Abstract
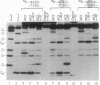
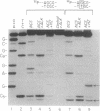
The diradical form of thiol-activated neocarzinostatin chromophore resides in the minor groove of DNA, where it has access to hydrogen atoms at the C-5', C-1', and C-4' positions of deoxyribose on each strand. In a dodecamer oligodeoxyribonucleotide containing the sequence AGC.GCT, a bistranded lesion staggered two nucleotides in the 3' direction, is generated that consists primarily of an abasic site (2'-deoxyribonolactone) at the C due to 1' chemistry and a direct strand break at the T due to 5' chemistry. Sequencing-gel analysis reveals that 72% of the damage at the C results from 1' chemistry with minor lesions consisting of a strand break due to 5' chemistry (15%) and 4' chemistry (less than 2%) and an abasic site (4'-hydroxylation product) (12%) due to 4' chemistry. Replacement of the G.C base pair 5' to the C by a G.T wobble mismatch results in a remarkable switching of the chemistry of damage at the C from C-1' to C-4'. The 1' chemistry is almost eliminated and replaced by 4' chemistry, so that the latter accounts for 64% of the damage, mainly in the form of the 4'-hydroxylation product (abasic site) and a smaller amount of the DNA fragment with a phosphoglycolate at the 3' end (strand break). Substitution of the radiation sensitizer misonidazole for dioxygen markedly enhances partitioning of the 4' chemistry in favor of the glycolate-containing product. On the complementary strand the G.T mismatch results in an increase in 4' chemistry at the T residue, but 5' chemistry remains the main mechanism. When a G.A mismatch is inserted 5' to the C, there is a marked decrease in all damage at this site without detectable switching of chemistry. These results show that the diradical form of thiol-activated neocarzinostatin chromophore acts as sensitive probe of DNA microheterogeneity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon P. C., Goldberg I. H. Influence of thiol structure on neocarzinostatin activation and expression of DNA damage. Biochemistry. 1992 Feb 25;31(7):1909–1917. doi: 10.1021/bi00122a003. [DOI] [PubMed] [Google Scholar]
- Dedon P. C., Goldberg I. H. Sequence-specific double-strand breakage of DNA by neocarzinostatin involves different chemical mechanisms within a staggered cleavage site. J Biol Chem. 1990 Sep 5;265(25):14713–14716. [PubMed] [Google Scholar]
- Dedon P. C., Jiang Z. W., Goldberg I. H. Neocarzinostatin-mediated DNA damage in a model AGT.ACT site: mechanistic studies of thiol-sensitive partitioning of C4' DNA damage products. Biochemistry. 1992 Feb 25;31(7):1917–1927. doi: 10.1021/bi00122a004. [DOI] [PubMed] [Google Scholar]
- Galat A., Goldberg I. H. Molecular models of neocarzinostatin damage of DNA: analysis of sequence dependence in 5'GAGCG:5'CGCTC. Nucleic Acids Res. 1990 Apr 25;18(8):2093–2099. doi: 10.1093/nar/18.8.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloni L., Takeshita M., Johnson F., Iden C., Grollman A. P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981 Aug 25;256(16):8608–8615. [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Wobble dG X dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kneale G., Anand N. N., Rabinovich D., Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J Biol Chem. 1987 Jul 25;262(21):9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Chen C. Q., Goldberg I. H. Atypical abasic sites generated by neocarzinostatin at sequence-specific cytidylate residues in oligodeoxynucleotides. Biochemistry. 1988 Jun 14;27(12):4331–4340. doi: 10.1021/bi00412a021. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Activation of neocarzinostatin chromophore and formation of nascent DNA damage do not require molecular oxygen. Nucleic Acids Res. 1985 Mar 11;13(5):1637–1648. doi: 10.1093/nar/13.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H., Frank B. L., Worth L., Jr, Christner D. F., Kozarich J. W., Stubbe J. Neocarzinostatin-induced hydrogen atom abstraction from C-4' and C-5' of the T residue at a d(GT) step in oligonucleotides: shuttling between deoxyribose attack sites based on isotope selection effects. Biochemistry. 1991 Feb 26;30(8):2034–2042. doi: 10.1021/bi00222a005. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Identification of 2-deoxyribonolactone at the site of neocarzinostatin-induced cytosine release in the sequence d(AGC). Biochemistry. 1989 Feb 7;28(3):1027–1032. doi: 10.1021/bi00429a016. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Kuwabara M., Yoon C., Goyne T., Thederahn T., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion: reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry. 1986 Nov 18;25(23):7401–7408. doi: 10.1021/bi00371a023. [DOI] [PubMed] [Google Scholar]
- Meschwitz S. M., Goldberg I. H. Selective abstraction of 2H from C-5' of thymidylate in an oligodeoxynucleotide by the radical center at C-6 of the diradical species of neocarzinostatin: chemical evidence for the structure of the activated drug-DNA complex. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3047–3051. doi: 10.1073/pnas.88.8.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Base substitution mutations induced in the cI gene of lambda phage by neocarzinostatin chromophore: correlation with depyrimidination hotspots at the sequence AGC. Nucleic Acids Res. 1986 Feb 11;14(3):1417–1426. doi: 10.1093/nar/14.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Houlgrave C. W., Han Y. H. Neocarzinostatin-induced DNA base release accompanied by staggered oxidative cleavage of the complementary strand. J Biol Chem. 1988 Dec 25;263(36):19263–19266. [PubMed] [Google Scholar]
- Steighner R. J., Povirk L. F. Bleomycin-induced DNA lesions at mutational hot spots: implications for the mechanism of double-strand cleavage. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8350–8354. doi: 10.1073/pnas.87.21.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H., Xu C., Murugesan N., Hecht S. M., van der Marel G. A., van Boom J. H. Chemistry of the alkali-labile lesion formed from iron(II) bleomycin and d(CGCTTTAAAGCG). Biochemistry. 1988 Jan 12;27(1):58–67. doi: 10.1021/bi00401a011. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Grollman A. P., Ohtsubo E., Ohtsubo H. Interaction of bleomycin with DNA. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5983–5987. doi: 10.1073/pnas.75.12.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]