Abstract
HIV-1 reverse transcriptase (RT) is a major drug target in the treatment of HIV-1 infection. RT inhibitors currently in use include non-nucleoside, allosteric RT inhibitors (NNRTIs), which bind to a hydrophobic pocket, distinct from enzyme's active site. We investigated RT-NNRTI interactions by solution 19F NMR, using singly 19F labeled RT proteins. Comparison of 19F chemical shifts of fluorinated RT and drug-resistant variants revealed that the fluorine resonance is a sensitive probe for identifying mutation-induced changes in the enzyme. Our data show that in the unliganded enzyme, the NNRTI-binding pocket is highly plastic and not locked into a single conformation. Upon inhibitor binding, the binding pocket rigidifies. In the inhibitor-bound state, the 19F signal of RT is similar to that of drug-resistant mutant enzymes, distinct from what is observe for the free state. Our results demonstrate the power of 19F NMR spectroscopy to characterize conformational properties using selectively 19F labeled protein.
Graphical Abstract
Introduction
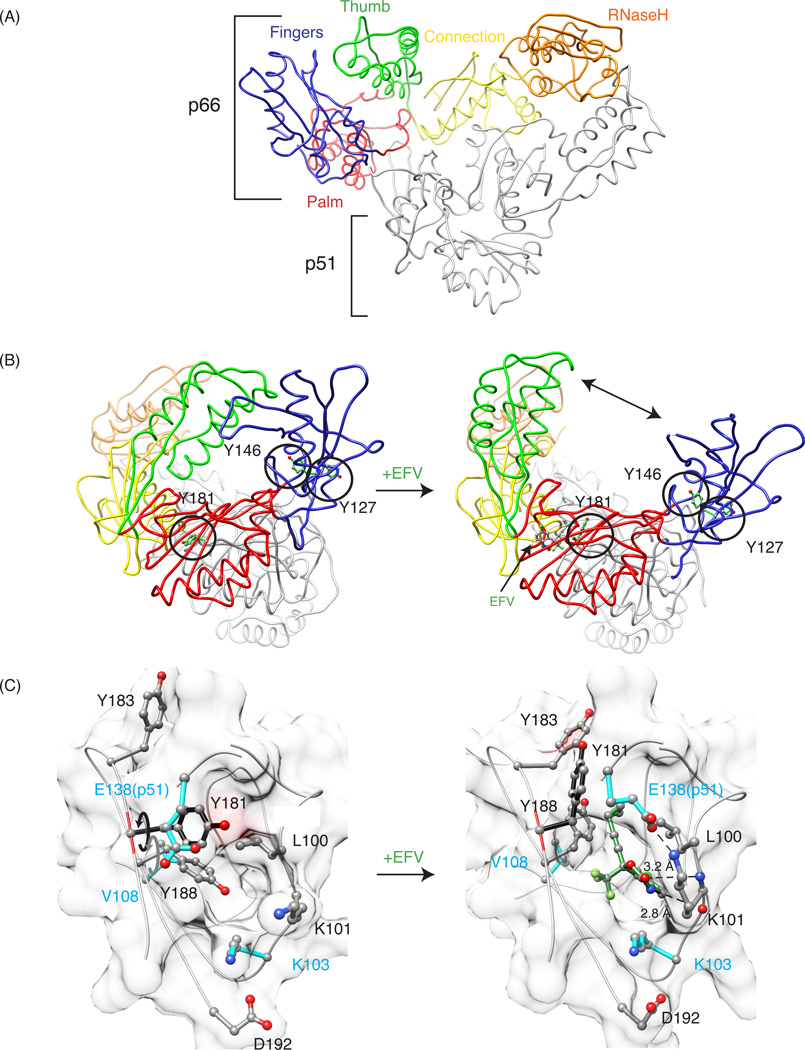
HIV-1 reverse transcriptase (RT) is an essential enzyme in the HIV-1 lifecycle and a major drug target in the treatment of HIV-1 infection. Current FDA approved RT inhibitors are effective, but continuous treatment can lead to the emergence of drug resistant strains.1 Understanding RT, its structure, and the mechanism of inhibitor action, is important for the development of novel inhibitors with more favorable resistance profiles. A large number of crystal structures of RT are available (wild-type and mutants), providing valuable information on the protein's conformations as well as drug interactions.2–9 Crystallographic studies have shown that RT is an asymmetric heterodimer that comprises two subunits p66 and p51. The p66 subunit contains two domains, a polymerase, and RNase H domain. The p51 subunit is identical in amino acid sequence to p66, apart from lacking the C-terminal RNase H domain. The polymerase domain of each subunit is further subdivided into fingers, palm, thumb, and connection subdomains.2 In the overall dimeric RT structure, the subdomains in the p51 and p66 subunits exhibit different relative orientations (Figure 1A).
Figure 1.
General description of RT structure, and comparison of apo and EFV-bound crystal structures of RT. (A) Tube representation of apo-RT (PDB: 1DLO), with the fingers, palm, thumb, connection, and RNase H domains in the p66 subunit colored in blue, pink, green, yellow and orange, respectively. The p51 subunit is colored grey. (B) Structural differences between apo-RT (left, PDB: 1DLO) and EFV-bound RT (right, PDB: 1FK9). A large conformational change, including the separation of the thumb and fingers domains (indicated by the arrow), is seen in the drug-bound structure. Tyrosine residues 127, 146 and 181 are depicted in ball and stick representation and encircled. (C) Details of the binding site in apo RT and the EFV-bound RT complex, illustrating the rotation of the Y181 (black arrow) and Y188 (grey arrow) side chains out of the binding pocket. The bound EFV molecule is shown in green and pertinent distances between the benzoxazin-2-one and the backbone carbonyl oxygen of K101 (2.8 Å), and the carbonyl group of the bexzoxazine-2-one and the backbone nitrogen of atom K101 (3.2 Å) are indicated.
Although highly effective as RT inhibitors and the first drugs to treat HIV-1 infection, nucleoside/nucleotide RT inhibitors, which act as chain terminators in the enzymatic reaction, are associated with numerous side effects. Therefore, non-competitive RT inhibitors were developed and have been in the clinic for almost 20 years.10–15 These non-nucleoside inhibitors (NNRTIs) include Nevirapine (NVP), Efavirenz (EFZ), Etravirine (ETR), and Rilpivirine (RPV). Although chemically diverse, they all bind to the same pocket, distinct from the polymerase active site, and inhibit RT allosterically.13,16 A comparison of the crystal structures of apo-RT and RT in the presence of NNRTIs reveals significant structural changes upon NNRTI binding. Apo-RT adopts a “closed” conformation, in which the p66 thumb subdomain folds down onto the fingers subdomain. In contrast, in the presence of NNRTIs, RT adopts an “open” conformation, in which the p66 thumb domain is ~30 Å away from the fingers subdomain (Figure 1B). Local conformational differences are also seen in the NNRTI-binding pocket in the p66 subunit. This pocket is not present in apo-RT, and the Y181 and Y188 side chains fill most of the cavity, which is occupied by the NNRTI in the NNRTI/RT complex (Figure 1C).2–5
Crystal structures are invaluable for pinpointing structural details of enzyme-inhibitor and substrate interactions, however, studies by other methods can offer complementary information. For RT, only a limited number of investigations in the absence of nucleic acid substrates have been reported, including EPR experiments and hydrogen exchange mass spectrometry (HXMS).17,18 Also, a few solution NMR studies, using [methyl-13C]-methionine or isoleucine labeled RT have been reported.19,20 In addition, several computational studies have been conducted to characterize RT dynamics and the effects of NNRTI binding.21–28 Yet, a general consensus on the mechanistic basis for NNRTI inhibition of RT has not been reached.2,12,29,30
Here, we used 19F solution NMR to study RT in solution by incorporating a single fluorine probe into the enzyme. Single site labeling prevents resonance overlap and enables simple and fast 1D NMR experiments. The fluorine nucleus was selected since it possesses a high gryomagnetic ratio, which results in excellent sensitivity (83% of 1H). In addition the 19F shielding is dominated by a large paramagnetic term, which makes it exquisitely sensitive to its local environment (the 19F chemical shift range is ~100 fold larger than that of 1H).31–33
In particular, we aimed to gain insight into the characteristic dynamics of the NNRTI-binding site, mechanism of action of NNRTIs, and the effects of drug resistant mutations. For this study, RT was labeled site-specifically with 4-trifluoromethylphenylalanine (tfmF) at positions 127, 146 and 181, producing three singly labeled RT proteins named RT127tfmF, RT146tfmF, and RT181tfmF, respectively. Comparisons of the 19F spectra of RT127tfmF, RT146tfmF, and RT181tfmF showed that distinctly different chemical shifts are observed for the trifluoromethyl group, demonstrating that the 19F probes in each RT protein are in distinct environments. Furthermore, linewidth analyses of these spectra suggest that the NNRTI-binding site is highly plastic in the ligand-free enzyme. Mutant, drug resistant proteins V108I, K103N, and E138K all modulate the conformational plasticity or average chemical environment of the NNRTI-binding site, with the K103N mutation producing the most prominent effect. In the presence of NVP, EFV, ETR, and RPV, the conformational plasticity of RT at the NNRTI-binding site is reduced, and the chemical shift of the NNRTI-bound signals depends on the identity of the inhibitor, regardless of the presence of drug resistant mutations. These data show that 19F NMR can be used as an effective tool for examining NNRTI -RT interactions.
Materials and Methods
Efavirenz (EFV), and Nevirapine (NVP) were kindly provided by Dr. Nicholas Sluis-Cremer. Etravirine (ETR) and Rilvipirine (RPV) were purchased from Selleckchem (Houston, TX). All NNRTIs were stored in DMSO at concentrations of 10 mM.
Cloning
Constructs for the codon optimized, C38V/C280S mutant p66 and p51 subunits of RT were prepared as previously described.34 The vectors encoding p66-127tfmF, p66-146tfmF, and p66-181tfmF proteins, were generated by replacing codons for tyrosines at positions 127, 146, or 181 with amber codons, using the appropriate TAG oligonucleotides as primers, and the p66 encoding vector as a template. The vectors encoding mutant p66-181tfmF, p66-146tfmF and p51 proteins (p66-181tfmF-K103N, p66-181tfmF-V108I, p66-146tfmF-K103N, p66-146tfmF-V108I, and the p51-V108I, p51-E138K, p51-K103N) were generated using the appropriate oligonucleotides as primers, and the p66-181tfmF, p66-146tfmF, and p51 encoding vectors as templates, respectively. All the p66 constructs contain a hexa-histidine tag at the C-terminus, and all the p51 constructs contain a Strep-tag at the N-terminus. The DNA sequences of all constructs were verified by DNA sequencing (Genewiz, South Plainfield, NJ).
Protein expression
The tfmF labeled p66 proteins (p66-127tfmF, p66-146tfmF, p66-181tfmF, p66-181tfmF-V108I, and p66-181tfmF-K103N) were produced using a protocol developed by the Mehl laboratory.33,35 Briefly, competent E. coli BL21 ai cells (Invitrogen) were co-transformed with the vector encoding the TAG containing constructs (above), and the pDule2 RS vector encoding the orthogonal amber tRNA/tRNA synthetase pair. Several transformants were screened for expression. All tfmF containing p66 proteins were produced at 27°C by growing cells for 24 h using auto-induction medium, containing tfmF at a final concentration of 1 mM. All p51 proteins (p51, p51-V108I, p51-K103N, p51-E138K) were transformed into E. coli BL21 (DE3) gold cells (Agilent Technologies, Santa Clara, CA). The p51 proteins were produced at 27°C by growing cells for 24 h using auto-induction media.33,36
RT protein purification
Cells were harvested by centrifugation, and re-suspended in lysis buffer, containing 25 mM sodium phosphate, 25 mM imidazole and 500 mM NaCl (pH 7.5). Equivalent amounts of cell pellets, containing the expressed p66 and p51 proteins, were mixed, and lysed using a microfluidizer. Cell debris was removed by centrifugation, and the supernatant was applied to a 5 mL HisTrap column (GE healthcare Life Sciences, Piscataway, NJ), equilibrated in lysis buffer. Proteins were eluted with a linear gradient of 0.025–0.5 M imidazole to separate fractions containing p66/p51 heterodimeric RT proteins from p51 monomer, and truncated unlabeled p66 proteins. RT-containing fractions were pooled and applied to a 5 mL StrepTrap column (GE healthcare), equilibrated in 25 mM sodium phosphate, 6 mM KCl, 280 mM NaCl (pH 7.5). RT proteins were eluted using the same buffer listed above supplemented with D-desthiobiotin to a final concentration of 3 mM (Sigma Aldrich, St. Louis, MO). This step was essential for the removal of p66 homodimer. Then, gel-filtration on a HiLoad 26/60 Superdex 200 column (GE Healthcare) was used as a final purification step. RT-containing fractions were pooled, and stored in 50% v/v glycerol at −80°C until use. A list of prepared RT proteins is provided in Table 1. A brief description of each RT protein is provided below. The RT127tfmF, RT146tfmF, and RT181tfmF proteins contain a single tfmF in the p66 subunit of RT at positions 127, 146 and 181, respectively. RT181tfmF-V108I and RT181tfmF-K103N proteins contain a single tfmF residue at position 181 in the p66 subunit, and amino acid changes (V108I or K103N) in both, the p51 and p66 subunits. The RT181tfmF-E138K(p51) proteins contain a single tfmF at position 181 in the p66 subunit, and the E138K amino acid change in the p51 subunit of RT.
Table 1.
RT proteins
| Name | RT subunits |
|---|---|
| RT127tmfF | p51/p66-127tfmF |
| RT146tfmF | p51/p66-146tfmF |
| RT181tfmF | p51/p66-181tfmF |
| RT181tfmF-V108I | p51-V108I/p66-181tfmF-V108I |
| RT181tfmF-K103N | p51-K103N/p66-181tfmF-K103N |
| RT181tfmF-E138K(p51) | p51-E138K/p66-181tfmF |
| RT146tfmF-V108I | p51-V108I/p66-146tfmF-V108I |
| RT146tfmF-K103N | p51-K103N/p66-146tfmF-K103N |
NMR experiments
Protein samples for NMR were buffer exchanged into 25 mM sodium phosphate buffer, 100 mM NaCl, 10% v/v D2O, pH 6.8 in an Amicon Ultra concentrator (EMD Millipore, Billerica, MA) to a final volume of 350 µL. All final protein concentrations were ~35 µM.19F 1D NMR spectra with 1H composite decoupling during acquisition were recorded on a 600 MHz Bruker AVANCE spectrometer, equipped with a CP TXO F/C-H-D triple-resonance z-axis gradient cryoprobe (Bruker Biospin, Billerica, MA). Spectra for the inhibitor-free proteins as well as samples containing NVP, EFV, ETR, and RPV at 1:1 and 1:5 RT: NNRTI inhibitor ratios were recorded using Topspin 3.1 (Bruker) and analyzed with MestReNova (Escondido, CA). Prior to Fourier transformation, the time-domain free-induction decays were apodized with an exponential function, using a line broadening factor of 30 Hz. Chemical shifts and linewidths were calculated using the peak deconvolution feature in MestReNova. An upper limit of uncertainty for the linewidths was qualitatively estimated by assuming that the fit error of each peak is associated with the linewidth error.
Results
Spectra of apo-RT127tfmF, apo-RT146tfmF, and apo-RT181tfmF
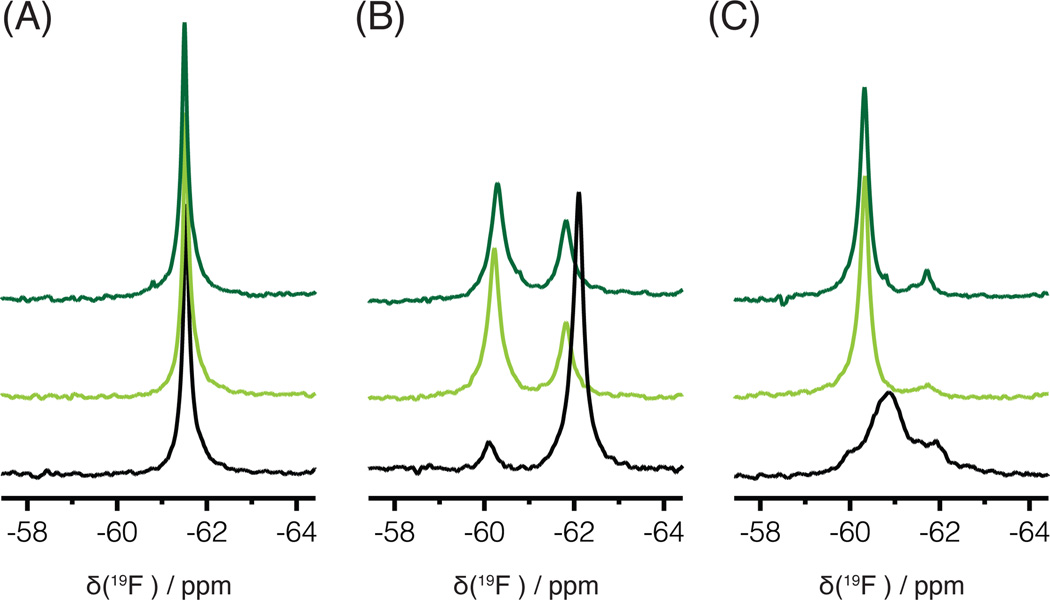
19F NMR spectra were recorded for RT127tfmF, RT146tfmF, and RT181tfmF, each containing a single tfmF at the indicated position in the p66 subunit of RT (Figure 1B). Different spectra were observed for the apo-proteins (Figure 2A–C, black traces), reflecting the distinct environment around the tfmF side chain of each RT protein. For apo-RT127tfmF, a single resonance signal is seen at −61.5 ppm (100 ± 2 Hz; Figure 2A), while for apo-RT146tfmF, a major signal is observed at −62.0 ppm (150 ± 2 Hz), and a small additional signal is seen at −60.2 ppm (150 ± 85 Hz), slightly downfield from the major resonance (Figure 2B). The spectrum of apo-RT181tfmF exhibits a very broad signal at −60.8 ppm (500 ± 5 Hz), much wider than those of apo-RT127tfmF and apo-RT146tfmF, and a smaller signal around −61.8 ppm (Figure 2C). In each spectrum, the major signal clearly originates from the p66/p51 heterodimeric RT. The very small signal in Figure 2C may originate from the small amount (< 8 %) of p66 monomer and/or homodimer in the sample.37 This was confirmed by comparing spectra of different RT samples (data not shown).
Figure 2.
1D 19F NMR spectra of RT with 4-trifluoromethyl-phenylalanines substituted for tyrosine residues at several positions in the p66 subunit, in the absence (black) and presence of EFV at 1:1 and 1:5 molar ratios (light and dark green, respectively). 19F spectra of (A) RT127tfmF, (B) RT146tfmF, and (C) RT181tfmF at 27°C are shown.
EFV binding to RT127tfmF, RT146tfmF, and RT181tfmF
The effect of EFV binding to the three RT variants, RT127tfmF, RT146tfmF, and RT181tfmF was investigated (Figure 2). At 1:1 RT:EFV molar ratio, no changes are observed for RT127tfmF (Figure 2A, light green trace). In contrast, for RT146tfmF, two new signals are observed, one very close to the one of the free protein (−62.0 ppm) and the other downfield, at −61.8 ppm (Figure 2B, light green trace). Both resonances, at −61.8 ppm and −62.0 ppm, exhibit comparable linewidths (180 ± 2 Hz). The broad asymmetric signal of RT181tfmF at −60.8 ppm (500 ± 2 Hz) disappears upon EFV binding (Figure 2C, light green trace) and a new, sharper resonance appears at −60.3 ppm (125 ± 2 Hz). Spectra were also recorded at a 1:5 RT: EFV molar ratio (Figure 2, dark green traces) to ensure saturation of the protein with ligand. For all proteins, the spectra at 1:5 are similar to spectra at 1:1 RT:EFV molar ratio, suggesting that saturation with EFV is essentially reached at the 1:1 molar ratio, consistent with the Kd value of 92 nM for EFV binding to the p66/p51 RT heterodimer.38
These spectral data are interpreted in light of the location of the three amino acid residues in RT crystal structures.5,39 Residue 127 is ~35 Å away from the NNRTI binding site, located on the fingers subdomain and pointing towards the solvent (Figure 1B), and no significant changes in the position of residue 127 in the absence or presence of EFV are noted when comparing crystal structures of apo-RT and the EFV/RT complex. The resonance frequency of RT127tfmF, which is essentially not affected by EFV binding, is consistent with this observation (Figure 2A). Residue 146 is ~30 Å away from the NNRTI binding site and is also located on the fingers subdomain. However, this residue points towards the thumb domain. In the RT146tfmF protein, the 19F probe at position 146 clearly senses EFV binding and splits into two resonances, at −60.2 and −61.8 ppm, in the EFV-bound form (Figure 2B). The presence of two resonances suggests that the 19F probe in RT146tfmF reports on two conformations, one of which is very similar to the free conformation, given that only a very small difference in frequency is involved, while the other one reflects a distinctly different conformation. The 181tfmF side chain is located in the NNRTI-binding pocket (Figure 1B,C) and EFV binding, by necessity, is expected to influence its conformation. The spectrum of RT181tfmF in the presence of EFV contains a substantially narrower resonance for the inhibitor bound state, which is shifted downfield compared to that of apo-RT181tfmF. The broad resonance observed in the spectrum of apo-RT181tfmF (500 ± 2 Hz) suggests that the ligand-free protein exhibits a substantial degree of conformational plasticity in the NNRTI binding site, which is reduced in the EFV/RT complex (Figure 2C), evidenced by the significantly narrower linewidth of the bound signal (125 ± 2 Hz).
Drug-resistant variants of RT
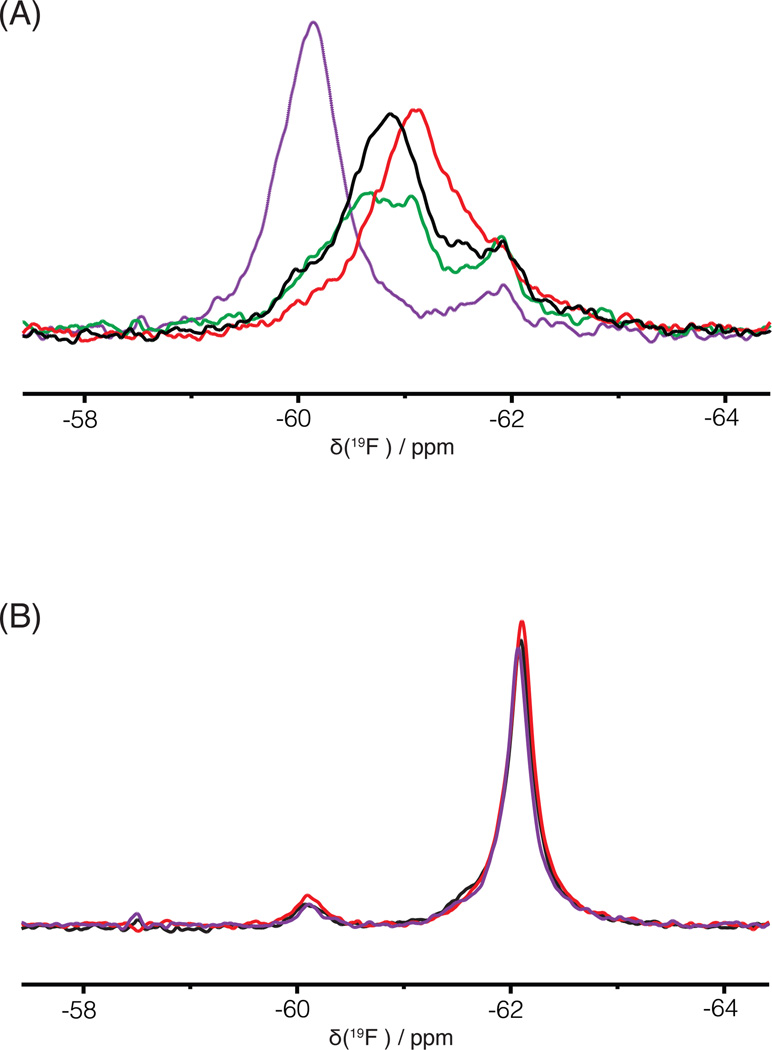
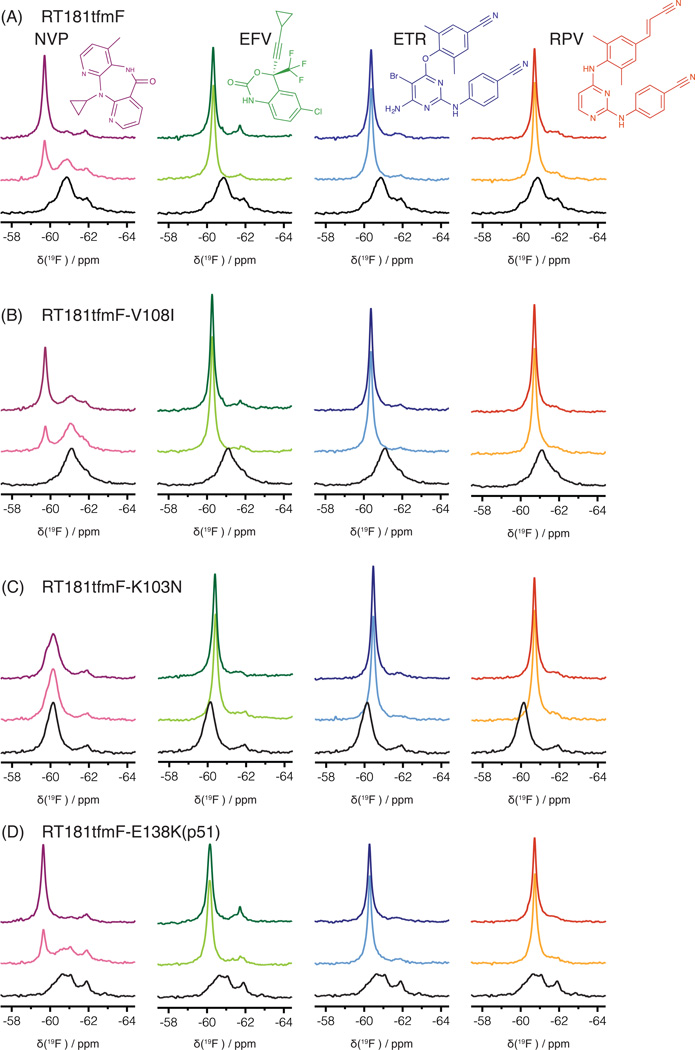
Three drug-resistant variants of RT were selected for investigation. V108I is associated with NVP resistance, K103N imparts NVP and EFV resistance,40–43 and E138K is connected with therapeutic failure of ETR and RPV.44–47 We evaluated these mutants in the context of RT181tfmF, since residue 181 resides in the NNRTI site and should report on possible effects of these mutations on the protein properties at this binding site. A superposition of the 19F spectra of all three apo RT181tfmF mutants is provided in Figure 3. The linewidth of the apo-RT181tfmF-V108I signal (red, 500 ± 5 Hz) is similar to that of the apo-RT181tfmF (black, 500± 5 Hz), although slightly upfield shifted, suggesting a minimal influence of this mutation on the NNRTI binding site. In contrast, the signal of apo-RT181tfmF-K103N is narrower and downfield shifted (purple; 300 ± 15 Hz, −60.1 ppm). This suggests that in this mutant a less plastic conformation is present in the binding site, compared to apo-RT181tfmF. The signal in the 19F spectrum of apo-RT181tfmF-E138K(p51) is also broad (green, 600 ± 20 Hz) and is similar to that of apo-RT181tfmF, indicating that only small changes are introduced into the flexible binding site. Collectively, these data suggest that the local environment around the 19F probe at the 181 position of RT is most prominently affected by the K103N mutation.
Figure 3.
1D 19F NMR spectra of RT181tfmF and several RT mutants at 27°C. (A) Superposition of the fluorine resonances of RT181tfmF (black), RT181tfmF-V108I (red), RT181tfmF-E138K(p51) (green), and RT181tfmF-K103N (purple). (B) Superposition of the fluorine resonances of RT146tfmF (black), RT146tfmF-V108I (red), and RT146tfmF-K103N (purple). All RT181tfmF and RT146tfmF variants contain amino acid changes in both the p51 and p66 domains, except for RT181tfmF-E138K in which the E138K change is only present in the p51 subunit.
NVP, EFV, ETR, and RPV binding to RT181tfmF and mutants associated with drug resistance
To examine the spectral perturbations of NNRTIs on the 19F spectrum of RT181tfmF, we recorded spectra in the presence of NVP, ETR, and RPV, in addition to EFV (Figure 4A; note the panel of Figure 2C is included as the 2nd panel in Fig. 4A for comparison). Spectra show that saturation with EFV, ETR and RPV is essentially complete at the 1:1 RT:NNRTI molar ratio, while, for NVP, much higher concentrations are needed to ensure saturation. These observations are consistent with results obtained in cell-based assays, that show the EC50 for NVP, 0.085 µM, is much higher than for EFV, ETR and RPV, which are 0.001, 0.002, and 0.0004 µM, respectively.48 Importantly, in the presence of each NNRTI, the defining characteristic of each spectrum is the significantly narrower NNRTI-bound signal (Figure 4A, colored traces), compared to the inhibitor-free signal of apo-RT181tfmF (500 ± 2 Hz) (Figure 4A, black trace) (Table 2). This suggests that NVP, EFV, ETR, and RPV all reduce the conformational plasticity of the NNRTI inhibitor binding site around position 181, as reflected in the narrower linewidth of the tfmF signal..
Figure 4.
1D 19F NMR spectra of RT181tfmF and several RT181tfmF mutants in the absence (black) and presence of NVP (pink), EFV (green), ETR (blue) and RPV (orange). (A) Superposition of the 19F spectra of apo-RT181tfmF and the 19F spectra of RT181tfmF in the presence of each NNRTI, (B) Superposition of the 19F spectra of apo-RT181tfmF-V108I and the 19F spectra of RT181tfmF in the presence of each NNRTI, (C) Superposition of the 19F spectra of apo-RT181tfmF-K103N and the 19F spectra of RT181tfmF-K103N in the presence of each NNRTI, (D) Superposition of the 19F spectra of apo-RT181tfmF-E138K(p51) and the 19F spectra of RT181tfmF-E138K(p51) in the presence of each NNRTI. The 19F spectra in the presence of each NNRTI at 1:1 and 1:5 molar ratios are shown in light and dark colors, respectively. Chemical formulae for each inhibitor are depicted in the individual panels.
Table 2.
19F Resonance frequencies and linewidthsa
| RT127tfmF | RT146tfmF | RT181tfmF |
RT181tfmF- V108I |
RT181tfmF- E138K(p51) |
RT181tfmF-K103N | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ppm | Hz | ppm | Hz | ppm | Hz | ppm | Hz | ppm | Hz | ppm | Hz | |
| Apo | −61.5 | 100 ± 2 | −60.2 −62.0 |
150 ± 85 150 ± 5 |
−60.8b | 500 ± 5 | −61.1b | 500 ± 5 | −60.8b | 600 ± 20 | −60.1 | 300 ± 15 |
| EFV | −61.5 | 100 ± 5 | −60.2 −61.8 |
180 ± 1 180 ± 2 |
−60.3 | 125 ± 2 | −60.3 | 125 ± 5 | −60.2 | 170 ± 5 | −60.4 | 150 ± 2 |
| ETR | −60.3 | 125 ± 2 | −60.3 | 125 ± 5 | −60.2 | 125 ± 2 | −60.4 | 145 ± 2 | ||||
| RPV | −60.7 | 140 ± 2 | −60.7 | 125 ± 5 | −60.7 | 125 ± 5 | −60.7 | 145 ± 2 | ||||
| NVP | −60.2 −61.9b |
180 ± 30 200 ± 25 |
−59.7 −60.8b |
180 ± 5 500 ± 10 |
−59.7 −61.1b |
125 ± 10 400 ± 10 |
−59.7 −60.9b |
125 ± 10 600 ± 15 |
−60.1b | 300 ± 20 | ||
the upper limit of uncertainty in the linewidth was qualitatively estimated as described in Materials and Methods.
although the signal is not symmetric, the resonance frequency and the linewidth were extracted assuming a single peak.
We also examined how NNRTI binding affects the different drug-resistant variants of RT181tfmF. In the presence of 5-fold excess of NVP, EFV, ETR and RPV, the spectra of RT181tfmF-V108I exhibit chemical shifts and linewidths similar to those of RT181tfmF (Figure 4A,B). All bound resonances are significantly sharper (~125 Hz) than in the apo form (~500 Hz). At 1:1 RT: NVP molar ratio, as expected, the ligand-free RT signal is still present, consistent with the 2-fold larger EC50 value, compared to wt-RT.49 RT181tfmF-K103N and RT181tfmF-E138K(p51) also exhibit similar changes in chemical shifts and linewidths, as seen with RT181tfmF upon NNRTI binding(Figure 4C,D; Table 2). Note that binding of NVP to RT181tfmF-K103N was not observed (Figure 4C). This agrees well with the much larger EC50 values reported in cell-based assays (> 1 µM).48 Taken together, upon NNRTI binding to RT, substantially narrower 19F signals are observed for the tfmF group at position 181, demonstrating that the NNRTI-binding pocket becomes confined and locked into a more rigid conformation.
NVP and EFV binding to RT146tfmF and mutants associated with drug resistance
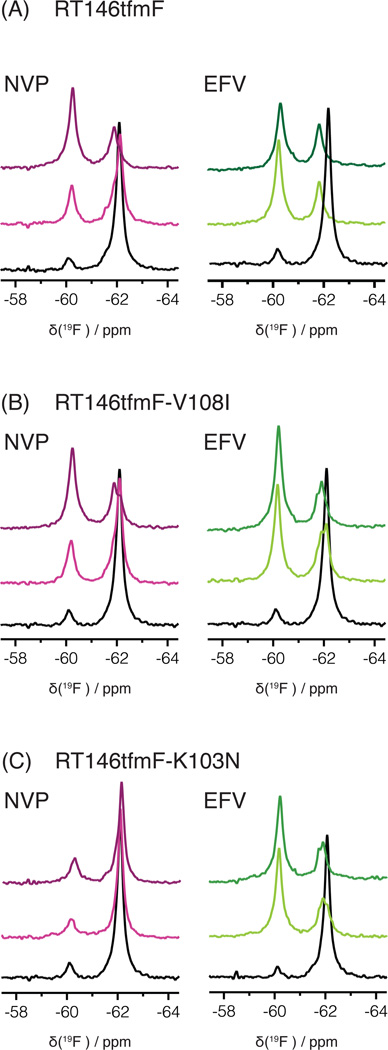
We also recorded 19F NMR spectra of RT146tfmF and its V108I and K103N mutants (Figure 3B). The spectra of these mutants in the apo form (without inhibitor) are essentially identical to each other, demonstrating that the tfmF group in position 146 is not affected by amino acid changes at positions 103 and 108 that cause drug resistance. From the 19F NMR spectra of RT146tfmF and the mutants in the presence of NVP or EFV, it can be appreciated that saturation with EFV is essentially complete at 1:1 RT:EFV molar ratio (Figure 5, note that the panel of Figure 2B is included as the 2nd panel in Figure 5A for comparison). In contrast, with NVP, much higher concentrations are needed to reach saturation. As expected,48 no NVP binding to RT146tfmF-K103N was detected. The spectra in the presence of NNRTIs exhibit two signals at approximately −62 ppm and −60 ppm, irrespective of the mutations. For RT146tfmF-V108I and RT146tfmF-K103N the EFV-bound signals (at 1:5 molar ratio) resonate at −62 ppm and are somewhat asymmetric, compared to that of RT146tfmF (Figure 5). However, these differences are small and may reflect minor differences in dynamics in these mutants. Overall, all available data for RT146tfmF show that no significant effect is seen in the spectra when the drug resistant mutational changes are introduced into the protein, both in the apo- and NNRTI-bound forms. This observation is consistent with the location of residue 146 in the RT structure on the fingers subdomain, ~30 Å away from the NNRTI binding site (Figure 1B).
Figure 5.
Superposition of 1D 19F NMR spectra of (A) RT146tfmF, (B) RT146tfmF-V108I, and (C) RT146tfmF-K103N, in the absence (black) and presence of NVP (pink) or EFV (green). The 19F spectra in the presence of each NNRTI at 1:1 and 1:5 molar ratios are shown in light and dark colors, respectively.
Sensing the NNRTI interaction in RT181tfmF and RT146tfmF using the tfmF probe
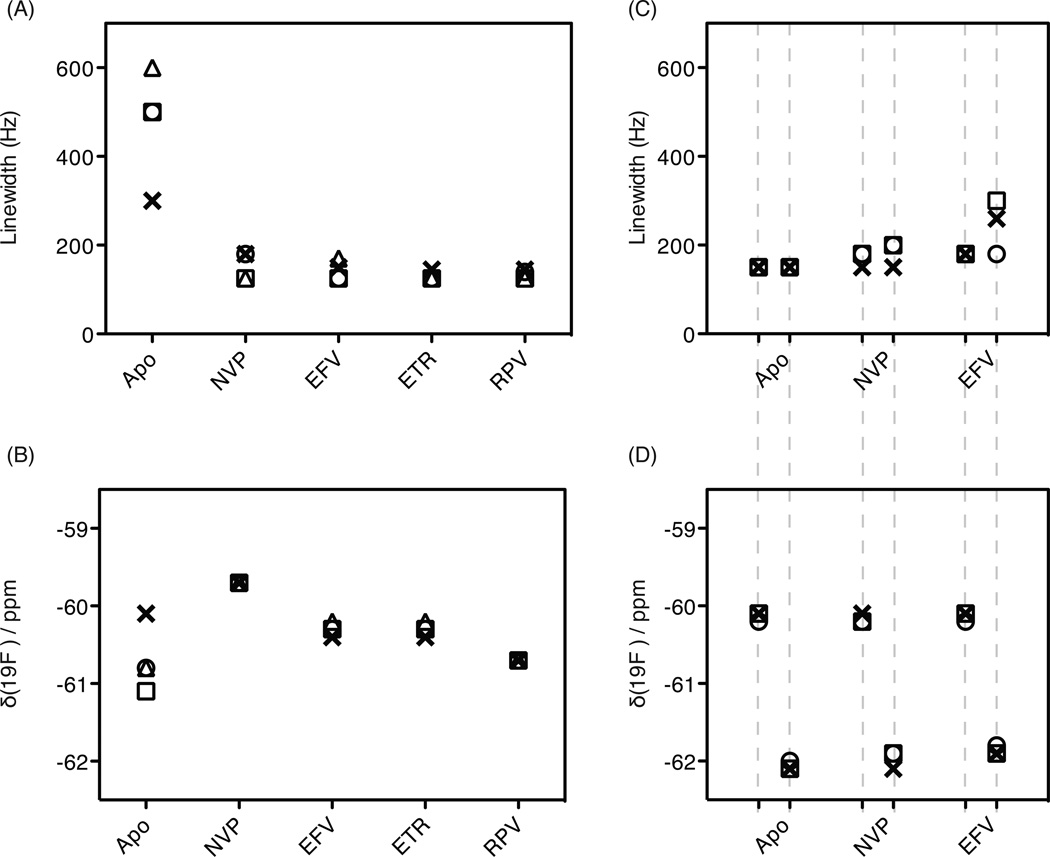
Given that residue 181 resides in the NNRTI binding site, extensive studies were carried out with different RT181tfmF variants. All chemical shifts and linewidths for these variants are summarized in Figure 6A,B. In the absence of inhibitors, the linewidths of the ligand-free signals are larger (Figure 6A) and the chemical shifts are diverse (Figure 6B). The chemical shifts of the NVP-, EFV-, ETR-, and RPV-bound RT181tfmF resonances are −59.7, −60.3, −60.3, −60.7 ppm, respectively, suggesting that different inhibitors create different chemical environments around the 181tfmF probe. Based on the linewidth data, although NNRTIs are chemically dissimilar, NVP, EFV, ETR, and RPV all seem to lock the NNRTI-binding pocket into a more rigid conformation (Figure 6A). For RT146tfmF no significant changes in resonance frequencies and linewidths in the apo- and NNRTI-bound forms among the variants are observed (Figure 6C and 6D).
Figure 6.
Plots of (A) and (C) linewidths and (B) and (D) chemical shifts of the signals in the 19F spectra of RT181tfmF and RT146tfmF and the drug-resistant variants, respectively, in the absence and presence of each NNRTI. In (A) and (B), plots are shown for apo- and NNRTI-bound signals of RT181tfmF (o), RT181tfmF-K103N (x), RT181tfmF-V108I (□), and RT181tfmF-E138K(p51) (△). In (C) and (D), plots are shown for apo- and NNRTI-bound signals of RT146tfmF (o), RT146tfmF-K103N (x), and RT146tfmF-V108I (□). Note that since the spectra of RT146tfmF and mutants thereof comprise two resonances at approximately at −62 and −60 ppm, two sets of points are contained in the plots presented in (C) and (D). Values were obtained from the spectra provided in Figures 4 and 5.
Furthermore, and most significantly, the inhibitor-bound chemical shifts of RT181tfmF resonances are specific for each inhibitor. Indeed, all RT variants, when complexed with NVP, exhibit the same chemical shift of −59.7 ppm. Likewise, the RPV-bound chemical shift is −60.7 ppm and EFV- and ETR-bound chemical shifts are −60.3 ± .1 ppm. These data suggest that the differences in the environment of the 19F probe at position 181 in the apo-RT proteins become diminished upon inhibitor binding, resulting in a chemical environment determined by the specific inhibitor, presumably by rigidifying the NNRTI-binding pocket around the inhibitor.
Discussion
19F solution NMR experiments on site-specifically 19F labeled RT variants were performed to assess the proteins' behavior in the absence and presence of several NNRTIs. The three singly fluorinated RT proteins, RT127tfmF, RT146tfmF and RT181tfmF exhibit distinct spectra, reflecting the different chemical environments surrounding the 19F probes. For two 19F-labeled proteins, RT146tfmF and RT181tfmF, EFV binding clearly can be monitored by the probe. Interestingly, the spectrum of free RT181tfmF (Figure 2C) exhibits a very broad signal, incompatible with a single, narrow conformation of the protein, and suggests a very plastic, mobile environment that is sensed by the 19F nucleus. In the crystal structures of apo-RT and NNRTI-bound RT, two different conformations of tyrosine 181 are observed, and rotation of the side chain out of the NNRTI-binding pocket in the complex is necessary to accommodate inhibitor binding.2–5,50 However, as described in the introduction, only limited data are available for apo-RT, in contrast to an abundance of data on ligand-bound RT. Since it is essential to have access to equivalent information for both, apo- and ligand-bound forms in order to evaluate conformational changes between the two states, we studied RT in solution. The spectrum of ligand-free RT suggests that the NNRTI-binding site is highly plastic in the free enzyme, and not confined to a single, narrow conformation. Thus, the solution NMR results offer different, but complimentary information to the crystallographic data.
We also investigated the effect of drug-induced mutations on the chemical environment around the 19F probe at position 181 (RT181tfmF-V108I, RT181tfmF-K103N and RT181tfmF-E138K(p51). Each mutation is located in a different position (Figure 1C). V108 is positioned behind residue Y188, and K103 (p51) and E138 (p51) are located at the entrance of the NNRTI-binding site. Our NMR data show that amino acid changes at these sites affect the chemical environment of the 19F probe at position 181, with the K103N change producing the greatest effects (Figure 3A). In particular, the narrow signal of RT181tfmF-K103N suggests that this mutation restricts the plasticity of the NNRTI-binding site. These results are consistent with previous studies, based on crystallographic and computational data, which suggest a “closed” form of the NNRTI-binding pocket in K103N RT.27,51 In contrast, 19F signals of the RT181tfmF-E138K and RT181tfmF-V108I variants are significantly broader, suggesting that they possess a more plastic NNRTI-binding site. Interestingly, the latter mutations are associated with the lower degree of NNRTI resistance.45,47,52
Our NMR data also revealed remarkable changes in chemical shifts and linewidths of the RT181tfmF signal upon interactions with NNRTIs. Importantly, the dynamic behavior of the 181 site is quenched upon EFV binding, consistent with previous results from HXMS experiments that showed higher protection for peptides 88–109 and 187–192 in the presence of EFV.18 A similar effect is seen for NVP, ETR and RPV binding. We find that all the NNRTI-bound signals are narrower, indicating a significant reduction in the flexibility of the inhibitor-bound forms, compared to the apo-form (Figure 6A). Most interestingly, the chemical shifts of the different NNRTI-bound variants revealed an intriguing pattern: while the resonance frequencies of the ligand-free RT181tfmF variants, V108I, K103N and E138K(p51), are all different, compared to RT181tfmF, once a particular inhibitor is bound, these differences disappear. For all four protein complexes, essentially the same chemical shifts are noted for the inhibitor-bound proteins; thus, the NNRTI-bound shifts are characteristic for a particular inhibitor, irrespective of the presence of mutations that are associated with drug resistance. This suggests that it is the identity of the inhibitor, which ultimately determines the resonance frequencies of the tfmF probe at position 181.
While it is possible that the fluorophenylalanine substitution for tyrosine at position 181 may influence the conformation in NNRTI-binding site, this effect has to be very small, given that the binding affinities of EFV and NVP to RT181tfmF are consistent with reports in the literature.48 In addition, the effects observed upon EFV binding when the tfmF probe resides at positions 127, 146, and 181 agree well with the distances between the 19F nucleus and the NNRTI binding site. Thus, observed 19F spectral changes upon NNRTI binding perhaps qualitatively reflect the native dynamics of individual sites. As described above, the 19F resonances of apo RT181tfmF and mutants thereof vary (Figure 3A).
In summary, our data provide new insights into the dynamics of the NNRTI-binding site and suggest a mechanism of action for NNRTIs. Our results clearly demonstrate that the NNRTI-binding site is highly plastic in the ligand-free enzyme, and that drug resistance mutants modulate this conformational plasticity. Importantly, NVP, EFV, ETR, and RPV all reduce the dynamics of RT in the NNRTI-binding site, and the NNRTI-bound chemical shifts are determined by the identity of the inhibitor. Furthermore, the present study demonstrates that 19F NMR can be used as an effective tool for examining ligand-protein interactions in cases where only small amounts of protein are available or limited solubility of protein or ligand exist.
Table 3.
19F Resonance frequencies and linewidths of mutants of RT146tfmFa
|
RT146tfmF- V108I |
RT146tfmF- K103N |
|||
|---|---|---|---|---|
| ppm | Hz | ppm | Hz | |
| Apo | −60.1 −62.1 |
150 ± 30 150 ± 5 |
−60.1 −62.1 |
150 ± 30 150 ± 5 |
| EFV | −60.1 −61.9b |
180 ± 5 300 ± 15 |
−60.1 −61.9b |
180 ± 5 260 ± 5 |
| NVP | −60.2 −61.9b |
180 ± 10 200 ± 25 |
−60.1 −62.1 |
150 ± 30 150 ± 5 |
the upper limit of uncertainty in the linewidth was qualitatively estimated as described in Materials and Methods.
although the signal is not symmetric, the resonance frequency and linewidth extracted assuming a single peak..
Acknowledgments
We thank Drs. Elena Matei and In-ja Byeon for help with 19F NMR spectroscopy, Mike Delk for NMR technical support, and Atticus Huberts for help with protein expression. Dr. Nicholas Sluis-Cremer is gratefully acknowledged for providing EFV and NVP, Dr. Teresa Brosenitsch for critical reading of the manuscript, and Drs. Stuart Le Grice, Mary Barkley, Ryan Mehl and Christopher Barnes for useful discussions.
Funding
This work was supported by National Institutes of Health grant P50GM082251, and N.G.S was the recipient of a Graduate Research Fellowship, 1247842, from the National Science Foundation.
Abbreviations
- tfmF
4-trifluoromethyl phenylalanine
- NNRTIs
non-nucleoside inhibitors
- RT
HIV-1 reverse transcriptase
- NVP
Nevirapine
- EFV
Efavirenz
- ETR
Etravirine
- RPV
Rilpivirine
References
- 1.Looney D, Ma A, Johns S. HIV therapy-the state of art. Curr. Top. Microbiol. Immunol. 2015;389:1–29. doi: 10.1007/82_2015_440. [DOI] [PubMed] [Google Scholar]
- 2.Kohlstaedt LA, Wang J, Friedman JM, Rice PA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. …. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 3.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold EE. Structure of unliganded HIV-1 reverse transcriptase at 2.7 å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 4.Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J. Med. Chem. 2010;53:4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]
- 5.Milton J, Weaver KL, Short SA. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure. 2000;8:1089–1094. doi: 10.1016/s0969-2126(00)00513-x. [DOI] [PubMed] [Google Scholar]
- 6.HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapkouski M, Tian L, Yang W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat. Struct. Mol. Biol. 2013;20:230–236. doi: 10.1038/nsmb.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das K, Feng JY, Tuske S, Clark AD, Jr, Boyer PL, Hou X, Jones RA, Hughes SH, Arnold EE. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J. Biol. Chem. 2009;284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu X, Bauman JD, Clark AD, Jr, Jones RA, Boyer PL, Hughes SH, Arnold EE. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antiviral Res. 1998;38:153–179. doi: 10.1016/s0166-3542(98)00025-4. [DOI] [PubMed] [Google Scholar]
- 11.Pauwels R. New non-nucleoside reverse transcriptase inhibitors (NNRTIs) in development for the treatment of HIV infections. Curr. Opin. Pharmacol. 2004;4:437–446. doi: 10.1016/j.coph.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Sluis-Cremer N. The emerging profile of cross-resistance among the nonnucleoside HIV-1 reverse transcriptase inhibitors. Viruses. 2014;6:2960–2973. doi: 10.3390/v6082960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold EE. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das K, Lewi PJ, Hughes SH, Arnold EE. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog. Biophys. Mol. Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 15.De Béthune M-P. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: A review of the last 20 years (1989–2009) Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Zhan P, Chen X, Li D, Fang Z, De Clercq E, Liu X. HIV-1 NNRTIs: structural diversity, pharmacophore similarity, and implications for drug design. Med Res Rev. 2013;33(Suppl 1):E1–E72. doi: 10.1002/med.20241. [DOI] [PubMed] [Google Scholar]
- 17.Kensch O, Restle T, Wöhrl BM, Goody RS, Steinhoff H-J. Temperature-dependent equilibrium between the open and closed conformation of the p66 subunit of HIV-1 reverse transcriptase revealed by site-directed spin labelling. J. Mol. Biol. 2000;301:1029–1039. doi: 10.1006/jmbi.2000.3998. [DOI] [PubMed] [Google Scholar]
- 18.Seckler JM, Barkley MD, Wintrode PL. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon Inhibitor Binding. Biophys. J. 2011;100:144–153. doi: 10.1016/j.bpj.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, Mueller GA, Derose EF, London RE. Solution characterization of [methyl-13C]methionine HIV-1 reverse transcriptase by NMR spectroscopy. Antiviral Res. 2009;84:205–214. doi: 10.1016/j.antiviral.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Perera L, Mueller GA, DeRose EF. Asymmetric conformational maturation of HIV-1 reverse transcriptase. Elife. 2015;4:e06359. doi: 10.7554/eLife.06359. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Madrid M, Evanseck JD, Madura JD. Effect of a bound non-nucleoside RT inhibitor on the dynamics of wild-type and mutant HIV-1 reverse transcriptase. J. Am. Chem. Soc. 2005;127:17253–17260. doi: 10.1021/ja053973d. [DOI] [PubMed] [Google Scholar]
- 22.Ivetac A, McCammon JA. Elucidating the inhibition mechanism of HIV-1 non-nucleoside reverse transcriptase inhibitors through multicopy molecular dynamics simulations. J. Mol. Biol. 2009;388:644–658. doi: 10.1016/j.jmb.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen L, Shen J, Luo X, Cheng F, Xu Y, Chen K. Steered molecular dynamics simulation on the binding of NNRTI to HIV-1 RT. Biophys. J. 2003;84:3547–3563. doi: 10.1016/S0006-3495(03)75088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahar I, Erman B, Jernigan RL, Atilgan AR, Covell DG. Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J. Mol. Biol. 1999;285:1023–1037. doi: 10.1006/jmbi.1998.2371. [DOI] [PubMed] [Google Scholar]
- 25.Seckler JM, Leioatts N, Miao H, Grossfield A. The interplay of structure and dynamics: insights from a survey of HIV-1 reverse transcriptase crystal structures. Proteins. 2013;81:1792–1801. doi: 10.1002/prot.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monroe JI, El-Nahal WG, Shirts MR. Investigating the mutation resistance of nonnucleoside inhibitors of HIV-RT using multiple microsecond atomistic simulations. Proteins. 2014;82:130–144. doi: 10.1002/prot.24346. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez-Barrios F, Gago F. Understanding the basis of resistance in the irksome Lys103Asn HIV-1 reverse transcriptase mutant through targeted molecular dynamics simulations. J. Am. Chem. Soc. 2004;126:15386–15387. doi: 10.1021/ja045409t. [DOI] [PubMed] [Google Scholar]
- 28.Wright DW, Hall BA, Kellam P, Coveney PV. Global conformational dynamics of HIV-1 reverse transcriptase bound to non-nucleoside inhibitors. Biology. 2012;1:222–244. doi: 10.3390/biology1020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das K, Ding J, Hsiou Y, Clark AD, Jr, Moereels H, Koymans L, Andries K, Pauwels R, Janssen PAJ, Boyer PL, Clark P, Smith RH, Jr, Kroeger Smith MB, Michejda CJ, Hughes SH, Arnold E. Crystal Structures of 8-Cl and 9-Cl TIBO Complexed with Wild-type HIV-1 RT and 8-Cl TIBO Complexed with the Tyr181Cys HIV-1 RT Drug-resistant Mutant. J. Mol. Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 30.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Mol. Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 31.Danielson MA, Falke JJ. Use of 19F NMR to probe protein structure and conformational changes. Annu. Rev. Biophys. Biomol. Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerig JT. Fluorine NMR of proteins. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:293–370. [Google Scholar]
- 33.Sharaf NG, Gronenborn AM. 19F-modified proteins and 19F-containing ligands as tools in solution NMR studies of protein interactions. Methods Enzymol. 2015;565:67–95. doi: 10.1016/bs.mie.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Sharaf NG, Poliner E, Slack RL, Christen MT, Byeon I-JL, Parniak MA, Gronenborn AM, Ishima R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014;82:2343–2352. doi: 10.1002/prot.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peeler JC, Mehl RA. Site-specific incorporation of unnatural amino acids as probes for protein conformational changes. Methods Mol. Biol. 2011;794:125–134. doi: 10.1007/978-1-61779-331-8_8. [DOI] [PubMed] [Google Scholar]
- 36.Studier FW. Protein production by auto-induction in high density shaking cultures. PREP. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry-Us. 2006;45:2779–2789. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 38.Holladay LA. Efavirenz binding to HIV-1 reverse transcriptase monomers and dimers. Biochemistry-Us. 2010;49:601–610. doi: 10.1021/bi901579y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold EE. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 40.Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, Sullivan J, Cheeseman S, Barringer K, Pauletti D. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuman EC, Rhee S-Y, Holmes SP, Shafer RW. Constrained patterns of covariation and clustering of HIV-1 non-nucleoside reverse transcriptase inhibitor resistance mutations. J. Antimicrob. Chemother. 2010;65:1477–1485. doi: 10.1093/jac/dkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bacheler L, Jeffrey S, Hanna G, D'Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 2001;75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee S-Y, Liu T, Ravela J, Gonzales MJ, Shafer RW. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob. Agents Chemother. 2004;48:3122–3126. doi: 10.1128/AAC.48.8.3122-3126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tambuyzer L, Vingerhoets J, Azijn H, Daems B, Nijs S, De Béthune M-P, Picchio G. Characterization of genotypic and phenotypic changes in HIV-1-infected patients with virologic failure on an etravirine-containing regimen in the DUET-1 and DUET-2 clinical studies. AIDS Res. Hum. Retroviruses. 2010;26:1197–1205. doi: 10.1089/aid.2009.0302. [DOI] [PubMed] [Google Scholar]
- 45.Tambuyzer L, Nijs S, Daems B, Picchio G, Vingerhoets J. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. J. Acquir. Immune Defic. Syndr. 2011;58:18–22. doi: 10.1097/QAI.0b013e3182237f74. [DOI] [PubMed] [Google Scholar]
- 46.Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J. Acquir. Immune Defic. Syndr. 2012;59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 47.Azijn H, Tirry I, Vingerhoets J, De Béthune M-P, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 2010;54:718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold EE. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Xu W, Koh Y-H, Shim JH, Girardet J-L, Yeh L-T, Hamatake RK, Hong Z. A novel nonnucleoside analogue that inhibits human immunodeficiency virus type 1 isolates resistant to current nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2007;51:429–437. doi: 10.1128/AAC.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodgers DW, Harris BA, Ray S, Hellmig B, Woolf DJ, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proceedings of the National Academy of Sciences. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsiou Y, Ding J, Das K, Clark AD, Jr, Boyer PL, Lewi P, Janssen PAJ, Kleim J-P, Rösner M, Hughes SH, Arnold E. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 2001;309:437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- 52.Geretti AM, Mackie N. Antiretroviral Resistance in Clinical Practice. Mediscript, London: 2006. Resistance to non-nucleoside reverse transcriptase inhibitors. [PubMed] [Google Scholar]