Abstract
Extraction of rare target cells from biosamples is enabling for life science research. Traditional rare cell separation techniques, such as magnetic activated cell sorting (MACS), are robust but perform coarse, qualitative separations based on surface antigen expression. We report a quantitative magnetic separation technology using high-force magnetic ratcheting over arrays of magnetically soft micro-pillars with gradient spacing, and use the system to separate and concentrate magnetic beads based on iron oxide content (IOC) and cells based on surface expression. The system consists of a microchip of permalloy micro-pillar arrays with increasing lateral pitch and a mechatronic device to generate a cycling magnetic-field. Particles with higher IOC separate and equilibrate along the miro-pillar array at larger pitches. We develop a semi-analytical model that predicts behavior for particles and cells. Using the system, LNCaP cells were separated based on the bound quantity of 1μm anti-EpCAM particles as a metric for expression. The ratcheting cytometry system was able to resolve a ±13 bound particle differential, successfully distinguishing LNCaP from PC3 populations based on EpCAM expression, correlating with flow cytometry analysis. As a proof of concept, EpCAM-labeled cells from patient blood were isolated with 74% purity, demonstrating potential towards a quantitative magnetic separation instrument.
Keywords: cell manipulation, equilibrium separation, circulating tumor cells
1. Introduction
Separating and concentrating cells from bulk solutions for analysis is a nontrivial task in life science research, diagnostics, and industrial processing. As such, various approaches based on physical[1] or biochemical[2] properties of cells have been developed to improve separation efficiency. Biochemical moieties on cell surfaces are most commonly used to distinguish cell populations, in which a specific receptor or protein is targeted with a recognition element (e.g. antibody, aptamer, ligand) to yield a fluorescent or magnetic label enabling downstream sorting.
The most widely used cell sorting techniques of this nature are fluorescence-activated cell sorting (FACS) and magnetic activated cell sorting (MACS). In FACS, multiple cell populations can be separated from heterogeneous mixtures based on the quantity of fluorophore associated with the cell. Though effective, the FACS process is performed serially and each cell is analyzed individually, increasing processing times for large sample volumes. Comparatively, magnetic based approaches are advantageous due to their simplicity and robustness, not requiring sophisticated fluid handling. These approaches are also able to operate on minimal cell quantities and/or process larger volumes more rapidly[3]. However, magnetic separation approaches remain less quantitative than FACS, which can gate on the relative quantity of a biomarker. This lack of quantification from traditional MACS is due to the fact that these approaches cannot discriminate effectively based on the number of bound magnetic particles. Additionally, some magnetic particles available today are not tightly controlled in size or magnetic content, further exacerbating efforts for quantification of biomarker levels as correlated to number of bound particles, emphasizing the need for techniques to purify particles based on magnetic content.
Several microfluidics approaches have been developed to quantitatively separate cells based on bound or internalized magnetic content[4],[5],[6],[7],[8][9][10]. In general, these techniques involve generating a magnetophoretic force orthogonal to a fluid flow direction, inducing cell deflection across streamlines and separation into different outlets depending on magnetic content. However, these “kinetic” based separations require precise tuning of flow rate, fluidic resistance, and magnetic field positioning. Additionally, many of these systems have low throughput as they rely on weaker bulk magnetic field gradients. Finally, the output from flow-through based systems often yields diluted solutions which may require concentration steps and is particularly challenging for isolating and locating rare cell types.
Magnetic ratcheting has the potential to achieve quantitative magnetic separations to both purify magnetic particle populations and separate cells based on bound number of particles. In magnetic ratcheting, arrays of magnetic micro-pillars combined with a directionally cycled magnetic field create dynamic potential energy wells that trap and manipulate superparamagnetic particles[11],[12],[13],[14] in a magnetic-content and particle-size dependent manner. However, previous ratcheting platforms have utilized thin film magnetic structures (height ≤ 200nm), which have minimal force capacities on the order of 10pN, due to the low aspect ratio of the structures used (SI Text). To compensate, larger particles are used (~3–10μm) to maximize the force envelope. However, larger particles have reduced magnetic labeling efficiency for cell separations due to slow diffusive motions. This slow diffusive motion results in inefficient binding of large particles to cell surface targets and the large increment in magnetic content per bound particle makes it difficult to relate bead binding to target expression levels. The use of smaller magnetic particles is necessary to increase labeling efficiency as well as provide a sensitive metric to relate bound particle numbers with cell surface expression, but is not practically compatible with current ratcheting technology. Additionally, previous ratcheting platforms[11] rely on velocity differences between particles to achieve magnetic based separation. But again this is a “kinetic” separation requiring initial sample concentration prior to process initiation, and time dependent collection functions. These challenges have limited use as a quantitative sorting tool. Ideally, an equilibrium separation could achieve reduced dependence on initial and final conditions of a sample yielding a more robust and quantitative separation.
To address these fundamental challenges we developed magnetic ratcheting arrays composed of 1:1 aspect ratio, electroplated magnetic structures, increasing the force capacity by 10 fold. Furthermore, the array was designed with increasing horizontal pitch enabling rapid magnetophoretic equilibrium separation of particles or cells, yielding concentrated “bands” which are quantized and proportional to magnetic content.
Our gradient magnetic ratcheting system (Movie S1) was able to rapidly separate and concentrate magnetic particles based on iron oxide (IO) content with high precision; yielding complete equilibrium separation of a particle batch in less than 60 seconds. The platform was then used to evaluate EpCAM expression on PC3 and LNCaP cells using the quantity of bound anti-EpCAM 1μm IO particles as a marker of expression. The system was able to successfully distinguish PC3 and LNCaP cell populations and was highly correlated with EpCAM expression as determined by flow cytometry of the two cell lines. We also demonstrated applicability as a rare cell cytometer achieving a capture efficiency of 26±4% and a purity of 67±35% of LNCaP cells spiked into 1mL of whole blood, which was comparable with other MACS-based systems[15],[16] but with a substantially higher separation purity. As an initial proof of concept, ratcheting cytometry was used to purify circulating tumor cells from blood samples obtained from three patients with metastatic castration resistant prostate cancer. In 2 of 3 patients, approximately 8–10 large nucleated CD45-negative cells were extracted and concentrated due to their large anti-EpCAM bound particle content with an average purity of 71%.
1.1 Theory
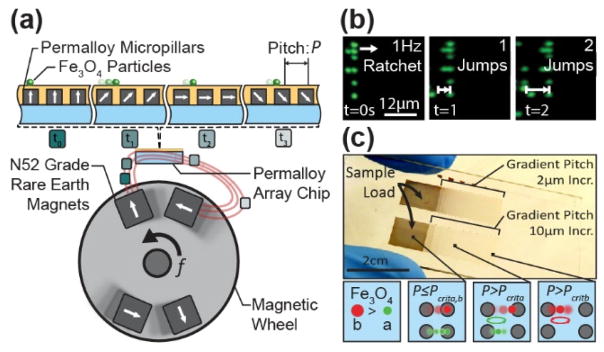
Transport of magnetic particles by ratcheting is achieved using arrays of magnetically soft micro-pillars combined with a directionally cycled magnetic field to dynamically modify the potential energy landscape. This creates translating potential wells that trap and manipulate magnetic particles (Figure S1). Upon application of a magnetic field from a mechatronically controlled magnetic system (Figure S2, Figure S3, & Movie S1), the micro-pillars magnetize in alignment to the bulk field, introducing potential wells in which superparamagnetic magnetic particles become trapped. As the magnetic wheel is cycled, particles follow the potential wells and ratchet through the pillars based on their size and magnetic properties (Figure 1a–b).
Figure 1.
Magnetic ratcheting utilizes arrays of electroplated permalloy micro-pillars, of pitch P, to create dynamic potential energy wells which can be used to trap and manipulate superparamagnetic particles. (a)The magnetic ratcheting system is comprised of a mechatronic instrument driving a rotating magnetic wheel as well as a microchip composed of permalloy micro-pillar arrays, with each pillar having a 1:1 aspect ratio. When proximal to the magnetic wheel, the micro-pillars magnetize in alignment to the bulk field; modifying the magnetic potential energy landscape and introducing potential wells in which superparamagnetic particles migrate. (b)As the wheel is cycled at frequency f, particles will follow the potential wells and ratchet through the pillars based on size and iron oxide content. Using a chip consisting of micro-pillar arrays with gradient horizontal pitch (c), particles will traverse the array until reaching their critical pitch, Pcrit, where they will collect and oscillate. Particles with increasing magnetic content will have correspondingly higher critical pitches and can therefore be separated.
Similar ratcheting transport , which has been previously studied in thin film uniform pitch ratcheting platforms[11],[12],[13],[14] , is characterized by a balance of the time averaged magnetic force, , with the time averaged drag force (Equations 1 & 2). The time averaged magnetic force is dependent on several parameters including magnetic particle volume VP, the particle susceptibility χP, the permeability of free space μ0, and the magnetic flux density B.
| Equation 1 |
Assuming a Stokes drag condition, the time averaged drag force can be described in terms of fluid viscosity μf, particle radius rp, and the time averaged particle speed . The time averaged particle speed can be further represented in terms of the total distance traversed over one ratcheting cycle, , and the ratcheting frequency f (Equation 2).
| Equation 2 |
Assuming the magnetic and drag forces equate, a particle ratcheted at a given frequency will be able to traverse a ratcheting array of pitch P given that and the average particle velocity becomes Pf (Movie S2). However, when the pitch reaches a critical value , the particle does not have sufficient migration time to reach the next pillar (and potential well) and will oscillate and become trapped (Equation 3).
| Equation 3 |
This bimodal behavior, which has also been observed in thin film ratcheting systems[11], is dependent on driving frequency, horizontal pitch between magnetic micro-pillars, particle size, and particle magnetic content. By designing a micro-pillar array with a gradient in horizontal pitch ( Figure 1c & Figure S4), particles with varying magnetic contents will equilibrate in different spatial locations and be separated from each other; as particles with higher magnetic content will have higher critical pitches. Furthermore, particles with similar magnetic content will concentrate into quantized bands at the critical pitch under a given driving frequency.
The net magnetic force on a magnetically labeled cell can be described as a summation of forces exerted by each bound particle NP. The magnetic gradient, as well as magnetic force, decays strongly with inter-pillar distance and is concentrated locally near the magnetized pillars. We quantified this empirically by recording particle speeds across the chip at various frequencies at each pitch and deriving the magnetic force (Figure S5). The magnetic force was best fit by a form αP−2, where α=550 pNμm2 with R2=0.85. Therefore the total magnetic force on a labeled cell becomes .
Equating the magnetic and drag forces on the cell and setting P = Pcritcell, a relationship between the number of bound particles and the critical pitch can be derived (Equation 4) where the cells critical pitch, Pcritcell , relates to Np1/3.
| Equation 4 |
Using Equation 4 as a predictive model, gradient ratcheting arrays can be intelligently designed to achieve quantitative and highly resolved equilibrium magnetic separation of particles and cells.
2. Results and Discussion
To realize a magnetic ratcheting based separation system we designed and fabricated micro-pillar arrays with gradients in horizontal pitch as well as a mechatronic system to generate the ratcheting field. As discussed theoretically, the pillar pitch and driving frequency can be modulated to control the trapping behavior of particles with varying magnetic content. The theoretical model informed the design of two gradient-pitch chips with linearly increasing pitch of either 10μm or 2μm increments for large dynamic range or high resolution separations.
2.1 Particle Equilibrium Separation Discretized by Iron Oxide Content
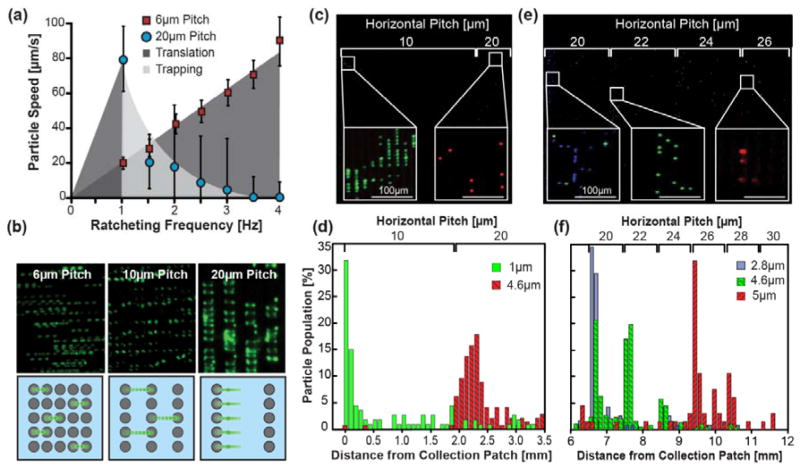
To characterize the system separation behavior, superparamagnetic particles with varying diameters and IO contents were separated under several driving frequencies. Particles ratcheted at a given frequency will traverse an array as long as the pitch, P, remains at or below that particle’s critical pitch value, Pcrit. In this regime the particle displays a linear relationship between particle speed and frequency. The particle will traverse the array until P > Pcrit, where it is unable to traverse to the next micro-pillar and will equilibrate and concentrate at the edge of this pitch region (Figure 2a–b).
Figure 2.
Equilibrium separation by ratcheting magnetophoresis on gradient-pitch permalloy arrays. (a) Ratcheting of 2.8μm diameter particles (IO content 10.1pg) within the chip demonstrate bimodal transport behavior dependent on the array pitch. (b) When P ≤ Pcrit the particle speed correlates linearly with frequency, but when P > Pcrit particles will trap and concentrate on the pillar edge. (c &d) Fluorescently labeled particle mixtures of green 1μm diameter and red 4.6μm diameter (IO contents of 0.98 and 11.1pg respectively) were separated on a 10μm increment chip ratcheted at 30Hz. 83% of the 1μm particles equilibrated at the 10μm pitch and 92% of the 4.6μm particles equilibrated at the 20μm increment patch, demonstrating a >90% separation purity. (e & f) Particle mixtures of blue 2.8μm, green 4.6μm and red 5μm (IO contents 10.1, 11.1 and 67.4 pg respectively) were separated on a 2μm pitch increment chip at 30Hz. 77% of the 2.8 and 41% of the 4.6μm particles equilibrated in the 20μm pitch region. An additional 41% of the 4.6μm particles occupied the 22μm pitch region while 85% of the 5μm particles collected at 26 and 28μm pitches.
Successful separation of mixed 1μm and 4.6μm particles was achieved using a 10μm incremented chip at 30Hz achieving ≥90% purity and 9 pg of IO resolution (Figure 2c–d). Though small, some inter-population overlap was observed which we suspect is due to 1μm particle aggregates or a lack of quality control in particle size. In addition to separation, each particle population was concentrated by 500 fold from bulk solution. As expected, the 2μm incremented chip leads to finer resolved separations with a mixture of three particle types demonstrating a resolution of 5.6 pg of IO (Figure 2e–f). The 2.8μm particles could be easily separated from 5μm particles with a >90% purity. Interestingly, the 4.6μm particles subdivided into three subpopulations at the 20, 22 and 24μm pitches which was unexpected as our model predicted a critical pitch of 24μm for this particle. We determined that this behavior was most likely due to variations in IO content as derived from the particle data sheet[17]. This suggests that the system can achieve resolutions bordering on 1pg of IO. However, these findings also demonstrate that the system is highly sensitive to variability in particle manufacturing which could be a potential limitation if the iron oxide content varies significantly between particles. To address this challenge, another potential application for the system is as a quality control tool for enriching particles with similar iron oxide content or assessing the distribution of IO content within a sample. Using this mode, we found that 1μm particles were highly homogenous in iron oxide content where > 90% of the injected particle population equilibrated at a single critical pitch. After characterizing separation behavior, we employed the magnetic ratcheting separation system to measure and concentrate cell populations based on surface expression level of Epithelial Cell Adhesion Molecule (EpCAM) using the 1μm particles.
2.2 Quantitative Magnetic Cell Separation Based on EpCAM Surface Expression
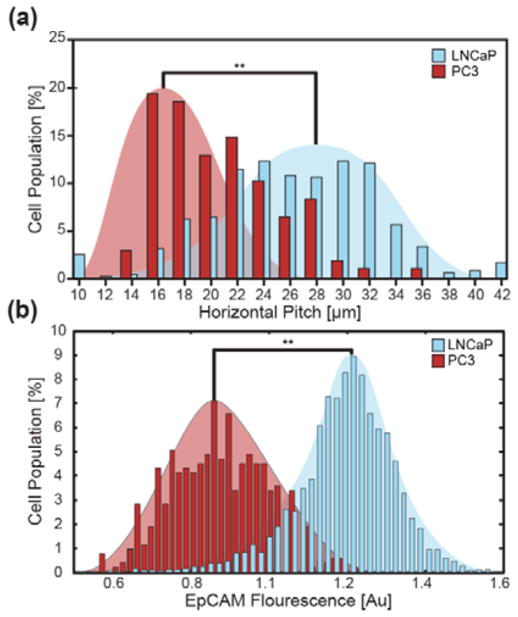
We first found that the quantity of 1 μm diameter, anti-EpCAM magnetic particle binding correlated with αEpCAM immunofluorescence on two prostate cancer lines with differential expression. LNCaP cells have been reported to have high but varying EpCAM expression (337,000 ± 37% molecules/cell[18]), which was in agreement with our flow cytometry analysis that demonstrated a variation from mean fluorescence of ± 27% (Figure S6). PC3 cells have comparatively lower EpCAM expression levels, ~ 52,000±78% molecules/cell which we confirmed with flow cytometry analysis. Quantity of bound magnetic particles followed a similar trend where PC3 cells ranged between 1–41 bound particles per cell while a majority of LNCaPs ranged between 21–103 particles per cell (NLNCaP=508, NPC3=57, p=5.7e-6).
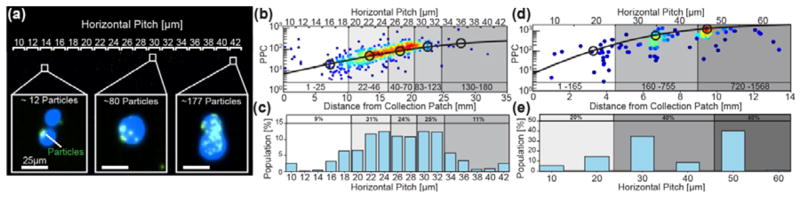
Once we confirmed that particle binding correlated positively with EpCAM expression, LNCaP cells were magnetically labeled and separated using the ratcheting system (Movie S3). Labeled LNCaPs driven with a 5Hz ratchet separated and equilibrated at critical pitches according to their bound particle quantity (Figure 3a–b). Using kmeans statistical analysis, cells were binned (unpaired two-tailed t-test, p≪0.05) into five subpopulations with particle binding quantities of 1–25, 22–46, 40–70, 83–123 &130–180 particles per cell (PPC) each equilibrating at their corresponding critical pitches. The average PPC’s for each subpopulation demonstrated the expected 1/3 power relationship between critical pitch and PPC, in agreement with the predictive (Equation 4, R2=0.91). Furthermore, the system was able to resolve the cells at high resolution resolving a 13 particle differential between the 10–24μm pitch regions. Additionally, the population distribution from the ratcheting cytometry chip (Figure 3c) was similar to flow cytometry data on the same LNCaP population (Figure 4a&b). The LNCaP distribution was centered on the 22μm pitch and had a coefficient of variation of 37%, which was close to the flow cytometry distribution with a 27% coefficient of variation.
Figure 3.
Equilibrium separation and analysis of LNCaP EpCAM distribution in a gradient pitch array. (a) A stitched image is shown of magnetically labeled LNCaP cells equilibrating at different pitches within a 2μm incremented chip driven at 5Hz frequency. Insets show cells equilibrating at pitches according to quantity of particles per cell (PPC). (b) The graph plots the PPC versus the on-chip location and critical pitch of individual cells (points). The theoretical 1/3 power relationship between PPC and pitch is shown as a solid line. Five statistically significant (p<0.05) populations were identified ranging from 1–25, 22–46, 40–70, 83–123 &130–180 particles per cell with corresponding averages (black circles). Each subpopulation equilibrated at increasing horizontal pitches, correlating well with the theoretical model (R2=0.91). (c) Independent of the attached particle number, the cell distribution as a function of pitch for both chips is also shown (N=508). (d)The PPC distribution of magnetically labeled LNCaP cells is also plotted for a 10μm incremented chip at 5Hz, again correlating with the predicted model (R2=0.89). More punctate trapping is observed due to the coarseness of the pitch gradient and additional populations of more strongly labeled cells are observed compared to the 2μm incremented chip. (e) The cell distribution as a function of pitch for this chip is also shown.
Figure 4.
Ratcheting cytometry of PC3 and LNCaP cells showed differential population distributions consistent with flow cytometry analysis. (a) PC3 cells demonstrate lower EpCAM expression and therefore equilibrate at lower critical pitches when ratcheted at 5Hz on a 2μm incremented chip compared to the LNCaP cells. (b) Flow cytometry analysis shows a similar trend in differential expression between the PC3 and LNCaP cell populations.
To accommodate separation and enrichment of cells with larger magnetic signatures, ratcheting cytometry with labeled LNCaPs was also performed on a 10μm incremented chip driven at 5Hz (Figure 3d & e). Using the same kmeans and t-test analysis, three PPC ranges of 1–165, 160–755 and 720–1598 PPC were resolved. This behavior on the 10μm incremented chip also correlated with the predictive model (R2=0.89) and separated a subpopulation of cells that was not detected in the 2μm increment. This population, with a PPC range of 720–1598, was not observed in the 2μm incremented chip most likely due insufficient pitch length, whereby this population may have ratcheted off the end of the chip.
In addition to LNCaP cells, ratcheting cytometry was also performed on PC3 cells to compare EpCAM expression profiles. Using a 2μm incremented chip, we were able to see differences in equilibrium pitches between LNCaP cells and PC3 cells ratcheted at 5Hz, in agreement with EpCAM expression levels (Figure 4a). PC3s exhibited PPC signatures of 1–23 and 19–41 (p=0.01), which equilibrated at 14–20μm and 22–26μm pitches respectively. In characterizing cell separation behavior on both the 2μm and 10μm incremented chips we determined that each chip design was optimal for different use cases. The 2μm incremented chip was best used for precise surface marker analysis and separation within a single cell population or between populations with similar expression. The 10μm incremented chip was best used in purifying a target cell population from heterogeneous cell solutions, such as blood, where the expression level of the target cells were significantly higher than the other populations.
2.3 Ratcheting Cytometry of Prostate Circulating Tumor Cells
The system was then assessed as a rare cell cytometer by spiking approximately 100 cancer cells into healthy blood, simulating prostate circulating tumor cell (CTCs) samples[19]. Our goal was to quantify surface expression profiles on CTCs but also address the major barrier of CTC purification which has limited MACS and other CTC capture systems. Using magnetic ratcheting cytometry, highly expressing CTCs can be quantitatively separated from the leukocyte background at high purity streamlining downstream precision assays. As a control, healthy blood was labeled with 1μm αEpCAM particles and ratcheted through a 10μm incremented chip to quantify contaminating leukocyte background. Most of non-specifically labeled leukocytes occupied a pitch range from 10–60μm and equilibrated mostly at the 20μm pitch corresponding to a PPC range of 1–25 (Figure S7). From this data we set a “cut-off” pitch at 60μm. Therefore, CTCs could be successfully purified as long as they equilibrated at a ≥70μm pitch.
To simulate patient samples LNCaP (~100 cells/mL) were spiked into 1mL volumes of whole blood from a healthy donor, diluted 5x in PBS, and then labeled with anti-EpCAM particles. The entirety of the sample was then flowed over the chip’s loading patch while magnetized to pull the labeled cells onto the loading patch. After accumulation onto the loading patch, the cells were separated on a 10μm incremented chip at 5Hz (Figure 5a). The spiked cells were successfully extracted from the leukocyte background and equilibrated mostly at the 80μm pitch (Figure 5b). Of the spiked cells approximately 24.9±1.94% were purified on the ratcheting chip. This capture efficiency is comparable to MACS based techniques targeting prostate cancer cells with EpCAM[15],[16]. However, compared to standard MACS techniques[20][21][22][23][24][25][26][10][27] the separation purity (Figure 5c) was significantly higher, the max being 51% [23], where a majority of the LNCaPs were successfully extracted from the leukocyte background. Note that for traditional MACS any amount of magnetic labeling leads to capture, ultimately resulting in lower purity which ranges between 51% to 0.1%.
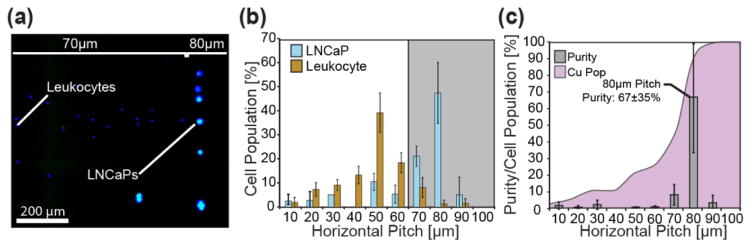
Figure 5.
Separation of LNCaP cells from leukocytes as a function of pitch. (a) After quantification of leukocyte background and setting a cut-off pitch at 60μm, LNCaP cells spiked into blood were separated and purified from the leukocyte background on a 10μm incremented chip under a 5Hz ratchet. (b) High purity separation of LNCaPs from leukocytes was achieved despite both populations shifting to higher pitches. The LNCaP majority equilibrated at the 70–100μm pitches peaking at 80μm while the leukocytes equilibrated between the 10–60μm pitches peaking at 50μm. (c) Purity, defined as the total number of LNCaPs to total cells, for each pitch was determined showing a maximum purity of 67±35% at the 80μm pitch. The cumulative population (Cu Pop) the spiked cells was determined by sequentially summing the spiked population beginning at the 10μm pitch and showed 26% of the spiked population equilibrated below the cut-off pitch.
Interestingly, the spiked LNCaPs and non-specifically-labeled leukocytes occupied higher pillar pitches than observed in buffer. A majority of the LNCaPs, 74%, resided at the 80μm pitch which we hypothesize is attributed to an effective particle concentration increase due to the excluded volume of the red blood cells. Additionally, the non-specifically labeled leukocyte population demonstrated a shift towards the higher pitches but a vast majority, ~90%, remained at or below the cutoff pitch of 60μm and therefore did not significantly affect the 70μm-100μm pitch purities. The system demonstrated minimal loss as shown in the cumulative distribution plots (Figure 5c). 74% of the extracted LNCaPs were cleared past the cut-off pitch and highly purified. The purity of the extracted cells above an 80μm pitch was 67±35% and contained 47±14% of the extracted population. In total, the ratcheting cytometry system was able to successfully concentrate and purify low concentrations of cancer cells from blood making it a tenable option as a clinical purification instrument for CTCs.
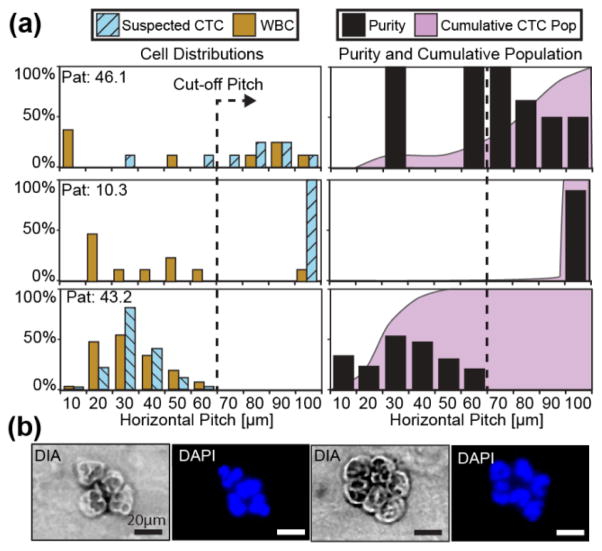
After validating the magnetic ratcheting cytometry system with spiked cancer cells, clinical blood samples of metastatic prostate cancer patients were run on the chip. We expected some prostate CTCs to exhibit high EpCAM expression (PPC 1700–3600) as observed in the spiking experiments, thereby equilibrating past the cutoff pitch under a 5Hz ratchet. Of note, we define suspected prostate CTCs as large nucleated cells (diameter≥9μm) with high EpCAM expression (PPC≥1700) and no detectable CD45 expression as quantified by florescence. Three patient samples were labeled and separated on the chip, two of which had several large, CD45 negative cells which equilibrated at the 70–100μm pitches (Figure 6a). These results were similar to our spiking experiments and, upon observation, demonstrated morphological characteristics consistent with CTC profiles including large nuclear to cytoplasmic ratio or multinucleated cells (Figure 6b). The CTC separation purity was high, where each binned purity for the 70–100μm pitches ranged between 50% to 100% for these two patients (Figure 6a Pat 46.1 & 10.3). Furthermore, the loss of suspicious cells was low as the cumulative populations of suspected prostate CTCs for these patients exhibited a skew towards the 70–100μm pitch values. Between 78% and 100% of the suspected CTCs were above the cut-off pitch for the two patients respectively, demonstrating high purity separation without significant loss of the captured cells.
Figure 6.
Blood biopsies (1ml volume) from prostate patients with metastatic castration resistant cancer were magnetically labeled with 1μm αEpCAM particles and cells were separated on the ratcheting cytometry system. (a) Patients 46.1 and 10.3 had several cells (N=7 and N=8 CTCs respectively) which equilibrated between the 70–100μm pitches with purity ranging from 50% to 100%. In contrast, patient 43.2 exhibited no cells in the 70–100μm pitches and a skewed cumulative population towards the lower pitches. Suspected prostate CTCs extracted from patients 10.3 (b) possessed morphological characteristics consistent with CTCs including large nuclear to cytoplasmic ratio, and large lobed or multiple nuclei in addition to being CD45 negative.
However, one patient exhibited a ratcheting profile more akin to the healthy controls where no cells occupied the 70–100μm pitch ranges (Figure 6 Pat 43.2). While many cells equilibrated at the lower pitches had little to no CD45 expression (N=188), they had small nuclei (< 9μm) and are therefore most likely non-specifically labeled leukocytes with low CD45 expression such as neutrophils or granulocytes [28].
In a parallel study with the same patient group, CTCs were captured using microfluidic vortex CTC isolation technology[29]. The findings from the ratcheting cytometry correlated with the vortex technology findings in two patients. The vortex chip detected high suspected CTC counts in Patient 46.1 and little to no CTCs in patient 43.1 which matched the findings of the ratcheting cytometer. Interestingly, ratcheting cytometry extracted several large suspected prostate CTCs from 10.3 where the vortex chip isolated only little to no CTCs. This discrepancy is most likely due to the clustered nature of the cells which prevented entrance into the vortex chip. Overall, the ratcheting cytometry system isolated EpCAM expressing suspected prostate CTCs from peripheral whole blood with an average purity of ~74%, a substantial improvement over traditional MACS-based techniques. The system shows promise as a high efficiency extraction method where high purity is also necessary, such as is the case for downstream sequencing for precision medicine.
3. Conclusion
Magnetic ratcheting cytometry enables widely used magnetic labeling techniques to be deployed for robust, efficient, and quantitative separations, while simultaneously concentrating target cells. Using 1:1 aspect ratio permalloy micro-pillar arrays we have increased the magnetophoretic force envelope 10 fold compared to thin film ratcheting systems. Increased force not only decreases processing time but enables the use of small particles which is advantageous due to their increased labeling efficiency attributed to large diffusion lengths. In developing a theoretical framework for high force magnetic ratcheting, arrays with gradient pitch can be rationally designed and constructed to achieve separation and concentration of magnetic particles and cells. Furthermore, these gradient pitch arrays achieve separation in a temporally stable, equilibrium based manner making them more robust than kinetic or flow based separation techniques which are sensitive to dynamic physical parameters such as flow rate. Additionally, magnetic ratcheting cytometry cleanly integrates separation and quantification (on the gradient pillar slide) into a single step assay, bringing a much needed quantitative aspect to MACS systems.
As demonstrated, ratcheting cytometry is an effective tool to capture and quantitatively separate rare cell types in both laboratory and clinical settings. Particularly, the system addresses the major challenge of purity that has plagued traditional MACS based systems. In future works separation resolution can be increased by using smaller magnetic particles to reduce the effective iron content per particle which will likely increase sample purity by reducing labeling time and off-target labeling. Another future system improvement could include a combined 10μm and 2μm incremented chip to enable both high dynamic range and high resolution separation. First cell populations can undergo a coarse separation in one axis and a fine separation in an orthogonal axis, thereby accommodating samples with a large PPC range without sacrificing separation resolution. Furthermore, ratcheting separation and manipulation can be combined with other size or deformability based techniques [30][29] to obtain thorough phenotypic profiles of enriched target cells. However, a significant challenge that needs to be addressed in transitioning this technology to a clinical instrument is the ability to extract the cells from the chip after separation. One option is to concentrate the separated cell populations by ratcheting vertically relative to the array to collect the cells at the bottom of each pitch zone (Movie S4). In this way, the concentrated cells could be extracted from the chip via fluid access wells aligned with the bottom of each pitch zone. Another option is to integrate all required analysis into the chip where cells, once separated, can be manipulated at high resolution into different modules directly on chip (Figure S8 & Movie S5). Indeed, ratcheting cytometry is not limited solely to separation but can be used as a manipulation platform to develop fully integrated lab-on-a chip systems enabling assays to be performed from whole samples down to single cells directly on the same chip. For example, target cells can first be quantitatively separated and concentrated using the gradient pitch arrays and then manipulated through fluid reservoirs to perform wash steps or various biochemical assays, or arrayed to enable single-cell interrogation. We envision the ratcheting system as a new platform for lab-on-a-chip automation, enabling sample-to-answer assays to be performed rapidly with single cell resolution.
4. Experimental
4.1 Automated Ratcheting System
The automated ratcheting system (Figure S2) consisted of a radial array of N52 grade rare earth neodymium ferrite magnets (KJ Magnetics), with a quasi halbach array arrangement (Figure S3, strength ranged from 20–200mT), driven by a custom designed mechatronic system and Labview® interface.
4.2 Chip Fabrication
Borosilicate glass (Fisher) slides were piranha cleaned and coated with a Ti-Cu-Ti metal seed layer via e beam evaporation. SPR220 resist was used to make electroplating molds, after which permalloy micro-pillars (1:1 aspect ratio ~4μm height) were electroplated. The chip was then sealed with silicon nitride and coated with spin-on polystyrene. Chips were soaked in 2% pluronic F127 solution before use. To create a fluidic chamber, PDMS (Dow-Corning) milli-channels were fabricated using scotch tape lithography[31] to create fluid access (Figure S4c–e). Solutions were added or injected onto the loading patch of the chip using a syringe See SI Text for details.
4.3 Particles and Particle Separations
Magnetic particles of 1μm, 2.8μm, 4.6μm and 5μm were made fluorescent through a variety of surface modifications (SI Text). Particle concentrations ranged between 0.5~1 x 106 particles/mL. Particle separation was observed by inverting the chip onto a PDMS milli-channel which was placed on the stage of a Nikon Eclipse Ti fluorescent microscope and positioning the mechatronic system above it. Of note that the Particles were injected onto the ratcheting chip loading patch, ratcheted at various frequencies, and imaged. Image analysis with ImageJ was used to identify particle distributions (Figure S9).
4.4 Cell Labeling
1μm iron oxide particles (Invitrogen) with anti-EpCAM (abcam) were diluted with PBS + 0.5% BSA to a concentration of 106 particles/mL and added to a solution of fixed LNCaP or PC3 cells (4% paraformaldehyde) at a 1:100 cell to particle ratio. Labeling was performed at room temperature with gentle mixing for at least 1 hour. Anti-mouse IgG Alexa Fluor-488 secondary (Invitrogen) was added to stain and visualize the particles. The solution was then washed and resuspended into 1mL of PBS. The quantity of particles per cell was determined using florescent image analysis, where the bound particle intensity was summed over the cell and normalized to the nuclear area. This metric was in turn used to determine PPC values (See SI Text and Figure. S 10).
4.5 Flow Cytometry
For flow cytometry experiments, the same fixed cells were used. ~105 cells of each type was labeled using anti-EpCAM PE (BD Biosciences) and incubated for an hour. The cells where then washed and resuspended in ~1mL of PBS. Flow cytometry was performed on a BD Accuri C6 flow cytometer with sampling of at least 2000 events.
4.6 Ratcheting Cytometry Separation based on EpCAM
1μm magnetically labeled LNCaP and PC3 cells (Hoechst stained) were injected at 50μL/min via a syringe/PEEK tube assembly onto the ratcheting chip’s loading patch. Note that chips with both 2μm and 10μm pitch increments where characterized. Simultaneously the mechatronic system was positioned over the loading patch to concentrate the cells. Ratcheting was initiated at a frequency of 5Hz for ~ 10 minutes. The entire chip was imaged under DAPI and FITC wavelengths.
4.7 Image Analysis and Aggregate Statistics
Stitched images of ratcheted cells and particles were analyzed using an automated MATLAB script to crop each cell, sum the intensity value of the particles (FITC), and normalize to the total cell area (DAPI). This numerical value was proportional to the number of bound particles per cell and was calibrated for each image analyzed to determine the PPC value (Figure. S 10).
Kmeans and t-test statistical analysis was used to characterize the relationship between bound particles and pillar pitch. Using a MATLAB script with a kmeans algorithm, the number of statistically significant populations (p≤0.05) and their mean PPC was determined.
4.8 Blood Spiking, Capture Efficiency and Purity Experiments
Peripheral whole blood collected from healthy donors, including 1 age matched, were aliquoted into 1mL volumes then spiked with ~100 FITC labeled LNCaPs. The blood was then diluted 5x with PBS and labeled with 1μm anti-EpCAM particles at 106 particles/mL (SI Text). Ratcheting separation was carried out at 5Hz and the entirety of the chip was imaged under DAPI and FITC filter sets. Capture efficiency was determined by counting the number of FITC positive cells and comparing to a control. Purity characterization was performed similarly where ~2000 FITC labeled LNCaP cells were spiked into 1mL of whole blood, labeled, then separated at 5Hz. Purity was defined as the ratio of spiked LNCaP cells to the total number of cells binned by pitch.
4.9 Ratcheting Cytometry with Prostate Cancer Patients
Blood was obtained from prostate patients with metastatic castration resistant cancer being treated at the UCLA medical center according to IRB approved protocol. Blood was aliquoted, stained with Hoechst & CD 45, diluted 5x in PBS and labeled with particles (SI Text). Similar to blood spiking experiments the cells were separated using a 5Hz ratchet and imaged under DAPI and TRITC filter sets.
Supplementary Material
Mechatronic ratcheting system can be mounted onto an inverted microscope for ratcheting of magnetic particles and cells across permalloy micro-pillar arrays. The system generates a cycling magnetic field using a wheel of rare earth magnets arranged in a halbach array orientation. The system can also be controlled to adjust the in plane ratcheting angle for separation and concentration functions. Ratcheting control can be achieved via a joystick or by an automation program for hands free manipulation and separations.
Green fluorescent 2.8μm superparamagnetic particles ratcheted on a permalloy micro-pillar array chip with gradient horizontal pitch. If the pitch is less than equal to the critical pitch, the particle will traverse the array. If the pitch exceeds the critical pitch, the particles will trap and coalesce. Critical pitch is dependent on the particle’s iron oxide content.
LNCaP cells magnetically labeled with αEpCAM 1μm diameter superparamagnetic particles can be quantitatively separated based on the quantity of bound particles. Cells will traverse the array until reaching their critical pitch where they will equilibrate and concentrate.
After quantitative separation, cells can be further concentrated for extraction off chip. By ratcheting orthogonally to the chip, cells can be transported to the base of the chip and collected.
Using the automated ratcheting system parallelized and high resolution manipulation of particles and cells can be achieved. Coarse mode manipulations of 2.8μm particles in a square trajectory can be achieved by translating particles between pillars and fine mode, nanometer scale manipulations can be achieved by piloting the particle radially and tangentially on a single pillar. In addition to particles, cells can also be piloted in an arbitrary trajectory in a parallelized and repeatable manner. Shown are cell piloted in a square trajectory and a square wave trajectory.
Figure. S1: Magnetic ratcheting utilizes arrays of electroplated permalloy micro-pillars to modify the magnetic potential energy landscape (Umag), creating dynamic potential energy wells to trap and manipulate superparamagnetic particles. When a bulk magnetic field is applied, the micro-pillars magnetize in alignment to the bulk field and modify the magnetic potential energy landscape introducing dynamic potential wells in which superparamagnetic particles become trapped. As the field orientation is cycled particles will follow the changing potential wells and ratchet through the pillars if the frequency and pitch between neighboring pillars is low enough and iron oxide content is high enough.
Figure. S2: Automated and scope mounted ratcheting system consists of an XYZ module for positioning and a ratcheting module for angular and ratcheting speed control (a). The chip is first inverted onto a PDMS well containing aqueous buffer, placed on the scope, and then the ratcheting system is positioned over it with an offset of 2mm (a & b). The system was able to operate a full cycle of the radial array at frequencies between 0.1 to 50 Hz. Note that the mechatronic system was aligned to the ratcheting chips such that the central axis of the ratcheting module was aligned with the centroid of the ratcheting chip. The centroid for both chip designs was approximately 14mm from the chip’s left edge and 0.5cm from the top edge. Alignment was performed as precisely as possible with a tolerance of approximately ±5mm. However, device performance is not highly sensitive to chip/mechatronic system alignment due to the fact that mm scale differences over the macroscale magnetic field de-amplify to nanometer scale changes of the micro-magnetic field within the permalloy micropillars.
Figure. S3: Magnetic wheel with a partial halbach array of N-52 grade rare earth magnets. As shown the peak flux density is 300mT with an average flux density of ~ 100mT with a max radial distance of 3mm from the surface of the drum.
Figure. S4: Schematic of gradient pitch arrays for ratcheting separation. Both chips consist of a 1cm x 1cm loading patch with 6μm horizontal and vertical pillar pitch where samples containing cells were loaded or flowed over. Going from left to right, the loading patch leads into pitch zones of incrementally increasing horizontal pitch but a constant 6 μm vertical pitch throughout. (a) The first chip consists of seventeen, 1mm x 1cm pitch zones, where the first pitch zone has a 10μm pitch (6μm edge to edge gap) and increments by 2μm with each zone; ending in a 42μm pitch. (b) The second chip consists of ten, 2mm x 1cm pitch zones, where the first pitch zone has a 10μm pitch, increments by 10μm, and ends in a 100μm pitch. (c) To achieve separation chips were inverted and placed onto an open air microfluidic PDMS chamber/channel produced using scotch tape lithography which was prefilled with PBS. The magnetic wheel from the mechatronic system was positioned directly above the inverted chip. (d–e) Sample was injected into the PDMS channel and the magnetically labeled cells were pulled up via the magnetic wheel and accumulated on the ratcheting array loading patch where they were separated after the entire sample volume was injected.
Figure. S5: Calculated magnetic force on a 1μm particle vs. horizontal pitch which was determined empirically by tracking ratcheting trajectories of 1μm Fe3O4 particles at various frequencies on 10μm incremented chip by balancing with drag force. A power law best fit the data, specifically αP−2, where α=550pN μm2. Pearson’s fit coefficient for this data was 0.85.
Figure. S6: Flow cytometry analysis for EpsCAM expression on LNCaP cells. The EpCAM expression shows a near normal distribution with approximately a 27% coefficient of variation.
Figure. S7: : Healthy control samples (N=3), including 1 age matched sample, was run through the chip to develop a healthy patient profile and determine cutoff pitch for leukocyte background. The non-specifically labeled leukocyte population occupied the 10–60μm pitches under a 5Hz ratchet, therefore the cut-off pitch was set as ≤60μm and cells equilibrating at ≥70μm under a 5Hz ratchet were past the leukocyte background.
Figure. S8: Using the automated ratcheting system parallelized and high resolution manipulation of particles and cells can be achieved. (a) Coarse mode manipulations of 2.8μm particles in a square trajectory can be achieved by translating particles between pillars and (b) fine mode, nanometer scale manipulations can be achieved by piloting the particle radially and tangentially on a single pillar. (c) Coarse mode manipulations have high consistency where 92% of particles were successfully piloted to a desired location. (d) Fine mode manipulations demonstrated nanometer scale error with 83% of the particles having a ±500 nm error and 93% of the particle population had a ±1μm error. (d) Cells can also be piloted in an arbitrary trajectory in a parallelized and repeatable manner.
Figure. S9: Shown above are distributions of several particle types (1μm, 2.8μm and 4.6μm diameter) driven at 30, 25, 10, 5 and 2.5 Hz ratcheting. As expected, particles with higher iron oxide content equilibrate at larger pillar pitches. As frequency decreases, the particle population migrates to higher pitches.
Figure. S10: The quantity of bound particles on magnetically labeled cells was determined using florescent image analysis. The intensity values from the bound particles (green) was summed over each cell and divided by the nuclear area (blue) to provide a cell area normalized particle intensity value. A calibration curve between the cell area normalized particle intensity value and particles per cell (counted manually in Image J) was generated to quantify the number of bound particles. This calibration curve was used to determine PPC values for separation on gradient pitch arrays.
Figure. S11: Predicted peak force on a 2.8μm magnetic particle ( Vp=1.95μm3, 17% Fe3O4, χ=0.65,μo=4π 1e–7) as a function of pillar aspect ratio was simulated using Comsol 4.2 Magnetostatics Module. In the simulation the pillars were set to have a relative magnetic permeability of 8500, a diameter of 4μm, an orthogonal pitch of 6μm and a height derived from the corresponding aspect ratio (a &b). Boundary conditions included a 304mT inward flux density with either a normal orientation (parallel to the pillar axis) or tangential (parallel to the pillar radii). The force density, defined as (B •∇)B, was numerically calculated at the surface of each pillar and the magnetic force derived for a 2.8μm magnetic particle (c).
Acknowledgments
This work was supported through the NIH Director’s New Innovator grant (1DP2OD007113). We thank Dr. Eric Diebold for flow cytometry analysis, Dr. Jere Harrison for help in microfabrication, and Brian McVerry for particle functionalization. Additional thanks to the California NanoSystems Institute for ISNC microfabrication facilities.
Footnotes
Contributions
C.M. and D.D. initiated the concept. P.T. developed the fabrication process. C.M and S.A. constructed the automated ratcheting system. C.M and E.P designed and executed the experiments. R.K and M. R. provided clinical guidance and patient blood samples. C.M., E.P., and D.D. wrote the manuscript.
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Weaver WM, Tseng P, Kunze A, Masaeli M, Chung AJ, Dudani JS, Kittur H, Kulkarni RP, Di Carlo D. Advances in high-throughput single-cell microtechnologies. Curr Opin Biotechnol. 2014 Feb;25:114–23. doi: 10.1016/j.copbio.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards BS, Sklar LA. Flow Cytometry: Impact on Early Drug Discovery. J Biomol Screen. 2015 Mar; doi: 10.1177/1087057115578273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Königsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, de Santis M, Zeillinger R, Hudec M, Dittrich C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011 Jun;50(5):700–10. doi: 10.3109/0284186X.2010.549151. [DOI] [PubMed] [Google Scholar]
- 4.Adams JD, Kim U, Soh HT. Multitarget magnetic activated cell sorter. Proc Natl Acad Sci U S A. 2008 Nov;105(47):18165–70. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci U S A. 2009 Mar;106(9):2989–94. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert D, Pamme N, Conjeaud H, Gazeau F, Iles A, Wilhelm C. Cell sorting by endocytotic capacity in a microfluidic magnetophoresis device. Lab Chip. 2011 Jun;11(11):1902. doi: 10.1039/c0lc00656d. [DOI] [PubMed] [Google Scholar]
- 7.Pamme N, Wilhelm C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip. 2006 Aug;6(8):974–80. doi: 10.1039/b604542a. [DOI] [PubMed] [Google Scholar]
- 8.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013 Apr;5(179):179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hejazian M, Li W, Nguyen N-T. Lab on a chip for continuous-flow magnetic cell separation. Lab Chip. 2014 Dec; doi: 10.1039/c4lc01422g. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Li P, Huang PH, Xie Y, Mai JD, Wang L, Nguyen NT, Huang TJ. Rare cell isolation and analysis in microfluidics. Lab Chip. 2014 Feb;14(4):626–45. doi: 10.1039/c3lc90136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yellen BB, Erb RM, Son HS, Hewlin R, Shang H, Lee GU. Traveling wave magnetophoresis for high resolution chip based separations. Lab Chip. 2007;7:1681–1688. doi: 10.1039/b713547e. [DOI] [PubMed] [Google Scholar]
- 12.Yellen BB, Virgin LN. Nonlinear dynamics of superparamagnetic beads in a traveling magnetic-field wave. Phys Rev E - Stat Nonlinear, Soft Matter Phys. 2009;80 doi: 10.1103/PhysRevE.80.011402. [DOI] [PubMed] [Google Scholar]
- 13.Gao L, Tahir MA, Virgin LN, Yellen BB. Multiplexing superparamagnetic beads driven by multi-frequency ratchets. Lab on a Chip. 2011;11:4214. doi: 10.1039/c1lc20683d. [DOI] [PubMed] [Google Scholar]
- 14.Tahir MA, Gao L, Virgin LN, Yellen BB. Transport of superparamagnetic beads through a two-dimensional potential energy landscape. Phys Rev E - Stat Nonlinear, Soft Matter Phys. 2011;84 doi: 10.1103/PhysRevE.84.011403. [DOI] [PubMed] [Google Scholar]
- 15.Dharmasiri U, Balamurugan S, Adams AA, Okagbare PI, Obubuafo A, Soper SA. Highly efficient capture and enumeration of low abundance prostate cancer cells using prostate-specific membrane antigen aptamers immobilized to a polymeric microfluidic device. Electrophoresis. 2009 Sep;30(18):3289–300. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, Lackner MR. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010 Jan;5(9):e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. [Accessed: 03-May-2015];COATED MAGNETIC MICROPARTICLES and NANOPARTICLES - Spherotech. [Online]. Available: http://www.spherotech.com/coa_mag_par.htm.
- 18.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Terstappen LWMM. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005 Jul;27(1):49–57. [PubMed] [Google Scholar]
- 19.Krebs MG, Hou JM, Ward TH, Blackhall FH, Dive C. Circulating tumour cells: their utility in cancer management and predicting outcomes. Ther Adv Med Oncol. 2010 Nov;2(6):351–65. doi: 10.1177/1758834010378414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen P, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014 Mar;9(3):694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004 Oct;32(10):891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Jatana KR, Balasubramanian P, Lang JC, Yang L, Jatana CA, White E, Agrawal A, Ozer E, Schuller DE, Teknos TN, Chalmers JJ. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Arch Otolaryngol Head Neck Surg. 2010 Dec;136(12):1274–9. doi: 10.1001/archoto.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009 Mar;106(10):3970–5. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilkenroth U, Taubert H, Riemann D, Rebmann U, Heynemann H, Meye A. Detection and enrichment of disseminated renal carcinoma cells from peripheral blood by immunomagnetic cell separation. Int J Cancer. 2001 May;92(4):577–82. doi: 10.1002/ijc.1217. [DOI] [PubMed] [Google Scholar]
- 25.Sbarbati R, De Boer M, Marzilli M, Scarlattini M, Rossi G, Van Mourik JA. Immunologic detection of endothelial cells in human whole blood. Blood. 1991;77(4):764–769. [PubMed] [Google Scholar]
- 26.Martin VM, Siewert C, Scharl A, Harms T, Heinze R, Ohl S, Radbruch A, Miltenyi S, Schmitz J. Immunomagnetic enrichment of disseminated epithelial tumor cells from peripheral blood by MACS. Exp Hematol. 1998 Mar;26(3):252–64. [PubMed] [Google Scholar]
- 27.Winer-Jones JP, Vahidi B, Arquilevich N, Fang C, Ferguson S, Harkins D, Hill C, Klem E, Pagano PC, Peasley C, Romero J, Shartle R, Vasko RC, Strauss WM, Dempsey PW. Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell. PLoS One. 2014 Jan;9(1):e86717. doi: 10.1371/journal.pone.0086717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuijpers TW, Tool AT, van der Schoot CE, Ginsel LA, Onderwater JJ, Roos D, Verhoeven AJ. Membrane surface antigen expression on neutrophils: a reappraisal of the use of surface markers for neutrophil activation. Blood. 1991 Aug;78(4):1105–11. [PubMed] [Google Scholar]
- 29.Sollier E, Go DE, Che J, Gossett DR, O’Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N, McCloskey S, Kulkarni RP, Di Carlo D. Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip. 2014 Jan;14(1):63–77. doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- 30.Loutherback K, Puchalla J, Austin RH, Sturm JC. Deterministic Microfluidic Ratchet. Phys Rev Lett. 2009 Jan;102(4):045301. doi: 10.1103/PhysRevLett.102.045301. [DOI] [PubMed] [Google Scholar]
- 31.Shrirao AB, Hussain A, Cho CH, Perez-Castillejos R. Adhesive-tape soft lithography for patterning mammalian cells: application to wound-healing assays. Biotechniques. 2012 Nov;53(5):315–18. doi: 10.2144/000113928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mechatronic ratcheting system can be mounted onto an inverted microscope for ratcheting of magnetic particles and cells across permalloy micro-pillar arrays. The system generates a cycling magnetic field using a wheel of rare earth magnets arranged in a halbach array orientation. The system can also be controlled to adjust the in plane ratcheting angle for separation and concentration functions. Ratcheting control can be achieved via a joystick or by an automation program for hands free manipulation and separations.
Green fluorescent 2.8μm superparamagnetic particles ratcheted on a permalloy micro-pillar array chip with gradient horizontal pitch. If the pitch is less than equal to the critical pitch, the particle will traverse the array. If the pitch exceeds the critical pitch, the particles will trap and coalesce. Critical pitch is dependent on the particle’s iron oxide content.
LNCaP cells magnetically labeled with αEpCAM 1μm diameter superparamagnetic particles can be quantitatively separated based on the quantity of bound particles. Cells will traverse the array until reaching their critical pitch where they will equilibrate and concentrate.
After quantitative separation, cells can be further concentrated for extraction off chip. By ratcheting orthogonally to the chip, cells can be transported to the base of the chip and collected.
Using the automated ratcheting system parallelized and high resolution manipulation of particles and cells can be achieved. Coarse mode manipulations of 2.8μm particles in a square trajectory can be achieved by translating particles between pillars and fine mode, nanometer scale manipulations can be achieved by piloting the particle radially and tangentially on a single pillar. In addition to particles, cells can also be piloted in an arbitrary trajectory in a parallelized and repeatable manner. Shown are cell piloted in a square trajectory and a square wave trajectory.
Figure. S1: Magnetic ratcheting utilizes arrays of electroplated permalloy micro-pillars to modify the magnetic potential energy landscape (Umag), creating dynamic potential energy wells to trap and manipulate superparamagnetic particles. When a bulk magnetic field is applied, the micro-pillars magnetize in alignment to the bulk field and modify the magnetic potential energy landscape introducing dynamic potential wells in which superparamagnetic particles become trapped. As the field orientation is cycled particles will follow the changing potential wells and ratchet through the pillars if the frequency and pitch between neighboring pillars is low enough and iron oxide content is high enough.
Figure. S2: Automated and scope mounted ratcheting system consists of an XYZ module for positioning and a ratcheting module for angular and ratcheting speed control (a). The chip is first inverted onto a PDMS well containing aqueous buffer, placed on the scope, and then the ratcheting system is positioned over it with an offset of 2mm (a & b). The system was able to operate a full cycle of the radial array at frequencies between 0.1 to 50 Hz. Note that the mechatronic system was aligned to the ratcheting chips such that the central axis of the ratcheting module was aligned with the centroid of the ratcheting chip. The centroid for both chip designs was approximately 14mm from the chip’s left edge and 0.5cm from the top edge. Alignment was performed as precisely as possible with a tolerance of approximately ±5mm. However, device performance is not highly sensitive to chip/mechatronic system alignment due to the fact that mm scale differences over the macroscale magnetic field de-amplify to nanometer scale changes of the micro-magnetic field within the permalloy micropillars.
Figure. S3: Magnetic wheel with a partial halbach array of N-52 grade rare earth magnets. As shown the peak flux density is 300mT with an average flux density of ~ 100mT with a max radial distance of 3mm from the surface of the drum.
Figure. S4: Schematic of gradient pitch arrays for ratcheting separation. Both chips consist of a 1cm x 1cm loading patch with 6μm horizontal and vertical pillar pitch where samples containing cells were loaded or flowed over. Going from left to right, the loading patch leads into pitch zones of incrementally increasing horizontal pitch but a constant 6 μm vertical pitch throughout. (a) The first chip consists of seventeen, 1mm x 1cm pitch zones, where the first pitch zone has a 10μm pitch (6μm edge to edge gap) and increments by 2μm with each zone; ending in a 42μm pitch. (b) The second chip consists of ten, 2mm x 1cm pitch zones, where the first pitch zone has a 10μm pitch, increments by 10μm, and ends in a 100μm pitch. (c) To achieve separation chips were inverted and placed onto an open air microfluidic PDMS chamber/channel produced using scotch tape lithography which was prefilled with PBS. The magnetic wheel from the mechatronic system was positioned directly above the inverted chip. (d–e) Sample was injected into the PDMS channel and the magnetically labeled cells were pulled up via the magnetic wheel and accumulated on the ratcheting array loading patch where they were separated after the entire sample volume was injected.
Figure. S5: Calculated magnetic force on a 1μm particle vs. horizontal pitch which was determined empirically by tracking ratcheting trajectories of 1μm Fe3O4 particles at various frequencies on 10μm incremented chip by balancing with drag force. A power law best fit the data, specifically αP−2, where α=550pN μm2. Pearson’s fit coefficient for this data was 0.85.
Figure. S6: Flow cytometry analysis for EpsCAM expression on LNCaP cells. The EpCAM expression shows a near normal distribution with approximately a 27% coefficient of variation.
Figure. S7: : Healthy control samples (N=3), including 1 age matched sample, was run through the chip to develop a healthy patient profile and determine cutoff pitch for leukocyte background. The non-specifically labeled leukocyte population occupied the 10–60μm pitches under a 5Hz ratchet, therefore the cut-off pitch was set as ≤60μm and cells equilibrating at ≥70μm under a 5Hz ratchet were past the leukocyte background.
Figure. S8: Using the automated ratcheting system parallelized and high resolution manipulation of particles and cells can be achieved. (a) Coarse mode manipulations of 2.8μm particles in a square trajectory can be achieved by translating particles between pillars and (b) fine mode, nanometer scale manipulations can be achieved by piloting the particle radially and tangentially on a single pillar. (c) Coarse mode manipulations have high consistency where 92% of particles were successfully piloted to a desired location. (d) Fine mode manipulations demonstrated nanometer scale error with 83% of the particles having a ±500 nm error and 93% of the particle population had a ±1μm error. (d) Cells can also be piloted in an arbitrary trajectory in a parallelized and repeatable manner.
Figure. S9: Shown above are distributions of several particle types (1μm, 2.8μm and 4.6μm diameter) driven at 30, 25, 10, 5 and 2.5 Hz ratcheting. As expected, particles with higher iron oxide content equilibrate at larger pillar pitches. As frequency decreases, the particle population migrates to higher pitches.
Figure. S10: The quantity of bound particles on magnetically labeled cells was determined using florescent image analysis. The intensity values from the bound particles (green) was summed over each cell and divided by the nuclear area (blue) to provide a cell area normalized particle intensity value. A calibration curve between the cell area normalized particle intensity value and particles per cell (counted manually in Image J) was generated to quantify the number of bound particles. This calibration curve was used to determine PPC values for separation on gradient pitch arrays.
Figure. S11: Predicted peak force on a 2.8μm magnetic particle ( Vp=1.95μm3, 17% Fe3O4, χ=0.65,μo=4π 1e–7) as a function of pillar aspect ratio was simulated using Comsol 4.2 Magnetostatics Module. In the simulation the pillars were set to have a relative magnetic permeability of 8500, a diameter of 4μm, an orthogonal pitch of 6μm and a height derived from the corresponding aspect ratio (a &b). Boundary conditions included a 304mT inward flux density with either a normal orientation (parallel to the pillar axis) or tangential (parallel to the pillar radii). The force density, defined as (B •∇)B, was numerically calculated at the surface of each pillar and the magnetic force derived for a 2.8μm magnetic particle (c).