Abstract
Active transport and localized translation of the ASH1 mRNA at the bud tip of the budding yeast Saccharomyces cerevisiae is an essential process that is required for the regulation of the mating type switching. ASH1 mRNA localization has been extensively studied over the past few years and the core components of the translocation machinery have been identified. It is composed of four localization elements (zipcodes), within theASH1 mRNA, and at least three proteins, She1p/Myo4p, She2p and She3p. Whereas the movement of the RNA can be attributed to direct interaction with myosin, the regulation of the RNA expression is less well understood. Recent insights have revealed a role for translation that might have a key function in the regulation of Ash1 protein sorting.
Introduction
Modulation of gene expression occurs at various steps, from initiation of transcription up to the final delivery of an active protein to its correct location within the cell. One powerful mechanism for control of gene expression involves localized translation of a specific mRNA at the place where the protein is needed. This cytoplasmic aspect of gene expression is a well-documented mechanism for regulating local protein expression in higher eukaryotes and patterning during embryonic development [1]. For instance, during Drosophila oogenesis, the future axes of the embryo are predetermined maternally by the specific localization of specific mRNAs to the two poles of the oocyte. Later in development, pair–rule mRNAs are actively transported and anchored to the cortex of the syncytium, preventing these morphogenes from diffusing along the embryo before cellularization occurs. Somatic cells use mRNA localization to locally produce proteins in the subcellular location where they are needed. For example, β-actin mRNA is localized to the leading edge of fibroblasts, where it is utilized for rapid responses to extracellular signals [2]. These examples confirm that specific mRNA subcellular localization generates the cellular protein asymmetry that is required for diverse cellular functions.
In the yeast Saccharomyces cerevisiae, ASH1 mRNA localization is used to control mating type switching. The localized translation of the transcriptional repressor, Ash1p, to the daughter cell specifies different patterns of gene expression between mother and daughter cells. Here, the most comprehensively understood mechanism of mRNA localization, that of ASH1 in budding yeast, will be discussed.
Budding yeast mating type switching
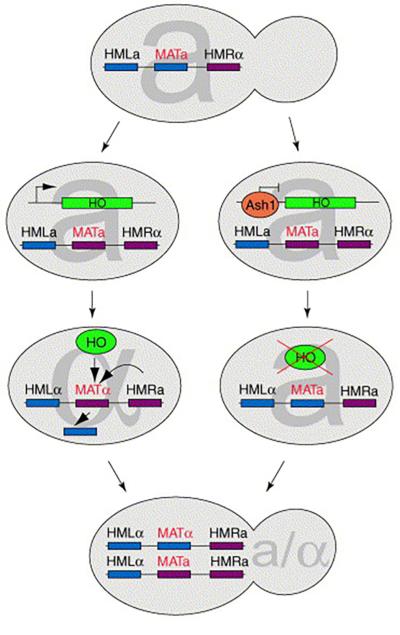
The budding yeast S. cerevisiae alternates between a diploid growing state and, under conditions of nutrient deprivation, a haploid growing state. The return to a diploid state is accomplished through the mating of two haploid cells of the opposite mating type (‘a’ or ‘α’). Mating type switching is unique to the mother cell. The daughter never switches, and thus mother and daughter cells are necessarily of opposite mating types. This ensures that an isolated spore will be able to form diploids cells through mating between its descendents. Mating type switching results from asymmetric (mother-specific) expression of the HO endonuclease. HO initiates a genomic rearrangement of the MAT locus, resulting in the conversion of an ‘a’ cell to an ‘α’ or vice–versa (Figure 1). The basis of asymmetric HO transcription comes from the restriction of the transcriptional repressor Ash1p to the daughter cell [3-5]; a result of the asymmetric distribution of its mRNA.
Figure 1.
Mating type switching in the yeast S. cerevisiae. The mating type of a cell is defined by the expression of ‘a’- or ‘α’- specific gene(s) from the active MAT locus. These specific factors are encoded by the transcriptionally silenced HMLa and HMRα loci. Switching to the opposite mating type occurs by gene conversion at the active MAT locus. The template used for this conversion is issued from the silenced HMLa or HMRα locus of the opposite type. HO endonuclease cuts the active MAT locus, initiating this replacement. In daughter cells, the transcriptional repressor Ash1p inhibits the expression of HO, thereby repressing mating type switching (see text for details).
ASH1 mRNA is packaged in a mRNP, the ‘locasome’, which is actively transported to the bud, ensuring the exclusive translation of Ash1p in the daughter cell [6]. ASH1 mRNA localization near to the daughter nucleus is the sole determinant of Ash1p sorting [7]. Ash1p asymmetry might not be restricted to mating type switching; it might also be required for pseudohyphal growth [8, 9]. Ash1p is restricted to the pseudohyphal cell-nucleus but the dependence of pseudohyphal growth on restricted expression of Ash1p has not been proven, thus far [9]. Together with daughter cell-specific ASH1 mRNA localization, the cell cycle-regulated transcription in late anaphase [4, 10] has a crucial role in mating type switching control. Ash1p also interacts with the promoters ofSGA1 and PCL1, which are implicated in sporulation control and cell cycle control, respectively [11]. Interestingly, Pcl1p is one of the cyclins associated with the Pho85 kinase complex, which has recently been demonstrated to phosphorylate Ash1p, regulating its stability [12]. In a PHO85Δ strain, Ash1p is stabilized so that its activity persists in the daughter cell nucleus during the cell cycle. This aberrantly represses mating type switching when the cell enters the next bud cycle [12].
The ASH1 mRNA transport machinery (the locasome)
ASH1 mRNA localization is mediated by different cis elements within the mRNA, termed ‘zipcodes’, that are recognized by trans-acting factors that mediate the directed movement to the bud tip (place of translation). ASH1 mRNA contains 4 cis-acting zipcodes, termed E1, E2a, E2b and E3 (Figure 2a) [13-16]. These zipcodes act synergistically, ensuring efficient transport. Each is able to direct an mRNA to the bud but all four together ensure that this happens more frequently [13, 14, 17]. The ASH1 zipcodes are contained within the coding sequence of the mRNA, with one of them overlapping the stop codon (Figure 2a). The sequence homology between the four elements is weak and no common sequence has yet been identified. Three of the four elements are entirely located in the coding region; this could introduce constraints in the evolution of such elements, making it difficult to identify clearly defined sequence/structure motifs. Additional functions of these elements, aside from their common role in localization, cannot be excluded.
Figure 2.
Ash1 mRNA localization. (a) Cartoon of the Ash1 mRNA core locasome transported on an actin filament. E1, E2a, E2b and E3 zipcodes are recognized by the She1–3p complex, providing the motor activity. (b)Schematic view of the different steps of ASH1 mRNA bud tip localization (see also Table1). 1 – Nuclear events: ASH1 transcription in late anaphase, association of ASH1 mRNA with She2p, interaction with Loc1p. 2 – Early cytoplasmic events: Assembly of the locasome by association of the newly exported ASH1mRNA-She2p RNP with She3p and She1p/Myo4p, recruitment to the actin network and interaction with Khd1p and She4p. 3 – Transport of the locasome to the bud tip. She1p/Myo4 provides the motor activity. Role of She4p as a translocation co-factor. 4 – Anchoring and translation at the bud tip: the mechanism of anchoring is still not understood; nevertheless it involves Khd1p and the nascent Ash1p polypeptide. Puf5p and Scp160p are not represented because their role remains largely unknown. (c) Upper image: phase image of a yeast budding cell in early anaphase. Middle image: fluorescent in situ hybridization (FISH) ofASH1 mRNA using 6 CY3-labeled probes in red and DAPI staining in blue. Lower image: overlay of the two previous images.
Each of the four localization elements is recognized by She2p, an RNA-binding protein, containing no known RNA-binding motifs [18, 19]. She2p interaction with the E3 zipcode in vitro can be demonstrated by UV cross-linking [13]. However, She2p in vitro binding to the full-length ASH1 mRNA is weak, showing a Kd in the micromolar range, indicating that an additional protein could be required for efficient binding in vivo (Hüttelmaier and Singer, unpublished). She2p shuttles from the nucleus to the cytoplasm in an RNA-dependent manner, indicating that newly transcribed ASH1 mRNA is predetermined for a cytoplasmic location before its nuclear export [20] (Figure 2c). Mutants in SHE2 were initially identified in a genetic screen for genes affecting HO expression [21]. This screen also identified two other genes that are directly involved inASH1 mRNA localization: SHE1 and SHE3. She1p, also called Myo4p, is a type V unconventional myosin, providing the motor activity of the locasome along the actin network [4, 13, 14, 17, 21, 22]. Motion driven from a single myosin V would be inefficient, due to the poor processivity of this type of motor. Redundancy of ASH1 mRNA zipcodes is therefore needed to provide a persistent motion of the cargo [23]. This is consistent with the presence of four localization elements in ASH1 mRNA, each presumably binding one myosin molecule (Figure 2a). The link between the RNA-binding protein She2p and the motor She1p/Myo4p is mediated by She3p [18, 24]. She1p/Myo4p and She3p are exclusively cytoplasmic proteins, and the process by which these two proteins recognize the newly exported mRNP containing She2p is yet to be determined. ASH1 mRNA, together with the trimeric complex She1p/Myo4p–She2p–She3p, initiates formation of the locasome — the RNA localization particle [18, 24]. Each of these components is absolutely necessary for ASH1 mRNA localization; their individual deletions result in the delocalization of ASH1 mRNA [13, 14]. The kinetics of the transport of the locasome to the bud tip has been demonstrated in vivo by tagging the mRNA with GFP reporters making ASH1 mRNA localization a powerful tool by which to study RNA movements in vivo [17, 25, 26]. (Figure 2).
Accessory factors involved in locasome targeting
She1p/Myo4p, She2p and She3p are core components of the locasome but additional proteins have been identified that affect ASH1 mRNA localization (Table 1). These proteins play an accessory role in the trafficking process and their deletion seems to affect general steps, such as cell polarization, motor-driven motility and translation [6].
Table 1.
Proteins involved in ASH1 mRNA localization.
| Protei n |
Altern ative names |
Function | Role in ASH1 mRNA localization |
Locaso me associ ation |
Subcell ular localiz ation |
Δ phenot ype bud/bu d tip |
References |
|---|---|---|---|---|---|---|---|
| She1 p |
Myo4p | Unconven tional myosin V type motor |
Locasome molecular motor |
Yes | C, bud enriche d |
Abolis hed |
13, 14, 17•, 21, 22. |
| She2 p |
RNA- binding |
Binds ASH1m RNA zipcodes |
Yes | N/C, bud enriche d |
Abolis hed |
13, 18, 19, 20•, 21, 22. | |
| She3 p |
Myosin binding protein |
Molecular link in between She2p and She1p |
Yes | C, bud enriche d |
Abolis hed |
13, 18, 19, 21, 22, 24. |
|
| She4 p |
Dim1p | Endocyto sis Enhance associatio n of Myo3p and Myo5p to the actin network Interacts with She1p |
Enhance association of She1p to the actin network? |
Yes | C, bud enriche d |
nd/nd bud neck |
13, 21, 28, 29. |
| She5 p |
Bni1p, Ppf3p |
Actin filament polymeriz ation at the bud tip |
Polarization and growth of the actin network |
No | C, bud tip |
nd/nd bud neck/b ud enrich ed |
13, 22, 26, 33. |
| Bud growth Establish ment of cell polarity |
Interacts with Bud6p (two- hybrid and synthetic lethality) |
||||||
| Bud6 p |
Aip3p | Actin filament organizati on at the bud tip |
Polarization and growth of the actin network |
No | C, bud tip |
nd/nd bud enrich ed |
26, 27, 34, 35. |
| Bud growth Establish ment of cell polarity |
Interacts with She5p (two- hybrid and synthetic lethality) |
||||||
| Loc1 p |
RNA binding rRNA processin g |
Binds ASH1zi pcodes, general translation defect? |
Yes (nucleu s) |
No | nd/13 | 36, 37. | |
| Khd1 p |
Hek2p | RNA binding, KH domain |
Involved in mRNA bud tip anchoring, translational repression |
Yes | C | 93/40 | 38••, 39. |
| Puf5p | Mpt5p, Htr1p, Uth4p |
Translatio nal regulation mRNA stability |
Involved in mRNA bud tip anchoring, Translational regulation? |
No | nd | 83/22 | 38••, 40. |
| Scp1 60p |
RNA binding, KH domain ER polysome s binding |
Involved in mRNA bud tip anchoring, Translational regulation? |
No | C | 84/23 | 38••, 42. |
C, Cytoplasmic; N, Nuclear; No, nucleolar; deletion phenotype is shown as: percentage of cells showing daughter cell localization of ASH1 mRNA/percentage of cells showing daughter cell bud tip localization ofASH1 mRNA. The wild type strain shows a phenotype of 99/87 [38••].
Two additional SHE genes (SHE4 and SHE5) were identified in the initial screen and, subsequently, were shown to affect ASH1 mRNA localization [13, 14, 17, 21, 26, 27]. SHE4 mutants show additional defects in endocytosis and actin polarization [28, 29]. She4p binds to the motor domains of unconventional myosins and enhances their binding to microfilaments [30, 31]. Interestingly, She4p interacts with She1p in a two-hybrid assay [31]. It has been proposed that She4p is required for the structural integrity or the regulation of the motor domain of unconventional myosins [31].
She5p/Bni1p is a formin that has recently been shown to promote nucleation of barbed-end actin polymerization [32, 33]. Deletion of SHE5/BNI1 leads to the accumulation of ASH1 mRNA at the bud neck [13], consistent with a defect in promoting polymerization of actin fibers at the bud tip. Bud6p binds to She5p/Bni1p and is also required for actin filament organization to the bud growth pole [34, 35]. Consistent with the SHE5Δ phenotype, in a BUD6Δ strain, the mRNA shows a directed motorized motion in the mother cell but mRNAs observed in the daughter cell are freely diffusing [26, 27].
In addition to the SHE gene mutations, four additional genes have been shown to affectASH1 mRNA localization: LOC1, KHD1, SCP160 and PUF5. Deletion of each of these four genes affects ASH1 mRNA localization to a lesser extent than the SHE mutants, which might explain why they were not identified in the original genetic screens. Loc1p was isolated by the three-hybrid system, due to its ability to bind to the E3 and E1 zipcodes. In a LOC1Δ strain the ASH1 mRNA is not correctly localized and restricted Ash1p localization to the daughter cell nucleus is strongly affected [36]. Loc1p is an exclusively nuclear protein and so might have a role in early ASH1 mRNP formation. Interestingly, Loc1p has been identified as part of the 66S pre-rRNA complex and has been shown to affect 60S rRNA processing [37]. It is actually unclear whether the ASH1mRNA delocalized phenotype is direct or if it involves a translational defect.
KHD1, SCP160 and PUF5 were identified in a systematic survey of yeast RNA-binding proteins that affect ASH1 mRNA localization [38]. Of these three proteins, only Khd1p co-localizes with the ASH1 mRNA locasome, indicating a direct role in mRNA localization. Interestingly, in a KHD1Δ, SCP160Δ or PUF5Δ strain, the strongest effect onASH1 mRNA localization is seen at the bud tip, meaning that the function of these genes might be more specific for anchoring of the mRNA to its translational location [38] (Table 1 and following sections).
Translation and localization
Sequential deletions of the four zipcodes of ASH1 mRNA progressively affect its correct localization, therefore, demonstrating that they all contribute in targeting the mRNA to the bud tip [7]. The number of zipcodes seems to be crucial for efficient ASH1 mRNA localization but their position is not. When inserted in the 3'UTR of a nonlocalizing mRNA, each one of these four elements is able to direct bud localization [7]. The shift to the 3'UTR of the four ASH1 zipcodes did not affect mRNA localization but it did affect Ash1p asymmetric localization. This demonstrates that the presence of localization elements within the coding region of ASH1 mRNA provide translational silencing while it is transported to the bud tip. Chartrand et al. [7] proposed that a reduced translation of theASH1 mRNA could balance the slow localization of ASH1 mRNA [17, 26]. Another interesting link between ASH1 mRNA translation and localization is the observation that, upon cycloheximide treatment, ASH1 mRNA is not properly localized to the bud tip [20]. Mutation of the ASH1 methionine initiation codon or insertion of a stop codon in the middle of the ASH1 coding sequence affects anchoring to the bud tip but doesn’t affect mRNA transport [15]. This phenotype cannot be rescued by the expression of the wild type Ash1p in trans, demonstrating that the C-terminus of Ash1p is implicated in anchoring as a cis-acting element. This finding introduces the interesting possibility that anchoring to a currently uncharacterized complex could be mediated co-translationally, via the nascent Ash1 polypeptide [15].
Recently, the KH domain containing protein Khd1p, has been reported to affect ASH1mRNA anchoring to the distal bud tip. Interestingly, overexpression of Khd1p reduces the cellular amounts of Ash1p, possibly due to a decrease in translation [38]. Khd1p was found in association with different translation initiation complexes in a proteomic survey [39]. Khd1p has been proposed to bind to the 5' region of ASH1 mRNA and so co-localize with the locasome [38], suggesting that Khd1 could silence ASH1 mRNA translation during transport. Khd1p could also have a more specific role at the bud tip, enabling the specific retention of ASH1 mRNA to the tip. The deletion of KHD1 [38] and the abolition of translation of the Ash1p C-terminus [15] appear to produce a similar phenotype on mRNA anchoring; we suggest that these two factors could be involved in the same process.
Puf5p belongs to a highly conserved family of RNA binding proteins with a well-characterized role in protein translation regulation [40]. Puf5p is a translational regulator of the HO endonuclease, functioning through binding to the 3'UTR of its mRNA [41]. Scp160p is known to interact with membrane-bound polysomes [42]. The role of SCP160 on ASH1 mRNA could be more general, as this protein was recently shown to interact with 69 mRNAs with diverse functions, among which, ASH1 mRNA was not found [43]. Aside from their general role in translation, the link between ASH1 mRNA localization and the function of these proteins remains unclear.
mRNAs actively transported to the daughter cell; a growing family
The function of the locasome is not restricted to the transport of ASH1 mRNA to the daughter cell bud tip. Immunoprecipitation experiments using the core locasome proteins (She1p–3p), followed by DNA microarray technology [44, 45] and validation in a living cell assay [45], identified a total of 22 mRNAs. All these mRNAs show a preferential localization to the bud in a SHE2-dependent manner, implying that their transport machinery is identical to the ASH1 locasome [45]. As is the case for ASH1 mRNA, all of these mRNAs contained localization elements within their coding sequence and for only three of them did the 3' UTR confer specific localization to a reporter mRNA, thus, demonstrating that at least some of them contain additional signals. Nevertheless, localization element redundancy is not a rule; IST2 mRNA seems to contain only one element [20]. Ist2p encodes an ion transporter that is restricted to the daughter cell membrane by the septin ring acting as a diffusion barrier [44]. In addition, eleven of the newly identified localized mRNAs code for proteins that are not localized to the daughter cell; moreover, eight additional mRNAs code for proteins that are able to localize to the daughter cell independently of their mRNA localization, indicating that mRNA localization is not a key determinant for the subcellular distribution of these 19 proteins. None of the 22 known localized mRNAs, nor the locasome components (she1–3), were essential in yeast. One hypothesis to explain the significance of these observations is that mRNA localization to the daughter cell is a dynamic event, ensuring enhanced distribution of mRNAs between mother and daughter cells. This could ensure the ability of the daughter cell to respond to environmental situations independently of transcription. Consistent with this, a fraction of mRNAs that are localized to the daughter cell code for proteins related to environmental sensing.
Conclusions
mRNA localization in S. cerevisiae has proven to be a complex system, involving more steps than just those for moving the RNA. Additional considerations must include the translational regulation of the mRNA during transport and anchoring at its final destination; the molecular mechanisms of these two crucial processes are yet to be fully elucidated. Clearly, the locasome has a role in this regulation, by sequestering the RNA away from the translational components. However, the mechanism by which it is disassembled at the bud tip, thereby enabling translation, remains elusive. Recent developments have also proved that generation of cell fate asymmetry could be one of several functions of mRNA localization in yeast. The discovery of localized mRNAs, not involved in protein asymmetry, and vice-versa, suggests that there might be other roles for mRNA localization.
Acknowledgements
We thank Kelly Shepard for sharing results before publication. The research carried in our laboratory is supported by the NIH grant GM57071.
Abbreviations
- 5'/3' UTR
5'/3' untranslated regions of a mRNA
- DAPI
4',6-diamidino-2-phenylindole
- FISH
fluorescent in situ hybridization
- Kd
dissociation constant
- mRNP
messenger RNA ribonucleoprotein complex
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 2.lathman V, Singer RH. Handbook of Cell Signaling. Vol. 3. (USA) ES; 2003. RNA localization and signal transduction; pp. 293–297. [Google Scholar]
- 3.Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 4.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in post anaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 5.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 6.Chartrand P, Singer RH, Long RM. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- 7.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. This paper shows that the position of Ash1 zipcodes within the coding region of the mRNA are required for the exclusive localization of Ash1p to the daughter cell. Ash1p translational efficiency is directly linked to the presence of the zipcodes within the coding region. Inefficient translation of ASH1 mRNA is required to prevent Ash1p from being prematurely synthesized in the mother cell. This paper also demonstrates that ASH1 mRNA localization is the only determinant of Ash1p daughter cell sorting. [DOI] [PubMed] [Google Scholar]
- 8.Pan X, Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandarlapaty S, Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 12.McBride HJ, Sil A, Measday V, Yu Y, Moffat J, Maxon ME, Herskowitz I, Andrews B, Stillman DJ. The protein kinase Pho85 is required for asymmetric accumulation of the Ash1 protein in Saccharomyces cerevisiae. Mol. Microbiol. 2001;42:345–353. doi: 10.1046/j.1365-2958.2001.02601.x. [DOI] [PubMed] [Google Scholar]
- 13.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 14.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 15•.Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. In addition to the mapping of ASH1 mRNA localization elements (see also [16]), this study demonstrates that specific synthesis of the C-terminal peptide through ASH1 mRNA translation is required for anchoring at the bud tip. This finding provided the first link between translation and anchoring. [DOI] [PubMed] [Google Scholar]
- 16.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 17•.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. This is the first report of the transport of a mRNP in vivo, using a GFP fusion reporter protein tethered to the mRNA. ASH1 mRNPs are transported at a speed compatible with a myosin motor. This work provided the first demonstration that myosin can directly localize mRNA. [DOI] [PubMed] [Google Scholar]
- 18.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, Ellenberg J, Jansen RP. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. This work demonstrated nucleo/cytoplasmic shuttling of She2p, dependent on its ability to bind to the mRNA. She2p binding to ASH1 mRNA in the nucleus implies that the cytoplasmic localization of ASH1mRNA is predetermined in the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 22.Munchow S, Sauter C, Jansen RP. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 1999;112:1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- 23.Peterson JN, Nassar R, Anderson PA, Alpert NR. Altered cross-bridge characteristics following haemodynamic overload in rabbit hearts expressing V3 myosin. J. Physiol. 2001;536:569–582. doi: 10.1111/j.1469-7793.2001.0569c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds ASH1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 27.Beach DL, Bloom K. ASH1 mRNA localization in three acts. Mol. Biol. Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berteaux-Lecellier V, Zickler D, Debuchy R, Panvier-Adoutte A, Thompson-Coffe C, Picard M. A homologue of the yeast SHE4 gene is essential for the transition between the syncytial and cellular stages during sexual reproduction of the fungus Podospora anserina. EMBO J. 1998;17:1248–1258. doi: 10.1093/emboj/17.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wesche S, Arnold M, Jansen RP. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr. Biol. 2003;13:715–724. doi: 10.1016/s0960-9822(03)00264-1. [DOI] [PubMed] [Google Scholar]
- 31.Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast,Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 33.Chang F, Peter M. Cell biology. Formins set the record straight. Science. 2002;297:531–532. doi: 10.1126/science.1074649. [DOI] [PubMed] [Google Scholar]
- 34.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 35.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 36.Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 38••.Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K, Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. This paper identifies three RNA binding proteins (Khd1p, Puf5p and Scp160p) that are specifically required for ASH1 mRNA bud tip anchoring, but not for transport. Whereas Puf5p and Scp160p function in ASH1 mRNA localization, Khd1p, interacting with the 5′ region of ASH1 mRNA, is a good candidate to participate in ASH1 mRNA translational regulation, as its overexpression reduces Ash1p levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 40.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 41.Tadauchi T, Matsumoto K, Herskowitz I, Irie K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey S, Pool M, Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 43.Li AM, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 45•.Shepard KA, Gerbert AP, Jambhekar A, Takizawa PA, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in S. cerevisiae: Identification of 20 new bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. Building on the analysis of Takizawa et al., 2000 [42], this study identifies 22 mRNAs transported to the daughter cell by She1–3p. Of particular interest is the finding that for most of these genes, mRNA localization by itself does not generate cellular asymmetry at the protein level; this opens the possibility that mRNA localization in yeast has additional or default functions. [DOI] [PMC free article] [PubMed] [Google Scholar]