Abstract
We previously showed that Aminaphtone, a drug used in the treatment of chronic venous insufficiency, modulates several vasoactive factors, such as endothelin-1 and adhesion molecules. Here, we provide data of time-course experiments about the effects of Aminaphtone on gene expression at the genome-wide level in human endothelial cells undergoing cytokine stimulation in vitro. ECV-304 endothelial cells were incubated with interleukin-1β (IL-1β) in the presence or absence of Aminaphtone for 1, 3, and 6 h. Gene expression profiles were analyzed by microarray. This article contains complete data on the genes significantly modulated by the drug over time. The data are supplemental to our original research article reporting detailed analysis of the actions of Aminaphtone on IL-1β stimulated endothelial cells at the molecular level, "Gene expression profiling reveals novel protective effects of Aminaphtone on ECV304 endothelial cells" (Salazar et al., 2016) [1].
Chemical compound studied in this article: Aminaphtone (PubChem CID: 84621)
Keywords: Endothelial cells, Transcriptome, Inflammation, Vasoactive drug
Specifications Table
| Subject area | Biology |
| More specific subject area | Cellular transcriptomics |
| Type of data | Tables, figure |
| How data was acquired | Affymetrix 7G Microarray Scanner (Affymetrix, Santa Clara, CA) |
| Data format | Filtered, analyzed |
| Experimental factors | Time-course experiments of gene expression responses of confluent human ECV304 endothelial cells, stimulated with recombinant IL-1β (100 IU/ml), to treatment with Aminaphtone (6 μg/ml) vs. medium alone |
| Experimental features | Whole-genome gene expression analysis performed at 1, 3, and 6-h time points using GeneChip Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA) |
| Data source location | Milan, Italy |
| Data accessibility | Filtered, analyzed data are reported within this article. Raw and normalized data are available via NCBI׳s GEO accession number GEO: GSE83297 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83297). |
Value of the data
-
•
Previously unreported transcriptional effects of Aminaphtone, a drug currently used for the treatment of chronic venous insufficiency.
-
•
Illustrate gene expression changes induced by Aminaphtone in a human endothelial cell line subjected to inflammatory stimulus.
-
•
May facilitate further experiments to unveil the still unknown mechanism of action of this drug.
-
•
May serve as a benchmark for comparison with data obtained in primary cells for further insight.
-
•
May stimulate further research on the clinical use of this drug in other disease conditions, in which inflammation and endothelial dysfunction are key pathophysiological elements.
1. Data
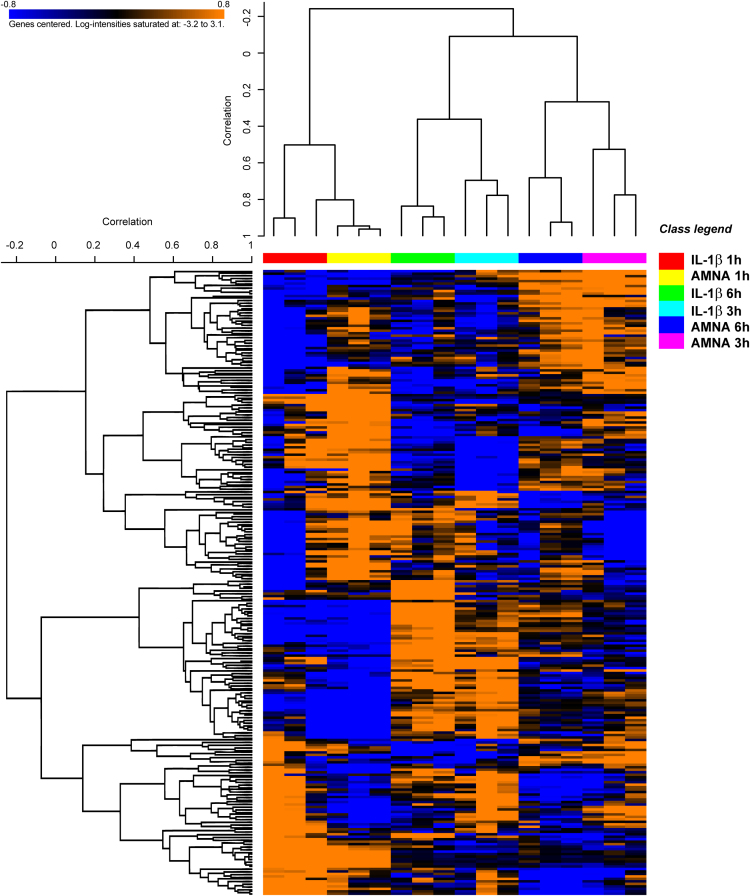
Transcriptional effects of Aminaphtone treatment on human ECV304 endothelial cells stimulated with IL-1β for 1, 3 and 6 h are shown in Table 1. The complete raw and normalized data are deposited in NCBI׳s Gene Expression Omnibus [2] and are accessible through GEO Series accession number GEO: GSE83297 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83297). A hierarchically clustered heatmap of the differentially expressed genes (Fig. 1) visualizes the arrangement of treatment groups into clusters. Table 2 illustrates data on the functional enrichment analysis of modulated genes, showing gene sets and molecular pathways affected by Aminaphtone in IL-1β stimulated ECV304 cells.
Table 1.
Differentially expressed genes in Aminaphtone-treated vs. untreated IL-1β-stimulated ECV304 endothelial cells.
| Symbol | Gene name | Gene ID | FDR | Ptreatment | Pinteraction |
FC AMNA/IL-1β |
Pairwise |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FC 1 h | FC 3 h | FC 6 h | 1 h | 3 h | 6 h | ||||||
| AAK1 | AP2 associated kinase 1 | 22848 | 9.13e-02 | 5.98e-03 | 7.26e-01 | 1.55 | 1.66 | 1.31 | * | ||
| ACAP1 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 | 9744 | 1.65e-02 | 2.07e-04 | 4.01e-01 | −1.50 | −1.23 | −1.36 | ** | * | |
| AGAP2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | 116986 | 1.73e-03 | 8.86e-05 | 4.53e-01 | −1.39 | −1.58 | −1.31 | ** | ** | * |
| AK4 | adenylate kinase 4 | 205 | 8.79e-03 | 4.27e-01 | 7.56e-04 | 1.41 | −1.01 | −1.64 | * | ** | |
| ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) | 27063 | 8.02e-03 | 2.75e-02 | 1.23e-02 | 1.26 | −1.46 | −1.78 | * | ** | |
| ANKRD13A | ankyrin repeat domain 13A | 88455 | 8.98e-02 | 2.37e-04 | 1.88e-01 | 1.23 | 1.47 | 1.18 | * | ** | |
| ANXA3 | annexin A3 | 306 | 4.39e-04 | 7.76e-02 | 7.05e-04 | 1.48 | −1.33 | −1.70 | ** | * | ** |
| AREG | amphiregulin | 374 | 7.88e-02 | 6.01e-03 | 6.32e-01 | −1.37 | −1.39 | −1.78 | * | ||
| ARHGEF26 | Rho guanine nucleotide exchange factor (GEF) 26 | 26084 | 7.10e-02 | 1.36e-03 | 9.35e-01 | 1.42 | 1.33 | 1.42 | * | * | |
| ARL4C | ADP-ribosylation factor-like 4C | 10123 | 2.31e-02 | 2.22e-04 | 8.41e-01 | −1.41 | −1.30 | −1.39 | ** | ** | |
| ATF7IP2 | activating transcription factor 7 interacting protein 2 | 80063 | 5.87e-02 | 5.59e-03 | 1.69e-01 | 1.77 | 1.17 | 1.22 | ** | ||
| BCL2L11 | BCL2-like 11 (apoptosis facilitator) | 10018 | 8.59e-04 | 7.06e-05 | 3.23e-02 | 1.60 | 1.48 | 1.09 | ** | ** | * |
| BCL3 | B-cell CLL/lymphoma 3 | 602 | 4.12e-03 | 3.18e-02 | 5.00e-03 | −1.98 | −1.05 | 1.13 | ** | ||
| BMP6 | bone morphogenetic protein 6 | 654 | 1.78e-02 | 3.25e-03 | 6.12e-02 | −1.00 | 1.65 | 1.47 | ** | * | |
| BNIP3 | BCL2/adenovirus E1B 19 kDa interacting protein 3 | 664 | 0.00e+00 | 3.64e-05 | 4.09e-06 | 1.11 | −1.12 | −2.54 | ** | ||
| BRCA1 | breast cancer 1, early onset | 672 | 7.46e-02 | 4.42e-01 | 1.64e-02 | 1.96 | −1.28 | −1.15 | ** | ||
| BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | 699 | 1.40e-02 | 5.76e-01 | 5.38e-03 | 1.93 | −1.28 | −1.26 | ** | ||
| BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | 701 | 5.11e-02 | 4.12e-01 | 1.69e-02 | 2.09 | −1.32 | −1.14 | ** | ||
| C14orf105 | chromosome 14 open reading frame 105 | 55195 | 6.13e-02 | 4.75e-03 | 5.28e-01 | −1.22 | −1.59 | −1.62 | * | * | |
| C15orf48 | chromosome 15 open reading frame 48 | 84419 | 5.83e-03 | 7.16e-05 | 4.51e-02 | −1.61 | −1.17 | −1.25 | ** | * | |
| C1orf63 | chromosome 1 open reading frame 63 | 57035 | 3.07e-03 | 1.82e-04 | 3.43e-01 | 1.61 | 1.27 | 1.38 | ** | * | |
| C2orf44 | chromosome 2 open reading frame 44 | 80304 | 9.87e-03 | 1.63e-03 | 2.02e-01 | 1.79 | 1.35 | 1.20 | ** | ||
| C3orf58 | chromosome 3 open reading frame 58 | 205428 | 1.08e-05 | 3.22e-01 | 1.35e-06 | 1.54 | 1.17 | −1.61 | ** | * | ** |
| C7orf53 | chromosome 7 open reading frame 53 | 286006 | 6.37e-05 | 1.06e-03 | 1.05e-02 | 2.19 | 1.05 | 1.27 | ** | ||
| C9orf131 | chromosome 9 open reading frame 131 | 138724 | 9.82e-03 | 4.39e-04 | 7.67e-02 | 1.67 | 1.21 | 1.19 | ** | ||
| CACHD1 | cache domain containing 1 | 57685 | 4.66e-02 | 2.02e-03 | 4.84e-01 | 1.31 | 1.63 | 1.29 | ** | ||
| CBLB | Cbl proto-oncogene, E3 ubiquitin protein ligase B | 868 | 8.71e-02 | 1.29e-02 | 2.19e-01 | 1.07 | 1.86 | 1.45 | * | ||
| CCL22 | chemokine (C–C motif) ligand 22 | 6367 | 0.00e+00 | 6.98e-06 | 6.53e-02 | −1.36 | −2.20 | −1.99 | * | ** | ** |
| CD70 | CD70 molecule | 970 | 9.39e-02 | 1.09e-03 | 2.37e-01 | −1.53 | −1.22 | −1.20 | ** | ||
| CD83 | CD83 molecule | 9308 | 1.63e-03 | 4.10e-04 | 8.35e-02 | 1.80 | 1.22 | 1.27 | ** | ||
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | 1052 | 6.06e-03 | 4.58e-01 | 6.34e-04 | −1.70 | 1.12 | 1.31 | ** | * | |
| CENPF | centromere protein F, 350/400 kDa | 1063 | 8.68e-02 | 6.92e-01 | 2.45e-02 | 1.90 | −1.75 | −1.31 | * | ||
| CEP55 | centrosomal protein 55 kDa | 55165 | 4.42e-02 | 7.14e-01 | 8.10e-03 | 1.68 | −1.40 | −1.35 | * | ||
| CEP76 | centrosomal protein 76 kDa | 79959 | 7.67e-02 | 2.29e-03 | 1.52e-01 | 1.57 | 1.32 | 1.09 | ** | * | |
| CHCHD7 | coiled-coil-helix-coiled-coil-helix domain containing 7 | 79145 | 6.62e-02 | 3.19e-03 | 4.05e-01 | 1.58 | 1.47 | 1.17 | * | * | |
| CITED2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 10370 | 7.93e-02 | 2.38e-01 | 3.69e-03 | −1.38 | 1.47 | 1.22 | * | ** | |
| CKAP2L | cytoskeleton associated protein 2-like | 150468 | 1.53e-02 | 1.45e-01 | 1.89e-02 | 1.57 | −1.97 | −1.57 | * | ||
| CLEC4E | C-type lectin domain family 4, member E | 26253 | 4.03e-03 | 8.42e-04 | 3.66e-01 | −1.25 | −1.53 | −1.71 | * | ** | |
| CNNM4 | cyclin M4 | 26504 | 1.79e-07 | 9.46e-05 | 4.77e-04 | −1.11 | 2.00 | 1.57 | ** | ** | |
| CSF2 | colony stimulating factor 2 (granulocyte-macrophage) | 1437 | 1.02e-03 | 8.79e-04 | 7.21e-01 | −1.68 | −1.88 | −1.48 | * | ** | |
| CTH | cystathionase (cystathionine gamma-lyase) | 1491 | 6.66e-03 | 1.90e-04 | 3.46e-01 | 1.29 | 1.59 | 1.31 | * | ** | * |
| CTSK | cathepsin K | 1513 | 2.91e-03 | 4.28e-04 | 9.47e-01 | 1.54 | 1.53 | 1.45 | * | * | * |
| CUZD1 | CUB and zona pellucida-like domains 1 | 50624 | 7.05e-02 | 3.72e-02 | 4.52e-02 | 1.95 | 1.02 | 1.05 | ** | ||
| CX3CL1 | chemokine (C-X3-C motif) ligand 1 | 6376 | 1.25e-02 | 3.11e-03 | 4.58e-01 | −1.94 | −1.48 | −1.33 | ** | ||
| CXCL3 | chemokine (C-X-C motif) ligand 3 | 2921 | 8.34e-02 | 1.65e-02 | 5.71e-02 | −1.70 | −1.34 | 1.07 | ** | ||
| CXCL6 | chemokine (C-X-C motif) ligand 6 | 6372 | 2.85e-03 | 1.69e-04 | 6.11e-01 | −1.39 | −1.34 | −1.58 | * | * | ** |
| CYB5D1 | cytochrome b5 domain containing 1 | 124637 | 2.40e-03 | 1.21e-03 | 7.39e-02 | 1.78 | 1.50 | 1.07 | ** | * | |
| CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | 1591 | 2.62e-05 | 4.54e-05 | 7.89e-01 | −1.52 | −1.69 | −1.51 | ** | ** | ** |
| DBR1 | debranching enzyme homolog 1 (Saccharomyces cerevisiae) | 51163 | 7.97e-02 | 1.06e-03 | 4.21e-01 | 1.52 | 1.26 | 1.24 | ** | ||
| DDIT4 | DNA-damage-inducible transcript 4 | 54541 | 5.57e-04 | 5.87e-04 | 5.05e-01 | −1.90 | −1.40 | −1.57 | ** | * | |
| DDR1 | discoidin domain receptor tyrosine kinase 1 | 780 | 1.43e-02 | 1.48e-03 | 8.30e-02 | −1.73 | −1.26 | −1.13 | ** | ||
| DENND2C | DENN/MADD domain containing 2C | 163259 | 3.14e-02 | 1.33e-03 | 9.57e-01 | 1.44 | 1.49 | 1.40 | * | * | * |
| DEPDC1 | DEP domain containing 1 | 55635 | 5.44e-02 | 9.05e-01 | 1.02e-02 | 1.75 | −1.26 | −1.44 | * | ||
| DKK1 | dickkopf 1 homolog (Xenopus laevis) | 22943 | 2.94e-05 | 5.83e-05 | 1.71e-02 | −1.25 | −1.25 | −1.94 | ** | ||
| DLEU1 | deleted in lymphocytic leukemia 1 (non-protein coding) | 10301 | 7.23e-02 | 5.02e-03 | 1.27e-01 | 1.71 | 1.19 | 1.14 | ** | ||
| DLGAP5 | discs, large (Drosophila) homolog-associated protein 5 | 9787 | 4.29e-03 | 1.58e-01 | 2.80e-03 | 1.51 | −1.58 | −1.42 | * | * | * |
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 3337 | 1.36e-02 | 4.99e-03 | 4.35e-02 | 1.82 | 1.31 | 1.00 | ** | ||
| DUSP8 | dual specificity phosphatase 8 | 1850 | 5.16e-03 | 1.05e-02 | 4.26e-03 | −1.87 | 1.06 | −1.07 | ** | ||
| EBI3 | Epstein-Barr virus induced 3 | 10148 | 0.00e+00 | 6.30e-06 | 6.87e-01 | −1.82 | −1.73 | −2.06 | ** | ** | ** |
| ECT2 | epithelial cell transforming sequence 2 oncogene | 1894 | 6.84e-03 | 5.43e-02 | 1.07e-02 | 1.36 | −1.84 | −1.45 | ** | ||
| EDN1 | endothelin 1 | 1906 | 0.00e+00 | 2.33e-06 | 8.60e-01 | −1.79 | −1.93 | −1.97 | ** | ** | ** |
| EFNA1 | ephrin-A1 | 1942 | 4.91e-03 | 1.20e-03 | 2.03e-02 | −1.74 | −1.02 | −1.31 | ** | * | |
| EFNB2 | ephrin-B2 | 1948 | 1.76e-02 | 3.27e-03 | 1.81e-01 | 1.81 | 1.40 | 1.13 | ** | ||
| EGR2 | early growth response 2 | 1959 | 7.52e-03 | 3.57e-03 | 6.43e-03 | −1.80 | −1.04 | −1.06 | ** | ||
| EIF4B | eukaryotic translation initiation factor 4B | 1975 | 3.63e-02 | 4.03e-01 | 9.48e-03 | 1.56 | −1.22 | −1.69 | * | * | |
| EIF4EBP2 | eukaryotic translation initiation factor 4E binding protein 2 | 1979 | 3.09e-04 | 5.53e-06 | 1.55e-01 | 1.25 | 1.56 | 1.42 | * | ** | ** |
| ELF3 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | 1999 | 6.05e-02 | 3.62e-02 | 8.45e-03 | −1.24 | 1.43 | 1.43 | * | * | |
| ELMSAN1 | ELM2 and Myb/SANT-like domain containing 1 | 91748 | 3.96e-05 | 1.34e-02 | 1.00e-05 | −1.86 | 1.19 | 1.08 | ** | * | |
| ELOVL4 | ELOVL fatty acid elongase 4 | 6785 | 8.64e-03 | 5.29e-04 | 3.98e-01 | 1.40 | 1.62 | 1.25 | * | ** | |
| ELOVL6 | ELOVL fatty acid elongase 6 | 79071 | 7.35e-02 | 1.86e-01 | 1.29e-02 | 1.84 | −1.08 | −1.15 | ** | ||
| ENO2 | enolase 2 (gamma, neuronal) | 2026 | 6.53e-05 | 1.50e-03 | 1.22e-02 | −1.27 | −1.06 | −2.31 | ** | ||
| ERCC6L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like | 54821 | 6.72e-02 | 8.17e-01 | 1.39e-02 | 1.75 | −1.58 | −1.21 | * | ||
| ERF | Ets2 repressor factor | 2077 | 1.91e-02 | 4.84e-04 | 1.05e-01 | −1.62 | −1.22 | −1.19 | ** | ||
| EVA1A | eva-1 homolog A (Caenorhabditis elegans) | 84141 | 1.25e-06 | 3.14e-07 | 4.49e-03 | −1.19 | −1.47 | −1.74 | * | ** | ** |
| FAM102A | family with sequence similarity 102, member A | 399665 | 3.29e-02 | 6.69e-02 | 5.48e-03 | −1.32 | 1.41 | 1.48 | * | * | |
| FAM111B | family with sequence similarity 111, member B | 374393 | 7.05e-02 | 5.89e-01 | 2.31e-02 | 2.18 | −1.58 | −1.06 | * | ||
| FAM46C | family with sequence similarity 46, member C | 54855 | 5.99e-04 | 5.82e-04 | 4.90e-03 | 1.87 | 1.20 | 1.03 | ** | ||
| FAM72D | family with sequence similarity 72, member D | 728833 | 2.54e-04 | 4.65e-01 | 1.25e-03 | 1.86 | −1.60 | −1.46 | ** | * | * |
| FBLIM1 | filamin binding LIM protein 1 | 54751 | 1.21e-03 | 1.31e-04 | 5.98e-01 | −1.61 | −1.38 | −1.39 | ** | * | * |
| FBXL20 | F-box and leucine-rich repeat protein 20 | 84961 | 1.87e-04 | 2.46e-04 | 6.47e-02 | 1.19 | 1.91 | 1.40 | ** | * | |
| FBXO33 | F-box protein 33 | 254170 | 4.21e-02 | 9.72e-04 | 7.70e-02 | 1.22 | 1.62 | 1.13 | ** | ||
| FLJ36840 | uncharacterized LOC645524 | 645524 | 2.12e-02 | 1.52e-01 | 2.64e-02 | 2.66 | −1.34 | 1.09 | ** | ||
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 2353 | 4.65e-06 | 4.26e-04 | 3.84e-04 | 2.13 | 1.01 | 1.08 | ** | ||
| FRMD6 | FERM domain containing 6 | 122786 | 4.02e-02 | 1.16e-02 | 3.29e-02 | 1.11 | −1.49 | −1.57 | * | ** | |
| FRMD8 | FERM domain containing 8 | 83786 | 3.02e-02 | 6.15e-03 | 2.25e-02 | −1.72 | −1.23 | 1.03 | ** | ||
| FST | follistatin | 10468 | 7.49e-02 | 1.12e-03 | 1.27e-01 | −1.07 | −1.44 | −1.42 | ** | ** | |
| FUT11 | fucosyltransferase 11 (alpha (1,3) fucosyltransferase) | 170384 | 5.23e-02 | 1.79e-03 | 6.02e-03 | −1.14 | 1.00 | −1.62 | ** | ||
| GAN | gigaxonin | 8139 | 4.82e-02 | 6.57e-03 | 1.16e-01 | 1.80 | 1.22 | 1.12 | ** | ||
| GAREM | GRB2 associated, regulator of MAPK1 | 64762 | 9.44e-02 | 1.61e-03 | 1.88e-02 | −1.00 | 1.54 | 1.23 | ** | * | |
| GAS2L3 | growth arrest-specific 2 like 3 | 283431 | 5.77e-02 | 2.78e-01 | 1.30e-02 | 1.92 | −1.14 | −1.17 | ** | ||
| GCLC | glutamate-cysteine ligase, catalytic subunit | 2729 | 3.29e-02 | 2.08e-03 | 7.58e-01 | 1.56 | 1.53 | 1.32 | * | * | |
| GCLM | glutamate–cysteine ligase, modifier subunit | 2730 | 1.37e-02 | 1.51e-03 | 5.69e-01 | 1.71 | 1.40 | 1.35 | ** | ||
| GLCCI1 | glucocorticoid induced transcript 1 | 113263 | 2.69e-02 | 1.03e-04 | 8.16e-01 | 1.37 | 1.38 | 1.28 | ** | ** | |
| GPR56 | G protein-coupled receptor 56 | 9289 | 1.49e-02 | 3.94e-03 | 7.47e-02 | −1.85 | −1.09 | −1.27 | ** | ||
| GPR89A | G protein-coupled receptor 89A | 653519 | 5.78e-02 | 3.12e-03 | 3.52e-01 | 1.67 | 1.32 | 1.22 | ** | ||
| GRB7 | growth factor receptor-bound protein 7 | 2886 | 1.09e-02 | 1.07e-03 | 5.88e-02 | −1.74 | −1.18 | −1.18 | ** | ||
| GTF2B | general transcription factor IIB | 2959 | 6.15e-04 | 8.75e-05 | 1.99e-01 | 1.69 | 1.35 | 1.29 | ** | * | * |
| HCAR1 | hydroxycarboxylic acid receptor 1 | 27198 | 9.99e-02 | 3.02e-03 | 7.95e-01 | −1.29 | −1.45 | −1.49 | * | * | |
| HILPDA | hypoxia inducible lipid droplet-associated | 29923 | 7.25e-05 | 9.73e-04 | 3.21e-03 | −1.20 | −1.01 | −2.09 | ** | ||
| HIST1H2AK | histone cluster 1, H2ak | 8330 | 3.09e-03 | 2.95e-03 | 1.11e-01 | −1.05 | −1.71 | −1.85 | * | ** | |
| HIST2H2BF | histone cluster 2, H2bf | 440689 | 4.80e-03 | 4.43e-05 | 6.62e-01 | 1.36 | 1.32 | 1.47 | ** | * | ** |
| HIST2H4A | histone cluster 2, H4a | 8370 | 0.00e+00 | 4.91e-07 | 1.85e-02 | 1.87 | 1.46 | 1.32 | ** | ** | ** |
| HK2 | hexokinase 2 | 3099 | 1.25e-06 | 4.59e-04 | 1.65e-03 | 1.06 | −1.37 | −2.25 | * | ** | |
| HMMR | hyaluronan-mediated motility receptor (RHAMM) | 3161 | 1.23e-02 | 2.21e-01 | 8.49e-03 | 1.58 | −1.43 | −1.71 | * | * | |
| HMOX1 | heme oxygenase (decycling) 1 | 3162 | 1.69e-02 | 3.20e-04 | 3.02e-01 | −1.51 | −1.19 | −1.41 | ** | ** | |
| ID2 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 3398 | 5.45e-03 | 1.51e-04 | 1.15e-02 | 1.38 | 1.58 | 1.03 | ** | ** | |
| IDI1 | isopentenyl-diphosphate delta isomerase 1 | 3422 | 6.89e-02 | 4.10e-03 | 2.47e-01 | 1.70 | 1.21 | 1.23 | ** | ||
| IGFL1 | IGF-like family member 1 | 374918 | 0.00e+00 | 1.06e-05 | 8.05e-01 | −2.11 | −2.05 | −2.41 | ** | ** | ** |
| IL1B | interleukin 1, beta | 3553 | 2.12e-03 | 1.50e-03 | 3.88e-01 | −1.29 | −1.92 | −1.69 | ** | * | |
| IL32 | interleukin 32 | 9235 | 3.56e-03 | 8.70e-05 | 1.94e-01 | −1.23 | −1.34 | −1.59 | ** | ** | |
| IL7R | interleukin 7 receptor | 3575 | 2.09e-04 | 7.60e-04 | 8.60e-02 | −1.11 | −1.95 | −1.69 | ** | ** | |
| IRF2BP2 | interferon regulatory factor 2 binding protein 2 | 359948 | 1.26e-02 | 7.17e-02 | 2.00e-04 | −1.65 | 1.11 | 1.13 | ** | ||
| ISCA1 | iron–sulfur cluster assembly 1 homolog (S. cerevisiae) | 81689 | 2.42e-02 | 2.41e-03 | 1.97e-01 | 1.60 | 1.54 | 1.10 | ** | * | |
| ISG20L2 | interferon stimulated exonuclease gene 20 kDa-like 2 | 81875 | 8.45e-02 | 4.01e-02 | 2.40e-03 | −1.62 | 1.08 | 1.03 | ** | ||
| KCTD11 | potassium channel tetramerisation domain containing 11 | 147040 | 4.26e-05 | 1.92e-04 | 3.55e-01 | −1.91 | −1.36 | −1.65 | ** | ** | |
| KDELC1 | KDEL (Lys-Asp-Glu-Leu) containing 1 | 79070 | 5.23e-02 | 7.58e-05 | 3.32e-01 | 1.20 | 1.43 | 1.30 | ** | ** | |
| KHDRBS3 | KH domain containing, RNA binding, signal transduction associated 3 | 10656 | 1.79e-03 | 2.61e-06 | 7.74e-04 | 1.05 | 1.57 | 1.32 | ** | ** | |
| KIAA0922 | KIAA0922 | 23240 | 1.60e-02 | 1.12e-03 | 3.47e-01 | 1.25 | 1.68 | 1.35 | ** | ||
| KIAA1524 | KIAA1524 | 57650 | 2.09e-02 | 3.06e-01 | 1.10e-02 | 1.64 | −1.68 | −1.44 | * | * | |
| KIF14 | kinesin family member 14 | 9928 | 1.42e-02 | 8.55e-01 | 1.29e-02 | 2.46 | −1.70 | −1.31 | * | ||
| KIF23 | kinesin family member 23 | 9493 | 7.31e-02 | 1.00e+00 | 1.64e-02 | 1.88 | −1.46 | −1.29 | * | ||
| KLRAP1 | killer cell lectin-like receptor subfamily A pseudogene 1 | 10748 | 9.99e-02 | 9.35e-01 | 2.08e-02 | 1.86 | −1.58 | −1.22 | * | ||
| KRT16 | keratin 16 | 3868 | 8.89e-04 | 7.06e-04 | 5.60e-01 | −1.73 | −1.76 | −1.37 | * | ** | |
| KRT17 | keratin 17 | 3872 | 1.35e-03 | 6.95e-04 | 2.03e-01 | −1.86 | −1.24 | −1.43 | ** | * | |
| KRT34 | keratin 34 | 3885 | 6.82e-02 | 5.18e-04 | 8.08e-01 | −1.27 | −1.37 | −1.40 | ** | ||
| LAMB3 | laminin, beta 3 | 3914 | 9.31e-03 | 7.05e-04 | 8.37e-01 | −1.57 | −1.44 | −1.39 | ** | * | * |
| LIF | leukemia inhibitory factor | 3976 | 1.04e-03 | 1.87e-02 | 1.93e-03 | −1.34 | 1.51 | 1.62 | * | ** | ** |
| LMCD1 | LIM and cysteine-rich domains 1 | 29995 | 7.13e-02 | 2.87e-04 | 8.53e-01 | −1.38 | −1.33 | −1.28 | ** | ||
| LOC100130713 | uncharacterized LOC100130713 | 100130713 | 8.73e-03 | 3.78e-02 | 1.06e-03 | −1.33 | 1.51 | 1.34 | * | ** | * |
| LOC440896 | uncharacterized LOC440896 | 440896 | 1.04e-02 | 7.89e-04 | 8.40e-01 | −1.55 | −1.49 | −1.37 | * | * | |
| LOX | lysyl oxidase | 4015 | 2.52e-02 | 2.34e-04 | 3.94e-02 | −1.15 | −1.19 | −1.60 | ** | ||
| LPIN1 | lipin 1 | 23175 | 1.40e-03 | 2.40e-04 | 4.71e-02 | 1.72 | 1.37 | 1.12 | ** | * | |
| MARCH8 | membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase | 220972 | 7.20e-02 | 2.41e-03 | 2.60e-01 | 1.12 | 1.54 | 1.42 | ** | * | |
| MEF2D | myocyte enhancer factor 2D | 4209 | 4.21e-03 | 9.11e-02 | 1.55e-03 | −1.84 | 1.24 | −1.00 | ** | ||
| MEGF9 | multiple EGF-like-domains 9 | 1955 | 1.01e-04 | 2.69e-04 | 9.48e-01 | 1.62 | 1.76 | 1.69 | * | ** | * |
| MERTK | c-mer proto-oncogene tyrosine kinase | 10461 | 6.83e-05 | 2.96e-05 | 6.07e-02 | 1.28 | 1.79 | 1.32 | * | ** | * |
| MINPP1 | multiple inositol-polyphosphate phosphatase 1 | 9562 | 4.12e-02 | 2.07e-03 | 6.87e-01 | 1.51 | 1.54 | 1.28 | * | * | |
| MIR29A | microRNA 29a | 407021 | 6.91e-05 | 9.06e-04 | 3.57e-01 | 1.59 | 1.74 | 2.69 | ** | ||
| MKI67 | antigen identified by monoclonal antibody Ki-67 | 4288 | 2.28e-02 | 6.57e-01 | 1.27e-02 | 2.18 | −1.58 | −1.13 | ** | ||
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 4312 | 1.40e-04 | 1.11e-03 | 3.61e-02 | −1.08 | −1.44 | −2.18 | * | ** | |
| MMP10 | matrix metallopeptidase 10 (stromelysin 2) | 4319 | 5.96e-08 | 1.29e-04 | 1.54e-03 | −1.02 | −1.38 | −2.37 | * | ** | |
| MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 4314 | 1.93e-05 | 1.63e-03 | 2.51e-02 | −1.01 | −1.63 | −2.57 | * | * | ** |
| MOB3C | MOB kinase activator 3C | 148932 | 2.40e-02 | 1.54e-02 | 7.70e-03 | −1.78 | 1.01 | −1.02 | ** | ||
| MYLIP | myosin regulatory light chain interacting protein | 29116 | 0.00e+00 | 9.67e-07 | 8.89e-05 | 2.84 | 1.52 | 1.12 | ** | ** | |
| NAV3 | neuron navigator 3 | 89795 | 7.05e-02 | 6.46e-01 | 6.03e-03 | 1.55 | −1.42 | −1.24 | * | * | |
| NCAPG | non-SMC condensin I complex, subunit G | 64151 | 8.61e-02 | 7.39e-01 | 1.66e-02 | 1.89 | −1.41 | −1.18 | * | ||
| NEK2 | NIMA-related kinase 2 | 4751 | 5.47e-02 | 2.39e-01 | 7.63e-03 | 1.78 | −1.23 | −1.04 | ** | ||
| NINJ1 | ninjurin 1 | 4814 | 6.07e-02 | 5.10e-03 | 8.39e-02 | −1.70 | −1.25 | −1.06 | ** | ||
| NIPAL4 | NIPA-like domain containing 4 | 348938 | 4.83e-03 | 6.92e-04 | 3.07e-01 | −1.74 | −1.31 | −1.32 | ** | ||
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 4929 | 2.09e-03 | 5.30e-01 | 1.08e-03 | −1.85 | 1.21 | 1.31 | ** | ||
| NUAK2 | NUAK family, SNF1-like kinase, 2 | 81788 | 1.41e-02 | 3.76e-04 | 1.70e-01 | −1.60 | −1.19 | −1.30 | ** | * | |
| NXF1 | nuclear RNA export factor 1 | 10482 | 1.95e-02 | 4.17e-04 | 7.68e-02 | 1.19 | 1.63 | 1.20 | ** | ||
| OLFM2 | olfactomedin 2 | 93145 | 9.51e-02 | 4.42e-03 | 7.98e-02 | −1.65 | −1.13 | −1.14 | ** | ||
| OSBPL11 | oxysterol binding protein-like 11 | 114885 | 9.51e-02 | 8.43e-03 | 4.90e-01 | 1.84 | 1.30 | 1.37 | * | ||
| OXTR | oxytocin receptor | 5021 | 1.11e-03 | 9.51e-06 | 5.52e-03 | −1.10 | −1.62 | −1.32 | ** | ** | |
| P4HA1 | prolyl 4-hydroxylase, alpha polypeptide I | 5033 | 1.04e-04 | 4.09e-01 | 3.61e-04 | 1.58 | −1.02 | −1.89 | ** | ** | |
| PCF11 | PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae) | 51585 | 2.42e-02 | 3.67e-02 | 3.25e-02 | 2.13 | −1.01 | 1.06 | ** | ||
| PDGFB | platelet-derived growth factor beta polypeptide | 5155 | 2.00e-04 | 6.09e-04 | 9.34e-02 | −2.09 | −1.26 | −1.36 | ** | ||
| PDK1 | pyruvate dehydrogenase kinase, isozyme 1 | 5163 | 1.91e-06 | 2.69e-02 | 4.56e-04 | 1.44 | −1.19 | −2.37 | * | ** | |
| PDZK1IP1 | PDZK1 interacting protein 1 | 10158 | 3.32e-02 | 2.18e-03 | 6.99e-02 | −1.60 | −1.03 | −1.39 | ** | * | |
| PER1 | period circadian clock 1 | 5187 | 1.43e-02 | 6.69e-02 | 3.66e-03 | −1.84 | 1.08 | 1.08 | ** | ||
| PFKFB4 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 | 5210 | 0.00e+00 | 3.32e-04 | 6.63e-05 | −1.06 | 1.04 | −2.67 | ** | ||
| PHLDA3 | pleckstrin homology-like domain, family A, member 3 | 23612 | 4.60e-02 | 7.89e-04 | 5.33e-01 | −1.52 | −1.27 | −1.31 | ** | * | |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | 5295 | 2.59e-03 | 1.84e-02 | 2.19e-02 | 2.40 | 1.14 | −1.02 | ** | ||
| PIK3R3 | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | 8503 | 1.86e-02 | 4.86e-04 | 5.59e-01 | 1.28 | 1.54 | 1.37 | ** | * | |
| PLAT | plasminogen activator, tissue | 5327 | 3.74e-02 | 1.90e-02 | 1.06e-01 | −2.20 | −1.18 | −1.12 | ** | ||
| PLCB4 | phospholipase C, beta 4 | 5332 | 7.45e-06 | 3.68e-02 | 2.37e-03 | 1.73 | −2.07 | −2.14 | * | ** | ** |
| PLEKHG2 | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | 64857 | 3.71e-02 | 3.43e-03 | 7.01e-02 | −1.72 | −1.23 | −1.08 | ** | ||
| PLEKHM1 | pleckstrin homology domain containing, family M (with RUN domain) member 1 | 9842 | 8.05e-02 | 9.03e-02 | 1.43e-02 | −1.28 | 1.65 | 1.26 | ** | ||
| PLEKHM3 | pleckstrin homology domain containing, family M, member 3 | 389072 | 3.51e-02 | 4.36e-03 | 9.12e-01 | 1.57 | 1.78 | 1.56 | * | ||
| PODXL | podocalyxin-like | 5420 | 1.55e-02 | 8.88e-04 | 9.91e-03 | −1.05 | −1.16 | −1.69 | ** | ||
| POLA1 | polymerase (DNA directed), alpha 1, catalytic subunit | 5422 | 9.69e-02 | 3.40e-01 | 2.19e-02 | 1.95 | −1.35 | −1.00 | ** | ||
| PPP1R18 | protein phosphatase 1, regulatory subunit 18 | 170954 | 2.09e-02 | 7.61e-03 | 1.02e-02 | −1.77 | −1.06 | −1.02 | ** | ||
| PPP1R3C | protein phosphatase 1, regulatory subunit 3C | 5507 | 1.19e-02 | 9.34e-03 | 7.03e-02 | 1.04 | −1.54 | −1.94 | * | ** | |
| PRDM1 | PR domain containing 1, with ZNF domain | 639 | 0.00e+00 | 1.29e-05 | 3.46e-06 | −2.47 | −1.04 | −1.01 | ** | ||
| PRKCA | protein kinase C, alpha | 5578 | 3.16e-03 | 6.04e-04 | 9.57e-01 | 1.48 | 1.59 | 1.54 | * | * | * |
| PRSS22 | protease, serine, 22 | 64063 | 2.23e-02 | 3.34e-04 | 8.39e-01 | −1.41 | −1.43 | −1.31 | ** | ** | |
| PTCH1 | patched 1 | 5727 | 8.87e-02 | 1.19e-02 | 6.15e-03 | −1.02 | 1.65 | 1.04 | ** | ||
| PTP4A3 | protein tyrosine phosphatase type IVA, member 3 | 11156 | 1.22e-04 | 2.90e-04 | 3.74e-01 | −1.51 | −1.95 | −1.41 | * | ** | |
| PXDC1 | PX domain containing 1 | 221749 | 3.90e-02 | 3.71e-01 | 2.02e-03 | −1.66 | 1.16 | 1.19 | ** | ||
| RAI14 | retinoic acid induced 14 | 26064 | 9.76e-02 | 7.26e-01 | 1.68e-02 | 1.87 | −1.31 | −1.26 | * | ||
| RASD2 | RASD family, member 2 | 23551 | 6.21e-02 | 7.34e-04 | 2.19e-02 | −1.58 | −1.08 | −1.15 | ** | ||
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | 83593 | 9.60e-02 | 2.47e-04 | 6.95e-01 | −1.29 | −1.39 | −1.25 | ** | ||
| RIPK4 | receptor-interacting serine–threonine kinase 4 | 54101 | 1.33e-03 | 3.07e-04 | 1.37e-02 | −1.79 | −1.10 | −1.23 | ** | ||
| RND1 | Rho family GTPase 1 | 27289 | 9.23e-03 | 1.56e-02 | 5.23e-02 | −1.12 | 1.59 | 2.06 | ** | ||
| RNF103 | ring finger protein 103 | 7844 | 6.68e-02 | 2.52e-03 | 9.20e-01 | 1.44 | 1.51 | 1.37 | * | * | |
| RRM2 | ribonucleotide reductase M2 | 6241 | 5.93e-02 | 3.12e-03 | 1.19e-02 | 1.59 | 1.22 | −1.04 | ** | ||
| S100A3 | S100 calcium binding protein A3 | 6274 | 0.00e+00 | 7.47e-06 | 5.36e-02 | −2.37 | −1.37 | −2.05 | ** | ** | |
| SAMD4A | sterile alpha motif domain containing 4A | 23034 | 7.19e-04 | 4.28e-06 | 7.70e-02 | 1.22 | 1.56 | 1.36 | * | ** | ** |
| SELE | selectin E | 6401 | 2.45e-03 | 2.50e-03 | 1.08e-02 | −1.01 | 1.24 | 1.89 | ** | ||
| SEMA4C | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | 54910 | 9.90e-02 | 1.27e-03 | 8.89e-02 | −1.53 | −1.26 | −1.08 | ** | * | |
| SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 12 | 2.93e-03 | 4.08e-03 | 1.68e-01 | −2.38 | −1.29 | −1.37 | ** | ||
| SERPINB9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 5272 | 4.70e-02 | 4.96e-03 | 1.48e-01 | −1.07 | −1.39 | −1.72 | ** | ||
| SERPINH1 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 871 | 6.23e-02 | 1.47e-03 | 4.28e-01 | −1.27 | −1.27 | −1.58 | ** | ||
| SERPINI1 | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 5274 | 2.82e-02 | 1.25e-03 | 6.61e-01 | 1.54 | 1.28 | 1.46 | * | * | |
| SLC2A1 | solute carrier family 2 (facilitated glucose transporter), member 1 | 6513 | 2.93e-03 | 1.28e-03 | 6.12e-02 | −1.23 | −1.19 | −1.91 | ** | ||
| SLC33A1 | solute carrier family 33 (acetyl-CoA transporter), member 1 | 9197 | 6.24e-02 | 6.09e-04 | 3.29e-01 | 1.42 | 1.42 | 1.16 | ** | ** | |
| SLC35D1 | solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 | 23169 | 5.29e-02 | 3.06e-03 | 6.97e-01 | 1.65 | 1.38 | 1.35 | * | ||
| SLC6A14 | solute carrier family 6 (amino acid transporter), member 14 | 11254 | 8.97e-02 | 5.67e-03 | 8.88e-02 | 1.01 | −1.46 | −1.50 | * | * | |
| SLC6A8 | solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | 6535 | 2.16e-03 | 1.14e-03 | 1.69e-02 | −1.03 | 1.70 | 1.47 | ** | ** | |
| SMIM14 | small integral membrane protein 14 | 201895 | 7.25e-02 | 3.85e-03 | 4.08e-01 | 1.19 | 1.64 | 1.41 | * | ||
| SNHG12 | small nucleolar RNA host gene 12 (non-protein coding) | 85028 | 2.55e-04 | 1.61e-05 | 6.39e-02 | 1.66 | 1.38 | 1.21 | ** | ** | * |
| SNORA61 | small nucleolar RNA, H/ACA box 61 | 677838 | 7.99e-04 | 6.54e-05 | 7.40e-02 | 1.62 | 1.45 | 1.15 | ** | ** | |
| SNORD105 | small nucleolar RNA, C/D box 105 | 692229 | 1.18e-02 | 5.73e-03 | 9.15e-02 | −1.82 | −1.58 | −1.00 | ** | * | |
| SNORD13P2 | small nucleolar RNA, C/D box 13 pseudogene 2 | 6077 | 1.60e-02 | 8.33e-02 | 5.74e-03 | −1.88 | 1.16 | 1.01 | ** | ||
| SNORD14E | small nucleolar RNA, C/D box 14E | 85391 | 1.44e-02 | 1.01e-02 | 9.15e-02 | 2.17 | 1.21 | 1.14 | ** | ||
| SNORD22 | small nucleolar RNA, C/D box 22 | 9304 | 1.49e-06 | 3.21e-04 | 3.16e-02 | 2.57 | 1.32 | 1.34 | ** | ||
| SNORD28 | small nucleolar RNA, C/D box 28 | 9300 | 4.12e-02 | 1.90e-03 | 1.47e-01 | 1.66 | 1.14 | 1.26 | ** | ||
| SNORD30 | small nucleolar RNA, C/D box 30 | 9299 | 1.45e-02 | 3.29e-03 | 4.36e-02 | 1.82 | 1.11 | 1.16 | ** | ||
| SNORD31 | small nucleolar RNA, C/D box 31 | 9298 | 5.13e-06 | 9.26e-05 | 3.20e-03 | 2.08 | 1.18 | 1.17 | ** | ||
| SNORD32A | small nucleolar RNA, C/D box 32A | 26819 | 6.96e-02 | 1.74e-02 | 3.10e-02 | −1.66 | −1.32 | 1.11 | ** | ||
| SPRY4 | sprouty homolog 4 (Drosophila) | 81848 | 8.04e-02 | 2.15e-03 | 1.03e-01 | −1.59 | −1.09 | −1.25 | ** | ||
| SRGN | serglycin | 5552 | 1.96e-02 | 1.14e-04 | 7.38e-02 | −1.15 | −1.28 | −1.55 | * | ** | |
| STAT5A | signal transducer and activator of transcription 5A | 6776 | 8.68e-02 | 6.33e-04 | 6.45e-01 | −1.22 | −1.38 | −1.40 | ** | ||
| STC1 | stanniocalcin 1 | 6781 | 8.00e-04 | 1.62e-03 | 1.37e-02 | −1.21 | −1.06 | −2.01 | ** | ||
| STC2 | stanniocalcin 2 | 8614 | 9.30e-04 | 1.85e-03 | 5.42e-03 | 1.01 | −1.19 | −1.93 | ** | ||
| STK17B | serine/threonine kinase 17b | 9262 | 1.23e-03 | 1.93e-03 | 1.18e-01 | 2.15 | 1.36 | 1.23 | ** | ||
| STX4 | syntaxin 4 | 6810 | 2.28e-03 | 1.23e-04 | 9.23e-01 | −1.49 | −1.40 | −1.43 | ** | * | ** |
| SYDE1 | synapse defective 1, Rho GTPase, homolog 1 (C. elegans) | 85360 | 9.00e-02 | 9.80e-04 | 4.81e-01 | −1.43 | −1.19 | −1.40 | ** | * | |
| TAGLN | transgelin | 6876 | 3.45e-03 | 3.65e-04 | 6.89e-01 | −1.43 | −1.37 | −1.61 | * | * | ** |
| TAS2R4 | taste receptor, type 2, member 4 | 50832 | 9.62e-02 | 1.10e-02 | 1.86e-01 | 1.83 | 1.15 | 1.22 | ** | ||
| TFRC | transferrin receptor (p90, CD71) | 7037 | 7.32e-02 | 3.99e-01 | 2.32e-02 | 2.18 | −1.21 | −1.23 | ** | ||
| TGFB1I1 | transforming growth factor beta 1 induced transcript 1 | 7041 | 8.92e-05 | 2.58e-05 | 9.24e-01 | −1.47 | −1.48 | −1.55 | ** | ** | ** |
| TGFB2 | transforming growth factor, beta 2 | 7042 | 1.50e-05 | 2.51e-03 | 3.22e-03 | 1.19 | −2.14 | −1.52 | ** | * | |
| TICAM1 | toll-like receptor adaptor molecule 1 | 148022 | 4.13e-03 | 8.38e-03 | 8.32e-04 | −1.79 | 1.05 | 1.01 | ** | ||
| TLE1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | 7088 | 3.37e-03 | 1.25e-02 | 3.72e-04 | −1.23 | 1.70 | 1.14 | * | ** | |
| TM4SF18 | transmembrane 4L six family member 18 | 116441 | 3.05e-02 | 6.72e-02 | 2.10e-03 | 1.31 | −1.24 | −1.55 | * | ** | |
| TMEM2 | transmembrane protein 2 | 23670 | 2.67e-02 | 5.08e-03 | 6.27e-01 | 2.00 | 1.44 | 1.53 | * | ||
| TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 7130 | 1.27e-02 | 4.06e-03 | 2.87e-01 | −1.16 | −1.70 | −1.84 | * | * | |
| TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b | 4982 | 6.74e-06 | 4.09e-04 | 2.27e-01 | −1.49 | −2.62 | −1.70 | ** | * | |
| TNFSF18 | tumor necrosis factor (ligand) superfamily, member 18 | 8995 | 0.00e+00 | 3.06e-06 | 3.10e-01 | −2.41 | −4.24 | −3.41 | ** | ** | ** |
| TOP2A | topoisomerase (DNA) II alpha 170 kDa | 7153 | 4.62e-02 | 8.29e-01 | 1.18e-02 | 1.90 | −1.42 | −1.24 | ** | ||
| TRAF1 | TNF receptor-associated factor 1 | 7185 | 2.49e-04 | 8.97e-05 | 2.32e-01 | −1.69 | −1.50 | −1.26 | ** | ** | |
| TRIP6 | thyroid hormone receptor interactor 6 | 7205 | 8.37e-02 | 9.63e-04 | 6.24e-01 | −1.48 | −1.30 | −1.26 | ** | * | |
| TSGA10 | testis specific, 10 | 80705 | 6.19e-02 | 6.67e-03 | 1.65e-02 | 1.68 | 1.07 | 1.04 | ** | ||
| TTLL3 | tubulin tyrosine ligase-like family, member 3 | 26140 | 3.38e-03 | 1.07e-03 | 1.29e-02 | −1.04 | 1.65 | 1.45 | ** | ** | |
| UBALD1 | UBA-like domain containing 1 | 124402 | 1.29e-02 | 9.81e-02 | 5.00e-03 | −1.88 | 1.23 | −1.03 | ** | ||
| ULBP3 | UL16 binding protein 3 | 79465 | 5.51e-02 | 3.20e-04 | 1.21e-01 | 1.30 | 1.50 | 1.12 | * | ** | |
| ULK1 | unc-51-like kinase 1 (C. elegans) | 8408 | 8.48e-02 | 4.11e-01 | 2.45e-03 | −1.44 | 1.33 | 1.27 | ** | * | |
| USP38 | ubiquitin specific peptidase 38 | 84640 | 8.54e-03 | 1.46e-04 | 4.15e-01 | 1.53 | 1.34 | 1.27 | ** | * | * |
| VAMP1 | vesicle-associated membrane protein 1 (synaptobrevin 1) | 6843 | 1.08e-03 | 3.77e-05 | 4.65e-01 | 1.49 | 1.48 | 1.28 | ** | ** | * |
| VLDLR | very low density lipoprotein receptor | 7436 | 4.04e-02 | 4.49e-04 | 2.18e-03 | −1.05 | −1.08 | −1.61 | ** | ||
| ZC3H7B | zinc finger CCCH-type containing 7B | 23264 | 6.99e-02 | 4.01e-04 | 8.97e-01 | −1.39 | −1.30 | −1.32 | ** | ||
| ZC3HAV1 | zinc finger CCCH-type, antiviral 1 | 56829 | 4.54e-02 | 3.89e-03 | 2.25e-01 | 1.74 | 1.29 | 1.17 | ** | ||
| ZCCHC14 | zinc finger, CCHC domain containing 14 | 23174 | 3.05e-02 | 1.36e-03 | 9.05e-02 | 1.11 | 1.66 | 1.27 | ** | ||
| ZNF253 | zinc finger protein 253 | 56242 | 5.52e-02 | 2.43e-02 | 1.07e-01 | 2.18 | 1.16 | 1.11 | ** | ||
| ZNF426 | zinc finger protein 426 | 79088 | 6.98e-02 | 3.42e-03 | 7.62e-01 | 1.55 | 1.51 | 1.30 | * | * | |
| ZNF48 | zinc finger protein 48 | 197407 | 5.05e-02 | 5.04e-03 | 1.14e-02 | −1.67 | −1.07 | −1.03 | ** | ||
| ZNF487P | zinc finger protein 487, pseudogene | 642819 | 5.33e-05 | 1.01e-03 | 5.62e-02 | 1.47 | 2.43 | 1.18 | ** | ||
| ZNF724P | zinc finger protein 724, pseudogene | 440519 | 3.69e-02 | 1.45e-01 | 1.73e-02 | 2.07 | −1.05 | −1.15 | ** | ||
| ZNF737 | zinc finger protein 737 | 100129842 | 2.08e-02 | 6.03e-03 | 3.48e-01 | 2.11 | 1.29 | 1.46 | ** | ||
| ZSWIM6 | zinc finger, SWIM-type containing 6 | 57688 | 4.63e-02 | 5.34e-03 | 4.44e-01 | 1.55 | 1.78 | 1.22 | * | ||
P-values in bold if significant at two-factor ANOVA (<0.05).
FDR: false discovery rate, according to BETR analysis. Ptreatment: P-value for treatment effect; and Pinteraction: P-value for interaction effect, at two-factor ANOVA. FC: fold-change in Aminaphtone-treated vs. untreated IL-1β-stimulated endothelial cells. AMNA: Aminaphtone. Pairwise: pairwise significant at two-tailed t-test.
P<0.05.
P<0.01 at two-tailed t-test.
Fig. 1.
Unsupervised hierarchical clustering of significant genes modulated over time by Aminaphtone in treated vs. untreated IL-1β stimulated ECV304 endothelial cells. The BETR algorithm identified 252 significantly modulated genes (FDR <0.1). Samples and genes were clustered using Pearson׳s correlation (centered) and average linkage method. Each combination of treatments and time points regrouped in distinct clusters. The log2 transformed, normalized, median-centered expression level of each gene is represented with a blue, black, and orange color scale: blue indicates below median, black, equal to, and orange above median. Class legends stand for: IL-1β (IL-1β without Aminaphtone); AMNA (IL-1β with Aminaphtone).
Table 2.
Gene sets significantly modulated by Aminaphtone in treated vs. untreated IL-1β-stimulated endothelial cells.
| GO Categorya | GO Ontology | GO term | #genes | LS | KS | Gene list |
|---|---|---|---|---|---|---|
| 0005975 | BP | carbohydrate metabolic process | 11 | 0.0805 | 0.0028 | DDIT4, ENO2, FUT11, GCLC, HK2, HMMR, PDGFB, PDK1, PFKFB4, PPP1R3C, SLC2A1 |
| 0007166 | BP | cell surface receptor signaling pathway | 46 | 0.0260 | 0.0020 | AAK1, BCL2L11, BMP6, CBLB, CITED2, CX3CL1, DDIT4, DDR1, DKK1, EBI3, ECT2, EDN1, EFNA1, EFNB2, EIF4B, EIF4EBP2, FOS, FST, GAREM, GPR56, GRB7, IL1B, IL7R, KCTD11, LIF, MERTK, OXTR, PDGFB, PIK3R1, PIK3R3, PLAT, PLEKHG2, PRDM1, PRKCA, PTCH1, SEMA4C, STAT5A, STC1, STC2, TGFB1I1, TGFB2, TICAM1, TLE1, TNFSF18, ULK1, VLDLR |
| 0022411 | BP | cellular component disassembly | 7 | 0.0042 | 0.8859 | BNIP3, DDIT4, DDR1, MMP1, MMP10, MMP3, TOP2A |
| 0048732 | BP | gland development | 11 | 0.0031 | 0.0022 | BCL2L11, CITED2, DDR1, ELF3, HK2, ID2, OXTR, PDGFB, PTCH1, STAT5A, TGFB2 |
| 0042592 | BP | homeostatic process | 26 | 0.0015 | 0.2886 | BCL2L11, BNIP3, CITED2, CTSK, EDN1, EGR2, GCLC, GCLM, GPR89A, HK2, HMOX1, ID2, IL1B, IL7R, MYLIP, OXTR, POLA1, PRKCA, PTCH1, SERPINA3, STAT5A, STC1, STC2, TFRC, TGFB2, TNFRSF11B |
| 0002376 | BP | immune system process | 47 | 0.0015 | 0.0284 | ANXA3, BCL2L11, BCL3, BMP6, BNIP3, CBLB, CCL22, CD70, CD83, CITED2, CLEC4E, CSF2, CTSK, CX3CL1, CXCL3, CXCL6, DDIT4, EBI3, EDN1, FOS, FST, GRB7, HMOX1, ID2, IL1B, IL32, IL7R, KIF23, LIF, MARCH8, MERTK, MMP1, PDGFB, PIK3R1, PIK3R3, PODXL, PRDM1, PRKCA, SELE, SERPINB9, STAT5A, TFRC, TGFB2, TICAM1, TNFSF18, ULBP3, ZC3HAV1 |
| 0006954 | BP | inflammatory response | 15 | 0.0131 | 0.0047 | BMP6, CCL22, CX3CL1, CXCL3, CXCL6, ELF3, FOS, HMOX1, IL1B, PRKCA, SELE, SERPINA3, STAT5A, TICAM1, TNFAIP6 |
| 0043066 | BP | negative regulation of apoptotic process | 20 | 0.0028 | 0.0677 | AGAP2, BCL3, BNIP3, CITED2, CSF2, CTH, CX3CL1, EDN1, GCLC, GCLM, HMOX1, IL1B, NR4A2, NUAK2, PIK3R1, PRKCA, SERPINB9, STAT5A, TLE1, TNFSF18 |
| 0007389 | BP | pattern specification process | 6 | 0.0137 | 0.0006 | CITED2, DKK1, EDN1, EGR2, FST, PTCH1 |
| 0010646 | BP | regulation of cell communication | 49 | 0.1744 | 0.0034 | AAK1, ACAP1, AGAP2, ANKRD1, ARHGEF26, BCL2L11, BCL3, BMP6, BNIP3, CBLB, CITED2, CSF2, CTH, DDIT4, DKK1, DUSP8, ECT2, EDN1, EFNA1, EGR2, FST, GAREM, GPR89A, HMOX1, IL1B, KCTD11, LIF, LMCD1, OXTR, PDGFB, PHLDA3, PLAT, PLEKHG2, PRDM1, PRKCA, PTCH1, RASD2, SEMA4C, SLC2A1, SPRY4, SYDE1, TGFB1I1, TGFB2, TICAM1, TLE1, TRAF1, TRIP6, ULK1, ZC3HAV1 |
| 0042127 | BP | regulation of cell proliferation | 23 | 0.0019 | 0.0322 | BMP6, BRCA1, CBLB, CSF2, DDR1, EBI3, EDN1, GAREM, HILPDA, HMOX1, ID2, IL1B, KCTD11, LIF, PDGFB, PRDM1, PRKCA, PTCH1, STAT5A, TGFB1I1, TGFB2, TICAM1, TNFSF18 |
| 0045619 | BP | regulation of lymphocyte differentiation | 5 | 0.0009 | 0.0034 | CD83, ID2, IL7R, PRDM1, STAT5A |
| 0051246 | BP | regulation of protein metabolic process | 38 | 0.0567 | 0.0032 | AGAP2, BCL3, BMP6, BRCA1, BUB1B, CBLB, CSF2, DDIT4, DKK1, DUSP8, EBI3, ECT2, EDN1, EFNA1, EIF4B, EIF4EBP2, GCLC, GRB7, HMOX1, IL1B, KRT17, LIF, LPIN1, MYLIP, PDGFB, PLAT, PRKCA, RASD2, SAMD4A, SERPINA3, SERPINB9, SERPINH1, SERPINI1, SPRY4, STAT5A, TGFB2, TICAM1, VLDLR |
| 0051090 | BP | regulation of sequence-specific DNA binding transcription factor activity | 10 | 0.0020 | 0.0058 | ANXA3, CTH, FOS, HMOX1, ID2, IL1B, PTCH1, TICAM1, TNFSF18, TRAF1 |
| 0006357 | BP | regulation of transcription from RNA polymerase II promoter | 30 | 0.0035 | 0.6000 | ANKRD1, BCL3, BMP6, BRCA1, CITED2, DKK1, EDN1, EFNA1, EGR2, ELF3, ERF, FOS, FST, HMOX1, ID2, IL1B, LIF, LMCD1, LPIN1, MEF2D, NR4A2, PER1, PIK3R1, POLA1, PRDM1, PTCH1, RRM2, STAT5A, TOP2A, VLDLR |
| 0036293 | BP | response to decreased oxygen levels | 14 | 0.0024 | 0.0002 | ANKRD1, BNIP3, CITED2, DDIT4, EDN1, HMOX1, NR4A2, OXTR, PDGFB, PDK1, PLAT, STC1, STC2, TGFB2 |
| 0009605 | BP | response to external stimulus | 32 | 0.0001 | 0.0071 | AGAP2, ANKRD1, BNIP3, CCL22, CITED2, CX3CL1, CXCL3, CXCL6, CYP24A1, EDN1, EFNB2, EGR2, ELOVL4, FOS, HMOX1, ID2, IL1B, NR4A2, NUAK2, PDGFB, PER1, PLAT, PRKCA, RND1, SELE, SLC2A1, STAT5A, STC1, STC2, TGFB2, TNFRSF11B, ULK1 |
| 0010033 | BP | response to organic substance | 48 | <0.0001 | 0.0168 | ANKRD1, BMP6, BRCA1, CD83, CITED2, CSF2, CTH, CX3CL1, CYP24A1, DKK1, DNAJB1, EBI3, EDN1, EGR2, EIF4B, EIF4EBP2, FOS, GAREM, GCLC, HCAR1, HMOX1, IDI1, IL1B, IL7R, LOX, LPIN1, MMP3, NR4A2, OXTR, PDGFB, PIK3R1, PIK3R3, PLAT, PRDM1, PRKCA, PTCH1, SELE, SERPINB9, SERPINH1, STAT5A, STC1, STC2, TGFB1I1, TGFB2, TICAM1, TNFRSF11B, TNFSF18, ZC3HAV1 |
| 0048511 | BP | rhythmic process | 6 | 0.0033 | 0.0194 | EGR2, ID2, OXTR, PER1, STAT5A, TGFB2 |
| 0046903 | BP | secretion | 20 | 0.0238 | 0.0008 | ANKRD1, ANXA3, BMP6, CLEC4E, DDR1, EDN1, FST, HK2, HMOX1, IL1B, LIF, MERTK, OXTR, PDGFB, PRKCA, SLC2A1, SRGN, STAT5A, TGFB2, VAMP1 |
| 0005125 | MF | cytokine activity | 15 | 0.0061 | 0.0047 | BMP6, CCL22, CD70, CSF2, CX3CL1, CXCL3, CXCL6, EBI3, EDN1, IL1B, IL32, LIF, TGFB2, TNFRSF11B, TNFSF18 |
| 0004175 | MF | endopeptidase activity | 6 | 0.0004 | 0.1037 | CTSK, MMP1, MMP10, MMP3, PLAT, PRSS22 |
| 0001071 | MF | nucleic acid binding transcription factor activity | 11 | 0.0006 | 0.0382 | BCL3, CEBPD, CITED2, EGR2, ELF3, ERF, FOS, MEF2D, NR4A2, PRDM1, STAT5A |
| 0000988 | MF | protein binding transcription factor activity | 11 | 0.0990 | 0.0032 | ANKRD1, BRCA1, CITED2, ELF3, ERF, LIF, LMCD1, LPIN1, PER1, TGFB1I1, TLE1 |
| 0019904 | MF | protein domain specific binding | 5 | 0.0031 | 0.0167 | ACAP1, CITED2, EGR2, IL1B, KHDRBS3 |
| 0001067 | MF | regulatory region nucleic acid binding | 6 | 0.0040 | 0.0378 | BRCA1, EGR2, FOS, MEF2D, PER1, STAT5A |
| 0031012 | CC | extracellular matrix | 9 | 0.0034 | 0.0040 | LAMB3, LMCD1, LOX, MMP1, MMP10, MMP3, TGFB1I1, TGFB2, TNFRSF11B |
| 0005615 | CC | extracellular space | 31 | 0.0003 | <0.0001 | BMP6, CCL22, CD70, CSF2, CTSK, CX3CL1, CXCL3, CXCL6, DKK1, EBI3, EDN1, HILPDA, HMOX1, IGFL1, IL1B, IL32, LIF, LMCD1, LOX, MERTK, MMP10, MMP3, PLAT, SELE, SERPINB9, SRGN, STC1, TGFB2, TNFRSF11B, TNFSF18, VLDLR |
| KEGG Pathwayb | Pathway description | #genes | LS | KS | Gene list |
|---|---|---|---|---|---|
| hsa04060 | Cytokine-cytokine receptor interaction | 13 | 0.0287 | 0.0095 | CCL22, CD70, CSF2, CX3CL1, CXCL3, CXCL6, IL1B, IL7R, LIF, PDGFB, TGFB2, TNFRSF11B, TNFSF18 |
| Broad MSigDB Curated Gene Setc | #genes | LS | KS | Gene list |
|---|---|---|---|---|
| HINATA_NFKB_TARGETS_KERATINOCYTE_UP | 13 | 0.0004 | 0.0095 | CD83, CSF2, CXCL3, CXCL6, EFNA1, IL1B, IL32, IL7R, MMP1, PLAT, STAT5A, TNFAIP6, TRAF1 |
| HINATA_NFKB_TARGETS_FIBROBLAST_UP | 6 | 0.0027 | 0.1123 | CD83, CXCL3, CXCL6, IL1B, MMP1, SELE |
| SCHOEN_NFKB_SIGNALING | 5 | 0.0192 | 0.0036 | CSF2, EDN1, IL1B, NUAK2, SERPINA3 |
| ELVIDGE_HYPOXIA_UP | 19 | 0.0017 | 0.0010 | AK4, BNIP3, CITED2, DDIT4, DDR1, ELF3, ENO2, FOS, HK2, LOX, P4HA1, PDGFB, PDK1, PRKCA, SAMD4A, SLC2A1, STC1, STC2, VLDLR |
| ELVIDGE_HIF1A_AND_HIF2A_TARGETS_DN | 14 | 0.0036 | 0.0001 | AK4, BNIP3, CITED2, ENO2, FOS, HK2, LOX, P4HA1, PDGFB, PDK1, SAMD4A, SLC2A1, STC1, VLDLR |
| ELVIDGE_HYPOXIA_BY_DMOG_UP | 17 | 0.0038 | 0.0023 | AK4, BNIP3, CITED2, DDIT4, ELF3, ENO2, FOS, HK2, LOX, P4HA1, PDK1, PRKCA, SAMD4A, SLC2A1, STC1, STC2, VLDLR |
| ELVIDGE_HIF1A_TARGETS_DN | 13 | 0.0114 | 0.0002 | AK4, BNIP3, CITED2, ENO2, FOS, HK2, LOX, P4HA1, PDGFB, PDK1, SLC2A1, STC1, VLDLR |
| WINTER_HYPOXIA_METAGENE | 18 | 0.0022 | 0.0376 | BNIP3, CITED2, DDIT4, EDN1, EFNA1, ELF3, FOS, HK2, HMOX1, ID2, LOX, P4HA1, PDGFB, PFKFB4, SLC2A1, SLC6A8, STC2, TFRC |
| PID_HIF1_TFPATHWAY | 9 | 0.0022 | 0.2078 | BNIP3, CITED2, EDN1, FOS, HK2, HMOX1, ID2, SLC2A1, TFRC |
| MENSE_HYPOXIA_UP | 10 | 0.0028 | 0.0004 | BNIP3, CEBPD, ENO2, HK2, LOX, P4HA1, PDK1, PFKFB4, PPP1R3C, STC2 |
| LEONARD_HYPOXIA | 11 | 0.0061 | 0.0007 | AK4, BNIP3, DDIT4, EFNA1, HK2, P4HA1, PDGFB, PFKFB4, PPP1R3C, SLC2A1, STC2 |
| JIANG_HYPOXIA_NORMAL | 15 | 0.0134 | 0.0009 | AK4, BNIP3, CITED2, CNNM4, EIF4B, ENO2, HMOX1, KHDRBS3, LOX, P4HA1, PFKFB4, PPP1R3C, RIPK4, SLC2A1, STC2 |
| KRIEG_HYPOXIA_VIA_KDM3A | 5 | 0.0182 | 0.0026 | C15orf48, CLEC4E, EDN1, HMOX1, LAMB3 |
| FARDIN_HYPOXIA_11 | 6 | 0.0200 | 0.0029 | AK4, BNIP3, DDIT4, FUT11, PDK1, PFKFB4 |
| NAGASHIMA_EGF_SIGNALING_UP | 8 | 0.0008 | 0.0007 | AREG, DNAJB1, EDN1, EGR2, FOS, LIF, NR4A2, TNFRSF11B |
| ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TNF_UP | 22 | 0.0021 | 0.0070 | BCL3, C15orf48, CD83, CXCL3, DDIT4, EDN1, EFNA1, GCLC, IGFL1, IL1B, IL32, IL7R, LAMB3, LIF, MMP10, NAV3, NINJ1, SAMD4A, SEMA4C, SERPINB9, TNFAIP6, TRAF1 |
| BASSO_CD40_SIGNALING_UP | 8 | 0.00498 | 0.0093 | CCL22, CD83, DDIT4, IL1B, PTP4A3, SRGN, STAT5A, TRAF1 |
| WIERENGA_STAT5A_TARGETS_GROUP2 | 5 | 0.0163 | 0.0036 | CCL22, CD83, CSF2, IL7R, TRAF1 |
GO ontology: BP=biological process, MF=molecular function, and CC=cellular component.
GO (Gene Ontology) categories,
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, and
MsigDB (Broad Institute Molecular Signature Database) curated gene sets found to be significant at the 0.005 significance level of either the LS or KS permutation tests. For each Gene Set, the table lists the unique identifier, the number of genes differentially modulated, the Fisher (LS) and Kolmogorov–Smirnov (KS) permutation P-values (P-values <0.005 are in bold), and the list of modulated genes.
2. Experimental design, materials and methods
2.1. Cell cultures and treatments
Human ECV304 endothelial cells (European Collection of Authenticated Cell Cultures, ECACC No. 92091712) were seeded at ×105/well in 6-well tissue-culture treated plates and grown to confluence in M199 complete medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT), at 37 °C in a humidified incubator with 5% CO2. Cells were serum starved (1% FBS) to synchronize the mitotic phase 24 h before treatment. Cells were then incubated for 1, 3, and 6 h with recombinant IL-1β 100 IU/ml (Sigma-Aldrich, St. Louis, MO) in the presence of Aminaphtone 6 μg/ml (Baldacci, Pisa, Italy) or an equal volume of medium alone. Experiments were performed in three independent replicates for each time point.
2.2. RNA isolation and whole-genome gene expression profiling
Total RNA extraction was performed with TRIzol (Life Technologies, Rockville, MD) directly added to the ECV304 culture plates. Removal of contaminating genomic DNA was done by treating RNA samples with RNase-free Turbo DNase (Life Technologies) for 15 min at room temperature. RNA quantity and quality were assessed respectively by micro-volume spectrophotometry on an Infinite 200 PRO plate reader (Tecan, Männedorf, Switzerland) and by on-chip capillary electrophoresis on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Absorbance ratio at 260 and 280 nm was ≥1.9 and the RNA integrity number (RIN) was >8 for all samples.
For each replicate, 100 ng of total RNA were amplified and labeled using the Whole-Transcript Sense Target Labeling Protocol by Affymetrix (Santa Clara, CA) without ribosomal RNA reduction. Affymetrix GeneChip Human Gene 1.0 ST arrays were hybridized with 11 µg of labeled sense DNA, washed, stained, and scanned on an Affymetrix 7G Scanner according to the manufacturer׳s protocols.
2.3. Data processing and probe mapping
Data were extracted using the Affymetrix Expression Console software. Background correction, log2 transformation, quantile normalization, and median polish probeset summarization was performed using the Robust Multi-array Average (RMA) method [3] implemented in the RMAExpress software v1.0.5. The resulting dataset consisted of 28869 probesets. The BRB-ArrayTools v4.3.2 (developed by Dr. Richard Simon and BRB-ArrayTools Development Team) and Bioconductor R packages v2.12 [4] were used for probe filtering and annotation. Probesets were deemed as non-informative and excluded from further analysis under any of the following conditions: P-value of the log-ratio variation greater than 0.01, i.e. genes showing minimal variation across samples; 17th percentile of intensity less than 10, i.e. genes with the lowest acceptable expression level at most in three samples. Multiple probesets were reduced to one per gene symbol by using the most variable probe measured by interquartile range across arrays. After applying these stringent quality control and gene filtering criteria, we analyzed the expression changes over time of 6461 genes. Project was annotated with the Bioconductor annotation package hugene10sttranscriptcluster.db v8.0.1.
2.4. Statistical and bioinformatics analysis
Differentially expressed genes were sought combining two statistics implemented in the software MultiExperiment Viewer (MeV) v4.9 [5]. To identify genes varying significantly between the two conditions across time points, we used the Bayesian Estimation of Temporal Regulation (BETR) method [6], which is a linear random-effect modeling framework that takes into account correlations within samples between sampling times. Genes assigned a False Discovery Rate (FDR) <0.1 were deemed significantly modulated. Then, to determine which genes were mainly influenced by the effect of Aminaphtone treatment per se (i.e. irrespective of the time response) and/or by the interaction effect of the two factors time and treatment, we applied a two-factor ANOVA to the gene list identified by BETR, given the balanced factorial design of the study. Genes were considered statistically significant if the P-values either for treatment and/or for interaction were <0.05. We finally performed post-hoc pairwise comparisons (2-tailed Student׳s t-test) to identify significant differences (P<0.05) between treatment classes at any time points.
Unsupervised hierarchical clustering was performed using the algorithms implemented in BRB-ArrayTools, to visualize similarities and differences in gene expression profiles that could discriminate treatment classes and/or changes over time. Log2 transformed, normalized gene expression values were median-centered, scaled, and clustered by Pearson׳s centered correlation and average linkage as distance metrics.
Functional analysis of significant genes identified by BETR was carried out by examining gene sets for differential expression between Aminaphtone treated and untreated samples. Gene sets were derived from the Gene Ontology (GO) database (http://www.geneontology.org) [7], the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html), and the curated gene sets of the Broad Institute Molecular Signature Database (MSigDB) [8]. LS/KS permutation tests were used to find gene sets with more genes differentially expressed among the phenotype classes than expected by chance. The threshold P-value was set at 0.005. Redundant GO terms were filtered out using the web-based tool REViGO [9], allowing a similarity threshold of 0.5.
Acknowledgments
The authors gratefully acknowledge the excellent technical assistance of Elena Grovetti (Department of Pathophysiology and Transplantation, Università degli Studi di Milano). The work was entirely supported by internal funds from the Fondazione IRCCS Ca׳ Granda Ospedale Maggiore Policlinico, Milano, Italy.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2016.06.051.
Transparency document. Supplementary material
Supplementary material
References
- 1.Salazar G., Bellocchi C., Todoerti K., Saporiti F., Piacentini L., Scorza R., Colombo G.I. Gene expression profiling reveals novel protective effects of Aminaphtone on ECV304 endothelial cells. Eur. J. Pharmacol. 2016;782:59–69. doi: 10.1016/j.ejphar.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 4.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 6.Aryee M.J., Gutierrez-Pabello J.A., Kramnik I., Maiti T., Quackenbush J. An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation) BMC Bioinform. 2009;10:409. doi: 10.1186/1471-2105-10-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Gene Ontology Consortium, Gene Ontology Consortium: going forward, Nucleic Acids Res., 43, 2015, pp. D1049–D1056. [DOI] [PMC free article] [PubMed]
- 8.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material