Abstract
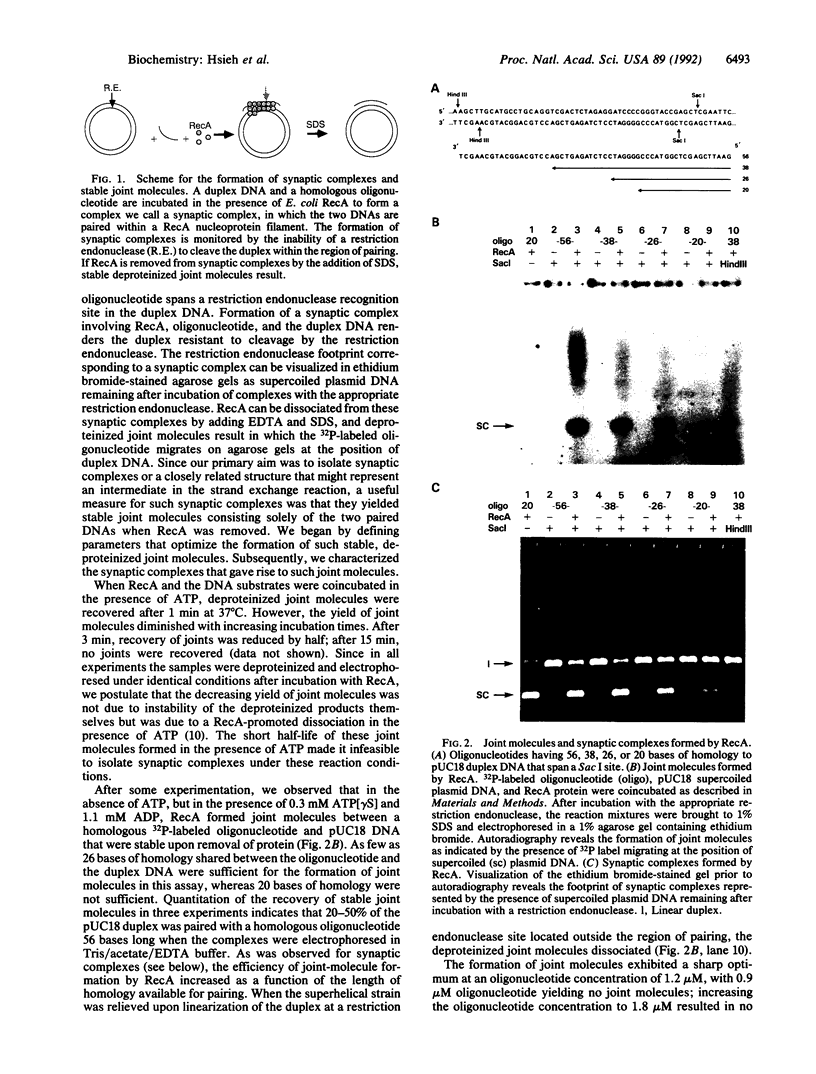
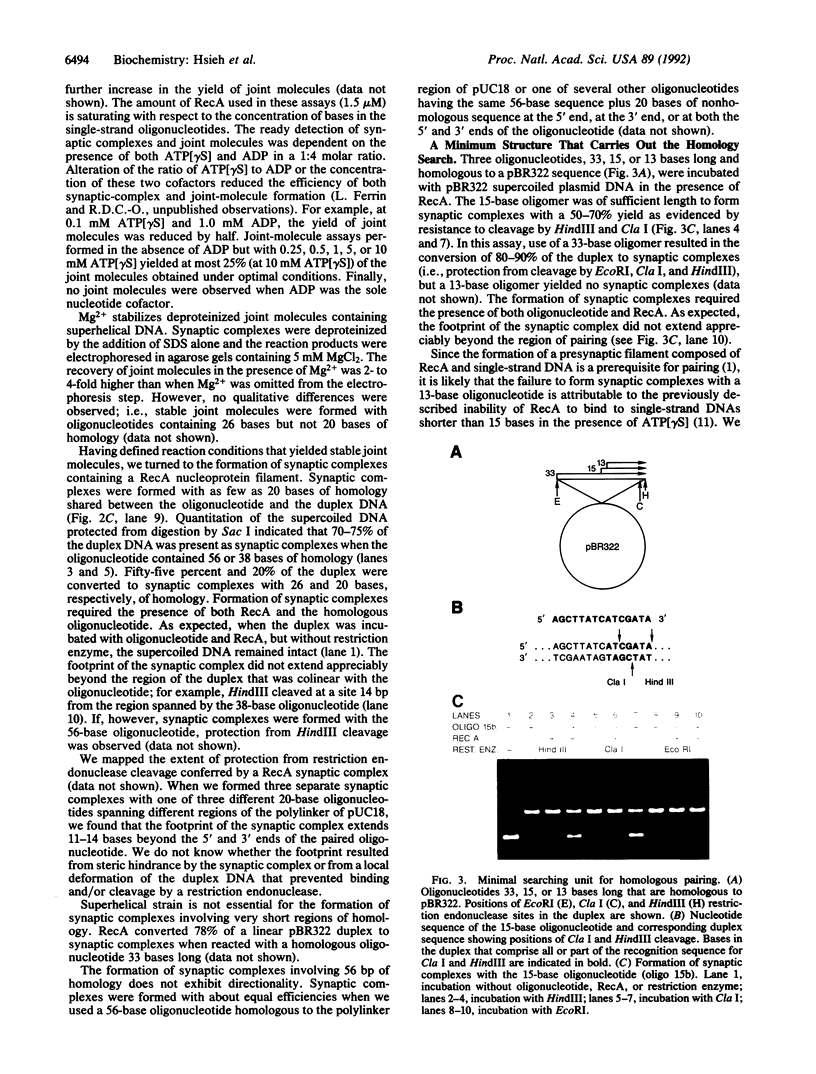
A key step in homologous recombination is the alignment and pairing of homologous DNAs. The Escherichia coli RecA protein initiates pairing by binding to single-strand DNA, forming a helical nucleoprotein filament. We demonstrate that in the presence of the nonhydrolyzable ATP analogue adenosine 5'-[gamma-thio]triphosphate and ADP, RecA can pair a homologous oligonucleotide 15 bases long with a duplex DNA to yield synaptic complexes consisting of the oligonucleotide and duplex DNA stabilized by RecA. RecA can pair as few as eight bases of homology to form such synaptic complexes. The homologous DNAs remain paired to each other upon removal of RecA provided that the length of shared homology is at least 26 base pairs. Based on our findings and the work of others, we propose that in vitro, one helical turn of a RecA nucleoprotein filament containing approximately six RecA monomers and 15 bases of single-strand DNA is the functional unit sufficient to carry out the homology search.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cheng S., Van Houten B., Gamper H. B., Sancar A., Hearst J. E. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC excinuclease. J Biol Chem. 1988 Oct 15;263(29):15110–15117. [PubMed] [Google Scholar]
- Cotterill S. M., Satterthwait A. C., Fersht A. R. recA protein from Escherichia coli. a very rapid and simple purification procedure: binding of adenosine 5'-triphosphate and adenosine 5'-diphosphate by the homogeneous protein. Biochemistry. 1982 Aug 31;21(18):4332–4337. doi: 10.1021/bi00261a023. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Egelman E. H., Stasiak A. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J Mol Biol. 1986 Oct 20;191(4):677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- Ferrin L. J., Camerini-Otero R. D. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991 Dec 6;254(5037):1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Heuser J., Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J Mol Biol. 1989 Dec 5;210(3):473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero C. S., Camerini-Otero R. D. Pairing of homologous DNA sequences by proteins: evidence for three-stranded DNA. Genes Dev. 1990 Nov;4(11):1951–1963. doi: 10.1101/gad.4.11.1951. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero R. D. Formation of joint DNA molecules by two eukaryotic strand exchange proteins does not require melting of a DNA duplex. J Biol Chem. 1989 Mar 25;264(9):5089–5097. [PubMed] [Google Scholar]
- Kowalczykowski S. C. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C. Interaction of recA protein with a photoaffinity analogue of ATP, 8-azido-ATP: determination of nucleotide cofactor binding parameters and of the relationship between ATP binding and ATP hydrolysis. Biochemistry. 1986 Oct 7;25(20):5872–5881. doi: 10.1021/bi00368a006. [DOI] [PubMed] [Google Scholar]
- Lauder S. D., Kowalczykowski S. C. Asymmetry in the recA protein-DNA filament. J Biol Chem. 1991 Mar 25;266(9):5450–5458. [PubMed] [Google Scholar]
- Leahy M. C., Radding C. M. Topography of the interaction of recA protein with single-stranded deoxyoligonucleotides. J Biol Chem. 1986 May 25;261(15):6954–6960. [PubMed] [Google Scholar]
- Menetski J. P., Bear D. G., Kowalczykowski S. C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci U S A. 1990 Jan;87(1):21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski J. P., Varghese A., Kowalczykowski S. C. Properties of the high-affinity single-stranded DNA binding state of the Escherichia coli recA protein. Biochemistry. 1988 Feb 23;27(4):1205–1212. doi: 10.1021/bi00404a021. [DOI] [PubMed] [Google Scholar]
- Menge K. L., Bryant F. R. ATP-stimulated hydrolysis of GTP by RecA protein: kinetic consequences of cooperative RecA protein-ATP interactions. Biochemistry. 1988 Apr 5;27(7):2635–2640. doi: 10.1021/bi00407a055. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Dutreix M., Radding C. M. Stable three-stranded DNA made by RecA protein. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):2984–2988. doi: 10.1073/pnas.88.8.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselli W., Stasiak A. Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J Mol Biol. 1990 Nov 20;216(2):335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- Rötig A., Cormier V., Blanche S., Bonnefont J. P., Ledeist F., Romero N., Schmitz J., Rustin P., Fischer A., Saudubray J. M. Pearson's marrow-pancreas syndrome. A multisystem mitochondrial disorder in infancy. J Clin Invest. 1990 Nov;86(5):1601–1608. doi: 10.1172/JCI114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Petes T. D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Ohtani T., Iwabuchi M., Ando T. D-loop cycle. A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by recA protein of Escherichia coli. J Biol Chem. 1982 Dec 10;257(23):13981–13986. [PubMed] [Google Scholar]
- Smith G. R. Conjugational recombination in E. coli: myths and mechanisms. Cell. 1991 Jan 11;64(1):19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Kahn R., DasGupta C., Radding C. M. Formation of nascent heteroduplex structures by RecA protein and DNA. Cell. 1982 Aug;30(1):37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]
- Zeviani M., Bresolin N., Gellera C., Bordoni A., Pannacci M., Amati P., Moggio M., Servidei S., Scarlato G., DiDonato S. Nucleus-driven multiple large-scale deletions of the human mitochondrial genome: a new autosomal dominant disease. Am J Hum Genet. 1990 Dec;47(6):904–914. [PMC free article] [PubMed] [Google Scholar]





