Abstract
Marijuana has drawn significant public attention and concern both for its medicinal and recreational use. Δ9-Tetrahydrocannabinol (THC), which is the main bioactive component in marijuana, has also been shown to possess potent anti-inflammatory properties by virtue of its ability to activate cannabinoid receptor-2 (CB-2) expressed on immune cells. In this study, we used RNA-seq to quantify the transcriptomes and transcript variants that are differentially regulated by THC in super antigen-activated lymph node cells and CD4+ T cells. We found that the expressions of many transcripts were altered by THC in both total lymph node cells and CD4+ T cells. Furthermore, the abundance of many miRNA precursors and long non-coding RNAs was dramatically altered in THC-treated mice. For example, the expression of miR-17/92 cluster and miR-374b/421 cluster was down-regulated by THC. On the other hand miR-146a, which has been shown to induce apoptosis, was up-regulated by THC. Long non-coding RNAs that are expressed from the opposite strand of CD27 and Appbp2 were induced by THC. In addition, THC treatment also caused alternative promoter usage and splicing. The functions of those altered transcripts were mainly related to immune response and cell proliferation.
Keywords: cannabinoid, gene expression, immunosuppression, inflammation, T helper cells
Introduction
Marijuana (Cannabis sativa) is the most widely used drug of abuse in the United States. Nonetheless, it is worth noting that its medicinal value has been known for many centuries and is getting increasing recognition in recent times. Thus, Δ9-tetrahydrocannabinol (THC),2 the major psychoactive principle in marijuana, is used for medicinal purposes such as for pain relief as well as an anti-emetic and appetite stimulant during cancer chemotherapy and in HIV/AIDS patients (1). In addition, marijuana is widely known anecdotally for its use to relieve pain, and prevent seizures in patients with epilepsy. Because of the potential therapeutic value of marijuana in a variety of clinical disorders, 23 states and the District of Columbia currently have laws legalizing marijuana for medicinal use. Four states and the District of Columbia have also legalized marijuana for recreational use. Clearly, legalization of marijuana for medicinal or recreational purposes raises significant questions on its long term use/abuse and the impact on human health.
THC has also been shown to modulate immune response primarily through activation of cannabinoid receptors (CB2) expressed primarily on cells of the immune system (2–4). In general, most studies have shown that THC has immunosuppressive properties mediated through multiple pathways (2, 5, 6). For example, it has been shown that THC suppresses Th1, whereas promoting Th2 cells (7). THC has also been shown to induce apoptosis in immune cells (8, 9), and induce myeloid-derived suppressor cells and Tregs, which are known to inhibit T cell proliferation (10–12). Moreover, induction of myeloid-derived suppressor cell by THC is associated with altered microRNA expression (13). Our previous studies have shown that epigenetic modifications such as histone methylation and acetylation play important roles in THC-mediated gene expression in immune cells (7).
In this study, we used RNA-seq to examine global gene expression in antigen-activated lymphocytes. RNA-seq is a next generation sequencing-based RNA quantification method (14). In this case, a population of RNA is isolated and converted to a library containing short cDNA fragments. Fragments with adapters to one or both ends are then sequenced using high-throughput sequencing technology. The fragments are then aligned to the genome to calculate the abundance of the transcript. Although RNA-seq is still under active development, it has shown many advantages over microarray (14). RNA-seq has a very low background signal because fragments can be unambiguously mapped to regions of the genome and unmapped sequences are filtered out. Because it is sequencing based, RNA-seq does not have an up or low limit for quantification. Therefore, it has a high sensitivity for genes expressed at very low or very high levels. The single-base resolution of RNA-seq can also detect the transcript boundaries that may lead to the discovery of alternative transcription start sites and splicing isoforms. Furthermore, novel transcripts including non-coding RNAs can be identified by RNA-seq.
Staphylococcal enterotoxin B (SEB), is a superantigen produced by Staphylococcus aureus and is responsible for causing food poisoning(15) and toxic shock (16). SEB binds to the non-polymorphic regions of MHC II on antigen presenting cells and the specific Vβ regions of the T-cell receptor (TCR) such as murine Vβ8, thereby activating a large population (up to 30%) of T cells that proliferate rapidly and trigger cytokine storm (17). Recent studies from our laboratory demonstrated that THC can attenuate SEB-mediated inflammation and toxicity to the lungs (18–20). In this study we wanted to examine the effect of THC on global gene expression in activated lymphocytes. We used the same animal model and SEB was used as the reagent to induce inflammation. We then used RNA-seq to analyze the lymph node (LN) cells as well as purified CD4+ T cells from mice immunized with SEB and treated with vehicle or THC to analyze RNA abundance. We found that the expression of many transcripts was altered by THC in both total lymph node cells and CD4+ T cells. Functional analysis revealed that those altered genes were mainly related to immune response and cell proliferation. Furthermore, the abundance of many miRNA precursors and long noncoding RNA (lncRNA) was dramatically altered in THC-treated mice, suggesting that noncoding RNA expression might play an important role in THC-mediated immune regulation.
Results
THC-mediated Suppression of Lymphocyte Proliferation and Inflammation
SEB is a potent superantigen that activates ∼30% of T cells particularly those expressing Vβ8 T cell receptor. Previous studies from our laboratory demonstrated that THC can attenuate SEB-induced inflammation (7, 21). However, to ensure that this model was working in the immune cells under current investigation, we used SEB to induce inflammation and tested the effect of THC. To that end, we immunized mice with SEB into the footpads so that SEB-specific T cell response can be studied in the draining popliteal LN as described (22, 23). To make sure that the THC effects were dependent on CB2 receptors, another group of mice received both THC and CB2 antagonist, SR144528 (24). The data shown in Fig. 1 demonstrated that THC treatment in SEB-immunized mice caused a significant decrease in total draining LN cellularity, CD4+ T cells as well as Ifn-γ+ CD4+ T cells when compared with vehicle controls (supplemental Fig. S1). Furthermore, in mice that received THC + SR144528, the immunosuppressive effects of THC were reversed (supplemental Fig. S1), thereby indicating that the THC effects in lymphocytes were mediated through CB2. These results were consistent with our previous report that THC suppresses cell proliferation and induces a shift in T cells from Th1 to Th2 (7).
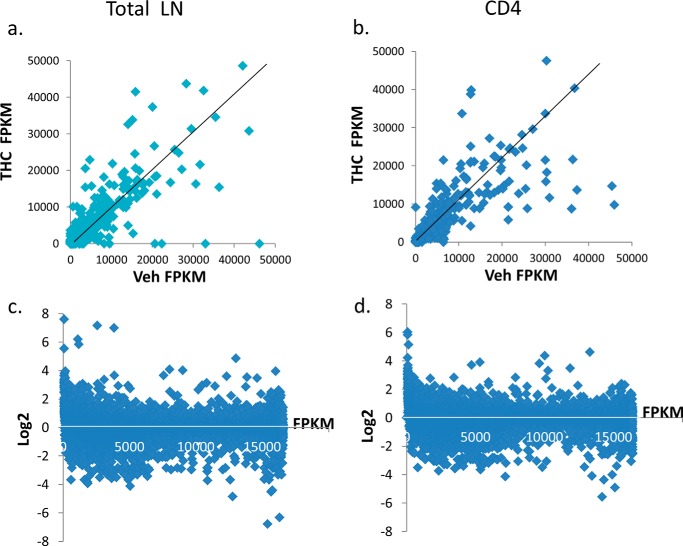
FIGURE 1.
Comparison of gene expression in vehicle- and THC-treated cells. C57BL/6J mice were treated with THC or vehicle as described under “Experimental Procedures.” Two hours after the second THC treatment, 10 μg of SEB was injected into each footpad. Three days after SEB challenge, draining popliteal lymph node cells (LN) of vehicle- or THC-treated mice (n = 5) were harvested. CD4+ cells were purified from the total LN cells. RNA was purified from those cells and the expression of each gene was quantified by RNA-seq. The expression level in total LN cells (a) and CD4+ cells (b) under the vehicle and THC treatment is presented as Fragments Per Kilobase per Million mapped reads (FPKM). The difference in gene expression is also presented as fold-change (c and d). The FPKM value of each gene in THC-treated LN cells (c) and CD4+ cells (d) was divided by its value in the vehicle-treated sample and expressed as log2.
THC-induced Differential Expression of Genes in Lymphocytes
To investigate the effect of THC treatment on SEB-induced gene expression, the abundance of each transcript was compared by RNA-seq between the SEB + vehicle- and SEB + THC-treated total lymph node cells as well as purified CD4+ cells. The rationale for using both purified CD4+ T cells and whole LN cells was as follows. 1) THC is well known to exert its effect on Th cell differentiation and therefore, studying CD4+ T cells would provide useful clues. 2) The cytokine storm induced by SEB can also activate other immune cells that can be studied using whole LN cells. 3) Our previous ChIP-seq study was performed in the total lymph node cells (7). Thus, comparing gene expression profile and histone modification profile would allow us to determine whether THC-induced alteration in gene expression is mediated by histone modification. We obtained 30 to 40 million reads in each sample, and 70–80% of reads were mapped to the mouse genome. In those mapped reads, 75–80% were uniquely mapped. Using Cufflinks and Cuffcompare, the expression level of each gene was calculated. Although most genes were expressed at a low level in both SEB + vehicle- and SEB + THC-treated samples, there were many genes that were differentially expressed under these two treatments (Fig. 1). In total LN cells, several genes were undetectable in SEB + THC-treated sample, whereas highly expressed in the SEB + vehicle-treated sample (Fig. 1a), thereby suggesting that THC was down-regulating the expression of genes induced by SEB. The differentially expressed genes were also presented as fold-changes (Fig. 1, c and d).
Genes that showed more than 2-fold difference and with an “OK” status as determined by Cuffcompare were further analyzed. Compared with SEB + THC group, 310 and 260 genes were highly expressed in the SEB + vehicle-treated total LN cells and CD4+ cells, respectively. Among those, 37 were common in total LN and CD4+ cells (Fig. 2a). On the other hand, the expression of 328 genes in the total LN cells and 333 genes in CD4+ cells were induced by THC + SEB treatment. Among them, 56 were common in two cell populations (Fig. 2b). A list of those differentially expressed genes is presented in supplemental Data S1–S3. Interestingly, among those differentially expressed genes, many of them were miRNA precursors and lncRNAs. There were 122 and 128 such transcripts that were highly expressed in the SEB + vehicle-treated total LN and CD4+ cells, respectively. On the other hand, there were 111 and 100 non-coding RNAs that were highly expressed in SEB + THC-treated LN and CD4+ cells, respectively. However, few of them were common in total LN and CD4+ cells (Fig. 2, c and d). The majority of these miRNAs had unknown functions (see supplemental Data S1–S3 for lists of these miRNAs).
FIGURE 2.
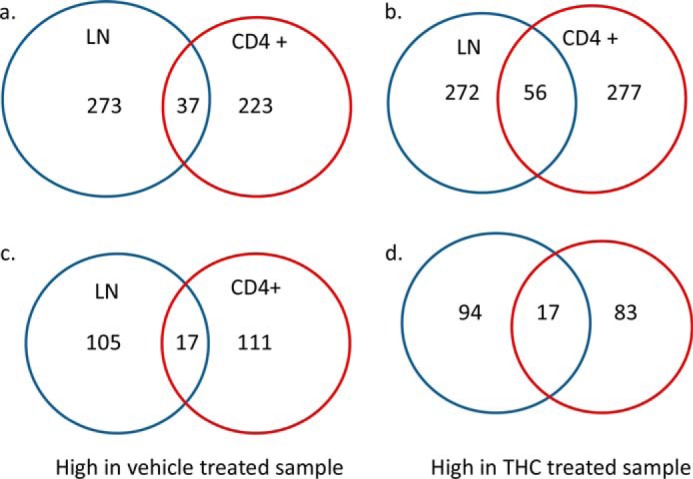
THC-induced differentially expressed genes. Mice were treated with SEB and vehicle or THC, and LN cells or CD4+ T cells were analyzed as described in the legend to Fig. 1. Genes with at least 2-fold differences in expression between the vehicle and THC-treated cells were identified. The Venn diagrams present the numbers of all genes (a), and miRNA precursors (c), whose expression was high in the vehicle-treated cells but suppressed by THC in the LN cells and CD4+ cells, and the numbers of all genes (b), and miRNA precursors (d), whose expression was high in THC-treated cells but low in vehicle-treated cells.
miRNA Precursors with Altered Expression after THC Treatment
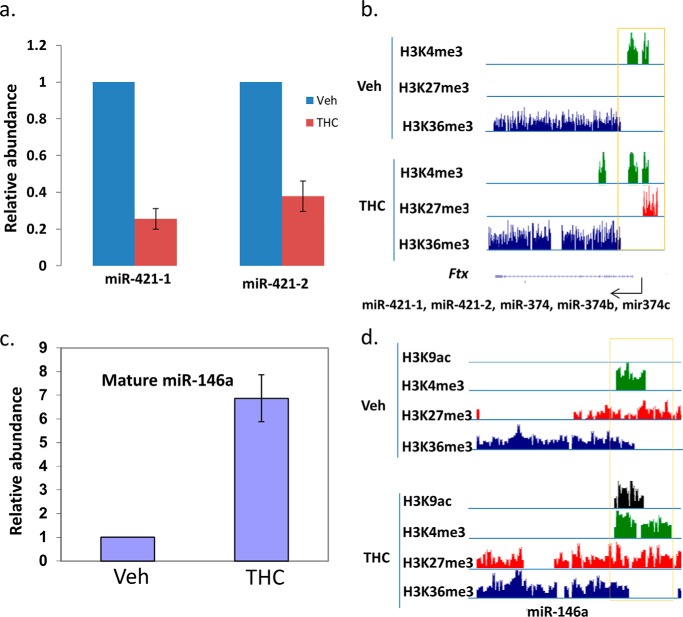
Because RNA-seq can only identify pri-miRNA, it was unclear whether the pri-miRNA level correlates with its mature miRNA expression. Suppression of miR-17/92 cluster by THC has been shown in another animal model (21). Indeed, the expression of precursor of this miRNA cluster was down-regulated by THC in this study. We confirmed the down-regulation of mature miRNAs of this cluster by real-time PCR (data not shown). This RNA-seq study also identified another miRNA cluster, miR-374b/421, whose precursor was also down-regulated by THC in total LN cells. pri-miR-421 can be processed into 2 mature miRNAs, miR-421-1 and miR-421-2. They are located at the 5′ and 3′ of pri-miR-421, respectively. Real-time PCR quantification of mature miR-421-1 and miR-421-2 confirmed that their expression levels were lower in THC-treated samples (Fig. 3a). In our previous ChIP-seq study, we have found that the promoter of certain miRNAs is differentially associated with histone marks in the THC-treated lymphocytes (7). In the promoter of miR-374b/421 cluster, histone H3 trimethylation at Lys-4 and Lys-36 (H3K4me3 and H3K36me3), which are associated with transcription activation, were present in both SEB + vehicle and SEB + THC samples. However, histone H3 trimethylaiton at Lys-27 (H3K27me3), which is associated with transcription suppression, was only present in the promoter area of this miRNA cluster in SEB + THC-treated sample, suggesting a possible histone methylation-mediated suppression (Fig. 3b). We also examined another miRNA, miR-146a. The expression of its precursor was induced in the LN cells by THC based on the RNA-seq data. Real-time PCR indeed showed that the mature miR-146a was significantly induced by THC (Fig. 3c). Again, we examined the association of histone marks under those two conditions. In this case, the association of H3K4me3, H3K4me27, and H3K36me3 did not differ significantly in the SEB + vehicle and SEB + THC-treated groups. However, histone H3 acetylation at Lys-9 (H3K9Ac) was only present in the promoter area of miR-146a in the THC-treated sample, suggesting histone acetylation might play a role in THC-induced expression of miR-146a (Fig. 3d).
FIGURE 3.
Examples of histone modification mediated miRNA expression. Mice were treated with SEB and vehicle or THC, and LN cells or CD4+ T cells were analyzed as described in the legend to Fig. 1. The expression of mature microRNA in vehicle-treated and THC-treated LN cells were quantified by real-time PCR (a and c). The amounts in the vehicle-treated cells were set as 1. The association of histone markers in their genes were identified by ChIP-seq and viewed in a genome browser (b and d). The area near the transcription start site is highlighted with a box.
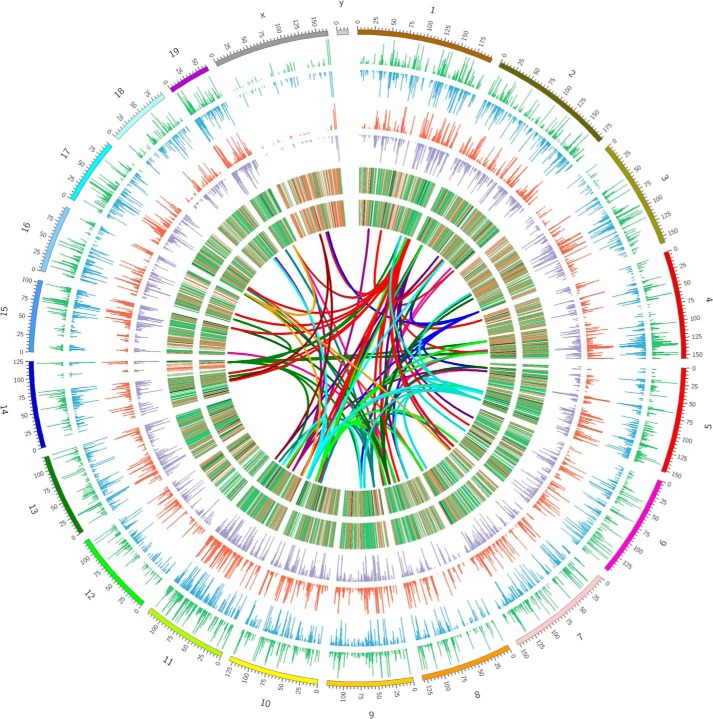
Fig. 4 is a Circos plot that shows genome-wide histone methylation, gene expression, and links between differentially expressed miRNAs and their target mRNAs. Two relatively well studied histone marks, H3K4me3 and H3K27me3, in total lymphocytes are presented as histograms. The differences in gene expression in both total lymphocytes and CD4+ cells are presented as heat maps. Genes in red and brown were those induced by THC and genes in blue and green are those suppressed by THC. Genes in yellow did not show significant difference. The links show the genomic positions of the differentially expressed miRNAs and their target mRNAs that were also differentially expressed. Cold colors (green, blue etc.) represent miRNAs that were down-regulated by THC, and hot colors (red, brown etc.) represent THC-induced miRNAs. A list of those miRNAs and their potential target mRNA genes are in supplemental Data S1–S3.
FIGURE 4.
Genome-wide histone modification and gene expression in THC-treated lymphocytes. Mice were treated with SEB and vehicle or THC, and LN cells or CD4+ T cells were analyzed as described in the legend to Fig. 1. Histone methylation marks H3K4me3 (blue and green) and H3K27me3 (red and purple) in lymphocytes are presented as histograms. The outward histograms are vehicle-treated samples and the inward ones are THC-treated samples. Differential gene expression in total lymphocytes (outer ring) and in CD4+ cells (inner ring) is presented as heat maps. The genomic positions of the differentially expressed miRNAs and their target mRNAs are shown as links. The graph was generated by Circos plot.
Pathway Analysis of Differentially Expressed Genes
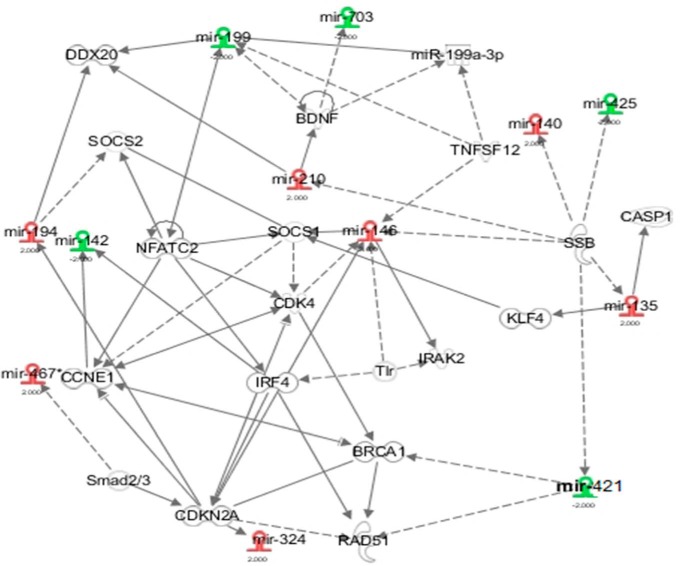
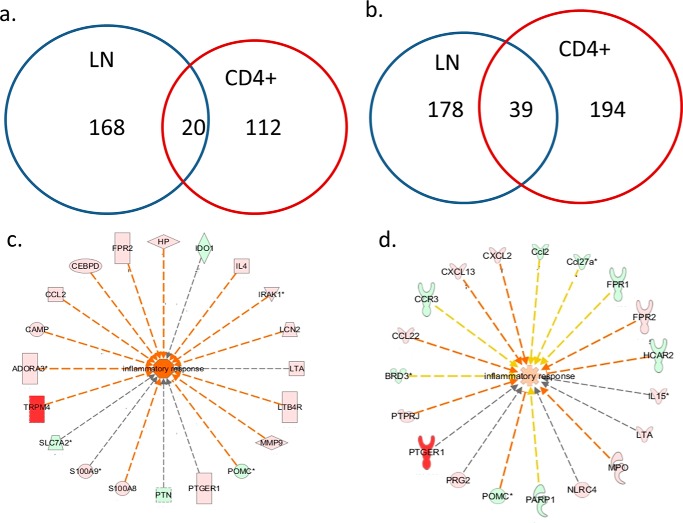
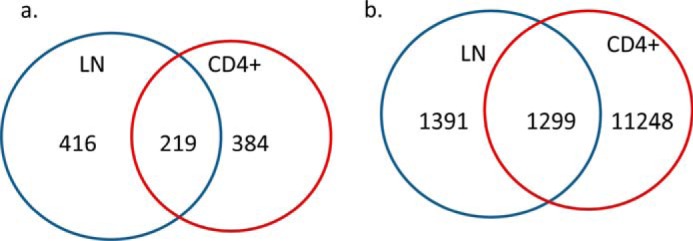
We further performed gene interaction analysis by Ingenuity pathway analysis on those differentially expressed genes. Most of those known miRNAs were involved in cell proliferation (Fig. 5). This was consistent with our previous study showing that THC promotes apoptosis in antigen-activated lymphocytes (7). For the protein-coding genes with more than 2-fold changes, THC decreased the expression of 188 genes in the total LN cells and 132 genes in CD4+ T cells. Among those, there were 20 genes that overlapped (Fig. 6a). On the other hand, 217 and 233 genes were up-regulated by THC in total LN and CD4+ cells, respectively, of which 39 genes overlapped (Fig. 6b). Functional analysis of those differentially expressed genes indicated that the major enriched pathway was inflammatory response (Fig. 6, c and d).
FIGURE 5.
Top network of THC-regulated miRNAs. Mice were treated with SEB and vehicle or THC, and LN cells or CD4+ T cells were analyzed as described in the legend to Fig. 1. Ingenuity pathway analysis indicated that cell death and survival was the most overrepresented network by miRNAs whose precursors were differentially expressed in the vehicle and THC-treated lymphocytes. THC up-regulated miRNAs are shown in red and THC down-regulated miRNAs are shown in green.
FIGURE 6.
Protein-coding genes that are differentially regulated by THC. Mice were treated with SEB and vehicle or THC, and LN cells or CD4+ T cells were analyzed as described in the legend to Fig. 1. Numbers of protein-coding genes whose expression was inhibited by THC (a) or induced by THC (b) in total LN cells and CD4+ cells are presented by Venn diagrams. Their differences under these two treatments were at least 2-fold. Functional analysis indicated that these differentially expressed genes, in both LN cells (c) and CD4+ cells (d), are involved in inflammatory response. THC-induced genes are in red, whereas THC suppressed genes are in green.
THC-induced Alteration in the Expression of Long Non-coding RNA
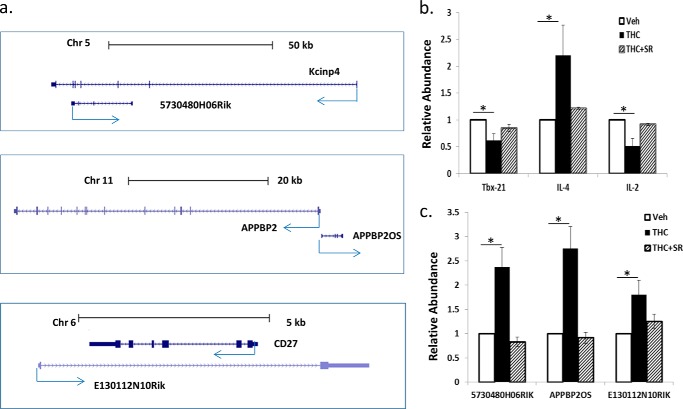
Besides alteration in miRNA and protein coding gene expression, THC also modulated the expression of several lncRNA (supplemental Data S1–S3). The functions of those lncRNAs are not yet clear. We selected 3 lncRNAs whose expression was induced by THC based on RNA-seq data. These lncRNAs are transcribed from the opposite strand of known genes, and may have a suppressive effect on those protein coding genes. 5730480H06Rik overlaps the 3′ end of Kv channel interacting protein 4 (KCNIP4). Appbp2OS overlaps the transcription start site of amyloid β precursor protein-binding protein 2. E130112N10Rik overlaps the whole transcript of Cd27 (Fig. 7a). To confirm that those non-coding RNAs were induced by THC through CB2 receptor, we examined their expression in an independent experiment by real-time PCR. First, we examined the marks of Th1 and Th2, which are known to be regulated by THC. Tbx-21, the transcription factor for Ifn-γ, and Il-2, the pro-inflammatory cytokine were suppressed, whereas Il-4, the anti-inflammatory cytokine was induced by THC (Fig. 7b). In the same set of samples, the expression of these non-coding RNAs was induced by THC, and the induction was reversed by SR144528 (Fig. 7c).
FIGURE 7.
Example of THC-induced lncRNA expression. Mice were treated as described in the legend to Fig. 1. The locations of 3 lncRNAs were obtained from UCSC genome browser (a). The expression of Th1 and Th2 marks (b) and lncRNAs (c) in vehicle-, THC-, or THC + SR-treated LN cells were quantified by real-time PCR. The amounts in the vehicle-treated cells were set as 1.
Transcripts Using Different Promoters or Having Different Splicing Forms after THC Treatment
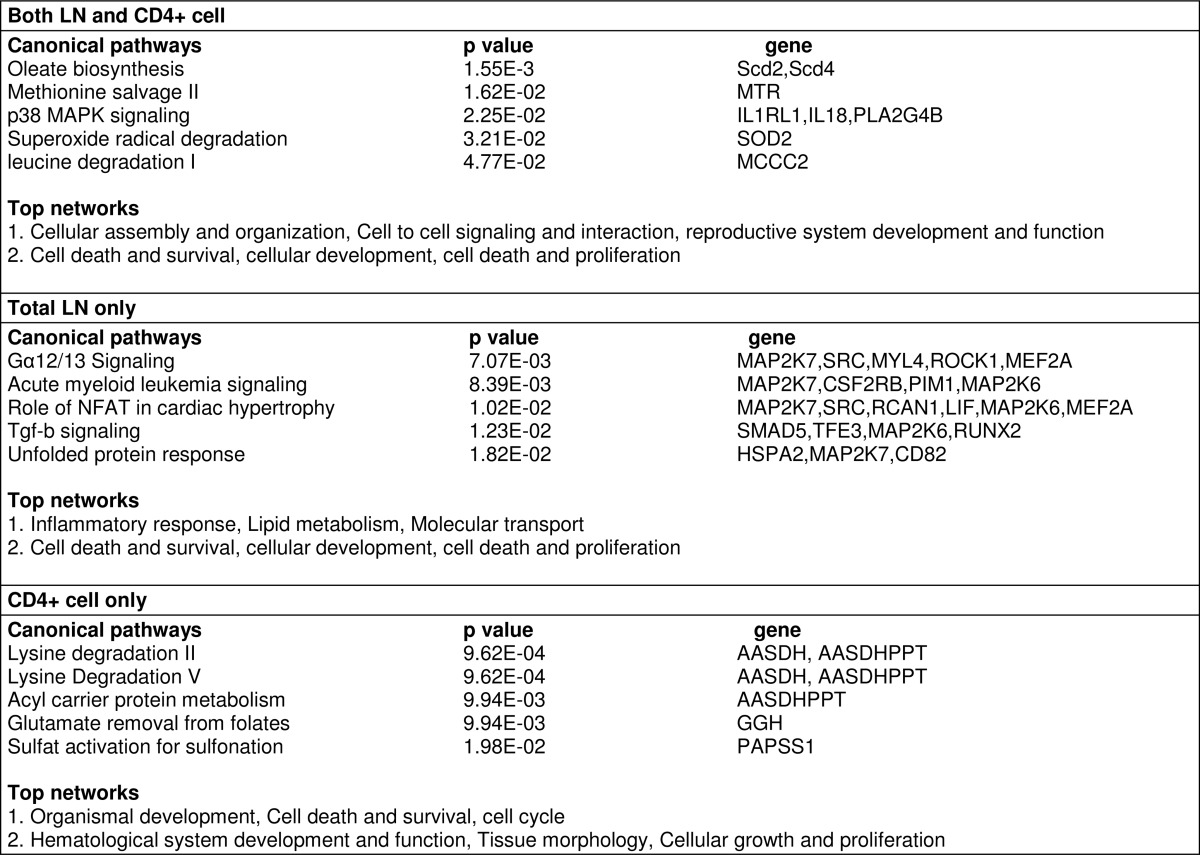
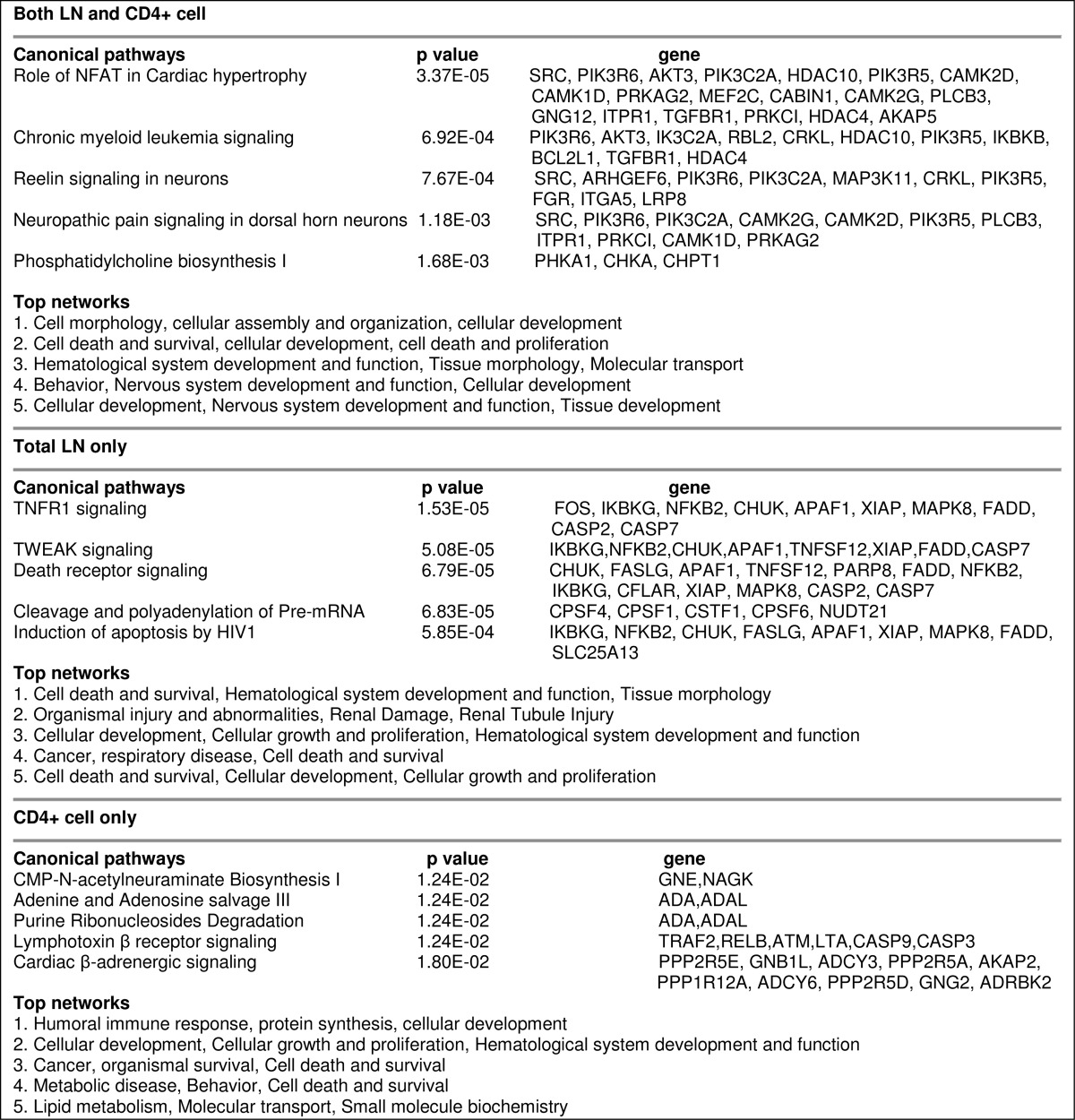
RNA-seq not only can identify differentially expressed genes but also transcripts with different promoter usage and splicing form. Approximately 600 transcripts in both total LN cells and CD4+ T cells used different promoters following SEB + vehicle and SEB + THC treatments. About half of those transcripts were common in both total LN and CD4+ T cells (Fig. 8a). As for alternative splicing, THC induced about 2500 different splicing and nearly half of them were common in both types of cells (Fig. 8b). Functional analysis of those transcripts with different promoters or splicing forms showed that those transcripts were involved in various pathways, whereas cell death and survival was the top network (Tables 1 and 2 and supplemental Data S1–S3).
FIGURE 8.
THC-induced alternative promoter usage and splicing. The transcripts that had alternative promoter usage (a), and splicing (b), after THC treatment were identified by Cuffcompare. The cutoff p value is 0.05. The overlap and unique transcripts in total LN and CD4+ cells are presented as Venn diagrams.
TABLE 1.
Functional analysis of genes with differential promoter usage
TABLE 2.
Functional analysis of genes with differential splicing
Discussion
Previous studies from our laboratory and elsewhere have shown that THC suppresses the expression of pro-inflammatory genes while inducing the expression of anti-inflammatory genes (6, 7, 25). To identify other genes whose expression might be regulated by THC, we used RNA-seq to compare the abundance in SEB-activated LN cells or CD4+ T cells, with and without THC treatment. Some genes such as Il-4 identified in this study have already been shown in our previous studies and those of others (7, 21). Interestingly, many genes such as Fpr2, which has been shown to have an anti-inflammatory role (26), but has never been shown previously to be induced by THC were also identified. However, the fact that only a small number of genes were either up-regulated or down-regulated by THC in total popliteal lymph node cells and CD4+ cells, suggested that THC may differentially regulate gene expression in different immune cells. This can be explained by the fact that although SEB activates primarily Vβ8+ T cells, the cytokine storm induced by this superantigen can also affect other immune cells. It is possible that THC may even have an opposite effect on the expression of certain genes in different subset of cells. It is, however, not clear whether this is the case. In the future, we will separate different types of cells from lymph nodes and compare their gene expression profiles. Despite few overlapping genes between total lymphocytes and CD4+ cells, most of those altered were related to cell survival and inflammatory response. This is consistent with our flow cytometry result using the same animal model in our previous study (7), which showed that THC promotes apoptosis in SEB-activated lymphocytes, and inhibits production of Th1 cytokines while inducing the production of Th2 cytokines (7).
A unique finding of this study is that the expression of many miRNA precursors was dramatically affected by THC. The majority of these miRNAs are newly identified with unknown functions. These data suggested that THC-mediated alteration in miRNA expression may play a critical role in cell proliferation and immune response. These novel miRNAs may have important roles in immune response. In the future, we will first validate the expression of these miRNAs in their mature miRNA form and then investigate their potential roles in the regulation of immune functions. Among those known miRNAs, the miR-17/92 cluster has been shown to be induced by SEB in an acute lung injury mouse model by regulating PIK3/Akt pathway (21). Because THC suppresses the expression of the miR-17/92 cluster in different tissues (21), it is likely that this effect is not cell type specific. This miRNA cluster has been shown to have many functions and THC may exert a broad spectrum of functions through this miRNA cluster. However, it is not clear how THC modulates the expression of miR-17/92 cluster. We examined the association of 5 histone modification marks (H3K4me3, H3K27me3, H3K36me3, H3K9me3, and H3K9ac) in its gene. No significant difference was found between the vehicle and THC-treated samples (data not shown), suggesting that these histone marks may not play a key role in THC-mediated modulation of the expression of miR-17/92 cluster.
The miRNA cluster 23, including miR-374b to miR-421, was also down-regulated by THC. In human cells, miR-421 suppresses the expression of Atm, a key kinase in the maintenance of genomic integrity, by targeting the 3′ UTR of the Atm transcript, and the expression of miR-421 is induced by oncogene N-Myc (27). This raises the possibility that THC may regulate Atm-dependent DNA damage response. miR-421 has been shown to be overexpressed in various types of cancer (28–31). A recent study suggested that miR-421 induces cell proliferation and apoptosis resistance by targeting Foxo4 (32). Down-regulation of miR-421 by THC is consistent with the notion that THC has a pro-apoptotic property. The miR-374b/421 cluster is located in the Ftx gene, which encodes a lncRNA. Another miRNA cluster, miR-545/374a, is also located in this lncRNA. This cluster is overexpressed in hepatitis B virus-induced hepatocellular carcinoma (33). It is unclear whether THC differentially regulates the expression of individual miRNAs in these clusters.
The expression of miR-146a is highly induced by THC at both the pri-miRNA level and mature miRNA level. This miRNA has been relatively well studied. Many studies indicated that miR-146a induces cell apoptosis and its abnormal expression has been found in various types of cancers (34–37). miR-146a also plays an important role in immune response. A study in miR-146a knock-out mice has shown that these mice produce an excessive amount of pro-inflammatory cytokines after bacterial challenge (38). A recent study showed that the expression of miR-146a and miR-146b is increased when monocytes differentiate into dendritic cells (39). Silencing of these two miRNAs enhances the expression of pro-inflammatory cytokines Il-12, Il-6, and Ifn-γ (39). In our model, THC-induced expression of miR-146a correlates with the decreased expression of these cytokines. We also find that several histone markers are associated with the miR-146a gene, suggesting that the expression of this miRNA may be regulated by the environmental stimuli through epigenetic mechanisms, which is supported by the presence of H3K9ac in the promoter area of its genes in THC-treated cells (Fig. 3d).
In this study, we also identified several THC-regulated lncRNAs. The functions of most lncRNAs are not known and they are involved in a variety of biological processes through different mechanisms. lncRNAs can serve as signals for transcription, guides for recruiting chromatin-modifying enzymes, or scaffolds for protein complex assembly (40). The biggest challenge of studying lncRNA functions is that it is very difficult to predict by which mechanism that a particular lncRNA may exert its regulatory function. A large proportion of lncRNAs is antisense to protein-coding transcripts. Antisense RNAs can suppress the expression of their sense counterparts through inhibiting transcription and translation (41). Here we show 3 examples of antisense lncRNA that overlap their sense protein-coding transcripts in different positions. Whether and how these lncRNAs suppress the expression of their counterparts needs further investigation. The purpose of this study is to screen for potentially interesting THC-regulated lncRNAs. KCNIP4 has been identified as an asthma gene (42). Marijuana has been linked to the cause as well as treatment for many respiratory diseases including asthma (9, 43).
Another interesting lncRNA is Appbp2os. Appbp2 is a microtubule-interacting protein and is involved in the transporting of amyloid precursor protein. It is not known whether Appbp2os regulates the expression or function of Appbp2 protein. However, THC can inhibit amyloid β-peptide aggregation (44). Cannabis-based medicine has been shown to reduce multiple pathological processes in an animal model of Alzheimer disease (45). Recently, CB2 receptor has been linked to amyloid-β processing and shows a minor therapeutic property in a mouse model (46). How THC affects the pathology of Alzheimer disease is worth further investigation.
THC-induced lncRNA E130112N10Rik is much longer than its counterpart sense transcript, Cd27, and covers the whole transcript of Cd27. CD27 is required for immune cells to maintain long-term antigen-specific T cell immunity (47). A study also showed that Tregs inhibit CD27-dependent Th1 priming (48). So far there are no reports regarding THC regulating CD27-mediated immune response. However, the role of THC in inhibiting Th1 is well documented. This lncRNA could add another possible mechanism of THC-induced immune suppression.
Pathway analysis of THC-induced differentially expressed genes, both protein-coding and nonprotein-coding genes, has indicated that THC has a pleotropic effect, from cell proliferation to metabolism. Therefore, it is necessary to investigate the role of THC in modulating various biological processes to weigh the benefits and risks of marijuana use. This study also shows that THC induces alternative promoter usage and splicing. More than half of human genes contain multiple promoters and have different splicing forms (49). Alternative transcription start sites can yield different pre-mRNAs. Alternative splicing of these pre-mRNAs leads to multiple transcript variants. As a result, one gene can produce various gene products under different conditions. Increasing evidence indicates that many diseases are associated with altered transcript isoforms. For example, it has been shown that using alternative transcription start and termination sites is a major source of transcriptome diversity in developing mouse cerebella (50). Moreover, that study indicates that H3K4me3 defines the use of alternative promoters that links a histone mark to promoter usage. The significance of THC-induced alternative promoter usage and splicing in immune cells is currently not clear.
In summary, our study has identified many protein coding and non-coding RNA transcripts whose expression is significantly altered by THC. This may provide information for future studies regarding the role of THC in various biological processes. It would also be interesting to investigate how histone modification, non-coding RNA, and alternative splicing coordinate each other for THC to exert its effects.
Experimental Procedures
Mice and Cell Isolation
Female C57BL/6J mice (6–7 weeks old) received intraperitoneal (i.p.) injection of THC (Sigma, 20 μg/kg of body weight) or the same amount of vehicle as described previously (51). The mice received the same treatment 24 h later. Two hours after the second treatment, 10 μg of SEB was injected in each hind footpads. To study the effect of CB2 antagonist, mice received SR144528 (Santa Cruz Biotechnology, 10 μg/kg of body weight) by i.p. route, at the same time as THC. Popliteal draining lymph nodes from the vehicle and THC-treated mice were collected 72 h after SEB injection and single cells were prepared using micro-Biomaster (Seward Lab Systems, West Sussex, UK). Samples from 4–7 mice in each group were pooled. Cells were filtered through a 70-μm mesh and counted. CD4+ cells were isolated using EasySep PE selection kit (StemCell Technologies, Vancouver, BC) according to the provided instruction. The antibody used was PE-anti-mouse CD4 (BioLegend, San Diego, CA).
Staining and FACS Analysis
For CD3 and CD4 staining, lymph node cells were stained with PE/Cy7-CD3 and PE-CD4 immediately after harvest. For intracellular staining, lymph node cells were cultured in complete RPMI in the presence of 1 nm PMA (Sigma), 1 μm calcium ionophore (Sigma), and 2 μm protein transport inhibitor monensin (Biolegend, San Diego, CA) for 4 h. Cells were washed and resuspended in FACS buffer (PBS containing 2% FBS and 0.1% sodium azide). Fc receptors were blocked by adding anti-mouse CD16/CD32 (10 μg/ml) followed by surface staining for PE-CD4 and intracellular staining with FITC-Ifn-γ. All antibodies were purchased from BD Biosciences. Cells were analyzed in a BC FC 500 flow cytometer.
RNA-seq
RNA-seq libraries were constructed using Illumina TruSeq RNA Sample Preparation kit. Briefly, total RNA was purified using Qiagen RNA easy kit. The oligo(dT) beads were added to 1 μg of total RNA and the purified mRNA was fragmented. The RNA fragments were then reverse transcribed into double-stranded cDNA fragments. The DNA fragments were repaired to generate blunt ends using T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. After DNA fragments were purified using a Qiagen PCR purification kit (Qiagen catalogue number 28004), an “A” base was added to the 3′ end of the blunt DNA fragment by Klenow fragment. Sequencing adapters were ligated to the ends of DNA fragments using DNA ligase. The libraries were then amplified by limited PCR (15 cycles) using primers provided by the kit. The PCR products were then separated in 2% agarose gel and fragments with size range from 250 to 400 bp were excised and purified using QIAquick Gel Extraction Kit (Qiagen number 28704). The concentration and distribution of the library were determined by a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). The library was sequenced by illumina HiSeq 2000 at Tufts University Genomic core facility. Raw sequencing reads (50 bp single-end) were mapped to mouse genome build mm9 using Tophat 2 (52). The accepted hits were used for assembling transcripts and estimating their abundances using Cufflinks (53). The differentially expressed gene, promoter usage, and splicing form were determined by Cuffdiff and Cuffcompare (53).
Quantitative Real-time PCR
Mature miRNA was quantified by real-time PCR using miScript SYBR Green PCR kit and miScript Primer Assays (Qiagen). Quantitative real-time PCR was performed according to the protocol provided by the manufacturer. Snord96a and Gapdh were used as the internal controls for miRNAs and mRNA, respectively. The amount in the vehicle-treated sample was set as 1.
Functional Enrichment
Ingenuity pathway analysis (Qiagen) was used for functional analysis. Ingenuity pathway analysis has tools to identify the canonical pathways and top networks in which a given set of genes are involved. We performed functional enrichment analysis with the transcriptome data by taking all the transcripts with a fold-change of 2 or more. Tables 1 and 2 were generated by such analysis. The software can also identify the interactions between miRNAs and target genes. Genes and the miRNAs that had more than 2-fold changes were analyzed together to see whether any of these genes are the targets of the dysregulated miRNAs. These interactions were presented as links in the Circos plot (54). The histogram of histone methylation marks and the heat map of gene expression were also generated by the Circos software.
Author Contributions
X. Y. and M. B. designed the study, performed the experiments, and wrote the paper. P. S. N. and M. N. conceived and coordinated the study, interpreted the results, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P01AT003961, P20GM103641, R01AT006888, R01ES019313, and R01MH094755. The authors declare that they have no conflict of interest with the contents of this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. S1 and Data S1–S3.
- THC
- Δ9-tetrahydrocannabinol
- SEB
- Staphylococcal enterotoxin B
- lncRNA
- long noncoding RNA
- LN
- lymph nodes.
References
- 1. Hill K. P. (2015) Medical marijuana for treatment of chronic pain and other medical and psychiatric problems: a clinical review. JAMA 313, 2474–2483 [DOI] [PubMed] [Google Scholar]
- 2. Do Y., McKallip R. J., Nagarkatti M., and Nagarkatti P. S. (2004) Activation through cannabinoid receptors 1 and 2 on dendritic cells triggers NF-κB-dependent apoptosis: novel role for endogenous and exogenous cannabinoids in immunoregulation. J. Immunol. 173, 2373–2382 [DOI] [PubMed] [Google Scholar]
- 3. Rao G. K., Zhang W., and Kaminski N. E. (2004) Cannabinoid receptor-mediated regulation of intracellular calcium by Δ9-tetrahydrocannabinol in resting T cells. J. Leukoc. Biol. 75, 884–892 [DOI] [PubMed] [Google Scholar]
- 4. Newton C. A., Klein T. W., and Friedman H. (1994) Secondary immunity to Legionella pneumophila and Th1 activity are suppressed by Δ9-tetrahydrocannabinol injection. Infect. Immun. 62, 4015–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sido J. M., Nagarkatti P. S., and Nagarkatti M. (2015) Δ9-Tetrahydrocannabinol attenuates allogeneic host-versus-graft response and delays skin graft rejection through activation of cannabinoid receptor 1 and induction of myeloid-derived suppressor cells. J. Leukoc. Biol. 98, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagarkatti P., Pandey R., Rieder S. A., Hegde V. L., and Nagarkatti M. (2009) Cannabinoids as novel anti-inflammatory drugs. Future Med. Chem. 1, 1333–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang X., Hegde V. L., Rao R., Zhang J., Nagarkatti P. S., and Nagarkatti M. (2014) Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J. Biol. Chem. 289, 18707–18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McKallip R. J., Nagarkatti M., and Nagarkatti P. S. (2005) Δ-9-Tetrahydrocannabinol enhances breast cancer growth and metastasis by suppression of the antitumor immune response. J. Immunol. 174, 3281–3289 [DOI] [PubMed] [Google Scholar]
- 9. Rieder S. A., Chauhan A., Singh U., Nagarkatti M., and Nagarkatti P. (2010) Cannabinoid-induced apoptosis in immune cells as a pathway to immunosuppression. Immunobiology 215, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hegde V. L., Nagarkatti M., and Nagarkatti P. S. (2010) Cannabinoid receptor activation leads to massive mobilization of myeloid-derived suppressor cells with potent immunosuppressive properties. Eur. J. Immunol. 40, 3358–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kusmartsev S. A., Li Y., and Chen S. H. (2000) Gr-1+ myeloid cells derived from tumor-bearing mice inhibit primary T cell activation induced through CD3/CD28 costimulation. J. Immunol. 165, 779–785 [DOI] [PubMed] [Google Scholar]
- 12. Bronte V., Apolloni E., Cabrelle A., Ronca R., Serafini P., Zamboni P., Restifo N. P., and Zanovello P. (2000) Identification of a CD11b+/Gr-1+/CD31+ myeloid progenitor capable of activating or suppressing CD8+ T cells. Blood 96, 3838–3846 [PMC free article] [PubMed] [Google Scholar]
- 13. Hegde V. L., Tomar S., Jackson A., Rao R., Yang X., Singh U., Singh U. P., Nagarkatti P. S., and Nagarkatti M. (2013) Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Δ9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer binding protein α by microRNA-690. J. Biol. Chem. 288, 36810–36826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z., Gerstein M., and Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Argudin M. Á., Mendoza M. C., and Rodicio M. R. (2010) Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kulhankova K., King J., and Salgado-Pabón W. (2014) Staphylococcal toxic shock syndrome: superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol. Res. 59, 182–187 [DOI] [PubMed] [Google Scholar]
- 17. Rieder S. A., Nagarkatti P., and Nagarkatti M. (2011) CD1d-independent activation of invariant natural killer T cells by staphylococcal enterotoxin B through major histocompatibility complex class II/T cell receptor interaction results in acute lung injury. Infect. Immun. 79, 3141–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rao R., Nagarkatti P., and Nagarkatti M. (2015) Role of miRNA in the regulation of inflammatory genes in staphylococcal enterotoxin B-induced acute inflammatory lung injury and mortality. Toxicol. Sci. 144, 284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rieder S. A., Nagarkatti P., and Nagarkatti M. (2012) Multiple anti-inflammatory pathways triggered by resveratrol lead to amelioration of staphylococcal enterotoxin B-induced lung injury. Br. J. Pharmacol. 167, 1244–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saeed A. I., Rieder S. A., Price R. L., Barker J., Nagarkatti P., and Nagarkatti M. (2012) Acute lung injury induced by Staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc. Microanal. 18, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao R., Nagarkatti P. S., and Nagarkatti M. (2015) Δ9-Tetrahydrocannabinol attenuates Staphylococcal enterotoxin B-induced inflammatory lung injury and prevents mortality in mice by modulation of miR-17–92 cluster and induction of T-regulatory cells. Br. J. Pharmacol. 172, 1792–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Busbee P. B., Nagarkatti M., and Nagarkatti P. S. (2014) Natural indoles, indole-3-carbinol and 3,3′-diindolymethane, inhibit T cell activation by Staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol. Appl. Pharmacol. 274, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camacho I. A., Nagarkatti M., and Nagarkatti P. S. (2002) 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces Fas-dependent activation-induced cell death in superantigen-primed T cells. Arch. Toxicol. 76, 570–580 [DOI] [PubMed] [Google Scholar]
- 24. Hegde V. L., Hegde S., Cravatt B. F., Hofseth L. J., Nagarkatti M., and Nagarkatti P. S. (2008) Attenuation of experimental autoimmune hepatitis by exogenous and endogenous cannabinoids: involvement of regulatory T cells. Mol. Pharmacol. 74, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaffal E., Cron M., Glodde N., and Tüting T. (2013) Anti-inflammatory activity of topical THC in DNFB-mediated mouse allergic contact dermatitis independent of CB1 and CB2 receptors. Allergy 68, 994–1000 [DOI] [PubMed] [Google Scholar]
- 26. Dufton N., Hannon R., Brancaleone V., Dalli J., Patel H. B., Gray M., D'Acquisto F., Buckingham J. C., Perretti M., and Flower R. J. (2010) Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J. Immunol. 184, 2611–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu H., Du L., Nagabayashi G., Seeger R. C., and Gatti R. A. (2010) ATM is down-regulated by N-Myc-regulated microRNA-421. Proc. Natl. Acad. Sci. U.S.A. 107, 1506–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Z., Guo J., Xiao B., Miao Y., Huang R., Li D., and Zhang Y. (2010) Increased expression of miR-421 in human gastric carcinoma and its clinical association. J. Gastroenterol. 45, 17–23 [DOI] [PubMed] [Google Scholar]
- 29. Hao J., Zhang S., Zhou Y., Liu C., Hu X., and Shao C. (2011) MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem. Biophys. Res. Commun. 406, 552–557 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Gong W., Dai S., Huang G., Shen X., Gao M., Xu Z., Zeng Y., and He F. (2012) Down-regulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Mol. Cancer Res. 10, 516–522 [DOI] [PubMed] [Google Scholar]
- 31. Zhong X. Y., Yu J. H., Zhang W. G., Wang Z. D., Dong Q., Tai S., Cui Y. F., and Li H. (2012) MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene 493, 44–51 [DOI] [PubMed] [Google Scholar]
- 32. Chen L., Tang Y., Wang J., Yan Z., and Xu R. (2013) miR-421 induces cell proliferation and apoptosis resistance in human nasopharyngeal carcinoma via down-regulation of FOXO4. Biochem. Biophys. Res. Commun. 435, 745–750 [DOI] [PubMed] [Google Scholar]
- 33. Zhao Q., Li T., Qi J., Liu J., and Qin C. (2014) The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS ONE 9, e109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumaraswamy E., Wendt K. L., Augustine L. A., Stecklein S. R., Sibala E. C., Li D., Gunewardena S., and Jensen R. A. (2015) BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene 34, 4333–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun Q., Zhao X., Liu X., Wang Y., Huang J., Jiang B., Chen Q., and Yu J. (2014) miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate 74, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 36. Sandhu R., Rein J., D'Arcy M., Herschkowitz J. I., Hoadley K. A., and Troester M. A. (2014) Overexpression of miR-146a in basal-like breast cancer cells confers enhanced tumorigenic potential in association with altered p53 status. Carcinogenesis 35, 2567–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao J. L., and Starczynowski D. T. (2014) Role of microRNA-146a in normal and malignant hematopoietic stem cell function. Front. Genet. 5, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boldin M. P., Taganov K. D., Rao D. S., Yang L., Zhao J. L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., Sun G., Tay J., Linsley P. S., and Baltimore D. (2011) miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park H., Huang X., Lu C., Cairo M. S., and Zhou X. (2015) miR-146a and miR-146b regulate human dendritic cell apoptosis and cytokine production by targeting of TRAF6 and IRAK1. J. Biol. Chem. 290, 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang K. C., and Chang H. Y. (2011) Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris K. V., and Vogt P. K. (2010) Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle 9, 2544–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Himes B. E., Sheppard K., Berndt A., Leme A. S., Myers R. A., Gignoux C. R., Levin A. M., Gauderman W. J., Yang J. J., Mathias R. A., Romieu I., Torgerson D. G., Roth L. A., Huntsman S., Eng C., et al. (2013) Integration of mouse and human genome-wide association data identifies KCNIP4 as an asthma gene. PLoS ONE 8, e56179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ocampo T. L., and Rans T. S. (2015) Cannabis sativa: the unconventional “weed” allergen. Ann. Allergy Asthma Immunol. 114, 187–192 [DOI] [PubMed] [Google Scholar]
- 44. Eubanks L. M., Rogers C. J., Beuscher A. E. 4th, Koob G. F., Olson A. J., Dickerson T. J., and Janda K. D. (2006) A molecular link between the active component of marijuana and Alzheimer's disease pathology. Mol. Pharm. 3, 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aso E., Sánchez-Pla A., Vegas-Lozano E., Maldonado R., and Ferrer I. (2015) Cannabis-based medicine reduces multiple pathological processes in AbetaPP/PS1 mice. J. Alzheimers Dis. 43, 977–991 [DOI] [PubMed] [Google Scholar]
- 46. Aso E., Andrés-Benito P., Carmona M., Maldonado R., and Ferrer I. (2016) Cannabinoid receptor 2 participates in amyloid-β processing in a mouse model of Alzheimer's disease but plays a minor role in the therapeutic properties of a cannabis-based medicine. J. Alzheimers Dis. 51, 489–500 [DOI] [PubMed] [Google Scholar]
- 47. Hendriks J., Gravestein L. A., Tesselaar K., van Lier R. A., Schumacher T. N., and Borst J. (2000) CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 1, 433–440 [DOI] [PubMed] [Google Scholar]
- 48. Dhainaut M., Coquerelle C., Uzureau S., Denoeud J., Acolty V., Oldenhove G., Galuppo A., Sparwasser T., Thielemans K., Pays E., Yagita H., Borst J., and Moser M. (2015) Thymus-derived regulatory T cells restrain pro-inflammatory Th1 responses by down-regulating CD70 on dendritic cells. EMBO J. 34, 1336–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kornblihtt A. R. (2005) Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 17, 262–268 [DOI] [PubMed] [Google Scholar]
- 50. Pal S., Gupta R., Kim H., Wickramasinghe P., Baubet V., Showe L. C., Dahmane N., and Davuluri R. V. (2011) Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21, 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandey R., Hegde V. L., Nagarkatti M., and Nagarkatti P. S. (2011) Targeting cannabinoid receptors as a novel approach in the treatment of graft-versus-host disease: evidence from an experimental murine model. J. Pharmacol. Exp. Ther. 338, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., and Marra M. A. (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.