ABSTRACT
Three wine estates (designated A, B, and C) were sampled in Sauternes, a typical appellation of the Bordeaux wine area producing sweet white wine. From those wine estates, 551 yeast strains were collected between 2012 and 2014, added to 102 older strains from 1992 to 2011 from wine estate C. All the strains were analyzed through 15 microsatellite markers, resulting in 503 unique Saccharomyces cerevisiae genotypes, revealing high genetic diversity and a low presence of commercial yeast starters. Population analysis performed using Fst genetic distance or ancestry profiles revealed that the two closest wine estates, B and C, which have juxtaposed vineyard plots and common seasonal staff, share more related isolates with each other than with wine estate A, indicating exchange between estates. The characterization of isolates collected 23 years ago at wine estate C in relation to recent isolates obtained at wine estate B revealed the long-term persistence of isolates. Last, during the 2014 harvest period, a temporal succession of ancestral subpopulations related to the different batches associated with the selective picking of noble rotted grapes was highlighted.
IMPORTANCE High genetic diversity of S. cerevisiae isolates from spontaneous fermentation on wine estates in the Sauternes appellation of Bordeaux was revealed. Only 7% of all Sauternes strains were considered genetically related to specific commercial strains. The long-term persistence (over 20 years) of S. cerevisiae profiles on a given wine estate is highlighted.
INTRODUCTION
Saccharomyces cerevisiae is widely distributed and associated with human-related fermentations, as well as with those from the natural environment (e.g., oak trees and fruits). The population genetic structure of S. cerevisiae has been shown to correlate with its ecological differentiation (1–4), as well as geographical distance (2, 5). Strains isolated from vineyards and wine-related environments constitute a genetically well-differentiated homogeneous group. In the last 20 years, many studies have described the genetic diversity of S. cerevisiae isolates from different grape varieties. Molecular methods, such as pulsed-field gel electrophoresis (PFGE) (6), mitochondrial DNA (mtDNA) restriction fragment length polymorphism (RFLP) analysis (7), inter-delta analysis (8), and microsatellite analysis (9), were used to describe the genetic diversity of vineyard-associated S. cerevisiae. Numerous factors, such as climate conditions, geographical locations of the vineyards, fungicide management, grape varieties, and winemaking practices, impact the natural yeast population's diversity (10). Grapes are thought to be the first source of S. cerevisiae strains involved in the winemaking process, and winery surfaces are probably the main microbial reservoir to carry out must spontaneous fermentation (11). Gayevskiy and Goddard were the first to show evidence for region-specific S. cerevisiae populations associated with vines and wines in New Zealand using microsatellite markers, and they also pointed out the exchange of strains among these regions (12). The presence of specific fermentative profiles with perennial persistence over successive years in a given wine-producing area has been highlighted by different authors (13). However, a recent study challenged the view of a stable population in a wine environment over time, showing that no S. cerevisiae strain was isolated in the same vineyard or cellar during three consecutive years (8). Until now, very few studies have reported long-term observations of the changes in the S. cerevisiae population over time (14).
The Sauternes region in France, similar to the Tokaj wine region in Hungary, is one of the most famous and highly esteemed areas for noble rot sweet wines. Musts are obtained from noble rotted grape cluster selective pickings (15). The noble rot development is subject to weather conditions that dictate the number of selective pickings each year, typically up to three or four. The resulting grape musts have specific characteristics, with high sugar, acid, glycerol, and mineral contents; nitrogen deficiency; special polysaccharides; and aroma composition, which provide extremely difficult nutritional and environmental conditions for yeast growth and fermentative metabolism (16). As a consequence, yeasts produce high levels of acetic acid during alcoholic fermentation, ranging from 0.56 to 1.50 g/liter, depending on the must (17), and fermentation can be a slow process. Thus, the use of selected yeast starters and subcultures, or “pied de cuve,” produced from fermenting must is generally recommended (18, 19).
Many previous studies reported the population dynamics on the surfaces of botrytized grapes and revealed a complex microbiota. Botrytis infection stimulates a high level of yeast diversity, and the community is likely enriched with fermentative and/or spoilage species (20). The presence of S. cerevisiae and Saccharomyces uvarum on Tokaj grapes has been described (21). The yeast microbial community of the grape must mirrors the grape microbiota and is highly diverse compared to that of traditional dry wines. Candida zemplinina, later renamed Starmerella bacillaris, could dominate fermentation during the first stages, and later, Kluyveromyces, Hanseniaspora, and Pichia were frequently isolated from midfermentation (22–25). Non-Saccharomyces yeast may contribute significantly to the fermentation of botrytized wines at early stages, but S. cerevisiae still dominates the fermentation process, frequently associated with S. uvarum (21, 23, 26, 27). The state of the damaged grape berries may impact the Saccharomyces yeast diversity and population level, since they may be very rich depositories of S. cerevisiae compared with sound berries (28). However, the S. cerevisiae population associated with the winemaking process of botrytized must has been poorly investigated until now. In a survey of wine estates in the southern region of Bordeaux, Frezier and Dubourdieu (13) described the existence of dominant S. cerevisiae profiles whenever white, red, and botrytized wine spontaneous fermentations were studied during two consecutive years. Later, Masneuf and Dubourdieu (29), using the PFGE method, established the karyotypes of 199 S. cerevisiae strains isolated from indigenous fermentation of botrytized must and reported high diversity in the profiles, with no dominant ones.
The occurrence of local and resident S. cerevisiae populations in a given viticultural region, and on a smaller scale in a given winery, is a recurring issue for the scientific community and winemakers. The microbial aspect of terroir was recently illustrated by different studies that suggested a link between vineyard environmental conditions and microbial inhabitation patterns and revealed the importance of microbial populations for the regional identity of wine (30, 31).
The objective of this study was to establish the population genetic structure of S. cerevisiae on a spatial (region/winery) and temporal (over 20 years) scale in the case of a fermentative system characterized by a highly complex microbiota and difficult nutritional and environmental conditions for yeast growth. For that purpose, we used a robust molecular method based on the analysis of 15 microsatellite markers. S. cerevisiae isolates were collected from spontaneous fermented must samples from three wine estates in the Sauternes appellation from 2012 to 2014. We aimed to gain deeper knowledge of cellar-associated S. cerevisiae ecology and possible exchanges between populations in the same appellation. We took advantage of having a large collection of S. cerevisiae isolates collected from spontaneously fermenting grape must since 1992 on one of the wine estates to survey the long-term diversity and population structure of cellar-associated S. cerevisiae and to test the hypothesis of the presence of specific wine cellar populations with perennial persistence in a given region or wine estate.
MATERIALS AND METHODS
Sample collection and processing.
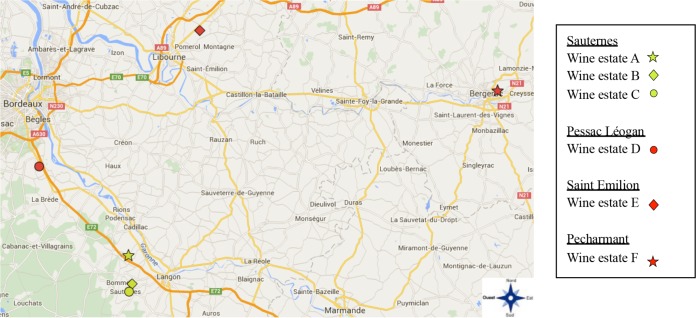
A total of 3 wine estates in the Sauternes appellation, which is one of the sweet-wine-producing areas in Gironde, part of the Aquitaine region in southwest France, were selected to conduct this study. The distance between wine estates A and B/C is 10 km, whereas the distance between wine estates B and C is 1.8 km. The three wine estates produce sweet wines from botrytized Sauvignon and Semillon grape varieties (Fig. 1). The initial sugar contents of the grape musts were between 350 and 450 g/liter. Wine estates A and B are managed according to organic practices, whereas wine estate C is managed conventionally. Briefly, sulfur and copper are used in both organic and conventional farming systems, whereas synthetic fungicides are also used in the conventional system. Alcoholic fermentation was stopped thanks to a massive addition of sulfur dioxide (200 to 300 mg/liter). Samples were taken at 75% of the alcoholic fermentation from barrels containing 225 liters of must. In the wine estates during harvest periods, several pickings were made in different crops, with different numbers of lots from different years for the study (Table 1). At wine estate A, sampling was performed for 2 years, in 2012 and 2014, from 2 and 4 different lots, respectively. At wine estate B, sampling was performed for 3 years, 2012, 2013, and 2014, with 3 lots in 2012 and 2013 and 2 in 2014. Finally, sampling was performed at wine estate C in 2014 in 5 different lots.
FIG 1.
Geographic localization of the wine estates in the appellations of the Bordeaux and Bergerac regions. The yellow labels represent wine estates in the white wine Sauternes appellation, and the red labels represent wine estates in red wine Pessac Léognan, Saint Emilion, and Pecharmant appellations. (Template map source, Google Maps.)
TABLE 1.
Summary of samples collected in the Bordeaux and Bergerac regions
| Wine estate | Yr | No. of samplings | No. of isolates |
|||
|---|---|---|---|---|---|---|
| Analyzed by microsatellites | With <4 missing markers | After removing all similar clones | After removing those with >75% similarity to commercial strains | |||
| A | 2012 | 2 | 55 | 54 | 52 | 47 |
| 2014 | 4 | 120 | 114 | 110 | 110 | |
| B | 2012 | 3 | 120 | 118 | 72 | 71 |
| 2013 | 3 | 48 | 46 | 35 | 19 | |
| 2014 | 2 | 60 | 55 | 55 | 49 | |
| C | 1992 | 5 | 43 | 43 | 43 | 43 |
| 1993 | 2 | 32 | 25 | 25 | 18 | |
| 2002 | NAa | 6 | 3 | 2 | 2 | |
| 2007 | NA | 2 | 2 | 2 | 2 | |
| 2011 | NA | 19 | 15 | 15 | 14 | |
| 2014 | 5 | 148 | 129 | 106 | 105 | |
| D | 2012 | 1 | 12 | |||
| E | 2013 | 1 | 9 | |||
| F | 2012 | 1 | 28 | |||
| Total | 653 | 604 | 517 | 529b | ||
NA, not applicable.
A total of 480 of these isolates were from estates A, B, and C.
Strain isolation.
Different dilutions (10−4, 10−5, and 10−6) of the collected samples were plated onto YPD (yeast extract, 1% [wt/vol]; peptone, 1% [wt/vol]; glucose, 2% [wt/vol]; agar, 2%, [wt/vol]) with 100 μg · ml−1 of chloramphenicol and 150 μg · ml−1 of biphenyl to delay bacterial and mold growth. A maximum of 40 randomly chosen colonies were collected after incubation (2 days at 26°C). After two subclonings on YPD plates, each colony was stored in glycerol (30% [vol/vol]) at −80°C.
Additional isolates.
For wine estate C, S. cerevisiae isolates collected since 1992 that were kept in the laboratory collection at −80°C were added to the data set collection, increasing the original data set sampled by 102 new isolates (Table 1).
As a possible external group, 49 new isolates collected from 3 red wine estates belonging to 3 different Bordeaux and Bergerac appellations were added to the data set. Appellation Saint Emilion was represented by wine estate D, Pessac Léognan by wine estate E, and Bergerac by wine estate F.
In addition to cellar samples, 33 yeast strains from diverse origins whose genomes have been sequenced (3, 4) (see Table S1 in the supplemental material) and 35 commercial wine strains (see Table S2 in the supplemental material) widely used as yeast starters were added to the data set.
Molecular methods.
Considering that all yeasts collected at the stage of wine must fermentations where 75% of the alcohol is fermented would very likely belong to the S. cerevisiae species and that this technique provides complete genotypes only for S. cerevisiae strains (32), all colonies were directly analyzed by microsatellites. For each of them, a small amount of fresh colony was suspended in 50 μl of Milli-Q water, and 7 μl of this suspension was dropped on an FTA card for DNA preservation. The samples were then genotyped using 2 multiplex PCRs of 8 and 7 microsatellite loci, respectively, for mixtures 1 and 2 (see Table S3 in the supplemental material) (32–37). Mixtures were prepared in a total volume of 84 μl for 8 samples, with 50 μl of 2× Qiagen Multiplex PCR master mix, 15.5 μl primers, and 18.5 μl water. Mixture 1 had 8 multiplexed primers, and mixture 2 had the other 7; each of them had a specific concentration as specified in Table S3 in the supplemental material. The PCRs were run in a final volume of 12 μl containing 10.5 μl mix and 2 μl of cell suspension. The following PCR program was used: initial denaturation at 95°C for 15 min, followed by 35 cycles of 95°C for 30 s, 57°C for 2 min, 72°C for 1 min, and a final extension at 60°C for 30 min. The PCR products were sized on an ABI3730 DNA analyzer (Applied Biosystems) using size standard 600LIZ (GeneScan).
Data analyses.
ABI3730 genotyping results were read using GeneMarker (V2.4.0, Demo). The presence of a missing value was allowed up to a maximum of 3 markers per sample. Estimation of population diversity was by rarefaction of 10,000 individuals repeated 10 times. The Shannon index (H′), the equitability index (J′), and the inverse Simpson diversity index (D − 1) were calculated with EstimateS (V9) (38) using the individual-based abundance method for intracellar analysis and sample-based abundance data for the whole-region sampling. H′ was determined with the following equation: , where S is the total number of genotypes in the population and Pi is the proportion of a specific genotype in the data set. D was determined with the following equation: . The term Pi was calculated as follows: Pi = Ni/N, where Ni is the number of individuals for a specific genotype and N is the total number of unique genotypes. GenClone (V2.0) software was used to remove strains with exactly similar profiles, resulting from potential clonal expansion, from our data set (39). Observed and expected heterozygosity, Fst (genetic distance), and analysis of molecular variance (AMOVA) were determined using Arlequin (V3.5.2.2) software (40).
SplitsTree V4.12.6 (41) was used to reconstruct a neighbor net phylogenetic network for S. cerevisiae using Bruvo's distance (42), calculated using the R program (43) with the following packages: ape (44) and poppr (45). The population structure was evaluated using a Bayesian clustering method with the software InStruct, which does not account for the Hardy-Weinberg equilibrium (46). Five chains of 150,000 iterations with a burn-in of 5,000 were run for a possible population number (K) of 1 to 25. The most likely number of ancestry populations was selected by choosing the lowest deviance information criterion (DIC). Ancestry profiles were drawn as bar plots from the Instruct output, using a different color for each inferred ancestral population under the R statistical environment. The contribution of each population was then evaluated with ObStruct software (47).
RESULTS
Cellar sample diversity.
To investigate S. cerevisiae population diversity in the typical appellation of Sauternes in the Bordeaux region, 3 wine estates, A, B, and C, were selected. Samples of spontaneously fermenting must were taken before mutage and at different times of the harvest corresponding to selective pickings. A total of 653 colonies were collected on the wine estates between 1992 and 2014 and analyzed by 15 microsatellite markers. Isolates with genotypes with missing values at more than 3 markers were removed from the data set. A summary of the samplings, years, and numbers of S. cerevisiae colonies with completed microsatellite genotypes collected is provided in Table 1. For wine estate C, S. cerevisiae strains from the laboratory collection that were isolated between 1992 and 2011 were included in the study (29). After microsatellite analyses, 43 additional S. cerevisiae isolates with full microsatellite genotypes were kept, giving a final data set of 604 S. cerevisiae isolates for further analysis (Table 1).
In order to compare the diversities of the yeast populations obtained from the three wine estates, we calculated three diversity indices using EstimateS: the Shannon index (H′), which measures the diversity within a population and takes into account both richness and evenness; the inverse Simpson index (1 − D), which gives more weight to common or dominant species; and the Pielou evenness index (J′). The different indices were evaluated on the basis of the number of different genotypes (Table 2) and on the standard deviations of H′ and 1 − D; all the results were significantly different. The Shannon index (H′) showed strong diversity in all 3 wine estates (over 4.50), with a slight decrease of diversity for wine estate B. The Pielou index (J′) was always close to 1, suggesting that genotypes have similar abundances within the population. The inverse Simpson index (1 − D) results were in accordance with the H′ and J′ indices, with high values over 0.97. When considering wine estates as a sample of the Sauternes appellation, the Pielou index value was even higher, reaching 100% diversity and again confirming the results from the inverse Simpson and diversity indices. The diversity index of the Sauternes S. cerevisiae population was similar to the diversity index obtained for the Merlot red wine cellar S. cerevisiae population (270 individuals), with H′ and J′ indices of 5.38 and 0.96, respectively (data not shown). The whole-appellation diversity, evaluated by the rarefaction analyses, estimated a number of unique genotypes on the whole-appellation scale greater than 72,533 (with 95% confidence limits of 7,010 to 138,057) and evaluated it with a sampling design including more than 1,000 S. cerevisiae samples throughout the region to achieve full diversity. The small number of genotypes shared between cellar sampling populations explains this high diversity.
TABLE 2.
Diversity analysis of the three wineries of the Sauterne appellation
| Parameter | Valuea |
|||
|---|---|---|---|---|
| A | B | C | Total | |
| No. of individuals | 168 | 219 | 217 | 604 |
| H′ | 5.04 | 4.57 | 5.11 | 6.22 |
| J′ | 0.995 | 0.90 | 0.99 | 1.00 |
| 1 − D | 0.993 | 0.97 | 0.992 | 0.998 |
The analyses are based on the 604 genotypes obtained after microsatellite typing of the isolates from the 3 different wine estates of the Sauternes appellation. The individual-method estimate was used for each cellar and the sample-based method for the estimates for the whole region.
Strain genetic relationships.
Due to clonal expansion of individual genotypes during fermentation, it was necessary to remove all identical genotypes within each sampling site before accessing the genetic relationships. From the initial 604 S. cerevisiae isolates, the GenClone software inferred 503 unique genotypes, grouping a total of 517 isolates from all 3 wine estates with 14 genotypes shared between the 3 wine estates (Table 1). Thirty-seven industrial S. cerevisiae strains widely used in the Bordeaux region and in Sauternes appellations specifically were then added to this data set in order to detect the potential presence of yeast starters within cellar populations, and 49 S. cerevisiae isolates from Bordeaux region Merlot must fermentations were also included in the analysis (Table 1). Finally, 33 S. cerevisiae strains of various origins whose genomes have been recently sequenced were also included as an outgroup in our data set.
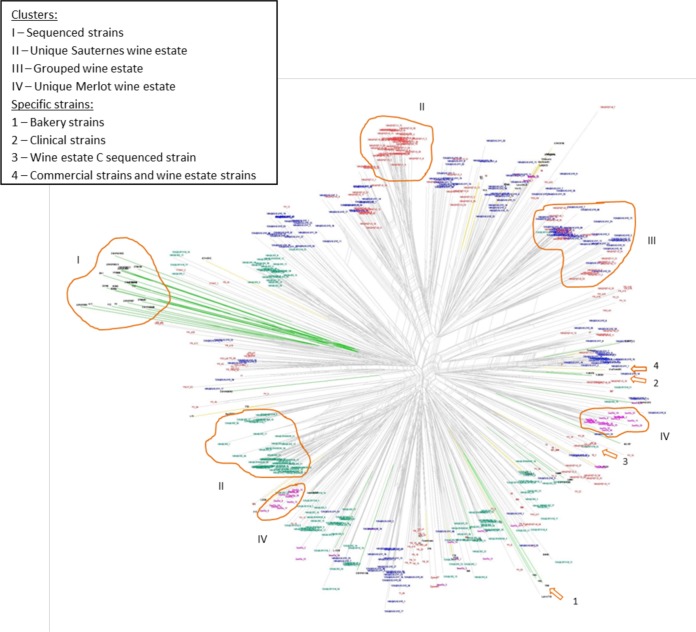
For the relationship between cellars and commercial strains, the 636 isolates were accessed from a phylogenic network built from the Bruvo's pairwise distance matrix (Fig. 2). As expected, one cluster gathered the sequenced strains of different origins (group I), and most of the sequenced strains originating from a wine environment clustered with our wine isolates, except for clinical strains (YJM978, -981, and -975) and baker strains (YS2, YS4, and YS9), which were grouped in the same branches, including Sauternes strains and commercial strains (arrows 2 and 1, respectively). Note that the sequenced strain YIIc17_E5, the genome for which has been sequenced, was isolated from the Sauternes region in 1992 and clustered with strains isolated on wine estate C in 1992 (arrow 3). Concerning the Sauternes cellar S. cerevisiae population, some branches clustered isolates from one wine estate with very close genetic relationships (group II), suggesting clonal variants. Those branches were mostly observed for wine estates A and C and to a lesser extent for wine estate B. Other branches were composed of clusters mixing wine estate B and C isolates, with only rare isolates from wine estate A (group III). Finally, there also appeared to be possible links between cellar strains and commercial strains (arrow 4), whichever wine estate was considered. Concerning Merlot isolates, all the strains from wine estate D and most from wine estate F clustered apart from the others (group IV), whereas a few isolates from wine estates E and F were spread over the network.
FIG 2.
Neighbor net network of 636 strains from the cellars of the 3 wine estates, 3 commercial strains, 33 strains from the S. cerevisiae sequenced database, and 49 strains from Merlot must fermentations. The network was constructed from Bruvo's distance between strains based on the polymorphism at 15 loci. Green, wine (red dots) and nonwine sequenced strains; yellow, commercial strains; green labels, domain A; blue labels, domain B; red labels, domain C; pink labels, wine estates D, E, and F.
To further compare the populations by wine estate, Fst statistics between all Sauternes and Merlot wine estates were calculated (Table 3). All the population comparisons indicated a significant difference (P < 0.001). As suggested by the individual network, there was higher differentiation between wine estates A and B (0.109) than between wine estates B and C (0.038), with the latter being more than twice as low as the comparison of A and C (0.145, the highest Sauternes pairwise Fst). Pairwise Fst statistics between Sauternes wine estates and Merlot wine estates ranged from 0.103 to 0.165, indicating a moderate but still notable differentiation. However, pairwise Fst statistics between Sauternes wine estates (A and C) could be higher than pairwise Fst statistics between Sauternes and Merlot wine estates (e.g., A and E or B and F), whereas the geographical distance was greater in these cases. Surprisingly, the pairwise Fst distances between Merlot wine estate D and any other Sauternes or Merlot wine estates were high (0.222 to 0.399), indicating strong differentiation, shown by the external positions of individuals in the network, which might also be explained by the lower number of strains from the sample.
TABLE 3.
Pairwise Fst statistic values between the 6 different Sauternes and Merlot wine estates after combining the different years of sampling
| Estate |
Fst or P value for estatea: |
|||||
|---|---|---|---|---|---|---|
| A (162) | B (162) | C (193) | D (12) | E (9) | F (28) | |
| A | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | |
| B | 0.109 | 0.001 | 0.001 | 0.001 | 0.001 | |
| C | 0.145 | 0.038 | 0.001 | 0.001 | 0.001 | |
| D | 0.222 | 0.299 | 0.325 | 0.001 | 0.001 | |
| E | 0.103 | 0.105 | 0.139 | 0.327 | 0.001 | |
| F | 0.147 | 0.120 | 0.165 | 0.399 | 0.138 | |
Fst values are in lightface, and P values are in boldface. All values are significant (P < 0.001). Italics indicate a comparison between Sauternes wine estates; underlining indicates a comparison between Merlot wine estates. The numbers of strains are in parentheses.
As highlighted in the network, some cellars' isolates appeared to be very close to commercial strains. Cellar isolates were further considered genetically related to the industrial strains when they shared at least 75% of the alleles. For example, one isolate from wine estate B was related to commercial strain VL3 and 4 isolates to strain X5. For commercial strain UvaFerm BC, 20 cellar isolates were related to the starter strain, 1 obtained from wine estate A, 18 from wine estate B, and 1 from wine estate C. Finally, 4 cellar isolates from wine estate A were related to commercial strain Levuline BRG. Commercial strains VL3, X5, UvaFerm BC, and Levuline BRG have been frequently used in the Sauternes region during the last 30 years, even by wine estates following organic agricultural practices. Nevertheless, only 7% of all Sauternes strains were considered genetically related to specific commercial strains, indicating a minor but substantial relationship between cellar and commercial strains (Table 1). In order to limit the potential impact of yeasts related to commercial starters on the detection of yeast population structures, they were removed from the data set, and differentiations between Sauternes and Merlot wine estates were estimated again, but the results did not change in a substantial manner (data not shown).
Population structure.
An AMOVA was then further performed in order to understand how genetic variations at these 15 microsatellite loci are structured (Table 4). For wine estate C, only samples from 2014, 1993, and 1992 were taken into account for the analysis, since isolates from several samples were available, and different groups were tested according to the year of sampling or the wine estate. The contributions of variation among individuals within groups (AIWP) always explained most of the global variation, ranging from 60 to 94% of the total variance. The percentage comparisons of variation among groups (AG) and among populations within a group (APWG) indicate different patterns. The comparison of genetic diversity from the different wine estates indicates that the wine estate has the highest impact on genetic diversity, as this factor explains from 11.7 to 31% of the genetic variability, according to the comparison. Wine estate A appeared to be more differentiated from wine estate C (31%) than from B (12.3%), whereas B and C were similarly differentiated (12.5%). Notably, these comparisons led to a moderate within-group variability (from 4.2 to 8.8%, except for the B-C comparison, with 14.1%) and a low variation among individuals within a population (AIWP). On the other hand, the vintage contributed less to the global variation for close vintages on a wine estate, with 0 to 6% global variation and a low intersample variation (5 to 7% variation in APWG) and the highest values for variation AIWP (88.6 to 94.5%). However, winery C presents a unique picture, as the differences between the most distant vintages explain the highest genetic variation (13.1 to 19.3%), whereas the differences between the 1992 and 1993 vintages were the lowest, similar to what can be observed for 2012 and 2013 and for 2013 and 2014 for wine estate B. Interestingly, this wine estate also shows the highest sample-to-sample variation (APWG explains 12 to 16% of the global variation).
TABLE 4.
AMOVA analyses, Fst values, and distribution of variance components based on microsatellite data for S. cerevisiae isolates obtained from the indicated groups of wine estates and vintages
| Fixed parametera | Variable parametera | % variation |
Fst | P (r < 0) | ||
|---|---|---|---|---|---|---|
| AG | APWG | AIWP | ||||
| A | 2012, 2014 | 6.23 | 5.14 | 88.62 | 0.113 | <0.000001 |
| B | 2012, 2013, 2014 | 1.66 | 5.06 | 93.26 | 0.063 | <0.000001 |
| 2012, 2013 | 3.75 | 5.16 | 91.08 | 0.089 | <0.000001 | |
| 2012, 2014 | 1.56 | 3.92 | 94.51 | 0.055 | <0.000001 | |
| 2013, 2014 | 0 | 7.17 | 93.21 | 0.067 | <0.000001 | |
| C | 2014, 1993, 1992 | 13.14 | 15.02 | 71.83 | 0.281 | <0.000001 |
| 1992, 1993 | 0 | 11.91 | 91.14 | 0.088 | <0.000001 | |
| 1992, 2014 | 13.91 | 15.51 | 70.57 | 0.294 | <0.000001 | |
| 1993, 2014 | 19.31 | 16.28 | 64.41 | 0.355 | <0.000001 | |
| 2012 | A, B | 11.69 | 4.19 | 84.11 | 0.158 | <0.000001 |
| 2014 | A, B, C | 22.42 | 8.68 | 68.89 | 0.311 | <0.000001 |
| A, B | 12.27 | 4.06 | 83.66 | 0.163 | <0.000001 | |
| A, C | 31.04 | 8.77 | 60.18 | 0.398 | <0.000001 | |
| B, C | 12.53 | 14.12 | 73.34 | 0.266 | <0.000001 | |
Vintages (years) or wine estates (A, B, and C).
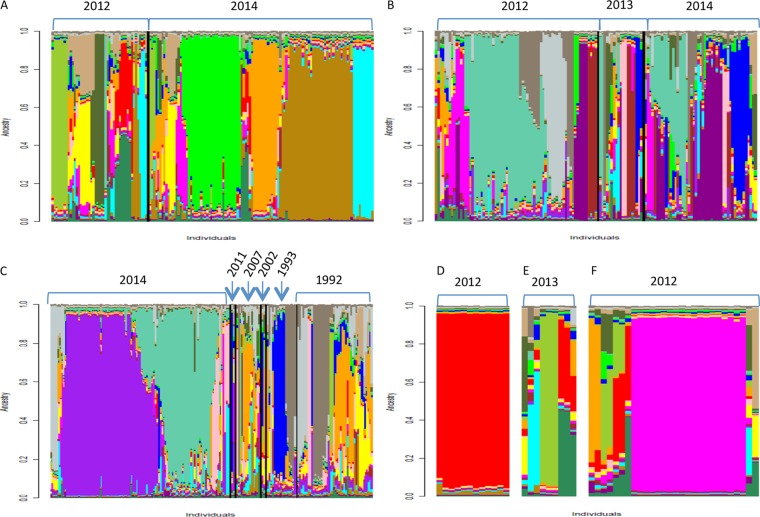
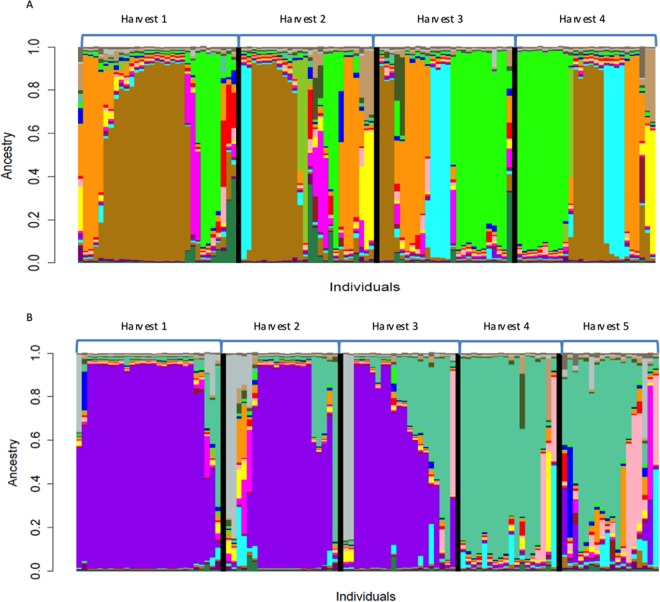
We used InStruct to evaluate the population structure from shared ancestry. The deviance information criterion indicated the most likely population number (K) to be a value of 19 (Fig. 3). In an overall view, whatever year or wine estate was considered, numerous strains were composed of a mosaic of ancestral subpopulations. Unique ancestral populations associated with a given wine estate were highlighted (A, D, and F in 2014). Strains from wine estate E presented mainly a mosaic ancestry, which may explain their dispersion over the individual network. Wine estate A shared few ancestral populations with wine estates B and C, whereas one of the main ancestral populations of wine estate C was shared with wine estate B, which also accounts for the mixed group seen in the network. The relationship between wine estates B and C was illustrated by shared ancestral populations in 2014 and, to a lesser extent, in 1992 and 1993. Except for wine estate B, where ancestral populations persisted through vintages (2012 to 2014), for wine estates A and C, only a few ancestral populations were shared from one year to another. Moreover, in 2014, 2 new ancestral subpopulations appeared to be predominant and absent from the former vintages for both wine estates A and C. When focusing on the population structures of wine estates A and C in 2014, a temporal succession of two ancestral subpopulations was clearly related to the different batches associated with the selective picking of noble rotted grapes (Fig. 4).
FIG 3.
Inference of populations using the InStruct program on the 604 S. cerevisiae cellar strains with the optimal K value of 19 classified according to years for each wine estate. (A) Wine estate A. (B) Wine estate B. (C) Wine estate C. (D) Wine estate D. (E) Wine estate E. (F) Wine estate F.
FIG 4.
(A) Inference of populations using the InStruct program on the 110 S. cerevisiae strains with the optimal K value of 19. The strains originated from wine estate A in 2014 and were classified according to the 4 different harvest batches. (B) Inference of populations using the InStruct program on the 105 S. cerevisiae strains with the optimal K value of 19. The strains originated from wine estate C in 2014 and were classified according to the 5 different harvest batches.
The ObStruct program permits an evaluation of the significance of different factors in the ancestry profile obtained from Instruct. Here, we can test the effect of the wineries on the population of Sauternes or Merlot wine estates (see Fig. S1 and Table S4a in the supplemental material). The two Merlot wine estates D and F had a strong influence on the global population structure. Sauternes wine estate B also contributed to this population structure shaping, but at a lower level. To focus on the Sauternes appellation, Merlot wine estates were removed from the data set. The ObStruct results for Sauternes wine estates only (see Table S4b in the supplemental material) showed that wine estate A still had the strongest influence on the shape of the Sauternes population structure, in agreement with the fact that it is clearly distinguished from the other 2 wine estates. Wine estates B and C had a lesser influence on the population structure, with B contributing slightly more than C.
DISCUSSION
Sauternes is a particular appellation of the Bordeaux region producing high-quality sweet wines. The development of noble rot on grapes results in the production of highly concentrated grape musts and typical wines (19). Fermentation conditions are highly stressful for wine yeast, mainly due to a high sugar content and a low level of assimilable nitrogen. A total of 653 isolates were collected over 3 consecutive years (2012, 2013, and 2014) on 3 different wine estates. Moreover, 102 additional strains collected from 4 to 23 years ago on wine estate C were added to our population sample. A highly discriminating method based on 15 microsatellite markers specific to S. cerevisiae was used for molecular typing at the strain level. In comparison to previous studies based on other methods, such as PFGE, mtDNA RFLP, and inter-delta analysis (6, 8, 48), this S. cerevisiae ecological study used deep sampling and relied on the robustness of the microsatellite marker method (36, 49–51), performed with 15 loci as microsatellite markers and providing more sensitivity than previous studies. Multilocus microsatellite analysis allowed us to evaluate the genetic diversity of our population. A total of 503 genotypes were revealed from an initial population of 653 S. cerevisiae isolates (77% of the different genotypes), indicating high genetic diversity, and 97% of the genotypes were wine estate specific. By sampling 21 different white and red ferments across three different regions in New Zealand, Gayevskiy and Goddard (12) obtained 353 S. cerevisiae isolates and 274 genotypes (78%) using 10 microsatellite markers, which is similar to our data but with lower diversity. However, our estimate of yeast diversity suggests that the Sauternes region is expected to contain an extremely high diversity of S. cerevisiae strains, with an underlying population of more than 72,533 unique genotype strains (with a wide confidence interval), a figure far higher than the estimate of 1,700 inferred for the New Zealand vineyard (52). The diversity index obtained for the Sauternes S. cerevisiae population was also similar to the diversity index obtained for the Merlot red wine cellar S. cerevisiae population. Several causes might explain these differences between our data and those obtained from New Zealand vineyards: the first obvious explanation might lie in the use of additional markers, enabling a deeper exploration of the diversity; however, as the most polymorphic markers are shared by the two studies, we doubt that this factor alone explains the differences. Because damaged grape berries may be very rich depositories of S. cerevisiae, in comparison to sound berries (28), we might expect to obtain higher diversity index values for botrytized ferment populations. However, on the contrary, the specific botrytized grape must composition, with a high sugar content, and the interaction with the action of Botrytis cinerea may constitute a highly selective medium, potentially limiting S. cerevisiae strain diversity. Finally, the recent origin of the New Zealand vineyard and its associated yeast population (M. R. Goddard, personal communication) may also provide another explanation. Indeed, the yeast diversity may carry the signature of the initial bottleneck associated with the founder effect of its introduction into New Zealand, resulting today in lower diversity despite its partial recovery.
The main objectives of the study were to define the population genetic structure and diversity of S. cerevisiae on both the Sauternes appellation and wine estate scales. The impact of commercial strains on the diversity of endogenous wine yeast strains is still controversial, since some authors have shown that the use of active dry yeasts reduced the variability of wine cellar strains (14), whereas other studies did not evidence any impact (6, 12, 53). In this study, only 7% of cellar strains were found to be related to 4 commercial strains usually used in sweet and dry white wine making in the Bordeaux region for over 25 years. Moreover, no significant variation in wine estate pairwise Fst values were obtained before and after removing strains genetically related to commercial starters. Despite the past or present use of yeast starters to inoculate dry white wines in the wine estates studied, this practice had a small impact on S. cerevisiae diversity and population genetic structure on the winery scale in the Sauternes region.
AMOVA, pairwise Fst, and ancestry profile and ObStruct analyses showed contrasting results concerning genetic differentiation between populations originating from different wine estates. While population differentiation between wine estate A and wine estates B and C was high, a much smaller differentiation was observed between wine estates B and C. Ancestry profile analysis provided evidence that wine estate B and C populations are mixed to a certain degree. Taking into account the short geographic distance between wine estates A, B, and C (less than 10 km), it is not realistic to postulate that the various degrees of genetic differentiation between wine estate populations are linked to their respective geographic distances. However, one of the possible explanations of the small differentiation between wine estates B and C in comparison to A is the short distance between B and C, which have juxtaposed vineyard plots. At such a short distance, insects like bees, wasps, and fruit flies, as well as birds, which are known to be vectors for yeasts, could have homogenized these yeast populations (51, 54, 55). Humans can also influence the yeast population structure and promote dispersal (49). Wine estates B and C shared seasonal staff and wine-growing equipment during the harvest and fermentation periods, which may also have facilitated exchanges between the S. cerevisiae populations of the two estates. On the very small scale of the appellation, this is an illustration of possible S. cerevisiae dispersion.
During a period of 23 years, strains from wine estate C were collected, and we could observe the systematic persistence of specific ancestral populations that were never dominant on wine estate C. The ancestral populations observed in 1992 and 1993 at winery C were also detected in the sampling performed during the 2012-2014 period on wine estate B but were absent on wine estate A. This result demonstrates, on the small scale of two wine estates, the existence of a local and stable group of strains with shared ancestry over 20 years, as well as the occurrence of multiple yeast population exchanges between the two wine estates over time. The phenotypic traits of this local and long-term stable group of strains would be interesting to investigate, in order to better understand to what extent those ancestral S. cerevisiae populations may contribute to the characteristics and typicality of the wine produced in this area.
Previous consecutive-year follow-up studies reported contrasting results concerning the possible establishment of strains as resident at a given winery (8, 14). The comparison by AMOVA of samples obtained from wine estate C over a long period revealed that the variation between the most distant years (1993 and 2014) provided more differences than a comparison of different samples from the same year or from successive years (1992 and 1993 or 2013 and 2014). From this preliminary analysis, we could hypothesize that time, over the long term, may be a key factor for genetic differentiation between cellar-resident S. cerevisiae populations at a given winery.
Finally, a cellar-associated S. cerevisiae population during the harvest period of 2014 for wine estates A and C was more closely explored. Ancestry profile analysis revealed a clear temporal succession of two main ancestral populations for wine estate C and, to a lesser extent, for wine estate A during the harvest campaign. A characteristic of both wine estates compared to wine estate B was the use of fermented batches to inoculate the others. This method, named “pied de cuve,” was shown to better acclimatize the yeast inoculum to the high sugar content of the fermentation medium. Such a stress factor provokes the upregulation of structural genes involved in glycerol synthesis and intracellular accumulation by S. cerevisiae in response to external osmolarity (56, 57), which results in the formation of acetic acid from acetaldehyde (58). The use of yeasts collected from already fermenting wine is advantageous, since the yeast cells had had the opportunity to acclimate to the high sugar content of the musts and produced less acetic acid than selected starters inoculated directly (19, 59). Our data indicated that the selection of specific ancestral S. cerevisiae populations through successive fermentations may also be favored by the use of subculture on wine estates A and C. Still, the factors that explain the selection of given ancestral populations remain to be elucidated. In the case of Sauternes winemaking, the sugar content of the musts, which is dramatically increased during harvest, with concentrations as high as 40 to 45% (wt/vol) at the end of the campaign, is probably a key parameter. In the case of wine estate C, this selection of one ancestral population during the harvest period was highlighted but raises the underlying question, to what extent does the increase of the must's sugar content explain this temporal succession?
Supplementary Material
ACKNOWLEDGMENTS
We thank Chateau Guiraud, Chateau Yquem, and Chateau Climens, who kindly provided fermented samples.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03627-15.
REFERENCES
- 1.Fay J, Benavides J. 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet 1:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legras J-L, Merdinoglu D, Cornuet J-M, Karst F. 2007. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 3.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Nguyen Ba AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. 2009. Comprehensive polymorphism survey elucidates population structure of S. cerevisiae. Nature 458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cromie G, Hyma K, Ludlow C, Garmendia-Torres C, Gilbert T, May P, Huang A, Dudley A, Fay J. 2013. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 3:2163–2171. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valero E, Cambon B, Schuller D, Casal M, Dequin S. 2007. Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Res 7:317–329. doi: 10.1111/j.1567-1364.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- 7.Cappello MS, Bleve G, Grieco F, Dellaglio F, Zacheo G. 2004. Characterization of Saccharomyces cerevisiae strains isolated from must of grape grown in experimental vineyard. J Appl Microbiol 97:1274–1280. doi: 10.1111/j.1365-2672.2004.02412.x. [DOI] [PubMed] [Google Scholar]
- 8.Vigentini I, De Lorenzis G, Fabrizio V, Valdetara F, Faccincani M, Panont CA, Picozzi C, Imazio S, Failla O, Foschino R. 2015. The vintage effect overcomes the terroir effect: a three year survey on the wine yeast biodiversity in Franciacorta and Oltrepò Pavese, two northern Italian vine-growing areas. Microbiology 161:362–373. doi: 10.1099/mic.0.000004. [DOI] [PubMed] [Google Scholar]
- 9.Schuller D, Casal M. 2007. The genetic structure of fermentative vineyard-associated Saccharomyces cerevisiae populations revealed by microsatellite analysis. Antonie Van Leeuwenhoek 91:137–150. doi: 10.1007/s10482-006-9104-8. [DOI] [PubMed] [Google Scholar]
- 10.Combina M, Elía A, Mercado L, Catania C, Ganga A, Martinez C. 2005. Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int J Food Microbiol 99:237–243. doi: 10.1016/j.ijfoodmicro.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Ciani M, Mannazzu I, Marinangeli P, Clementi F, Martini A. 2004. Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Antonie Van Leeuwenhoek 85:159–164. doi: 10.1023/B:ANTO.0000020284.05802.d7. [DOI] [PubMed] [Google Scholar]
- 12.Gayevskiy V, Goddard MR. 2012. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J 6:1281–1290. doi: 10.1038/ismej.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frezier V, Dubourdieu D. 1992. Ecology of yeast strains Saccharomyces cerevisiae during spontaneous fermentation in Bordeaux winery. Am J Enol Vitic 43:375–380. [Google Scholar]
- 14.Beltran G, Torija MJ, Novo M, Ferrer N, Poblet M, Guillamón JM, Rozès N, Mas A. 2002. Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst Appl Microbiol 25:287–293. doi: 10.1078/0723-2020-00097. [DOI] [PubMed] [Google Scholar]
- 15.Magyar I. 2011. Botrytized wines. Adv Food Nutr Res 63:147–206. doi: 10.1016/B978-0-12-384927-4.00006-3. [DOI] [PubMed] [Google Scholar]
- 16.Ribereau-Gayon P, Lafon-Lafourcade S, Dubourdieu D, Lucmaret V, Larue F. 1979. Métabolisme de Saccharomyces cerevisiae dans le moût de raisins parasités par Botrytis cinerea. Inhibition de la fermentation, formation d'acide acétique et de glycérol. C R Hebd Seances Acad Sci 289:441–444. [Google Scholar]
- 17.Bely M, Masneuf-Pomarède I, Dubourdieu D. 2005. Influence of physiological state of inoculum on volatile acidity production by Saccharomyces cerevisiae during high sugar fermentation. J Int Sci Vigne Vin 39:191–197. [Google Scholar]
- 18.Ribereau-Gayon P, Glories Y, Maujean A, Dubourdieu D. 2000. Micro-oxygenation presents promise with potential peril for quality winemaking, p 83–88. In Rieger T. (ed), The chemistry of wine and stabilization and treatments. John Wiley & Sons, New York, NY. [Google Scholar]
- 19.Bely M, Masneuf I, Dubourdieu D. 2005. Influence of physiological state of inoculum on volatile acidity production by “Saccharomyces cerevisiae” during high sugar fermentation. J Int Sci Vigne Vin Int J Vine Wine Sci 39:191–198. [Google Scholar]
- 20.Nisiotou A., Nychas GJE. 2007. Yeast populations residing on healthy or Botrytis-infected grapes from a vineyard in Attica, Greece. Appl Environ Microbiol 73:2765–2768. doi: 10.1128/AEM.01864-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naumov GI, Naumova ES, Antunovics Z, Sipiczki M. 2002. Saccharomyces bayanus var. uvarum in Tokaj wine-making of Slovakia and Hungary. Appl Microbiol Biotechnol 59:727–730. doi: 10.1007/s00253-002-1077-6. [DOI] [PubMed] [Google Scholar]
- 22.Sipiczki M. 2003. Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int J Syst Evol Microbiol 53:2079–2083. doi: 10.1099/ijs.0.02649-0. [DOI] [PubMed] [Google Scholar]
- 23.Fleet GH, Lafon-Lafourcade S, Ribéreau-Gayon P. 1984. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl Environ Microbiol 48:1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills DA, Johannsen EA, Cocolin L. 2002. Yeast diversity and persistence in botrytis-affected wine fermentations. Appl Environ Microbiol 68:4884–4893. doi: 10.1128/AEM.68.10.4884-4893.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nisiotou A, Spiropoulos A, Nychas G-JE. 2007. Yeast community structures and dynamics in healthy and botrytis-affected grape must fermentations. Appl Environ Microbiol 73:6705–6713. doi: 10.1128/AEM.01279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antunovics Z, Csoma H, Sipiczki M. 2003. Molecular and genetic analysis of the yeast flora of botrytized Tokaj wines. Bull OIV 76:380–397. [Google Scholar]
- 27.Naumov GI, Masneuf I, Naumova ES, Aigle M, Dubourdieu D. 2000. Association of Saccharomyces bayanus var. uvarum with some French wines: genetic analysis of yeast populations. Res Microbiol 151:683–691. doi: 10.1016/S0923-2508(00)90131-1. [DOI] [PubMed] [Google Scholar]
- 28.Mortimer R, Polsinelli M. 1999. On the origins of wine yeast. Res Microbiol 150:199–204. doi: 10.1016/S0923-2508(99)80036-9. [DOI] [PubMed] [Google Scholar]
- 29.Masneuf I, Dubourdieu D. 2000. Rôle de la souche de levures sur les combinaisons du dioxyde de soufre des vins issus de raisins botrytisés et passerillés. J Int Sci Vigne Vin Int J Vine Wine Sci 34:27–32. [Google Scholar]
- 30.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight S, Klaere S, Fedrizzi B, Goddard MR. 2015. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep 5:14233. doi: 10.1038/srep14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legras J-L, Ruh O, Merdinoglu D, Karst F. 2005. Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int J Food Microbiol 102:73–83. doi: 10.1016/j.ijfoodmicro.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Bradbury JE, Richards KD, Niederer HA, Lee SA, Dunbar PR, Gardner RC. 2006. A homozygous diploid subset of commercial wine yeast strains. Antonie Van Leeuwenhoek 89:27–37. doi: 10.1007/s10482-005-9006-1. [DOI] [PubMed] [Google Scholar]
- 34.Field D, Wills C. 1998. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci U S A 95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González Techera A, Jubany S, Carrau FM, Gaggero C. 2001. Differentiation of industrial wine yeast strains using microsatellite markers. Lett Appl Microbiol 33:71–75. doi: 10.1046/j.1472-765X.2001.00946.x. [DOI] [PubMed] [Google Scholar]
- 36.Hennequin C, Thierry A, Richard GF, Lecointre G, Nguyen HV, Gaillardin C, Dujon B. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J Clin Microbiol 39:551–559. doi: 10.1128/JCM.39.2.551-559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez MA., Gallego FJ, Martínez I, Hidalgo P. 2001. Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett Appl Microbiol 33:461–466. doi: 10.1046/j.1472-765X.2001.01032.x. [DOI] [PubMed] [Google Scholar]
- 38.Colwell R. 2004. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology 85:2717–2727. [Google Scholar]
- 39.Arnaud-Haond S, Belkhir K. 2007. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7:15–17. [Google Scholar]
- 40.Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 41.Huson D, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. [DOI] [PubMed] [Google Scholar]
- 42.Bruvo R, Michiels NK, D'Souza TG, Schulenburg H. 2004. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- 43.R Development Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 44.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 45.Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer J 2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Williamson S, Bustamante C. 2007. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gayevskiy V, Klaere S, Knight S, Goddard M. 2014. ObStruct: a method to objectively analyse factors driving population structure using Bayesian ancestry profiles. PLoS One 9:e85196. doi: 10.1371/journal.pone.0085196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuller D, Alves H, Dequin S, Casal M. 2005. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol Ecol 51:167–177. doi: 10.1016/j.femsec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Goddard M, Anfang N, Tang R, Gardner R, Jun C. 2010. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol 12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- 50.Schuller D, Cardoso F, Sousa S, Gomes P, Gomes AC, Santos MAS, Casal M. 2012. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS One 7:e32507. doi: 10.1371/journal.pone.0032507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francesca N, Canale D, Settanni L, Moschetti G. 2012. Dissemination of wine-related yeasts by migratory birds. Environ Microbiol Rep 4:105–112. doi: 10.1111/j.1758-2229.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- 52.Knight S, Goddard MR. 2015. Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J 9:361–370. doi: 10.1038/ismej.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cordero-Bueso G, Arroyo T, Serrano A, Valero E. 2011. Remanence and survival of commercial yeast in different ecological niches of the vineyard. FEMS Microbiol Ecol 77:429–437. doi: 10.1111/j.1574-6941.2011.01124.x. [DOI] [PubMed] [Google Scholar]
- 54.Shihata AME-TA, Mrak EM. 1952. Intestinal yeast floras of successive populations of Drosophila. Evolution 6:325–332. doi: 10.2307/2405417. [DOI] [Google Scholar]
- 55.Stefanini I, Dapporto L, Legras J-L, Calabretta A, Di Paola M, De Filippo C, Viola R, Capretti P, Polsinelli M, Turillazzi S, Cavalieri D. 2012. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci U S A 109:13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blomberg A, Adler L. 1989. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol 171:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varela JC, Mager WH. 1996. Response of Saccharomyces cerevisiae to changes in external osmolarity. Microbiology 142:721–731. doi: 10.1099/00221287-142-4-721. [DOI] [PubMed] [Google Scholar]
- 58.Erasmus D, van der Merwe G, van Vuuren HJ. 2003. Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res 3:375–399. doi: 10.1016/S1567-1356(02)00203-9. [DOI] [PubMed] [Google Scholar]
- 59.Ciani M, Comitini F, Mannazzu I, Domizio P. 2010. Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res 10:123–133. doi: 10.1111/j.1567-1364.2009.00579.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.