Abstract
Oligomeric protein nanopores with rigid structures have been engineered for the purpose of sensing a wide range of analytes including small molecules and biological species such as proteins and DNA. We chose a monomeric β-barrel porin, OmpG, as the platform from which to derive the nanopore sensor. OmpG is decorated with seven flexible loops that move dynamically to create a distinct gating pattern when ionic current passes through the pore. Biotin was chemically tethered to the most flexible one of these loops. The gating characteristic of the loop’s movement in and out of the porin was substantially altered by analyte protein binding. The gating characteristics of the pore with bound targets were remarkably sensitive to molecular identity – even providing the ability to distinguish between homologues within an antibody mixture. A total of five gating parameters were analyzed for each analyte to create a unique fingerprint for each biotin binding protein. Our exploitation of gating noise as a molecular identifier may allow more sophisticated sensor design while OmpG’s monomeric structure greatly simplifies nanopore production.
Keywords: nanopore, protein sensor, single-molecule detection, OmpG
Protein nanopores have become powerful single-molecule analytical tools that enable the study of fundamental problems in chemistry and biology,1, 2 including protein folding3 and unfolding,4-8 enzymatic activity,9-11 chemical reactions12, 13 and stability of complex formation.14 Beyond basic research, nanopores also hold tremendous promise in biotech applications such as DNA sequencing11, 15-17 and biosensing.1 Molecular detection using a single nanopore works by observing modulations in ionic current flowing through the pore during an applied potential. Typically, binding (or translocation) of an analyte within (or through) the pore’s lumen partially blocks the flow of current and provides information about a molecule’s size, concentration and affinity18. Protein nanopores based on protein toxins, especially α-hemolysin (αHL), have been used to detect metal ions,19, 20 organic molecules,21-23 oligonucleotides11, 17, 24 and measure the size of polymers.25, 26
Although αHL works well for small analyte detection, molecules larger than 27Å in diameter cannot fit in the pore’s lumen. Direct protein detection with nanopores is therefore problematic, though some strategies have been developed to transmit the binding signal from solution to the pore’s interior.27-30 For example, binding of a kinase was performed using an αHL pore modified with an inhibitor peptide attached to its stem side.28 The binding of lethal factor to the PA63 pores of the anthrax toxin orients the N-terminal leader sequence towards the pore’s lumen.31, 32 In both cases, analyte docking to the binding site on the sensor pore manifest as a current blockage.28, 32 In addition to direct current blockade, target analytes may also be detected indirectly through current modulation. A common strategy involves a nanopore-permeable molecule, e.g. a small chemical ligand or ligand-modified polymer whose partitioning into or translocation through the nanopore was altered after analyte binding. Following this scheme, the detection of streptavidin or avidin was demonstrated by tethering biotin via a PEG polymer to αHL30 or monitoring the translocation of biotinylated poly nucleic acids through αHL.33-35
Another strategy is to use larger nanopores for analyte detect. For example, the bacterial toxin ClyA, with a 70Å diameter, was modified at one end with an aptamer specific to thrombin.36 So far, ClyA represents the largest protein pore for sensing. Although there are many proteins that form larger pores in nature,37 e.g. perfringolysin O (~15 nm in diameter),38 their application as sensors has yet to be realized. Synthetic nanopores do not have the size limitation and are more robust39-41 and have been applied to identify proteins either during translocation40-42 or via capture by specific receptors immobilized on the wall of the pore.39, 43-45 However, synthetic nanopores lack the well-controlled geometry common to their protein pore counterparts.
Unlike other multimeric proteinaceous nanopores such as αHL and ClyA,27, 36 outer membrane protein G (OmpG) from Escherichia coli (E. coli) is monomeric.46 Thus, complex and asymmetric alterations by chemical or genetic modifications are straightforward, making OmpG an attractive nanopore platform for developing nanopore-based sensing technology. OmpG is composed of 14 β-strands connected by seven flexible loops on the extracellular side and seven short turns on the periplasmic side (Fig. 1a).47-49 The extracellular opening is 8 Å in diameter and the periplasmic side is 14 Å.50 Wild-type OmpG spontaneously gates during an applied potential as revealed by planar bilayer studies.46, 50 Pore gating is attributed to loop 6 which flops in and out of the pore, intermittently blocking the current (Fig. 1a, b).50, 51 To reduce gating, a disulfide bond or lipid anchor was introduced into OmpG’s structure which effectively pinned the flexible loop 6 in place.50, 51 The resulting quiet OmpG was used to sense ADP in the presence of a cyclodextrin adapter.50
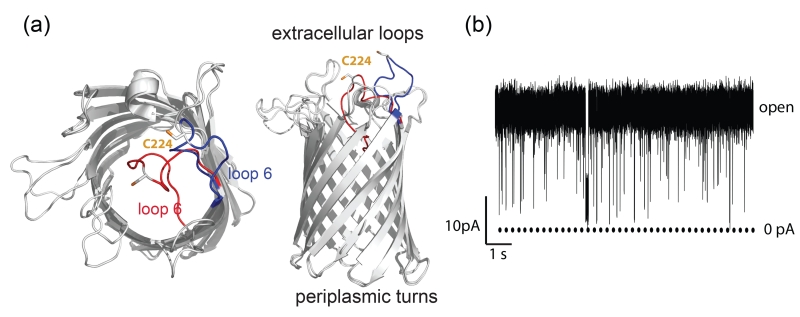
Figure 1.
Structures of OmpG and its gating activity. (a) The top view (left) and side view (right) of the structural alignment of the open (2IWV) and closed (2IWW) states. Loop 6 is highlighted in blue in the open state and red in the closed state. The D224C mutation is shown in ball and stick model. (b) Single channel recording trace of a wild type OmpG pore. The data was obtained in buffer 10 mM Tris·HCl, pH 8.0, 150 mM KCl at +50 mV.
So far, a rigid and stable structure is usually sought for protein pores for sensing.2, 52 The protein pores with demonstrated sensing applications include αHL,30 MspA,53 ClyA,36 aerolysin54 and phi29 DNA packaging motor55 all of which are homo-oligomers that possess a rigid structure. Two monomeric outer membrane porins, OmpG50 and FhuA56 with flexible loops have also been used for sensing purposes. However in both cases the flexible loops were considered as the major obstacle for sensing. These loops were either fixed or removed to stabilize a single open conformation by protein engineering.50, 56 Here, we directly exploit loop dynamics instead of pore blockage to detect protein interactions. Our results demonstrate that the flexibility of OmpG’s structure represents a unique feature, which can be used for resolving subtle differences between the surface properties of highly homologous protein analytes. This capability has not been demonstrated with other nanopores.
Results and discussion
Detection of Streptavidin by OmpG-PEG11-biotin pore
To detect proteins, we designed an OmpG nanopore with a ligand tethered to loop 6 to “fish” for target proteins. We hypothesized that target binding would alter the flexibility of loop 6 and therefore alter the gating pattern as a recognizable signal to indicate detection. To validate the concept of the OmpG sensor, we first chose biotin and streptavidin as the model ligand and target protein because of its very low dissociation constant of ~10−15 M57. A single cysteine mutation was introduced to the D224 residue of OmpG by site-directed mutagenesis (Fig.1). The OmpG D224C was expressed in E. coli as inclusion bodies and purified by ion-exchange chromatography. Purified OmpG D224C proteins were labeled with maleimide-(PEG)11-biotin and the resulting OmpG-PEG11-biotin construct was refolded to its native structure (Fig. S1). The biotin group could extend out from the OmpG pore by approximately 60Å to facilitate the capture of the analyte proteins (Fig. 2a). Single-channel recording of OmpG-D224C and OmpG-PEG11-biotin revealed that neither the mutation nor the tethered biotin group induced a measurable change in the unitary conductance or gating pattern of OmpG when compared to the wild type protein (Fig. S2). Addition of 3 nM streptavidin to the OmpG-PEG11-biotin pore induced an irreversible change in its gating pattern, i.e. a marked increase in gating frequency from 111±30 s−1 to 199 ±27 s−1 (n=3) was observed for OmpG-PEG11-biotin pore at pH 5.7 (Fig. 2b).
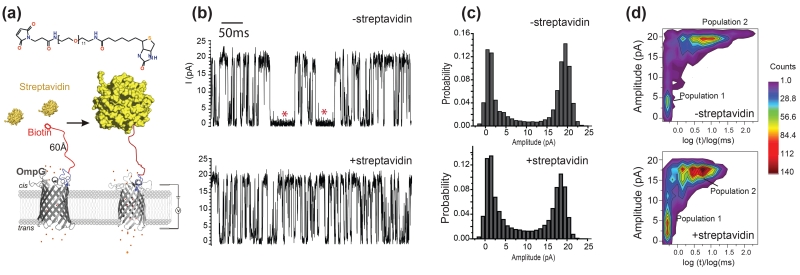
Figure 2.
Detection of streptavidin by OmpG-PEG11-biotin pore. (a) Schematic model showing the OmpG nanopore chemically modified with maleimide-PEG11-biotin. The model was generated in Pymol using PDB files of OmpG (2IWV) and streptavidin (3RY1). The streptavidin was placed approximately 60Å away from the OmpG pore in the model of the bound state. (b) Representative traces of the OmpG pores before and after the addition of the streptavidin (3 nM). The measurements were performed in buffer 10 mM sodium phosphate, pH5.7, 150 mM KCl at +50 mV. The gating event frequency increases from 75 s−1 to 97 s−1 after the addition of streptavidin. (c) All current histogram of the corresponding traces in Fig 2b. (d) Two dimensional histogram of the gating events. Gating events collected from a 15s recording trace were distributed based on their intensity versus duration. The color scale indicates the number of events.
We plot all the gating events according to their gating amplitude and duration in a two-dimensional (2D) event distribution plot (Fig. 2d). From the 2D plot analysis, we observe two population of events. Population 1 only partially blocks the pore with amplitudes between 0 to 7.5 pA and dwell time between 0-0.4 ms (Fig. S3); population 2 almost fully blocks the pore with amplitudes larger than 10 pA (10-20pA) and dwell time longer than 1ms (1-50 ms) (Fig. S3). From previous studies and known structures of OmpG,47, 50 we expect that loop 6 cannot fully block the pore on its own as it cannot occupy sufficient space within the lumen. For complete blockage, we expect that as much as one third of strand 12 must also unfold so that loop 6 is long enough to completely occlude the opening. We give the term “flickering” and “bending” to describe partial vs complete blockages, respectively. This distinction is important when considering the behavior observed in the 2D plots. For example, flickering events (population 1) seem relatively constant in the presence or absence of target, while the bending events (population 2) shorten considerably when the target binds (Fig. 2d). By contrast, the average dwell time of the bending events decreased from 5.1±0.14 ms to 3.8±0.15 ms (n=3) (Fig. S4) when streptavidin was bound. In particular, those bending events of especially long duration (>10 ms), indicated with red asterisks, were eliminated during the streptavidin-bound state (Figs. 2b, d). We hypothesize that the bending events are shortened by bound streptavidin by destabilizing the closed state. However, due to the increased gating frequency, the open probability of the OmpG pore actually reduced slightly from 0.58±0.09 to 0.51±0.10 (n= 3) upon streptavidin binding as revealed by the decrease of the open state peak (Fig 2c). As controls, streptavidin has also been added to the unmodified OmpG-D224C pores (15 pores tested), we have not observed any change in the gating pattern (Fig. S5). Thus, specific binding of streptavidin to the tethered biotin induces a clear but slight change in the gating properties of OmpG-PEG11-biotin pore.
Shortening the ligand linker to strengthen signal
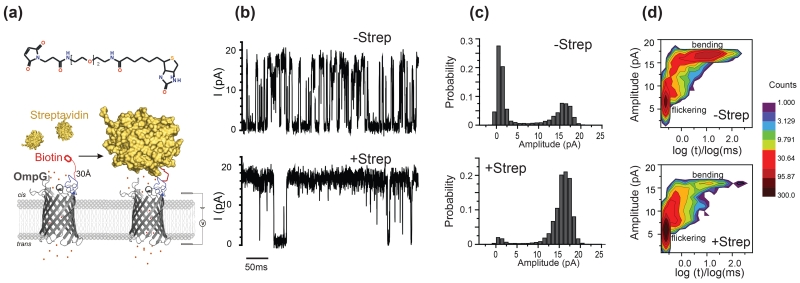
Since the binding between the OmpG-PEG11-biotin and streptavidin produced a relatively small effect on the gating, we hypothesized that the polyethylene linker was too long to effectively restrict the dynamic movement of loop 6. Therefore, we shortened the length of the PEG linker to just two units, creating the OmpG-PEG2-biotin construct where the biotin could extend ~30 Å into solution (Fig. 3a). The shortened linker did not affect the gating pattern when compared to OmpG D224C (Fig. S2). By shortening the linker, the effect of streptavidin binding was much more pronounced, permanently reducing the frequency and amplitude of gating events (Fig. 3b, c). Quantitative analysis of three OmpG-PEG2-biotin pores showed that the gating event frequency was reduced by more than 6 fold from 104 ± 6 s−1 to 16 ± 2 s−1 (n=3). Comparison of the two-dimensional plots of all events reveals that the occurrence of bending events with long duration time (>0.1ms) and high intensity (>10 pA) were mostly eliminated due to streptavidin binding (Fig. 3d). Gating events of transient duration time (<0.1 μs) and low intensity (<10 pA) still persist albeit with greatly reduced frequency. The data indicate that streptavidin bound to the PEG2 linker can strongly restrict bending but not the flickering of loop 6. As a control, streptavidin was added to OmpG D224C pores and no change was observed (10 pores tested). Adding excess BSA (1μM) to the OmpG-PEG2-biotin pore also did not show any effect (Fig. S6). These observations confirmed that the alteration of the gating pattern is caused by the specific interaction between the streptavidin and the tethered biotin ligand. In summary, binding of streptavidin to the OmpG-PEG2-biotin nanopore can be detected via reduction in gating behavior.
Figure 3.
Detection of streptavidin by OmpG-PEG2-biotin pore. (a) Schematic model showing the OmpG nanopore chemically modified with a maleimide-PEG2-biotin. The streptavidin is placed around 30Å away from the OmpG pore in the model of the bound state. (b) Representative single channel recording traces of the OmpG pores before and after the addition of the streptavidin (3 nM). The measurements were performed in buffer 10 mM sodium phosphate, pH 5.7, 150 mM KCl at +50 mV. (c) All current histogram of the corresponding traces in (b). (d) Two dimensional histogram of the gating events. Total number of 4000 gating events collected from ~220 s and ~40s recording traces of OmpG pore with and without streptavidin bound were distributed based on their intensity versus duration. The color scale indicates the number of the events.
Detection of reversible antibody binding
The biotin-streptavidin interaction is effectively irreversible, thus only one binding event can be detected with the nanopore sensor. Here, we introduce proteins with weaker dissociation constants to look at reversible interactions. Mouse monoclonal anti-biotin antibodies (mAb) were added to a recording chamber with a single OmpG-PEG2-biotin (Fig. 4a). The electrical trace showed that the presence of biotin-antibody induced a dose-dependent gating pattern that was distinct from the streptavidin bound state (Fig. 4b). During antibody binding, the pore shifted to more closed conformation as revealed by the larger closed state peak in the all current histograms (Fig. 4b, c). Indeed the calculated open probability was reduced from 0.73±0.04 at the no binding to 0.52±0.04 at the bound state (n=6). In addition, although the current fluctuates between open and closed states during both the antibody-free and antibody-bound states, the current of the pore in the fully open conformation was slightly reduced by 3.5±0.86 pA (13.6±3.8%, n=6) during the antibody bound state compared to the unbound state (Fig. 4c). This effect was not observed during the experiments using streptavidin and might suggest that the antibody is in closer proximity to the pore opening when bound. As a control, addition of mAb to OmpGwt and unmodified OmpG D224C pores did not induce any detectable binding signal (Fig. S7a). Neither were mouse anti-histag nor anti-Glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH) monoclonal antibodies (20 nM) detected by current recording with OmpG-PEG2-biotin pores (Fig. S7b). Thus, these gating events resulted from the specific mAb binding to the tethered biotin.
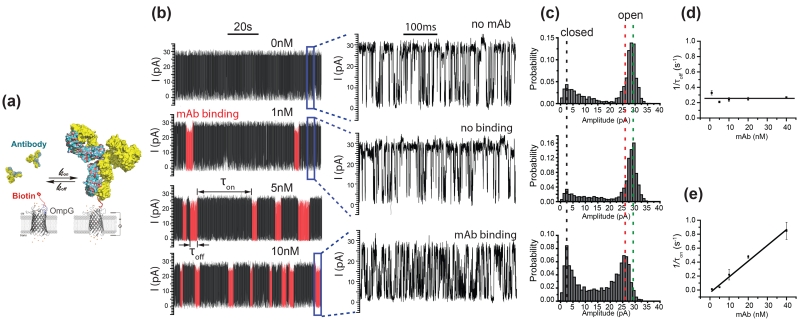
Figure 4.
Detection of monoclonal anti-biotin antibody by OmpG-PEG2-biotin pore. (a) Schematic model showing the reversible binding of monoclonal anti-biotin antibody to OmpG-PEG2-Biotin pore. The model is generated in Pymol using pdb files of OmpG (2IWV) and a mouse monoclonal anti-phenobarbital antibody (1IGY). The antibody was placed approximately 30Å away from the OmpG pore in the captured model. (b) Representative single channel recording traces at various mAb concentrations. The mAb binding regions in the recording traces are highlighted in red. Increase of the mAb binding frequency was observed with increasing concentration of mAb. The measurements were performed in buffer 10 mM sodium phosphate, pH 6.0, 300 M KCl at +50 mV. (c) All current histogram of the corresponding traces in (b). The green and red dashed lines emphasize the shift of the fully open states in current at unoccupied and mAb bound states respectively. (d) (e) Concentration dependence of the 1/τoff and 1/τon. Error bars represent the standard deviations from the measurements of at least three independent pores.
Next, the dwell time (τoff) and inter-event intervals (τon) of mAb binding was calculated (Fig. S8). The average dissociation rate constant (koff=1/τoff) of the mAb binding events was 0.25±0.04 s−1 (n=4) which was independent of the antibody concentration (Fig. 4d). The observed association constant (kon’=1/τon) increased linearly with the increasing concentration of antibody (Fig. 4e). The association rate constant kon of antibody binding was 2.30±0.43×107M−1·s−1 (n=4). The equilibrium dissociation constant (Kd) of the mouse monoclonal antibody to biotin was 1.12±0.28 ×10−8M−1 (n=4). At the lowest mAb concentration tested (1 nM), the mean inter-event interval was 74.5 ± 31 s meaning the OmpG-PEG2-biotin sensor can detect 1nM anti-biotin mAb within tens of min.
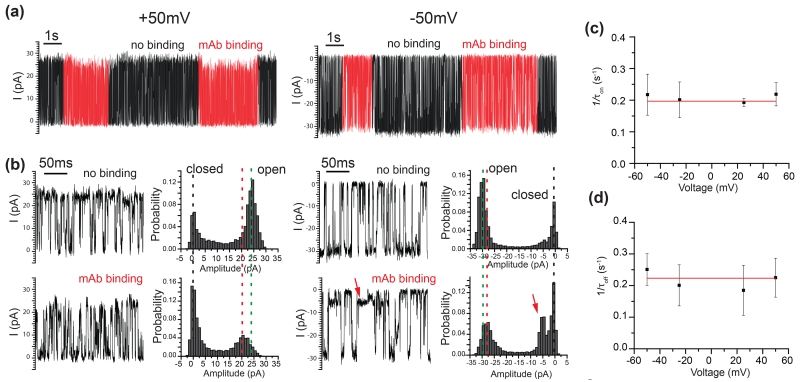
Influence of voltage on the mAb binding
OmpG exhibits asymmetrical gating pattern at positive and negative voltages. Therefore, we were interested to see if the polarity of the voltage could similarly affect the dynamic motion of loop 6 during the mAb bound state. Figure 5a shows that mAb binds to OmpG-PEG2-biotin at both +50 mV and −50 mV. The open probability of the mAb bound state at +50 mV and −50 mV is 0.52±0.04 (n=6) and 0.40±0.09 (n=6) respectively, in comparison to 0.73±0.04 (n=6) and 0.71±0.01 (n=3) of the non-binding state. Thus in the mAb bound state, the pore switched to a slightly more closed state at the negative potential (Fig. 5b). OmpG pore also showed a decreased current by 1.2±0.4 pA (n=3) at its fully open state at −50 mV. This decrease is ~5.4% of the current of a no binding state in comparison to the 13.6% decrease at the positive potential. Moreover it had a partial closure state with 6 pA of residual current as indicated by the red arrow in the recording trace and all current histograms (Fig. 5b). This result shows that the loop gating during the mAb bound state is still strongly influenced by the polarity of the applied potential. This is a useful feature that can be used for sensing because the asymmetric response of OmpG to target protein binding adds one more parameter for specific analyte protein recognition.
Figure 5.
Effect of voltage on the mAb binding. (a) Representative single channel recording trace of OmpG-PEG2-biotin showing reversible binding of the mAb at both +50 mV and −50 mV. We define the positive potential as the potential of the chamber where the loops are located is positive as indicated in Fig. 4a. The measurement was performed in buffer, 10 mM sodium phosphate buffer, pH 6.0, 300 mM KCl in the presence of 10 nM mAb. (b) Representative single channel recording traces of OmpG-PEG2-biotin at the unoccupied or mAb bound states at +50 mV and −50 mV. All current histogram of the corresponding current recording traces are also shown. The green and red dashed lines emphasize the shift of the fully open states in current at unoccupied and mAb bound states respectively. The positive potential caused a larger shift of the open state current than the negative potential. (c) (d) Voltage independence of 1/τon and 1/τoff. The measurements were performed in buffers 10 mM sodium phosphate buffer, pH 6.0, 300 mM KCl in the presence of 10 nM mAb at various applied voltages ranging from −50 mV to +50 mV.
Previous studies on nanopore detection have shown that voltage could alter analyte binding kinetics.28, 43, 58 Therefore, single channel recording was performed at applied voltages ranging from −50 mV to +50 mV in the presence of mouse mAbs. The voltage-dependent gating of OmpG prevented us from testing higher potentials as OmpG tends to close completely at ±75 mV.46 Neither τon nor τoff exhibited a strong dependence on voltages (Fig. 5c, d) (Fig. S9). Thus, we concluded that the mAb binding to biotin is not affected at applied potentials ranging from −50mV to 50 mV. The independence of binding from voltage at this range is advantageous since proteins can be analyzed regardless of the applied potential.
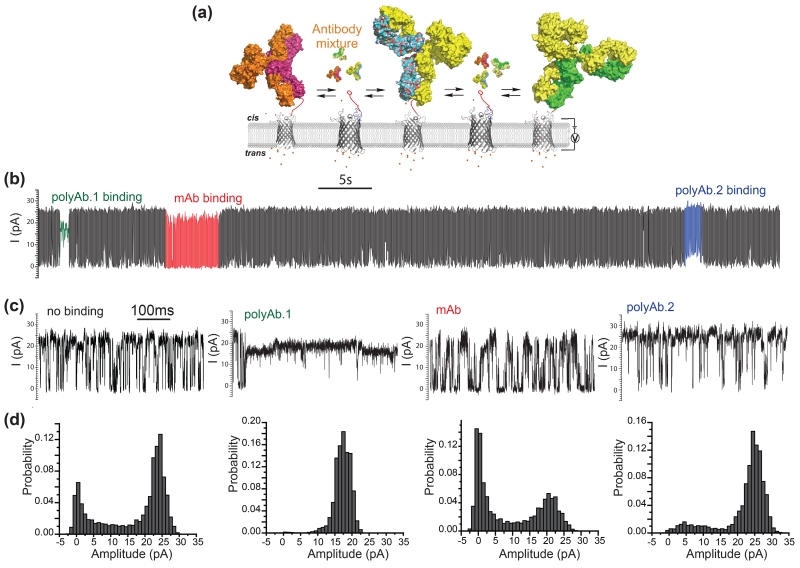
Simultaneous detection of mouse mAb and goat polyclonal anti-biotin antibody
Although the antibody and streptavidin both bound the biotinylated OmpG, they produced remarkably unique gating patterns. We wondered how sensitive the OmpG sensor would be to various factors such as a protein’s size, shape, surface charge or rigidity. In an attempt to distinguish between these, we analyzed the binding of a polyclonal anti-biotin antibody derived from goat. To our surprise, the polyclonal antibody produced gating patterns distinct from the structurally very similar mAb tested earlier (Fig. 6a, b). These specific gating patterns were not observed when adding polyclonal antibody to unmodified OmpG D224C pore (Fig. S10). Furthermore, the polyclonal sample showed clear evidence that at least two readily distinguished populations of antibody were present (Fig. 6b, c). We categorized their gating activities into two classes, called type I and type II.
Figure 6.
Detection of mouse mAb and polyclonal Ab binding by OmpG-PEG2-biotin pore. (a) Schematic model of simultaneous detection of multiple target proteins by OmpG nanopore. (b) Representative current trace of a single OmpG-biotin. The measurement was performed in the presence of mouse mAb (1 nM) and goat pAb (72 nM) in 10 mM sodium phosphate, pH 6.0, 300 mM KCl at applied potential of +50 mV. The mAb and polyAb binding events were highlighted in colors, i.e. mAb in red, pAb. 1 in green and pAb.2 in blue. (c) Representative current recording trace of OmpG-PEG2-biotin pore at the unoccupied and mAb and polyAb bound states. (d) All current histogram of the corresponding current recording traces.
During type I binding (pAb.1) the current decreased by 50% and contained few gating events. During type II (pAb.2) binding events, the open state conductance was unchanged but the gating frequency was slightly reduced (Fig. 6c, d). Mouse mAb was then added to the chamber already containing pAb and the binding was observed (Fig. 6b). All types of antibodies bound with their respective characteristics regardless of the presence of the other antibodies. These gating patterns were not seen when antibodies and streptavidin when added to unmodified OmpG D224C (Fig. S11). This is the first example of a nanopore than can distinguish between three antibodies with virtually identical shape in a complex mixture.
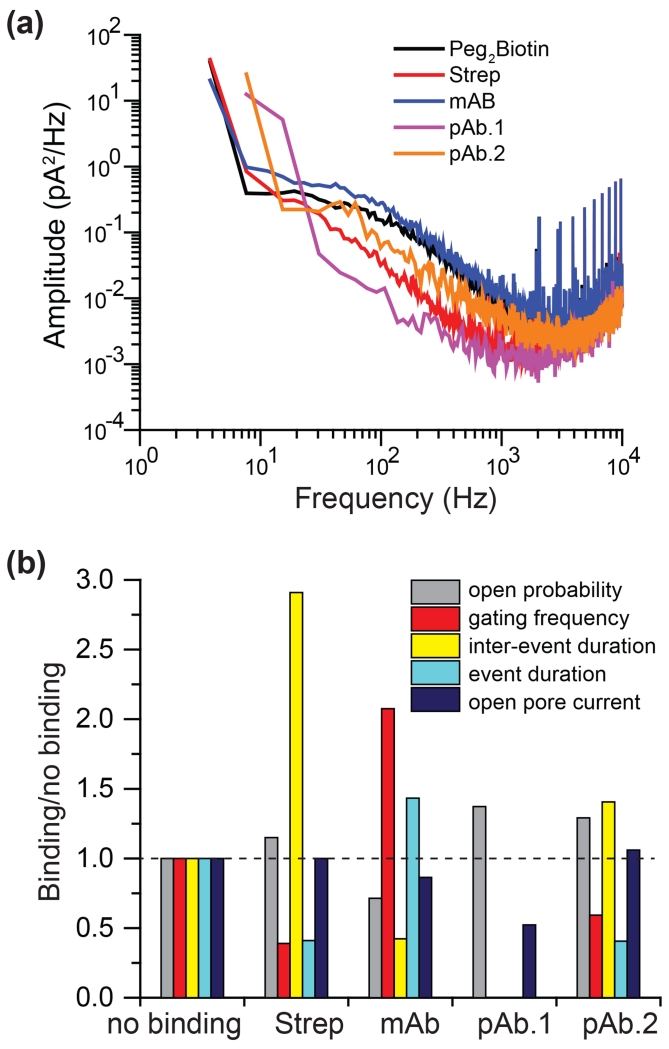
Power spectrum analysis and fingerprint of analyte protein binding signal
Nanopore sensing often relies on blockade amplitude and/or the mean duration time of binding to discriminating target molecules. In OmpG nanopore, the binding of analyte not necessarily induced a current blockage. Instead, alteration of the gating/noise of OmpG was indicative of the interaction. Noise spectral density analysis of each biotin-protein bound state revealed that mAb showed a slightly higher noise than the unbound state (Fig. 7a, S12). pAb. 1 exhibited the lowest noise while the level of streptavidin and pAb. 2 was between that of pAb. 1 and the unbound state. Thus, the noise analysis contributes key information for distinguishing analytes. Noise spectral density analysis of all OmpG pores were also obtained and showed very small changes in noise (Fig. S13)
Figure 7.
Comparison of the gating patterns of OmpG-PEG2-biotin at analyte binding states. (a) Power spectra of the protein-binding states for four biotin-binding proteins: streptavidin, mAb, pAb.1 and pAb.2. To compare the streptavidin binding with other biotin-binding proteins under the same condition, experiments were performed in 10 mM sodium phosphate, pH 6.0, 300 mM KCl (Fig. S12). Electrical traces under this condition were used to derive the power spectra and the fingerprint characteristic shown Table 1 and Fig. 7b. (b) Fingerprints of the biotin binding proteins. The gating events of different analyte protein binding states were characterized by five parameters, i.e. open probability, gating frequency, inter-event duration, event duration and the conductance of the open pore state. Changes of these parameters relative to the no binding state generate the fingerprint unique for each antibody.
However, noise analysis alone is insufficient for analyte identification. For example, the mAb-bound state was similar to the unbound state and the open pore current cannot be seen by noise analysis. To thoroughly analyze the characteristics of the traces at the analyte binding state, we analyze five parameters: (i) open probability, (ii) gating events frequency, (iii) inter gating event duration, (iv) duration of gating events and (v) the open state conductance to identify the protein(Fig. S14). Table 1 summarizes these parameters which provide a fingerprint for each analyte (Fig. 7b). The OmpG-biotin sensor can unambiguously detect and discriminate between 4 biotin-binding proteins including three antibody species (two pAb and one mAb) that share highly homologous structures.
Table 1.
Fingerprint of each type of gating events
| Open probability |
Event frequency (s−1) |
Inter-event duration (ms) |
Gating duration (ms) |
Relative conductance of open state (%) |
|
|---|---|---|---|---|---|
|
| |||||
| No binding | 0.73±0.04* | 97±3.6 | 8.68±2.14 | 2.93±0.52 | 100 |
| Streptavidin | 0.95±0.08 | 45±7 | 22.75±3.2 | 0.62±0.19 | 100±4.2 |
| mAb | 0.52±0.04 | 201±103 | 3.67±1.57 | 4.20±1.90 | 86.4±1.3 |
| pAb.1 | 0.99±0.01 | 7±1 | n/a | n/a | 52.3±4.7 |
| pAb.2 | 0.94±0.02 | 57.5±2 | 12.2±1.3 | 1.09±0.21 | 106.5±6.5 |
Values were calculated from at least three independent experiments. The errors indicate the standard deviation.
Discussion
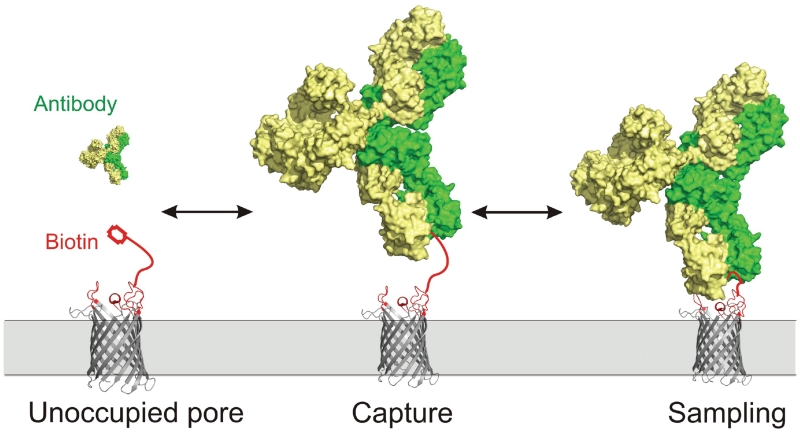
Since proteins of the same size and shape produce unique signals, we hypothesize that the OmpG sensor recognizes unique targets based on other factors such as charge, hydrophobicity or perhaps post-translational modification of the surface. The structure of OmpG, along with the data presented here, sheds some light on the possible mechanism of protein detection via the nanopore strategy. Although the four biotin-binding proteins trigger a characteristic gating pattern upon binding to the OmpG nanopore, they can be categorized into two groups. In pAb.2 and streptavidin cause a decreased gating frequency, which suggests binding to the PEG2 tethered biotin hindered the dynamics of the loop 6. According to the crystal structure, the D224 residue traverses approximately 7.5 Å between the open and closed states47. However, a recent NMR study of OmpG shows this residue may migrate as far as 30 Å between the fully open and closed conformers.49, 59 Our results suggest that such a large conformational change is strongly hindered by streptavidin binding and moderately by pAb.2 binding. The results also suggest minimal interaction between the streptavidin and pAb.2 with the loops at the opening of OmpG. In another category, mAb and pAb.1 both caused a decrease of current in the fully open state. This observation suggests that the two antibodies obstruct the current flow at the entrance, presumably by partially docking to the extracellular loops of OmpG. Because all seven loops at OmpG’s entrance are negatively charged, the two antibodies are likely positively charged or have a positively charged patch near the biotin-binding site that mediate this interaction. This speculation is supported by the observation of mAb’s asymmetrical behavior under an applied potential. Namely, a positive potential might push the mAb closer towards the OmpG pore to cause 13.6% partial block of the current (Fig. 5). In contrast, at negative potential, the electric field would repel mAb away from the pore entrance. Indeed, we observed that the open pore conductance was less affected, only ~5.4% blockage seen. These results suggest that not only the ligand-tethered loop, but all the loops on the extracellular entrance may be involved in interacting and sampling the target proteins, which explains its ability to discriminate between highly structurally homologous proteins. We expect the interaction between the loops and the two antibodies is weak because mAb and pAb.1 did not induce noticeable changes to the gating of unlabeled OmpG D224C. However these interactions may be enhanced after the antibodies bind the tethered biotin ligand. In summary, our data suggests a novel mechanism underlying OmpG nanopore sensing that contains two steps (Fig. 8). First, OmpG captures the target protein via its tethered high-affinity ligand. Consequently, the bound protein interferes with the movement of loop 6 generating its characteristic gating pattern. Second, the extracellular loops of OmpG may sample the target protein via unspecific interactions which further alters the ionic current providing additional readout. While further study is required to delve deeper into the precise mechanism of protein detection, our sensor’s ability to discriminate between structurally homologous antibodies within a multi-component mixture represents a powerful advance over previous approaches.
Figure 8.
Schematic model illustrating the principle of OmpG nanopore detection.
Previously, the detection of streptavidin and anti-biotin antibody was demonstrated using αHL that contained a PEG biotin group tethered to its vestibule.30 In the absence of the target, the PEG polymer traversed through the constriction site from the cis and trans chamber and back through the pore. This movement was manifest as rapid gating. Analyte protein binding of the biotin group eliminated the gating and provided the readout signal for protein sensing. In contrast to our OmpG nanopore, αHL-biotin sensor did not differentiate binding events derived from streptavidin and mAb which differ greatly in size, shape and surface properties. Two features of OmpG may contribute to its higher resolution compared to αHL (Fig. S15). The first is the location of the constriction site which is the narrowest part of the pore that determines the conductance. The constriction site of OmpG-PEG2-biotin is located at the entrance to the pore next to the ligand interaction site while the location of the constriction site of αHL is in the middle making it inaccessible for large folded analyte protein. Because of this, analyte protein binding at the pore entrance directly affected the conductance of OmpG but not αHL. Secondly OmpG has flexible loops at the binding site which allows conformational changes to occur in response to analyte protein binding. Instead, αHL possesses a rather stable and rigid structure at the two ends.60 Although the biotin-binding proteins might also interact with the two entrances of αHL nanopore, the rigidity of the αHL structure does not allow large conformational changes to occur, so the interaction of different target proteins with the entrance did not induce noticeable changes in the current flow that passed through the constriction site.
Our study points out an alternative design in the architecture of nanopore sensors. By creating a nanopore with a dynamic structure that changes upon analyte binding, new regions of data may be interpreted that give a greater sensitivity and selectivity for detecting protein analytes. We have shown that even protein isoforms in a mixture can be clearly distinguished using this new sensing scheme. These features are not available in other nanopore sensing strategies, making the OmpG sensor particularly useful. Further, monomeric proteins such as OmpG are ready to use after refolding and require no further assembly and purification steps compared to other oligomeric nanopores.
Conclusion
We have shown that binding of target protein to an OmpG-biotin nanopore can be deduced from changes in the gating activity of OmpG. The principle of the OmpG nanopore relies on detecting the modulation of loop dynamics upon target protein binding rather than the occupation in the pore lumen. More importantly, the OmpG nanopore exhibited the ability to resolve protein homologues that share the same high-affinity ligand, making this sensing approach well suitable for screening for homologous disease markers in complex mixtures. In the future, this principle may be extended to a broader spectrum of analytes, such as proteins, viruses, or bacteria without the need to use a far larger nanopore.
Methods
Single channel recording of OmpG proteins
Single channel recording of OmpG was similar to the previous study.50 Briefly, experiments were performed in an apparatus containing two chambers separated by a 25 μm thick Teflon film. An aperture of approximately 100 μm diameter had been made near the center of the film with an electric spark. The aperture was pretreated with a hexadecane/pentane (10% v/v) solution before each chamber was filled with buffers as indicated specifically. An Ag/AgCl electrode was immersed in each chamber with the cis chamber grounded. 1,2-Diphytanoyl-sn-glycerol-3-phosphocholine (Avanti Polar Lipids, USA) dissolved in pentane (10mg/ml) was deposited on the surface of the buffer in both chambers and monolayers formed after the pentane evaporated. The lipid bilayer was formed by raising the liquid level up and down across the aperture. OmpG proteins (~1 nM, final concentration) were added to the cis chamber and +200mV was applied to facilitate OmpG insertion. After a single OmpG pore inserted, the applied voltage was lowered to 50 mV for recording. OmpG proteins inserted in the planar lipid bilayer bi-directionally with its extracellular loops located at either cis or trans side. After 10 min recording, the orientation of the OmpG pore in the lipid bilayer was determined by analyzing the asymmetrical gating pattern at positive and negative potentials.61 Streptavidin or antibodies were added to the cis or trans chamber depending on the pore orientation and the solution was stirred for 10 s. We define a positive potential as the potential of the chamber where the extracellular loops were exposed to is positive. Current was amplified with an Axopatch 200B integrating patch clamp amplifier (Axon Instruments, Foster City, CA). Signals were filtered with a Bessel filter at 2 kHz (unless otherwise stated) and then acquired by a computer (sampling at 50 μs) after digitization with a Digidata 1320A/D board (Axon Instruments).
Single-channel current analysis
For power spectra analysis, data were recorded with a Bessel filter at 50 kHz and acquired at 250 kHz. Power spectra were calculated from a 20 s recording trace in Clampfit using segment lengths of 32768 samples (spectra resolution 7.62 Hz) by applying Hamming window. Data shown were derived from averaged spectra segments with 50% window overlap. To analyze the mAb and polyclonal antibody binding, power spectra of multiple binding events from a total of 20 s recording time were calculated and averaged. The power spectra densities for all traces were plotted in OriginPro 9.1.
Supplementary Material
ACKNOWLEDGMENTS
M.F was supported in part by a Fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface training grant (T32 GM08515).
Footnotes
ASSOCIATED CONTENT
Full documentation of materials; mutagenesis of OmpG; preparation of biotin-labelled OmpG pore; Analysis of the duration of bending events before and after streptavidin binding; details of fingerprint analysis of antibody binding; comparison of αHL and OmpG. This information is available free of charge via the Internet at http://pubs.acs.org
Reference
- 1.Howorka S, Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 2.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of Biological Pores in Nanomedicine, Sensing, and Nanoelectronics. Curr Opin Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefureac RI, Lee JS. Nanopore Analysis of the Folding of Zinc Fingers. Small. 2008;4:1646–1650. doi: 10.1002/smll.200800585. [DOI] [PubMed] [Google Scholar]
- 4.Merstorf C, Cressiot B, Pastoriza-Gallego M, Oukhaled A, Betton JM, Auvray L, Pelta J. Wild Type, Mutant Protein Unfolding and Phase Transition Detected by Single-Nanopore Recording. ACS Chem Biol. 2012;7:652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 5.Nivala J, Marks DB, Akeson M. Unfoldase-Mediated Protein Translocation through an Alpha-Hemolysin Nanopore. Nat Biotechnol. 2013;31:247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oukhaled G, Mathe J, Biance AL, Bacri L, Betton JM, Lairez D, Pelta J, Auvray L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys Rev Lett. 2007;98:158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 7.Payet L, Martinho M, Pastoriza-Gallego M, Betton JM, Auvray L, Pelta J, Mathe J. Thermal Unfolding of Proteins Probed at the Single Molecule Level Using Nanopores. Anal Chem. 2012;84:4071–4076. doi: 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Larrea D, Bayley H. Multistep Protein Unfolding During Nanopore Translocation. Nat Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majd S, Yusko EC, MacBriar AD, Yang J, Mayer M. Gramicidin Pores Report the Activity of Membrane-Active Enzymes. J Am Chem Soc. 2009;131:16119–16126. doi: 10.1021/ja904072s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q, de Zoysa RS, Wang D, Jayawardhana DA, Guan X. Real-Time Monitoring of Peptide Cleavage Using a Nanopore Probe. J Am Chem Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luchian T, Shin SH, Bayley H. Single-Molecule Covalent Chemistry with Spatially Separated Reactants. Angew Chem Int Ed Engl. 2003;42:3766–3771. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- 13.Shin SH, Luchian T, Cheley S, Braha O, Bayley H. Kinetics of a Reversible Covalent-Bond-Forming Reaction Observed at the Single-Molecule Level. Angew Chem Int Ed Engl. 2002;41:3707–3709. 3523. doi: 10.1002/1521-3773(20021004)41:19<3707::AID-ANIE3707>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Arnaut V, Langecker M, Simmel FC. Nanopore Force Spectroscopy of Aptamer-Ligand Complexes. Biophys J. 2013;105:1199–1207. doi: 10.1016/j.bpj.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The Potential and Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanunu M. Nanopores: A Journey Towards DNA Sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc Natl Acad Sci U S A. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayley H, Cremer PS. Stochastic Sensors Inspired by Biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 19.Braha O, Gu LQ, Zhou L, Lu X, Cheley S, Bayley H. Simultaneous Stochastic Sensing of Divalent Metal Ions. Nat Biotechnol. 2000;18:1005–1007. doi: 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- 20.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. Designed Protein Pores as Components for Biosensors. Chem Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 21.Cheley S, Gu LQ, Bayley H. Stochastic Sensing of Nanomolar Inositol 1,4,5-Trisphosphate with an Engineered Pore. Chem Biol. 2002;9:829–838. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 22.Wu HC, Bayley H. Single-Molecule Detection of Nitrogen Mustards by Covalent Reaction within a Protein Nanopore. J Am Chem Soc. 2008;130:6813–6819. doi: 10.1021/ja8004607. [DOI] [PubMed] [Google Scholar]
- 23.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic Sensing of Organic Analytes by a Pore-Forming Protein Containing a Molecular Adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 24.Howorka S, Cheley S, Bayley H. Sequence-Specific Detection of Individual DNA Strands Using Engineered Nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 25.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JW. Theory for Polymer Analysis Using Nanopore-Based Single-Molecule Mass Spectrometry. Proc Natl Acad Sci U S A. 2010;107:12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson JW, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ. Single-Molecule Mass Spectrometry in Solution Using a Solitary Nanopore. Proc Natl Acad Sci U S A. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotem D, Jayasinghe L, Salichou M, Bayley H. Protein Detection by Nanopores Equipped with Aptamers. J Am Chem Soc. 2012;134:2781–2787. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie H, Braha O, Gu LQ, Cheley S, Bayley H. Single-Molecule Observation of the Catalytic Subunit of Camp-Dependent Protein Kinase Binding to an Inhibitor Peptide. Chem Biol. 2005;12:109–120. doi: 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Howorka S, Nam J, Bayley H, Kahne D. Stochastic Detection of Monovalent and Bivalent Protein-Ligand Interactions. Angew Chem Int Ed Engl. 2004;43:842–846. doi: 10.1002/anie.200352614. [DOI] [PubMed] [Google Scholar]
- 30.Movileanu L, Howorka S, Braha O, Bayley H. Detecting Protein Analytes That Modulate Transmembrane Movement of a Polymer Chain within a Single Protein Pore. Nat Biotechnol. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Udho E, Wu Z, Collier RJ, Finkelstein A. Protein Translocation through Anthrax Toxin Channels Formed in Planar Lipid Bilayers. Biophys J. 2004;87:3842–3849. doi: 10.1529/biophysj.104.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halverson KM, Panchal RG, Nguyen TL, Gussio R, Little SF, Misakian M, Bavari S, Kasianowicz JJ. Anthrax Biosensor, Protective Antigen Ion Channel Asymmetric Blockade. J Biol Chem. 2005;280:34056–34062. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 33.Henrickson SE, DiMarzio EA, Wang Q, Stanford VM, Kasianowicz JJ. Probing Single Nanometer-Scale Pores with Polymeric Molecular Rulers. J Chem Phys. 2010;132:135101. doi: 10.1063/1.3328875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA Transport into an Asymmetric Nanometer-Scale Pore. Phys Rev Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 35.Kasianowicz JJ, Henrickson SE, Weetall HH, Robertson B. Simultaneous Multianalyte Detection with a Nanometer-Scale Pore. Anal Chem. 2001;73:2268–2272. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 36.Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. An Engineered Clya Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012;12:4895–4900. doi: 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, et al. The Macpf/Cdc Family of Pore-Forming Toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olofsson A, Hebert H, Thelestam M. The Projection Structure of Perfringolysin O (Clostridium Perfringens Theta-Toxin) FEBS Lett. 1993;319:125–127. doi: 10.1016/0014-5793(93)80050-5. [DOI] [PubMed] [Google Scholar]
- 39.Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. Protein Biosensors Based on Biofunctionalized Conical Gold Nanotubes. J Am Chem Soc. 2005;127:5000–5001. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 40.Han A, Schürmann G, Mondin G, Bitterli RA, Hegelbach NG, de Rooij NF, Staufer U. Sensing Protein Molecules Using Nanofabricated Pores. Appl Phys Lett. 2006;88:093901. [Google Scholar]
- 41.Li W, Bell NA, Hernandez-Ainsa S, Thacker VV, Thackray AM, Bujdoso R, Keyser UF. Single Protein Molecule Detection by Glass Nanopores. ACS Nano. 2013;7:4129–4134. doi: 10.1021/nn4004567. [DOI] [PubMed] [Google Scholar]
- 42.Fologea D, Ledden B, McNabb DS, Li J. Electrical Characterization of Protein Molecules by a Solid-State Nanopore. Appl Phys Lett. 2007;91:539011–539013. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic Sensing of Proteins with Receptor-Modified Solid-State Nanopores. Nat Nanotechnol. 2012;7:257–263. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- 44.Ding S, Gao C, Gu LQ. Capturing Single Molecules of Immunoglobulin and Ricin with an Aptamer-Encoded Glass Nanopore. Anal Chem. 2009;81:6649–6655. doi: 10.1021/ac9006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Controlling Protein Translocation through Nanopores with Bio-Inspired Fluid Walls. Nat Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conlan S, Zhang Y, Cheley S, Bayley H. Biochemical and Biophysical Characterization of Ompg: A Monomeric Porin. Biochemistry. 2000;39:11845–11854. doi: 10.1021/bi001065h. [DOI] [PubMed] [Google Scholar]
- 47.Yildiz O, Vinothkumar KR, Goswami P, Kuhlbrandt W. Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subbarao GV, van den Berg B. Crystal Structure of the Monomeric Porin Ompg. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 49.Liang B, Tamm LK. Structure of Outer Membrane Protein G by Solution Nmr Spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M, Khalid S, Sansom MS, Bayley H. Outer Membrane Protein G: Engineering a Quiet Pore for Biosensing. Proc Natl Acad Sci U S A. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhuang T, Tamm LK. Control of the Conductance of Engineered Protein Nanopores through Concerted Loop Motions. Angew Chem Int Ed Engl. 2014 doi: 10.1002/anie.201400400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Movileanu L. Interrogating Single Proteins through Nanopores: Challenges and Opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-Molecule DNA Detection with an Engineered Mspa Protein Nanopore. Proc Natl Acad Sci U S A. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, Betton J-M, Pelta J. Dynamics of Unfolded Protein Transport through an Aerolysin Pore. J Am Chem Soc. 2011;133:2923–2931. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 55.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Translocation of Double-Stranded DNA through Membrane-Adapted Phi29 Motor Protein Nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J Am Chem Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green NM. Avidin and Streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 58.Gu LQ, Bayley H. Interaction of the Noncovalent Molecular Adapter, Beta-Cyclodextrin, with the Staphylococcal Alpha-Hemolysin Pore. Biophys J. 2000;79:1967–1975. doi: 10.1016/S0006-3495(00)76445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang T, Chisholm C, Chen M, Tamm LK. Nmr-Based Conformational Ensembles Explain Ph-Gated Opening and Closing of Ompg Channel. J Am Chem Soc. 2013;135:15101–15113. doi: 10.1021/ja408206e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 61.Chen M, Li QH, Bayley H. Orientation of the Monomeric Porin Ompg in Planar Lipid Bilayers. Chembiochem. 2008;9:3029–3036. doi: 10.1002/cbic.200800444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.