Abstract
The genetic diversity and identification of slow- and fast-growing soybean root nodule bacterial isolates from different agro-climatic regions in Mpumalanga, Limpopo and Gauteng Provinces of South Africa were evaluated. The 16S-rDNA-RFLP analysis of 100 rhizobial isolates and eight reference type strains placed the isolates into six major clusters, and revealed their site-dependent genomic diversity. Sequence analysis of single and concatenated housekeeping genes (atpD, glnII and gyrB), as well as the symbiotic gene nifH captured a considerably higher level of genetic diversity and indicated the dominance of Bradyrhizobium diazoefficiens and Bradyrhizobium japonicum in Mpumalanga, Limpopo and Gauteng Provinces. Gene sequence similarities of isolates with type strains of Bradyrhizobium ranged from 97.3 to 100% for the 16S rDNA, and 83.4 to 100% for the housekeeping genes. The glnII gene phylogeny showed discordance with the other genes, suggesting lateral gene transfer or recombination events. Concatenated gene sequence analysis showed that most of the isolates did not align with known type strains and might represent new species from South Africa. This underscores the high genetic variability associated with soybean Bradyrhizobium in South African soils, and the presence of an important reservoir of novel soybean-nodulating bradyrhizobia in the country. In this study, the grouping of isolates was influenced by site origin, with Group I isolates originating from Limpopo Province and Groups II and III from Mpumlanga Province in the 16S rDNA-RFLP analysis.
Keywords: Colony morphology, 16S rDNA-RFLP, Horizontal gene transfer, Housekeeping genes
Introduction
Soybean (Glycine max [L.] Merrill) is a grain legume belonging to the family Leguminosae (Fabaceae) and sub-family Papilionoideae [16], [23], [65]. It originated from North-eastern China and is currently cultivated worldwide under various climatic conditions [6], [46]. It may have been introduced to Africa in the 19th century by Chinese traders along the east coast of Africa [50]. Its mature grain contains about 40% protein, 30% carbohydrate (cellulose, pectin and phytic acid), 10% digestible fibre, the vitamins E, K, riboflavin, thiamine, niacin and choline and minerals such as K, Mg, Ca, Zn, Fe and Cu, as well as anti-oxidants [16], [37]. Soybean alone accounts for 80% of the land area used for legume cultivation in the world and about 68% of the world's legume production [23]. About 109 980 000 ha of land are under soybean production worldwide [59]. However, soybean production in Africa is constrained by several factors, including the lack of compatible rhizobia in many African soils [41]. Promiscuous soybean varieties, which nodulate with indigenous soil rhizobia, have been bred to overcome the nodulation problem [1], [41].
Diverse groups of bacteria (fast- and slow-growers) belonging to the genera Bradyrhizobium, Sinorhizobium (Ensifer) and Mesorhizobium are responsible for establishing effective N2-fixing symbiosis with soybean [57], [62]. The slow-growers are distributed across different species, which include Bradyrhizobium japonicum [28], Bradyrhizobium elkanii [31], Bradyrhizobium liaoningense [68], and Bradyrhizobium yuanmingense [70]. The fast-growers consists of Sinorhizobium fredii and Sinorhizobium xinjiangense [12], [24], [29], [44], while Mesorhizobium tianshanese [11], with varying generation time, has also been reported to be a soybean microsymbiont. The B. japonicum and B. elkanii species have been found in various climatic regions across the world, while B. liaoningense, B. yuanmingense and Bradyrhizobium canariense have not yet been surveyed worldwide. The alkaline soils of temperate to subtropical climates of South and South-east Asia are home to B. liaoningense [2], [6], [22], [33], while the warm tropical climates of India, Kenya and Nepal favour B. yuanmingense [6], [46], [64]. The acid-tolerant B. canariense was found in North-east China [69]. In contrast, the fast- growing S. fredii and S. xinjiangense species were isolated from saline-alkaline soils in China, Vietnam, and Japan [7], [22], [33], [35], [44], [54]. Species of M. tianshanense were also isolated from China [11].
Characterisation and classification of rhizobia into different groups has been achieved using phenotypic, physiological and/or molecular methods. Phenotypic characterisation deals with traits exhibited by the rhizobial colonies, which include colony shape, colour and size [67]. Molecular characterisation, on the other hand, involves studying genome of the rhizobial cell. The molecular techniques commonly used to study rhizobial diversity include restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP) and rapid amplified polymorphic DNA (RAPD) [9], [32], [34]. Other methods such as DNA–DNA hybridisation, sequence analysis of the 16S rDNA gene and multilocus sequence analysis (MLSA) have also been used to study rhizobial taxonomy and diversity [15], [36], [39].
MLSA involves the study of sequences of the more conserved housekeeping genes such as atpD, glnII and gyrB genes. The sequencing of the 16S rDNA gene, on the other hand, is accurate at the genus level but poor at inter- and intra-species discrimination due to its low variation in sequences [36], [60]. MLSA is currently considered a more reliable alternative for studying genomic relationships between rhizobia [42], [58]. The genes used in MLSA have a higher degree of sequence divergence, hence MLSA is good for inter- and intra-species identification [36]. Additionally, housekeeping genes are more discriminatory and can therefore identify rhizobial strains from closely related lineages [36]. The housekeeping genes analysed in this study were reported previously as good taxonomic markers [36], [72].
Despite the growing cultivation of soybean in South Africa, and Africa at large, there is very limited information in the literature on the type of rhizobia nodulating this legume crop in South African soils. Studies of the type of rhizobia preferred by the soybean in South Africa, the dominant rhizobial strains in local soils, and their genetic diversity are needed to serve as guide for the production of soybean inoculants. The aim of this study was to determine the genetic diversity and phylogenetic identification of bacteria nodulating soybean in the different agro-climatic regions of South Africa.
Materials and methods
Sampling of root nodules and soils for rhizobial isolation
Two sources of root-nodule bacteria were used in this study: i) rhizobia isolated from soybean nodules collected from inoculated and uninoculated farmers’ fields in different areas of Mpumalanga Province in South Africa, and ii) rhizobia trapped in pot experiments using surface-sterilised soybean seed and uncultivated soils (10–20 cm depth) collected from different locations in Mpumalanga, Gauteng and Limpopo Provinces. The sites of nodule and soil collection are indicated in Table S1. Soil samples were processed and used to trap rhizobia, as described by Somasegaran and Hoben [51].
Rhizobia isolation and morpho-physiological characterisation
Rhizobia from soybean root nodules were isolated following standard procedures [61], [67]. Colony morphology and acid/alkaline reactions were evaluated on YMA containing bromothymol blue (25 μg/ml) as indicator. Pure single colony of each isolate was used for host-nodulation test with soybean variety PAN 1666, in fulfilment of Koch's postulate. Reference strains such as Rhizobium leguminosarum USDA 2370T, B. japonicum USDA 6T, B. diazoefficiens USDA 110T, B. elkanii USDA 76T, Ensifer meliloti (Sinorhizobium meliloti) USDA 1002T and Ensifer medicae (S. medicae) USDA 103T used in this study, were obtained from the National Rhizobium Germplasm Resource Collection USDA-ARS-SGIL, Beltsvile, USA.
Rhizobial DNA isolation and PCR amplification of 16S rDNA gene region
Rhizobial genomic DNA was extracted using GenElute Bacterial Genomic DNA kit according to the manufacturer's instructions (Sigma Aldrich, USA) and examined using 1% agarose gel containing ethidium bromide. The 16S-rDNA regions of bacterial genomic DNA was amplified in 25 μL reaction mixture containing 3 μl (5×) My Taq PCR buffer, 0.1 μl Taq polymerase (5 U) (Bioline, USA), 1 μl (10 pM) of each forward and reverse primer and 1 μL (40–50 ng) DNA as template. DNA amplifications were performed by Thermal cycle (T100 BIORAD, USA) with respective primers and standard temperature profile and examined on horizontal gel electrophoresis in 1.2% agarose gel (Table S2 [43], [52]).
Restriction fragment length polymorphism (RFLP) analysis of PCR-amplified 16S rDNA region
Products of the PCR-amplified 16S rDNA region were digested with three different four base-cutting restriction enzymes (namely, MspI, RsaI and HaeIII), following the procedure recommended by the manufacturer (Thermo Scientific, Lithuania) and digested products were separated by electrophoresis in 2.5% agarose gel.
RFLP cluster analysis of 16S rDNA region
The digested fragments of restriction enzymes were scored as “1” for presence, and “0” for absence of homologous bands. A dendrogram was constructed from the distance matrix using the un-weighted pair group method with arithmetic mean algorithm (UPGMA) with the help of NTSYSpc 2.1 software [47]. The Shannon–Wiener diversity index (H′) [49] was estimated based on the number of isolates of each province belonging to each cluster of the 16S rDNA-RFLP dendrogram.
PCR amplification, sequencing and phylogenetic analysis of housekeeping (atpD, glnII and gyrB) and symbiotic (nifH) genes
The PCR amplification of atpD, glnII, gyrB and nifH genes of rhizobial genome was done as described above for 16S rDNA. The primers used and thermal cycle conditions are listed in Table S2. The PCR amplified products of 16S rDNA, symbiotic gene (nifH) and housekeeping loci (atpD, glnll and gyrB) were purified using GeneJET PCR purification kit (Thermo Scientific, Lithuania). The purified samples were sequenced (Macrogen, Netherlands), and the quality of all sequences checked using BioEdit 7.0.0 software [21]. NCBI GenBank databases were used to identify species that were closely related to our test strains by means of the BLASTn program, and sequences submitted to the NCBI GenBank to get accession numbers (Table S3). Reference type sequences were selected to align with sequences of the test strains using MUSCLE [17] for construction of phylogenetic trees using MEGA 6.0 program [56]. Phylogenetic trees were generated by kimura 2-parameter method to calculate evolutionary distances [30], and evolutionary history was inferred using the Maximum-Likelihood method with 1000 bootstrap support [18]. Nucleotide information was obtained from conserved, variable, parsimony-informative, and singleton regions using consensus sequences. Tajima neutral mutation test was done by pair-wise comparison of analysis of the genetic variation by observing the number of nucleotide differences. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated.
Results
A total of 110 pure single-colony isolates were obtained and tested for plant nodulation under glasshouse conditions in conformity with Koch's postulate. Out of the 110 test isolates, 100 were identified as soybean rhizobia, given their ability to nodulate non-promiscuous soybean variety PAN1666 in plant nodulation assays.
Colonies of soybean nodule isolates appeared on YMA plates over variable time periods (2–14 days), exhibited different colony shapes, and failed to absorb the red colour of Congo red dye on YMA plates (Table S4). Isolates TUTMP16.2, TUTMP14.10.1, TUTMP14.10.3, TUTMP14.10.3b, TUTGP10.10.1, TUTLI1.10.3, TUTLI2.20.7, TUTLI4.20.5, TUTLI4.20.16, TUTLI8.20.1, TUTLI8.20.5 and TUTLI8.20.5b formed colonies before 4 days after inoculation on YMA plates. PCR amplification of 16S rDNA gene region of each isolate's genome produced a single band of approximately 1500 bp. The isolates exhibited variation in the banding pattern of the 16S rDNA gene when digested with 4-base cutter MspI, RsaI and HaeIII restriction endonucleases. All tested restriction endonucleases were able to produce polymorphic banding pattern with the test isolates. Restriction endonucleases MspI, RsaI and HaeIII yielded 16(A-P), 12(A-L) and 19 (A-S) restriction banding patterns, respectively (Table S4). On the basis of combined digested banding patterns, the dendrogram clustered isolates and reference strains into six main groups by the binary matrix 0/1 (0 for absence and 1 for presence of the restriction type (see Fig. S1). Group I consisted of 15 rhizobial isolates with a similarity coefficient of 0.22. No reference strain clustered with this group. Group II consisted of 22 rhizobial isolates that clustered with reference Bradyrhizobium strains USDA 6, H1, USDA 76 and USDA 110 with a similarity coefficient of 0.20. Groups III and IV respectively consisted of 20 and eight rhizobial isolates with a similarity coefficient of 0.12 and 0.08. No reference strain clustered with these groups. Group V had two rhizobial isolates (7 and 8) with a similarity coefficient of 0.50 and clustered with reference strains Rhizobium leguminosarum (USDA 2370), Sinorhizobium meliloti (USDA 1002) and Sinorhizobium medicae (USDA 1037). Group VI was the largest cluster with 31 isolates from Mpumalanga and Limpopo Provinces. Two rhizobial isolates (namely, 58 and 70) did not cluster with any reference strain or other test isolates, and stood independently with a 0.00 similarity coefficient to the rest of the isolates.
The Shannon–Weiner diversity index was highest (1.47) in Mpumalanga Province, followed by Limpopo (1.37), and then Gauteng Province (0.69) (Table S1).
Phylogenetic analysis of 16S rDNA, nifH, atpD, gyrB and glnII genes
A total of 22 bradyrhizobial isolates were selected from 16S rDNA-RFLP analysis as representative of each cluster to get the phylogenetic position. The PCR amplification of nifH (dinitrogenase reductase), and housekeeping genes [atpD (ATP synthase beta chain), gyrB (DNA gyrase) and glnII (glutamine synthase II)] yielded a single band of about 800 bp for nifH, atpD and gyrB, and 750 bp for glnII products.
Based on partial 16S rDNA gene sequence comparisons with the NCBI GenBank references, all the tested isolates were identified as B. diazoefficiens, B. japonicum, B. liaoningense and B. yuanmingense. The phylogenetic tree of the 16S rDNA gene grouped isolates into four distinct clusters (I–IV). Isolates TUTLl2.20.6, TUTLl4.10.7, TUTLl2.20.7, TUTLl4.10.9, TUTMP7.7, TUTMP2.9 and TUTMP9.7 grouped with reference strains of B. liaoningense, B. yuanmingense and B. japonicum in Cluster I with all isolates having 97–100% sequence identity with Bradyrhizobium type strains. Isolates TUTGP10.10.2, TUTMP18.11 and TUTMP18.6 aligned with B. diazoefficiens USDA 110 in Cluster II with a low bootstrap support of 60%. Only isolate TUTLl2.10.7 grouped with Bradyrhizobium iriomotense, Bradyrhizobium huanghuaihaiense, Bradyrhizobium denitrificans and Bradyrizobium ganzhouense in Cluster III. Surprisingly, isolate TUTMP16.11 clustered with Rhizobium lusitanum P1-7 with a high 99% bootstrap support and with sequence identity of 99.1%, while showing 91.0–92.2% identity with a type strain of Bradyrhizobium (tree not shown).
Analysis of individual housekeeping genes
Sequence analysis of the housekeeping genes atpD, glnII and gyrB respectively revealed 83.4–100%, 88.4–100% and 89.9–100% similarities with type strains of Bradyrhizobium. The lowest level of conservation (54.57%), the highest variable (45.43%) and the highest singleton sequences (19.25%) were observed for gyrB. The levels of conservation were 65.26% for atpD and 61.74% for glnII, while the highest parsimony-informative sequence information (32.21%) was observed in glnII (Table S5). In Tajima neutral mutation test, the highest (208) segregation sites was observed in gyrB locus, with atpD and gyrB showing negative D values (Table S6).
Phylogenetic trees were constructed using the maximum likelihood method for each housekeeping gene and this resulted in different Bradyrhizobium groupings. Isolates that were not clearly delineated at genus or species level by 16S rDNA gene, were distinctly separated into B. elkanii, B. japonicum, B. yuanmingense, B. iriomotense and B. diazoefficiens in the housekeeping gene phylogeny. Isolates with difficulties in PCR amplification or poor sequence quality were excluded in tree construction. In each of the housekeeping gene's phylogenetic tree, most of the test isolates were consistently grouped into a major clade with B. diazoefficiens (atpD and gyrB) and B. elkanii (glnII).
The phylogenetic tree from the atpD, glnII and gyrB gene analysis formed different clusters with B. diazoefficiens, B. japonicum, Bradyrhizobium jicamae, B. yuanmingense and B. elkanii (Figs. 1, S2 and S3). The single gene phylogenetic study showed incongruency of the isolates with similar type strains in all studied phylograms. Isolates TUTLl4.10.7, TUTMP18.11, TUTMP16.11, TUTMP14.10.3 and TUTLl4.10.5b showed similar species level cluster with B. diazoefficiens in atpD and gyrB phylograms, but aligned with B. elkanii in glnII phylogeny. There was a major shift of the isolates between clusters, with the relative positions of a few strains varying between atpD, glnII and gyrB genes trees.
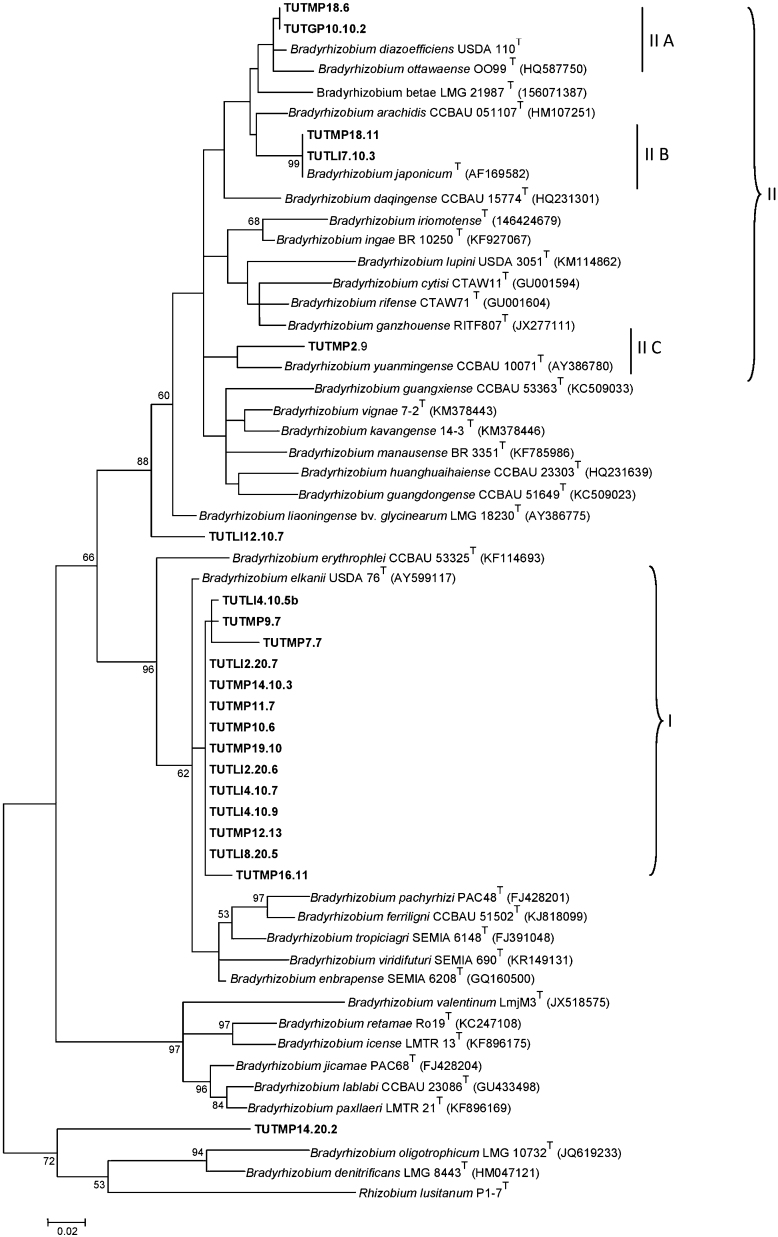
Fig. 1.
Phylogenetic tree based on glnII sequences generated by Maximum-Likelihood method. Bootstrap values (1000 replicates) are indicated above the branches.
All isolates from Limpopo Province, (except for isolate TUTLl2.20.7) clustered with B. diazoefficiens and B. japonicum in atpD and gyrB phylogenies, while Mpumlanga isolates clustered with B. diazoefficiens, B. japonicum, Bradyrhizobium guangxiense, Bradyrhizobium guangdongense, B. yuanmingense, B. jicamae and B. elkanii (Figs. S2 and S3). Isolate TUTGP10.10.2 from Gauteng Province also grouped with B. diazoefficiens in glnII and 16S rDNA phylograms.
Reconstruction of the phylogenetic tree of glnII sequences revealed that all isolates were grouped into two major clusters (I, II). Unlike atpD and gyrB phylogenetic analysis, most of the isolates proximally related with B. elkanii in Cluster I (Fig. 1), while Cluster II was divided into three subclusters (IIA, IIB, IIC). Subcluster IIA had isolates TUTMP18.6 and TUTGP10.10.2 which delineated with B. diazoefficiens, whereas isolates from subcluster IIB and IIC grouped with B. japonicum and B. yaunmingense, respectively (see Fig. 1). Isolate TUTLl2.10.7 distantly linked with Cluster II as an outgroup, while TUTMP14.20.2 stood alone with Bradyrhizobium oligotrophicum and B. denitrificans type strains (Fig. 1).
Phylogenetic analysis based on the combined atpD and gyrB gene sequences
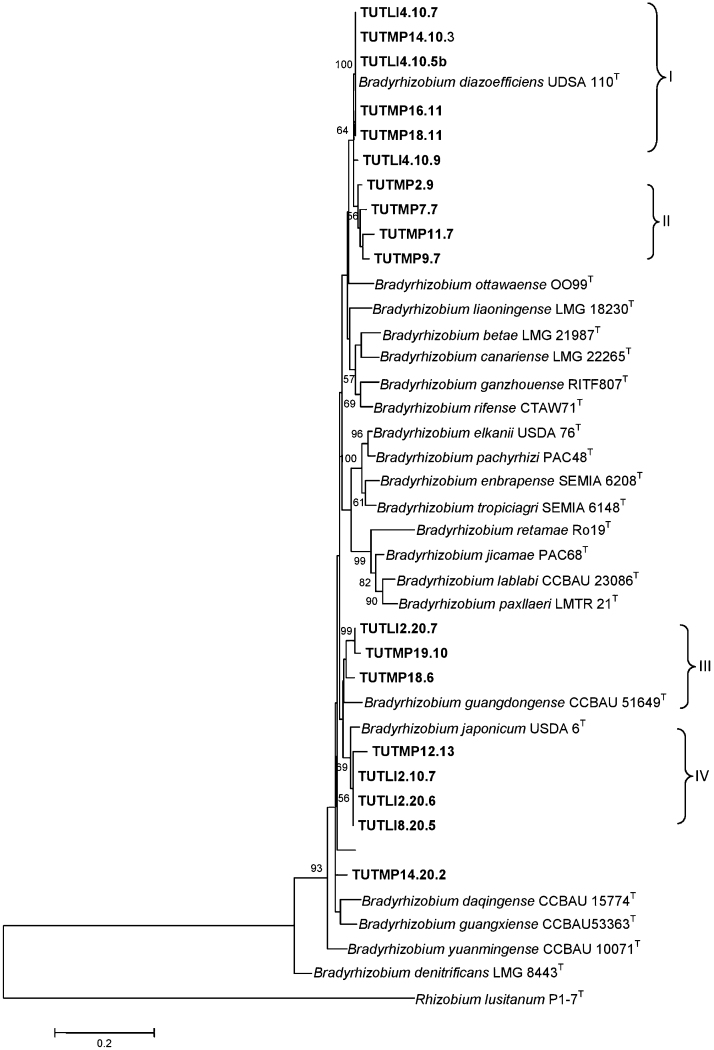
A phylogenetic tree based on concatenated genes confirmed the presence of diverse and novel type of Bradyrhizobium isolates nodulating soybean in South Africa. With the discordance observed in test isolates from glnII and 16S rDNA phylograms in comparison to those of atpD and gyrB housekeeping genes, we constructed concatenated tree with atpD + gyrB sequences (Fig. 2). The concatenated phylogeny was based on average consensus sequences of 843 analysed sites containing 509 (60.6%), 331 (39.4%), 170 (20.24%) and 161 (19.17%) of conserved, variable, parsimony-informative and singleton information sites, respectively (Table S5). The constructed concatenated tree revealed four large clusters (Clusters I–IV) with Bradyrhizobium species corresponding to B. diazoefficiens, Bradyrhizobium guangdongense and B. japonicum (Fig. 2). Five test strains in Cluster I grouped with B. diazoefficiens with high bootstrap support (100%) and 100% sequence identity, while isolate TUTLl4.10.9 was linked to the Cluster I outgroup with 64% bootstrap support and 99.1% sequence identity (Fig. 2; Table S7). In the atpD phylogram, the same isolate TUTLl4.10.9 grouped with B. japonicum in Cluster II.
Fig. 2.
Phylogenetic tree based on atpD + gyrB sequences generated by Maximum-Likelihood method. Bootstrap values (1000 replicates) are indicated above the branches.
Cluster II consisted of four isolates that failed to group with any type strains even though they were aligned with B. diazoefficiens in the gyrB phylogenetic tree. Although the four isolates converged in Cluster II in the concatenated phylogeny, they behaved differently in individual gene phylogenies. In the atpD tree, TUTMP2.9 grouped with B. yuanmingense, isolate TUTMP11.7 aligned with B. elkanii, TUTMP7.7 closed to B. jicamae, while isolate TUTMP9.7 stood alone. Monophyletic Cluster IV also had four isolates (TUTMP12.13, TUTLl2.20.6, TUTLl2.10.7 and TUTLl8.20.5) that grouped with B. japonicum in the same way as in the gyrB phylogram with a 69% bootstrap support and 97.1 to 98.3% sequence identity. Cluster III comprised three isolates that grouped without any reference type strains, but close to B. guangdongense, while isolate TUTMP14.20.2 stood alone in the tree (Fig. 2).
Phylogenetic analysis based on the nifH gene sequence
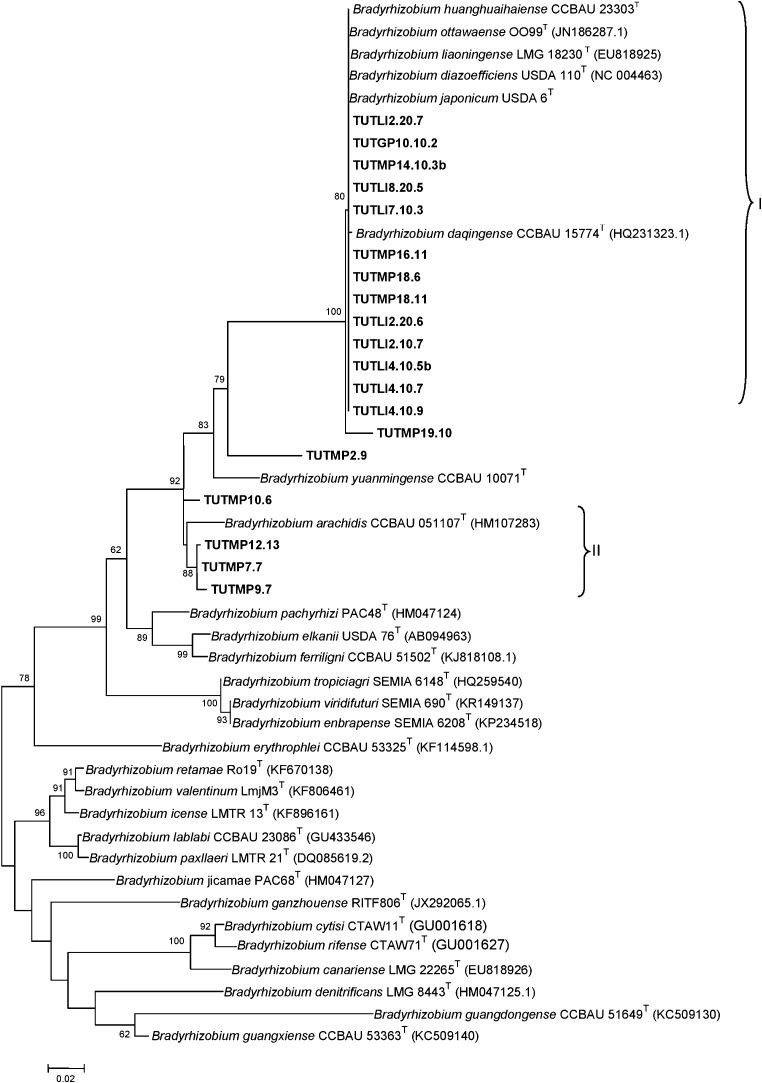
The reconstructed phylogenetic tree based on nifH was congruent with the tree built with atpD gene. Isolates TUTMP11.7, TUTMP14.20.2 and TUTMP14.10.3 were not included in the nifH study due to poor sequence results. In the nifH phylogenetic tree, all isolates from Limpopo Province, isolate TUTGP.10.10.2 from Gauteng, and some isolates from Mpumalanga were tightly clustered in one clade along with type strains such as Bradyrhizobium daqingense CCBAU 15774T, Bradyrhizobium ottawaense OO99T, Bradyrhizobium huanghuaihaiense CCBAU 23303T, B. liaoningense LMG 18230T, B. japonicum USDA 6T and B. diazoefficiens USDA 110T with a high 80 bootstrap support and 100% pair-wise sequence identity (Fig. 3). Isolates TUTMP2.9 and TUTMP19.10 were located on the outgroups of Cluster I with bootstrap support 79 and 100, respectively, and pairwise sequence similarity of 91.6% and 98.3% with the Bradyrhizobium type strains present in Cluster I. Cluster II consisted of only Mpumalanga isolates TUTMP10.6, TUTMP9.7, TUTMP7.7 and TUTMP12.13 without any type or reference strains (Fig. 3). In the atpD phylogram (Fig. S2), isolate TUTMP10.6 was closely grouped with B. elkanii USDA 76 with a 56% bootstrap support, while TUTMP12.13, TUTMP9.7 and TUTMP7.7 clustered together without any reference strains.
Fig. 3.
Phylogenetic tree based on nifH sequences generated by Maximum-Likelihood method. Bootstrap values (1000 replicates) are indicated above the branches.
Discussion
Phenotypic variation among bacterial symbionts nodulating soybean
In this study, we analysed rhizobial isolates from soybean root nodules obtained from different agro-climatic zones in South Africa. Out of 110 isolates, 100 formed root nodules with soybean variety PAN 1666 in fulfilment of Koch's postulate. Of the 100 rhizobial isolates, 66 were trapped from soils that had no recent history of crop cultivation, while 34 were isolated from farmers’ fields, previously inoculated with B. japonicum inoculants. Although it has been reported that some N2-fixing isolates can lose their symbiotic genes and thus fail to nodulate the homologous host during nodulation assay [5], [25], that was not the case in this study. The 100 bacterial symbionts comprised both fast- and slow-growers, and respectively produced acid and alkali reaction on BTB-containing YMA plates (Table S4).
Soybean-nodulating bacteria in the genus Bradyrhizobium are usually slow-growers (growth rate of ≥5 days) and alkali-producing with smaller colonies (<2 mm). However, strains from the genus Sinorhizobium, which also nodulates soybean, are fast-growers (growth rate of ≤5 days), and acid-producing with larger colonies (>2 mm) [51], [61]. The data in Table S4 clearly show that soybean-nodulating isolates from South Africa exhibited a huge diversity, as evidenced by the proportions of colony characteristics which included 87% dome shape, 5% conical, 6% flat and 2% oval isolates. Various studies [3], [13], [38] have similarly found isolate diversity in relation to cell growth rate (82% slow-growers vs. 18% fast-growers), as well as bromothymol blue reaction (83% alkali and 17% acid-producing). Some fast-growers (TUTMP15.3 and TUTGP10.10.1) showed alkali reaction, while slow-growers (TUTMP5.16, TUTMP11.1 and TUTMP19.10) showed acid reaction with BTB on YMA plates.
Molecular analysis of microsymbiont diversity of soybean-nodulating bacteria
PCR-RFLP has become a potent tool for classifying bradyrhizobial isolates due to the restriction site variation in certain amplified regions of the genome of different rhizobia [26], [40]. Soybean is nodulated by diverse bacterial symbionts that can undergo differentiation, resulting in new rhizobial types [1], [60], [69]. In this study, 16S rDNA-RFLP was used to molecularly characterise bradyrhizobial isolates from soybean root nodules. Based on the patterns of digested fragments, the number and distance of restriction sites recognised by a particular restriction endonuclease were found to differ among the different rhizobial isolates [4]. In fact, some of the isolates showed similar fingerprint after restriction digestion. This supported the findings of Amarger [4] which showed that similar banding pattern of bacterial isolates, when separately digested by more than two restriction enzymes, is an indication of genetic relatedness.
Some test isolates also showed variable RFLP digestion profiles of the 16S rDNA genomic region, which confirmed the presence of diverse bradyrhizobial isolates in the local microsymbiont population. This is inconsistent with the findings of Germano et al. [19] which found no diversity reflection in the16S rDNA sequence region of rhizobia. Furthermore, a dendrogram constructed from the RFLP digestion profiles clustered the bacterial isolates into six major groups. Except for the isolates in Group II, the RFLP patterns of the isolates in Groups I, III, IV and VI were clearly different and showed that they are genetically distinct from all the reference strains. However, Group II isolates were highly related to reference strains B. japonicum (USDA 6T, H1), B. diazoefficiens (USDA 11 T), and B. elkanii (USDA 76T), a finding consistent with the RFLP results of Abaidoo et al. [1]. In other previous studies, RFLP-PCR analysis of the 16S rRNA gene clearly differentiated Bradyrhizobium from Rhizobium, and even B. japonicum from B. elkanii [19], [32], [63]. In this study, the grouping of isolates was influenced by the site of origin. For example, the isolates in Group I originated from Limpopo Province, while Group III and IV were isolated from Mpumalanga Province. Our finding contrasted those of Yang & Zhou [69] and Grossman et al. [20], who showed that the clustering of soybean isolates, in their studies, was not influenced by the site of origin.
In this study, the phylogeny of soybean rhizobia was studied using sequence analysis of the 16S rDNA, nifH, atpD, glnII and gyrB genes. Based on the 16S rDNA gene analysis, the estimated similarity percentage between test isolates and type reference strains of Bradyrhizobium from NCBI GenBank ranged from 97.3 to 100%, except for isolate TUTMP16.11, which showed 89.8–92.2% identity. Thus, even though the 16S rDNA gene is highly conserved in members of the genus Bradyrhizobium [39], [66], our analysis showed that many isolates stood alone in a branch separate from other species of Bradyrhizobium and also revealed incongruency with studied housekeeping genes, a finding inconsistent with the results of Zilli et al. [74].
In this study, considerable phylogenetic congruency was found with the sequencing of atpD and gyrB genes, but not glnII, which produced discordant results. The observed phylogenetic incongruences could be attributed to the different evolutionary histories of the genes studied [73], intra-genomic re-arrangements and vertical/horizontal gene transfer, as well as subsequent recombinations that probably occurred in the genes [10], [14], [27], [45], [48], [71]. Gene recombination is known to play a major role in the evolution of soybean rhizobia. Li et al. [33] have reported significant gene recombination in chromosomal housekeeping genes of soybean rhizobia, just as they also observed both lateral and vertical gene transfer among soybean-nodulating microsymbionts. Symbiotic genes such as the nifH gene are easily transmitted through genetic re-arrangement [53].
In the Tajima neutral mutation test, the D values for glnII and nifH were positive, which suggests natural selection by increasing genetic variation [55]. In contrast, the D values for atpD and gyrB were negative, possibly indicating insertion or deletion of nucleotides in the region. Such insertions/deletions of nucleotides could account for the distant, not-so-close, alignment of test isolates with known Bradyrhizobium species in the phylogram.
Isolate groupings based on single gene phylogenies and concatenated phylogeny were compared and found to be interesting. Isolates TUTLl2.20.7, TUTMP18.6 and TUTMP19.10 delineated with B. daqingense, B. ottawaense, B. huanghuaihaiense, B. liaoningense, B. japonicum and B. diazoefficiens in the nifH phylogeny, whereas in the concatenated atpD + gyrB gene phylogeney, they failed to group with any reference strains, thus indicating a lack of correlation between the core and symbiotic gene phylogeny [8]. The results further revealed that the test soybean isolates clustered with the type/reference strains from the genus Bradyrhizobium, with B. japonicum and B. diazoefficiens dominating in all phylogenetic trees. These findings are consistent with other reports which showed that B. japonicum is the most dominant soybean microsymbiont in some soils [33], [71].
In the concatenated atpD + gyrB gene phylogeny, some of the isolates trapped from soils without cropping history grouped together with B. japonicum and B. diazoefficiens, while some isolates (TUTLl2.20.7, TUTMP19.10, TUTMP18.6) trapped from soil stood alone without any reference strains (Cluster II); this suggests that our bacterial isolates are very diverse, and probably related to different phylogenetic groups of Bradyrhizobium. There is therefore the strong possibility that some of these soybean-nodulating bradyrhizobia in South African soils, are unique and novel, especially those trapped from soils without any history of cultivation (e.g. TUTLl2.20.7, TUTMP19.10, TUTMP18.6). But to ascertain that would require further studies to unravel their uniqueness. It was however also interesting to note the grouping of inoculant strains isolated from farmers’ fields specifically with B. japonicum and B. diazoefficiens.
In conclusion, rhizobial isolates from this study exhibited varying phenotypic, physiological and molecular characteristics, which clearly indicated that the bacterial symbionts isolated from uncultivated soils and from farmers’ fields in Mpumalanga, Limpopo and Gauteng Provinces of South Africa consisted of different bradyrhizobia. These isolates, especially those trapped from soils without any history of cultivation, should be further studied to detail their identity and diversity, while identifying microsymbionts with greater N2-fixing ability for use as inoculants for soybean cultivation in Africa.
Footnotes
New sequences: Submitted to NCBI GenBank and accession numbers have been provided.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.syapm.2016.05.009.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Abaidoo R.C., Keyser H.H., Singleton P.W., Borthakur D. Bradyrhizobium spp. (TGx) isolates nodulating the new soybean cultivars in Africa are diverse and distinct from bradyrhizobia that nodulate North American soybean. Int. J. Syst. Evol. Microbiol. 2000;50:225–234. doi: 10.1099/00207713-50-1-225. [DOI] [PubMed] [Google Scholar]
- 2.Adhikari D., Kaneto M., Itoh K., Suyama K., Pokharel B.B., Gaihre Y.K. Genetic diversity of soybean-nodulating rhizobia in Nepal in relation to climate and soil properties. Plant Soil. 2012;357:131–145. [Google Scholar]
- 3.Alberton O., Kaschuk G., Hungria M. Sampling effects on assessment of genetic diversity of rhizobia associated with soybean and common bean. Soil Biol. Biochem. 2006;38:1298–1307. [Google Scholar]
- 4.Amarger N. Rhizobia in the field. Adv. Agron. 2001;73:109–168. [Google Scholar]
- 5.Angelini J., Ibanez F., Taurian T., Tonelli M.L., Valetti L., Fabra A. A study of the prevalence of bacteria that occupy nodules within single pea nut plants. Curr. Microbiol. 2011;62:1752–1759. doi: 10.1007/s00284-011-9924-2. [DOI] [PubMed] [Google Scholar]
- 6.Appunu C., Angele N., Laguerre G. Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural–ecological–climatic regions of India. Appl. Environ. Microbiol. 2008;74:5991–5996. doi: 10.1128/AEM.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appunu C., Sasirekha N., Prabavathi V.R., Nair S. A significant proportion of indigenous rhizobia from India associated with soybean (Glycine max L.) distinctly belong to Bradyrhizobium and Ensifer genera. Biol. Fertil. Soils. 2009;46:57–63. [Google Scholar]
- 8.Aserse A.A., Rasanen L.A., Assefa F., Hailemariam A., Lindstrom K. Phylogenetic and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 2012;35(2):120–131. doi: 10.1016/j.syapm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Botha W.J., Jaftha J.B., Bloem J.F., Habig J.H., Law I.J. Effect of soil bradyrhizobia on the success of soybean inoculant strain CB 1809. Microbiol. Res. 2004;159:219–231. doi: 10.1016/j.micres.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Charles L., Carbone I., Davies K.G., Bird D., Burke M., Kerry B.R., Opperman C.H. Phylogenetic analysis of Pasteuria penetrans by use of multiple genetic loci. J. Bacteriol. 2005;187:5700–5708. doi: 10.1128/JB.187.16.5700-5708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W.X., Wang E.T., Li Y.B., Chen X.Q., Li Y. Characterisation of Rhizobium tianshanense sp. nov. moderately and slowly growing nodule bacterium isolated from an acid saline environment in Xinjiang, People's Republic of China. Int. J. Syst. Bacteriol. 1995;45:153–159. doi: 10.1099/00207713-45-1-153. [DOI] [PubMed] [Google Scholar]
- 12.Chen L.S., Figueredo A., Pedrosa F.O., Hungria M. Genetic characterization of soybean rhizobia in Paraguay. Appl. Environ. Microbiol. 2000;66:5099–5103. doi: 10.1128/aem.66.11.5099-5103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L., Figueredo A., Villani H., Michajluk J., Hungria M. Diversity and symbiotic effectiveness of rhizobia isolated from field-grown soybean nodules in Paraguay. Biol. Fertil. Soils. 2002;35(6):448–457. [Google Scholar]
- 14.Christensen H., Kuhnert P., Olsen J.E., Bisgaard M. Comparative phylogenies of the housekeeping genes atpD, infB and rpoB and the 16S rRNA gene within the Pasteurellaceae. Int. J. Syst. Evol. Microbiol. 2004;54:1601–1609. doi: 10.1099/ijs.0.03018-0. [DOI] [PubMed] [Google Scholar]
- 15.Coenye T., Gevers D., van de Peer Y., Vandamme P., Swings J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 2005;29:147–167. doi: 10.1016/j.femsre.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Dugje I.Y., Omoigui L.O., Ekeleme F., Bandyopadyay R., Kumar L.P., Kamara A.Y. 2009. Farmers’ guide to soybean production in northern Nigeria. http://www.iita.org/c/document_library/get_file?p_l_id=25368&folderId=25529&name=DLFE-192.pdf. [Google Scholar]
- 17.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Germano M.G., Menna P., Mostasso F.L., Hungria M. RFLP analysis of the rRNA operon of a Brazilian collection of bradyrhizobial strains from 33 legume species. Int. J. Syst. Evol. Microbiol. 2006;56(1):217–229. doi: 10.1099/ijs.0.02917-0. [DOI] [PubMed] [Google Scholar]
- 20.Grossman J.M., Schipanski M.E., Sooksanguan T., Seehaver S., Drinkwater L.E. Diversity of rhizobia in soybean [Glycine max (Vinton)] nodules varies under organic and conventional management. Appl. Soil Ecol. 2011;50:14–20. [Google Scholar]
- 21.Hall T. 2004. BioEdit Version 7.0.0. Distributed by the author, website: www.mbio.ncsu.edu/BioEdit/bioedit.html. [Google Scholar]
- 22.Han L.L., Wang T.T., Han T.X., Liu J., Sui X.H., Chen W.F., Chen W.X. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil. 2009;324:291–305. [Google Scholar]
- 23.Herridge D.F., Peoples M.B., Boddey R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil. 2008;311:1–18. [Google Scholar]
- 24.Hungria M., de O., Chueire LgM., Coca R.G., Megías M. Preliminary characterization of fast growing rhizobial strains isolated from soyabean nodules in Brazil. Soil Biol. Biochem. 2001;33:1349–1361. [Google Scholar]
- 25.Ibanez F., Angelini J., Taurian T., Tonelli L., Fabra A. Endophytic occupation of pea nut root nodules by opportunistic Gammaproteobacteria. Syst. Appl. Microbiol. 2009;32:49–55. doi: 10.1016/j.syapm.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Jaiswal S.K., Anand A., Dhar B., Vaishampayan A. Genotypic characterization of phage-typed indigenous soybean bradyrhizobia and their host range symbiotic effectiveness. Microb. Ecol. 2012;63:116–126. doi: 10.1007/s00248-011-9950-4. [DOI] [PubMed] [Google Scholar]
- 27.Jaiswal S.K., Beyan S.M., Dakora F.D. Distribution, diversity and population composition of soybean-nodulating bradyrhizobia from different agro-climatic regions in Ethiopia. Biol. Fertil. Soils. 2016 [Google Scholar]
- 28.Jordan D. Notes: Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 1982;32:136–139. [Google Scholar]
- 29.Keyser H.H., Bohlool B.B., Hu T., Weber D.F. Fast-growing rhizobia isolated from root nodules of soybean. Science. 1982;215:1631–1632. doi: 10.1126/science.215.4540.1631. [DOI] [PubMed] [Google Scholar]
- 30.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 31.Kuykendall L., Saxena B., Devine T., Udell S. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 1992;38:501–505. [Google Scholar]
- 32.Laguerre G., Mavingui P., Allard M.R., Charnay M.P., Louvrier P., Mazurier S.I., Rigottier-Gois L., Amarger N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q.Q., Wang E.T., Zhang Y.Z., Zhang Y.M., Tian C.F., Sui X.H., Chen W.F., Chen W.X. Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei Province, China. Microb. Ecol. 2011;61:917–931. doi: 10.1007/s00248-011-9820-0. [DOI] [PubMed] [Google Scholar]
- 34.Lin D.X., Man C.X., Wang E.T., Chen W.X. Diverse rhizobia that nodulate two species of Kummerowia in China. Arch. Microbiol. 2007;188:495–507. doi: 10.1007/s00203-007-0271-4. [DOI] [PubMed] [Google Scholar]
- 35.Man C.X., Wang H., Chen W.F., Sui X.H., Wang E.T., Chen W.X. Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil. 2008;310:77–87. [Google Scholar]
- 36.Martens M., Dawyndt P., Coopman R., Gillis M., de Vos P. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium) Int. J. Syst. Evol. Microbiol. 2008;58:200–214. doi: 10.1099/ijs.0.65392-0. [DOI] [PubMed] [Google Scholar]
- 37.Meghvansi M.K., Prasad K., Mahna S.K. Symbiotic potential, competitiveness and compatibility of indigenous Bradyrhizobium japonicum isolates to three soybean genotypes of two distinct agro-climatic regions of Rajasthan, India. Saudi J. Biol. Sci. 2010;17:303–310. doi: 10.1016/j.sjbs.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menna P., Hungria M., Barcellos F.G., Bangel E.V., Hess P.N., Martínez-Romero E. Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst. Appl. Microbiol. 2006;29(4):315–332. doi: 10.1016/j.syapm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Menna P., Barcellos F.G., Hungria M. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int. J. Syst. Evol. Microbiol. 2009;59:2934–2950. doi: 10.1099/ijs.0.009779-0. [DOI] [PubMed] [Google Scholar]
- 40.Milagre S.T., Macielb C.D., Shinodac A.A., Hungria M., Almeida J.R.B. Multidimensional cluster stability analysis from a Brazilian Bradyrhizobium sp. RFLP/PCR data set. J. Comput. Appl. Math. 2009;227:30–319. [Google Scholar]
- 41.Mpepereki S., Javaheri F., Davis P., Giller K.E. Soybean and sustainable agriculture: promiscuous soyabeans in Southern Africa. Field Crop Res. 2000;65:137–149. [Google Scholar]
- 42.Naser S.M., Thompson F.L., Hoste B., Gevers D., Dawyndt P., Vancanneyt M., Swings J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology. 2005;151:2141–2150. doi: 10.1099/mic.0.27840-0. [DOI] [PubMed] [Google Scholar]
- 43.Nzoué A., Miché L., Klonowska A., Laguerre G., de Lajudie P., Moulin L. Multilocus sequence analysis of bradyrhizobia isolated from Aeschynomene species in Senegal. Syst. Appl. Microbiol. 2009;32(6):400–412. doi: 10.1016/j.syapm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Peng G.X., Tan Z.Y., Wang E.T., Reinhold-Hurek B., Chen W.F., Chen W.X. Identification of isolates from soybean nodules in Xinjiang Region as Sinorhizobium xinjiangense and genetic differentiation of S. xinjiangense from Sinorhizobium fredii. Int. J. Syst. Evol. Microbiol. 2002;52:457–462. doi: 10.1099/00207713-52-2-457. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro R.A., Barcellos F.G., Thompson F.L., Hungria M. Multilocus sequence analysis of Brazilian rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res. Microbiol. 2009;160:297–306. doi: 10.1016/j.resmic.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Risal C.P., Yokoyama T., Ohkama-Ohtsu N., Djedidi S., Sekimoto H. Genetic diversity of native soybean bradyrhizobia from different topographical regions along the southern slopes of the Himalayan Mountains in Nepal. Syst. Appl. Microbiol. 2010;33:416–425. doi: 10.1016/j.syapm.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Rohlf F.J. Setauket; New York: 2009. NTSYSpc: Numerical Taxonomy System, Ver. 2.21c. Exeter Software. [Google Scholar]
- 48.Rokas A., King N., Finnerty J., Carroll S.B. Conflicting phylogenetic signals at the base of the metazoan tree. Evol. Dev. 2003;5:346–359. doi: 10.1046/j.1525-142x.2003.03042.x. [DOI] [PubMed] [Google Scholar]
- 49.Saeki Y., Minami M., Yamamoto A., Akao S. Estimation of the bacterial community diversity of soybean-nodulating bradyrhizobia isolated from Rj-genotype soybeans. Soil Sci. Plant Nutr. 2008;54(5):718–724. [Google Scholar]
- 50.Shurtleff W., Aoyagi A. Soyinfo Center; 2009. History of Soybeans and Soyfoods in Africa (1857–2009): Extensively Annotated Bibliography and Sourcebook. [Google Scholar]
- 51.Somasegaran P., Hoben H.J. Springer-Verlag, NifTAL Publications, Library of Congress; New York: 1994. Handbook for Rhizobia. [Google Scholar]
- 52.Stępkowski T., Żak M., Moulin L., Króliczak J., Golińska B., Narożna D., Mądrzak C.J. Bradyrhizobium canariense and Bradyrhizobium japonicum are the two dominant Rhizobium species in root nodules of lupin and serradella plants growing in Europe. Syst. Appl. Microbiol. 2011;34(5):368–375. doi: 10.1016/j.syapm.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan J.T., Ronson C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K., Oguro H., Yamakawa T., Yamamoto A., Akao S., Saeki Y. Diversity and distribution of indigenous soybeannodulating rhizobia in the Okinawa islands, Japan. Soil Sci. Plant Nutr. 2008;54:237–246. [Google Scholar]
- 55.Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan Z.Y., Xu X.D., Wang E.T., Gao J.L., Martínez-romero E., Chen W.X. Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int. J. Syst. Bact. 1997;47:874–879. doi: 10.1099/00207713-47-3-874. [DOI] [PubMed] [Google Scholar]
- 58.Thompson F.L., Gevers D., Thompson C.C., Dawyndt P., Naser S., Hoste B., Munn C.B., Swings J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.USDA . Government Printer; Washington: 2013. World Agricultural Production. [Google Scholar]
- 60.van Berkum P., Fuhrmann J.J. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 2000;50:2165–2172. doi: 10.1099/00207713-50-6-2165. [DOI] [PubMed] [Google Scholar]
- 61.Vincent J.M. Blackwell Scientific; Oxford: 1970. A Manual for the Practical Study of Root Nodule Bacteria. [Google Scholar]
- 62.Vinuesa P., Rojas-Jiménez K., Contreras-Moreira B., Mahna S.K., Prasad B.N., Moe H., Selvaraju S.B., Thierfelder H., Werner D. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 2008;74:6987–6996. doi: 10.1128/AEM.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang E.T., Rogel M.A., GarćIa-de Los Santos A., Martinez-Romero J., Cevallos M.A., Martinez-Romero E. Rhizobium etli bv. mimosae, a novel biovar isolated from Mimosa affinis. Int. J. Syst. Bact. 1999;49:1479–1491. doi: 10.1099/00207713-49-4-1479. [DOI] [PubMed] [Google Scholar]
- 64.Wasike V.W., Lesueur D., Wachira F.N., Mungai N.W., Mumera L.M., Sanginga N., Mburu H.N., Mugadi D., Wango P., Vanlauwe B. Genetic diversity of indigenous Bradyrhizobium nodulating promiscuous soybean [Glycine max (L) Merr.] varieties in Kenya: impact of phosphorus and lime fertilization in two contrasting sites. Plant Soil. 2009;322:151–163. [Google Scholar]
- 65.Werner D., Newton W.E., editors. Nitrogen Fixation in Agriculture, Forestry, Ecology, and the Environment. Springer; Dordrecht: 2005. [Google Scholar]
- 66.Willems A., Doignon-Bourcier F., Goris J., Coopman R., deLajudie P., De Vos P., Gillis M. DNA–DNA hybridization study of Bradyrhizobium strains. Int. J. Syst. Evol. Microbiol. 2001;51:1315–1322. doi: 10.1099/00207713-51-4-1315. [DOI] [PubMed] [Google Scholar]
- 67.Woomer P.L., Karanja N., Kisamuli S.M., Murwira M., Bala A. 2011. A Revised Manual for Rhizobium Methods and Standard Protocols.http://www.n2africa.org [Google Scholar]
- 68.Xu L.M., Ge C., Cui Z., Li J., Fan H. Bradyrhizobium liaoningense sp. nov. isolated from the root nodules of soybeans. Int. J. Syst. Bact. 1995;45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 69.Yang J.K., Zhou J.C. Diversity, phylogeny and host specificity of soybean and Peanut bradyrhizobia. Biol. Fert. Soils. 2008;44:843–851. [Google Scholar]
- 70.Yao Z.Y., Kan F.L., Wang E.T., Wei G.H., Chen W.X. Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int. J. Syst. Evol. Microbiol. 2002;52:2219–2230. doi: 10.1099/00207713-52-6-2219. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X.X., Guo H.J., Wang R., Sui X.H., Zhang Y.M., Wang E.T., Tian C.F., Chen W.X. Genetic divergence of Bradyrhizobium strains nodulating soybeans as revealed by multilocus sequence analysis of genes inside and outside the symbiosis island. Appl. Environ. Microbiol. 2014;80(10):3181–3190. doi: 10.1128/AEM.00044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y.M., Tian C.F., Sui X.H., Chen W.F., Chen W.X. Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS ONE. 2012;7(9):e44936. doi: 10.1371/journal.pone.0044936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao L., Fan M., Zhang D., Yang R., Zhang F., Xu L., Wei X., Shen Y., Wei G. Distribution and diversity of rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in Northwest China. Syst. Appl. Microbiol. 2014;37:449–456. doi: 10.1016/j.syapm.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 74.Zilli J.E., Baraúna A.C., da Silva K., De Meyer S.E., Farias E.N., Kaminski P.E., O’Hara G. Bradyrhizobium neotropicale sp. nov., isolated from effective nodules of Centrolobium paraense. Int. J. Syst. Evol. Microbiol. 2014;64:3950–3957. doi: 10.1099/ijs.0.065458-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.