ABSTRACT
We previously showed that modeled microgravity conditions alter the physiological characteristics of Escherichia coli O157:H7. To examine how microgravity conditions affect bacterial heat stress responses, D values, membrane fatty acid composition, and heat stress-related gene expression (clpB, dnaK, grpE, groES, htpG, htpX, ibpB, and rpoH), E. coli O157:H7 ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895 were cultured under two different conditions: low-shear modeled microgravity (LSMMG, an analog of spaceflight conditions) and normal gravity (NG, Earth-like conditions). When 24-h cultures were heated to 55°C, cells cultured under LSMMG conditions showed reduced survival compared with cells cultured under NG conditions at all time points (P < 0.05). D values of all tested strains were lower after LSMMG culture than after NG culture. Fourteen of 37 fatty acids examined were present in the bacterial membrane: nine saturated fatty acids (SFA) and five unsaturated fatty acids (USFA). The USFA/SFA ratio, a measure of membrane fluidity, was higher under LSMMG conditions than under NG conditions. Compared with control cells grown under NG conditions, cells cultured under LSMMG conditions showed downregulation of eight heat stress-related genes (average, −1.9- to −3.7-fold). The results of this study indicate that in a simulated space environment, heat resistance of E. coli O157:H7 decreased, and this might be due to the synergistic effects of the increases in membrane fluidity and downregulated relevant heat stress genes.
IMPORTANCE Microgravity is a major factor that represents the environmental conditions in space. Since infectious diseases are difficult to deal with in a space environment, comprehensive studies on the behavior of pathogenic bacteria under microgravity conditions are warranted. This study reports the changes in heat stress resistance of E. coli O157:H7, the severe foodborne pathogen, under conditions that mimic microgravity. The results provide scientific clues for further understanding of the bacterial response under the simulated microgravity conditions. It will contribute not only to the improvement of scientific knowledge in the academic fields but also ultimately to the development of a prevention strategy for bacterial disease in the space environment.
INTRODUCTION
Bacteria have evolved numerous strategies that enable them to survive when exposed to environmental stresses (1, 2), including alterations in temperature, pH, and osmotic pressure (3–5). Similarly, bacteria exhibit survival responses under modeled microgravity conditions, which simulate a space-like environment. Recently, we showed that low-shear modeled microgravity (LSMMG) conditions alter the physiological characteristics of the foodborne pathogen Escherichia coli O157:H7 (6). Bacteria actively grew in minimal medium despite a reduced pH. In addition, bacterial cells cultured under LSMMG conditions were larger than those cultured under normal gravity (NG) conditions. Since bacterial resistance to stress changes upon exposure to various environmental conditions (7–9), we can hypothesize that reduced gravity affects stress responses and physiological characteristics.
Heat treatment is the most common method for reducing bacterial populations in foodstuffs. It is therefore important to understand bacterial heat resistance mechanisms, because these determine the effectiveness of thermal interventions (10, 11). Some foods consumed by astronauts are heated prior to consumption (12); therefore, understanding the bacterial heat resistance under microgravity conditions may improve food safety in the space environment. The D value estimates bacterial heat resistance. It is the time required to destroy 90% (1 log cycle) of the target microorganism at a specific temperature. Under NG conditions, E. coli O157:H7 has a large D value because it can survive relatively high temperatures (13). The D value of E. coli O157:H7 under modeled microgravity conditions is not known.
The mechanisms underlying bacterial stress responses, including exposure to microgravity (14), are still not fully understood. There is, however, a relationship between heat stress resistance and membrane fatty acid composition (15). For example, bacterial cells with high membrane fluidity (represented by a lower ratio of unsaturated fatty acids to saturated fatty acids [USFA/SFA]) are more susceptible to heat (16, 17).
Previous studies have sought to explain bacterial stress responses by examining changes in gene expression (15, 18–21). The primary mechanism of cellular protection against heat stress involves the expression of heat shock proteins (HSPs), which act as molecular chaperones (22). Molecular chaperones play essential roles in protein folding, degradation, assembly, and transport; thus, they help thermally damaged proteins regain biological function after heat stress (23, 24). The well-studied heat shock sigma factor σH provides protection against cytoplasmic heat stress by regulating the transcription of HSPs (22, 25). In E. coli, the DnaK-DnaJ-GrpE and GroEL-GroES systems are the most extensively characterized molecular chaperones (26). Others, including the Clp ATPases (ClpB, ClpX, and ClpY), the Hsp90 homolog HtpG, and the small HSPs IbpA and IbpB are also believed to function as molecular chaperones in vivo (27, 28). The transcription levels of relevant genes provide insights into related protein translation; however, it is still not known if there are any differences in the expression of genes encoding HSPs and σH in E. coli O157:H7 under LSMMG conditions. Therefore, changes in the gene expression should be considered alongside changes in membrane fatty acid composition when assessing the bacterial heat stress response.
Although bacterial stress responses under simulated microgravity conditions have been reported (14, 29–34), none of those studies have examined the severe foodborne pathogen E. coli O157:H7. Moreover, it is technically difficult to identify the mechanisms that underlie bacterial heat resistance under microgravity conditions. Here, we examined the effects of the simulated microgravity on heat resistance, membrane fatty acid composition, and gene expression of σH (rpoH) and seven HSP-encoding genes (clpB, dnaK, gorES, grpE, htpG, htpX, and ibpB) in E. coli O157:H7 in a simulated space environment.
MATERIALS AND METHODS
Preparation of bacterial strains.
E. coli O157:H7 ATCC 35150 (acid resistant; human feces isolate), ATCC 43889 (acid adaptable; human feces isolate), ATCC 43890 (acid sensitive; human feces isolate), and ATCC 43895 (acid resistant; ground beef isolate) were used for all experiments (35). The strains were obtained from the Food Microbiology Culture Collection at Korea University (Seoul, South Korea) and stored at −20°C in Luria-Bertani medium (LB; Difco, Sparks, MD, USA) containing 20% glycerol. Prior to use in the experiments, each strain was grown overnight in LB medium at 37°C in a shaking incubator at 225 rpm (VS-8480S; Vision Scientific Co., Ltd., Seoul, South Korea). The initial turbidity of the stationary-phase cultures was measured in a SmartSpec Plus spectrophotometer (Bio-Rad, Hercules, CA, USA).
Generation of normal and microgravity conditions.
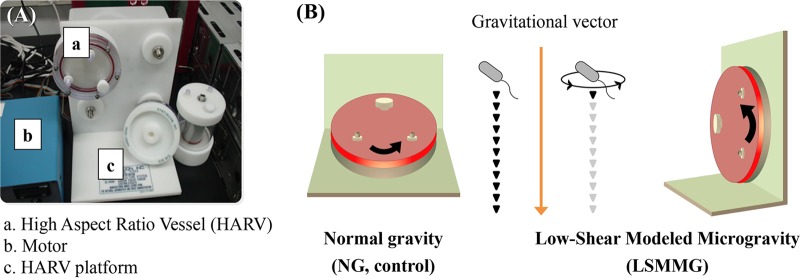
A rotary cell culture system (RCCS; Synthecon, Houston, TX, USA) and a high-aspect-ratio vessel (HARV; Synthecon) were used to generate LSMMG and NG conditions, as previously described (6) (Fig. 1). The HARV apparatus was filled with diluted (1:500) overnight cultures (50 ml) with zero headspace that were then maintained in a constant state of suspension. To simulate LSMMG conditions, the vessels were rotated at 25 rpm around a horizontal axis to offset gravitational forces with hydrodynamic forces (centrifugal, Coriolis, and shear forces) (29). NG conditions were provided by rotating the vessels around a vertical axis. All incubations were performed at 37°C. A gas-permeable membrane on the back of the HARV bioreactor allowed air exchange throughout the experiments.
FIG 1.
(A) High-aspect-ratio vessel-rotary cell culture system used to generate simulated microgravity conditions in ground-based investigations. (B) Differential rotation of the vessel perpendicular or parallel to the gravitational vector generates NG (Earth-like) or LSMMG (spaceflight analog) conditions, respectively.
Survival of E. coli O157:H7 during heat treatment.
Cells cultured under LSMMG or NG conditions for 6 h (exponential phase) and 24 h (stationary phase) were removed through a port on the front plate of the HARV and added to 10 ml of fresh LB medium (1:100 dilution) in a glass culture tube (18-mm internal diameter by 150 mm in length). Prior to use, each fresh-medium-containing culture tube was prewarmed by immersion in a water bath (Vision Science Co., Incheon, South Korea). The survival of E. coli O157:H7 was examined at 55°C. After heat treatment, culture tubes were removed from the water bath at the appropriate time intervals (10, 20, or 30 min), and samples were serially diluted 10-fold in M9 minimal medium with no glucose. The diluted samples were then spread plated on LB agar (Difco) and incubated for 24 h at 37°C. Bacterial counts were expressed as log CFU per milliliter. All experiments were repeated six times.
Determination of D values.
D values (time to inactivate 90% of the bacterial population at a specific temperature) were determined by plotting the mean values of the number of survivors (log CFU per milliliter) against time at 55°C. This was done for each bacterial cell culture exposed to both gravity treatments for 6 and 24 h. The linear fit for survivor plots was determined by linear regression in the Sigma Plot 10.0 software (Systat Software, Richmond, CA, USA), and the D values were calculated as the negative reciprocal of the slope for each regression line.
Determination of membrane fatty acid composition.
Bacterial pellets (about 50 mg) were collected by centrifugation (13,000 × g, 10 min) after culture under LSMMG or NG conditions for 24 h. Fatty acid methyl esters were then extracted as described in MIDI Technical note no. 101, with a slight modification (15). Fatty acid composition was analyzed using an Agilent gas chromatograph (model 7890A; Agilent Technologies, Santa Clara, CA, USA) equipped with a flame-ionizing detector. Fatty acids were separated on a DB-Wax column (25 m length by 0.2 mm inner diameter [i.d.] by 0.33-μm film thickness; Agilent Technologies). The conditions for gas chromatography were the following: sample (1.0 μl; split ratio, 20) was injected at an injector temperature of 250°C; the initial oven temperature was 50°C, held for 1 min; and the oven temperature was subsequently increased to 200°C at a rate of 25°C/min for 5 min, and then to 230°C at a rate of 3°C/min for 20 min. The detector temperature was held at 280°C. Helium was used as the carrier gas. The data were expressed as percentage of composition, calculated as the molar ratio of each individual fatty acid to the total fatty acid content. All experiments were conducted in triplicate.
RNA extraction and cDNA synthesis.
After cell culture under LSMMG or NG conditions for 24 h, total RNA was extracted using TRIzol reagent (Invitrogen, Grand Island, NY, USA), according to the manufacturer's instructions. The total RNA concentration was determined using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The quality of the RNA samples was deemed acceptable if the A260/280 ratio was 1.9 to 2.1 (36).
RNA was reverse transcribed using high-capacity cDNA reverse transcription kits (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. In brief, 0.5 μg of RNA was reverse transcribed with 10× random primers, 25× dinucleoside triphosphate (dNTP) mix (100 mM), 10× buffer, 1.0 μl of MultiScribe reverse transcriptase, and 1.0 μl of RNase inhibitor (total reaction volume, 20 μl). cDNA was synthesized in a thermocycler (Thermo Fisher Scientific, Rockford, IL, USA) under the following cycling conditions: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min, followed by a 4°C hold to stop the reaction.
Real-time RT-PCR for heat stress-related gene expression.
Expression of the stress-related genes in E. coli O157:H7 under LSMMG and NG conditions was evaluated by real-time reverse transcription-PCR (RT-PCR). Each 25-μl reaction mixture contained 2 μl of reverse-transcribed cDNA, 12.5 μl of Maxima SYBR green/ROX quantitative PCR (qPCR) master mix (Thermo Scientific, Hampton, NH, USA), a concentration of 0.2 μM of each primer, and 5.5 μl of nuclease-free water. The Maxima SYBR green/ROX qPCR master mix contained the following components: Maxima Hot Start Taq DNA polymerase, dNTPs, SYBR green I dye, ROX passive reference dye, and optimized PCR buffer. The DNA sequences of the primers used in this study are listed in Table S1 in the supplemental material (19, 37).
Real-time RT-PCR was performed on a Bio-Rad iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA), under the following conditions: pretreatment at 50°C for 2 min; initial denaturation at 95°C for 10 min; and 40 cycles of denaturation, annealing, and extension at 95°C for 15 s, 63°C for 1 min, and 72°C for 30 s, respectively. Fluorescence data were collected at the end of each cycle. A no-template control was included, and no threshold cycle (CT) values were obtained for the negative controls (data not shown). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included to normalize the input amounts of RNA. Relative gene expression levels were determined using the comparative critical threshold (2−ΔΔCT) method (38), whereby the normalized LSMMG value was divided by the NG value. All experiments were repeated 10 times.
Statistical analysis.
Statistical analysis was performed using SAS software version 9.1 (SAS Institute, Inc., Cary, NC, USA). Data were evaluated using a general linear model for analysis of variance. Tukey's test was used to determine the significance of differences in bacterial survival between E. coli O157:H7 cultures grown under LSMMG conditions and those grown under NG conditions. Statistical significance was regarded at a P value of <0.05.
RESULTS
Survival of E. coli O157:H7 under heat stress conditions.
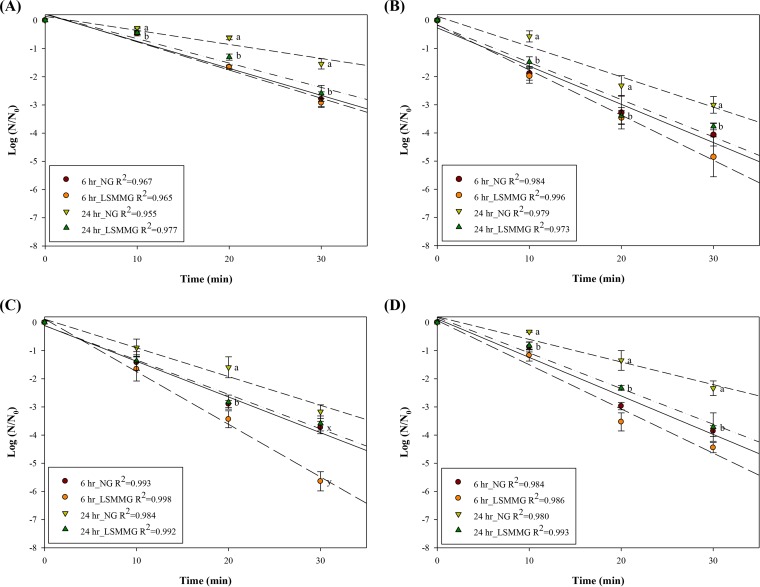
Survival curves for E. coli O157:H7 at 55°C were generated by fitting a log linear model to the data. All survival models fitted the data well (R2 > 0.95). In Fig. 2, the y axis represents the log ratio of the number of viable cells remaining after heat treatment (N) relative to the initial number of cells (N0), where N0 ranged from 6.8- to 7.4-log CFU/ml.
FIG 2.
Heat resistance at 55°C of E. coli O157:H7 ATCC 35150 (A), ATCC 43889 (B), ATCC 43890 (C), and ATCC 43895 (D) cultured under LSMMG and NG conditions for 6 and 24 h. Log (N/N0) values represent the mean ± standard error (SE) (n = 6). The lines denote predictions from linear regression (R2 > 0.95). a and b, means of 24-h NG and LSMMG culture within each time point are significantly different when letters are different (P < 0.05). x and y, means of 6-h NG and LSMMG culture within each time point are significantly different when letters are different (P < 0.05).
When E. coli O157:H7 was cultured under LSMMG or NG conditions for 6 h and then heated to 55°C, there was no significant difference between the survival rates of LSMMG and NG cultures (P > 0.05), except for ATCC 43890. When 6-h cultures of ATCC 43890 were heated for 30 min, the log reductions were greater in LSMMG cultures than in NG cultures (5.6 and 3.7 log CFU/ml, respectively; P < 0.05). LSMMG had a significant impact on the survival of 24-h cultures of E. coli O157:H7 subjected to heat stress. Cells cultured under LSMMG conditions showed reduced survival compared with cells cultured under NG conditions at all time points (P < 0.05), except ATCC 43890 during 10- and 30-min heat treatment (P > 0.05). When fitting a log linear model, survival in the stationary phase (24 h) was higher than that in the exponential phase (6 h) for all cultures. Thus, in general, the smallest log reduction was observed in 24-h NG cultures, followed by 24-h LSMMG cultures and 6-h NG and LSMMG cultures.
Among the four strains tested, E. coli O157:H7 ATCC 35150 was most resistant. When cells cultured under NG conditions for 24 h were heated to 55°C for 30 min, log reductions of 1.5, 3.0, 3.2, and 2.3 log CFU/ml were observed for ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895, respectively. The log reductions were greater in LSMMG cultures than in NG cultures (2.6, 3.8, 3.6, and 3.7 log CFU/ml, respectively).
Differences in D values between LSMMG and NG cultures.
D values at 55°C were obtained from the linear portion of the survival curves under each gravity treatment and cultivation time (Table 1). The D values for E. coli O157:H7 cultured under LSMMG conditions for 24 h were generally lower than those seen under NG conditions (P < 0.05). The 24-h D values for E. coli O157:H7 ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895 were 21.8, 9.6, 10.1, and 13.5 min under NG conditions, and 12.3, 7.6, 7.7, and 8.5 min under LSMMG conditions, respectively. The D values for most of the 6-h cultures were not significantly different (P > 0.05), with the exception of ATCC 43890 (NG, 7.7 min; LSMMG, 5.5 min; P < 0.05). Among the four strains tested, ATCC 35150 showed the greatest heat resistance under all treatments (P < 0.05).
TABLE 1.
D values (minutes at 55°C) for E. coli O157:H7 strains cultured under LSMMG and NG conditions for 6 and 24 h
| Strain | D values (mean ± SE) (min)a |
|||
|---|---|---|---|---|
| 6-h culture |
24-h culture |
|||
| LSMMG | NG | LSMMG | NG | |
| ATCC 35150 | 10.7 ± 0.5 A | 10. 8 ± 0.9 A | 12.3 ± 1.2 A C | 21.8 ± 3.1 A D |
| ATCC 43889 | 6.9 ± 1.0 B | 7.8 ± 0.8 B | 7.6 ± 0.3 B C | 9.6 ± 0.8 B D |
| ATCC 43890 | 5.5 ± 0.4 B C | 7.7 ± 0.4 B D | 7.7 ± 0.8 B | 10.1 ± 0.7 B |
| ATCC 43895 | 6.7 ± 0.3 B | 7.5 ± 0.3 B | 8.5 ± 0.7 B C | 13.5 ± 1.7 B D |
D values were calculated by linear regression (n = 6 replicates). Values denoted by different letters of A and B are significantly different (cells cultured for the same time under each condition) (P < 0.05). Values denoted by different letters of C and D are significantly different (cells cultured under the same gravity conditions) (P < 0.05).
Differences in membrane fatty acid composition in cells cultured under LSMMG and NG conditions.
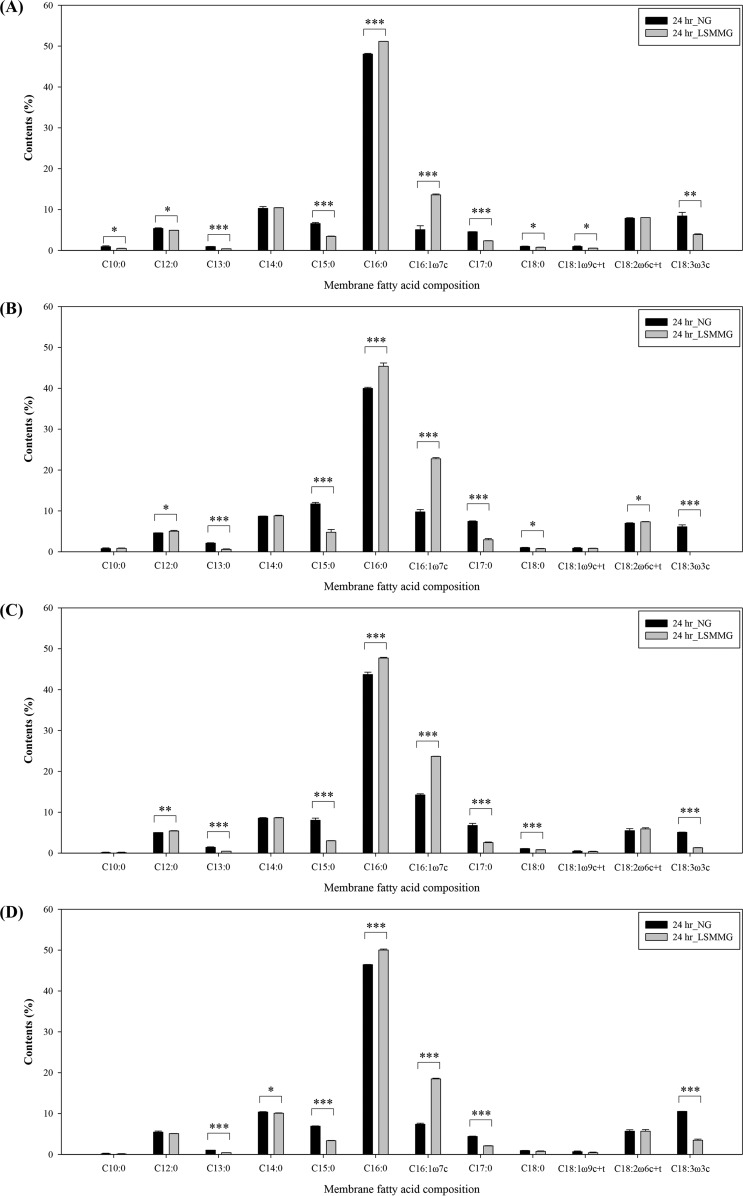
The fatty acid composition of the E. coli O157:H7 membrane after culture under LSMMG and NG conditions is shown in Fig. 3. Fourteen of the tested 37 fatty acids were identified in the bacterial membrane. The nine predominant fatty acids were lauric acid (C12:0), myristic acid (C14:0), pentadecylic acid (C15:0), palmitic acid (C16:0), palmitoleic acid (C16:1ω7c), margaric acid (C17:0), linoleic acid (C18:2ω6c), linolelaidic acid (C18:2ω6t), and α-linolenic acid (C18:3ω3c). The other five (present at levels <1.0%) were capric acid (C10:0), tridecylic acid (C13:0), stearic acid (C18:0), oleic acid (C18:1ω9c), and elaidic acid (C18:1ω9t).
FIG 3.
Membrane fatty acid composition of E. coli O157:H7 ATCC 35150 (A), ATCC 43889 (B), ATCC 43890 (C), and ATCC 43895 (D) cultured under LSMMG and NG conditions for 24 h. The data represent the mean ± standard error (SE) (n = 3). Asterisk indicates significant differences between NG and LSMMG (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). On the x axis, ω denotes the location of the first double bond of unsaturated fatty acid, c and t denote cis and trans forms, respectively, and c + t means the sum of the cis and trans fatty acid amounts.
In all E. coli O157:H7 strains tested, the membranes of cells cultured under LSMMG conditions contained mostly C16:0 (45.4 to 51.2%), followed by C16:1ω7c (13.6 to 23.7%) and C14:0 (8.6 to 10.4%). The membrane fatty acid content of cells cultured under NG conditions showed no particular trend, although C16:0 was predominant (40.0 to 48.0%). Among the fatty acids identified, the amounts of C15:0, C16:0, C16:1ω7c, C17:0, and C18:3ω3c were significantly different under LSMMG conditions (P < 0.05). Overall, growth under LSMMG conditions resulted in less C15:0, C17:0, and C18:3ω3c but more C16:0 and C16:1ω7c. The largest difference was observed in membranes of E. coli O157:H7 ATCC 43889, which contained 5.4% more C16:0 and 13% more C16:1ω7c under LSMMG conditions than under NG conditions.
Changes in membrane fluidity under LSMMG conditions.
Total membrane fatty acid composition, including minor fatty acids, SFA, USFA, and the USFA/SFA ratio (which is indicative of membrane fluidity), is shown in Table 2. Overall, the total amount of each fatty acid under LSMMG conditions was significantly higher than under NG conditions (P < 0.05). The USFA/SFA ratio increased when bacterial cells were cultured under LSMMG conditions, reaching maximum values of 0.46 for E. coli O157:H7 ATCC 43890, 0.45 for ATCC 43889, 0.39 for ATCC 43895, and 0.35 for ATCC 35150. Lower USFA/SFA ratios were observed in NG cultures (approximately 0.3); the lowest ratio was observed in ATCC 35150.
TABLE 2.
Total membrane fatty acid composition and USFA/SFA ratio in E. coli O157:H7 strains cultured under LSMMG and NG conditions for 24 h
| Strain | Membrane fatty acid | Content (mean ± SE) (%)a |
|
|---|---|---|---|
| LSMMG | NG | ||
| ATCC 35150 | Total minor fatty acidsb | 2.2 ± 0.1 A | 3.8 ± 0.5 B |
| Total SFA | 72.3 ± 0.1 A | 74.9 ± 0.4 B | |
| Total USFA | 25.5 ± 0.1 A | 21.4 ± 0.1 B | |
| USFA/SFA | 0.35 | 0.29 | |
| ATCC 43889 | Total minor fatty acids | 3.0 ± 0.1 A | 4.8 ± 0.3 B |
| Total SFA | 66.9 ± 0.1 A | 72.4 ± 0.3 B | |
| Total USFA | 30.1 ± 0.2 A | 22.8 ± 0.0 B | |
| USFA/SFA | 0.45 | 0.31 | |
| ATCC 43890 | Total minor fatty acids | 1.8 ± 0.0 A | 3.1 ± 0.2 B |
| Total SFA | 67.4 ± 0.3 A | 72.1 ± 0.6 B | |
| Total USFA | 30.9 ± 0.3 A | 24.8 ± 0.8 B | |
| USFA/SFA | 0.46 | 0.34 | |
| ATCC 43895 | Total minor fatty acids | 1.7 ± 0.2 A | 2.8 ± 0.1 B |
| Total SFA | 70.6 ± 0.3 A | 73.6 ± 0.6 B | |
| Total USFA | 27.7 ± 0.5 A | 23.6 ± 0.6 B | |
| USFA/SFA | 0.39 | 0.32 | |
n = 3 per group. Values with different of each strain are significantly different (P < 0.05).
Minor fatty acids were capric (C10:0), tridecylic (C13:0), stearic (C18:0), oleic (C18:1ω9c), and elaidic (C18:1ω9t) acid; composition values were all <1.0%.
Relative expression of heat stress-related genes during growth under LSMMG conditions.
E. coli O157:H7 ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895 were examined for variation in gene expression using real-time RT-PCR. The relative LSMMG/NG expression levels of the heat stress-related genes, clpB, dnaK, grpE, groES, htpG, htpX, ibpB, and rpoH, are shown in Fig. 4. Values below −1 indicate decreased gene expression (downregulation) under LSMMG conditions compared to that under NG conditions. In all E. coli O157:H7 strains tested, the expression of all genes was reduced under LSMMG conditions, from an average −1.9- to −3.7-fold. Downregulation of clpB, dnaK, htpX, and rpoH in response to LSMMG conditions was conspicuous, with average fold changes of −2.9, −3.5, −3.2, and −3.7, respectively. Among the four strains, E. coli O157:H7 ATCC 43889 showed the most marked downregulation of clpB, dnaK, and htpX, with fold changes of −4.5, −6.5, and −6.9, respectively (see Table S2 in the supplemental material). In the case of rpoH, the strongest response to LSMMG conditions was shown in E. coli O157:H7 ATCC 43890, followed by ATCC 43889 (−4.8- and −4.4-fold, respectively).
FIG 4.
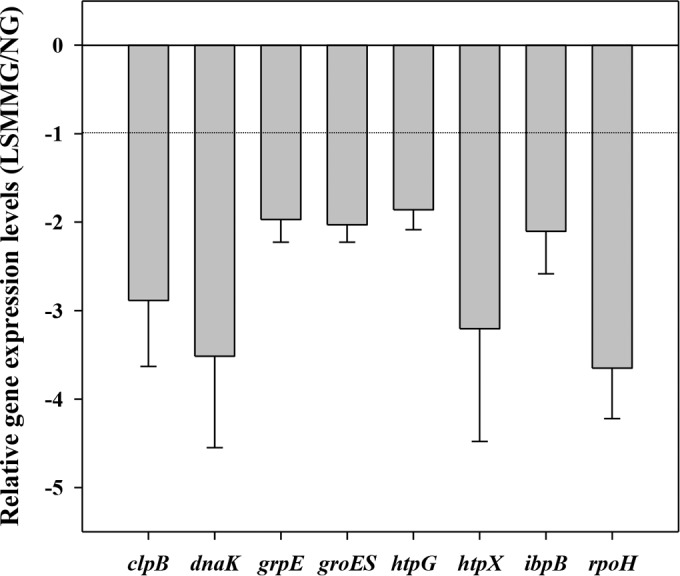
Relative expression levels of heat stress-related genes under LSMMG conditions in E. coli O157:H7 ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895. The data represent the mean ± standard error (SE) (n ≥ 10).
DISCUSSION
Bacterial heat resistance is influenced by many factors, including growth temperature (16, 39), medium composition (40), and pH (16). Likewise, microgravity conditions can induce changes in bacterial heat stress responses. The present study shows that the heat resistance of E. coli O157:H7 decreased when cells were cultured under LSMMG conditions. Heat resistance was greater when cells were in the stationary phase. For all strains, D values at 55°C decreased in the order 24-h NG, 24-h LSMMG, 6-h NG, and 6-h LSMMG. After 24 h, the D values of LSMMG cultures were 1.3- to 1.8-fold lower than those for NG cultures (in terms of log population size). These data differ from those published by Wilson et al. (14), who observed that growth for 10 h under LSMMG conditions increased the heat resistance of Salmonella enterica serovar Typhimurium 8.6-fold (in terms of percentage survival). On the other hand, the heat resistance of Streptococcus pneumoniae strain TIGR4 was diminished after growth under LSMMG conditions (41), indicating that bacterial heat resistance under LSMMG conditions might be species specific. Nevertheless, we have shown that low-gravity conditions do affect the heat stress response of E. coli O157:H7.
Cell membrane fluidity can be roughly measured by membrane polarization; a high polarization value indicates low membrane fluidity. England et al. (42) reported that no differences were observed in membrane polarization values of Pseudomonas aeruginosa grown under LSMMG and NG conditions. To investigate the possible mechanisms underlying changes in the heat resistance of E. coli O157:H7, we examined the fatty acid composition of the membranes of cells grown under LSMMG and NG conditions using gas chromatography. As mentioned earlier, membrane fluidity is affected by fatty acid composition. For instance, membranes containing higher levels of SFA are more densely packed and lack fluidity, whereas membranes containing higher levels of USFA are looser and more fluid (43). We found that the membranes of E. coli O157:H7 consisted primarily of one SFA (C16:0; palmitic acid; average, 48.6% under LSMMG and 44.5% under NG) and one USFA (C16:1ω7c; palmitoleic acid; average, 19.6% under LSMMG and 9.1% under NG). Under LSMMG conditions, the total amount of USFA increased, while the total amount of SFA decreased, yielding a higher USFA/SFA ratio that is indicative of increased membrane fluidity. This change was mainly due to a marked increase in C16:1ω7c and a decrease in C15:0. For example, we observed a >2-fold increase in the amount of C16:1ω7c in most strains cultured under LSMMG conditions, with the exception of ATCC 43890 (∼1.7-fold increase). The levels of C15:0 in LSMMG cultures were 1.9- to 2.7-fold higher than those in NG cultures.
Changes in membrane fluidity are related to bacterial adaptation to a variety of environmental stresses (43, 44). For example, changes in growth temperature (45–47) and acidity (48–50) lead to changes in membrane fatty acid composition. Heat-adapted E. coli O157:H7 grown at 42°C and 45°C shows a reduced USFA/SFA ratio, whereas low-temperature bacterial growth increases the amount of unsaturated fatty acids in the membrane (51). Also, acid adaptation by E. coli O157:H7 is accompanied by an increase in the amount of SFA and a decrease in the amount of USFA, resulting in reduced membrane fluidity (50). Here, we found that exposure to LSMMG conditions induced changes in heat resistance and membrane fatty acid composition similar to those observed after low-temperature growth (reduced heat resistance and increased membrane fluidity).
Membrane fatty acid composition and heat resistance are clearly related. Indeed, D values are higher for cells with a low USFA/SFA ratio, which results in low membrane fluidity in general (52). Here, the USFA/SFA ratio of bacterial cells varied according to strain and microgravity conditions, which in turn affected heat resistance. We found that the USFA/SFA ratio in E. coli O157:H7 under LSMMG conditions increased 1.2- to 1.5-fold, indicating greater membrane fluidity. The USFA/SFA ratios increased in the following order: ATCC 35150 (0.35), ATCC 43895 (0.39), ATCC 43889 (0.45), and ATCC 43890 (0.46). Membrane rigidity (the converse of membrane fluidity) decreased in the following order: ATCC 35150, ATCC 43895, and ATCC 43890 and ATCC 43889. The same trend was observed for heat resistance in LSMMG cultures: D values decreased in the order ATCC 35150 (12.3 min), ATCC 43895 (8.5 min), ATCC 43890 (7.7 min), and ATCC 43889 (7.6 min). Therefore, changes in membrane fatty acid composition at least partly explain the decreased heat resistance of E. coli O157:H7 cultured under LSMMG conditions.
To understand the genetic mechanisms of the heat stress response under LSMMG conditions, we examined the relative expression levels (LSMMG/NG) of the genes encoding HSPs (clpB, dnaK, grpE, groES, htpG, htpX, and ibpB) and the global transcription regulator σH (rpoH). The major heat shock protein, DnaK, assists in the refolding of misfolded proteins (53) and, in combination with ClpB, plays a role in nondestructive protein disaggregation (54). HtpX also plays a major role in the proteolytic quality control of membrane proteins (55). After cultivation under LSMMG conditions, the expression of clpB, dnaK, and htpX was largely downregulated compared to that of other HSP genes (average, −2.9-, −3.5-, and −3.2-fold, respectively; Fig. 4). In contrast, Chopra et al. (34) reported upregulation of dnaK in S. Typhimurium under LSMMG conditions. Otherwise, Vukanti et al. (32) reported downregulation of genes in the unfolded protein response of E. coli under LSMMG conditions, including htpG, ibpB, dnaK, clpB, Lon, dnaJ, and ibpA (−2.0- to −6.5-fold). This is in accordance with our result, which might explain the decreased heat resistance of E. coli O157:H7 under LSMMG conditions. The other HSP genes (grpE, groES, htpG, htpX, and ibpB) were also downregulated (average, −1.9- to −2.1-fold) under LSMMG conditions, which is also consistent with the heat stress response of the LSMMG culture.
The global transcription regulator σH directs the transcription of genes encoding heat shock chaperones and consequently makes bacteria more resistant to heat (22). Here, the transcription of rpoH decreased under LSMMG conditions (−2.5- to −4.8-fold; average, −3.7-fold). Similarly, the expression of rpoH is reduced when cells are cultured at low temperatures (15). The downregulation of rpoH may thus be a possible explanation for the downregulation of HSP genes and the reduced heat resistance of cells cultured under LSMMG conditions.
Overall, the heat resistance of LSMMG cultures was lower than that of NG cultures due to the complex effects of increases in membrane fluidity and downregulation of relevant heat stress genes (clpB, dnaK, grpE, groES, htpG, htpX, ibpB, and rpoH). The present study is the first to report altered heat stress responses in E. coli O157:H7 cultures under LSMMG conditions and to elucidate the most likely underlying mechanisms. Since changes in membrane fatty acid composition probably affect antibiotic resistance (56) and protein secretion through the bacterial membrane (57), further studies should examine whether the observed changes in membrane composition affect E. coli O157:H7 antibiotic resistance and verotoxin secretion. Testing an isogenic mutant of downregulated heat stress-related genes in E. coli O157:H7 also might provide insight into the function of genes under LSMMG conditions.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Korea Research Foundation (grant NRF-2011-0012712).
We thank the School of Life Sciences and Biotechnology of Korea University for BK 21 PLUS and the Institute of Biomedical Science and Food Safety, CJ-Korea University Food Safety Hall, for providing the equipment and facilities.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00050-16.
REFERENCES
- 1.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. 2004. Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev 68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 3.Hengge-Aronis R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol 4:341–346. [PubMed] [Google Scholar]
- 4.Hecker M, Völker U. 2001. General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol 44:35–91. doi: 10.1016/S0065-2911(01)44011-2. [DOI] [PubMed] [Google Scholar]
- 5.Audia JP, Webb CC, Foster JW. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol 291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 6.Kim H, Matin A, Rhee M. 2014. Microgravity alters the physiological characteristics of Escherichia coli O157:H7 ATCC 35150, ATCC 43889, and ATCC 43895 under different nutrient conditions. Appl Environ Microbiol 80:2270–2278. doi: 10.1128/AEM.04037-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazzotta AS, Gombas DE. 2001. Heat resistance of an outbreak strain of Listeria monocytogenes in hot dog batter. J Food Prot 64:321–324. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins D, Schultz J, Matin A. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol 170:3910–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leenanon B, Drake M. 2001. Acid stress, starvation, and cold stress affect poststress behavior of Escherichia coli O157:H7 and nonpathogenic Escherichia coli. J Food Prot 64:970–974. [DOI] [PubMed] [Google Scholar]
- 10.Davidson PM, Critzer FM. 2012. Interventions to inhibit or inactivate bacterial pathogens in foods, p 189–202. In Oyarzabal OA, Backert S (ed), Microbial food safety: an introduction. Springer, New York, NY. [Google Scholar]
- 11.Sergelidis D, Abrahim A. 2009. Adaptive response of Listeria monocytogenes to heat and its impact on food safety. Food Control 20:1–10. doi: 10.1016/j.foodcont.2008.01.006. [DOI] [Google Scholar]
- 12.Perchonok M, Bourland C. 2002. NASA food systems: past, present, and future. Nutrition 18:913–920. doi: 10.1016/S0899-9007(02)00910-3. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. 2011. Enterohaemorrhagic Escherichia coli (EHEC). Fact sheet no. 125. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs125/en/.
- 14.Wilson JW, Ott CM, Ramamurthy R, Porwollik S, McClelland M, Pierson DL, Nickerson CA. 2002. Low-shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS-independent manner. Appl Environ Microbiol 68:5408–5416. doi: 10.1128/AEM.68.11.5408-5416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Khoo WJ, Zheng Q, Chung H-J, Yuk H-G. 2014. Growth temperature alters Salmonella Enteritidis heat/acid resistance, membrane lipid composition and stress/virulence related gene expression. Int J Food Microbiol 172:102–109. doi: 10.1016/j.ijfoodmicro.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Álvarez-Ordóñez A, Fernández A, López M, Bernardo A. 2009. Relationship between membrane fatty acid composition and heat resistance of acid and cold stressed Salmonella senftenberg CECT 4384. Food Microbiol 26:347–353. doi: 10.1016/j.fm.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Yuk H-G, Marshall DL. 2003. Heat adaptation alters Escherichia coli O157:H7 membrane lipid composition and verotoxin production. Appl Environ Microbiol 69:5115–5119. doi: 10.1128/AEM.69.9.5115-5119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White-Ziegler CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23oC) increases expression of biofilm-, cold-shock-and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 19.Trudeau K, Vu KD, Déziel É, Shareck F, Lacroix M. 2014. Effect of γ-irradiation on gene expression of heat shock proteins in the foodborne pathogen Escherichia coli O157:H7. Int J Radiat Biol 90:268–273. doi: 10.3109/09553002.2014.859766. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y, He Y, Irwin PL, Jin T, Shi X. 2011. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl Environ Microbiol 77:2325–2331. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen KJ, Lepp D, McKellar RC, Griffiths MW. 2008. Examination of stress and virulence gene expression in Escherichia coli O157:H7 using targeted microarray analysis. Foodborne Pathog Dis 5:437–447. doi: 10.1089/fpd.2008.0100. [DOI] [PubMed] [Google Scholar]
- 22.Chung H, Bang W, Drake M. 2006. Stress response of Escherichia coli. Compr Rev Food Sci Food Saf 5:52–64. doi: 10.1111/j.1541-4337.2006.00002.x. [DOI] [Google Scholar]
- 23.Hartl FU. 1996. Molecular chaperones in cellular protein folding. Nature 381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 24.Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 25.Spector MP, Kenyon WJ. 2012. Resistance and survival strategies of Salmonella enterica to environmental stresses. Food Res Int 45:455–481. doi: 10.1016/j.foodres.2011.06.056. [DOI] [Google Scholar]
- 26.Thomas JG, Baneyx F. 1998. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG in vivo. J Bacteriol 180:5165–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross CA. 1996. Function and regulation of the heat shock proteins, p 1382–1399. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 28.Squires C, Squires CL. 1992. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol 174:1081–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch SV, Brodie EL, Matin A. 2004. Role and regulation of σS in general resistance conferred by low-shear simulated microgravity in Escherichia coli. J Bacteriol 186:8207–8212. doi: 10.1128/JB.186.24.8207-8212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacello F, Rotilio G, Battistoni A. 2012. Low-shear modeled microgravity enhances Salmonella enterica resistance to hydrogen peroxide through a mechanism involving KatG and KatN. Open Microbiol J 6:53–64. doi: 10.2174/1874285801206010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosado H, Doyle M, Hinds J, Taylor PW. 2010. Low-shear modelled microgravity alters expression of virulence determinants of Staphylococcus aureus. Acta Astronaut 66:408–413. doi: 10.1016/j.actaastro.2009.06.007. [DOI] [Google Scholar]
- 32.Vukanti R, Mintz E, Leff L. 2008. Changes in gene expression of E. coli under conditions of modeled reduced gravity. Microgravity Sci Technol 20:41–57. doi: 10.1007/s12217-008-9012-9. [DOI] [Google Scholar]
- 33.Wilson JW, Ramamurthy R, Porwollik S, McClelland M, Hammond T, Allen P, Ott CM, Pierson DL, Nickerson CA. 2002. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc Natl Acad Sci U S A 99:13807–13812. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chopra V, Fadl A, Sha J, Chopra S, Galindo C, Chopra A. 2006. Alterations in the virulence potential of enteric pathogens and bacterial-host cell interactions under simulated microgravity conditions. J Toxicol Environ Health A 69:1345–1370. doi: 10.1080/15287390500361792. [DOI] [PubMed] [Google Scholar]
- 35.Berry ED, Cutter CN. 2000. Effects of acid adaptation of Escherichia coli O157:H7 on efficacy of acetic acid spray washes to decontaminate beef carcass tissue. Appl Environ Microbiol 66:1493–1498. doi: 10.1128/AEM.66.4.1493-1498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain M, Nijhawan A, Tyagi AK, Khurana JP. 2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 37.Carey CM, Kostrzynska M, Thompson S. 2009. Escherichia coli O157:H7 stress and virulence gene expression on Romaine lettuce using comparative real-time PCR. J Microbiol Methods 77:235–242. doi: 10.1016/j.mimet.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 39.Martínez S, López M, Bernardo A. 2003. Thermal inactivation of Enterococcus faecium: effect of growth temperature and physiological state of microbial cells. Lett Appl Microbiol 37:475–481. doi: 10.1046/j.1472-765X.2003.01431.x. [DOI] [PubMed] [Google Scholar]
- 40.de Matos RE. 1998. Heat resistance of Listeria monocytogenes in dairy products as affected by the growth medium. J Appl Microbiol 84:234–239. doi: 10.1046/j.1365-2672.1998.00334.x. [DOI] [PubMed] [Google Scholar]
- 41.Allen C, Galindo C, Williams N, Pandya U, Chopra A, Niesel D. 2007. A global transcriptional analysis of Streptococcus pneumoniae in response to low-shear modeled microgravity. Gravit Space Res 19:143–144. [Google Scholar]
- 42.England L, Gorzelak M, Trevors J. 2003. Growth and membrane polarization in Pseudomonas aeruginosa UG2 grown in randomized microgravity in a high aspect ratio vessel. Biochim Biophys Acta 1624:76–80. doi: 10.1016/j.bbagen.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Moorman MA, Thelemann CA, Zhou S, Pestka JJ, Linz JE, Ryser ET. 2008. Altered hydrophobicity and membrane composition in stress-adapted Listeria innocua. J Food Prot 71:182–185. [DOI] [PubMed] [Google Scholar]
- 44.Yoon Y, Lee H, Lee S, Kim S, Choi K-H. 2015. Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res Int 72:25–36. doi: 10.1016/j.foodres.2015.03.016. [DOI] [Google Scholar]
- 45.Casadei M, Mañas P, Niven G, Needs E, Mackey B. 2002. Role of membrane fluidity in pressure resistance of Escherichia coli NCTC 8164. Appl Environ Microbiol 68:5965–5972. doi: 10.1128/AEM.68.12.5965-5972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suutari M, Laakso S. 1994. Microbial fatty acids and thermal adaptation. Crit Rev Microbiol 20:285–328. doi: 10.3109/10408419409113560. [DOI] [PubMed] [Google Scholar]
- 47.Annous BA, Becker LA, Bayles DO, Labeda DP, Wilkinson BJ. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 63:3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampathkumar B, Khachatourians GG, Korber DR. 2004. Treatment of Salmonella enterica serovar Enteritidis with a sublethal concentration of trisodium phosphate or alkaline pH induces thermotolerance. Appl Environ Microbiol 70:4613–4620. doi: 10.1128/AEM.70.8.4613-4620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Jonge R, Ritmeester W, van Leusden F. 2002. Adaptive responses of Salmonella enterica serovar Typhimurium DT104 and other S. Typhimurium strains and Escherichia coli O157 to low pH environments. J Appl Microbiol 94:625–632. [DOI] [PubMed] [Google Scholar]
- 50.Yuk H-G, Marshall DL. 2004. Adaptation of Escherichia coli O157:H7 to pH alters membrane lipid composition, verotoxin secretion, and resistance to simulated gastric fluid acid. Appl Environ Microbiol 70:3500–3505. doi: 10.1128/AEM.70.6.3500-3505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marr AG, Ingraham JL. 1962. Effect of temperature on the composition of fatty acids in Escherichia coli. J Bacteriol 84:1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Álvarez-Ordóñez A, Fernández A, López M, Arenas R, Bernardo A. 2008. Modifications in membrane fatty acid composition of Salmonella Typhimurium in response to growth conditions and their effect on heat resistance. Int J Food Microbiol 123:212–219. doi: 10.1016/j.ijfoodmicro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Acebrón SP, Martín I, del Castillo U, Moro F, Muga A. 2009. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett 583:2991–2996. [DOI] [PubMed] [Google Scholar]
- 54.Barends TR, Werbeck ND, Reinstein J. 2010. Disaggregases in 4 dimensions. Curr Opin Struct Biol 20:46–53. doi: 10.1016/j.sbi.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 55.Kornitzer D, Teff D, Altuvia S, Oppenheim A. 1991. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J Bacteriol 173:2944–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishra S, Pierson D. 1992. Space flight: effects on microorganisms. In Schaechter M. (ed), Desk encyclopedia of microbiology. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 57.Wai SN, Takade A, Amako K. 1995. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol Immunol 39:451–456. doi: 10.1111/j.1348-0421.1995.tb02228.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.