Abstract
The Hippo pathway is crucial for not only normal growth and apoptosis but also cell fate specification during development. What controls Hippo pathway activity during cell fate specification is incompletely understood. In this article, we identify the insulator protein BEAF-32 as a regulator of Hippo pathway activity in Drosophila photoreceptor differentiation. Though morphologically uniform, the fly eye is composed of two subtypes of R8 photoreceptor neurons defined by expression of light-detecting Rhodopsin proteins. In one R8 subtype, active Hippo signaling induces Rhodopsin 6 (Rh6) and represses Rhodopsin 5 (Rh5), whereas in the other subtype, inactive Hippo signaling induces Rh5 and represses Rh6. The activity state of the Hippo pathway in R8 cells is determined by the expression of warts, a core pathway kinase, which interacts with the growth regulator melted in a double-negative feedback loop. We show that BEAF-32 is required for expression of warts and repression of melted. Furthermore, BEAF-32 plays a second role downstream of Warts to induce Rh6 and prevent Rh5 fate. BEAF-32 is dispensable for Warts feedback, indicating that BEAF-32 differentially regulates warts and Rhodopsins. Loss of BEAF-32 does not noticeably impair the functions of the Hippo pathway in eye growth regulation. Our study identifies a context-specific regulator of Hippo pathway activity in post-mitotic neuronal fate, and reveals a developmentally specific role for a broadly expressed insulator protein.
KEY WORDS: Color vision, Photoreceptor, Cell fate, Insulator, Drosophila retina, RNAi screen, Hippo pathway, Regulatory networks, Warts tumor suppressor, Rhodopsin
Summary: During cell fate specification in the Drosophila eye, BEAF-32 regulates expression of Hippo pathway components and acts downstream of Warts to control photoreceptor identity.
INTRODUCTION
The Hippo signaling pathway is a crucial regulator of growth and apoptosis in organ size control (Irvine and Harvey, 2015; Yu et al., 2015; Zhao et al., 2011). However, proliferation-independent roles for the pathway during animal development have also been discovered. The identification of the Hippo pathway as a regulator of R8 photoreceptor subtype specification in Drosophila was among the first examples of a mitosis-independent role for the Hippo pathway in determining cell fate (Mikeladze-Dvali et al., 2005). More recently, the pathway has been shown to regulate dendritic field tiling in neurons (Emoto et al., 2006), cell differentiation in pre-implantation embryos (Cockburn et al., 2013; Nishioka et al., 2009), neuroblast differentiation upon cell cycle exit (Reddy et al., 2010), and hematopoiesis (Milton et al., 2014), among others. Because R8 photoreceptors are post-mitotic neurons and are not competent to divide, they are an excellent system in which to elucidate context-specific mechanisms of Hippo pathway function (Hsiao et al., 2013; Jukam and Desplan, 2011; Jukam et al., 2013). How the Hippo pathway is regulated differently in division and differentiation is incompletely understood. Here, we describe the insulator protein BEAF-32 as a regulator of Hippo pathway activity in cell fate specification in the developing Drosophila retina.
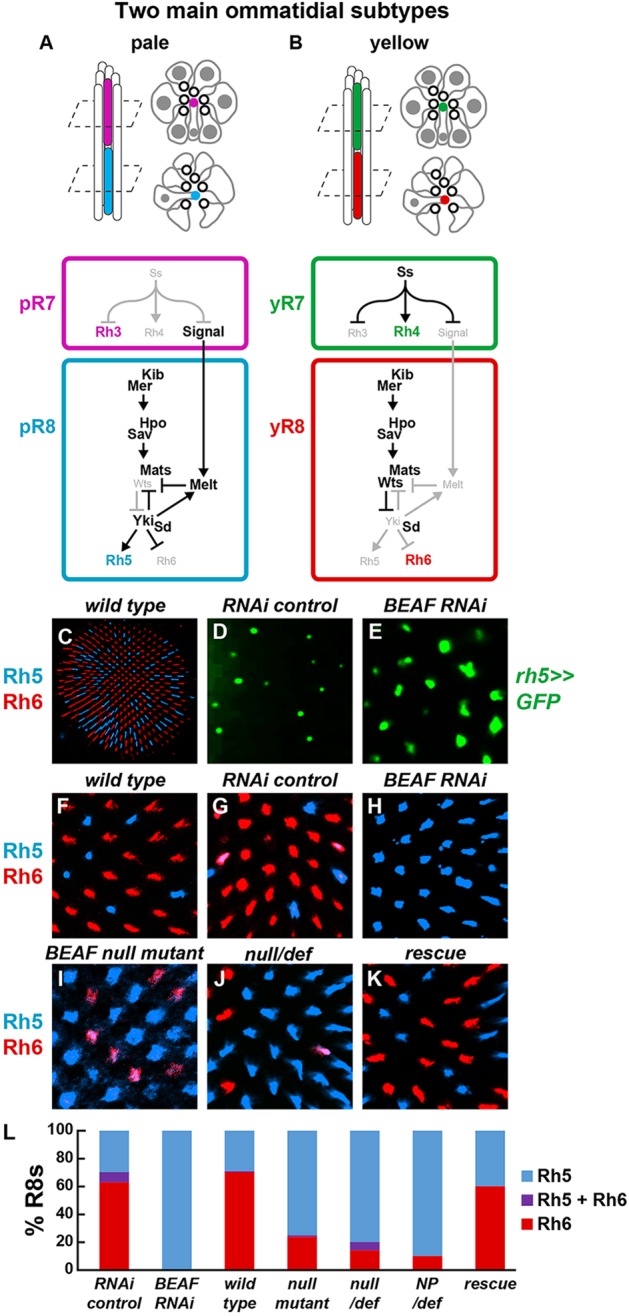
The fly eye is composed of ∼800 ommatidia (unit eyes); each ommatidium contains eight photoreceptors named R1-R8 (Hardie, 1985). The outer photoreceptors, R1-R6, express the broad spectrum-detecting Rhodopsin 1 (Rh1; also known as NinaE) and function in motion detection (Heisenberg and Buchner, 1977; Yamaguchi et al., 2008; Wardill et al., 2012). The inner photoreceptors, R7 and R8, are specialized for color vision, with some contribution from R1-R6 (Schnaitmann et al., 2013). Though morphologically uniform, the fly eye is composed of two main types of ommatidia defined by expression of color-sensing Rhodopsins (Rhs) in the inner photoreceptors (Rister et al., 2013). In the ‘pale’ (p) subtype, pR7s express Rhodopsin 3 (Rh3) and pR8s express Rhodopsin 5 (Rh5) (Fig. 1A). In the ‘yellow’ (y) subtype, yR7s express Rhodopsin 4 (Rh4) and yR8s express Rhodopsin 6 (Rh6) (Fig. 1B). The ommatidial subtypes are stochastically distributed throughout the eye in a p:y ratio of ∼35:65 (Fig. 1C,F).
Fig. 1.
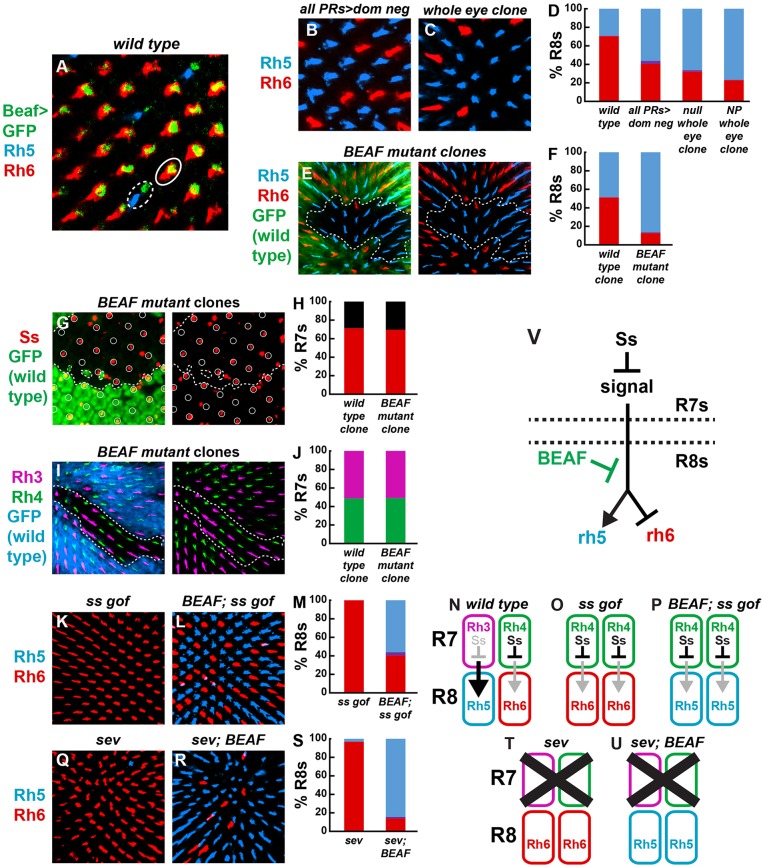
BEAF is required for yR8 subtype specification. (A,B) Schematic showing eight photoreceptors and a cross-section of their rhabdomeres, the membranous structures containing Rhodopsin (Rh) proteins, within an ommatidium. Gray indicates cell bodies and nuclei. White circles with black outlines indicate outer photoreceptor rhabdomeres. Colored rhabdomeres indicate R7 (top) and R8 (bottom). Below is the regulatory network controlling Rh expression in R7 (top) and R8 (bottom). (A) Pale ‘p’ ommatidial subtype. (B) Yellow ‘y’ ommatidial subtype. (C) Retina showing Rh5 and Rh6 expression in stochastic and mutually exclusive R8 subsets. R8 subtypes are visualized by Rh5 (pR8, blue) and Rh6 (yR8, red) antibodies in all panels unless otherwise noted. (D) rh5≫GFP was expressed in a subset of R8s in RNAi controls. Visualized by water immersion (see Materials and Methods). (E) rh5≫GFP was expressed in most R8s when BEAF was knocked down by RNAi. (F,G) Rh5 and Rh6 were expressed in subsets of R8s in wild-type (F) or RNAi Gal4 control (G) retinas. (H-J) Most R8s contained Rh5, and few contained Rh6, in retinas expressing BEAF RNAi (H) or homozygous mutant for BEAFAB-KO (I) or BEAFAB-K0 over a deficiency covering the BEAF locus (J). (K) A BEAF genomic fragment restored normal Rh5 and Rh6 expression in BEAF homozygous null mutants. (L) Quantification of phenotypes.
The specification of ommatidial subtypes is determined in R7s by the stochastic ON/OFF expression of the PAS-bHLH transcription factor Spineless (Ss) (Johnston and Desplan, 2014; Wernet et al., 2006). The ON/OFF state of Ss determines R8 subtype fate through an inductive signal (Chou et al., 1996; Papatsenko et al., 1997) that results in mutually exclusive R8 expression of the Hippo pathway kinase Warts (Wts) and the growth regulator Melted (Melt). In pR7s lacking Ss, Rh3 is expressed in R7s and a signal from R7s triggers activation of melt and repression of wts, leading to Rh5 expression in pR8s (Fig. 1A) (Mikeladze-Dvali et al., 2005). In yR7s expressing Ss, Rh4 is expressed and the signal is repressed, causing the default state of melt repression and wts activation leading to Rh6 expression (Fig. 1B). The double-negative feedback loop between wts and melt controls the presence or absence of Wts downstream of the constitutively active upstream Hippo pathway (Jukam and Desplan, 2011). Wts negatively regulates Yorkie (Yki), which acts with a network of photoreceptor-specific transcription factors to transduce Hippo pathway output into expression of Rh5 or Rh6 (Jukam et al., 2013).
Here, we identify the insulator protein BEAF-32 as a regulator of Wts and Hippo pathway activity in R8 subtype specification. BEAF-32 is required for the expression of wts and also functions downstream of Wts to regulate Rhodopsins, but does not noticeably affect growth. Finally, we demonstrate that BEAF-32 is differentially required for Hippo pathway positive feedback and Rh expression. The role of BEAF-32 in post-mitotic determination of photoreceptor subtypes suggests that insulators have highly specific functions in development.
RESULTS AND DISCUSSION
BEAF-32 is a regulator of the Hippo pathway controlling Rhodopsin expression
To identify transcription factors that regulate the Hippo pathway and R8 subtype specification, we conducted an in vivo RNAi screen for genes for which knockdown caused a change in the proportion of R8s expressing a rh5-LexA, lexAOP-GFP transcriptional reporter (rh5≫GFP) (Vasiliauskas et al., 2011) (i.e. low Hippo pathway activity) (Fig. 1D). We screened 652 lines targeting transcription factor genes, which resulted in 113 lethal phenotypes, 155 eye morphology phenotypes, and one line with a dramatic increase in Rh5.
In the screen, we identified BEAF-32 as a positive regulator of the Hippo pathway. RNAi knockdown of BEAF-32 caused a dramatic increase in the proportion of R8s that express rh5≫GFP (Fig. 1E). BEAF-32 RNAi also caused an increase in R8s that express Rh5 protein and a decrease in R8s expressing Rh6 protein (Fig. 1G,H,L).
BEAF-32 (Boundary element-associated factor of 32 kD; hereafter referred to as BEAF) is one of several known Drosophila insulator proteins, including CTCF, GAGA factor (also known as Trl), Su(Hw), Zw5 (Dwg), CP190 and Mod(mdg4). BEAF binds preferentially near promoters at several thousand sites in the genome (Emberly et al., 2008; Jiang et al., 2009; Negre et al., 2011; Yang et al., 2012) and generally promotes gene expression. Two BEAF isoforms, BEAF-32A and BEAF-32B, are identical except for the 80 amino acid DNA-binding domain; however, BEAF-32B appears to be the dominant isoform (Jiang et al., 2009; Roy et al., 2007). Though BEAF binds throughout the genome, homozygous BEAF mutants, null for both isoforms (BEAFAB-KO), are viable, suggesting that BEAF is required for specific developmental processes such as R8 subtype specification.
Similar to BEAF RNAi, homozygous null BEAF mutants (BEAF-32AB-KO) (Roy et al., 2007) (Fig. 1I,L) and flies with the BEAF null mutant allele over a 105 kb deficiency completely lacking the BEAF locus (Fig. 1J,L) displayed an increase in Rh5 and decrease in Rh6 expression. An independent BEAF mutant allele caused by a P-element insertion (BEAF-32NP6377) placed over the deficiency showed similar changes in the Rh5:Rh6 ratio (NP/def; Fig. 1L). All three BEAF mutant conditions and BEAF RNAi displayed significant increases in Rh5 and decreases in Rh6, and any phenotypic variability among these is likely due to differences in genetic background. A genomic fragment containing the BEAF gene locus (Roy et al., 2007) rescued the mutant phenotype, restoring the normal Rh5:Rh6 ratio (Fig. 1K,L), indicating that the Rh phenotype is specifically due to loss of BEAF. RNAi-mediated knockdown of other insulator genes [CTCF, Cp190, mod(mdg4), su(Hw) and GAGA factor] did not significantly increase Rh5 in the retina (Fig. S1). Therefore, the regulation of Rhodopsins in R8s by insulators is likely to be restricted to BEAF and not a general property of insulator function.
BEAF acts in R8s downstream of R7 signaling to control Rhodopsins
We next determined the cellular focus of BEAF activity. Consistent with previous reports that BEAF is expressed in all cells of the fly (Roy et al., 2007), a 900 bp BEAF promoter drove a BEAF-GFP transgene in all photoreceptors, including all R8s (Fig. 2A). Photoreceptor-specific expression of a dominant-negative BEAF protein lacking the DNA-binding domain and containing only the protein-binding BED domain (all PRs>dom neg) (Gilbert et al., 2006) induced an increase in Rh5 and decrease in Rh6 (Fig. 2B,D). Whole-retina clones of BEAF null or P-element insertion mutants displayed changes in Rh expression (Fig. 2C,D) similar to those in viable whole-animal BEAF mutants. BEAF null mutant clones displayed upregulation of Rh5 and loss of Rh6 compared with wild-type clones (Fig. 2E,F). Thus, BEAF is required for proper expression of Rh5 and Rh6 in R8 photoreceptor neurons of the eye.
Fig. 2.
BEAF acts in R8s downstream of R7 signaling to control Rhodopsins. (A) BEAF-GFP under control of the BEAF promoter was expressed in both R8 subtypes. Rh5-expressing pR8 (dashed oval); Rh6-expressing yR8 (solid oval). (B) Rh5-expressing R8s increased and Rh6-expressing R8s decreased when a BEAF dominant-negative construct was expressed specifically in photoreceptors. (C) A similar phenotype was observed in whole eye BEAF null mutant clones. (D) Quantification of the data shown in B,C. (E,F) BEAF null mutant clones (GFP−) contained more R8s expressing Rh5 compared with wild-type or heterozygous tissue (GFP+). Dashed lines represent clone boundary in all panels unless otherwise noted. (G,H) Ss was expressed stochastically with similar frequency in BEAF null (GFP−) and control (GFP+) tissue in pupal retinas. R7 cells are circled. Red indicates percentage of R7s expressing Ss; black indicates percentage of R7s lacking Ss. (I,J) The Rh3 and Rh4 expression ratio was normal in BEAF null mutant clones (GFP−). (K) Ectopic Ss expression in all R7s induced Rh6 and inhibited Rh5 expression in nearly all R8s. (L) Ectopic Ss expression in the absence of BEAF resulted in increased Rh5- and decreased Rh6-expressing R8s. (M) Quantification of the data shown in K,L. (N-P) Schematics depicting wild type, K and L. (Q) Genetic ablation of R7s (and hence the signal to R8) in sev mutants caused expression of Rh6 and loss of Rh5 in nearly all R8s. (R) sev; BEAF null double mutants displayed upregulation of Rh5 and downregulation of Rh6. (S) Quantification of the data shown in Q,R. (T,U) Schematics describing the observations shown in Q,R. (V) Model for how BEAF acts in R8s, downstream of R7 signaling to control Rhodopsins.
The most upstream trigger for R8 subtype fate is the stochastic ON/OFF expression of Ss in R7s. Expression of Ss represses an unknown signal to R8s, resulting in Wts expression, active Hippo signaling, and Rh6 expression (Fig. 1B). In the absence of Ss, the signal induces repression of wts, leading to inactive Hippo signaling and Rh5 expression (Fig. 1A). Ss was expressed at a similar frequency in BEAF null mutant clones as in wild-type clones (Fig. 2G,H), indicating that BEAF is not required for Ss expression. Rh3 and Rh4, targets of Ss regulation in R7s (Thanawala et al., 2013; Wernet et al., 2006), were expressed at similar ratios in BEAF null mutant and wild-type clones (Fig. 2I,J). Thus, BEAF is not required for Ss expression or R7 subtype specification.
We next showed that BEAF acts downstream of Ss and the signal to control R8 subtypes. Ectopic expression of Ss in all R7s from a BAC transgene (Johnston and Desplan, 2014) repressed the signal to R8s, causing nearly all R8s to adopt yR8 fate and express Rh6 (Fig. 2K,M,O). Ectopic Ss expression in R7s in BEAF null mutants displayed increased Rh5 and decreased Rh6 expression (Fig. 2L,M,P), showing that Ss requires BEAF activity to control R8 subtype. Genetic ablation of R7s in sevenless (sev) mutants removed the signal from R7s to R8s, causing nearly all R8s to acquire yR8 fate and express Rh6 (Fig. 2Q,S,T). sev; BEAF null double mutant R8s primarily expressed Rh5 (Fig. 2R,S,U), showing that the default Hippo activity ON state of R8s requires BEAF to activate Rh6 and repress Rh5. Altogether, these data indicate that BEAF acts in R8s downstream of the signal from R7s to control R8 subtype specification (Fig. 2V).
BEAF binds genes encoding the Hippo pathway members and melt
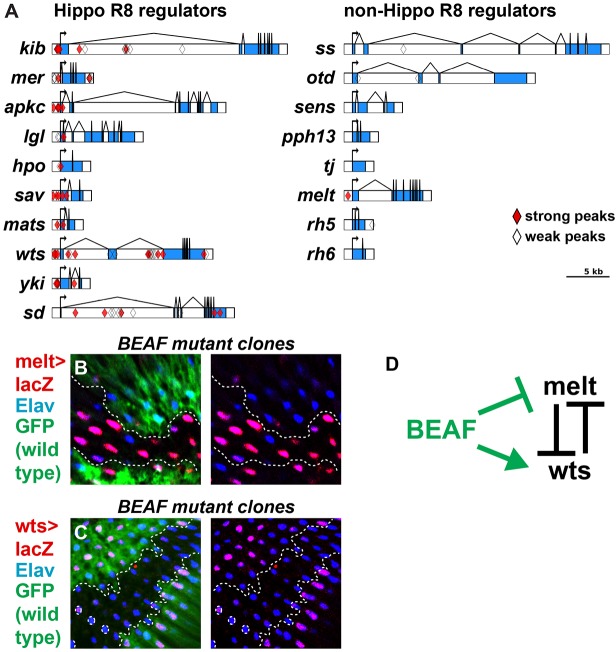
To explore how BEAF regulates the R8 regulatory network, we examined five independent ChIP datasets (four ChIP-chip, one ChIP-seq) available from the modENCODE consortium (Negre et al., 2011). We identified strong peaks (Fig. 3A, red diamonds) that are likely to be direct binding sites for BEAF and weak peaks (Fig. 3A, unfilled diamonds) that may be the result of DNA looping and insulator-insulator interaction (Liang et al., 2014). Strong BEAF binding peaks were present for all the core members of the Hippo pathway (hpo, sav, mats, wts), upstream regulators known to function in R8 {kib (kibra), mer, aPKC, lgl [l(2)gl]}, and output regulators (yki, sd) (Fig. 3A). BEAF also bound melt, part of the melt-wts bistable feedback loop (Fig. 3A). Although BEAF weakly bound ss, the fate trigger in R7s, and otd (also known as oc), a general activator of Rh3 and Rh5 and repressor of Rh6, we did not detect any defects indicative of changes in ss (Fig. 2G-J) or otd (Fig. 1D-J; Fig. 2E) expression in BEAF mutants. Additionally, BEAF does not bind at loci of the other photoreceptor-restricted transcription factors that regulate R8 Rhodopsins (sens, pph13 and tj). The absence of the BEAF consensus DNA binding sequence (CGATA) in the rh5 and rh6 promoter regions, which are sufficient to induce their subtype specific expression, is consistent with a model wherein BEAF does not regulate Rh5 or Rh6 expression through direct binding. Together, these binding profiles suggest that BEAF could regulate R8 subtype fate by controlling aspects of the Hippo pathway or expression of melt and wts.
Fig. 3.
BEAF regulates R8 subtypes by promoting warts expression and preventing melt expression. (A) Summary of BEAF modEncode ChIP binding data at loci of R8 Hippo pathway genes (left) and non-Hippo R8 subtype regulators (right). Diamonds are ChIP-chip peak centers (red diamonds are strong peaks; unfilled diamonds are weak peaks); exons are blue; non-coding sequence is white. (B) melt-lacZ was upregulated in BEAF null mutant clones (GFP−) compared with control tissue (GFP+). (C) wts-lacZ was lost in BEAF null mutant clones (GFP−) compared with control tissue (GFP+). (D) Model for BEAF regulation of wts and melt expression. Dashed lines represent clone boundary.
BEAF is required for repression of melt and activation of wts
Since BEAF bound melt and wts, which are in a transcriptional double-negative feedback loop crucial for R8 subtype specification, we examined the role of BEAF in their regulation. In pR8s, melt represses wts expression to activate Rh5 and repress Rh6 (Fig. 1A) (Mikeladze-Dvali et al., 2005). In yR8s, wts is expressed to repress Rh5 and induce Rh6 (Fig. 1B). BEAF null mutants display loss of Rh6 and gain of Rh5 expression, suggesting that melt is upregulated and wts expression is downregulated. Indeed, melt (melt-lacZ) is de-repressed (Fig. 3B) and warts expression (warts-lacZ) is lost (Fig. 3C) in R8s in BEAF null mutant clones, indicating that BEAF is required for the repression of melt and activation of warts expression (Fig. 3D).
BEAF is required downstream of Wts and Melt for regulation of Rhodopsins
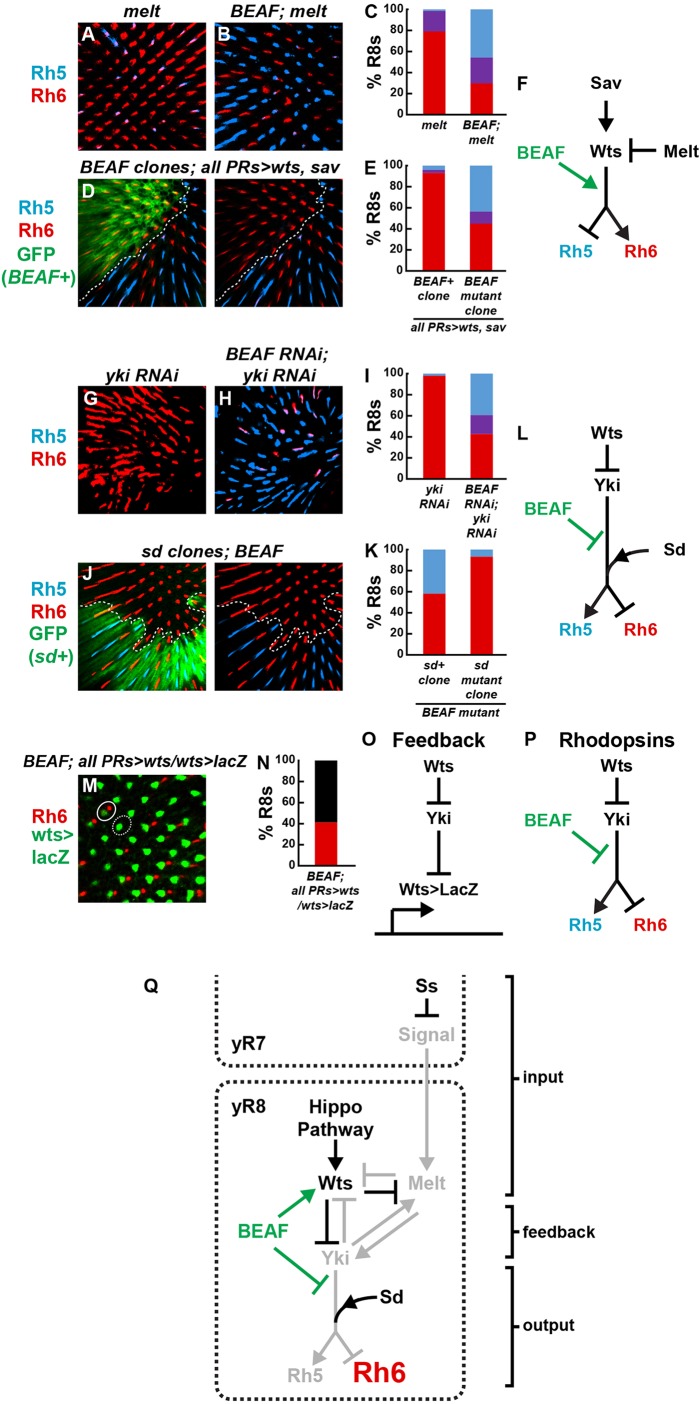
The Rh phenotype observed in BEAF mutants could be caused simply by de-repression of melt and loss of wts expression (Fig. 3B-D). Alternatively, BEAF could play other roles in the pathway and the BEAF mutant phenotype could be due to misregulation of additional downstream genes. In melt mutants, wts is expressed and the Hippo pathway is active, inducing nearly all R8s to express Rh6 and lose Rh5 (Fig. 4A,C) (Mikeladze-Dvali et al., 2005). Removing BEAF in melt mutants caused upregulation of Rh5 and downregulation of Rh6 (Fig. 4B,C) compared with melt single mutants, suggesting that BEAF acts downstream of or in parallel with melt to control Hippo pathway activity and Rh expression state (Fig. 4F).
Fig. 4.
BEAF is essential for Hippo pathway regulation of Rhodopsins, but not positive feedback, in R8 subtype specification. (A) melt mutants contained Rh6 in almost all R8s. (B) Rh5 was upregulated in BEAF; melt null double mutants. (C) Quantification of the data shown in A,B. (D) 0GMR-wts+sav induced Rh6 and inhibited Rh5 expression in otherwise wild-type R8s (GFP+). BEAF null mutant clones (GFP−) in GMR-wts+sav retinas showed increased Rh5 and decreased Rh6 expression. (E) Quantification of the data shown in D. (F) Schematic showing how BEAF acts downstream of wts and melt to regulate Rhodopsins. (G) yki-RNAi retinas displayed Rh6 expression in all R8s. (H) BEAF-RNAi+yki-RNAi resulted in upregulation of Rh5 and downregulation of Rh6. (I) Quantification of the data shown in G,H. (J) sd mutant clones (GFP−) in whole-eye BEAF null mutant background (GFP+) showed Rh6 in almost all R8s. (K) Quantification of the data shown in J. (L) Model for BEAF regulation of Rhodopsin output downstream of Yki and upstream of Sd. (M) wts-lacZ (green) was expressed in all R8s in BEAF null;GMR-wts retinas. Circle shows Rh6 and wts-lacZ co-expressed; dotted circle shows R8 expressing wts-lacZ but not Rh6. Note: R8 nuclei are in a different cell region than Rh-containing rhabdomeres. (N) Quantification of the data shown in M. Red indicates percentage of R8s expressing Rh6; black indicates percentage of R8s lacking Rh6 (presumably expressing Rh5). (O,P) BEAF is dispensable for Hippo pathway feedback, but not Hippo pathway regulation of Rhodopsins. (Q) Working model for BEAF regulation of R8 subtypes. Arrows indicate genetic regulation.
We next tested whether wts and the Hippo pathway require BEAF to regulate Rh expression. Misexpression of Wts and Salvador (Sav, an upstream positive regulator of Wts) in wild-type clones (BEAF+) induced Rh6 in all R8s (Fig. 4D,E). BEAF null mutant clones generated in retinas simultaneously misexpressing Wts and Sav in all photoreceptors resulted in the upregulation of Rh5 and loss of Rh6 (Fig. 4D,E). BEAF null mutant clones in retinas with misexpression of Wts alone displayed similar phenotypes (data not shown). Thus, BEAF is required for Hippo pathway activity to promote the Rh6 R8 fate. Altogether, our epistasis analysis indicates that BEAF acts downstream of or in parallel to wts and melt to regulate Rh5 and Rh6 expression (Fig. 4F), as well as upstream of wts and melt to regulate their expression (Fig. 3D).
Yki but not Sd requires BEAF function to regulate Rhodopsins
The transcription factors Yorkie (Yki) and Scalloped (Sd) are heterodimerization partners that regulate Hippo pathway target genes downstream of Wts (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). In pR8s with Hippo pathway OFF, Yki and Sd are active and induce Rh5 and repress Rh6 (Fig. 1A). In yR8s with active Hippo signaling, Yki is inactive and Rh6 is expressed, whereas Rh5 is repressed (Fig. 1B). yki null mutant cells are eliminated via apoptosis and cannot be examined in adult eyes, but strong expression of RNAi can effectively knockdown yki function in the retina (Jukam et al., 2013). RNAi knockdown of yki caused a loss of Rh5 and gain of Rh6 in all R8s (Fig. 4G,I). Retinas with simultaneous RNAi knockdown of yki and BEAF displayed upregulation of Rh5 and loss of Rh6 (Fig. 4H,I) relative to yki RNAi alone. With the caveats inherent to RNAi-based epistasis analysis, we conclude that BEAF is required downstream of or in parallel with yki to control Rh5 and Rh6 (Fig. 4L).
Sd is present in all R8s and appears to play a permissive role in Rh regulation. sd mutants display Rh6 in all R8s and completely lose Rh5 (Jukam et al., 2013). Whereas wild-type clones (sd+) in homozygous BEAF null mutant tissue upregulated Rh5 and lost Rh6, sd mutant clones in BEAF mutant tissue expressed Rh6 and lost Rh5 in most R8s (Fig. 4J,K), suggesting that Sd acts downstream of BEAF to regulate Rhs. These data are consistent with a model wherein Yki, but not Sd, requires BEAF to regulate Rhodopsins, suggesting that Yki and Sd may have separable roles in R8 subtype specification (Fig. 4L). Given the strong but incomplete phenotypic suppression in the above epistasis, however, we cannot exclude more complicated models.
Positive-feedback regulation of wts expression is independent of BEAF
We next tested BEAF for a role in the positive network-level feedback that is a feature of the R8 Hippo pathway, but not the Hippo growth pathway (Jukam et al., 2013). Wts and Yki cross-regulate in a double-negative feedback loop, in which Wts phosphorylates Yki to inactivate it in yR8s and Yki downregulates transcription of wts in pR8s (Fig. 1A,B). Thus, Wts activates its expression by inhibiting its repressor Yki (Fig. 4O). In wild-type R8s, wts (i.e. wts-lacZ) is expressed in yR8s to generate an active Hippo pathway and Rh6 expression. Ectopic expression of wts (GMR-wts) caused all R8s to express wts (Mikeladze-Dvali et al., 2005). Since BEAF is required for wts expression in otherwise wild-type yR8s, we predicted that BEAF would be required for positive-feedback regulation of wts when wts was ectopically expressed. However, retinas with ectopic wts expression in BEAF null mutants displayed wts-lacZ expression in all R8s (Fig. 4M), suggesting that BEAF is not required for the positive feedback regulation of wts (Fig. 4O,P). One interpretation of this result is that BEAF is required only for initiation of wts expression and not its maintenance. Alternatively, the wild-type wts/yki feedback loop could be near a threshold that is highly sensitive to BEAF regulation, whereas ectopic expression of wts biases the regulation strongly towards wts expression overcoming the absence of BEAF. Consistent with a role for BEAF downstream of wts and yki to regulate opsins, we still observe loss of Rh6 in most BEAF mutant cells that express wts-lacZ (Fig. 4M,N).
Conclusions
We have shown that the insulator protein BEAF is required for a post-mitotic neuronal fate decision in Drosophila photoreceptors. BEAF regulates Hippo pathway activity to control R8 subtype fate and Rhodopsin expression (Fig. 4Q). First, BEAF regulates wts and melt expression by acting upstream. Second, BEAF is required for the Hippo pathway to promote Rh6 and repress Rh5. We also demonstrate that BEAF acts downstream of or in parallel with Yki for regulation of Rhodopsins. Finally, we show that this regulation of Rhodopsins is independent of wts feedback. It appears likely that BEAF regulates cell specification by permissively promoting Hippo pathway activity and wts expression to specify the default yR8 fate.
Despite its role in regulating the Hippo pathway in post-mitotic neuronal fate, BEAF appears to be dispensable for Hippo growth signaling in the eye because homozygous null mutants are viable, exhibit no gross external morphological defects, and show no dramatic differences in eye clone size or pupal interommatidial cell number compared with wild-type tissue (Fig. 2E,G,I; Fig. 3B,C; Fig. 4D; Fig. S2). Additionally, BEAF depletion did not suppress the under-proliferation in yki-RNAi eyes despite suppressing yki-RNAi Rhodopsin phenotypes (Fig. 4G,H). This differential regulation of the Hippo pathway in R8s compared with growth is consistent with other transcriptional regulators of R8 subtypes (ewg, tj) having minimal or no proliferation defects (Hsiao et al., 2013; Jukam et al., 2013). It is possible that BEAF regulates the Hippo pathway indirectly in R8s, by acting on yet-to-be-discovered R8-specific Hippo pathway regulators. Alternatively, BEAF may play a larger role in R8s because of Hippo pathway positive feedback, and have less effect in Hippo growth signaling where homeostatic regulation through negative feedback may compensate for the absence of BEAF. The compensation of ectopic Wts on wts expression, but not opsin control, in BEAF mutants is consistent with such a model.
Non-CTCF insulators appear to be restricted to arthropods, and among several insect species examined (Anopheles gambiae, Apis mellifera or Tribolium castaneum), BEAF was present exclusively in the Drosophila genus (Heger et al., 2013; Schoborg and Labrador, 2010). We speculate that conserved signaling modules of the Hippo pathway in growth control may be co-opted for cell fate specification by regulatory factors such as BEAF that are unique to dipterans.
Insulators were classically defined as proteins that bind particular DNA sequences to either interfere with promoter-enhancer interactions or prevent chromatin-state position effects from affecting transgenes (Gaszner and Felsenfeld, 2006). This definition has expanded to include proteins that mediate chromosomal interactions to regulate 3D chromatin organization and global gene expression (Bushey et al., 2009; Phillips-Cremins and Corces, 2013; Wood et al., 2011). Despite these studies, surprisingly few roles for insulator proteins in specific biological processes in flies have been characterized, including the regulation of oogenesis (Hsu et al., 2015; Roy et al., 2007; Soshnev et al., 2013) and spermatogenesis (Soltani-Bejnood et al., 2007; Thomas et al., 2005). Our result that BEAF regulates Hippo pathway activity for terminal differentiation of R8 neuronal subtypes, but has no observed effect on general growth control or other photoreceptor fate, indicates that broadly expressed insulators can have exquisitely specific functions in development.
MATERIALS AND METHODS
Drosophila genotypes and stocks
See Table S1 for details of Drosophila genotypes and stocks.
Drosophila genetics and transgene descriptions
Flies were raised on corn meal-molasses-agar medium under standard laboratory conditions. y1, w67;+;+ flies were considered ‘wild type’ and used as a control for Rhodopsin gene expression. All experiments were conducted at 25°C unless otherwise noted.
lGMR-Gal4 (long Glass Multiple Reporter) contains a pentamerized 38 bp Glass binding site and is expressed in all photoreceptors and some other retina cells posterior to the morphogenetic furrow (Wernet et al., 2003). ey-Gal4 drives transgene expression in eye primordium and eye imaginal discs. UAS-Dicer2(Dcr2) is co-expressed to increase RNAi processing efficiency (Dietzl et al., 2007). warts-lacZ contains a P-element inserted into the warts locus (Justice et al., 1995). melt-lacZ contains the first intron of melted cloned upstream of nls:lacZ (Mikeladze-Dvali et al., 2005).
BEAF-32AB-KO is semi-viable (Roy et al., 2007). The original BEAF-32AB-KO stock possibly contained a second-site mutation that caused rhabdomere defects, and the chromosome was cleaned during recombination to FRT42D. After recombination, the resulting FRT42D BEAF-32AB-KO flies contained normal rhabdomeres. BEAF-32NP6377 is a null allele caused by a P-element insertion (Gurudatta et al., 2012). We also recombined this allele onto an FRT42D chromosome, which removed the lethality and growth defects previously described on the chromosome (Gurudatta et al., 2012). The deficiency Df(2R)BSC429 (Bloomington stock #24933) contains a 105 kb FLP/FRT-derived deletion that completely removes the BEAF coding sequence, in addition to several other genes. Placing BEAF-32AB-KO trans-heterozygous over a second, ∼250 kb deficiency (Df(2R)BSC858; stock#27928) gave similar results.
Homozygous mutant adult eyes (whole eye mutant clones) were generated using the FLP/FRT system (Xu and Rubin, 1993). FLP recombinase expressed under control of the eyeless (ey) promoter (ey-FLP) (Newsome et al., 2000) induced recombination of FRT chromosomes containing a cell lethal mutation and GMR-hid to remove all non-mutant eye tissue (Stowers et al., 2000). Mutant clones were made using FRT-FLP-mediated recombination between the mutant chromosome and an otherwise wild-type chromosome containing P[w+, ubi-GFP].
RNAi screen
Transcription factors were defined according to the FlyTF database (Adryan and Teichmann, 2006), which includes manual curation from the literature and computationally generated structure homologies. The data set identifies 1052 candidate DNA-binding proteins, including 753 proposed as transcription factors (∼450 site specific). Another 299 genes did not meet their criteria, but had transcription-related Gene Ontology annotations (Adryan and Teichmann, 2006, 2007).
UAS-RNAi fly lines were obtained from a genome-wide library of Drosophila RNAi at the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al., 2007). Each transgenic line contained a 300-500 bp inverted hairpin construct under the control of a 10× multimerized UAS promoter. We tested several Gal4 drivers, including lGMR-Gal4 (strongly expressed in all photoreceptors after the morphogenetic furrow), sens-Gal4 (strongly expressed in R8 and weakly and variably expressed in other photoreceptors), and ey-Gal4+lGMR-Gal4 (ey-Gal4 is strongly expressed in the early eye primordium and disc), with or without co-expression of UAS-Dicer2 (Dcr-2). Dcr-2 is thought to enhance RNAi-processing efficiency in cell types more refractory to UAS-RNAi such as adult neurons (Dietzl et al., 2007).
UAS-Dcr2; ey-Gal4, lGMR-Gal4 was determined to be the optimal Gal4 driver because it induced RNAi phenotypes very similar to phenotypes of a gene's respective loss-of-function mutant for seven out of seven known R7 and R8 subtype regulators: Rh6 was lost with warts, merlin, mats and sav RNAi, whereas Rh5 was lost with melt RNAi. spineless RNAi flies had expansion of Rh3 into all R7 photoreceptors. In addition, dve RNAi resulted in an increase in Rh5 expression, in the outer photoreceptors. An independent paper describing a role for Dve in restricting R8 Rhodopsins from R1-R6 was in preparation during the screen and has since been published (Johnston et al., 2011). otd RNAi completely removed rh5 expression, consistent with a requirement for Otd for rh5 transcription and direct binding to the rh5 promoter. These positive controls demonstrate that expression of UAS-RNAi under control of UAS-Dcr2; ey-Gal4, lGMR-Gal4 is an effective tool to induce loss-of-function developmental phenotypes detectable in adult photoreceptors.
The screen was performed by crossing UAS-RNAi males to virgin female reporter-driver lines. To maximize virgin collection, the female driver stock contained an hs-hid transgene on the Y chromosome (Dietzl et al., 2007). We performed two 30 min heat-shocks spaced 8 h apart at 37°C in late larval and early pupal stages to eliminate males.
R8 subtypes were assessed for defects in F1 progeny by examining for a change in the proportion of R8s expressing a rh5-LexA::VP16, lexAOP::GFP (rh5≫GFP) transcriptional reporter. The LexA/lexAOP binary expression system was used to amplify GFP levels while keeping the reporter Gal4 independent (Lai and Lee, 2006; Vasiliauskas et al., 2011).
Water immersion protocol for visualizing rh5≫GFP
GFP was visualized in living adult flies by neutralizing the cornea using a water immersion technique (Pichaud and Desplan, 2001). Ten to twelve flies of the appropriate F1 genotype were placed on a streak of clear nail polish (Wet n Wild) perpendicular to the straight edge with one retina facing up, in the middle of a 10 cm Petri dish. The dish was placed on the microscope stage and water added to submerge flies. Images of rh5≫GFP were taken with a SPOT camera mounted on a fluorescence microscope with a Nikon Plan Fluor 40× objective lens immersed in water. About 30-40 ommatidia are visible in the same focal plane. If rh5≫GFP appeared in >60% of ommatidia of at least two flies, the genotype was later dissected and Rh5 and Rh6 visualized with antibodies. During the pilot screen it was discovered that the ey-Gal4+lGMR-Gal4 flies exhibited, on occasion, an increase in the Rh6:Rh5 ratio (from 70:30 to 85:15). Therefore we only assayed subtype phenotypes that increased the Rh5 R8 proportion.
BEAF binding analysis
BEAF-32 ChIP data for Drosophila melanogaster were obtained from five available studies from modENCODE (http://www.modencode.org). For the purpose of this analysis, the pre-processed peak calls available in the modENCODE's ‘dmel-interpreted-1’ FTP directory were used. The midpoint of each identified ChIP peak regions was used to annotate the gene diagrams, based on the FlyBase dm3 gene annotation on the UCSC Genome Browser (genome.ucsc.edu). Strong peaks were defined as having >2.5-fold enrichment. Weak peaks were defined as having <2.5-fold enrichment. The scripts used for analysis can be found at https://github.com/pdeford/beaf32-hippo-chip.
Antibodies
Antibodies and dilutions were as follows: mouse anti-Rh3 (1:10; gift from S. Britt, University of Colorado, CO, USA), rabbit anti-Rh4 (1:100; gift from C. Zuker, Columbia University, NY, USA), mouse anti-Rh5 (1:200; Chou et al., 1996), rabbit anti-Rh6 (1:2000; Tahayato et al., 2003), guinea pig anti-Ss 2.21 (1:200; gift from Y.N. Jan, University of California, San Francisco, CA, USA) (Kim et al., 2006), rat anti-ElaV (1:50; Developmental Studies Hybridoma Bank), sheep anti-GFP (1:500; AbD Serotec, 4745-1051), mouse anti-Dlg (1:75; Developmental Studies Hybridoma Bank) and goat anti-β-gal (1:50; Biogenesis, 4600-1409). All secondary antibodies were Alexa Fluor 488-, 555- or 647-conjugated antibodies (1:400) made in donkey (Molecular Probes).
Antibody staining and imaging
Adult or staged pupal retinas were dissected as described (Hsiao et al., 2012). Briefly, retinas were dissected and immediately fixed for 15 min with 4% paraformaldehyde at room temperature. After lamina removal, retinas were rinsed twice in PBX (PBS+0.2% Triton X-100) then washed in PBX for >2 h. Retinas were incubated overnight with primary antibodies diluted in PBX at room temperature, rinsed twice in PBX and then washed in PBX for >4 h. Retinas were incubated 4-6 h or overnight with secondary antibodies diluted in PBX at room temperature, rinsed twice in PBX and then washed in PBX for >2 h. Adult retinas were mounted in SlowFade (Molecular Probes) and pupal retinas in Vectashield (Vector Laboratories) on glass slides with coverslip. Images were acquired using a Leica TCS SP5, Zeiss 710 or Zeiss 780 confocal microscope. Objectives were 10×, 20× or 60×. Images were processed in Photoshop (Adobe) or ImageJ. Brightness or contrast adjustments, if any, were simple linear adjustments made to the entire image, in accordance with journal guidelines. Figures were prepared using Illustrator (Adobe).
Quantification of photoreceptor subtypes
Confocal images were taken and the number of R8 cells that expressed Rh5, Rh6, both, or neither were counted. The percentage of R8s expressing Rh5 (%Rh5) was calculated for each retina, and mean %Rh5 of all retinas within a genotype was used to compare across genotypes. Retinas were scored if there were 75 or more ommatidia present in a single focal plane. Most retinas contained ∼200-300 ommatidia in a single image. For all genotypes, more retinas were observed than quantified to confirm a particular phenotype. Means and standard deviations for all experiments can be found in Table S2.
Acknowledgements
We are grateful to Steve Britt, Steve Cohen, Barry Dickson, Georg Halder, Iswar Hariharan, Craig Hart, Ken Irvine, Jin Jiang, D. J. Pan, Jessica Treisman, Tian Xu, Charles Zuker, the Bloomington Stock Center, the Kyoto Stock Center and the Vienna Drosophila RNAi Center (VDRC) for generously providing published fly stocks and antibodies. We thank Pam Geyer and Judy Kassis for helpful comments on the manuscript. We thank Vince Lau for technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.J. and R.J.J. designed the research and RNAi screen. D.J. performed the RNAi screen. D.J., K.V., C.A., C.Z., J.Y., J.C., and R.J.J. performed genetic experiments and analyzed the data. P.D. analyzed the BEAF modEncode ChIP data. D.J. and R.J.J. wrote the manuscript with input from all authors.
Funding
This work was supported by a Pew Scholar Award from Pew Charitable Trusts [00027373 to R.J.J.]; a Basil O'Connor Scholar Award from the March of Dimes Foundation [5-FY15-21 to R.J.J.]; and a grant from the National Institutes of Health [R01EY025598 to R.J.J.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.134700.supplemental
References
- Adryan B. and Teichmann S. A. (2006). FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics 22, 1532-1533. 10.1093/bioinformatics/btl143 [DOI] [PubMed] [Google Scholar]
- Adryan B. and Teichmann S. A. (2007). Computational identification of site-specific transcription factors in Drosophila. Fly 1, 142-145. 10.4161/fly.4571 [DOI] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E. and Corces V. G. (2009). Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23, 1338-1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W.-H., Hall K. J., Wilson D. B., Wideman C. L., Townson S. M., Chadwell L. V. and Britt S. G. (1996). Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17, 1101-1115. 10.1016/S0896-6273(00)80243-3 [DOI] [PubMed] [Google Scholar]
- Cockburn K., Biechele S., Garner J. and Rossant J. (2013). The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195-1201. 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Emberly E., Blattes R., Schuettengruber B., Hennion M., Jiang N., Hart C. M., Käs E. and Cuvier O. (2008). BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 6, e327 10.1371/journal.pbio.0060327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., Parrish J. Z., Jan L. Y. and Jan Y.-N. (2006). The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443, 210-213. 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Gaszner M. and Felsenfeld G. (2006). Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7, 703-713. 10.1038/nrg1925 [DOI] [PubMed] [Google Scholar]
- Gilbert M. K., Tan Y. Y. and Hart C. M. (2006). The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173, 1365-1375. 10.1534/genetics.106.056002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J. and Zider A. (2008). SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435-441. 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- Gurudatta B. V., Ramos E. and Corces V. G. (2012). The BEAF insulator regulates genes involved in cell polarity and neoplastic growth. Dev. Biol. 369, 124-132. 10.1016/j.ydbio.2012.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie R. C. (1985). Functional organization of the fly retina. In Sensory Physiology (ed. Ottoson D.), pp. 1-79. New York, NY: Springer-Verlag. [Google Scholar]
- Heger P., George R. and Wiehe T. (2013). Successive gain of insulator proteins in arthropod evolution. Evolution 67, 2945-2956. 10.1111/evo.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. and Buchner E. (1977). The role of retinula cell types in visual behavior of Drosophila melanogaster . J. Comp. Physiol. 117, 127-162. 10.1007/BF00612784 [DOI] [Google Scholar]
- Hsiao H. Y., Johnston R. J., Jukam D., Vasiliauskas D., Desplan C. and Rister J. (2012). Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J. Vis. Exp. 69, e4347 10.3791/4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao H.-Y., Jukam D., Johnston R. and Desplan C. (2013). The neuronal transcription factor erect wing regulates specification and maintenance of Drosophila R8 photoreceptor subtypes. Dev. Biol. 381, 482-490. 10.1016/j.ydbio.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-J., Plata M. P., Ernest B., Asgarifar S. and Labrador M. (2015). The insulator protein Suppressor of Hairy wing is required for proper ring canal development during oogenesis in Drosophila. Dev. Biol. 403, 57-68. 10.1016/j.ydbio.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K. D. and Harvey K. F. (2015). Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb. Perspect Biol. 7, a019224 10.1101/cshperspect.a019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Emberly E., Cuvier O. and Hart C. M. (2009). Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 29, 3556-3568. 10.1128/MCB.01748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J. Jr and Desplan C. (2014). Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science 343, 661-665. 10.1126/science.1243039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J. Jr, Otake Y., Sood P., Vogt N., Behnia R., Vasiliauskas D., McDonald E., Xie B., Koenig S., Wolf R. et al. (2011). Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell 145, 956-968. 10.1016/j.cell.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D. and Desplan C. (2011). Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev. Cell 21, 874-887. 10.1016/j.devcel.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D., Xie B., Rister J., Terrell D., Charlton-Perkins M., Pistillo D., Gebelein B., Desplan C. and Cook T. (2013). Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342, 1238016 10.1126/science.1238016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R. W., Zilian O., Woods D. F., Noll M. and Bryant P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534-546. 10.1101/gad.9.5.534 [DOI] [PubMed] [Google Scholar]
- Kim M. D., Jan L. Y. and Jan Y. N. (2006). The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 20, 2806-2819. 10.1101/gad.1459706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.-L. and Lee T. (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9, 703-709. 10.1038/nn1681 [DOI] [PubMed] [Google Scholar]
- Liang J., Lacroix L., Gamot A., Cuddapah S., Queille S., Lhoumaud P., Lepetit P., Martin P. G. P., Vogelmann J., Court F. et al. (2014). Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol. Cell 53, 672-681. 10.1016/j.molcel.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., Wernet M. F., Pistillo D., Mazzoni E. O., Teleman A. A., Chen Y.-W., Cohen S. and Desplan C. (2005). The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775-787. 10.1016/j.cell.2005.07.026 [DOI] [PubMed] [Google Scholar]
- Milton C. C., Grusche F. A., Degoutin J. L., Yu E., Dai Q., Lai E. C. and Harvey K. F. (2014). The Hippo pathway regulates hematopoiesis in Drosophila melanogaster. Curr. Biol. 24, 2673-2680. 10.1016/j.cub.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., Wagner U., Kheradpour P., Eaton M. L., Loriaux P., Sealfon R. et al. (2011). A cis-regulatory map of the Drosophila genome. Nature 471, 527-531. 10.1038/nature09990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome T. P., Asling B. and Dickson B. J. (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851-860. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K.-I., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N. et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Papatsenko D., Sheng G. and Desplan C. (1997). A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development 124, 1665-1673. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins J. E. and Corces V. G. (2013). Chromatin insulators: linking genome organization to cellular function. Mol. Cell 50, 461-474. 10.1016/j.molcel.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F. and Desplan C. (2001). A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development 128, 815-826. [DOI] [PubMed] [Google Scholar]
- Reddy B. V. V. G., Rauskolb C. and Irvine K. D. (2010). Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development 137, 2397-2408. 10.1242/dev.050013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J., Desplan C. and Vasiliauskas D. (2013). Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development 140, 493-503. 10.1242/dev.079095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Gilbert M. K. and Hart C. M. (2007). Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics 176, 801-813. 10.1534/genetics.106.068056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitmann C., Garbers C., Wachtler T. and Tanimoto H. (2013). Color discrimination with broadband photoreceptors. Curr. Biol. 23, 2375-2382. 10.1016/j.cub.2013.10.037 [DOI] [PubMed] [Google Scholar]
- Schoborg T. A. and Labrador M. (2010). The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J. Mol. Evol. 70, 74-84. 10.1007/s00239-009-9310-x [DOI] [PubMed] [Google Scholar]
- Soltani-Bejnood M., Thomas S. E., Villeneuve L., Schwartz K., Hong C.-S. and McKee B. D. (2007). Role of the mod(mdg4) common region in homolog segregation in Drosophila male meiosis. Genetics 176, 161-180. 10.1534/genetics.106.063289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., Baxley R. M., Manak J. R., Tan K. and Geyer P. K. (2013). The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 140, 3613-3623. 10.1242/dev.094953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers R. S., Garza D., Rascle A. and Hogness D. S. (2000). The L63 gene is necessary for the ecdysone-induced 63E late puff and encodes CDK proteins required for Drosophila development. Dev. Biol. 221, 23-40. 10.1006/dbio.2000.9685 [DOI] [PubMed] [Google Scholar]
- Tahayato A., Sonneville R., Pichaud F., Wernet M. F., Papatsenko D., Beaufils P., Cook T. and Desplan C. (2003). Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev. Cell 5, 391-402. 10.1016/S1534-5807(03)00239-9 [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D. and Hariharan I. K. (2002). salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467-478. 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Thanawala S. U., Rister J., Goldberg G. W., Zuskov A., Olesnicky E. C., Flowers J. M., Jukam D., Purugganan M. D., Gavis E. R., Desplan C. et al. (2013). Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev. Cell 25, 93-105. 10.1016/j.devcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. E., Soltani-Bejnood M., Roth P., Dorn R., Logsdon J. M. and McKee B. D. (2005). Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123, 555-568. 10.1016/j.cell.2005.08.043 [DOI] [PubMed] [Google Scholar]
- Vasiliauskas D., Mazzoni E. O., Sprecher S. G., Brodetskiy K., Johnston R. J. Jr, Lidder P., Vogt N., Celik A. and Desplan C. (2011). Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature 479, 108-112. 10.1038/nature10451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardill T. J., List O., Li X., Dongre S., McCulloch M., Ting C.-Y., O'Kane C. J., Tang S., Lee C.-H., Hardie R. C. et al. (2012). Multiple spectral inputs improve motion discrimination in the Drosophila visual system. Science 336, 925-931. 10.1126/science.1215317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet M. F., Labhart T., Baumann F., Mazzoni E. O., Pichaud F. and Desplan C. (2003). Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115, 267-279. 10.1016/S0092-8674(03)00848-1 [DOI] [PubMed] [Google Scholar]
- Wernet M. F., Mazzoni E. O., Çelik A., Duncan D. M., Duncan I. and Desplan C. (2006). Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440, 174-180. 10.1038/nature04615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. M., Van Bortle K., Ramos E., Takenaka N., Rohrbaugh M., Jones B. C., Jones K. C. and Corces V. G. (2011). Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol. Cell 44, 29-38. 10.1016/j.molcel.2011.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J. and Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Xu T. and Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A. and Yu W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053-1063. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Wolf R., Desplan C. and Heisenberg M. (2008). Motion vision is independent of color in Drosophila. Proc. Natl Acad. Sci. USA 105, 4910-4915. 10.1073/pnas.0711484105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ramos E. and Corces V. G. (2012). The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 22, 2199-2207. 10.1101/gr.142125.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B. and Guan K.-L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811-828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B. and Jiang J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Tumaneng K. and Guan K.-L. (2011). The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 13, 877-883. 10.1038/ncb2303 [DOI] [PMC free article] [PubMed] [Google Scholar]