Abstract
Accumulating evidences have indicated that the functional -94 ins/del ATTG polymorphism in the promoter region of human nuclear factor-kappa B1 (NFKB1) gene may be associated with cancer risk. However, some studies yielded conflicting results. To clarify precise association, we performed a comprehensive meta-analysis of 42 case-control studies involving 43,000 subjects (18,222 cases and 24,778 controls). The overall results suggested that the -94 ins/del ATTG polymorphism had a decreased risk for cancer, reaching significant levels in five genetic models (dominant model: OR = 0.86, 95% CI = 0.79–0.95, P = 0.002; recessive model: OR = 0.84, 95% CI = 0.74–0.94, P = 0.003; homozygous model: OR = 0.77, 95% CI = 0.66–0.90, P = 0.001; heterozygous model: OR = 0.90, 95% CI = 0.83–0.98, P = 0.011; allelic model: OR = 0.89, 95% CI = 0.83–0.96, P = 0.002). Furthermore, the -94 ins/del ATTG polymorphism could confer a decreased or increased risk for cancer development among Asians and Caucasians, respectively. Additionally, the stratification analysis revealed a significant association between the variant and decreased risk of oral, ovarian, and nasopharyngeal cancer in Asians. After we adjusted p values using the Benjamini-Hochberg false discovery rate method to account for multiple comparisons, these associations remained.
Characterized by angiogenesis and inflammation, cancer is a major public health problem and a complex terminal disease worldwide, which is injurious to human health and lives1,2. With the aging population together with increasing formation of cancer-causing habits, it is estimated that there would be more than 22 million new cases suffering from cancer in the next decades. Therefore, the clinical and economic burden of cancer on healthcare will be heavier2,3. Although the mechanism of carcinogenesis has not been completely clarified, abundant evidences indicated that a combination of genetic predisposition and environmental exposures is believed to contribute to pathogenesis of cancer4. With numerous recent advances in genetic research, many genes that are related to malignancy susceptibility have been identified as candidates in multiple populations, especially nuclear factor-kappa B 1 (NFKB1)5,6,7,8,9,10.
There is mounting evidence that inflammatory cytokines and carcinogens closely related to cancer development are involved in activating the common cell survival signaling pathways11,12. NF-κB serves as a major cell survival signal and is involved in multiple steps in carcinogenesis and in cancer cell’s resistance to chemo- and radiotherapy12,13. It was identified as an inducible transcriptional factor binding to the intronic enhancer of the kappa light chain gene in every cell type14,15. Subsequently, NF-κB acts as an important regulator of more than 200 genes known to be involved in cell adhesion, cell survival, inflammation, apoptosis, and growth16,17. The NF-κB family is composed of five members in mammal: p50/p105, p65/RelA, c-Rel, RelB, and p52/p100 (NFKB2), which can form different dimeric combinations. NFKB1 gene, mapped to chromosome 4q24, encodes p105/p50 isoforms of NF-κB, involved in cancer initiation and progression. The -94 insertion/deletion (ins/del) ATTG polymorphism within the promoter region of the NFKB1 gene could potentially affect the transcription of the gene and the function of NF-κB protein, sequentially leading to loss of binding capacity to nuclear proteins and reduced promoter activity18.
The -94 ins/del ATTG (rs28362491) polymorphism that involves deletion of four nucleotides in the promoter region of NFKB1 gene consists of three genotypes: homozygote insertion or wild-type (WW), homozygote deletion or variant (DD), and heterozygous ins/del (DW)7,18. According to published data, several search groups have investigated the relationship between the polymorphism and cancers risk7,10,19,20,21,22,23,24,25,26,27. However, these results obtained remains inconsistent. For instance, numerous studies reported that the del/del genotype of the polymorphism could contribute to an increased colorectal cancer (CRC)28,29,30, papillary thyroid carcinoma (PTC)24 and lung cancer (LC)13 in Chinese or Caucasian populations, while some other studies showed that the ins/ins genotype of the polymorphism could increase the risk of CRC20,31, LC26 and bladder cancer (BC)32 in different populations. In addition, several studies showed a lack of relationship between the polymorphism and CRC risk7.
Up to now, some earlier meta-analyses based on different strategies have tried to investigate the link between the polymorphism and cancer risk. Yang et al.33 and Nian et al.34 reported that the -94 ins/del ATTG polymorphism was significantly associated with decreased cancer risk in Asian populations, but not in Caucasian populations. Unfortunately, the two previous meta-analyses included some studies, in which the genotype distributions of control groups were not consistent with Hardy–Weinberg equilibrium (HWE). In addition, obviously high heterogeneity was detected under all five genetic models in their studies, but they only conducted subgroup analysis based on ethnicity to investigate the origin of the high heterogeneity. Furthermore, the authors did not perform tests to access statistical significance of multiple null hypotheses35. Henceforth, some new studies further assessed the relationship between the polymorphism and cancer risk among multiple ethnic populations. However, the association remains inconclusive due to the inconsistent results from the published publications23,24,25,26,27,30,36,37,38,39,40,41,42. Therefore, the data need to be updated and more reliable evaluations of the polymorphism with cancer risk are warranted.
Due to the inconsistency of past studies and the critical role of the variant in the pathogenesis of cancer, we conducted an updated meta-analysis to investigate the association of NFKB1 -94 ins/del ATTG promoter polymorphism and cancer risk by precise results.
Materials and Methods
Study selection
A systematic search of PubMed, Embase, ISI Web of Science, and Chinese National Knowledge Infrastructure (CNKI) database was conducted by two independent reviewers to identify relevant articles, which had investigated the association between NFKB1 -94 ins/del ATTG polymorphism and susceptibility to cancer (latest electronic search was updated on March 10, 2016). The search terms were used as follows: ‘cancer’ or ‘carcinoma’ or ‘tumor’ or ‘carcinogenesis’ or ‘neoplasm’ And ‘NF-κB’ or ‘Nuclear factor-κB 1’ or ‘NFKB1” And ‘polymorphism’ or ‘variant’ or ‘mutation’ or ‘polymorphisms’ or ‘variants’ or ‘mutations’. No publication date or languages restrictions were imposed. Studies eligible for this meta-analysis must fulfill the following inclusion criteria: (a) studies using a case-control design; (b) studies evaluating the association between NFKB1 -94 ins/del ATTG polymorphism and cancer risk; (c) genotype distributions should be available for estimating the odds ratios (OR) with 95% confidence interval (CI); (d) the genotype distributions among control groups must be consistent with HWE. For multiple reports based on overlapping samples, only the most recent study or the study with the largest number of subjects were retained. Exclusion criteria were defined as follows: (a) abstracts, conferences, reviews, letters, case reports, and non-human studies; (b) repeated or overlapping publications; (c) studies reporting neither genotype distributions nor allele frequency; (d) the studies that do not follow HWE. We also inspected the references list of reviews or previous meta-analyses for potentially relevant publications. Two investigators independently screened all abstracts and citations to extract potentially eligible studies.
Data extraction
Two investigators independently collected the information of each eligible study based on the inclusion and exclusion criteria. First author, publication year, country, ethnicity, control source, genotyping technology, genotype and allele distributions, total number of cases and controls, and cancer type were extracted. In case of discrepancies, we checked the collected data and reached a consensus through discussion to ensure accuracy of the data. The information is shown in Tables 1 and 2.
Table 1. Characteristics of 42 included studies in this meta-analysis.
| Author | Year | Country | Ethnicity | Cases/Control | Sample size | Control source | Cancer type | Genotyping method | Quality score |
|---|---|---|---|---|---|---|---|---|---|
| Lin | 2006 | China | Asian | 212/201 | 413 | HB | OSCC | PCR-PAGE | 6 |
| Riemann | 2006 | Germany | Caucasian | 139/307 | 446 | HB | CRC | pyrosequencing | 7 |
| Riemann | 2006 | Germany | Caucasian | 72/307 | 379 | HB | BCCLL | pyrosequencing | 7 |
| Riemann | 2006 | Germany | Caucasian | 140/307 | 447 | HB | RCC | pyrosequencing | 7 |
| Riemann | 2007 | Germany | Caucasian | 242/307 | 549 | HB | BC | pyrosequencing | 7 |
| Lehnerdt | 2008 | Germany | Caucasian | 364/307 | 671 | HB | HNSCC | PCR-RFLP | 7 |
| Lo | 2009 | China | Asian | 182/116 | 298 | HB | GC | PCR-RFLP | 6 |
| He | 2009 | China | Asian | 202/404 | 606 | HB | HCC | PCR-RFLP | 7 |
| Zhang | 2009 | China | Asian | 117/143 | 260 | HB | PC | PCR-PAGE | 8 |
| Zhou | 2010 | China | Asian | 233/365 | 598 | Mixed | CSCC | PCR-PAGE | 7 |
| Andersen | 2010 | Denmark | Caucasian | 378/756 | 1134 | PB | CRC | TaqMan | 8 |
| Tang | 2010 | China | Asian | 207/228 | 435 | HB | BC | PCR-PAGE | 7 |
| Song | 2011 | China | Asian | 1,001/1,005 | 2006 | PB | CRC | PCR-RFLP | 8 |
| Fan | 2011 | China | Asian | 179/223 | 402 | PB | OC | PCR-CGE | 7 |
| Lin | 2012 | China | Asian | 462/520 | 982 | HB | OSCC | TaqMan | 8 |
| Ungerback | 2012 | Sweden | Caucasian | 348/622 | 966 | PB | CRC | TaqMan | 8 |
| Huang | 2013 | China | Asian | 1056/1056 | 2112 | HB | LC | TaqMan | 7 |
| Huang | 2013 | China | Asian | 503/623 | 1126 | HB | LC | TaqMan | 7 |
| Cheng | 2013 | China | Asian | 135/520 | 655 | HB | HCC | TaqMan | 8 |
| Li | 2013 | China | Asian | 609/640 | 1249 | HB | BC | TaqMan | 8 |
| Oltulu | 2014 | Turkey | Caucasian | 95/99 | 194 | NA | LC | PCR-RFLP | 7 |
| Kopp | 2013 | Denmark | Caucasian | 334/334 | 668 | PB | PC | TaqMan | 8 |
| Huo | 2013 | China | Asian | 187/221 | 408 | HB | OC | MassARRAY | 7 |
| Vangsted | 2012 | Denmark | Caucasian | 328/1696 | 2024 | PB | MM | Taqman | 6 |
| Zhou | 2009 | China | Asian | 163/203 | 366 | HB | NPC | PCR-PAGE | 8 |
| Han | 2015 | China | Asian | 936/936 | 1872 | PB | PC | PCR-RFLP | 8 |
| Li | 2015 | China | Asian | 220/222 | 442 | HB | Osteosarcoma | PCR-RFLP | 7 |
| Liu | 2015 | China | Asian | 906/1072 | 1978 | HB | NPC | TaqMan | 7 |
| Liu | 2015 | China | Asian | 684/907 | 1591 | HB | NPC | TaqMan | 7 |
| Wang | 2015 | China | Asian | 421/425 | 846 | HB | LC | PCR-RFLP | 8 |
| Lu | 2015 | China | Asian | 687/687 | 1374 | HB | OC | PCR-RFLP | 7 |
| Kopp | 2015 | Denmark | Caucasian | 1,010/1,829 | 2634 | PB | CRC | PCR-KASP | 6 |
| Chen | 2015 | China | Asian | 411/438 | 852 | HB | OC | MassARRAY | 7 |
| Zhang | 2014 | China | Asian | 208/1606 | 2230 | HB | HCC | PCR-RFLP | 6 |
| Hua | 2014 | China | Asian | 401/433 | 834 | HB | GC | MassARRAY | 8 |
| Wang | 2015 | China | Asian | 352/459 | 811 | HB | PTC | PCR-PAGE | 8 |
| Li | 2015 | China | Asian | 730/780 | 1510 | HB | BC | TaqMan | 8 |
| Li | 2015 | China | Asian | 1216/1588 | 2804 | HB | RCC | TaqMan | 8 |
| Li | 2015 | China | Asian | 820/945 | 1765 | HB | PC | TaqMan | 8 |
| Rybka | 2016 | Poland | Caucasian | 62/126 | 188 | NA | AML | PCR-CGE | 6 |
| Eskandari | 2016 | Iran | Asian | 236/203 | 439 | PB | BC | AS-PCR | 9 |
| Zhang | 2015 | China | Asian | 718/718 | 1436 | PB | LC | PCR-RFLP | 8 |
AS-PCR, Allele-Specific polymerase chain reaction; PCR-CGE, polymerase chain reaction with capillary gel electrophoresis; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-PAGE, polymerase chain reaction-polyacrylamide gel electrophoresis; HB, hospital-based study; PB, population-based study; NA, not available; BC, bladder cancer; PC, prostate cancer; OC, ovarian cancer; LC, lung cancer; HCC, hepatocellular carcinoma; MM, multiple myeloma; OSCC, oral squamous cell carcinoma; CSCC, cervical squamous cell carcinoma; CRC, colorectal cancer; NPC, nasopharyngeal carcinoma; GC, gastric cancer; GNT, gastroenteropancreatic neuroendocrine tumor; HNSCC, squamous cell carcinomas of the head and neck region; BCLL, B cell chronic lymphocytic leukemia; RCC, renal cell carcinoma; ESCC, esophageal squamous cell carcinoma; BRC, breast cancer; PTC, papillary thyroid carcinoma; AML, acute myeloid leukaemia.
Table 2. Distributions of NF-κB1-94 ins/del ATTG promoter polymorphism allele and genotypes in different groups.
| Author | Case |
Control |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DD | DW | WW | D (%) | W (%) | DD | DW | WW | D (%) | W (%) | HWE | |
| Lin | 50 | 103 | 59 | 203 (47.9) | 221 (52.1) | 58 | 100 | 43 | 216 (53.7) | 186 (46.3) | 0.993 |
| Riemann | 27 | 58 | 54 | 112 (40.3) | 166 (59.7) | 48 | 141 | 118 | 237 (38.6) | 377 (61.4) | 0.586 |
| Riemann | 13 | 41 | 18 | 67 (46.5) | 77 (53.5) | 48 | 141 | 118 | 237 (38.6) | 377 (61.4) | 0.586 |
| Riemann | 17 | 76 | 47 | 110 (39.3) | 170 (60.7) | 48 | 141 | 118 | 237 (38.6) | 377 (61.4) | 0.586 |
| Riemann | 30 | 124 | 88 | 184 (38) | 300 (62) | 48 | 141 | 118 | 237 (38.6) | 377 (61.4) | 0.586 |
| Lehnerdt | 53 | 179 | 132 | 285 (39.2) | 443 (60.8) | 48 | 141 | 118 | 237 (38.6) | 377 (61.4) | 0.58 |
| Lo | 31 | 89 | 62 | 151 (41.5) | 213 (58.5) | 34 | 62 | 20 | 130 (56) | 102 (44) | 0.361 |
| He | 35 | 84 | 83 | 154 (38.1) | 250 (61.9) | 124 | 183 | 97 | 431 (53.3) | 377 (46.7) | 0.07 |
| Zhang | 14 | 57 | 46 | 85 (36.3) | 149 (63.7) | 31 | 68 | 44 | 130 (45.5) | 156 (54.5) | 0.624 |
| Zhou | 20 | 105 | 108 | 145 (31.1) | 321 (68.9) | 64 | 166 | 135 | 294 (40.3) | 436 (59.7) | 0.297 |
| Andersen | 62 | 195 | 121 | 319 (42.2) | 437 (57.8) | 102 | 347 | 307 | 551 (36.4) | 961 (63.6) | 0.801 |
| Tang | 26 | 92 | 89 | 144 (34.8) | 270 (65.2) | 46 | 108 | 74 | 200 (43.9) | 256 (56.1) | 0.565 |
| Song | 138 | 500 | 363 | 776 (38.8) | 1226 (61.2) | 186 | 522 | 297 | 894 (44.5) | 1116 (55.5) | 0.102 |
| Fan | 17 | 84 | 78 | 118 (33) | 240 (67) | 44 | 103 | 76 | 191 (42.8) | 255 (57.2) | 0.396 |
| Lin | 100 | 246 | 116 | 446 (48.3) | 478 (51.7) | 168 | 271 | 81 | 607 (58.4) | 433 (41.6) | 0.099 |
| Ungerback | 43 | 187 | 114 | 273 (39.7) | 415 (60.3) | 96 | 270 | 256 | 462 (37.1) | 782 (62.9) | 0.079 |
| Huang | 225 | 459 | 372 | 909 (43) | 1203 (57) | 210 | 491 | 355 | 911 (43.1) | 1201 (56.9) | 0.089 |
| Huang | 104 | 230 | 169 | 438 (43.5) | 568 (56.5) | 145 | 289 | 189 | 579 (46.5) | 667 (53.5) | 0.092 |
| Cheng | 29 | 64 | 42 | 122 (45.2) | 148 (54.8) | 168 | 271 | 81 | 607 (58.4) | 433 (41.6) | 0.099 |
| Li | 151 | 269 | 189 | 571 (46.9) | 647 (53.1) | 93 | 324 | 223 | 510 (39.8) | 770 (60.2) | 0.156 |
| Oltulu | 16 | 44 | 35 | 76 (40) | 114 (60) | 6 | 47 | 46 | 59 (29.8) | 139 (70.2) | 0.18 |
| Kopp | 54 | 152 | 128 | 260 (38.9) | 408 (61.1) | 64 | 161 | 109 | 289 (43.3) | 379 (56.7) | 0.741 |
| Huo | 22 | 82 | 83 | 126 (33.7) | 248 (66.3) | 47 | 103 | 71 | 197 (44.6) | 245 (55.4) | 0.399 |
| Vangsted | 55 | 163 | 110 | 273 (41.6) | 383 (58.4) | 253 | 778 | 665 | 1284 (37.9) | 2108 (62.1) | 0.303 |
| Zhou | 22 | 67 | 74 | 111 (34.1) | 215 (62.9) | 42 | 90 | 71 | 174 (42.9) | 232 (57.1) | 0.177 |
| Han | 534 | 339 | 63 | 1407 (75) | 465 (25) | 567 | 331 | 38 | 1465 (78.3) | 407 (21.7) | 0.23 |
| Li | 46 | 114 | 60 | 206 (46.8) | 234 (53.2) | 66 | 106 | 50 | 238 (53.6) | 206 (46.4) | 0.55 |
| Liu | 152 | 438 | 316 | 742 (41) | 1070 (59) | 224 | 512 | 336 | 960 (44.8) | 1184 (55.2) | 0.262 |
| Liu | 117 | 331 | 236 | 565 (41) | 803 (59) | 195 | 438 | 274 | 828 (45.6) | 986 (54.4) | 0.169 |
| Wang | 89 | 219 | 113 | 397 (47.2) | 445 (52.8) | 131 | 205 | 89 | 467 (54.9) | 383 (45.1) | 0.595 |
| Lu | 221 | 351 | 115 | 793 (57.7) | 581 (42.3) | 253 | 339 | 95 | 845 (61.5) | 529 (38.5) | 0.271 |
| Kopp | 146 | 449 | 320 | 741 (40.5) | 1089 (59.5) | 253 | 787 | 679 | 1293 (37.6) | 2145 (62.4) | 0.311 |
| Chen | 95 | 195 | 120 | 385 (47) | 435 (53) | 122 | 235 | 85 | 479 (54.2) | 405 (45.8) | 0.136 |
| Zhang | 107 | 312 | 205 | 526 (42.2) | 722 (57.8) | 274 | 790 | 542 | 1338 (41.7) | 1874 (58.3) | 0.631 |
| Hua | 127 | 182 | 92 | 436 (54.4) | 366 (45.6) | 83 | 230 | 120 | 396 (45.7) | 470 (54.3) | 0.144 |
| Wang | 60 | 186 | 106 | 306 (43.5) | 398 (56.5) | 79 | 209 | 171 | 367 (40) | 551 (60) | 0.273 |
| Li | 187 | 316 | 227 | 690 (47.3) | 770 (52.7) | 124 | 395 | 261 | 643 (41.2) | 917 (58.8) | 0.208 |
| Li | 188 | 577 | 451 | 953 (39.2) | 1479 (60.8) | 225 | 781 | 582 | 1231 (38.8) | 1945 (61.2) | 0.152 |
| Li | 144 | 377 | 299 | 665 (40.6) | 975 (59.4) | 136 | 462 | 347 | 734 (38.8) | 1156 (61.2) | 0.371 |
| Rybka | 7 | 30 | 25 | 44 (35.5) | 80 (64.5) | 14 | 69 | 43 | 97 (38.5) | 155 (61.5) | 0.08 |
| Eskandari | 18 | 122 | 96 | 158 (33.5) | 314 (66.5) | 35 | 106 | 62 | 176 (43.3) | 230 (56.7) | 0.37 |
| Zhang | 32 | 252 | 434 | 316 (22) | 1120 (78) | 76 | 290 | 352 | 442 (30.8) | 994 (69.2) | 0.16 |
HWE, Hardy-Weinberg equilibrium.
WW, homozygous insertion or wild-type; DD, homozygous deletion or variant; DW, heterozygous ins/del.
Quality score assessment
The quality of included studies was estimated according to the Newcastle–Ottawa Scale (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The scale use a ‘-’ rating system to evaluate quality based on three points including the selection of the study groups, comparability of the groups, and the ascertainment of the exposure or outcome of interest in the case-control study. Total scores range from 0 to 9 points. A study with scores of more than 7 points was considered to be a high quality study (Table 1). Discrepancies would be resolved by discussion with a third author until a consensus was reached.
Statistical method
In this meta-analysis, all of the data in eligible studies were calculated as odds ratios (OR) with 95% confidence interval (CI) to assess the strength of association between NFKB1 -94 ins/del ATTG promoter polymorphism and the risk of cancer. The -94 ins/del ATTG polymorphism consists of three genotypes: homozygous insertion or wild-type (WW), homozygous deletion or variant (DD), and heterozygous ins/del (DW). The pooled OR was estimated in five genetic models: (1) dominant model: del/del + del/ins versus ins/ins (DD+DW vs. WW); (2) recessive model: del/del versus del/ins + ins/ins (DD vs. DW+WW); (3) homozygous model: del/del versus ins/ins (DD vs. WW); (4) heterozygous del/ins versus ins/ins (DW vs. WW); and (5) allelic model: deletion allele versus insertion allele (D vs. W), which was considered statistically significant when the P value of the Z test was less than 0.05.
Heterogeneity among studies was evaluated by Chi-squared-based Q-test and I2 statistics, which confirmed the heterogeneity at P-value < 0.10. If no or low heterogeneity existed (P < 0.10), the random-effects model (REM) was applied to estimate pooled OR. Otherwise, the fixed-effects model (FEM) was used. Moreover, HWE was recalculated in control groups by Pearson’s χ2 test before this meta-analysis was conducted. To explore the potential heterogeneity among studies, we conducted subgroup analysis based on ethnicity (Asian and Caucasian), control source (hospital-based (HB) and population-based (PB) population), genotyping method (TaqMan and others), quality score of studies, and type of cancer. In addition, sensitivity analyses were also conducted through sequentially excluded individual studies to investigate the potential origin of heterogeneity and to assess the stability of the results.
Potential publication bias was tested by several methods. Visual inspection of asymmetry in funnel plots was carried out. Furthermore, Egger’ regression and Begg’s test were also utilized to detect publication bias, and P < 0.05 was considered statistically significant. Benjamini–Hochberg false discovery rate (FDR) test was used to correct for multiple comparisons yielding Pcorr value less than 0.05 was considered as significant. All statistical analyses were performed with STATA 12.0 software (Stata Corp LP, College Station, TX).
Results
Study inclusion and characteristics
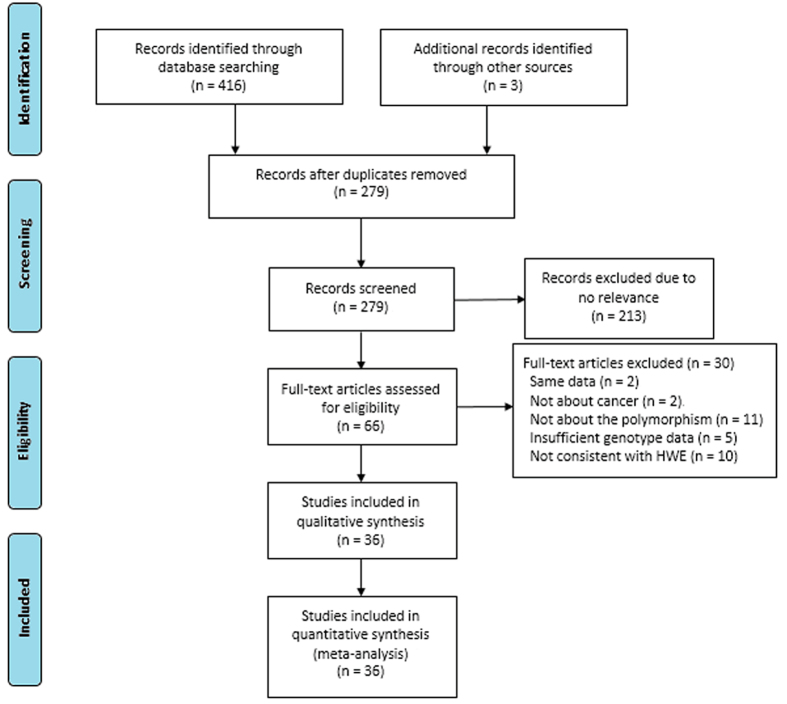
This meta-analysis design was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) prospectively. The process of study selection was showed in Fig. 1. We totally identified 419 articles with our search strategy. After removing duplications, scanning titles and abstracts and reading the full-text, 36 articles were eligible based on our inclusion and exclusion criteria in this meta-analysis. Among these selected articles, Riemann et al.43 and Li et al.39 investigated the link between the -94 ins/del ATTG polymorphism and the risk of three different cancer types, respectively. Therefore, we treated them as six separate comparisons. In addition, Huang et al.44 and Liu et al.41 conducted their research in two different places (eastern and southern china), which were treated as four individual comparisons. Therefore, a total of 42 studies from 36 articles were identified, including 43,000 subjects (18,222 cases and 24,778 controls)8,10,13,19,20,21,22,23,24,25,26,27,28,29,30,32,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55. Of the 42 included studies, five studies involving 7,186 subjects reported on colorectal cancer (CRC). In the remaining studies, there were two studies on renal cell carcinoma (RCC), four studies on bladder cancer (BC), two studies on gastric cancer (GC), three studies on hepatocellular carcinoma (HCC), four studies on prostate cancer (PC), four studies on ovarian cancer (OC), five studies on lung cancer (LC), three studies on nasopharyngeal carcinoma (NPC), two studies on oral squamous cell carcinoma (OSCC). Other cancers, such as multiple myeloma, breast cancer, cervical squamous cell carcinoma, acute myeloid leukaemia, were relatively less investigated and thereby merged into “other cancer” category. In addition, there were 30 studies for the Asian population and 12 studies for the Caucasian population. As for control source, 10 studies employed PB control, while 29 studies conducted their studies applying HB control. Moreover, the estimated quality of each study was in the range of 6–9 points. The genotype distributions for control populations in all studies were conformed to HWE. Characteristics of included studies are shown in Tables 1 and 2.
Figure 1. Flow diagram of literature selection and data extraction for meta-analysis.
Quantitative data synthesis
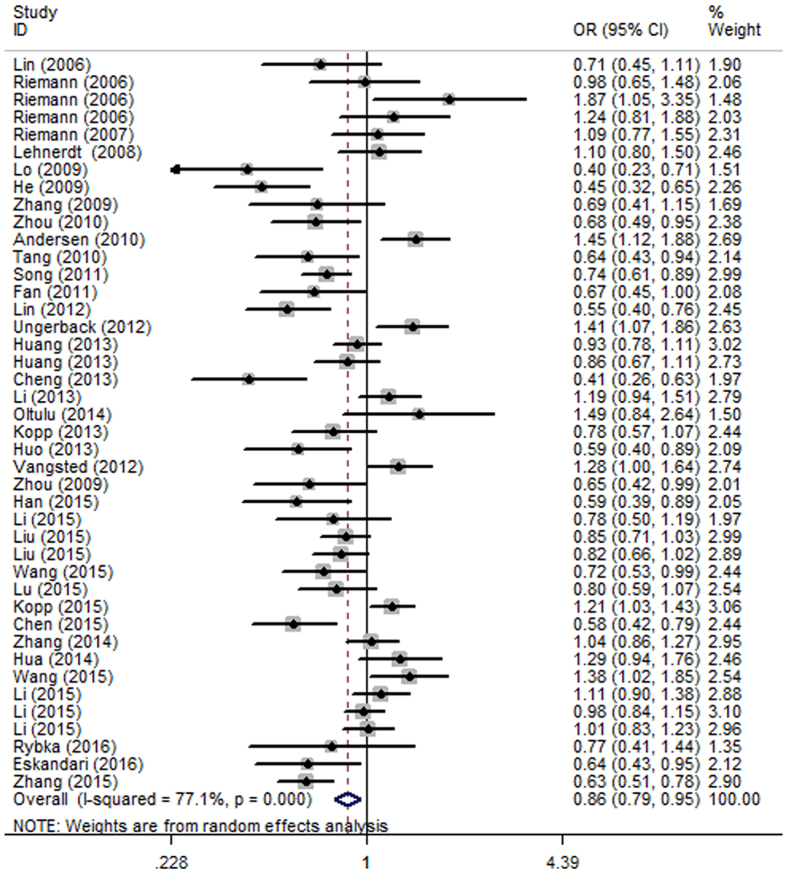
The meta-analysis results have been summarized in Table 3. For the presence of moderate heterogeneity across all levels of analysis, random-effects model (REM) was applied to calculate the summary odds ratios (OR) in all five genetic models (Table 4). Overall, a significantly decreased risk of cancer was associated with NFKB1 -94 ins/del ATTG polymorphism in all five genetic models (DD+DW vs. WW: OR = 0.86, 95% CI = 0.79–0.95, P = 0.002; DD vs. DW+WW: OR = 0.84, 95% CI = 0.74–0.94, P = 0.003; DD vs. WW: OR = 0.77, 95% CI = 0.66–0.90, P = 0.001; DW vs. WW: OR = 0.90, 95% CI = 0.83–0.98, P = 0.011; D vs. W: OR = 0.89, 95% CI = 0.83–0.96, P = 0.002) (Fig. 2). In addition, corrected P values for multiple testing remained significant (Table 4).
Table 3. Summary of overall results and subgroup for the association between the NF-κB1-94 ins/del ATTG promoter polymorphism and cancer risk.
| No. | Sample size (N) |
DD+DI vs. II |
DD vs. DI+II |
DD vs. II |
DI vs. II |
D vs. I |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | POR a | OR (95% CI) | POR a | OR (95% CI) | POR a | OR (95% CI) | POR a | OR (95% CI) | POR a | ||
| Overall | 42 | 18,222 | 24,778 | 0.86 (0.79–0.95) | 0.002 | 0.84 (0.74–0.94) | 0.003 | 0.77 (0.66–0.90) | 0.001 | 0.90 (0.83–0.98) | 0.011 | 0.89 (0.83–0.96) | 0.002 |
| Control source | |||||||||||||
| PB | 10 | 5,369 | 8,212 | 0.90 (0.72–1.13) | 0.354 | 0.77 (0.62–0.96) | 0.02 | 0.72 (0.52–1.01) | 0.053 | 0.96 (0.78–1.17) | 0.689 | 0.89 (0.76–1.04) | 0.15 |
| HB | 29 | 12,463 | 15,976 | 0.85 (0.77–0.95) | 0.003 | 0.86 (0.74–0.99) | 0.04 | 0.78 (0.65–0.93) | 0.007 | 0.88 (0.81–0.96) | 0.004 | 0.89 (0.81–0.97) | 0.006 |
| Ethnicity | |||||||||||||
| Caucasian | 12 | 3,413 | 6,887 | 1.20 (1.1–1.31) | <0.0001 | 1.03 (0.92–1.16) | 0.583 | 1.15 (1.01–1.31) | 0.03 | 1.22 (1.11–1.34) | <0.0001 | 1.11 (1.04–1.17) | 0.001 |
| Asian | 30 | 14,809 | 17,891 | 0.77 (0.70–0.86) | <0.0001 | 0.77 (0.67–0.90) | 0.001 | 0.66 (0.55–0.80) | <0.0001 | 0.85 (0.81–0.90) | <0.0001 | 0.82 (0.76–0.90) | <0.0001 |
| Genotyping methods | |||||||||||||
| PCR-RFLP | 11 | 5,450 | 6,525 | 0.75 (0.62–0.90) | 0.002 | 0.72 (0.60–0.87) | 0.001 | 0.62 (0.47–0.82) | 0.001 | 0.81 (0.70–0.94) | 0.005 | 0.87 (0.80–0.95) | 0.001 |
| MassARRAY | 3 | 998 | 1,096 | 0.77 (0.44–1.32) | 0.334 | 0.93 (0.43–2.01) | 0.862 | 0.77 (0.29–2.07) | 0.608 | 0.75 (0.53–1.07) | 0.111 | 0.87 (0.80–0.95) | 0.609 |
| TaqMan | 14 | 8,505 | 12,059 | 0.95 (0.83–1.09) | 0.446 | 0.99 (0.83–1.21) | 0.977 | 0.95 (0.76–1.19) | 0.654 | 0.95 (0.84–1.07) | 0.375 | 0.87 (0.80–0.95) | 0.685 |
| PCR-PAGE | 6 | 1,284 | 1,599 | 0.08 | 0.65 (0.50–0.85) | 0.002 | 0.58 (0.39–0.86) | 0.008 | 0.86 (0.66–1.12) | 0.257 | 0.87 (0.80–0.95) | 0.015 | |
| PCR-CGE | 2 | 241 | 349 | 0.70 (0.50–0.98) | 0.037 | 0.61 (0.26–1.40) | 0.239 | 0.51 (0.23–1.13) | 0.096 | 0.78 (0.55–1.12) | 0.174 | 0.87 (0.80–0.95) | 0.017 |
| Pyrosequencing | 4 | 593 | 1,228 | 1.18 (0.94–1.48) | 0.146 | 0.96 (0.72–1.28) | 0.756 | 1.06 (0.78–1.45) | 0.693 | 1.23 (0.94–1.60) | 0.125 | 0.87 (0.80–0.95) | 0.364 |
| Quality score | |||||||||||||
| <7 points | 24 | 8,849 | 13,548 | 0.013 | 0.81 (0.71–0.91) | 0.001 | 0.75 (0.63–0.89) | 0.001 | 0.91 (0.82–1.01) | 0.076 | 0.87 (0.80–0.95) | 0.002 | |
| >7 points | 18 | 9,373 | 11,230 | 0.87 (0.74–1.01) | 0.066 | 0.87 (0.70–1.08) | 0.205 | 0.79 (0.60–1.04) | 0.086 | 0.89 (0.78–1.01) | 0.073 | 0.91 (0.80–1.03) | 0.126 |
| Cancer Type | |||||||||||||
| OSCC | 2 | 674 | 721 | 0.60 (0.46–0.77) | <0.0001 | 0.63 (0.49–0.81) | <0.0001 | 0.49 (0.33–0.72) | <0.0001 | 0.67 (0.51–0.88) | 0.004 | 0.70 (0.60–0.82) | <0.0001 |
| CRC | 5 | 2,777 | 4,409 | 1.12 (0.85–1.48) | 0.409 | 0.98 (0.76–1.26) | 0.849 | 1.06 (0.74–1.52) | 0.768 | 1.14 (0.87–1.49) | 0.336 | 1.06 (0.88–1.27) | 0.558 |
| BC | 4 | 1,788 | 1,955 | 0.884 | 1.16 (0.68–1.97) | 0.578 | 1.12 (0.64–1.96) | 0.692 | 0.95 (0.81–1.11) | 0.48 | 1.05 (0.82–1.36) | 0.686 | |
| GC | 2 | 583 | 549 | 0.74 (0.24–2.31) | 0.603 | 1.00 (0.26–3.85) | 0.996 | 0.78 (0.12–5.12) | 0.799 | 0.72 (0.33–1.57) | 0.41 | 0.90 (0.34–2.24) | 0.814 |
| HCC | 3 | 961 | 2,530 | 0.59 (0.30–1.15) | 0.119 | 0.67 (0.40–1.10) | 0.114 | 0.50 (0.21–1.16) | 0.106 | 0.65 (0.37–1.15) | 0.137 | 0.69 (0.44–1.09) | 0.114 |
| PC | 4 | 2,207 | 2,358 | 0.79 (0.61–1.02) | 0.074 | 0.89 (0.66–1.20) | 0.447 | 0.72 (0.45–1.16) | 0.177 | 0.84 (0.71–1.00) | 0.05 | 0.88 (0.74–1.05) | 0.149 |
| OC | 4 | 1,463 | 1,573 | 0.67 (0.56–0.79) | <0.0001 | 0.68 (0.52–0.89) | 0.005 | 0.54 (0.40–0.73) | <0.0001 | 0.73 (0.61–0.87) | 0.001 | 0.75 (0.65–0.86) | <0.0001 |
| LC | 5 | 2,793 | 2,921 | 0.83 (0.67–1.03) | 0.092 | 0.82 (0.54–1.26) | 0.368 | 0.77 (0.48–1.26) | 0.302 | 0.84 (0.74–0.96) | 0.008 | 0.88 (0.70–1.09) | 0.246 |
| NPC | 3 | 1,573 | 2,182 | 0.82 (0.72–0.93) | 0.003 | 0.74 (0.63–0.88) | <0.0001 | 0.63 (0.49–0.81) | <0.0001 | 0.63 (0.49–0.81) | 0.067 | 0.83 (0.76–0.91) | <0.0001 |
| RCC | 2 | 1,356 | 1,895 | 1.01 (0.87–1.17) | 0.892 | 1.01 (0.72–1.40) | 0.973 | 1.06 (0.85–1.31) | 0.625 | 1.07 (0.78–1.47) | 0.688 | 1.02 (0.92–1.13) | 0.71 |
| Other cancers | 8 | 1,867 | 3,685 | 0.99 (0.78–1.27) | 0.962 | 0.79 (0.59–1.05) | 0.1 | 0.81 (0.55–1.21) | 0.307 | 1.07 (0.86–1.31) | 0.553 | 0.93 (0.77–1.13) | 0.468 |
OR, odds ratio; CI, confidence interval.
P values in bold denotes significance.
aThe P values of Z test for odds ratios test.
Table 4. Summary of the corrected P value for multiple testing and heterogeneity test in this meta-analysis.
| No. | Sample size (N) |
DD+DI vs. II |
DD vs. DI+II |
DD vs. II |
DI vs. II |
D vs. I |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | PCorra | I2(%)b | PHet c | PCorra | I2(%)b | PHet c | PCorra | I2(%)b | PHet c | PCorra | I2(%)b | PHet c | PCorra | I2(%)b | PHetc | ||
| Overall | 42 | 18,222 | 24,778 | 0.005 | 77.1 | <0.0001 | 0.0037 | 80.2 | <0.0001 | 0.005 | 83.8 | <0.0001 | 0.011 | 64.1 | <0.0001 | 0.005 | 84 | <0.0001 |
| Control source | ||||||||||||||||||
| PB | 10 | 5,369 | 8,212 | 0.443 | 86.6 | <0.0001 | 0.1 | 81.5 | <0.0001 | 0.133 | 83.1 | <0.0001 | 0.689 | 71.3 | <0.0001 | 0.25 | 87.8 | <0.0001 |
| HB | 29 | 12,463 | 15,976 | 0.015 | 73.3 | <0.0001 | 0.04 | 77.5 | <0.0001 | 0.0088 | 79.1 | <0.0001 | 0.01 | 53.9 | <0.0001 | 0.01 | 81.2 | <0.0001 |
| Ethnicity | ||||||||||||||||||
| Caucasian | 12 | 3,413 | 6,887 | <0.001 | 35.3 | 0.108 | 0.583 | 21.8 | 0.229 | 0.0375 | 31.9 | 0.136 | <0.001 | 32.8 | 0.128 | 0.0017 | 32 | 0.135 |
| Asian | 30 | 14,809 | 17,891 | <0.001 | 74 | <0.0001 | 0.001 | 84.5 | <0.0001 | <0.001 | 86.3 | <0.0001 | <0.001 | 46.5 | 0.003 | <0.001 | 81.1 | <0.0001 |
| Genotyping methods | ||||||||||||||||||
| PCR-RFLP | 11 | 5,450 | 6,525 | 0.0025 | 73.6 | <0.0001 | 0.003 | 71.9 | <0.0001 | 0.005 | 78.9 | <0.0001 | 0.005 | 55.7 | 0.012 | 0.002 | 80.6 | <0.0001 |
| MassARRAY | 3 | 998 | 1,096 | 0.835 | 86.7 | 0.001 | 0.862 | 89 | <0.0001 | 1.000 | 89.1 | <0.0001 | 0.555 | 65 | 0.057 | 0.761 | 88 | <0.0001 |
| TaqMan | 14 | 8,505 | 12,059 | 1.000 | 78.4 | <0.0001 | 0.977 | 84.2 | <0.0001 | 1.000 | 85.1 | <0.0001 | 1.000 | 69 | <0.0001 | 0.856 | 85.2 | <0.0001 |
| PCR-PAGE | 6 | 1,284 | 1,599 | 0.1 | 70.1 | 0.005 | 0.01 | 39.5 | 0.142 | 0.02 | 67.1 | 0.01 | 0.257 | 58.6 | 0.034 | 0.025 | 72.1 | 0.003 |
| PCR-CGE | 2 | 241 | 349 | 0.0925 | 0 | 0.722 | 0.239 | 55.7 | 0.133 | 0.16 | 43.6 | 0.183 | 0.218 | 0 | 0.879 | 0.085 | 13.4 | 0.283 |
| Pyrosequencing | 4 | 593 | 1,228 | 0.365 | 12.4 | 0.331 | 0.756 | 6.9 | 0.358 | 0.866 | 0 | 0.402 | 0.625 | 27.7 | 0.246 | 0.607 | 0 | 0.472 |
| Quality score | ||||||||||||||||||
| <7 points | 24 | 8,849 | 13,548 | 0.016 | 71.6 | <0.0001 | 0.005 | 61 | <0.0001 | 0.003 | 74.5 | <0.0001 | 0.076 | 57.5 | <0.0001 | 0.0033 | 76.7 | <0.0001 |
| >7 points | 18 | 9,373 | 11,230 | 0.33 | 78.9 | <0.0001 | 0.205 | 82 | <0.0001 | 0.143 | 85.3 | <0.0001 | 0.183 | 67.5 | <0.0001 | 0.158 | 87 | <0.0001 |
| Cancer Type | ||||||||||||||||||
| OSCC | 2 | 674 | 721 | <0.001 | 0 | 0.377 | <0.001 | 3.9 | 0.308 | <0.001 | 33 | 0.222 | 0.004 | 0 | 0.57 | <0.001 | 6.9 | 0.3 |
| CRC | 5 | 2,777 | 4,409 | 1.000 | 85 | <0.0001 | 0.849 | 68.3 | 0.013 | 0.96 | 81.4 | <0.0001 | 1.000 | 82.1 | <0.0001 | 0.93 | 84.4 | <0.0001 |
| BC | 4 | 1,788 | 1,955 | 0.884 | 60.8 | 0.054 | 1.000 | 88.4 | <0.0001 | 0.865 | 87.1 | <0.0001 | 1.000 | 11.3 | 0.336 | 0.966 | 85.7 | <0.0001 |
| GC | 2 | 583 | 549 | 1.000 | 84.4 | <0.0001 | 0.996 | 90.3 | <0.0001 | 1.000 | 91.4 | <0.0001 | 1.000 | 80.9 | 0.022 | 1.000 | 91.6 | <0.0001 |
| HCC | 3 | 961 | 2,530 | 0.149 | 92.2 | <0.0001 | 0.285 | 82.4 | 0.003 | 0.53 | 89.9 | <0.0001 | 0.137 | 85.5 | <0.0001 | 0.19 | 89.8 | <0.0001 |
| PC | 4 | 2,207 | 2,358 | 0.185 | 57.6 | 0.07 | 0.447 | 70 | 0.019 | 0.221 | 78.2 | 0.003 | 0.25 | 9.6 | 0.345 | 0.248 | 69.5 | 0.02 |
| OC | 4 | 1,463 | 1,573 | <0.001 | 0 | 0.462 | 0.005 | 51.6 | 0.102 | <0.001 | 39.8 | 0.173 | 0.0012 | 0 | 0.416 | <0.001 | 38.5 | 0.181 |
| LC | 5 | 2,793 | 2,921 | 0.23 | 69.1 | 0.012 | 0.368 | 86.3 | <0.0001 | 0.378 | 86.9 | <0.0001 | 0.04 | 12 | 0.337 | 0.41 | 83.2 | <0.0001 |
| NPC | 3 | 1,573 | 2,182 | 0.0038 | 0 | 0.504 | <0.001 | 0 | 0.728 | <0.001 | 0 | 0.561 | 0.067 | 0 | 0.633 | <0.001 | 0 | 0.426 |
| RCC | 2 | 1,356 | 1,895 | 1.000 | 1.9 | 0.313 | 0.973 | 34.2 | 0.218 | 1.000 | 0 | 0.583 | 1.000 | 53.6 | 0.142 | 1.000 | 0 | 0.945 |
| Other cancers | 8 | 1,867 | 3,685 | 0.962 | 71.8 | 0.001 | 0.500 | 53.2 | 0.046 | 0.768 | 76.4 | 0.002 | 0.691 | 56.4 | 0.025 | 0.78 | 77.6 | <0.0001 |
P values in bold denotes significance.
aP Values were corrected to adjust for multiple testing.
bThe value of I2 statistics for heterogeneity test.
cP value of the Q-test for heterogeneity test.
Figure 2. Forest plot of cancer risk associated with NFKB1 -94 ins/del ATTG polymorphism in dominant model (DD+DW vs. WW) for the overall population.
Subgroup analysis
To investigate the moderate heterogeneity in the outcomes and the inference of the studies, we pooled the odds ratios (OR) and 95% confidence interval (CI) from further subgroup analyses of ethnicity, control source, genotyping method, cancer type, and quality score. When stratified by control source, REM was utilized due to the presence of heterogeneity in all above genetic models (Table 4). Consistently, NFKB1 -94 ins/del ATTG polymorphism was significantly associated with decreased cancer susceptibility among HB population in all genetic models, among which the most obvious is for dominant model (DD+DW vs. WW: OR = 0.85, 95% CI = 0.77–0.95, P = 0.003), and weakest for recessive model (DD vs. DW+WW: OR = 0.86, 95% CI = 0.74–0.99, P = 0.04). Corrected P values for multiple testing remained significant. However, no such significant relationship was detected for PB population in any genetic model (Table 3).
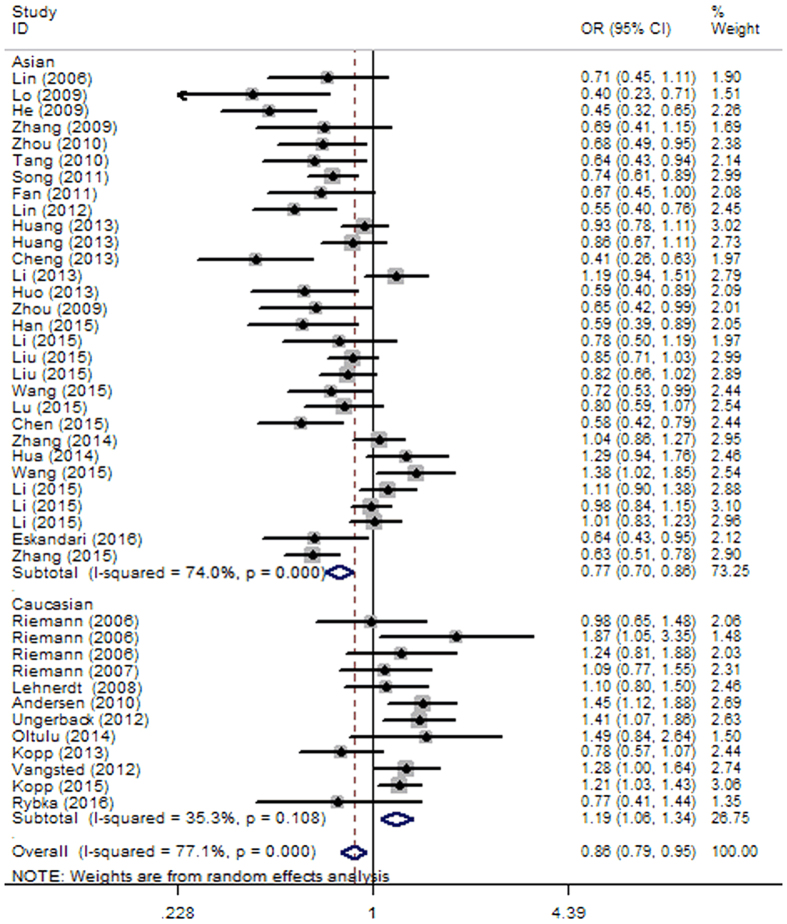
Subgroup analysis based on ethnicity was also conducted in this analysis. Since moderate heterogeneity was observed in all genetic models, we used the REM of analysis. The overall results showed that a significantly strong association between NFKB1 -94 ins/del ATTG polymorphism and decreased cancer risk was seen for Asian population in all genetic models (DD+DW vs. WW: OR = 0.77, 95% CI = 0.70–0.86, P < 0.0001; DD vs. DW+WW: OR = 0.77, 95% CI = 0.67–0.90, P = 0.001; DD vs. WW: OR = 0.66, 95% CI = 0.55–0.80, P < 0.0001; DW vs. WW: OR = 0.85, 95% CI = 0.81–0.90, P < 0.0001; D vs. W: OR = 0.82, 95% CI = 0.76–0.90, P < 0.0001) (Fig. 3). In contrast, the Caucasian population exhibit NFKB1 -94 ins/del ATTG promoter variant associated increased susceptibility to cancer in dominant (DD+DW vs. WW: OR = 1.20, 95% CI = 1.10–1.31, P < 0.0001), homozygous (DD vs. WW: OR = 1.15, 95% CI = 1.01–1.31, P = 0.03), heterozygous (DW vs. WW: OR = 1.22, 95% CI = 1.11–1.34, P < 0.0001), and allelic (D vs. W: OR = 1.11, 95% CI = 1.04–1.17, P = 0.001) models (Table 3), with low heterogeneity found in Caucasians (Table 4). Moreover, no significant variables were identified after corrections for multiple comparisons.
Figure 3. Forest plot of cancer risk associated with NFKB1 -94 ins/del ATTG polymorphism in dominant model (DD+DW vs. WW) for the subgroup analysis by ethnicity (Caucasian and Asian).
Subgroup analysis was conducted among 6 methods for genotyping. REM was used since moderate heterogeneity was identified in this subgroup analysis. In the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) group and polymerase chain reaction-polyacrylamide gel electrophoresis (PCR-PAGE) group, significantly decreased association between NFKB1 -94 ins/del ATTG promoter variant and cancer risk were identified, among which the most obvious is for recessive model in the two groups (PCR-RFLP group: DD vs. DW+WW: OR = 0.72, 95% CI = 0.60–0.87, P = 0.001; PCR-PAGE group: DD vs. DW+WW: OR = 0.65, 95% CI = 0.50–0.85, P = 0.002), respectively. In contrast, no relationship was detected in TaqMan, MassARRAY, PCR-CGE, and Pyrosequencing group (Table 3). When stratified by quality score, significant decreased association was only identified in the group whose quality score ≤ 7 points in dominant (DD+DW vs. WW: OR = 0.86, 95% CI = 0.76–0.97, P = 0.013), recessive (DD vs. DW+WW: OR = 0.81, 95% CI = 0.71–0.91, P = 0.001), homozygous (DD vs. WW: OR = 0.75, 95% CI = 0.63–0.89, P = 0.001), and allelic (D vs. W: OR = 0.87, 95% CI = 0.80–0.95, P = 0.002) model (Table 3).
In addition, stratification analysis by cancer type was also performed. Further analysis showed that the substitution was significantly related to decreased risk of OSCC (DD+DW vs. WW: OR = 0.60, P < 0.0001; DD vs. DW+WW: OR = 0.63, P < 0.0001; DD vs. WW: OR = 0.49, P < 0.0001; DW vs. WW: OR = 0.67, P = 0.004; D vs. W: OR = 0.70, P < 0.0001), OC (DD+DW vs. WW: OR = 0.67, P < 0.0001; DD vs. DW+WW: OR = 0.68, P = 0.005; DD vs. WW: OR = 0.54, P < 0.0001; DW vs. WW: OR = 0.73, P = 0.001; D vs. W: OR = 0.75, P < 0.0001), and NPC (DD+DW vs. WW: OR = 0.82, P = 0.003; DD vs. DW+WW: OR = 0.74, P < 0.0001; DD vs. WW: OR = 0.63, P < 0.0001; D vs. W: OR = 0.83, P < 0.0001) in genetic models. Moreover, significantly decreased relationship between the polymorphism and LC risk was only found in heterozygous model (DW vs. WW: OR = 0.84, 95% CI = 0.74–0.96, P = 0.008). In addition, corrected p values for multiple testing remained significant (Table 4). Nevertheless, there was no significant relationship between the polymorphism and the remaining cancer types in any genetic model, including CRC, RCC, BC, GC, HCC, PC and other cancer (Table 3).
Publication bias
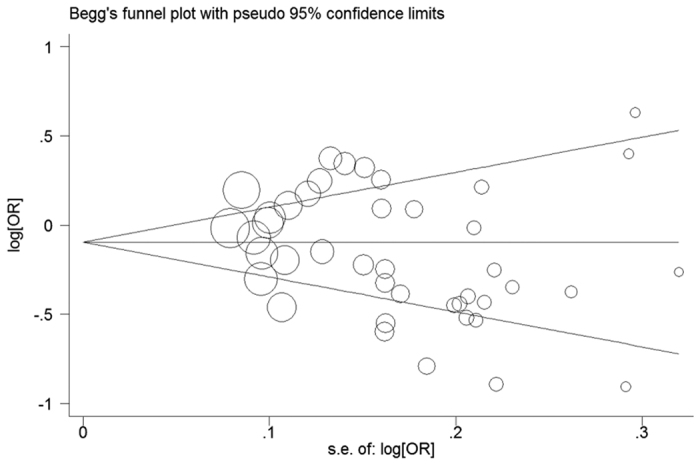
Publication bias was assessed through the Begg’s funnel plot and Egger’s regression intercept tests. The shape of the Begg’s funnel plot did not reveal basically asymmetric distribution in all comparisons of the overall population. Moreover, the result of Egger’s test (dominant model: P = 0.269; recessive model: P = 0.202; homozygous model: P = 0.133; heterozygous model: P = 0.471; allelic model: P = 0.245) further provided no evidence of significant publication bias. Our observation of symmetric funnel plots and non-significant statistical tests confirmed no publication bias (Fig. 4).
Figure 4. Begg’s funnel plot of publication bias test for the association between NFKB1 -94 ins/del ATTG polymorphism and cancer risk under dominant model (DD+DW vs. WW).
Sensitivity analyses
To validate our results, we further conducted sensitivity analyses to evaluate the stability of these results in this meta-analysis. Firstly, the random-effects model was compared with the fixed-effects model, and the statistically similar results were obtained in all genetic models. Secondly, we applied leave-one-out method by sequentially excluding each study to assess the influence of individual study on the obtained conclusions. The conclusions remained unchanged in all comparisons, suggesting the stability of our meta-analysis. Furthermore, due to the higher number of studies of Asian population, we conducted the analysis with randomly selecting 50% of the case-control studies for many times to determine the sensitivity. However, the corresponding pooled ORs were generally similar in each comparison (data not shown). Therefore, our conclusion in this meta-analysis was stable and credible.
Discussion
The NF-κB family constitutes a class of pleiotropic transcription factors, which seems to contribute to tumor angiogenesis and progression by regulating the transcription of genes involved in immune response, cell proliferation, apoptosis, and angiogenesis17,56. The NF-κB family members can form homo-dimmers and hetero-dimmers to produce more than 15 distinct active complexes, among which p50/p65 heterodimer is one of the most sufficient forms and is present in almost all cell types57. NF-κB complexes are constitutively active in various cancer cell lines and tissues, indicating the involvement of NF-κB in tumorigenesis58,59. In addition, various inflammatory cytokines, such as TNF-a, IL-6, and IL-8, regulated their biological effects by activating NF-κB. In consequence, the abnormality and disturbance of NF-κB function can reduce diverse biological processes, including uncontrolled cell proliferation and suppression of apoptosis, thereby rendering a cell more prone to malignant transformation31.
A functional -94 ins/del ATTG (rs28362491) polymorphism in the promoter region of the p50/p105-encoding NFKB1 gene located between two received key promoter regulatory elements modulates its transcription and then affects the production of protein subunits18,31. Previous studies have shown that the -94del allele may result in significantly decreased NFKB1 transcript level and hence, reduce p50/p105 protein production18,20. Therefore, the different level of the protein subunits between the carriers of -94del allele and -94ins allele may be expected to contribute to inter-individual variations in cancer risk.
In this meta-analysis, we investigated the -94 ins/del ATTG on NFKB1 gene locus with 42 separate case-control studies (18,222 cases and 24,778 controls) focusing on the relationship of this variant to cancer risk. The overall results revealed a significant association between -94 del/del variant genotype carriers and decreased cancer risk. Despite the mechanism remains unclear, considering the critical role of NF-κB in various biological pathways, we could hypothesize that ATTG deletion causes the loss of binding to nuclear proteins, which leads to reduced promoter activity13. The promoter sequence with -94 del allele results in lower transcriptional activity and thereby causes decreased levels of p50/p105 in the cytoplasm, which serves a critical function in trans-activating anti-apoptosis genes and inhibiting cell apoptosis, thereby promoting cellular proliferation. In addition, the p50 in -94 del carriers can form lesser heterodimers with p65 to mediate the inflammatory pathway compared to the -94 ins allele carriers. Therefore, the -94 del allele may play a protective role in cancer risk.
However, we must treat these results cautiously when referring to the findings, because moderate heterogeneity was detected in this meta-analysis, which may be attributed to different characteristics of the cohort, environmental exposure, clinical information and intrinsic complexity of cancer architecture. In addition, there are 30 studies for the Asian population and 12 studies for the Caucasian population. The high proportion of over-represented studies of the Asian population in the combined analysis clearly resulted in a bias, which affected the overall results. Therefore, we conducted a subgroup analysis to identify the origin of heterogeneity by ethnicity, control source, genotyping method, cancer type, and quality score.
When analysis was restricted to ethnicity, the variant del allele could confer a decreased or increased risk of developing cancer to its carriers among Asians and Caucasians, respectively. Natural selection and balance to other genetic variants may result in different genotype frequencies in different ethnicities. In addition, accumulated evidence demonstrated that individuals with the same genotype living in the different areas with different environments and life style might have a different cancer risk7,20,31. Therefore, this discrepancy between Caucasians and Asians could be attributed to the possible differences in genetic background, gene-environment, and non-genetic risk factors, such as smoking and high intake of salted foods. In populations where the -94del allele was identified to increase cancer risk, a plausible interpretation for such observation could be that the p50 protein lacks the C-terminal transactivation domain and may form inhibitory homo-dimers, which can serve as a role in repressing pro-inflammatory genes and then resulting in decreased levels of p50/p50 repressive homo-dimers in -94 del allele carriers60,61. Consequently, -94 del allele carriers may be genetically with a higher inflammatory response. Interestingly, the results were contrary to the results of two previous meta-analyses33,34 that no significant association was seen between NFKB1 -94 ins/del ATTG polymorphism and the risk of cancer in Caucasians. The contradictory results are interpreted by the following reasons: (1) the small sample was included in the previous two meta-analyses. Therefore, the results may reduce the power to reveal a reliable relationship. There were twelve case-control studies eligible for this meta-analysis among Caucasians, so the result may be closer to the real value; (2) the previous studies included many studies, which do not follow HWE. These studies may make the result incredible; (3) the sensitivity analysis was conducted through three different methods in this meta-analysis, and the results were consistent with the previous results, suggesting the results of this study were stable; (4) different clinicopathological features of cases included, different experimental designs and genotyping methods utilized in these studies; (5) we used Benjamini–Hochberg procedures to adjust for multiple comparisons in setting the level of significance. There were no significant variables after corrections for multiple comparisons.
The stronger and significantly decreased risk of cancer when analysis was restricted to hospital-based (HB) group was observed in all genetic model, but such association was not seen in population-based (PB) group. Interestingly, it was noted that moderate heterogeneity appeared in the HB group, which may result from selection bias. Additionally, HB controls were selected from hospital, which may not validly represent the exposure situation of the overall population. Therefore, results from the PB controls were thought to be more reliable.
Cancer was an overly broad collection of different types, which may have confounding factors to not reveal a reliable association. Therefore, further stratification analysis by cancer type was also conducted. The decreased risk of cancer also remained in the subgroups of oral, ovarian, and nasopharyngeal carcinoma in Asian populations. Moreover, the clinical and demographic information are essential factors affecting the overall results. Zhang et al.26 and Wang et al.25 reported that the increased risk of -94 ins/del ATTG polymorphism remained when analyses were done according to gender and smoking status in lung cancer among Asian population. The study by Oltulu et al.13 demonstrated that gender and age associated with lung cancer among Caucasian population. Additionally, Han et al.38 demonstrated that the impact of the -94 ins/del ATTG polymorphism was not present in nonsmokers and younger subjects in prostate cancer. Consequently, only increasing the number of populations, without taking into clinical and demographic information into account, will not guarantee the validity of the results. We tried to collect some subsets of clinical information and demographic data from the eligible publications to conduct further analysis. However, on one side, the relatively small sample size could reduce the statistical power of this study and result in unreliable association. On the other side, these data were collected from various cancers and ethnicities, which may give rise to confounding variables to reveal an unreliable association. Therefore, in my opinion, a better result can be given exclusively from a case-control study of large sample size, the same cancer type, and the same ethnicity.
We note several potential limitations of this meta-analysis when interpreting these results. First, we were unable to perform further analysis to investigate other risk factors of cancer risk for insufficient original data, such as age, tumor location, and gene-environment/gene-gene interaction. Second, moderate between-study heterogeneity was checked in some genetic comparisons. Further subgroup analysis showed that cancer type and control source might be the source of heterogeneity. Nevertheless, there is other inexplicable heterogeneity affecting the results. Third, publication bias might have occurred for only including published articles. Some relevant published studies or unpublished studies with null results were missed, which may affect our results. Fourth, many studies included in this meta-analysis were performed in Asian and Caucasian populations, so the association needs to be verified in other ethnicities. Despite these limitations, we created a strict protocol, and performed study selection, data identification and statistical analysis to reduce potential bias through the whole process. Thus, the objectivity and reliability of the results are guaranteed.
In conclusion, to the best of our knowledge, this is a comprehensive study to assess the relationship between NFKB1 -94 ins/del ATTG polymorphism and cancer risk. Our results indicated that the -94 ins/del ATTG polymorphism was significantly associated with decreased cancer risk in Asian populations, but increased cancer risk in Caucasian populations. However, due to the limitations of this current meta-analysis, the conclusion might be not conclusive. Therefore, in the future, there will be a need for larger sample size and more ethnic groups to further confirm the results of this meta-analysis.
Additional Information
How to cite this article: Wang, D. et al. Genetic association between NFKB1 −94 ins/del ATTG Promoter Polymorphism and cancer risk: a meta-analysis of 42 case-control studies. Sci. Rep. 6, 30220; doi: 10.1038/srep30220 (2016).
Footnotes
Author Contributions Z.Z. and F.P. conceived and designed this study; D.W., H.W. and J.X. searched databases and collected the data; D.W., J.X. and T.X. performed the statistical analysis; J.X., W.Z., K.Z. and S.R. prepared Tables 1,2–3 and supplemental Tables 1–2; H.W., S.R., W.Z. and T.X. prepared Figures 1,2,3–4; K.Z., D.W. and J.X. wrote the manuscript. All authors reviewed the final manuscript.
References
- Hammam O. et al. A possible role for TNF-α in coordinating inflammation and angiogenesis in chronic liver disease and hepatocellular carcinoma. Gastrointestinal cancer research: GCR 6, 107 (2013). [PMC free article] [PubMed] [Google Scholar]
- Vineis P. & Wild C. P. Global cancer patterns: causes and prevention. The Lancet 383, 549–557 (2014). [DOI] [PubMed] [Google Scholar]
- Bray F., Jemal A., Grey N., Ferlay J. & Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. The lancet oncology 13, 790–801 (2012). [DOI] [PubMed] [Google Scholar]
- Foulkes W. D. Inherited susceptibility to common cancers. The New England journal of medicine 359, 2143–2153, 10.1056/NEJMra0802968 (2008). [DOI] [PubMed] [Google Scholar]
- Jiao J., Wu J., Huang D. & Liu L. Lack of association of the iNOS gene polymorphism with risk of cancer: a systematic review and Meta-Analysis. Scientific reports 5, 9889, 10.1038/srep09889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Zhu M., Wang Y., He J. & Zheng L. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta-analysis. Scientific reports 5, 7986, 10.1038/srep07986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewander A. et al. Polymorphism in the promoter region of the NFKB1 gene increases the risk of sporadic colorectal cancer in Swedish but not in Chinese populations. Scand J Gastroenterol 42, 1332–1338, 10.1080/00365520701396026 (2007). [DOI] [PubMed] [Google Scholar]
- Lehnerdt G. et al. No Association of the NF-κB1− 94INS/Delattg Promoter Polymorphism with Relapse-Free and Overall Survival in Patients with Squamous Cell Carcinomas of the Head and Neck Region. International journal of immunopathology and pharmacology 21, 827–832 (2008). [DOI] [PubMed] [Google Scholar]
- Burnik F. S. & Yalcin S. NFKB1 -94 insertion/deletion ATTG polymorphism in gastroenteropancreatic neuroendocrine tumors. Chemotherapy 55, 381–385, 10.1159/000237744 (2009). [DOI] [PubMed] [Google Scholar]
- Lo S. S., Chen J. H., Wu C. W. & Lui W. Y. Functional polymorphism of NFKB1 promoter may correlate to the susceptibility of gastric cancer in aged patients. Surgery 145, 280–285, 10.1016/j.surg.2008.11.001 (2009). [DOI] [PubMed] [Google Scholar]
- Chen W., Li Z., Bai L. & Lin Y. NF-kappaB in lung cancer, a carcinogenesis mediator and a prevention and therapy target. Frontiers in bioscience (Landmark edition) 16, 1172–1185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koti M. et al. Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC cancer 13, 549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltulu Y. M. et al. Investigation of NF-kappaB1 and NF-kappaBIA gene polymorphism in non-small cell lung cancer. Biomed Res Int 2014, 530381, 10.1155/2014/530381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi G., Sung B. & Aggarwal B. B. Nuclear factor-κB activation: from bench to bedside. Experimental Biology and Medicine 233, 21–31 (2008). [DOI] [PubMed] [Google Scholar]
- Salim P. et al. Interleukin‐10 Gene Promoter and NFKB1 Promoter Insertion/Deletion Polymorphisms in Systemic Sclerosis. Scandinavian journal of immunology 77, 162–168 (2013). [DOI] [PubMed] [Google Scholar]
- Lin S. C. et al. Areca (betel) nut extract activates mitogen-activated protein kinasesand NF-κB in oral keratinocytes. International journal of cancer 116, 526–535 (2005). [DOI] [PubMed] [Google Scholar]
- Hayden M. S. & Ghosh S. Shared principles in NF-κB signaling. Cell 132, 344–362 (2008). [DOI] [PubMed] [Google Scholar]
- Karban A. S. et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Human molecular genetics 13, 35–45 (2004). [DOI] [PubMed] [Google Scholar]
- Zhou B. et al. Relationship between NFKB1 -94 insertion/deletion ATTG polymorphism and susceptibility of cervical squamous cell carcinoma risk. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 21, 506–511, 10.1093/annonc/mdp507 (2010). [DOI] [PubMed] [Google Scholar]
- Song S. et al. NFkappaB1 and NFkappaBIA polymorphisms are associated with increased risk for sporadic colorectal cancer in a southern Chinese population. PloS one 6, e21726, 10.1371/journal.pone.0021726 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangsted A. J. et al. A functional polymorphism in the promoter region of the IL1B gene is associated with risk of multiple myeloma. British journal of haematology 158, 515–518, 10.1111/j.1365-2141.2012.09141.x (2012). [DOI] [PubMed] [Google Scholar]
- Kopp T. I., Friis S., Christensen J., Tjonneland A. & Vogel U. Polymorphisms in genes related to inflammation, NSAID use, and the risk of prostate cancer among Danish men. Cancer Genet 206, 266–278, 10.1016/j.cancergen.2013.06.001 (2013). [DOI] [PubMed] [Google Scholar]
- Lu Z. H. et al. Association between genetic polymorphisms of inflammatory response genes and the risk of ovarian cancer. J Formos Med Assoc, 10.1016/j.jfma.2015.01.002 (2015). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. A functional insertion/deletion polymorphism in the promoter region of the NFKB1 gene increases the risk of papillary thyroid carcinoma. Genet Test Mol Biomarkers 19, 167–171, 10.1089/gtmb.2014.0271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen L., Pan L., Xue J. & Yu H. The association between NFKB1-94ins/del ATTG polymorphism and non-small cell lung cancer risk in a Chinese Han population. Int J Clin Exp Med 8, 8153–8157 (2015). [PMC free article] [PubMed] [Google Scholar]
- Zhang J. W., Chen Q. S., Zhai J. X., Lv P. J. & Sun X. Y. Polymorphisms in NF-kappaB pathway genes & their association with risk of lung cancer in the Chinese population. Pakistan journal of medical sciences 31, 1411–1416, 10.12669/pjms.316.7935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybka J. et al. Variations in genes involved in regulation of the nuclear factor - kappaB pathway and the risk of acute myeloid leukaemia. International journal of immunogenetics 43, 101–106, 10.1111/iji.12255 (2016). [DOI] [PubMed] [Google Scholar]
- Andersen V., Christensen J., Overvad K., Tjonneland A. & Vogel U. Polymorphisms in NFkB, PXR, LXR and risk of colorectal cancer in a prospective study of Danes. BMC Cancer 10, 484, 10.1186/1471-2407-10-484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerback J. et al. Genetic variation and alterations of genes involved in NFkappaB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis 33, 2126–2134, 10.1093/carcin/bgs256 (2012). [DOI] [PubMed] [Google Scholar]
- Kopp T. I., Andersen V., Tjonneland A. & Vogel U. Polymorphisms in NFKB1 and TLR4 and interaction with dietary and life style factors in relation to colorectal cancer in a Danish prospective case-cohort study. PloS one 10, e0116394, 10.1371/journal.pone.0116394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Suzairi M. S. et al. The functional -94 insertion/deletion ATTG polymorphism in the promoter region of NFKB1 gene increases the risk of sporadic colorectal cancer. Cancer epidemiology 37, 634–638, 10.1016/j.canep.2013.05.007 (2013). [DOI] [PubMed] [Google Scholar]
- Tang T. et al. Insertion/deletion polymorphism in the promoter region of NFKB1 gene increases susceptibility for superficial bladder cancer in Chinese. DNA Cell Biol 29, 9–12, 10.1089/dna.2009.0937 (2010). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. Association between NFKB1 -94ins/del ATTG Promoter Polymorphism and Cancer Susceptibility: An Updated Meta-Analysis. International journal of genomics 2014, 612972, 10.1155/2014/612972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nian X. et al. Meta-analysis of studies on the association between the NF-kappaB1-94ins/del ATTG promoter polymorphism and cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 35, 11921–11931, 10.1007/s13277-014-2470-3 (2014). [DOI] [PubMed] [Google Scholar]
- Efird J. T. & Nielsen S. S. A method to compute multiplicity corrected confidence intervals for odds ratios and other relative effect estimates. International journal of environmental research and public health 5, 394–398 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 25, 2413–2419, 10.1093/annonc/mdu451 (2014). [DOI] [PubMed] [Google Scholar]
- Chen L. P., Cai P. S. & Liang H. B. Association of the genetic polymorphisms of NFKB1 with susceptibility to ovarian cancer. Genet Mol Res 14, 8273–8282, 10.4238/2015.July.27.15 (2015). [DOI] [PubMed] [Google Scholar]
- Han X. et al. Polymorphisms in NFKB1 and NFKBIA Genes Modulate the Risk of Developing Prostate Cancer among Han Chinese. Med Sci Monit 21, 1707–1715, 10.12659/msm.893471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. The relationship between functional promoter -94 ins/del ATTG polymorphism in {NF-kappa B1} gene and the risk of urinary cancer. Cancer biomarkers: section A of Disease markers, 10.3233/cbm-150536 (2015). [DOI] [PubMed] [Google Scholar]
- Li X., Zhang C., Qiao W., Zhou X. & Sun M. NFKB1 -94ins/del ATTG polymorphism increases osteosarcoma risk in a Chinese Han population. Int J Clin Exp Med 8, 1420–1423 (2015). [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Polymorphisms of NFkappaB1 and IkappaBalpha and Their Synergistic Effect on Nasopharyngeal Carcinoma Susceptibility. Biomed Res Int 2015, 362542, 10.1155/2015/362542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari-Nasab E., Hashemi M., Ebrahimi M. & Amininia S. The functional 4-bp insertion/deletion ATTG polymorphism in the promoter region of NF-KB1 reduces the risk of BC. Cancer biomarkers: section A of Disease markers 16, 109–115, 10.3233/cbm-150546 (2016). [DOI] [PubMed] [Google Scholar]
- Riemann K. et al. No association of the NFKB1 insertion/deletion promoter polymorphism with survival in colorectal and renal cell carcinoma as well as disease progression in B-cell chronic lymphocytic leukemia. Pharmacogenet Genomics 16, 783–788, 10.1097/01.fpc.0000230414.74726.f6 (2006). [DOI] [PubMed] [Google Scholar]
- Huang D. et al. Functional polymorphisms in NFkappaB1/IkappaBalpha predict risks of chronic obstructive pulmonary disease and lung cancer in Chinese. Hum Genet 132, 451–460, 10.1007/s00439-013-1264-9 (2013). [DOI] [PubMed] [Google Scholar]
- Lin S. C. et al. Functional polymorphism in NFKB1 promoter is related to the risks of oral squamous cell carcinoma occurring on older male areca (betel) chewers. Cancer letters 243, 47–54, 10.1016/j.canlet.2005.11.019 (2006). [DOI] [PubMed] [Google Scholar]
- Riemann K. et al. Insertion/deletion polymorphism in the promoter of NFKB1 as a potential molecular marker for the risk of recurrence in superficial bladder cancer. Int J Clin Pharmacol Ther 45, 423–430 (2007). [DOI] [PubMed] [Google Scholar]
- He Y. et al. IkappaBalpha gene promoter polymorphisms are associated with hepatocarcinogenesis in patients infected with hepatitis B virus genotype C. Carcinogenesis 30, 1916–1922, 10.1093/carcin/bgp226 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. et al. A functional insertion/deletion polymorphism in the promoter region of the NFKB1 gene increases susceptibility for prostate cancer. Cancer Genet Cytogenet 191, 73–77, 10.1016/j.cancergencyto.2009.01.017 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou B. et al. A functional insertion/deletion polymorphism in the promoter region of NFKB1 gene increases susceptibility for nasopharyngeal carcinoma. Cancer letters 275, 72–76, 10.1016/j.canlet.2008.10.002 (2009). [DOI] [PubMed] [Google Scholar]
- Fan Y. et al. NFKB1 insertion/deletion promoter polymorphism increases the risk of advanced ovarian cancer in a Chinese population. DNA Cell Biol 30, 241–245, 10.1089/dna.2010.1107 (2011). [DOI] [PubMed] [Google Scholar]
- Lin C. W. et al. Effects of NFKB1 and NFKBIA gene polymorphisms on susceptibility to environmental factors and the clinicopathologic development of oral cancer. PloS one 7, e35078, 10.1371/journal.pone.0035078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. W. et al. Effects of NFKB1 and NFKBIA gene polymorphisms on hepatocellular carcinoma susceptibility and clinicopathological features. PloS one 8, e56130, 10.1371/journal.pone.0056130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Z. H., Zhong H. J., Zhu Y. S., Xing B. & Tang H. Roles of functional NFKB1 and beta-TrCP insertion/deletion polymorphisms in mRNA expression and epithelial ovarian cancer susceptibility. Genet Mol Res 12, 3435–3443, 10.4238/2013.March.11.6 (2013). [DOI] [PubMed] [Google Scholar]
- Li P. et al. Functional promoter -94 ins/del ATTG polymorphism in NFKB1 gene is associated with bladder cancer risk in a Chinese population. PloS one 8, e71604, 10.1371/journal.pone.0071604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua T. et al. Nuclear factor-kappa B1 is associated with gastric cancer in a Chinese population. Medicine (Baltimore) 93, e279, 10.1097/md.0000000000000279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. & Verma I. M. NF-κB regulation in the immune system. Nature Reviews Immunology 2, 725–734 (2002). [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A. & Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harbor perspectives in biology 1, a000034, 10.1101/cshperspect.a000034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai D. I. et al. Constitutive activation of nuclear factor kappaB in hepatocellular carcinoma. Cancer 89, 2274–2281 (2000). [PubMed] [Google Scholar]
- Chen C. D. & Sawyers C. L. NF-kappa B activates prostate-specific antigen expression and is upregulated in androgen-independent prostate cancer. Molecular and cellular biology 22, 2862–2870 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkawy A. M. et al. The NF-kappaB p50:p50:HDAC-1 repressor complex orchestrates transcriptional inhibition of multiple pro-inflammatory genes. Journal of hepatology 53, 519–527, 10.1016/j.jhep.2010.03.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih V. F., Tsui R., Caldwell A. & Hoffmann A. A single NFkappaB system for both canonical and non-canonical signaling. Cell research 21, 86–102, 10.1038/cr.2010.161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]