. Identification and characterization of three PeSHRs and one PeSCR involved in adventitious root development of Populus. Plant Cell Tissue Organ C. 2014;117(2):253–264. doi: 10.1007/s11240-014-0437-0. [DOI] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, Kamiya Y, Yamaguchi S, Sun TP. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci. 2011;108(5):2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]


