Abstract
In higher plants, L-galactono-1,4-lactone dehydrogenase (GLDH) plays important roles in ascorbic acid (AsA) biosynthesis and assembly of respiration complex I. Here we report three homoeologous genes (TaGLDH-A1, -B1 and -D1) encoding common wheat GLDH isozymes and a unique allelic variant (TaGLDH-A1b) associated with enhanced drought tolerance. TaGLDH-A1, -B1 and -D1 were located on chromosomes 5A, 5B and 5D, respectively, and their transcripts were found in multiple organs. The three homoeologs each conferred increased GLDH activity when ectopically expressed in tobacco. Decreasing TaGLDH expression in wheat significantly reduced GLDH activity and AsA content. TaGLDH-A1b differed from wild type allele TaGLDH-A1a by an in-frame deletion of three nucleotides. TaGLDH-A1b was biochemically less active than TaGLDH-A1a, and the total GLDH activity levels were generally lower in the cultivars carrying TaGLDH-A1b relative to those with TaGLDH-A1a. Interestingly, TaGLDH-A1b cultivars showed stronger water deficiency tolerance than TaGLDH-A1a cultivars, and TaGLDH-A1b co-segregated with decreased leaf water loss in a F2 population. Finally, TaGLDH-A1b cultivars generally exhibited smaller leaf stomatal aperture than TaGLDH-A1a varieties in control or water deficiency environments. Our work provides new information on GLDH genes and function in higher plants. TaGLDH-A1b is likely useful for further studying and improving wheat tolerance to drought stress.
Drought is a major abiotic stress decreasing global crop production and food security1,2. It may become even more severe owing to global warming3. Consequently, understanding and improving crop tolerance to drought is a top target in current plant biology and breeding research1,2,4,5. Drought tolerance is a complex trait controlled by polygenes, involving multiple physiological and developmental processes and affected by the environment1,2. The advent of molecular genetic and functional genomic studies has substantially improved understanding on drought response in model plants (e.g., Arabidopsis thaliana and rice)5,6,7. However, much less is known about the genetic and molecular basis of drought tolerance in crop species1,2. This is particularly evident for common wheat (Triticum aestivum, 2n = 6x = 42, AABBDD), which carries a large and complex hexaploid genome and has only limited functional genomic resources for efficiently studying the molecular basis of drought tolerance and other agronomic traits8,9.
L-galactono-1,4-lactone dehydrogenase (GLDH) (EC 1.3.2.3) is a mitochondrion located enzyme found in many eukaryotic organisms10,11,12,13. In higher plants, GLDH is essential for the synthesis of ascorbic acid (AsA), a vital and abundant antioxidant, through the D-mannose/L-galactose pathway14. GLDH acts in the final step of this pathway, converting L-galactono-1,4-lactone (L-GalL) into AsA14. GLDH has also been found required for the assembly and accumulation of plant respiratory complex I (RCI)10,15. Through transgenic studies in multiple plant species, GLDH activities have been shown required for the normal growth and development of plant cells and organs and their efficient response to adverse environmental factors. For example, in both tobacco and tomato, suppression of GLDH function results in decreased cell and organ growth16,17. In rice, inhibition of GLDH activity leads to decreased AsA content, with the transgenic plants exhibiting reductions in seed set and tiller number18,19. In Arabidopsis, our study revealed that partial reduction of GLDH expression through RNA interference not only lowers AsA content, but also causes a substantial decrease of foliar water loss rate and potential tolerance to drought stress20. Overexpression of GLDH confers enhanced tolerance to oxidative stress in tobacco cells21 and to salt stress in tobacco plants22. In the studies above, changes in GLDH expression and activity levels are mainly rendered using transgenic technology. To date, there is still no published information on natural molecular variations of GLDH genes and their potential influences on agronomic traits. One study has revealed that GLDH protein and activity levels differ significantly between two common wheat cultivars with high (cv Buck Chambergo, BCH) or low (cv Cooperativa Maipún, CM) AsA content, and that drought treatment can substantially up-regulate the amounts of GLDH protein and activity in CM13. However, the molecular basis underlying GLDH expression difference between the two varieties is still unclear, and the functional importance of such difference in wheat drought tolerance requires further study.
The findings described by Bartoli and colleagues13 and the observation that partial suppression of Arabidopsis GLDH activity led to enhanced drought tolerance20 stimulated us to further study the potential involvement of GLDH in common wheat response to water deficiency stress. Towards this end, we characterized the three homoeologous genes (designated TaGLDH-A1, -B1 and -D1, respectively) encoding common wheat GLDH isozymes, tested their GLDH activity through viral vector mediated ectopic expression in Nicotiana benthamiana, analyzed their function in AsA biosynthesis using virus induced gene silencing (VIGS), and examined their molecular variations in common wheat germplasm lines. On the basis of these experiments, we comparatively analyzed two alleles of TaGLDH-A1, the major wild type (WT) allele TaGLDH-A1a and a rare allele TaGLDH-A1b, in more detail, and found that TaGLDH-A1b was associated with the enhanced drought tolerance of a number of varieties cultivated in arid regions. Our work provides new information on GLDH encoding genes and their function in a polyploid crop species, and demonstrates the association between GLDH and plant tolerance to drought stress.
Results
Characteristics of TaGLDH-A1, -B1 and -D1 homoeologs
To investigate the genes encoding TaGLDH-A1, -B1 and -D1 isozymes in common wheat, a bacterial artificial chromosome (BAC) library prepared for the bread wheat variety Xiaoyan 54 was screened by PCR (see Methods). A total of seven unique positive BAC clones containing TaGLDH sequence were identified. Three similar but not identical TaGLDH genomic open reading frames (ORFs) were amplified from the seven BAC clones, and completely sequenced. The genes represented by the three ORFs were designated as TaGLDH-A1, -B1 and -D1 based on their chromosomal locations (see below). The complete genomic ORF of TaGLDH-A1, -B1 and -D1 (from start to stop codons) was 5498, 5685 or 5526 bp, and each was composed of six exons and five introns (Fig. 1). This exon-intron pattern was also found for the GLDH genes in other monocot (e.g., rice and Brachypodium distachyon) and dicot (e.g., Arabidopsis thaliana) plants (Fig. 1). The ORF sequences of TaGLDH-A1, -B1 and -D1 were 97% identical, with nucleotide differences found mainly in the introns. The coding region sequences of TaGLDH-A1, -B1 and -D1, as deduced from their cDNA clones, were all 1752 bp (excluding the stop codon) and 99% identical. The deduced proteins of the three homoeologs all contained 584 residues, and were 98–99% identical. Comparison with previously characterized GLDH proteins from Arabidopsis, cauliflower, sweet potato and tobacco indicated the presence of a mitochondrial targeting signal peptide (78 residues) and conservation of the residues involved in FAD-binding and those taking part in substrate recognition and catalysis in TaGLDH-A1, -B1 and -D1 (Fig. 2). The calculated molecular mass was 66.1 kD for TaGLDH-A1, -B1 and -D1 preproteins, and 57.5 kD for their putative mature proteins (after removing the putative mitochondrial targeting signal peptide).
Figure 1. Conservation of exon and intron structure among TaGLDH-A1, -B1 and -D1 homoeologs and orthologous GLDH genes in rice (OsGLDH1 and OsGLDH2), B. distachyon (BdGLDH) and Arabidopsis (AtGLDH).
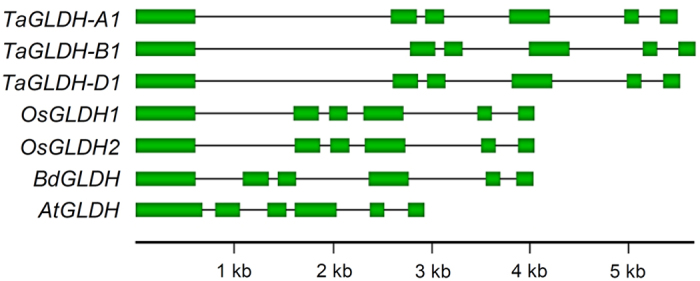
The chromosomal loci of OsGLDH1, OsGLDH2, BdGLDH and AtGLDH are Os11g0143500, Os12g0139600, Bradi4g43070 and At3g47930, respectively. The GenBank accession numbers for TaGLDH-A1, -B1 and -D1 are KU695146, KU695147 and KU695148, respectively.
Figure 2. Multiple alignment of the deduced amino acid sequences of TaGLDH-A1, -B1 and -D1 homoeologs and orthologous GLDH proteins from Arabidopsis (AtGLDH), cauliflower (BoGLDH), sweet potato (IbGLDH) and tobacco (NtGLDH).
Arrow indicates the likely cleavage site of the mitochondrial targeting peptide. The underlined sequence element is required for FAD binding. The conserved cysteine residue labeled in purple is involved in regulating the enzyme activity of plant GLDH proteins, whereas the E-R pair of residues marked in blue are required for efficient substrate binding and catalysis. The glutamic acid (E) marked in red is deleted in a novel allele (TaGLDH-A1b) of TaGLDH-A1. Asterisks denote residues conserved among the compared GLDHs. The UniProtKB numbers for AtGLDH, BoGLDH, IbGLDH and NtGLDH are Q8GY16, O47881, Q9ZWJ1 and Q9FXL9, respectively.
To find the chromosome locations of TaGLDH-A1, -B1 and -D1, PCR mapping was carried out using the amplicons yielded by primer sets PS3 and PS4 (Table S1, Figure S1). In the PCR with the genomic DNA of Xiaoyan 54 or Chinese Spring (CS) as templates, both PS3 and PS4 gave rise to three amplicons with variable size diagnostic for TaGLDH-A1, -B1 and -D1 (Figure S2). Through examining amplicons from the nulli-tetrasomic (NT) lines of CS23, TaGLDH-A1, -B1 and -D1 were assigned to 5A, 5B and 5D chromosomes, respectively (Figure S2). Consistent with this finding, three different GLDH genes (Traes_5AL_E7806F3C1, Traes_5BL_E8D36EEA8 and Traes_5DL_696F6D975) have been annotated in the draft genome sequence of CS24. These three genes are located on the long arms of 5A, 5B and 5D, respectively. The deduced amino acid sequences of Traes_5BL_E8D36EEA8 and Traes_5DL_696F6D975 are identical to those of TaGLDH-B1 and -D1, respectively. However, the gene model annotated for Traes_5AL_E7806F3C1 is partial because the polypeptide deduced from it contains only 400 amino acids. Nevertheless, this hypothetical polypeptide is nearly identical to the last 400 residues of TaGLDH-A1.
Expression pattern of TaGLDH in common wheat organs
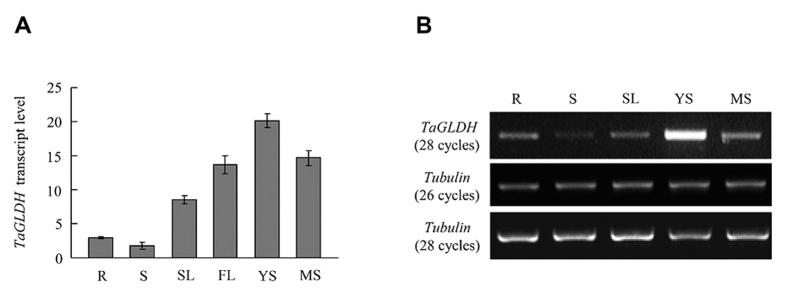
As revealed by qRT-PCR (Fig. 3A), the expression level of TaGLDH was highest in the young spikelets, intermediate in the seedling leaves, flag leaves and mature spikelets, and comparatively low in the roots and stems. The relatively high expression level of TaGLDH in young spikelets was also found by semi-quantitative RT-PCR assay comparing TaGLDH transcript levels in five vegetative and reproductive organs of common wheat (Fig. 3B).
Figure 3.
Analysis of relative transcript levels of TaGLDH in different common wheat organs using qRT-PCR (A) and semi-quantitative RT-PCR (B). R: root; S: stem; SL: seedling leaf; FL: flag leaf; YS: young spikelet; MS: mature spikelet. The values shown in (A) are means ± SD of three biological replicates. In (B), a wheat tubulin gene (GenBank accession U76745) was used as the internal control. Three separate semi-quantitative RT-PCR assays were conducted with highly similar results obtained.
GLDH activity of TaGLDH-A1, -B1 and -D1
To examine if TaGLDH-A1, -B1 and -D1 may possess GLDH activity, they were individually expressed in N. benthamiana using the viral vector based on pea early browning virus (PEBV, see Methods). Four recombinant viruses (PEBV:GUS, PEBV:A1, PEBV:B1 and PEBV:D1, Figure S3a) were constructed and introduced into N. benthamiana plants. Two weeks after the inoculation of PEBV:GUS, abundant β-glucuronidase (GUS) staining signals were detected in the inoculated leaves (Figure S3b), thus confirming the capacity of PEBV vector to mediate high level of ectopic expression of heterologous coding sequence in N. benthamiana. Subsequently, the inoculated leaves in the plants infected by PEBV:GUS, PEBV:A1, PEBV:B1 or PEBV:D1 were collected and examined for total GLDH enzyme activity. Additionally, the mock-inoculated leaves in the control plants were also analyzed for GLDH activity. From Fig. 4A, it is clear that GLDH activity level was significantly higher in the leaves infected by PEBV:A1, PEBV:B1 or PEBV:D1 than in those infected by PEBV:GUS, but there was no significant difference in GLDH activity level between PEBV:GUS infected leaves and the mock-inoculated controls. However, the total AsA content did not differ significantly among the examined leaf samples (Fig. 4B).
Figure 4. Ectopic expression of TaGLDH-A1, -B1 and -D1 homoeologs in N. benthamiana using PEBV viral vector.
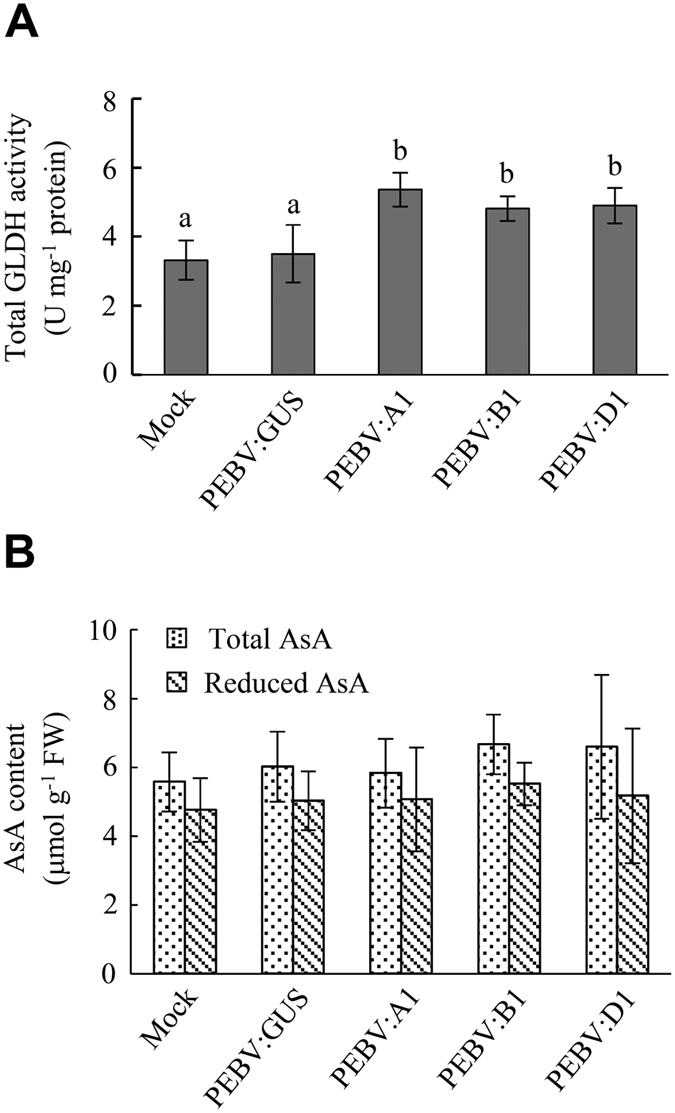
PEBV:A1, PEBV:B1 and PEBV:D1 are three recombinant viruses expressing TaGLDH-A1, -B1 and -D1, respectively. PEBV:GUS is another recombinant virus expressing a bacterial GUS gene (encoding β-glucuronidase). (A) Total GLDH activity levels in mock controls and the plants infected by different recombinant viruses. The values displayed are means ± SD of three measurements for three different plants. Different letters above the histograms indicate statistical difference (P < 0.05). (B) Total and reduced AsA contents in mock controls and the plants infected by different recombinant viruses. The contents shown are means ± SD of three measurements of three different plants. Two independent experiments were performed with similar results obtained.
Function of TaGLDH in AsA biosynthesis
To investigate the involvement of TaGLDH in AsA biosynthesis in common wheat, the effects of decreasing TaGLDH expression on AsA content were analyzed. The reduction of TaGLDH expression was achieved by VIGS with the vector derived from barley stripe mosaic virus (BSMV) (see Methods). Among the three recombinant BSMVs (BSMV:GFP, BSMV:PDSas and BSMV:GLDHas, Figure S4a) used in this analysis, BSMV:GLDHas was developed to specifically silence all three TaGLDH homoeologs in common wheat. A 431 bp sequence element strictly conserved among TaGLDH-A1, -B1 and -D1 was employed to construct BSMV:GLDHas. This element recognized only the three GLDH genes (Traes_5AL_E7806F3C1, Traes_5BL_E8D36EEA8 and Traes_5DL_696F6D975, see above) in the draft genome sequence of CS. BSMV:GFP expresses green fluorescent protein (GFP) in wheat cells, and is useful for monitoring virus spread in the inoculated seedlings. BSMV:PDSas, silencing wheat phytoene desaturase (PDS) gene and causing leaf bleaching, is valuable for checking the progress of VIGS.
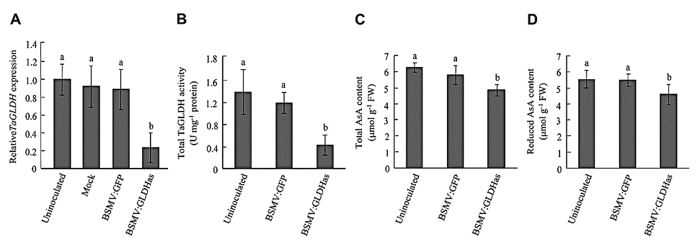
Two weeks after virus inoculation, strong GFP fluorescence was observed in the leaf tissues of the seedlings infected with BSMV:GFP (Figure S4b), and apparent leaf bleaching appeared in those infected by BSMV:PDSas (Figure S4c), thus verifying the effectiveness of BSMV VIGS in this work. In the seedlings infected by BSMV:GLDHas, the expression level of TaGLDH was significantly reduced (by >75%) relative to that detected for the controls (uninoculated, mock-inoculated or BSMV:GFP infected seedlings, Fig. 5A). In line with this finding, total TaGLDH activity level in BSMV:GLDHas-infected seedlings was substantially decreased (by >70%) relative to that in the controls (mock-inoculated or infected by BSMV:GFP, Fig. 5B). Silencing TaGLDH expression caused significant reductions of total AsA (Fig. 5C) and reduced AsA (Fig. 5D) contents in the plants infected by BSMV:GLDHas, but for both parameters the reduction was only about 20%.
Figure 5. Function of TaGLDH in AsA biosynthesis as investigated using BSMV mediated VIGS.
BSMV:GFP, expressing a green fluorescence protein, was used for monitoring viral spread in infected wheat plants, whereas BSMV:GLDHas was for silencing TaGLDH transcripts. (A) Comparison of relative TaGLDH expression levels among different plant samples, with TaGLDH expression in the uninoculated plants set as 1. TaGLDH expression level was assayed by qRT-PCR. Significant decrease of TaGLDH expression was detected in the plants infected by BSMV:GLDHas but not in the mock controls or those infected by BSMV:GFP. (B–D) Significant reductions of TaGLDH activity and the contents of total and reduced AsA in the plants infected by BSMV:GLDHas but not in those infected by BSMV:GFP. The values depicted are all means ± SD of three measurements for three different plants. Different letters above the histograms mark statistical difference (P < 0.05). The assays were repeated at least twice with similar results found.
Past studies have shown that exogenous application of L-GalL can substantially stimulate AsA biosynthesis in plant tissues25,26. Thus, the small decrease of AsA content in the plants infected by BSMV:GLDHas might be due to an insufficient amount of L-GalL available for AsA biosynthesis in the cells. To investigate this possibility, we exogenously applied 15 mM L-GalL to the leaf segments excised from BSMV:GLDHas infected seedlings and the controls, followed by measuring total and reduced AsA contents. For the controls (mock-inoculated or infected with BSMV:GFP), the leaf segments treated with L-GalL exhibited dramatically increased total and reduced AsA contents (by 6.32–6.63 times) relative to those without exogenous application of the substrate (Figure S5). For the plants infected by BSMV:GLDHas, treatment with L-GalL also resulted in increases of total and reduced AsA contents but at much lower levels (by 3.83–3.98 times, Figure S5). In the presence of 15 mM L-GalL, the total and reduced AsA contents in BSMV:GLDHas infected leaf tissues were reduced by about 42% relative to those of the controls (Figure S5). This level of reduction was substantially higher than that found for the BSMV:GLDHas infected leaf tissues (~20%, Fig. 5C,D) in the absence of artificially supplied L-GalL.
Identification and characterization of TaGLDH-A1b
Potential molecular variations in the coding region of TaGLDH-A1, -B1 or -D1 in the micro-core collection (MCC) of Chinese wheat germplasm were examined by genomic PCR with the primer sets PS3 and PS4 (Table S1, Figure S1), respectively. The 262 MCC lines, including both land races and improved varieties, represent 70% of the genetic diversity of Chinese common wheat27. No length variation was detected in the amplicons produced by PS3. However, in the amplicons by PS4, length variation was found for TaGLDH-A1 in four unique cultivars (i.e., Hongdongmai, Kashi 1, Kashibaipi and Tutoumai, Figure S6). The size of TaGLDH-A1 amplicons in the four lines was 452 bp, which was 3 bp shorter than those from Xiaoyan 54 and the remaining MCC lines (Figure S6). Sequencing the full-length genomic ORF and cDNA coding region of TaGLDH-A1 in the four cultivars revealed an in-frame deletion of three nucleotides (GAG) compared to TaGLDH-A1 sequence in Xiaoyan 54. However, the four lines did not differ from Xiaoyan 54 in either the coding sequence of TaGLDH-B1 or that of TaGLDH-D1. To facilitate further characterization, the TaGLDH-A1 sequence in Xiaoyan 54 was designated as the WT allele TaGLDH-A1a, whereas that in Hongdongmai, Kashi 1, Kashibaipi and Tutoumai was named as TaGLDH-A1b. Comparison of deduced proteins showed that the glutamic acid residue at position 501 of TaGLDH-A1a was absent in TaGLDH-A1b (Fig. 2). This glutamic acid was 42 residues away from the conserved Glu-Arg pair (Fig. 2). These two residues have been suggested to reside in the active site of plant GLDH proteins, and are thus essential for optimal function of these enzymes28.
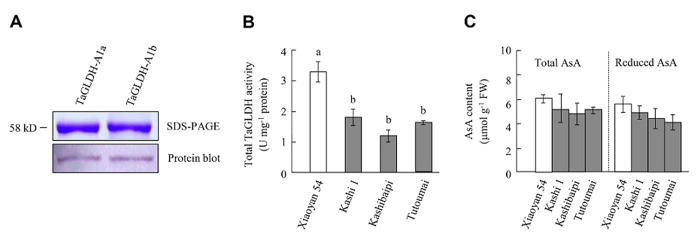
Potential biochemical differences between the proteins specified by TaGLDH-A1a or TaGLDH-A1b were examined. TaGLDH-A1a and TaGLDH-A1b, lacking the sequence coding for the putative mitochondrion targeting peptide, were individually expressed in Escherichia coli using the bacterial expression vector pET-30a (see Methods), with the aim to produce recombinant TaGLDH-A1a and TaGLDH-A1b proteins without the mitochondrial targeting peptide but with a histidine tag at the N-terminus. The recombinant proteins were purified, and verified by protein blot analysis with an antibody specific for the histidine tag (Fig. 6A). Subsequently, the kinetic properties of purified TaGLDH-A1a and TaGLDH-A1b were investigated using L-GalL as a substrate. Based on their Michaelis-Menten kinetic characteristics, the mean Km and Vmax of TaGLDH-A1a were estimated to be 61 μM and 11.43 μmol. mg−1 protein.min−1, respectively, and the corresponding values for TaGLDH-A1b were 181 μM and 0.16 μmol.mg−1protein.min−1, respectively (Table 1). These data indicated that TaGLDH-A1b had significantly reduced kinetic activities than TaGLDH-A1a towards the substrate tested. In line with the biochemical difference between TaGLDH-A1a and TaGLDH-A1b, total TaGLDH activity levels in the TaGLDH-A1b lines were generally and significantly lower than that found for Xiaoyan 54 carrying TaGLDH-A1a (Fig. 6B). Under normal growth conditions, total AsA contents in the seedling leaves of the three TaGLDH-A1b lines tended to be lower than that of Xiaoyan 54, but the differences were not statistically significant (Fig. 6C).
Figure 6. Analysis of TaGLDH-A1a and TaGLDH-A1b alleles.
(A) SDS-PAGE and protein blot assays of TaGLDH-A1a and TaGLDH-A1b recombinant proteins. The two alleles were expressed in E. coli cells without the putative mitochondrial targeting peptide but with a histidine tag at the N-terminal end, followed by purification with nickel affinity chromatography and the protein blot assay using an antibody specific for histidine tag. (B,C) Comparison of total TaGLDH activity levels and the contents of total and reduced AsA in Xiaoyan 54 (carrying TaGLDH-A1a) and the three Xinjiang wheat cultivars (Kashi 1, Kashibaipi and Tutoumai, with TaGLDH-A1b). The values are means ± SD of three measurements for three different plants. Different letters above the histograms indicate statistical difference (P < 0.05). For both (B,C), the data displayed are typical of three independent assays.
Table 1. Kinetic parameters of recombinant TaGLDH-A1a and -A1b proteinsa.
| Recombinant protein | Km (μM) | Vmax (μmol.mg−1protein.min−1) |
|---|---|---|
| TaGLDH-A1a | 61.0 ± 5.4 | 11.43 ± 0.88 |
| TaGLDH-A1b | 181.0 ± 67.1* | 0.16 ± 0.04*** |
aValues are means ± SD of three independent assays. Statistical analysis between the Km or Vmax values was conducted with ANOVA. *Indicates significant difference from the Km of TaGLDH-A1a at P < 0.05. ***Indicates significant difference from the Vmax of TaGLDH-A1a at P < 0.001.
Association of TaGLDH-A1b with enhanced tolerance to water deficiency stress
The four cultivars carrying TaGLDH-A1b all came from the Xinjiang Uygur Autonomous Region (Xinjiang hereafter) in Northwest China29. The common wheat lines cultivated in Xinjiang generally have enhanced drought tolerance because this region has a strong continental climate with an annual rainfall varying from about 40 mm (in the south) to 192 mm (towards the north)29,30. Therefore, we tested if TaGLDH-A1b might be found in an additional set of 11 wheat cultivars from Xinjiang (Table S2) with a newly developed molecular marker CAP-A1b (Figure S7a). The PCR fragments derived from TaGLDH-A1a (538 bp) and TaGLDH-A1b (535 bp) were digested using the restriction enzyme BsaJI, which cleaved the amplicon of TaGLDH-A1a once (yielding two fragments of 266 and 272 bp, respectively) but not that of TaGLDH-A1b (Figure S7a). Among the 11 cultivars, three lines (Jiuchun 1, Redstar and Xinchun 3) were detected to have TaGLDH-A1b, with the remaining ones carrying TaGLDH-A1a (Table S2). Together, seven cultivars carrying TaGLDH-A1b were found among a total of 273 germplasm lines examined in this study. The frequency of TaGLDH-A1b was thus 2.56%. The seven TaGLDH-A1b lines are all known to have enhanced tolerance to abiotic stresses (Table S3), and four of them (Hongdongmai, Kashibaipi, Redstar, and Xinchun 3) have been extensively cultivated in Xinjiang29. One of the TaGLDH-A1b lines, Redstar, was originally from the former Soviet Union29, whereas the remaining ones were native to Xinjiang (Table S3).
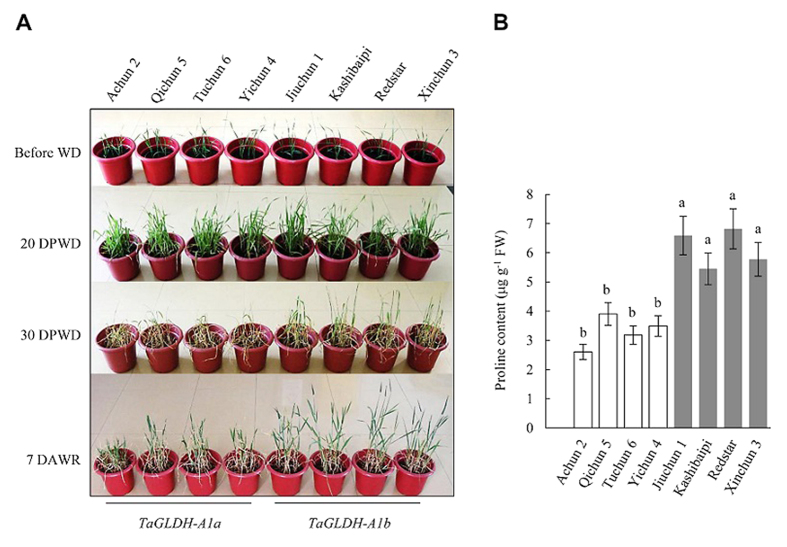
Next, we performed three independent pot trials for comparing the drought tolerance of eight Xinjiang cultivars, four carrying TaGLDH-A1a and four with TaGLDH-A1b. The plants of the eight lines were deprived of water for 30 days followed by rewatering, with their growth behavior recorded at specific time points. At 20 days post water deprivation (DPWD), the four lines with TaGLDH-A1b were apparently greener than those carrying TaGLDH-A1a (Fig. 7A). At 30 DPWD (just before rewatering), the four TaGLDH-A1a lines exhibited severe wilting, but the wilting was much less severe in the four TaGLDH-A1b lines (Fig. 7A). At the 7th day after rewatering, the TaGLDH-A1b lines were obviously taller and had more green leaves than the ones with TaGLDH-A1a (Fig. 7A), indicating that the former lines recovered from water deficiency stress more efficiently than the latter ones. Free proline content is an important indicator of plant drought tolerance31. At 20 DPWD, free proline contents in the leaf tissues of TaGLDH-A1b lines were generally and significantly higher than those of TaGLDH-A1a lines (Fig. 7B). The plants of the eight cultivars, whether carrying TaGLDH-A1a or TaGLDH-A1b, all survived under the above drought treatment conditions; no difference was observed in the survival rate between the two types of lines in the three pot trials.
Figure 7. Different responses to water deprivation between the wheat cultivars carrying TaGLDH-A1a or TaGLDH-A1b.
Among the eight Xinjiang wheat cultivars tested, Achun 2, Qichun 5, Tuchun 6 and Yichun 4 had TaGLDH-A1a whereas Jiuchun 1, Kashibaipi, Redstar and Xinchun 3 possessed TaGLDH-A1b. (A) Growth performance of the two types of Xinjiang wheat cultivars before water deprivation (WD), at 20 and 30 days post water deprivation (DPWD), and at 7 days after water resumption (DAWR). The data shown are typical of three separate growth trials. (B) Free proline contents in the leaf tissues of the eight cultivars at 20 DPWD. Each value is the mean ± SD of five measurements using five different plants. Different letters above the histograms indicate statistical difference (P < 0.05). The data are typical of two independent assays.
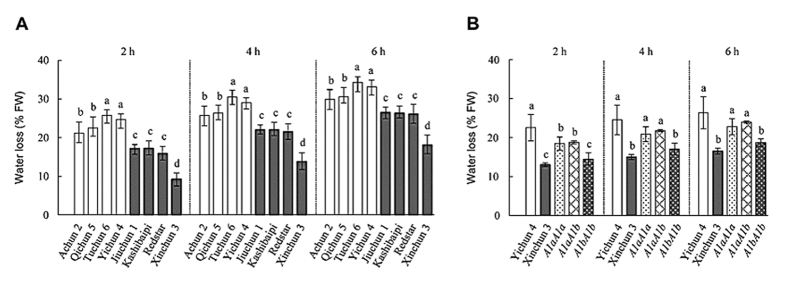
Lastly, we compared the degree of leaf water loss between TaGLDH-A1a and TaGLDH-A1b lines because this parameter is another valuable indicator of plant response to water deficiency stress2,5. Water loss behavior of the leaves was monitored at 2, 4 and 6 h post detachment, respectively, for the two groups of lines carrying TaGLDH-A1a or TaGLDH-A1b. At all three time points, the levels of water loss recorded for the TaGLDH-A1b lines were consistently and significantly lower than those found for the TaGLDH-A1a lines (Fig. 8A). Furthermore, we investigated if there would be differences in water loss behavior among the different genotypes identified in a F2 population developed using two Xinjiang cultivars Yichun 4 (carrying TaGLDH-A1a) and Xinchun 3 (with TaGLDH-A1b) (Table S2). After genotyping 134 F2 individuals with the DNA marker CAP-A1b (Figure S7b), the individuals homozygous for TaGLDH-A1a or TaGLDH-A1b were 35 and 30, respectively, and those heterozygous at TaGLDH-A1 locus were 69. The levels of water loss determined for Xinchun 3 or TaGLDH-A1b/TaGLDH-A1b leaves were highly similar, both being significantly lower than those obtained for Yichun 4, TaGLDH-A1a/TaGLDH-A1a or TaGLDH-A1a/TaGLDH-A1b leaves (Fig. 8B). The water loss levels of Yichun 4, TaGLDH-A1a/TaGLDH-A1a and TaGLDH-A1a/TaGLDH-A1b differed only at 2 h post detachment (Fig. 8B).
Figure 8. Effects of TaGLDH-A1a and TaGLDH-A1b on leaf water loss value.
Among the eight Xinjiang wheat cultivars used, Achun 2, Qichun 5, Tuchun 6 and Yichun 4 had TaGLDH-A1a whereas Jiuchun 1, Kashibaipi, Redstar and Xinchun 3 possessed TaGLDH-A1b. (A) Water loss values of the eight cultivars determined at 2, 4 and 6 h after leaf detachment, respectively. The four cultivars with TaGLDH-A1b exhibited significantly reduced water loss (filled bars) than those carrying TaGLDH-A1a (open bars). The data shown are representative of three independent assays. (B) Water loss values scored at three different time points after leaf detachment (2, 4 and 6 h) for Yichun 4, Xinchun 3 and the F2 seedlings with A1aA1a (homozygous for TaGLDH-A1a), A1aA1b (heterozygous) or A1bA1b (homozygous for TaGLDH-A1b) genotype. The data depicted are representative of two independent assays. In both (A,B), each water loss value is the mean ± SD of three measurements using a pool of eight fully expanded leaves detached from eight separate plants. The histograms labelled by unidentical letters are statistically different (P < 0.05).
Comparison of stomatal aperture between TaGLDH-A1a and TaGLDH-A1b cultivars
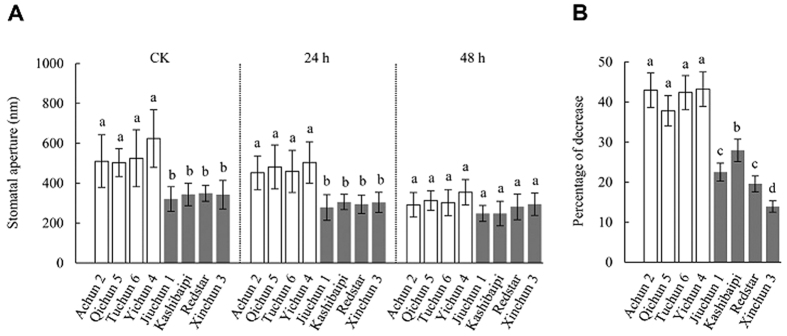
The drought tolerance exhibited by TaGLDH-A1b cultivars, especially their comparatively lower water loss levels, prompted us to compare stomatal aperture size between TaGLDH-A1a and TaGLDH-A1b cultivars. Under control conditions, the mean stomatal aperture size of TaGLDH-A1b lines (320–349 nm) was substantially lower than that of TaGLDH-A1a lines (503–623 nm) (Fig. 9A). Water deficiency stress, imposed by 13% polyethylene glycol 6000 (PEG 6000), caused decreases of stomatal aperture size for both types of lines, with the percentages of the decrease being much stronger for TaGLDH-A1a lines (Fig. 9B). After 48 h PEG 6000 treatment, the percentages of stomatal aperture size reduction shown by TaGLDH-A1b lines (13.8–27.9%) were generally and significantly lower than those of TaGLDH-A1a lines (37.8–43.2%) (Fig. 9B).
Figure 9. Effects of TaGLDH-A1a and TaGLDH-A1b on leaf stomatal aperture.
Among the eight Xinjiang wheat cultivars examined, Achun 2, Qichun 5, Tuchun 6 and Yichun 4 carried TaGLDH-A1a whereas Jiuchun 1, Kashibaipi, Redstar and Xinchun 3 had TaGLDH-A1b. (A) Stomatal aperture values measured for control plants (CK) and those treated with PEG 6000 for 24 and 48 h, respectively. The four cultivars with TaGLDH-A1b showed significantly reduced stomatal aperture (filled bars) than those carrying TaGLDH-A1a (open bars) under control conditions and after PEG 6000 treatment for 24 h. Each stomatal aperture value is the mean ± SD of the measurements of at least 200 stomata in six epidermal peels from three plants. The histograms labeled by unidentical letters are statistically different (P < 0.05). The data shown are typical of three independent experiments. (B) Percentages of stomatal aperture decrease after PEG 6000 treatment for 48 h in the eight cultivars. The percentages were calculated using the data depicted in (A). Relative to TaGLDH-A1a lines, the extents of stomatal aperture decrease in those with TaGLDH-A1b (filled bars) were generally and significantly lower.
Discussion
Although understanding and enhancing drought tolerance represent a major task in current plant biology and crop improvement research, only a few genes have so far been molecularly characterized for their involvement in wheat drought tolerance9,32,33,34 Here, we investigated the function of TaGLDH homoeologs and explored the association of a previously undescribed TaGLDH-A1 allele (TaGLDH-A1b) with enhanced drought tolerance by combining molecular, biochemical and physiological approaches. The new information obtained is discussed below.
Molecular features of TaGLDH-A1, -B1 and -D1 homoeologs
Prior to this work, little is known about TaGLDH homoeologs and their protein products at the molecular level, although there is evidence for TaGLDH protein expression and enzyme activity in common wheat13. Based on the data gathered here, several suggestions can be made on the molecular features of TaGLDH homoeologs. First, TaGLDH-A1, -B1 and -D1 are located on wheat group 5 chromosomes. This finding is consistent with the annotation of three TaGLDH homoeologous genes on 5A, 5B and 5D chromosomes, respectively, by the draft genome sequence of CS24. Second, the exon and intron pattern in TaGLDH-A1, -B1 and -D1 is identical to that found for orthologous GLDH genes in rice, B. distachyon and Arabidopsis, suggesting the preservation of exon and intron structure in GLDH genes in higher plants. Third, the deduced primary structure and mitochondrial targeting of TaGLDH-A1, -B1 and -D1 are highly similar to those of orthologous GLDHs in Arabidopsis, cauliflower, sweet potato and tobacco, highlighting the conservation of GLDH protein structure and subcellular location in plant cells. Fourth, TaGLDH-A1, -B1 and -D1 all have GLDH activity, and may not differ substantially from each other in the level of such activity. This is supported by similar increases of total GLDH activity in the N. benthamiana plants ectopically expressing TaGLDH-A1, -B1 or -D1. Fifth, the recombinant protein of TaGLDH-A1a (lacking the putative mitochondrial targeting peptide) is biochemically active towards the substrate L-GalL, with a Km value very similar to that reported for tobacco GLDH enzyme35. Lastly, TaGLDH transcripts are present in multiple vegetative and reproductive organs of common wheat, with relatively high transcript abundance in the leaf and spike tissues.
The observation of a higher TaGLDH transcript level in the leaves by this work agrees well with the finding of abundant TaGLDH proteins in wheat leaf tissues by a previous study13. However, it is interesting to note that TaGLDH transcripts are highly abundant in the spikelets, particularly in the young spikelet tissues. It is well known that the development of cereal spikelets involves intensive cell growth and differentiation and enhanced metabolic activity36, and that the level of reactive oxygen species in this developmental process needs to be tightly controlled especially under stress conditions37. Furthermore, AsA has been found to regulate flowering20,38. Therefore, the high TaGLDH transcript level detected in the immature spikelets by this work may reflect the function of TaGLDH in active AsA biosynthesis during common wheat spikelet development. The importance of having sufficient GLDH proteins during cereal spikelet development has recently been demonstrated by the finding that silencing rice GLDH expression led to substantially reduced seed set19.
Function of TaGLDH in AsA biosynthesis in common wheat
In Arabidopsis, rice and several other plant species, the function of GLDH in AsA biosynthesis is well supported by genetic evidence obtained using defined mutants and transgenic lines with altered expression GLDH level17,19,20,21,22. But in common wheat genetic evidence for the function of GLDH in AsA biosynthesis is not available before this work. From the data of our BSMV VIGS experiment, it is clear that normal TaGLDH expression is required for efficient AsA biosynthesis in common wheat seedlings. In agreement with previous studies39,40,41,42, our BSMV VIGS was reliable and effectively lowered the expression of TaGLDH. However, in the leaf tissues undergoing TaGLDH silencing, the reductions of total and reduced AsA contents were considerably below the decreases in TaGLDH expression and GLDH activity. The lack of correlation in between GLDH expression level, GLDH activity and AsA content has been observed in a number of plant species13,43, and several possibilities have been suggested to explain such a phenomenon13,25. Here we found that exogenous application of 15 mM L-GalL strongly increased AsA contents in both controls and the plants infected by BSMV:GLDHas, and substantially enlarged the degree of AsA content reduction by silencing TaGLDH expression. Therefore, in common wheat leaf tissues, the function of TaGLDH in AsA biosynthesis is at least partly affected by the amount of substrate.
One concern on VIGS data is that their reliability may be affected by off target silencing39,41. In this work, the chance of off target silencing in the VIGS initiated by BSMV:GLDHas is likely to be very small because the fragment employed for developing BSMV:GLDHas recognized only the three TaGLDH homoeologs and no additional GLDH paralogous sequences were found in the draft genome sequence of CS. Therefore, the reductions of TaGLDH expression, TaGLDH activity and AsA contents in BSMV:GLDHas infected plants are due to specific silencing of the expression of TaGLDH-A1, -B1 and -D1. Nevertheless, a more detailed understanding of the function of TaGLDH-A1, -B1 and -D1 in AsA biosynthesis (and other processes) would require the preparation and examination of wheat mutants lacking one or more of the three homoeologs. We are now in the process of developing such mutants.
Association of TaGLDH-A1b with enhanced drought tolerance
Natural allelic variation is a fundamental mechanism in the genetic control of plant traits44. Mining allelic variations at the molecular level has contributed enormously to understanding and improving the genetic basis of valuable traits in higher plants45. The cloning of three TaGLDH homoeologs in this work enabled us to examine their molecular variations in common wheat germplasm, leading to the discovery of a novel allele TaGLDH-A1b. Compared to WT allele (TaGLDH-A1a), TaGLDH-A1b missed one glutamic acid residue, and exhibited a significant reduction in its catalytic activity towards the tested substrate (Table 1). To our knowledge, this type of allele has not been described previously for either plant or animal GLDH proteins. Further, our data suggest that there is very likely a positive association between TaGLDH-A1b and enhanced drought tolerance in common wheat. This proposition is supported by four lines of evidence. First, the seven cultivars carrying TaGLDH-A1b were all from Xinjiang that has a limited rainfall in wheat production season29,30. Second, our tests showed that, relative to TaGLDH-A1a lines, those with TaGLDH-A1b generally displayed better tolerance to water deficiency stress in growth trails (Fig. 7A), had a higher foliar proline content (Fig. 7B), and showed lower leaf water loss rate (Fig. 8A). We did not observe any difference in the survival rate between the cultivars carrying TaGLDH-A1a or TaGLDH-A1b probably because the drought treatment applied was not sufficiently severe. Third, TaGLDH-A1b co-segregated with lower water loss in a F2 population (Fig. 8B). Lastly, under either normal or water deficiency conditions, TaGLDH-A1b cultivars generally had smaller stomatal aperture (Fig. 9), which is known to contribute directly to reduction of water loss and to stronger drought tolerance in water limited environments2,5. Interestingly, our previous study showed that partial suppression of GLDH activity also resulted in enhanced drought tolerance in Arabidopsis20. In both our previous study20 and this work, partial reduction of GLDH activity did not cause a drastic decrease of AsA content, and yet drought tolerance was significantly increased. This suggests that, in both monocot and dicot plant species, GLDH activity may be partially lowered to enhance drought tolerance.
Because the frequency of TaGLDH-A1b (2.56%) was relatively low, we deduce that TaGLDH-A1b is a rare allele that has so far not gained intensive use in wheat breeding. However, TaGLDH-A1b may have at least two different geographical origins because one of its carriers (i.e., Redstar) is not native to Xinjiang29. There is now growing interest in characterizing rare alleles of plant genes because many of them have been found to confer positive effects on agronomic traits44,45. For example, a rare allele of the crtRB1 gene enhances β-carotene content in maize grains46. The rare alleles of rice OsPPKL1 and GS2/OsGRF4 genes are associated with larger grains and higher yield47,48. Considering the overwhelming importance of wheat as a major staple food and the urgent need to develop drought tolerant wheat cultivars in both China and the world9,33,49, it is highly worthwhile to test if TaGLDH-A1b may confer improved drought tolerance when transferred to high yielding common wheat varieties with more diverse genetic backgrounds. Aided by the CAP-A1b marker developed in this work (Figure S7a), we have now embarked on this test.
Mechanism underlying the function of TaGLDH-A1b in enhanced drought tolerance
From the data gathered in this study (Figs 8 and 9), it is clear that decrease of stomatal aperture and reduction of leaf water loss are two physiological events closely linked with the enhanced drought tolerance by TaGLDH-A1b. However, further efforts are needed to reveal the additional physiological processes and molecular interactions involved. A previous study has shown that reducing AsA content in transgenic tobacco plants led to enhanced water stress tolerance through accumulation of a higher concentration of H2O2 in the guard cells and an increased stomatal closure in water limiting environment50. But in common wheat water deficiency treatment, although increasing GLDH activity in certain cultivar, did not cause substantial changes in AsA content13. We also found that water deficiency stress did not alter AsA content considerably in the common wheat plants carrying TaGLDH-A1b (Figure S8). Furthermore, although the cultivars carrying TaGLDH-A1b generally exhibited substantial reductions in TaGLDH activity, their foliar AsA contents did not differ significantly from those of the cultivars with TaGLDH-A1a (Fig. 6C). These findings suggest that the overall foliar AsA content may not be directly related to the smaller stomatal aperture observed for TaGLDH-A1b plants. However, there may be a difference in the guard cell AsA content between TaGLDH-A1a and TaGLDH-A1b plants. This is possible because H2O2 accumulation in the guard cells has been found vital for stomata closing51,52, and AsA, being the most abundant water-soluble antioxidant in plant cells, plays a critical role in regulating cellular level of H2O214,53. Consequently, further work will be directed to compare if the guard cells of TaGLDH-A1a and TaGLDH-A1b leaves may differ in the levels of AsA and H2O2 under normal or water deficiency conditions.
Because it has been shown that GLDH regulates the assembly and function of plant RCI10,15, it will also be interesting to investigate if RCI activity may differ between TaGLDH-A1a and TaGLDH-A1b plants, and whether such difference might be involved in the enhanced drought tolerance associated with TaGLDH-A1b. Two recent studies have shown that partial reduction of RCI activity can lead to constitutive enhancement of drought tolerance in tobacco plants through reducing stomatal conductance and aperture54,55. Thus, it is worthy to analyze if the RCI activity of TaGLDH-A1b plants may be altered as compared to those carrying TaGLDH-A1a, and if so, its potential impact on stomatal aperture control and leaf water loss behavior.
Finally, owing to the difficulty in designing homoeolog specific primers, we were unable to compare if TaGLDH-A1a and TaGLDH-A1b may differ in their expression level by qRT-PCR. The resolving of this question is clearly important for further studying the functional difference between the two alleles. Therefore, we have started a RNA sequencing experiment, which should reveal any potential difference between the transcript levels of TaGLDH-A1a and TaGLDH-A1b. Because we observed a co-segregation between TaGLDH-A1b and reduced leaf water loss rate (Fig. 8B), and the occurrence of smaller stomatal aperture in TaGLDH-A1b plants (Fig. 9), it becomes necessary to examine if TaGLDH-A1b may co-segregate with decreased stomatal aperture. However, the investigation of this question using segregating F2 plants is time consuming. The development of defined mutants for TaGLDH homoeologs (see above) or near isogenic lines for TaGLDH-A1a and TaGLDH-A1b should facilitate this analysis in further research.
In summary, this work has improved our understanding on plant GLDH genes and their function through characterizing TaGLDH homoeologs and discovering an association between TaGLDH-A1b and enhanced drought tolerance. The new information obtained may prompt further research on GLDH genes and their application in improving the drought tolerance and other traits of crop plants.
Methods
Plant materials, nucleic acid extraction, oligonucleotide primers and chemicals
Four sets of common wheat varieties were used in this study. The first set included Xiaoyan 54, CS and the NT lines derived from CS. The second set was the mini-core collection (MCC) of Chinese wheat germplasm composed of 262 landraces and improved varieties27. The third set included 11 cultivars from Xinjiang (Table S2). The fourth set consisted of 134 F2 seedlings developed in this work through crossing two Xinjiang varieties (i.e., Yichun 4 × Xinchun 3). Unless specifically stated, wheat plants were grown in a greenhouse at 25 °C (day)/20 °C (night) and with a 16 h light/8 h dark photoperiod. A vernalization treatment (5 weeks at 4 °C) was applied to Xiaoyan 54 seedlings to promote flowering. Genomic DNA and total RNA samples were prepared from desired wheat tissues as described previously40. The oligonucleotide primer sets (PS1 to PS11) used in this work are listed in Table S1, and their positions are given in Figure S1.
Cloning TaGLDH homoeologs and expression analysis of TaGLDH
The BAC library of Xiaoyan 54 was screened by PCR with the conserved primer set PS1 (Table S1, Figure S1), and following the procedure reported previously56. TaGLDH ORFs, corresponding to TaGLDH-A1, -B1 and -D1, respectively, were amplified from the positive BAC clones using the primer set PS2 (Table S1, Figure S1). PS2 was also used to clone the cDNA coding region of TaGLDH-A1, -B1 and -D1 by RT-PCR with the cDNAs derived from Xiaoyan 54 leaf tissues as described previously40. Subsequently, two primer sets (PS3 and PS4, Table S1, Figure S1), both of which could yield amplicons with different lengths for TaGLDH-A1, -B1 and -D1, were developed for chromosome assignment of the three homoeologs. The assignment was facilitated using CS and associated NT lines and by fragment analysis of fluorescently labeled PCR amplicons57.
Because the coding sequences of TaGLDH-A1, -B1 and -D1 were nearly identical, it was difficult to design copy specific PCR primers to study the expression patterns of individual homoeologs. Therefore, two primer sets PS5 and PS6 (Table S1, Figure S1), capable of recognizing all three TaGLDH homoeologs, were designed for detecting the transcript level of TaGLDH by qRT-PCR and semi-quantitative RT-PCR, respectively. Total RNA samples were extracted from vegetative (root, stem, seedling leaf and flag leaf) and reproductive (young and mature spikelets) organs. The cDNAs derived from the different organs were used for qRT-PCR with the 26S rRNA gene (GenBank accession Z11889) as an internal reference and following the cycling conditions reported previously40. The semi-quantitative RT-PCR assay was performed as reported in a previous study57, with the wheat beta-tubulin gene 2 (GenBank accession U76745) as an internal reference gene.
Ectopic expression of TaGLDH homoeologs in N. benthamiana
The coding sequence of TaGLDH-A1, -B1 or -D1 was each expressed in N. benthamiana using the PEBV vector. This vector has been used successfully for ectopically expressing cloned genes in N. benthamiana58,59. For constructing PEBV:GUS, GUS coding sequence was excised from the plasmid pJIT16660 using NcoI and EcoRI digestions, followed by cloning into PEBV vector. The coding sequence of TaGLDH-A1, -B1 and -D1 was each cloned into PEBV vector using PCR fragment amplified with the primer sets PS7 (for A1) or PS8 (for B1 and D1) to form PEBV:A1, PEBV:B1 and PEBV:D1, respectively. The information on PS7 and PS8 is given in Table S1 and Figure S1. The inoculation of N. benthamiana plants with PEBV:GUS, PEBV:A1, PEBV:B1 or PEBV:D1 was conducted as detailed in our previous publication59. Two weeks after inoculation, the leaves infected by PEBV:GUS were collected for histochemical staining of GUS activity61, whereas those infected by PEBV:A1, PEBV:B1 or PEBV:D1 were harvested for determining total GLDH activity and AsA content (see below).
VIGS analysis
Among the three recombinant BSMVs used in this analysis (Figure S4a), BSMV:GFP and BSMV:PDSas were developed previously40,42. BSMV:GLDHas was constructed using a 431 bp sequence element conserved among TaGLDH-A1, -B1 and -D1 coding sequence. This element was amplified from leaf cDNAs as a NheI fragment using the primer set PS9 (Table S1, Figure S1). It was cloned into BSMV vector and gave rise to BSMV:GLDHas. The genome wide target specificity of the 431 bp fragment was checked by searching the draft genome sequence of CS at http://plants.ensembl.org/Triticum_aestivum/Info/Index.
RNA transcripts were prepared for the three recombinant BSMVs, and introduced into Xiaoyan 54 seedlings (at two leaf stage) as detailed previously40,42. For each virus, 60 seedlings were inoculated. The same number of seedlings was buffer-inoculated as mock controls, and another 60 seedlings were kept as uninoculated controls. Three weeks after inoculation, the fourth leaves of the plants infected by BSMV:GLDHas were collected for examining TaGLDH silencing by qRT-PCR and alterations of total GLDH activity and AsA content (see below). For testing the effect of exogenous application of L-GalL on AsA biosynthesis, the fourth leaves were collected from mock controls and the plants infected by BSMV:GFP or BSMV:PDSas. They were cut into 1 mm segments, followed by incubation in a 15 mM solution of L-GalL for 24 h at 25 °C under a light intensity of 150 μmol/m2·s. Afterwards, the segments were collected for measuring total and reduced AsA contents (see below).
Measurement of total GLDH activity and AsA content in leaf tissues
The assay of total GLDH activity in leaf tissues was carried out using a crude mitochondrial fraction that was prepared at 4 °C following a previous study11. Measurement of GLDH activity level in the crude mitochondrial fraction was conducted in 0.5 ml volume consisting of 50 mM Tris-HCl, pH 8.0, 60 μM cytochrome c, 0.15% (w/v) Triton X-100 and 25 μg sample protein. The reaction was started by adding 4 μM L-GalL. After the reaction, the assay mixtures (150 μl per sample) were transferred to a 96 well microtiter plate, and the absorbance at 550 nm was recorded for each well using a Thermo Multiscan Scientific Spectrophotometer (Thermo Scientific, Waltham, USA). One unit of GLDH activity was defined as the formation of 2 nm reduced cytochrome c per minute.
To determine AsA content in a leaf sample, about 40 mg foliar materials were homogenized, and suspended in 6% trichloroacetic acid. The suspension was centrifuged at 13,000 g for 5 min at 4 °C, and the supernatant was kept for measuring the contents of total and reduced AsA following the method detailed previously62.
Screening of TaGLDH variants and tagging of TaGLDH-A1b
The screening was executed by fragment analysis of fluorescently labeled PCR amplicons. Briefly, genomic DNA samples were extracted from 262 MCC lines, and used in two series of genomic PCR assays with the primer sets PS3 and PS4 (Table S1, Figure S1), respectively. The resultant amplicons were subjected to fragment analysis as described above. For tagging TaGLDH-A1b allele, a cleaved amplified polymorphic marker CAP-A1b was developed (Figure S7a). A portion of the genomic ORF of TaGLDH-A1 (538 bp for TaGLDH-A1a and 535 bp for TaGLDH-A1b) was specifically amplified by PCR with the primer set PS11 (Table S1, Figure S1). The amplicons were digested with BsaJI, which cut the amplicons from TaGLDH-A1a but not those of TaGLDH-A1b. CAP-A1b was employed to identify TaGLDH-A1b in 11 Xinjiang common wheat lines, and in the F2 individuals derived from Yichun 4 × Xinchun 3 (see below).
Bacterial expression and biochemical study of TaGLDH-1a and -A1b
TaGLDH-A1a and TaGLDH-A1b sequences were amplified from the cDNAs of Xiaoyan 54 and Kashibaipi, respectively, using the primer set PS10 (Table S1, Figure S1). The resultant TaGLDH-A1a and TaGLDH-A1b fragments lacked the nucleotide sequence encoding the first 78 residues (the putative mitochondrial targeting peptide). These fragments were cloned into the plasmid vector pET-30a (Novagen, Madison, WI, USA), yielding the constructs pET-A1a and pET-A1b, respectively. Induction of TaGLDH-A1a and TaGLDH-A1b expression and purification of the two recombinant proteins by combining nickel chelate affinity chromatography and fast performance liquid chromatography followed the practices detailed previoulsy59. The purified TaGLDH-A1a and TaGLDH-A1b were checked by 10% SDS-PAGE and a protein blot assay with an anti-histidine antibody (Roche Diagnostic GmbH, Mannheim, Germany).
For investigating the kinetic properties of recombinant TaGLDH-A1a and TaGLDH-A1b, the assay was performed as described above for testing GLDH activity with minor modifications. Each assay was conducted in 0.2 ml volume consisting of 25 mM Tris-HCl, pH 8.0, 25 mM NaCl, 60 μM cytochrome c, 0.167 μg recombinant protein (TaGLDH-A1a or TaGLDH-A1b) and a specific concentration of L-GalL. A total of 11 concentrations of L-GalL, varying from 0.015625 to 5 mM, were used. The reaction was started by addition of TaGLDH-A1a or TaGLDH-A1b. After the reaction, the absorbance at 550 nm was read for each assay. The obtained readings were then employed to calculate the kinetic parameters (Km and Vmax) with the SigmaPlot Enzyme Kinetics Module for Windows (SPSS Inc., Chicago, IL, USA).
Drought tolerance growth trial and determination of free proline content
The trial was conducted essentially as described by Zhang and coauthors33. In brief, each trail started with the seedlings at three-leaf stage, and a total of 40 uniform seedlings (5 individuals per pot) were used for each variety. The plants were placed in a growth chamber set at 23 °C (day)/20 °C (night) and with a 16 h light/8 h dark photoperiod, but without watering for 30 days. They were re-watered afterwards. The volumetric water content (VWC) of each pot was checked every two days, and the plants at the same VWC were used for comparison. The trial was repeated three times. At 20 days post water deprivation, free proline content was determined for each variety using the leaves collected from 5 individual plants in five different pots. The measurement of proline content followed a procedure detailed previoulsy63. The proline content assay was conducted in two of the three drought tolerance trails.
Water loss assay
The assay was carried out using wheat plants (each with 3 tillers) cultured in 1 × Hoagland solution in a growth chamber with a temperature regime of 15 °C (day)/10 °C (night) and a photoperiod of 16 h light/8 h dark. In each assay and for each cultivar, 7 fully expanded leaves from 7 plants were pooled and subjected to water loss measurement33. For investigating water loss behavior of the F2 plants derived from Yichun 4 × Xinchun 3, the F2 individuals (at tillering stage) were first genotyped with the marker CAP-A1b (see above). This divided the F2 progenies into three groups, homozygous for TaGLDH-A1a (i.e., A1aA1a) or TaGLDH-A1b (A1bA1b) and heterozygous at TaGLDH-A1 locus (A1aA1b) (Figure S7b). The water loss assay was then conducted for the three types of plants and their parents as described above.
Measurement of stomatal aperture
Wheat plants were cultured as described above for the water loss assay. For each of the eight wheat varieties under examination, 9 uniform plants (each with 3 tillers) were selected and divided into 3 batches (3 in each batch). The first batch of plants was transferred into double distilled water (as controls), with 6 epidermal peels immediately prepared from the 3 plants. The second and third batches of plants were transferred into 13% PEG 6000, and treated for 24 and 48 h, respectively. Subsequently, 6 epidermal peels were immediately prepared from the second and third batches of plants, respectively. The epidermal peels were stained with 2′,7′-dichlorofluorescin diacetate64, which facilitated the examination of guard cells and stomatal aperture under a confocal laser scanning microscope (LSM710; Carl Zeiss AG, Oberkochen, Germany). Ten photos were taken for each epidermal peel, and used for measuring stomatal aperture with the ZEN lite 2012 software. This experiment was repeated three times.
Statistical analysis
Statistical analysis of the experimental data (presented as mean ± SD) was performed by ANOVA with the SPSS program (SAS Institute Inc., Cary, NC, USA).
Additional Information
Accession codes: The genomic sequences of TaGLDH-A1, -B1 and -D1 have been deposited in the GenBank under the accession codes KU695146, KU695147 and KU695148, respectively.
How to cite this article: Zhang, J. et al. A novel allele of L-galactono-1,4-lactone dehydrogenase is associated with enhanced drought tolerance through affecting stomatal aperture in common wheat. Sci. Rep. 6, 30177; doi: 10.1038/srep30177 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012AA10A308) and the National Natural Science Foundation of China (31371611). We thank Dr. Hui Liang for assistance in fragment analysis and confocal microscopy.
Footnotes
Author Contributions J.Z., B.L. and D.W. designed the study and wrote the manuscript. J.Z., B.L. and Y.Y. accomplished most of the experimental work. L.D., X.L. and H.L. helped the screen of BAC library. W.Q., K.Z. and H.Q. assisted the experiments involving PEBV and BSMV vectors. P.M. contributed to the screen of Xinjiang wheat cultivars. All authors read and approved the manuscript.
References
- Shanker A. K. et al. Drought stress responses in crops. Funct & Integr Genomics 14, 11–22 (2014). [DOI] [PubMed] [Google Scholar]
- Mir R. R., Zaman-Allah M., Sreenivasulu N., Trethowan R. & Varshney R. K. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125, 625–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenberth K. E. et al. Global warming and changes in drought. Nat Clim Change 4, 17–22 (2014). [Google Scholar]
- Mickelbart M. V., Hasegawa P. M. & Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16, 237–251 (2015). [DOI] [PubMed] [Google Scholar]
- Hu H. & Xiong L. Genetic engineering and breeding of drought-resistant crops. Annu Plant Biol 65, 715–741 (2014). [DOI] [PubMed] [Google Scholar]
- Singh D. & Laxmi A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6, 895 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Kim Y. S., Han S. H., Lee B. D. & Paek N. C. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang S. & Xia G. From genome to gene: a new epoch for wheat research? Trends Plant Sci 20, 380–387 (2015). [DOI] [PubMed] [Google Scholar]
- Fleury D., Jefferies S., Kuchel H. & Langridge P. Genetic and genomic tools to improve drought tolerance in wheat. J Exp Bot 61, 3211–3222 (2010). [DOI] [PubMed] [Google Scholar]
- Pineau B., Layoune O., Danon A. & De Paepe R. L-Galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J Biol Chem 283, 32500–32505 (2008). [DOI] [PubMed] [Google Scholar]
- Bartoli C. G., Pastori G. M. & Foyer C. H. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123, 335–343 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard J., Persiau G., Davey M. W., Bauw G. & Van Montagu M. Isolation of a cDNA coding for L-galactono-gamma-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Purification, characterization, cDNA cloning, and expression in yeast. J Biol Chem 272, 30009–30016 (1997). [DOI] [PubMed] [Google Scholar]
- Bartoli C. G. et al. Ascorbate content of wheat leaves is not determined by maximal L-galactono-1,4-lactone dehydrogenase (GalLDH) activity under drought stress. Plant Cell Environ 28, 1073–1081 (2005). [Google Scholar]
- Smirnoff N., Conklin P. L. & Loewus F. A. Biosynthesis of ascorbic acid in plants: A renaissance. Annu Rev Plant Physiol Plant Mol Biol 52, 437–467 (2001). [DOI] [PubMed] [Google Scholar]
- Schertl P. et al. L-Galactono-1,4-lactone dehydrogenase (GLDH) forms part of three subcomplexes of mitochondrial complex I in Arabidopsis thaliana. J Biol Chem 287, 14412–14419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhagdow M. et al. Silencing of the mitochondrial ascorbate synthesizing enzyme L-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol 145, 1408–1422 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K., Oba K., Suzuki K. & Esaka M. Generation and properties of ascorbic acid-deficient transgenic tobacco cells expressing antisense RNA for L-galactono-1,4-lactone dehydrogenase. Plant J 27, 139–148 (2001). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Tiller number is altered in the ascorbic acid-deficient rice suppressed for L-galactono-1,4-lactone dehydrogenase. J Plant Physiol 170, 389–396 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Y. H., Yu L. & Wang R. Z. Level of ascorbic acid in transgenic rice for l-galactono-1,4-lactone dehydrogenase overexpressing or suppressed is associated with plant growth and seed set. Acta Physiol Plant 33, 1353–1363 (2011). [Google Scholar]
- Li B. et al. Partial suppression of L-galactono-1,4-lactone dehydrogenase causes significant reduction in leaf water loss through decreasing stomatal aperture size in Arabidopsis. Plant Growth Regul 72, 171–179 (2014). [Google Scholar]
- Tokunaga T., Miyahara K., Tabata K. & Esaka M. Generation and properties of ascorbic acid-overproducing transgenic tobacco cells expressing sense RNA for L-galactono-1,4-lactone dehydrogenase. Planta 220, 854–863 (2005). [DOI] [PubMed] [Google Scholar]
- Liu W., An H. M. & Yang M. Overexpression of Rosa roxburghii L-galactono-1,4-lactone dehydrogenase in tobacco plant enhances ascorbate accumulation and abiotic stress tolerance. Acta Physiol Plant 35, 1617–1624 (2013). [Google Scholar]
- Sears E. R. The aneuploids of common wheat. Univ Missouri Agric Exp Stn Bull 572, 1–58 (1954). [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 (2014). [DOI] [PubMed]
- Bartoli C. G. et al. Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J Exp Bot 57, 1621–1631 (2006). [DOI] [PubMed] [Google Scholar]
- Davey M. W. et al. Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol 121, 535–543 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C. Y. et al. Genetic diversity and construction of core collection in Chinese wheat genetic resources. Chinese Sci Bull 53, 1518–1526 (2008). [Google Scholar]
- Leferink N. G., Jose M. D., van den Berg W. A. & van Berkel W. J. Functional assignment of Glu386 and Arg388 in the active site of L-galactono-gamma-lactone dehydrogenase. FEBS Lett 583, 3199–3203 (2009). [DOI] [PubMed] [Google Scholar]
- He Z. H., Rajaram S., Xin Z. Y. & Huang G. Z. A history of wheat breeding in China. (CIMMYT, 2001). [Google Scholar]
- Sun B. Q., Zhang A. M. & Bonjean A. P. In The World Wheat Book (eds Bonjean A. P. & Angus W. J. ) 667–701 (Lavoisier Publishing, 2001). [Google Scholar]
- Szabados L. & Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci 15, 89–97 (2010). [DOI] [PubMed] [Google Scholar]
- Shavrukov Y., Baho M., Lopato S. & Langridge P. The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol J 14, 313–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Y. et al. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol 196, 1155–1170 (2012). [DOI] [PubMed] [Google Scholar]
- Xue G. P. et al. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4, 697–712 (2011). [DOI] [PubMed] [Google Scholar]
- Yabuta Y., Yoshimura K., Takeda T. & Shigeoka S. Molecular characterization of tobacco mitochondrial L-galactono-gamma-lactone dehydrogenase and its expression in Escherichia coli. Plant Cell Physiol 41, 666–675 (2000). [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N. & Schnurbusch T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci 17, 91–101 (2012). [DOI] [PubMed] [Google Scholar]
- Barnabas B., Jager K. & Feher A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31, 11–38 (2008). [DOI] [PubMed] [Google Scholar]
- Kotchoni S. O., Larrimore K. E., Mukherjee M., Kempinski C. F. & Barth C. Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiol 149, 803–815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S. R. & Brandt A. S. Virus-induced gene silencing in hexaploid wheat using barley stripe mosaic virus vectors. Methods Mol Biol 894, 93–112 (2012). [DOI] [PubMed] [Google Scholar]
- Wang G. F. et al. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol 191, 418–431 (2011). [DOI] [PubMed] [Google Scholar]
- Scofield S. R. & Nelson R. S. Resources for virus-induced gene silencing in the grasses. Plant Physiol 149, 152–157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. B. et al. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J 52, 420–434 (2007). [DOI] [PubMed] [Google Scholar]
- Loscos J., Matamoros M. A. & Becana M. Ascorbate and homoglutathione metabolism in common bean nodules under stress conditions and during natural senescence. Plant Physiol 146, 1282–1292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C. et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21, 1877–1896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss-Fels K. & Snowdon R. J. Understanding and utilizing crop genome diversity via high-resolution genotyping. Plant Biotechnol J 14, 1086–1094 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. B. et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat Genet 42, 322–327 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang X. J. et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109, 21534–21539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8, 1455–1465 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang J. H. China’s success in increasing per capita food production. J Exp Bot 62, 3707–3711 (2011). [DOI] [PubMed] [Google Scholar]
- Chen Z. & Gallie D. R. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16, 1143–1162 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. & Song C.-P. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178, 703–718 (2008). [DOI] [PubMed] [Google Scholar]
- Huang X.-Y. et al. A previoulsy unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23, 1805–1817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. Ascobic acid: metabolism and fucntions of a multi-facetted molecule. Curr Opin Plant Biol 3, 229–235 (2000). [PubMed] [Google Scholar]
- Rzigui T., De Paepe R., Cornic G. & Streb P. In the mitochondrial CMSII mutant of Nicotiana sylvestris photosynthetic activity remains higher than in the WT under persisting mild water stress. Plant Sci 205–206, 20–28 (2013). [DOI] [PubMed] [Google Scholar]
- Djebbar R. et al. Respiratory complex I deficiency induces drought tolerance by impacting leaf stomatal and hydraulic conductances. Planta 235, 603–614 (2012). [DOI] [PubMed] [Google Scholar]
- Dong L. L. et al. New insights into the organization, recombination, expression and functional mechanism of low molecular weight glutenin subunit genes in bread wheat. PLoS One 5, e13548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. M. et al. Molecular analysis of phosphomannomutase (PMM) genes reveals a unique PMM duplication event in diverse Triticeae species and the main PMM isozymes in bread wheat tissues. BMC Plant Biol 10, 214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin G. D. et al. Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J 40, 622–631 (2004). [DOI] [PubMed] [Google Scholar]
- Qian W. Q. et al. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J 49, 399–413 (2007). [DOI] [PubMed] [Google Scholar]
- Niu Y. et al. Molecular and functional characterization of sphingosine-1-phosphate lyase homolog from higher plants. J Integr Plant Biol 49, 323–335 (2007). [Google Scholar]
- Gallagher S. R. GUS Protocols: Using the GUS gene as a reporter of gene expression. (Academic Press, 1992). [Google Scholar]
- Gillespie K. M. & Ainsworth E. A. Measurement of reduced, oxidized and total ascorbate content in plants. Nat Protoc 2, 871–874 (2007). [DOI] [PubMed] [Google Scholar]
- Hu C. A., Delauney A. J. & Verma D. P. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA 89, 9354–9358 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua D. et al. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24, 2546–2561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.