Abstract
Extremophiles are expected to represent a source of enzymes having unique functional properties. The hypothetical protein NIS_0185, termed NitAly in this study, was identified as an alginate lyase-homolog protein in the genomic database of ϵ-Proteobacteria Nitratiruptor sp. SB155-2, which was isolated from deep-sea hydrothermal vents at a water depth of 1,000 m. Among the characterized alginate lyases in the polysaccharide lyase family 7 (PL-7), the amino acid sequence of NitAly showed the highest identity (39%) with that of red alga Pyropia yezoensis alginate lyase PyAly. Recombinant NitAly (rNitAly) was successfully expressed in Escherichia coli. Purified rNitAly degraded alginate in an endolytic manner. Among alginate block types, polyM was preferable to polyG and polyMG as a substrate, and its end degradation products were mainly tri-, tetra-, and penta-saccharides. The optimum temperature and pH values were 70 °C and around 6, respectively. A high concentration of NaCl (0.8–1.4 m) was required for maximum activity. In addition, a 50% loss of activity was observed after incubation at 67 °C for 30 min. Heat stability was decreased in the presence of 5 mm DTT, and Cys-80 and Cys-232 were identified as the residues responsible for heat stability but not lyase activity. Introducing two cysteines into PyAly based on homology modeling using Pseudomonas aeruginosa alginate lyase PA1167 as the template enhanced its heat stability. Thus, NitAly is a functional alginate lyase, with its unique optimum conditions adapted to its environment. These insights into the heat stability of NitAly could be applied to improve that of other PL-7 alginate lyases.
Keywords: algae, alginate lyase, bacteria, carbohydrate, carbohydrate processing, alginate, brown algae, extremophile, heat stability
Introduction
Alginate consists of two uronic acids, β-d-mannuronic acid (M) and its C5 epimer α-l-guluronic acid (G) (1, 2). These units are linearly linked by a 1–4-glycosidic bond and are arranged in an M-consecutive sequence (M-block), G-consecutive sequence (G-block), and M/G random sequence (MG-block). Only a limited number of organisms produce alginate in nature. As a typical example, brown algae biosynthesize alginate as a cell wall component (3), although some types of mucoid bacteria, such as Pseudomonas spp. and Azotobacter vinelandii, produce alginate as an exopolysaccharide (4).
Alginate lyase enzymes catalyze the cleavage of the 1–4-glycosidic bond of alginate via a β-elimination mechanism (5). These enzymes are found in organisms that have either alginate-metabolizing systems or alginate biosynthesis systems. In alginate metabolization, endo-type alginate lyase(s) attack alginates and degrade them to oligosaccharides. Then, exo-type alginate lyase(s) release monosaccharides, and 4-deoxy-l-erythro-5-hexoseulose uronic acid is generated by non-enzymatic conversion. 4-Deoxy-l-erythro-5-hexoseulose uronic acid is reduced to 2-keto-3-deoxygluconate by a 4-deoxy-l-erythro-5-hexoseulose uronic acid-specific reductase (6–8), with 2-keto-3-deoxygluconate possibly being metabolized by the Entner-Doudoroff pathway (9, 10). The ability to degrade alginate is required to synthesize the alginate polymer because Pseudomonas aeruginosa does not produce alginate when inactive alginate lyase is expressed (11). In addition, alginate-producing bacteria use this ability to escape from biofilms containing alginate (12). Although a similar alginate biosynthesis process may be present in brown algae, candidate genes for alginate lyases have not been found, even following the completion of genomic analysis (13).
To date, dozens of alginate lyases have been isolated and characterized. According to the CAZy database (14), alginate lyases are structurally classified into seven polysaccharide lyase families, PL-5, -6, -7, -14, -15, -17, and -18. At present, PL-7 alginate lyases are the best characterized of all classified alginate lyases and are well documented. In alginate-producing bacteria, the alginate lyases PA1167 from P. aeruginosa (15) and AlyA1, AlyA2, and AlyA3 from A. vinelandii (16) have been characterized. Many PL-7 alginate lyases have also been identified in bacteria, i.e. Flavobacterium sp. (17, 18), Sphingomonas sp. (19–21), Vibrio sp. (22–26), Streptomyces sp. (27–29), Corynebacterium sp. (30), Agarivorans sp. (31, 32), Pseudoalteromonas sp. (33), Photobacterium sp. (34), Klebsiella pneumoniae (35), Zobellia galactanivorans (36), Saccharophagus degradans (37), and an uncultured bacterium (38). Within the last year, a novel alginate lyase, PyAly belonging to the PL-7 family, was identified from the red alga Pyropia yezoensis (39). This discovery was unexpected because P. yezoensis is not considered to have any alginate utilization systems. Although its physiological role remains obscure, PyAly showed alginate degradation activity in an endolytic manner, and its gene was confirmed to be derived from eukaryotic cells, not from contaminating prokaryotic bacteria. Moreover, we noticed that the amino acid sequence of PyAly showed a significant degree of identity with that of the hypothetical protein NIS_0185 (GenBankTM accession no. YP_001355656) from Nitratiruptor sp. SB155-2, a bacterium of the ϵ-Proteobacteria phylum that was isolated from deep-sea hydrothermal vents at a water depth of 1,000 m (40, 41). In such extreme environments, alginate producers, such as brown algae, are not present, and alginate-utilizing organisms have yet to be found. This finding of amino acid sequence similarity led us to question whether the product of NIS_0185 could have alginate degradation activity.
In this study, the enzymatic properties of the Nitratiruptor sp. SB155-2 hypothetical protein NIS_0185, which was termed NitAly, were investigated using recombinant NitAly protein (rNitAly).2 Our results are expected to reveal the residues responsible for the heat stability of NitAly, which could be used to improve the heat tolerance of PyAly.
Results
Cloning and Sequencing of NitAly
NitAly was identified by the analysis of the genomic sequence of Nitratiruptor sp. SB155-2, as a protein homolog of the red alga alginate lyase PyAly. This gene was annotated as a hypothetical protein, NIS_0185, in the genomic sequence of Nitratiruptor sp. SB155-2 from the NCBI Genome database. The gene was amplified with a set of specific primers (Table 1) using DNA isolated from Nitratiruptor sp. SB155-2 as the template. The amplified product was estimated to have a length of about 700 bp, based on agarose gel electrophoresis results, and was sequenced. The nucleotide sequence was completely identical to that of NIS_0185, with 243 amino acids being deduced. Twenty two residues at the N terminus were predicted to comprise the secretion signal using the SignalP 3.0 software program (42), and the mature protein was considered to consist of 221 residues with a total molecular mass of 26,692 Da.
TABLE 1.
Primers used in this study
Introduced restriction sites are underlined. Mutated sequences are double underlined. F indicates forward; R indicates reverse.
| Primer | Sequence |
|---|---|
| For nitaly cloning | |
| Nit-F | 5′-ATGCACCAACTAAAAGTTTTG-3′ |
| Nit-R | 5′-CTATTTCATTGTTAATGAATC-3′ |
| For rNitAly expression | |
| Nit-BamF | 5′-GCGGATCCCACGATGCGCCCTACGCTAT-3′ |
| Nit-XbaR | 5′-CGTCTAGACTATTTCATTGTTAATGAAT-3′ |
| For rNitAlyC80A, rNitAlyC232A, or rNitAlyC80A/C232A expression | |
| Nit-C80A-F | 5′-ACCTTTTTCATGGCCGGGAAAAAACAT-3′ |
| Nit-C80A-R | 5′-ATGTTTTTTCCCGGCCATGAAAAAGGT-3′ |
| Nit-C232A-F | 5′-CAAGGAGACGGAGCCGCAAAAGTTTT-3′ |
| Nit-C232A-R | 5′-AAAACTTTTGCGGCTCCGTCTCCTTG-3′ |
| For rPyAlyG79C, rPyAlyD230C, or rPyAlyG79C/D230C expression | |
| Py-G79C-F | 5′-GTGTTTGTCATGTGTGGCGACTCAC-3′ |
| Py-G79C-R | 5′-GTGAGTCGCCACACATGACAAACAC-3′ |
| Py-D230C-F | 5′-GAGGGCAGCCCCTGCGCGAGGGTAGTG-3′ |
| Py-D230C-R | 5′-CACTACCCTCGCGCAGGGGCTGCCCTC-3′ |
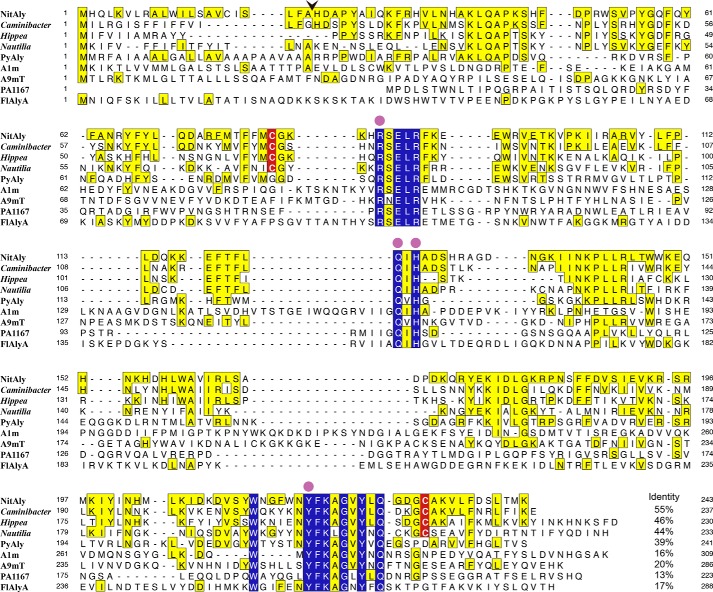
A BLAST search showed that the deduced amino acid sequence of NitAly shared a significant degree of identity with Caminibacter mediatlanticus hypothetical protein (55%), Hippea jasoniae hypothetical protein (46%), and Nautilia profundicola predicted alginate lyase (44%) (Fig. 1). Interestingly, these bacteria were also isolated at deep-sea hydrothermal vents (43–45). Among the characterized PL-7 alginate lyases, the amino acid sequence of NitAly showed the highest identity with that of PyAly (39%) (Fig. 1). Lower identities were detected for alginate lyases from deep-sea sediments, i.e. Agarivorans sp. JAM-A1m alginate lyase A1m (16%) (31) and Vibrio sp. alginate lyase A9mT (20%) (25). Other characterized PL-7 alginate lyases were found to have a relatively low degree of identity with NitAly, e.g. 17% for FlAlyA from Flavobacterium sp. UMI-01 and 13% for PA1167 from P. aeruginosa. Among the listed sequences in Fig. 1, 13 residues were fully conserved, namely Arg-85, Glu-87, Leu-88, Arg-89, Gln-123, His-125, Trp-214, Tyr-220, Phe-221, Lys-222, Gly-224, Tyr-226, and Gln-228 in NitAly. Of these, Arg-85, Gln-123, His-125, and Tyr-226 corresponded to the catalytic residues proposed to exist in some PL-7 alginate lyases, due to their location at an active cleft based on the resolved crystal structures of PL-7 alginate lyases (15, 21).
FIGURE 1.
Comparison among the amino acid sequences of NitAly, its homologous proteins, and several characterized family PL-7 alginate lyases. NitAly, Nitratiruptor sp. SB155-2 hypothetical protein NIS_0185 (GenBankTM accession no. WP_012081562); Caminibacter, C. mediatlanticus hypothetical protein (GenBankTM accession no. WP_007473671); Hippea, H. jasoniae hypothetical protein (GenBankTM accession no. WP_051904545); Nautilia, N. profundicola predicted alginate lyase (GenBankTM accession no. WP_015901805); PyAly, P. yezoensis alginate lyase PyAly (GenBankTM accession no. BAI66416) (39); PA1167, P. aeruginosa PAO1 alginate lyase PA1167 (GenBankTM accession no. AAG04556) (15); A1m, Agarivorans sp. JAM-A1m alginate lyase A1m (GenBankTM accession no. AB426616) (31); A9mT, Vibrio sp. alginate lyase A9mT (GenBankTM accession no. AB473598) (25); FlAlyA, Flavobacterium sp. UMI-01 alginate lyase FlAlyA (18). Residues invariant among all listed proteins are indicated with blue boxes. Residues identical with those of NitAly are indicated with yellow boxes. Cys-80 and Cys-232 in NitAly and their corresponding conserved Cys residues in other proteins are highlighted with red. An arrowhead shows the cleavage site of the signal peptide in NitAly predicted by the SignalP 3.0 software program. Catalytic residues proposed in PL-7 alginate lyases (15, 21, 65) are shown by solid circles. The level of identity of each other protein with NitAly is shown at the end of each sequence, respectively.
Alginate Degradation Activity of NitAly
rNitAly was successfully expressed in Escherichia coli using the pCold expression system and purified (Fig. 2, A and B). The yield was about 1.2 mg of protein per 1 liter of culture. The molecular mass of purified rNitAly was estimated at ∼29 kDa under reducing conditions using SDS-PAGE. This value corresponded reasonably well with the calculated mass of rNitAly (29,636 Da) (Fig. 2B, left lane). Under non-reducing conditions, rNitAly appeared at a lower position, at ∼27 kDa (Fig. 2B, right lane). This result indicated that it has a compact conformation, due to an internal disulfide bond forming via the two Cys residues (Cys-80 and Cys-232) in rNitAly. Therefore, we used PEG-Mal to evaluate the formation of the disulfide bond. PEG-Mal showed that rNitAly migrated less after pretreatment with DTT compared when no pretreatment was used (Fig. 2C). The number of free thiol groups in rNitAly detected by the 4,4′-dithiodipyridine method was 0.01 ± 0.01 and 1.4 ± 0.2/rNitAly molecules under non-reducing and reducing conditions, respectively. Thus, Cys-80 and Cys-232 primarily form an internal disulfide bond in rNitAly.
FIGURE 2.

Bacterial expression and purification of rNitAly. A, schematic drawing of rNitAly. B, SDS-PAGE of purified rNitAly. Reduced and non-reduced indicate that the samples were prepared in the presence and absence of 20 mm DTT, respectively. C, SDS-PAGE of PEG-Mal-labeled rNitAly pretreated in the presence and absence of 20 mm DTT.
We subsequently investigated nine commercially available substrates for polysaccharide lyases that might be degraded by rNitAly, namely alginate, pectin, xanthan gum, polygalacturonic acid, heparin, hyaluronic acid, chondroitin sulfate A, chondroitin sulfate B, and chondroitin sulfate C. The substrates were incubated in a solution containing 10 mm sodium phosphate (pH 6.0), 1 m NaCl, 0.1 mg/ml BSA, 0.1 mg/ml rNitAly, and 0.25% (w/v) polysaccharide for 24 h at 50 °C. Then, each mixture was analyzed by TLC, with only alginate producing degradation products (data not shown). Therefore, we used alginate as the substrate for rNitAly. Next, the alginate degradation activity of rNitAly was evaluated based on the rate of decrease in the viscosity of an alginate solution produced by the enzyme reaction. As shown in Fig. 3, the relative viscosity rapidly decreased within 1 h and declined gradually thereafter. Along with the decrease in viscosity, the absorbance of the reaction mixture at 235 nm increased linearly. Thus, rNitAly might degrade alginate endolytically, with the lyase reaction introducing the double bonds to the degradation products.
FIGURE 3.
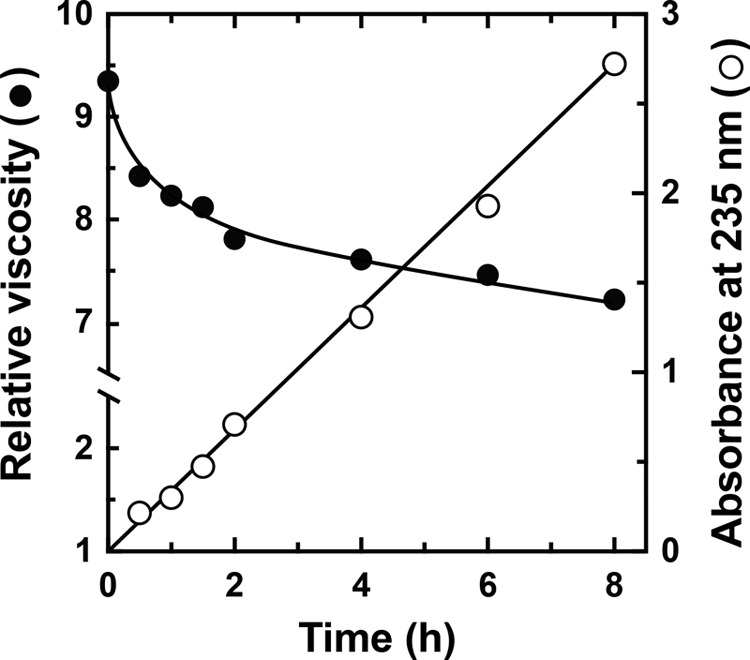
Evaluation of an alginate lyase activity of rNitAly. Relative viscosity (closed circles) and absorbance at 235 nm (open circles) were measured in a mixture of 10 mm sodium phosphate (pH 6.0), 1 m NaCl, 0.1 mg/ml BSA, 1% (w/v) sodium alginate, and 0.005 mg/ml rNitAly at 30 °C at the indicated time points.
The substrate preference of rNitAly was assayed using polyM, polyG, polyMG, and sodium alginate as the substrates (Fig. 4A). PolyM was the most preferable substrate for rNitAly, whereas polyG was the least preferable. The activity level of rNitAly against polyMG and alginate was about one-third of that against polyM. Thus, rNitAly may preferably attack consecutive mannuronic residues of alginate.
FIGURE 4.
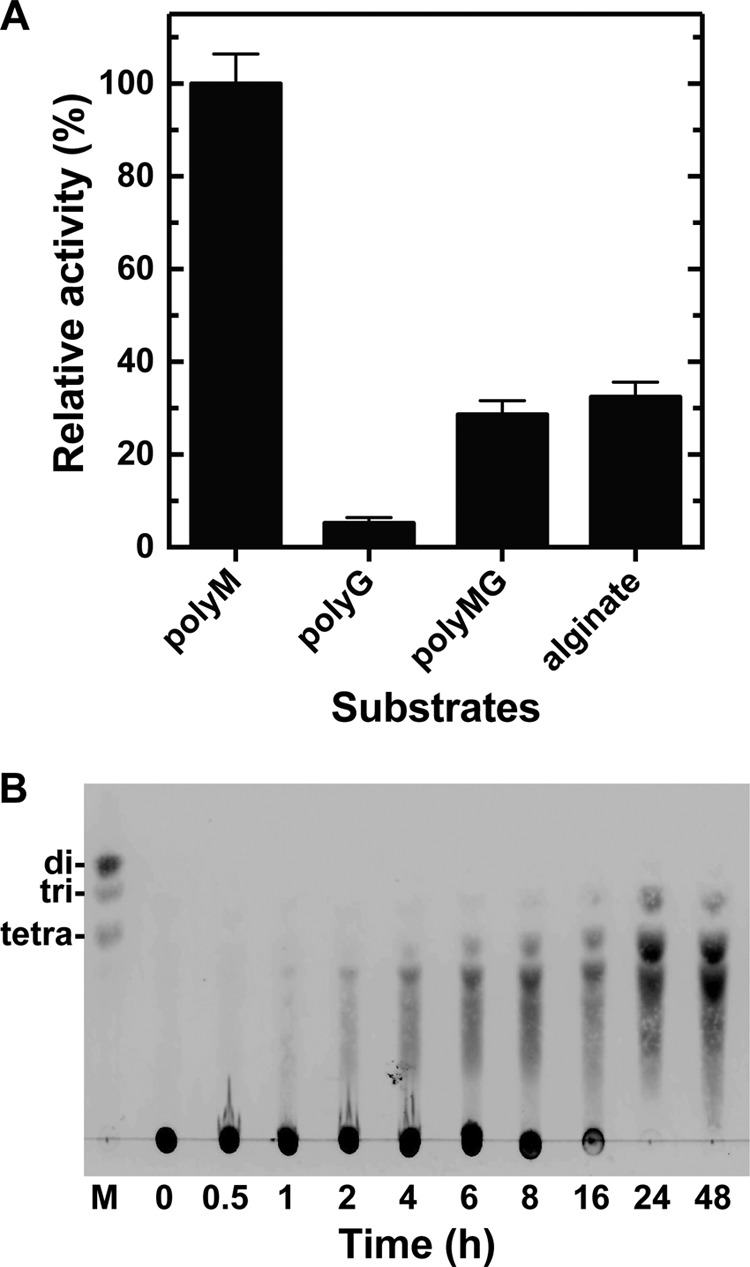
Substrate preferences of rNitAly and degradation products. A, substrate preferences of rNitAly. Enzyme reactions were conducted in a solution containing 10 mm sodium phosphate buffer (pH 6.0), 1 m NaCl, 0.1 mg/ml BSA, 0.005 mg/ml rNitAly, and 0.5% (w/v) substrate (polyM, polyG, polyMG, or sodium alginate) at 50 °C for 15 min. A relative activity of 100% was equivalent to 1,290 units/mg. Assays were done three times, and the data were shown as mean ± S.D. B, thin layer chromatography analysis of the degradation products of polyM following incubation with rNitAly. The enzyme reaction was conducted using polyM as the substrate under the same conditions described for A, and was stopped by the addition of 4 volumes of ice-cold ethanol at the indicated time point.
The degradation products of polyM produced by digestion using rNitAly were analyzed by TLC (Fig. 4B). After 8 h, oligosaccharides with a degree of polymerization of 3 and above were generated; however, most substrates remained undigested at this time point. The original substrate spot disappeared, and the main degradation products that were detected included tri-, tetra-, and penta-saccharides after 24 h and later. Disaccharides were not detected, even after 48 h; therefore, the smallest end products of alginate digestion by rNitAly were identified as tri-saccharides, whereas the major end products were tetra- and penta-saccharides.
Enzymatic Properties of NitAly
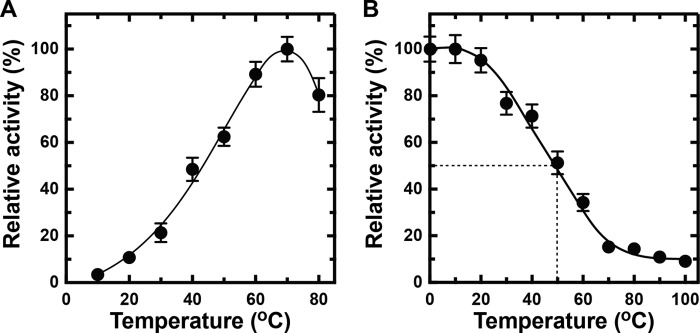
To determine the optimum conditions for rNitAly, its alginate lyase activity was assayed using polyM as the substrate under different conditions. rNitAly activity gradually increased as the temperature rose from 10 to 70 °C (Fig. 5A). Maximum activity (1,620 units/mg) was observed at 70 °C.
FIGURE 5.
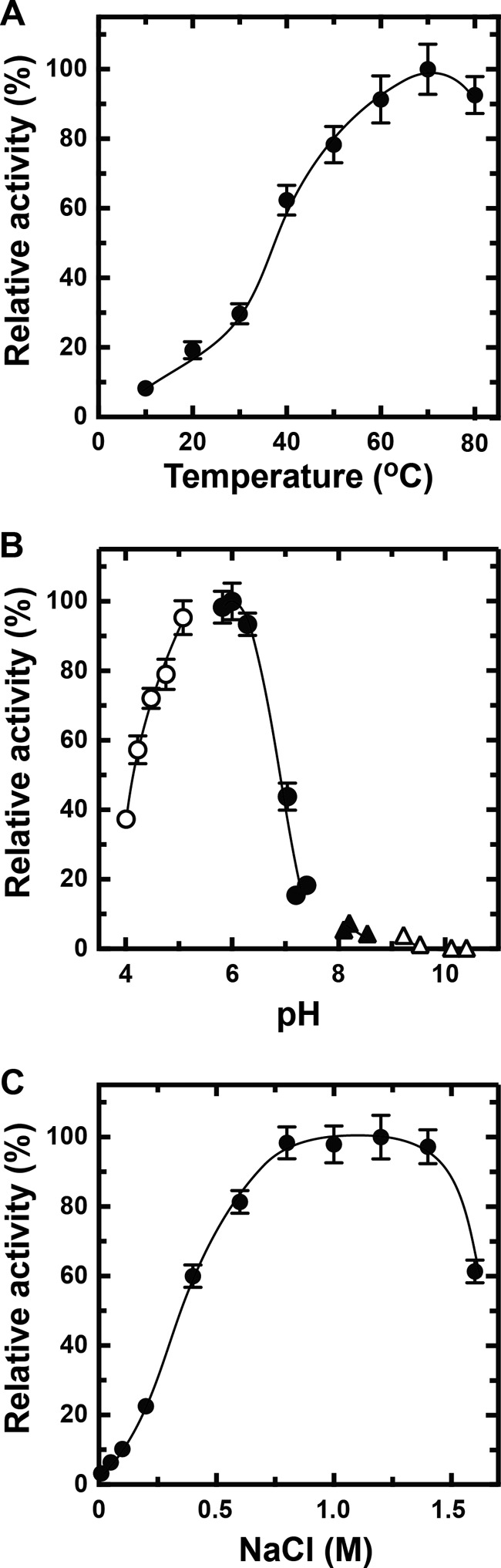
Effects of temperature, pH, NaCl, and divalent metals on rNitAly activity. A, temperature dependence of alginate lyase activity of rNitAly. Enzyme reactions were conducted in a solution containing 10 mm sodium phosphate (pH 6.0), 1 m NaCl, 0.1 mg/ml BSA, 0.005 mg/ml rNitAly, and 0.5% (w/v) polyM for 15 min at the indicated temperatures. Relative activity of 100% was equivalent to 1,620 units/mg. B, pH dependence of the alginate lyase activity of rNitAly. Assays were performed as described for A, except that reaction mixtures were incubated at 50 °C for 15 min in 10 mm sodium acetate for pH 4.0–5.1 (open circles), 10 mm sodium phosphate for pH 5.7–7.4 (closed circles), 10 mm Tris-HCl for pH 8.1–8.5 (closed triangles), and 10 mm glycine-NaOH for pH 9.2–10.4 (open triangles). Relative activity at 100% was equivalent to 1,230 units/mg. C, NaCl dependence of the alginate lyase activity of rNitAly. Assays were performed as described for A at 50 °C, except that different concentrations of NaCl were used as indicated. Relative activity at 100% was equivalent to 1,430 units/mg.
The effect of pH on rNitAly activity was also assayed. Optimum pH was around 6, and relative activities of 80% or above were observed between pH 5.0 and 6.5 (Fig. 5B). The activity of rNitAly noticeably decreased at pH 7 and above. Thus, rNitAly preferred acid conditions to neutral or alkaline conditions for alginate degradation.
Next, the effect of NaCl concentration on rNitAly activity was investigated (Fig. 5C). Alginate degradation activity was fully activated in the presence of 0.8–1.4 m NaCl; however, its activity was extremely low in the presence of 0.15 m NaCl. The effects of other monovalent cations were investigated. All tested cations (K+, Li+, and Cs+) showed similar activation with Na+ (Table 2). These finding showed that rNitAly requires a high concentration of monovalent cations for maximum activity.
TABLE 2.
Effects of monovalent cations on the alginate lyase activity of NitAly
| Relative activitya (%) |
|||
|---|---|---|---|
| 0.1 m | 0.5 m | 1.0 m | |
| NaCl | 9.2 ± 0.5 | 74.2 ± 2.1 | 100 ± 4.3b |
| KCl | 8.3 ± 0.4 | 71.6 ± 1.8 | 113 ± 6.1 |
| LiCl | 10.3 ± 0.4 | 77.3 ± 3.2 | 101 ± 3.0 |
| CsCl | 8.7 ± 0.3 | 72.4 ± 1.2 | 114 ± 7.4 |
a Enzyme reactions were conducted in a solution containing 10 mm sodium phosphate (pH 6.0), 0.1 mg/ml BSA, 0.005 mg/ml rNitAly, 0.5% (w/v) polyM, and the indicated concentration of monovalent cation at 50 °C for 15 min.
b Relative activity at 100% was equivalent to 1,350 units/mg.
The effects of divalent cations, trivalent cations, and chelating reagents were examined (Table 3). Among the tested reagents, Sn2+, Pb2+, Fe2+, and Fe3+ inhibited rNitAly activity by 84.6, 81.6, 68.8, and 86.1%, respectively. Ni2+, Zn2+, Mn2+, Cd2+, Hg2+, Mg2+, Ca2+, Sr2+, Co2+, Al3+, EDTA, and EGTA had no significant effect on rNitAly activity. Although Hg2+ is a blocking agent for cysteine thiol groups, it did not inhibit rNitAly activity. Thus, the disulfide bond in rNitAly (Figs. 1 and 2) is not responsible for alginate degradation through the lyase reaction.
TABLE 3.
Effects of divalent cations, trivalent cations, and chelate reagents on the alginate lyase activity of NitAly
| Relative activitya% | |
|---|---|
| Controlb | 100 ± 2.7c |
| Divalent cation (1 mm) | |
| NiCl2 | 110 ± 6.3 |
| ZnCl2 | 109 ± 4.5 |
| MnCl2 | 109 ± 5.3 |
| CdCl2 | 107 ± 3.2 |
| HgCl2 | 107 ± 2.2 |
| MgCl2 | 105 ± 6.2 |
| CaCl2 | 104 ± 7.8 |
| SrCl2 | 97.3 ± 4.3 |
| CoCl2 | 95.1 ± 5.6 |
| SnCl2 | 84.6 ± 3.2 |
| PbCl2 | 81.6 ± 4.1 |
| FeCl2 | 68.8 ± 3.2 |
| Trivalent cation (1 mm) | |
| AlCl3 | 97.2 ± 4.3 |
| FeCl3 | 86.1 ± 5.2 |
| Chelate reagent (5 mm) | |
| EDTA | 95.8 ± 6.2 |
| EGTA | 101 ± 3.8 |
a Enzyme activity in this experiment was measured by TBA method (62), due to the increment of absorbance at 235 nm in the absence of rNitAly through the addition of some metal ions, possibly as a result of gelation. Enzyme reactions were conducted in a solution containing 10 mm sodium phosphate (pH 6.0), 1 m NaCl, 0.1 mg/ml BSA, 0.005 mg/ml rNitAly, and 0.5% (w/v) polyM for the specified divalent cations, trivalent cations, or chelate reagents at 50 °C for 15 min.
b Control activity was measured in the absence of metal ions or chelate reagents.
c Relative activity at 100% was equivalent to 1,320 units/mg.
Effects of Temperature and pH on NitAly Stability
The heat stability of rNitAly was investigated by measuring the residual alginate lyase activity of rNitAly after incubation at different temperatures for 30 min (Fig. 6A). Heat treatment did not affect rNitAly activity between 0 and 30 °C. The loss of activity was observed after incubation at 40 °C and above, and it reached 50% after incubation at 67 °C for 30 min. Interestingly, rNitAly showed residual alginate lyase activity of about 10% even after treatment at 100 °C for 30 min.
FIGURE 6.
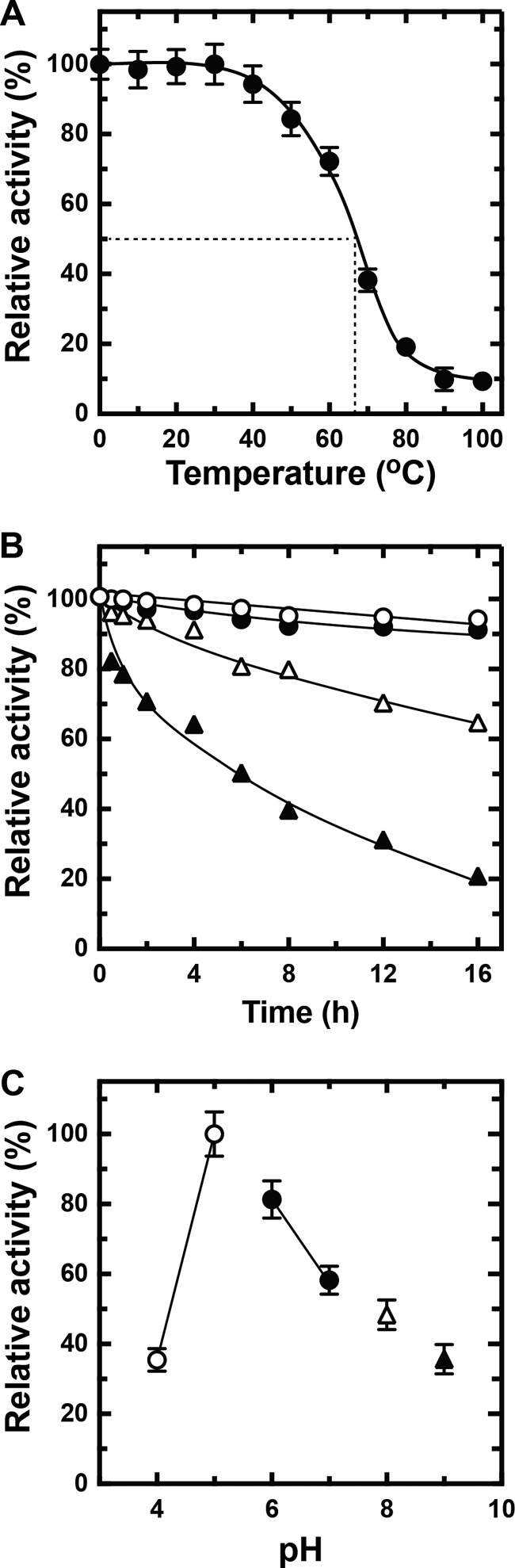
Effects of temperature and pH on rNitAly stability. A, heat stability of rNitAly. For this assay, 0.2 mg/ml rNitAly in 10 mm sodium phosphate (pH 6.0) and 1 m NaCl were incubated for 30 min at the indicated temperatures and placed on ice. Enzyme activity was then assayed as described in Fig. 5A at 50 °C. Relative activity at 100% was equivalent to 1,230 units/mg. B, effects of time extension of heat treatment on the heat stability of rNitAly. For this assay, 0.2 mg/ml rNitAly in 10 mm sodium phosphate (pH 6.0) and 1 m NaCl were incubated for the indicated time at 20 °C (open circles), 30 °C (closed circles), 40 °C (open triangles), and 50 °C (closed triangles) and placed on ice. Assays were performed as described for A. Relative activity at 100% was equivalent to 1,210 units/mg. C, pH stability of rNitAly. For this assay, 0.2 mg/ml rNitAly in 10 mm sodium phosphate (pH 6.0) and 1 m NaCl was dialyzed against 1,000 volumes of 1 m NaCl and 10 mm sodium acetate (pH 4.0 or 5.0) (open circles), 10 mm sodium phosphate (pH 6.0 or 7.0) (closed circles), 10 mm Tris-HCl (pH 8.0) (open triangle), and 10 mm glycine-KOH (pH 9.0) (closed triangle) at 4 °C for 8 h. Assays were performed as described for A. Relative activity at 100% was equivalent to 1,410 units/mg. All assays were repeated three times, and the data are shown as mean ± S.D.
Extending the duration of heat treatment of rNitAly at 20 and 30 °C to 16 h slightly decreased its residual activity (Fig. 6B). After incubation for 16 h under these conditions, rNitAly still showed over 90% of its original activity, although its residual activity was reduced to about 70 and 20% by incubation at 40 and 50 °C, respectively, for 16 h.
The pH stability of rNitAly was investigated by measuring its residual activity after incubation under different pH conditions for 8 h at 4 °C (Fig. 6C). rNitAly exhibited the highest stability at pH 5. The residual activity of rNitAly significantly decreased at pH 4 and 7–10.
Effects of DTT on the Optimum Temperature and Heat Stability of NitAly
While measuring the alginate lyase activity of NitAly at 30 °C, we noticed that the absorbance of the reaction solution at 235 nm did not increase linearly. The slope of the increase in absorbance over time decreased after 1.5 h, unlike that shown in Fig. 2, when DTT was added to the reaction mixture to reduce the protein disulfide bonds. In comparison, incubation at 30 °C in the absence of DTT did not significantly affect rNitAly activity (Fig. 6B). Thus, we investigated how DTT affected the optimum temperature and heat stability of rNitAly. Fig. 7A shows the temperature dependence of rNitAly activity in the presence of 5 mm DTT. Under this condition, the activity of rNitAly between 10 and 50 °C was slightly lower in comparison with its activity in the absence of DTT (Fig. 5A). Yet, the optimum temperature for the activity of rNitAly in the presence of DTT remained at 70 °C, corresponding to that observed without DTT (Fig. 5A). In addition, the specific activity of rNitAly at 70 °C was not significantly different in the absence (1,620 units/mg) and presence (1,590 units/mg) of DTT. Yet, the presence of DTT markedly decreased the heat stability of rNitAly. rNitAly activity fell to 78% of its original activity after incubation at 30 °C for 30 min in the presence of DTT (Fig. 7B). In comparison, incubation at 20 °C or below had no effect on rNitAly activity. Compared with the decrease in rNitAly activity in the absence of DDT, it showed a marked further decline in the presence of DTT at incubation temperatures of 40–70 °C. rNitAly activity declined to 50% after incubation at 50 °C, which was 17 °C lower than the temperature that produced the same decrease in activity in the absence of DTT (Fig. 6A). Thus, DTT tended to have no effect on the optimum temperature and specific activity of rNitAly. In contrast, DTT had a major effect on the heat stability of rNitAly, probably due to the reduction of disulfide bond(s).
FIGURE 7.
Effects of DTT on the temperature dependence and heat stability of NitAly. A, temperature dependence of alginate lyase activity of rNitAly in the presence of DTT. Assays were performed as described for Fig. 5A, except that DTT was added to the reaction mixture at a final concentration of 5 mm. Relative activity at 100% was equivalent to 1,590 units/mg. B, heat stability of rNitAly in the presence of DTT. Samples were preincubated as described for Fig. 6A, except that DTT was added at a final concentration of 5 mm. Assays were performed as described in A at 50 °C. Relative activity at 100% was equivalent to 1,080 units/mg. All assays were repeated three times, and the data are shown as mean ± S.D.
Identification of the Residues Responsible for the Heat Stability of rNitAly
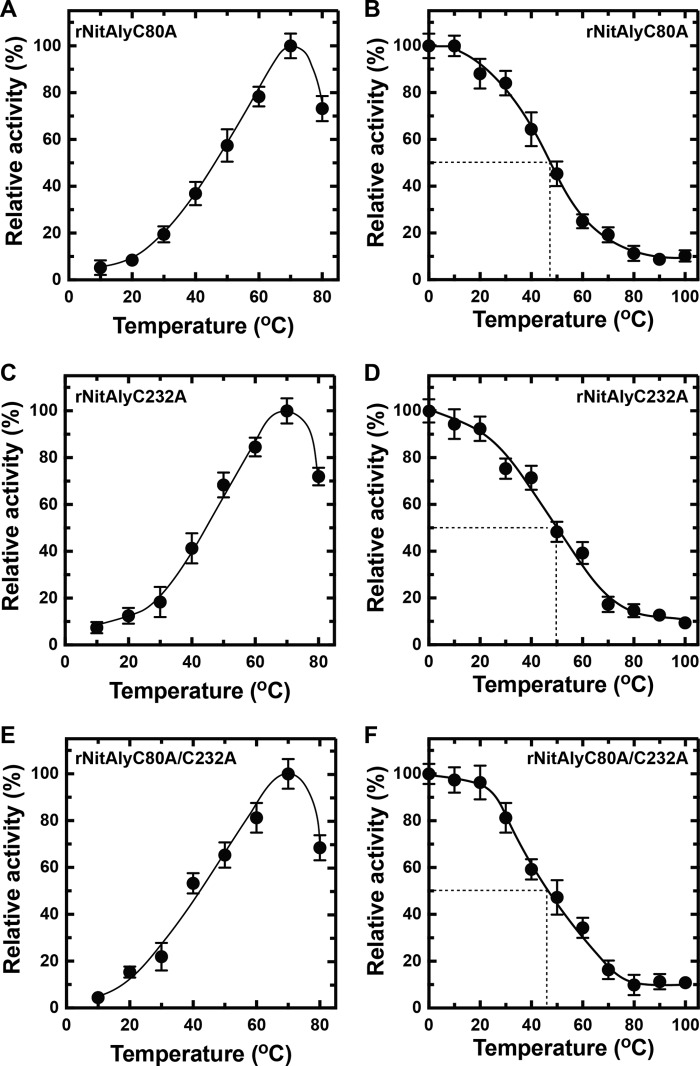
In the amino acid sequence of NitAly, three cysteines were found at the positions of 17, 80, and 232 (Fig. 1). Because Cys-17 was located in the predicted signal sequence region, rNitAly did not contain this amino acid. Hence, we focused on the other two cysteines that we suggested form an internal disulfide bond (Fig. 2). Homology modeling of NitAly also supported that Cys-80 and Cys-232 are physically located close to one another in the folded protein (Fig. 8 A).Therefore, Cys-80 and/or Cys-232 of NitAly were replaced with Ala by point mutation to produce the mutant proteins rNitAlyC80A, rNitAlyC232A, and rNitAlyC80A/C232A. Then, the optimum temperature and heat stability of each mutant were investigated in the absence of DTT. All of these mutants showed maximum activity at 70 °C, with specific activities of 1,520, 1,630, and 1,480 units/mg for rNitAlyC80A, rNitAlyC232A, and rNitAlyC80A/C232A, respectively (Fig. 9, A, C, and E). In addition, the effect of temperature on the activity of all mutants was similar to that of wild-type NitAly. Thus, the replacement of Cys-80 and/or Cys-232 with Ala had no significant effect on the activity and optimum temperature of rNitAly. Further experiments revealed that alterations to the residual activity of each mutant after incubation at different temperatures in the presence of DTT were indistinguishable from that of the wild-type rNitAly protein (Fig. 9, B, D, and E). A loss of 18–24% activity was observed in all mutants after incubation at 30 °C in the absence of DTT, whereas the wild-type protein showed no loss of activity under the same conditions. Incubation temperatures of 48, 50, and 46 °C caused a 50% loss of activity for rNitAlyC80A, rNitAlyC232A, and rNitAlyC80A/C232A, respectively. Overall, the replacement of Cys-80 and/or Cys-232 caused the heat stability of NitAly to decline. Thus, the disulfide bond that forms between Cys-80 and Cys-232 is responsible for the heat stability of NitAly.
FIGURE 8.
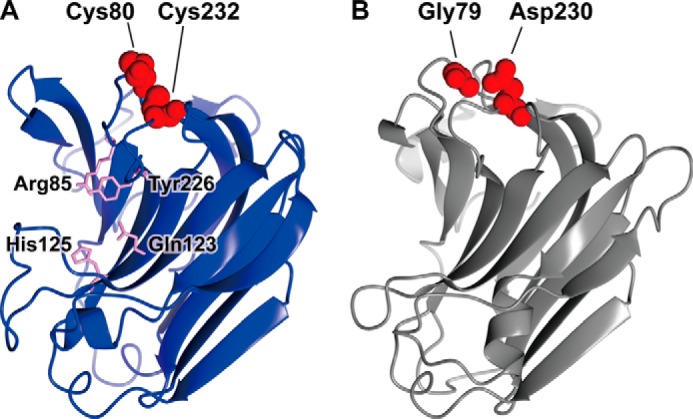
Homology modeling of NitAly and PyAly. The structures of NitAly (A, residues 34–242) and PyAly (B, residues 13–216) were predicted using the PHYRE2 program (63) and P. aeruginosa alginate lyase PA1167 (Protein Data Bank code 1vav) as the template (15) with 100% confidence, respectively. A, Cys-80 and Cys-232 are shown by red globules. The residues Arg-85, Gln-123, His-125, and Tyr-226, which correspond to the suggested catalytic residues in PA1167 (15, 21), are shown by pink sticks. B, Gly-79 and Asp-230 are shown by red globules.
FIGURE 9.
Effects of cysteine residue mutations in rNitAly on temperature dependence and heat stability. A, C, and E, temperature dependence of alginate lyase activity of rNitAlyC80A (A), rNitAlyC232A (C), and rNitAlyC80A/C232A (E). Assays were performed as described for Fig. 5A. Relative activities at 100% were equivalent to 1,520 units/mg (A), 1,630 units/mg (C), and 1,480 units/mg (E). B, D, and F, heat stability of rNitAlyC80A (B), rNitAlyC232A (D), and rNitAlyC80A/C232A (F). Assays were performed as described for Fig. 6A. Relative activity at 100% was equivalent to 970 units/mg (B), 1,060 units/mg (D), and 960 units/mg (F). All assays were repeated three times, and the data are shown as mean ± S.D.
Improvement to the Heat Stability of rPyAly
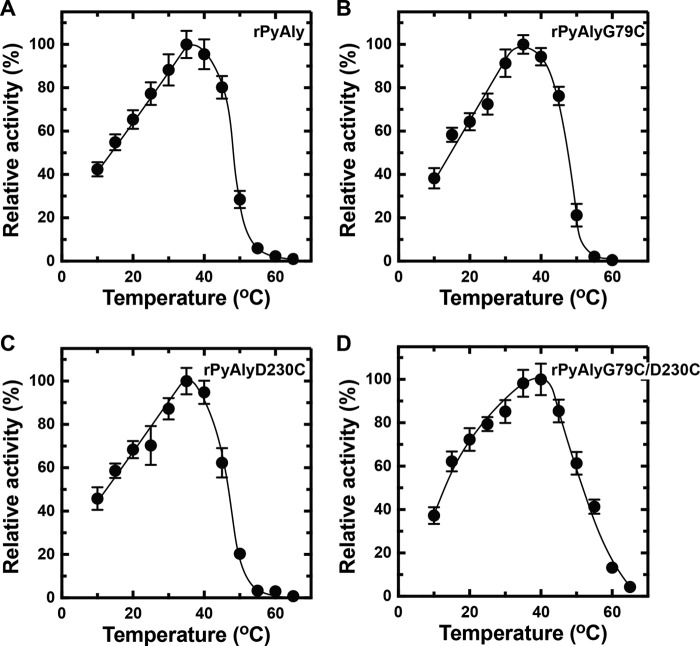
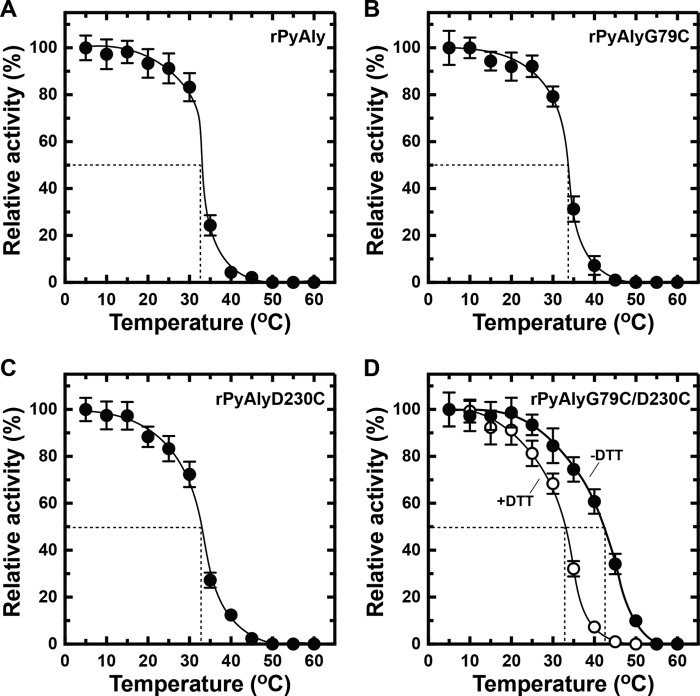
NitAly showed the highest amino acid sequence identity with that of the red alga alginate lyase PyAly among the characterized family PL-7 alginate lyases (Fig. 1). The heat stability of PyAly appeared to be inferior to that of NitAly. For instance, PyAly showed residual activity of 50% after incubation at 32.5 °C for 30 min, and all activity ceased at incubation temperatures above 50 °C (39). Alignment of the amino acid sequences of PyAly and NitAly (Fig. 1) suggested that two residues in PyAly, Gly-79 and Asp-230, correspond to the Cys-80 and Cys-232 residues, respectively, in NitAly. These two residues of PyAly may be positioned close to each other based on the molecular structure obtained by homology modeling (Fig. 8B). In addition, Cys residues were not found in the whole sequence of PyAly. Thus, Gly-79 and/or Asp-230 in PyAly were replaced with Cys (single Cys mutant, rPyAlyG79C or rPyAlyD230C; double Cys mutant, rPyAlyG79D230C). These mutant proteins and the wild-type protein (rPyAlyWT) were expressed and purified. First, the effect of temperature on the alginate lyase activity of these mutant proteins was assayed. Maximum activity was observed at 35 °C for PyAlyG79C and PyAlyD230C and at 40 °C for PyAlyD230C. Specific activity was 1,820 units/mg for rPyAlyWT, 1,740 units/mg for PyAlyG79C, 1,710 units/mg for PyAlyD230C, and 1,780 units/mg for PyAlyG79C/D230C (Fig. 10). Therefore, Cys mutation(s) at Gly-79 and/or Asp-230 in PyAly had no significant effect on the optimum temperature and activity of NitAly. Interestingly, rPyAlyG79C/D230C activity significantly declined to 61% of its original activity after incubation at 50 °C for 30 min. In comparison, the relative activity of the other mutant and wild-type PyAly enzymes shown declined to 20–28%. Thus, the dual replacement of Gly-79 and Asp-230 residues in PyAly with Cys residues improved the heat stability of the enzyme at 50 °C.
FIGURE 10.
Effects of replacing the residues of rPyAly with cysteine on its temperature dependence. Enzyme reactions were conducted in a solution containing 10 mm sodium phosphate (pH 8.0), 0.1 m NaCl, 0.1 mg/ml BSA, 0.25% (w/v) polyM, and 0.005 mg/ml rPyAly (A), rPyAlyG79C (B), rPyAlyD230C (C), or rPyAlyG79C/D230C (D) for 15 min at the indicated temperatures. Relative activity at 100% was equivalent to 1,820 units/mg (A), 1,740 units/mg (B), 1,710 units/mg (C), and 1,780 units/mg (D). All assays were repeated three times, and the data are shown as mean ± S.D.
The heat stability of the mutant and wild-type PyAly enzymes was then evaluated. The heat stability of both PyAlyG79C and PyAlyD230C was mostly identical to that of rPyAlyWT (Fig. 11). A 50% loss and a complete loss of activity occurred at about 33 °C and above 50 °C, respectively, in both mutants. However, the introduction of the two Cys residues to PyAly enhanced heat stability (Fig. 11D). A temperature of 45 °C caused a 50% loss of activity in PyAlyG79C/D230C, which was more than 10 °C higher than the temperatures causing similar losses of activity in rPyAlyWT, PyAlyG79C, and PyAlyD230C. Furthermore, PyAlyG79C/D230C showed residual activity of 25% after incubation at 50 °C, whereas rPyAlyWT, PyAlyG79C, and PyAlyD230C showed no residual activity. Moreover, the enhanced heat stability of PyAlyG79C/D230C was removed in the presence of DTT (Fig. 11D). Thus, the disulfide bond that formed by introducing the two Cys residues to PyAly improved the heat stability of the enzyme without decreasing its alginate lyase activity.
FIGURE 11.
Effects of replacing residue(s) of rPyAly with cysteine on its heat stability. Enzyme solution containing 10 mm sodium phosphate (pH 8.0), 0.1 m NaCl, and 0.15 mg/ml rPyAly (A), rPyAlyG79C (B), rPyAlyD230C (C), or rPyAlyG79C/D230C (D, −DTT) was incubated for 30 min at the indicated temperatures and placed on ice. For rPyAlyG79C/D230C, 5 mm DTT was also added during incubation (D, +DTT). Enzyme activity was then assayed as described for Fig. 10 at 30 °C. Relative activity at 100% was equivalent to 1,730 units/mg (A), 1,620 units/mg (B), 1,540 units/mg (C), 1,600 units/mg (D, −DTT), and 1,420 units/mg (D, +DTT). All assays were repeated three times, and the data are shown as mean ± S.D.
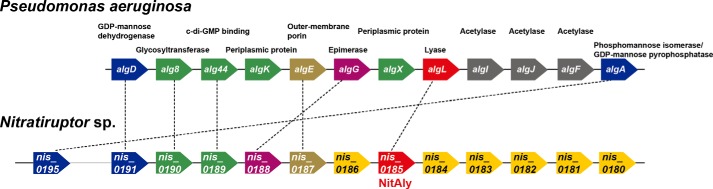
Location of the NitAly Gene in the Alginate Biosynthetic Cluster of Nitratiruptor sp. SB155-2 Genome
BLAST searches of a series of predicted proteins from Nitratiruptor sp. SB155-2 against P. aeruginosa proteins located within the alginate biosynthesis operon revealed that seven genes encoding the Nitratiruptor sp. SB155-2 homologs were adjacent to one another in the Nitratiruptor sp. SB155-2 genome, namely the P. aeruginosa proteins AlgD (GDP-mannose dehydrogenase), Alg8 (glycosyltransferase), Alg44 (c-di-GMP binding), AlgE (outer membrane porin), AlgG (mannuronic acid C5-epimerase), AlgL (alginate lyase), and AlgA (phosphomannose isomerase/GDP-mannose pyrophosphatase). Thus, these genes may constitute an operon system in Nitratiruptor sp. SB155-2, supporting that NitAly performs alginate biosynthesis in its natural habitat. Yet the candidate genes for AlgK and AlgX, which are involved in alginate polymerization, were not found. These proteins are needed to form the alginate biosynthesis machinery complex (46), whereby AlgK interacts with AlgE, Alg44, and AlgX. Furthermore, AlgX interacts with Alg44 and the specifically interacting serine protease MucD (47). In addition, AlgI, AlgJ, and AlgF were not found, despite being involved in the O-acetylation of alginate. Thus, proteins with these functions may have low homology with known proteins and may be the currently unidentified proteins encoded by genes such as nis_0180–nis_0184 and nis_0186 located near the nitaly (nis_0185) gene.
Discussion
This study identified NitAly as a functional alginate lyase belonging to the PL-7 family. Compared with other characterized PL-7 alginate lyases, NitAly activity was maximum at 1,620 units/mg, 70 °C, and pH 6. This activity was comparable with the maximum activity of PyAly at 35 °C and pH 8 (39), and it was ∼2.5% that of FlAlyA at 30 °C and pH 8 (48). When comparing the maximum activity of rNitAly with other PL enzymes, abalone alginate lyase HdAly belonging to PL-14 has about 1.3-fold higher activity at 35 °C and pH 8 (49).
One important finding of this study was that a disulfide bond between Cys-80 and Cys-232 is critical for the high heat stability of NitAly (Figs. 7–9). The breaking of this bond, by a reducing reagent or point mutation, caused heat stability to noticeably decline. These Cys residues are fully conserved among alginate lyases, showing significant homologies to NitAly that are produced by bacteria isolated from deep-sea hydrothermal vents but are not fully conserved among alginate lyases from terrestrial bacteria, marine bacteria isolated from deep-sea sediments, and red algae (Fig. 1). C. mediatlanticus (43), H. jasoniae (44), and N. profundicola (45) are thermophilic bacteria isolated from deep-sea hydrothermal vent sites. These species produce uncharacterized proteins that show homology with NitAly (Fig. 1). If these uncharacterized proteins have alginate lyase activity, we could confirm that the two conserved Cys residues are important for their high heat stability.
Interestingly, rNitAly did not completely lose its activity, regardless of the presence of DTT, even after incubation at 100 °C for 30 min (Fig. 7B). This result was observed in the wild-type as well as the C80A, C232A, and C80A/C232A mutants of rNitAly (Fig. 9). Thus, other heat tolerant structure(s) may exist in NitAly, separate from the disulfide bond between Cys-80 and Cys-232. Although there is no firm explanation for this additional heat tolerance yet, regions with a high conservation of residues between NitAly and its homologous proteins from thermophilic bacteria (including proteins other than PL-7 alginate lyases) may be involved. For example, the Asp-49–Phe-59, His-152–Ser-165, and Lys-198–Asn-202 regions of NitAly represent possible candidates. Thus, further studies are required to understand the relationship between the structure of NitAly and several of its properties, including its heat stability. At present, we are attempting to resolve the crystal structure of NitAly.
Although the high optimum temperature and heat stability of NitAly were expected, considering the habitat occupied by Nitratiruptor sp. SB155-2, other unexpected unique properties were also found. NitAly optimally functions under acidic conditions, rather than neutral and alkaline conditions. To date, the characterized PL-7 alginate lyases have shown maximum activity at pH 7–9. In contrast, NitAly showed a markedly decreased activity at this higher pH range, with its optimum pH being around 6 (Fig. 5B). This mild acidophilicity may reflect the natural habitat of Nitratiruptor sp. SB155-2, where deep-sea hydrothermal vent fluids (typical pH ∼4) mix with seawater (typical pH ∼8). It is not possible to determine the exact pH in such an environment due to local and steep fluctuations in pH. However, in general, the environment should be more acidic than that at the sea surface. Consequently, NitAly is adapted to acidic conditions. Thus, NitAly may be secreted to function outside of the cell, rather than in the periplasmic space.
Our results imply that bacterial communities inhabiting deep-sea hydrothermal vents use alginate. Previous reports on the existence or utilization of alginate in this type of environment are not available. One of the best characterized alginate-producing bacteria, P. aeruginosa, is a member of microbiota that reside in the soil, water, plants, and animals. The exocellular matrix produced by P. aeruginosa contains alginate as a component (4). This matrix functions as a biofilm to protect the bacterium from attack and to physically attach it to target sites (50). If Nitratiruptor sp. SB155-2 produces alginate, which is a component of exopolysaccharides matrices in nature, it might have the capacity to adhere to biotic and abiotic surfaces, as well as the cells of other Nitratiruptor sp. SB155-2. The adherent capacity of alginate stems from its ability to bind divalent metals (such as Ca2+, Sr2+, and Ba2+) to form a cross-linked sticky gel, in accordance with the egg-box molecular model (51–54). Alginate gels show high heat stability compared with gels consisting of other polysaccharides (e.g. agar or carrageenan) and are stable at 0–100 °C (55). The heat resistance property of alginate gels means that they could be used around deep-sea hydrothermal vents where cold seawater associates with hydrothermal vent fluids emerging at temperatures of 300–400 °C (56). In addition, Nitratiruptor sp. SB155-2 thrives at a water depth of 1,000 m, where the pressure is 101 atm (=10.1 × 105 Pa). Under such high pressure, alginate gels are stable. A previous study reported no significant change in the strength of an alginate gel after treatment at a pressure of 10.5 × 107 Pa (57).
In conclusion, we showed that Nitratiruptor sp. SB155-2 has a gene encoding a functional alginate lyase enzyme, NitAly, that is mildly acidophilic and heat stable. Because the nitaly gene appears to be a member of an alginate biosynthetic operon (Fig. 12), this bacterium may be able to produce alginate. If this is the case, NitAly might be involved in alginate biosynthesis to form and decompose an exocellular matrix (such as a biofilm).
FIGURE 12.
Assignment of genes in the alginate biosynthesis operon of P. aeruginosa and their homologs in Nitratiruptor sp. SB155-2. Genes encoding proteins with homology between P. aeruginosa and Nitratiruptor sp. SB155-2 are connected by dotted lines. Each pentagon is classified by a different color based on known function in P. aeruginosa as follows: blue, precursor biosynthesis; green, alginate polymerization; brown, alginate export; violet, epimerization of mannuronic acid to guluronic acid; red, alginate degradation; gray, O-acetylation. Yellow pentagons show genes with unidentified functions in Nitratiruptor sp. SB155-2.
Experimental Procedures
Host Strain and Vectors
E. coli DH5α (Nippon Gene, Tokyo, Japan) and pTac-1 vector plasmid (BioDynamics, Tokyo, Japan) were used for DNA cloning. E. coli BL21(DE3) (Nippon Gene) and pCold I plasmid vector (Takara Bio, Shiga, Japan) were used for recombinant protein expression.
Reagents
Sodium alginate with a viscosity of 80–120 centipoise was purchased from Wako Pure Chemical Industries (Osaka, Japan). PolyM, polyG, and polyMG blocks were prepared by the method of Gacesa and Wusteman (58). Restriction enzymes and DNA-modifying enzymes were purchased from Nippon Gene. All other chemical reagents were purchased from Wako Pure Chemical Industries, unless otherwise stated.
DNA Cloning and Sequencing of NitAly
Genomic DNA was isolated from Nitratiruptor sp. SB155-2, as described previously (41). A primer set of Nit-F and Nit-R (Table 1) was used in combination with Platinum® TaqDNA polymerase (Thermo Fisher Scientific, Waltham, MA) to amplify the full-length gene encoding the hypothetical protein NIS_0185 (GenBankTM accession no. YP_001355656). Polymerase chain reaction (PCR) was conducted using temperature settings of 95 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s. The final step for extension was performed at 72 °C for 7 min. Amplified DNA was ligated into the pTac-1 vector and was sequenced using a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA).
Construction of Expression Plasmid for rNitAly
Restriction sites of BamHI and XbaI were introduced into the 5′ and 3′ termini of the nitaly gene using the Nit-BamF and Nit-XbaR primer sets (Table 1), respectively, by PCR. The reaction conditions were the same as those described above. Amplified DNA was digested with BamHI and XbaI after subcloning in the pTac-1 vector. DNA fragments were isolated and recovered from agarose gel following agarose gel electrophoresis and were ligated in the pCold I vector digested with BamHI and XbaI. After DNA sequencing, BL21(DE3) cells were transformed with the recombinant pCold I construct.
Expression and Purification of rNitAly
Transformed cells were cultured in lysogeny broth medium supplemented with 50 μg/ml ampicillin at 37 °C for 12 h. Then, the medium was cooled at 15 °C for 1 h and isopropyl β-d-1-thiogalactopyranoside was added at a final concentration of 0.1 mm. After incubation at 15 °C for 12 h, cells were harvested by centrifugation at 5,000 × g for 15 min. Pellets were suspended and sonicated in a buffer containing 10 mm imidazole-HCl (pH 7.4), 0.5 m NaCl, 1% (v/v) TritonTM X-100, and 0.05 mg/ml lysozyme. After centrifugation at 10,000 × g for 15 min, the supernatant was mixed with 250 μl of nickel-nitrilotriacetic acid-agarose in a conical tube, and the tube was rotated at 4 °C for 30 min. Resins were washed with a 20-fold volume of 30 mm imidazole (pH 7.4) and 0.5 m NaCl. Protein elution was conducted with a solution containing 250 mm imidazole (pH 7.4) and 0.5 m NaCl. Fractions containing the targeted proteins were combined and dialyzed against 20 mm sodium acetate (pH 5.0) and 0.1 m NaCl at 4 °C for 8 h. Purified proteins were stored on ice and were assayed within 3 days. Protein concentration was determined by the method of Bradford (59), using bovine serum albumin (BSA) fraction V as the protein standard.
Site-directed Mutagenesis of Alginate Lyase Proteins
The listed primers (Table 1) were designed for the site-directed mutation of rNitAly and rPyAly. Mutagenesis was conducted using recombinant plasmid rNitAly-pCold I (in this study) or rPyAly-pCold I (39) as a template with a Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA). The mutant proteins of NitAly (rNitAlyC80A, rNitAlyC232A, and rNitAlyC80A/C232A) were purified using the same method as that described for rNitAly in the previous section. Wild-type rPyAly and its mutant proteins (rPyAlyG79C, rPyAlyD230C, and rPyAlyG79C/D230C) were purified using the same method as that used for rPyAly, as reported previously (39).
SDS-PAGE
SDS-PAGE was conducted on a 10 or 12% (v/v) polyacrylamide gel using the method of Porzio and Pearson (60). After electrophoresis, the gel was stained with 0.1% (w/v) Coomassie Brilliant Blue R-250 in 50% (v/v) methanol, 10% (v/v) acetic acid, and the background of the gel was destained with 5% (v/v) methanol, 7% (v/v) acetic acid. Protein Molecular Weight Marker (Broad) (Takara Bio) was used as the molecular weight marker.
Labeling of Cys Residues in rNitAly
Purified rNitAly (0.1 mg) was incubated in 20 mm Tris-HCl (pH 7.5) and 2.5% SDS in the presence and absence of 20 mm DTT at 25 °C for 5 h. Then, the proteins were precipitated by 10% trichloroacetic acid and were dissolved in 20 mm Tris-HCl (pH 7.5), 3% SDS, and 10 mm PEG-Mal (PEG number average molecular weight of 5,000, Sigma). After incubation at 25 °C for 30 min, the samples were analyzed by SDS-PAGE.
Determination of the Number of Free Thiol Groups in rNitAly
Purified rNitAly (0.1 mg) was incubated in 10 mm Tris-HCl (pH 7.5) and 6 m guanidinium chloride in the presence and absence of 5 mm DTT at 25 °C for 2 h. Then, the number of free thiol groups was determined using 4,4′-dithiodipyridine by measuring absorbance at 324 nm using the method of Le and Means (61).
Assay for Alginate Lyase Activity
The decrease in the viscosity of alginate solutions produced by the alginate lyase enzyme reactions was measured at 30 °C using an Ostwald-type viscometer. The alginate lyase activities of rNitAly and its mutants were assayed in a solution containing 10 mm sodium phosphate (pH 6.0), 1.0 m NaCl, 0.25% (w/v) sodium alginate, 0.1 mg/ml BSA, and 0.005 mg/ml enzyme, unless otherwise stated. For PyAly and its mutants, the reaction mixture contained 10 mm sodium phosphate (pH 8.0), 0.1 m NaCl, 0.25% (w/v) sodium alginate, 0.1 mg/ml BSA, and 0.005 mg/ml enzyme. Alginate lyase activity was measured by an increase in absorbance at 235 nm, due to the formation of a double bond between C-4 and C-5 at the non-reducing end by the cleavage of alginate. Enzyme reactions were monitored using a U-3010 spectrophotometer (Hitachi, Tokyo, Japan) equipped with an SP-12R thermal control unit (TAITEC, Koshigaya, Japan). One unit of alginate lyase was defined as the amount of enzyme that increased absorbance at 235 nm by 0.01 in 1 min.
Alginate lyase activity of rNitAly in the presence of divalent cations, trivalent cations, or chelator reagent was measured by the 2-thiobarbituric acid method (62). Assay conditions were the same as those described above. One unit of activity was defined as the amount of enzyme required to liberate 1 μmol of β-formyl-pyruvic acid per min at 50 °C. Because 1 unit in this assay was experimentally determined to be equivalent to an increase of 0.62 in the absorbance at 235 nm in 1 min, the calculated activity was converted to activity comparable with that measured by the absorbance at 235 nm.
Thin Layer Chromatography (TLC)
After each enzyme reaction, ethanol was added to the mixture at a final concentration of 80% (v/v), and the resulting solution was stored at −20 °C for 2 h. Pellets were produced by centrifugation at 12,000 × g for 15 min, and were washed with ethanol 3 times. Dried pellets were dissolved with 10 mm sodium phosphate (pH 8.0), and were analyzed by TLC using a silica gel 60 plate (Merck, Darmstadt, Germany). The resulting solvent contained ethyl acetate, acetic acid, and water (2:2:1, v/v/v). Sugars developed on the TLC plate were detected by spraying 10% (v/v) sulfuric acid in ethanol followed by heating at 130 °C for 10 min.
Computational Analysis of Proteins
The presence of a signal peptide was predicted using the program SignalP 3.0 (42). Homology modeling of protein structures was carried out using the PHYRE2 protein fold recognition server (63). The predicted protein structures were visualized using the program CCP4mg (64).
Author Contributions
A. I. designed the study and wrote the paper. A. I., M. A., and T. O. carried out DNA cloning, the construction of the protein expression system, and enzymatic characterization. S. N. performed the culture of Nitratiruptor sp. SB155-2 and the extraction of genomic DNA.
This work was supported by the Program for Constructing “Tohoku Marine Science Bases” supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- rNitAly
- recombinant NitAly
- PEG-Mal
- methoxypolyethylene glycol maleimide.
References
- 1. Pawar S. N., and Edgar K. J. (2012) Alginate derivatization: a review of chemistry, properties and applications. Biomaterials 33, 3279–3305 [DOI] [PubMed] [Google Scholar]
- 2. Remminghorst U., and Rehm B. H. (2006) Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 28, 1701–1712 [DOI] [PubMed] [Google Scholar]
- 3. Michel G., Tonon T., Scornet D., Cock J. M., and Kloareg B. (2010) The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in eukaryotes. New Phytol. 188, 82–97 [DOI] [PubMed] [Google Scholar]
- 4. Boyd A., and Chakrabarty D. A. (1995) Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15, 162–168 [DOI] [PubMed] [Google Scholar]
- 5. Wong T. Y., Preston L. A., and Schiller N. L. (2000) Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54, 289–340 [DOI] [PubMed] [Google Scholar]
- 6. Takase R., Ochiai A., Mikami B., Hashimoto W., and Murata K. (2010) Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim. Biophys. Acta 1804, 1925–1936 [DOI] [PubMed] [Google Scholar]
- 7. Takase R., Mikami B., Kawai S., Murata K., and Hashimoto W. (2014) Structure-based conversion of the coenzyme requirement of a short-chain dehydrogenase/reductase involved in bacterial alginate metabolism. J. Biol. Chem. 289, 33198–33214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inoue A., Nishiyama R., Mochizuki S., and Ojima T. (2015) Identification of a 4-deoxy-l-erythro-5-hexoseulose uronic acid reductase, FlRed, in an alginolytic bacterium Flavobacterium sp. strain UMI-01. Mar. Drugs 13, 493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Preiss J., and Ashwell G. (1962) Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 237, 309–316 [PubMed] [Google Scholar]
- 10. Preiss J., and Ashwell G. (1962) Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J. Biol. Chem. 237, 317–321 [PubMed] [Google Scholar]
- 11. Albrecht M. T., and Schiller N. L. (2005) Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187, 3869–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costerton J. W., Stewart P. S., and Greenberg E. P. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 13. Michel G., Tonon T., Scornet D., Cock J. M., and Kloareg B. (2010) Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: insights into the origin and evolution of storage carbohydrates in Eukaryotes. New Phytol. 188, 67–81 [DOI] [PubMed] [Google Scholar]
- 14. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamasaki M., Moriwaki S., Miyake O., Hashimoto W., Murata K., and Mikami B. (2004) Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a β-sandwich fold. J. Biol. Chem. 279, 31863–31872 [DOI] [PubMed] [Google Scholar]
- 16. Gimmestad M., Ertesvåg H., Heggeset T. M., Aarstad O., Svanem B. I., and Valla S. (2009) Characterization of three new Azotobacter vinelandii alginate lyases, one of which is involved in cyst germination. J. Bacteriol. 191, 4845–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang L., Zhou J., Li X., Peng Q., Lu H., and Du Y. (2013) Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. J. Ind. Microbiol. Biotechnol. 40, 113–122 [DOI] [PubMed] [Google Scholar]
- 18. Inoue A., Takadono K., Nishiyama R., Tajima K., Kobayashi T., and Ojima T. (2014) Characterization of an alginate lyase, FlAlyA, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs. 12, 4693–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyake O., Ochiai A., Hashimoto W., and Murata K. (2004) Origin and diversity of alginate lyases of families PL-5 and -7 in Sphingomonas sp. strain A1. J. Bacteriol. 186, 2891–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoon H.-J., Hashimoto W., Miyake O., Murata K., and Mikami B. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J. Mol. Biol. 307, 9–16 [DOI] [PubMed] [Google Scholar]
- 21. Yamasaki M., Ogura K., Hashimoto W., Mikami B., and Murata K. (2005) A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 352, 11–21 [DOI] [PubMed] [Google Scholar]
- 22. Sugimura I. I., Sawabe T., and Ezura Y. (2000) Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar. Biotechnol. 2, 65–73 [DOI] [PubMed] [Google Scholar]
- 23. Han F., Gong Q.-H., Song K., Li J.-B., and Yu W.-G. (2004) Cloning, sequence analysis and expression of gene alyVI encoding alginate lyase from marine bacterium Vibrio sp. QY101. DNA Seq. 15, 344–350 [DOI] [PubMed] [Google Scholar]
- 24. Kawamoto H., Horibe A., Miki Y., Kimura T., Tanaka K., Nakagawa T., Kawamukai M., and Matsuda H. (2006) Cloning and sequencing analysis of alginate lyase genes from the marine bacterium Vibrio sp. O2. Mar. Biotechnol. 8, 481–490 [DOI] [PubMed] [Google Scholar]
- 25. Uchimura K., Miyazaki M., Nogi Y., Kobayashi T., and Horikoshi K. (2010) Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 12, 526–533 [DOI] [PubMed] [Google Scholar]
- 26. Zhu B., Tan H., Qin Y., Xu Q., Du Y., and Yin H. (2015) Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 75, 330–337 [DOI] [PubMed] [Google Scholar]
- 27. Kim H. K., Lee J. C., Kang N. H., Kim S. H., Kim J. G., and Chung K. C. (2007) Purification and characterization of the extracellular alginate lyase from Streptomyces sp. MET 0515. J. Life Sci. 17, 625–633 [Google Scholar]
- 28. Kim D. E., Lee E. Y., and Kim H. S. (2009) Cloning and characterization of alginate lyase from a marine bacterium Streptomyces sp. ALG-5. Mar. Biotechnol. 11, 10–16 [DOI] [PubMed] [Google Scholar]
- 29. Kim H. S. (2010) Characterization of recombinant polyG-specific lyase from a marine bacterium, Streptomyces sp. M3. J. Life Sci. 20, 1582–1588 [Google Scholar]
- 30. Matsubara Y., Iwasaki K., and Muramatsu T. (1998) Action of poly(α-l-guluronate)lyase from Corynebacterium sp. ALY-1 strain on saturated oligoguluronates. Biosci. Biotechnol. Biochem. 62, 1055–1060 [DOI] [PubMed] [Google Scholar]
- 31. Kobayashi T., Uchimura K., Miyazaki M., Nogi Y., and Horikoshi K. (2009) A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 13, 121–129 [DOI] [PubMed] [Google Scholar]
- 32. Li S., Yang X., Zhang L., Yu W., and Han F. (2015) Cloning, expression, and characterization of a cold-adapted and surfactant-stable alginate lyase from marine bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 25, 681–686 [DOI] [PubMed] [Google Scholar]
- 33. Duan G., Han F., and Yu W. (2009) Cloning, sequence analysis, and expression of gene alyPI encoding an alginate lyase from marine bacterium Pseudoalteromonas sp. CY24. Can. J. Microbiol. 55, 1113–1118 [DOI] [PubMed] [Google Scholar]
- 34. Brown B. J., Preston J. F., and Ingram L. O. (1991) Cloning of alginate lyase gene (alxM) and expression in Escherichia coli. Appl. Environ. Microbiol. 57, 1870–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baron A. J., Wong T. Y., Hicks S. J., Gacesa P., Willcock D., and McPherson M. J. (1994) Alginate lyase from Klebsiella pneumoniae, subsp. aerogenes: gene cloning, sequence analysis and high-level production in Escherichia coli. Gene 143, 61–66 [DOI] [PubMed] [Google Scholar]
- 36. Thomas F., Lundqvist L. C., Jam M., Jeudy A., Barbeyron T., Sandström C., Michel G., and Czjzek M. (2013) Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 288, 23021–23037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim H. T., Ko H.-J., Kim N., Kim D., Lee D., Choi I.-G., Woo H. C., Kim M. D., and Kim K. H. (2012) Characterization of a recombinant endo-type alginate lyase (Alg7D) from Saccharophagus degradans. Biotechnol. Lett. 34, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 38. Sim S.-J., Baik K. S., Park S. C., Choe H. N., Seong C. N., Shin T.-S., Woo H. C., Cho J.-Y., and Kim D. (2012) Characterization of alginate lyase gene using a metagenomic library constructed from the gut microflora of abalone. J. Ind. Microbiol. Biotechnol. 39, 585–593 [DOI] [PubMed] [Google Scholar]
- 39. Inoue A., Mashino C., Uji T., Saga N., Mikami K., and Ojima T. (2015) Characterization of an eukaryotic PL-7 alginate lyase in the marine red alga Pyropia yezoensis. Curr. Biotechnol. 4, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakagawa S., Takai K., Inagaki F., Hirayama H., Nunoura T., Horikoshi K., and Sako Y. (2005) Distribution, phylogenetic diversity and physiological characteristics of ϵ-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7, 1619–1632 [DOI] [PubMed] [Google Scholar]
- 41. Nakagawa S., Takaki Y., Shimamura S., Reysenbach A.-L., Takai K., and Horikoshi K. (2007) Deep-sea vent ϵ-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U.S.A. 104, 12146–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 43. Voordeckers J. W., Starovoytov V., and Vetriani C. (2005) Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 55, 773–779 [DOI] [PubMed] [Google Scholar]
- 44. Flores G. E., Hunter R. C., Liu Y., Mets A., Schouten S., and Reysenbach A. L. (2012) Hippea jasoniae sp. nov., and Hippea alviniae sp. nov., thermoacidophilic members of the class Deltaproteobacteria isolated from deep-sea hydrothermal vent deposits. Int. J. Syst. Evol. Microbiol. 62, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 45. Smith J. L., Campbell B. J., Hanson T. E., Zhang C. L., and Cary S. C. (2008) Nautilia profundicola sp. nov., a thermophilic, sulfur-reducing ϵ-proteobacterium from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 58, 1598–1602 [DOI] [PubMed] [Google Scholar]
- 46. Fata Moradali M., Donati I., Sims I. M., Ghods S., and Rehm B. H. (2015) Alginate polymerization and modification are linked in Pseudomonas aeruginosa. MBio. 6, e00453–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gutsche J., Remminghorst U., and Rehm B. H. (2006) Biochemical analysis of alginate biosynthesis protein AlgX from Pseudomonas aeruginosa: purification of an AlgX-MucD (AlgY) protein complex. Biochimie 88, 245–251 [DOI] [PubMed] [Google Scholar]
- 48. Inoue A., Nishiyama R., and Ojima T. (2016) The alginate lyases FlAlyA, FlAlyB, FlAlyC, and FlAlex from Flavobacterium sp. UMI-01 have distinct roles in the complete degradation of alginate. Algal Res. 10.1016/j.algal.2016.03.008 [DOI] [Google Scholar]
- 49. Inoue A., Mashino C., Kodama T., and Ojima T. (2011) Protoplast preparation from Laminaria japonica with recombinant alginate lyase and cellulase. Mar. Biotechnol. 13, 256–263 [DOI] [PubMed] [Google Scholar]
- 50. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., and Lappin-Scott H. M. (1995) Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 [DOI] [PubMed] [Google Scholar]
- 51. Haug A., and Smidsrod O. (1967) Strontium–calcium selectivity of alginates. Nature 215, 757–757 [DOI] [PubMed] [Google Scholar]
- 52. Haug A., Smidsrød O., Högdahl B., Øye H. A., Rasmussen S. E., Sunde E., and Sørensen N. A. (1970) Selectivity of some anionic polymers for divalent metal ions. Acta Chem. Scand. 24, 843–854 [Google Scholar]
- 53. Grant G. T., Morris E. R., Rees D. A., Smith P. J., and Thom D. (1973) Biological interactions between polysaccharides and divalent cations: the egg-box model. FEBS Lett. 32, 195–198 [Google Scholar]
- 54. Smidsrød O. (1974) Molecular basis for some physical properties of alginates in the gel state. Farad. Discuss. 57, 263–274 [Google Scholar]
- 55. Oates C. G., and Ledward D. A. (1990) Studies on the effect of heat on alginates. Food Hydrocoll. 4, 215–220 [Google Scholar]
- 56. Kelley Deborah S., Baross John A., and Delaney J. R. (2003) Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu. Rev. Earth Planet. Sci. 30, 385–491 [Google Scholar]
- 57. Schwertfeger M. (1999) in Advances in High Pressure Bioscience and Biotechnology (Ludwig H., ed) pp. 337–340, Springer, Berlin [Google Scholar]
- 58. Gacesa P., and Wusteman F. S. (1990) Plate assay for simultaneous detection of alginate lyases and determination of substrate specificity. Appl. Environ. Microbiol. 56, 2265–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 60. Porzio M. A., and Pearson A. M. (1977) Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim. Biophys. Acta 490, 27–34 [DOI] [PubMed] [Google Scholar]
- 61. Le M., and Means G. E. (1995) A procedure for the determination of monothiols in the presence of dithiothreitol–an improved assay for the reduction of disulfides. Anal. Biochem. 229, 264–271 [DOI] [PubMed] [Google Scholar]
- 62. Weissbach A., and Hurwitz J. (1959) The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. J. Biol. Chem. 234, 705–709 [PubMed] [Google Scholar]
- 63. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McNicholas S., Potterton E., Wilson K. S., and Noble M. E. (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Osawa T., Matsubara Y., Muramatsu T., Kimura M., and Kakuta Y. (2005) Crystal structure of the alginate (poly α-l-guluronate) lyase from Corynebacterium sp. at 1.2Å resolution. J. Mol. Biol. 345, 1111–1118 [DOI] [PubMed] [Google Scholar]