Abstract
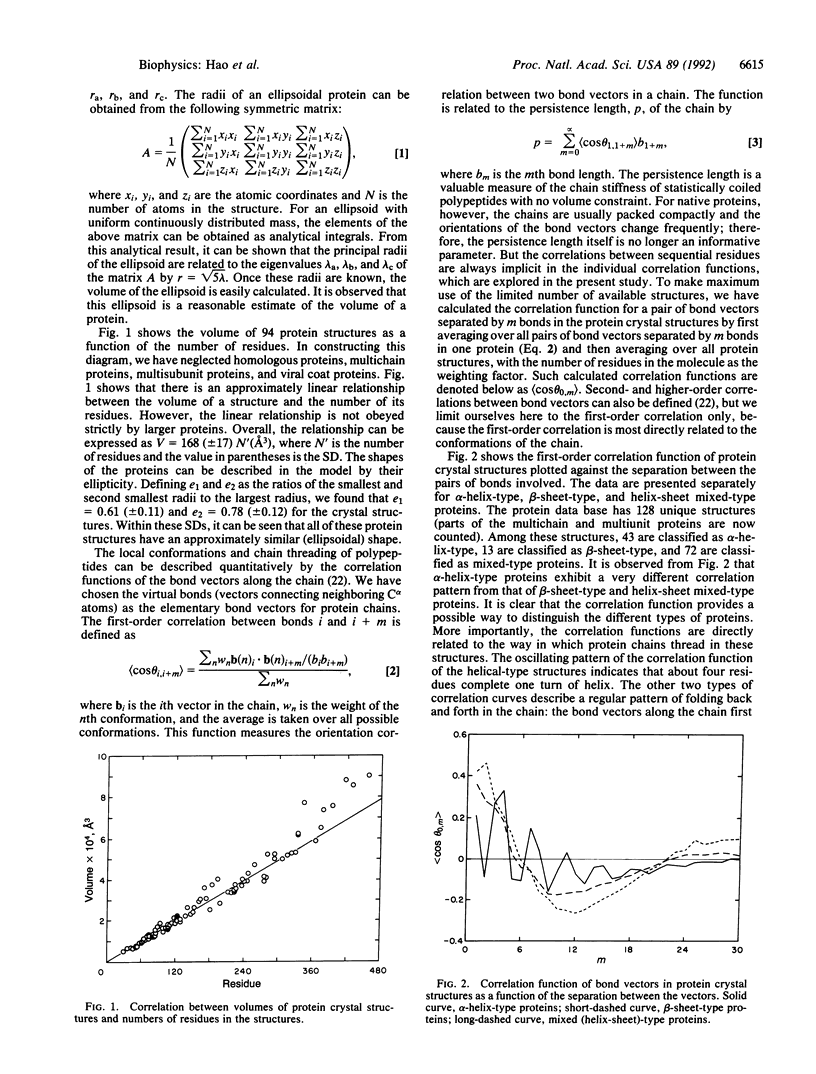
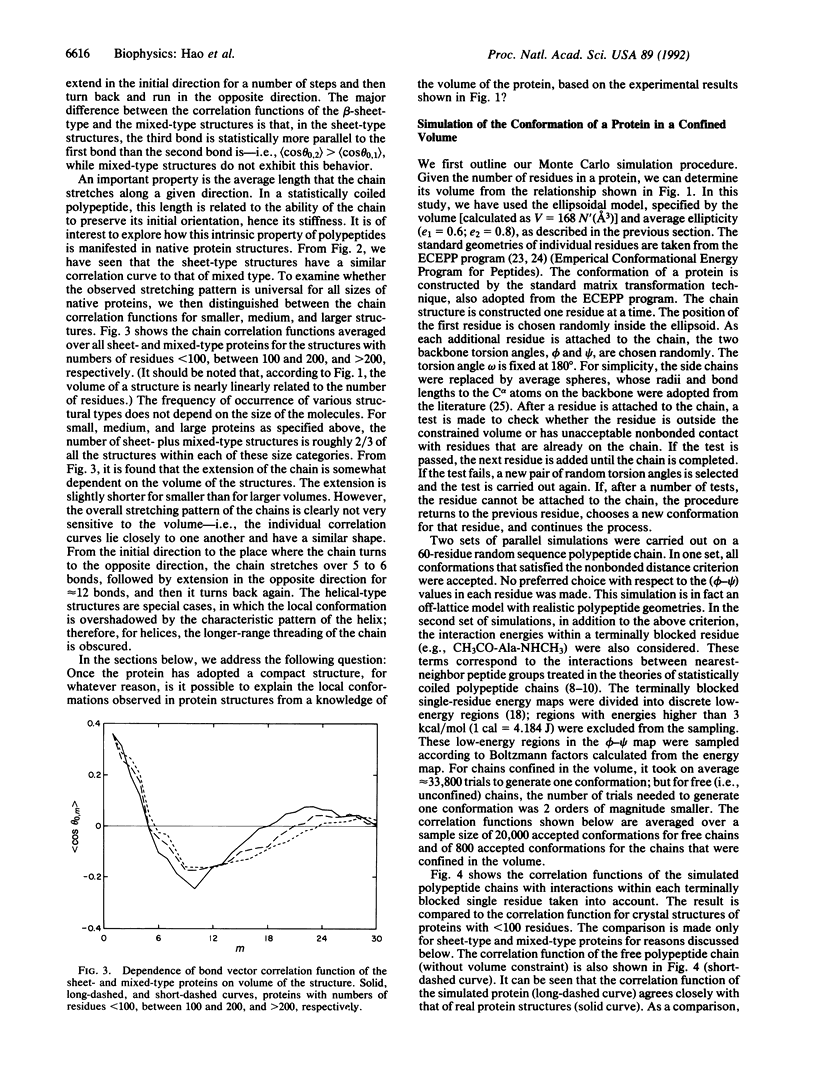
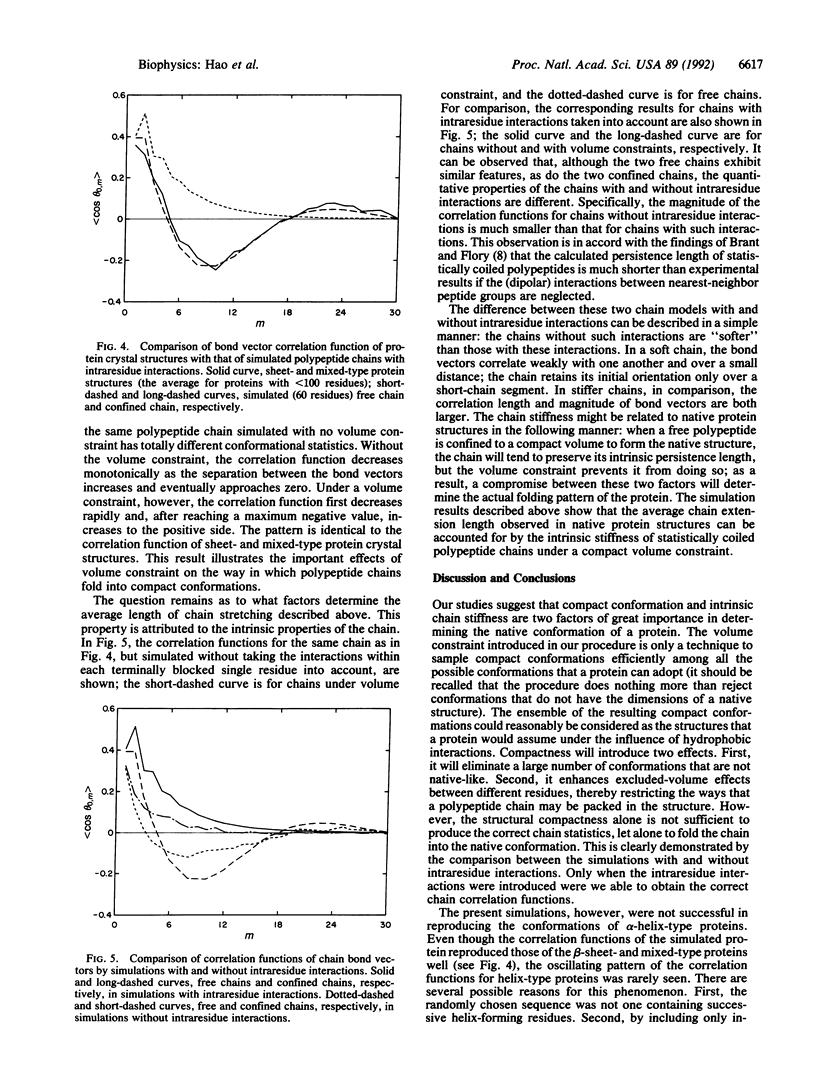
An investigation of the statistical properties of the native conformations of proteins, observed from crystal structures, is reported. Protein conformations were analyzed in terms of a bond vector correlation function and molecular volume. It was observed that, while the volume of a protein structure varies nearly linearly with the number of residues, the bond vector correlation function exhibits a universal feature for all sizes of proteins. To interpret the nature of the bond vector correlation function of native protein structures quantitatively, Monte Carlo simulations of realistic polypeptide chains of specific but arbitrary amino acid sequence were carried out. The molecule was constrained in an ellipsoidal volume determined by its chain length, and conformations with unacceptable nonbonded contacts between different amino acid residues were excluded. The interactions within a terminally blocked single residue, which correlate two nearest-neighbor peptide groups in a chain, were taken into account by an energetically biased sampling of its phi-psi space. The simulated chain correlation functions were found to be in good agreement with those of the crystal structures of beta-sheet-type and mixed-type (alpha+beta) proteins of similar length. On the basis of these calculations, it is concluded that the observed conformations of these native proteins may arise from two basic factors: the compactness of structures under hydrophobic interactions and the intrinsic stiffness of polypeptide chains due to the interactions within each terminally blocked residue.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Covell D. G., Jernigan R. L. Conformations of folded proteins in restricted spaces. Biochemistry. 1990 Apr 3;29(13):3287–3294. doi: 10.1021/bi00465a020. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry. 1990 Aug 7;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Ptitsyn O. B. Why do globular proteins fit the limited set of folding patterns? Prog Biophys Mol Biol. 1987;50(3):171–190. doi: 10.1016/0079-6107(87)90013-7. [DOI] [PubMed] [Google Scholar]
- Flory P. J. Moments of the End-to-End Vector of a Chain Molecule, Its Persistence and Distribution. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1819–1823. doi: 10.1073/pnas.70.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory P. J., Schimmel P. R. Dipole moments in relation to configuration of polypeptide chains. J Am Chem Soc. 1967 Dec 20;89(26):6807–6813. doi: 10.1021/ja01002a001. [DOI] [PubMed] [Google Scholar]
- Gregoret L. M., Cohen F. E. Protein folding. Effect of packing density on chain conformation. J Mol Biol. 1991 May 5;219(1):109–122. doi: 10.1016/0022-2836(91)90861-y. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kneller D. G., Cohen F. E., Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990 Jul 5;214(1):171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Flory P. J. Conformational energy and configurational statistics of poly-L-proline. Proc Natl Acad Sci U S A. 1967 Jul;58(1):52–59. doi: 10.1073/pnas.58.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. Interfacial free energy and the hydrophobic effect. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4175–4176. doi: 10.1073/pnas.76.9.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz D. H., Scheraga H. A. Influence of water on protein structure. An analysis of the preferences of amino acid residues for the inside or outside and for specific conformations in a protein molecule. Macromolecules. 1978 Jan-Feb;11(1):9–15. doi: 10.1021/ma60061a002. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. S., Pottle M. S., Némethy G., Scheraga H. A. Conformational analysis of the 20 naturally occurring amino acid residues using ECEPP. Macromolecules. 1977 Jan-Feb;10(1):1–9. doi: 10.1021/ma60055a001. [DOI] [PubMed] [Google Scholar]


