ABSTRACT
Fungi, including the yeast Saccharomyces cerevisiae, lack ferritin and use vacuoles as iron storage organelles. This work explored how plant ferritin expression influenced baker's yeast iron metabolism. Soybean seed ferritin H1 (SFerH1) and SFerH2 genes were cloned and expressed in yeast cells. Both soybean ferritins assembled as multimeric complexes, which bound yeast intracellular iron in vivo and, consequently, induced the activation of the genes expressed during iron scarcity. Soybean ferritin protected yeast cells that lacked the Ccc1 vacuolar iron detoxification transporter from toxic iron levels by reducing cellular oxidation, thus allowing growth at high iron concentrations. Interestingly, when simultaneously expressed in ccc1Δ cells, SFerH1 and SFerH2 assembled as heteropolymers, which further increased iron resistance and reduced the oxidative stress produced by excess iron compared to ferritin homopolymer complexes. Finally, soybean ferritin expression led to increased iron accumulation in both wild-type and ccc1Δ yeast cells at certain environmental iron concentrations.
IMPORTANCE Iron deficiency is a worldwide nutritional disorder to which women and children are especially vulnerable. A common strategy to combat iron deficiency consists of dietary supplementation with inorganic iron salts, whose bioavailability is very low. Iron-enriched yeasts and cereals are alternative strategies to diminish iron deficiency. Animals and plants possess large ferritin complexes that accumulate, detoxify, or buffer excess cellular iron. However, the yeast Saccharomyces cerevisiae lacks ferritin and uses vacuoles as iron storage organelles. Here, we explored how soybean ferritin expression influenced yeast iron metabolism, confirming that yeasts that express soybean seed ferritin could be explored as a novel strategy to increase dietary iron absorption.
INTRODUCTION
Iron is an essential micronutrient for most living organisms because it participates as a redox cofactor in many metabolic pathways, including respiration, lipid biosynthesis, translation, DNA replication, and photosynthesis. Despite its abundance, iron bioavailability is very low because Fe3+ forms ferric hydroxides that tend to precipitate at a physiological pH. In humans, iron deficiency anemia (IDA) is the most extended and common nutritional disorder worldwide. The World Health Organization estimates that IDA affects approximately one-fourth of the world's population, especially women and children. Consequences of IDA include diminished learning ability in infants, fatigue and reduced physical capacity and work productivity in adults, and poor pregnancy outcomes (reviewed in references 1 and 2). Iron supplementation with ferrous salts is one of the most widely applied strategies to combat IDA. However, such treatment can cause gastric problems like vomiting, faintness, constipation, or diarrhea. Furthermore, inorganic iron induces oxidative changes, and its absorption in the intestine is vastly altered by diet composition. Many studies have demonstrated that the iron contained in ferritin is soluble and bioavailable to alleviate iron deficiency (3–5). Thus, an emerging strategy to fight IDA consists of supplementing iron in the form of ferritin, which protects iron from chelators like the phytates and polyphenols present in the diet and reduces oxidative damage and the risk of gastric problems, and its iron is properly absorbed via receptor-mediated endocytosis (reviewed in references 6–10).
Ferritin is a multimeric iron storage and detoxification protein that is ubiquitously distributed in animals, plants, and bacteria but is absent in fungi. Ferritin complexes consist of 24-subunit heteropolymers that assemble as spherical shells and can store up to 4,500 Fe3+ ions each (reviewed in references 11 and 12). In vertebrates, ferritin localizes to the cytoplasm and is composed of two distinct subunits, H and L, whose ratio varies depending on the tissue. The H subunit contains ferroxidase centers, whereas the L subunit facilitates nucleation of the mineral core. Some studies have shown that human ferritin H is also found in cell nuclei (13). A human ferritin encoded in the nuclear genome, expressed as a ferritin H-like precursor, is targeted to mitochondria and processed to a functional subunit that assembles into typical ferritin shells with ferroxidase activity (14). Despite animal and plant ferritins having evolved from a common ancestor and possessing a conserved primary structure (15), plant ferritins (phytoferritins) exhibit distinctive features but with some parallels to human mitochondrial ferritin (reviewed in references 8, 9, 16, and 17). Phytoferritins localize to nongreen plastids and possess only H-type subunits with both a ferroxidase center and a nucleation site. In addition to a four-helix bundle (helices A, B, C, and D) and a fifth short helix (helix E) at their amino terminus, plant ferritin subunits contain a transit peptide (TP) that is responsible for their translocation to the plastid and a specific domain known as the extension peptide (EP) (18, 19) (Fig. 1A). The plant ferritin EP, which localizes to the exterior protein surface, helps stabilize the oligomer and acts as a second ferroxidase center for iron binding and oxidation (20–24). In vitro studies have suggested that the soybean seed ferritin H1 (SFerH1) EP displays serine protease-like activity that catalyzes its autodegradation by facilitating iron release from ferritin during seed germination (25). Studies in Arabidopsis thaliana have indicated that phytoferritin functions mainly in the buffering of iron needed for metabolic processes and in protecting cells against oxidative stress (26). However, in pea seeds, >90% of iron is stored in phytoferritin, which is required for plant germination and early growth (27, 28). In vitro work has shown that the SFerH1 and SFerH2 subunits synergistically interact during iron mineralization by exhibiting greater iron oxidation activity than separate homopolymers (29, 30). Soybean ferritin has been used to enrich cereals with iron (31–33). Nonetheless, successful plant iron biofortification requires a complex combinatorial approach that simultaneously modifies iron acquisition, transport, and accumulation in its edible part (34, 35).
FIG 1.
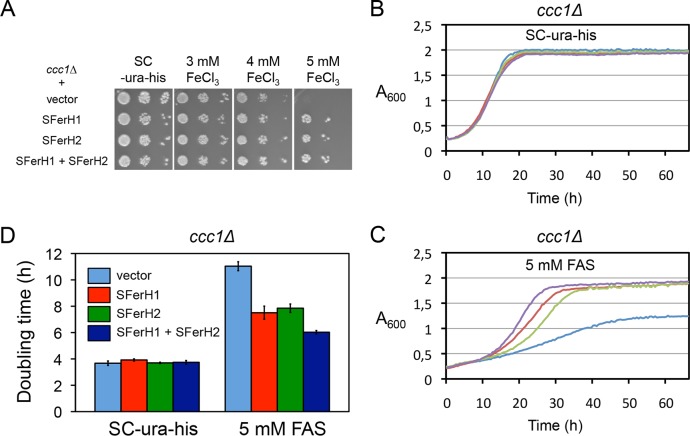
Soybean seed ferritin expression in Saccharomyces cerevisiae. (A) Schematic representation of the most relevant SFerH1 and SFerH2 domains. Numbers indicate amino acid positions. TP, transit peptide; EP, extension peptide. (B) Soybean ferritin levels obtained with different yeast expression vectors. Wild-type BY4741 yeast cells transformed with plasmid p416ADH-SFerH1, p416TEF-SFerH1, p416GPD-SFerH1, p413ADH-SFerH2, p413TEF-SFerH2, or p413GPD-SFerH2 were cultivated in SC−Ura (SFerH1) or SC−His (SFerH2) medium to the exponential growth phase; total protein was extracted; and soybean seed ferritin levels were determined by Western blotting with an anti-SFerH1 antibody. Pgk1 protein levels were used as a loading control. ADH represents p413ADH or p416ADH, TEF represents p413TEF or p416TEF, and GPD represents p413GPD or p416GPD.
The yeast Saccharomyces cerevisiae is used as a model organism to decipher the mechanisms that regulate the response of eukaryotic organisms to changes in iron bioavailability. In response to iron limitation, the yeast Aft1 transcription factor coordinately activates the expression of a set of genes, collectively known as the iron regulon, that function in inorganic iron uptake, siderophore iron acquisition, and the remodeling of iron-dependent metabolism (36–38). Iron regulon members include the cuproferroxidase Fet3, required for high-affinity iron uptake (39); the cell wall-associated mannoprotein Fit3, involved in siderophore iron transport (40); and the RNA-binding protein Cth2, which rearranges cellular iron metabolism to optimize iron utilization (41). Elevated iron concentrations accelerate the formation of reactive oxygen species (ROS) via Fenton redox reactions, which leads to oxidative damage in cellular membranes, proteins, and nucleic acids. In response to high iron concentrations, yeast cells activate the expression of Ccc1, a protein that protects cells from iron toxicity by transporting iron from the cytoplasm to the vacuole, where it is stored bound to polyphosphates (42–44). Thus, ccc1Δ yeast strains are sensitive to elevated iron concentrations due to detoxification defects (42).
The baker's yeast S. cerevisiae is a GRAS (generally recognized as safe) organism used to obtain multiple fermented foods and beverages, livestock feed for fish and poultry, and food supplements for human consumption because it is rich in vital amino acids, fiber, and B-type vitamins. Iron-enriched yeasts are currently being evaluated and appear to be promising iron sources to prevent and palliate iron deficiency in humans and animals (45). In fact, recent reports have indicated that iron-enriched baker's yeasts efficiently help animals recover from iron deficiency while maintaining their fermentative and bakery properties (46, 47). Previous studies have reported that the expression of animal or human ferritin genes in yeast cells leads to increased iron accumulation (48–50). In this report, we characterized how yeast cells that express two phytoferritin genes, SFerH1 and SFerH2, behave in response to elevated iron concentrations with regard to gene expression, iron accumulation, iron resistance, and the cellular redox state.
MATERIALS AND METHODS
Yeast strains and growth conditions.
In this study, S. cerevisiae wild-type strain BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) and the ccc1Δ strain (BY4741 ccc1::KanMX) were used. Yeasts were cultivated at 30°C in synthetic complete (SC) medium that lacked uracil (SC−Ura medium), histidine (SC−His medium), or both (SC−Ura−His medium). Iron was added to autoclaved SC medium at the desired final concentration from either a 250 mM FeCl3 (ferric chloride; Sigma) or a 100 mM Fe(NH4)2(SO4)2 (ferrous ammonium sulfate [FAS]; Sigma) stock solution. Both solutions were prepared in 0.1 M HCl to facilitate iron solubility and were used fresh. A control with the equivalent HCl concentration was always prepared but did not alter growth. To cultivate yeast cells in liquid media, precultures grown overnight in SC medium were reinoculated at an absorbance at 600 nm (A600) of 0.2 and then incubated for 24 h at 30°C at 190 rpm in 50-ml flasks that contained 5 ml of SC−Ura−His medium supplemented with the corresponding iron salt concentration (51). Depending on the experiment, these cultures were employed to determine the A600, cell concentration, dry weight, and endogenous iron level. For growth in 96-well plates, exponentially growing yeast cells were reinoculated at an A600 of 0.2 in 260 μl of liquid SC−Ura−His medium, and the A600 was determined with a SPECTROstar Nano microplate reader (BMG Labtech) every 30 min for 3 days at 28°C. The doubling time in the exponential phase was determined. To perform growth assays in solid media, cells from precultures grown overnight were diluted at an A600 of 0.1 and then spotted onto plates directly and after 1:10 and 1:100 dilutions in sterile water. For the oxidative stress experiments, methylene blue (Sigma) was added to plates containing SC−Ura−His medium or SC−Ura−His medium with FeCl3 from a 1% stock solution. Plates were incubated at 30°C for 3 days and then photographed. For Prussian blue staining, yeast transformants were cultivated for 8 h in SC−Ura−His medium with 5 mM ferric citrate to the exponential phase.
Plasmids.
Plasmids pSFerH1 and pAT-T7-SFerH2 (kindly provided by Taro Masuda) (29), which contained soybean (Glycine max) seed ferritin H1 and H2 cDNAs, respectively, were used as the templates to PCR amplify the coding sequences of ferritins H1 and H2, which lacked their transit peptide (amino acids 1 to 48 in SFerH1 and amino acids 1 to 47 in SFerH2) (Fig. 1A). Oligonucleotides SSFH1-BamHI-F and SSFH1-SalI-R were used to amplify ferritin H1, whereas oligonucleotides SSFH2-BamHI-F and SSFH2-SalI-R were used to obtain ferritin H2 (Table 1). The SFerH1 PCR product was cloned into centromeric vectors p416ADH, p416TEF, and p416GPD, and SFerH2 was cloned into centromeric vectors p413ADH, p413TEF, and p413GPD (52). The URA3 selection marker was used for SFerH1 plasmids, and HIS3 was used for SFerH2 plasmids. In all cases, BamHI and SalI restriction sites were utilized for cloning. All PCR amplifications were performed with Phusion high-fidelity DNA polymerase (Thermo Scientific), and cloned inserts were sequenced. The Escherichia coli One Shot TOP10 strain (Invitrogen) was used for plasmid propagation and isolation.
TABLE 1.
Oligonucleotides used in this work

Western blot analyses.
Total protein extracts were obtained by the alkali method (53). Equal amounts of protein were resolved in SDS-PAGE gels and transferred onto nitrocellulose membranes. Ponceau staining was used to assess protein transfer. Both soybean ferritin H1 and H2 protein levels were determined with an anti-soybean ferritin H1 primary antibody (kindly provided by Taro Masuda), whereas Pgk1 protein levels were determined with an anti-Pgk1 antibody (catalog number 22C5D8; Invitrogen). Immunoblots were developed with horseradish peroxidase (HRP)-labeled secondary antibodies and the ECL Select Western blotting detection kit (GE Healthcare Life Sciences). Immunoblot images were obtained with an ImageQuant LAS 4000 Mini biomolecular imager (GE Healthcare Life Sciences).
Prussian blue staining.
To determine whether soybean ferritin expressed in yeast assembled into multimeric complexes that stored iron, staining with Prussian blue was performed as previously described (49). Briefly, yeast cells that corresponded to a total optical density at 600 nm (OD600) of 60 were harvested and washed twice with distilled water and once with extraction buffer (20 mM Tris-HCl [pH 7.4]). The cells were then disrupted with 200 μl of extraction buffer and 200 μl of glass beads in a bead beater (Biospec Products Inc.). Cell lysates were centrifuged for 3 min at 10,000 × g at 4°C, and the supernatant was transferred to a fresh tube. Since ferritin is a thermostable protein, it was partially purified from yeast cell extracts by heating at 75°C for 15 min. Cell extracts were centrifuged for 10 min at 16,000 × g, the supernatant was transferred to a fresh tube, and protein concentrations were determined by the Bradford method (54). Ten micrograms of proteins was resolved by 7% native PAGE under nondenaturing conditions. Two identical gels were prepared: one was stained for protein by using 0.2% Coomassie brilliant blue, and the other was stained for iron loading with a fresh 1:1 (vol/vol) mixture of 2% K4Fe(CN)6 (ferrocyanide or Prussian blue; Sigma) and 2% 11.6 M HCl (0.116 M final HCl concentration). Horse spleen ferritin (Sigma) was included as a control (data not shown).
Endogenous iron measurements.
Yeast cells that corresponded to a total OD600 of 5.0 were collected by centrifugation and washed twice with 1 mM EDTA and once with MilliQ water. Cells were dissolved in 500 μl of 3% HNO3 and incubated at 98°C for 16 h with agitation. After digestion, extracts were used to determine iron levels. For yeast cells grown in SC medium, iron levels were determined by inductively coupled plasma mass spectrometry (ICP-MS) at the Servei Central d'Instrumentació Científica of the Universitat Jaume I (Castellón, Spain). We used a colorimetric assay for the other samples (55). We previously confirmed that both methods provided similar results (51).
RNA analyses.
To determine the expression levels of the specific mRNAs, total RNA was extracted, and quantitative reverse transcription-PCR (qPCR) was performed as previously described (56). For each analyzed gene, a standard curve was made with serial dilutions of the cDNA sample. ACT1 mRNA values were used to normalize data. Data and error bars represent the averages and standard deviations of results from at least three independent biological samples. All the primers used in this study are listed in Table 1.
Cell concentration and yeast dry weight.
Yeast cells incubated in liquid cultures were properly diluted and sonicated before their concentration (cells per milliliter) was determined with a Beckman Coulter Z counter. To establish yeast dry weight, cells that corresponded to a total OD600 of 30 were collected by centrifugation and washed with distilled water. The cells were then transferred to a previously weighed Eppendorf tube and dried at 50°C for 3 days. Finally, the yeast dry weight was obtained by subtracting the total weight from the Eppendorf tube weight. At least three independent biological samples were used to determine the yeast cellular concentration and dry weight.
Statistical analyses.
To evaluate statistical significance, the data shown in Fig. 5B were analyzed by the multiple-comparison Tukey test using SigmaPlot 12.5 software. In this case, the endogenous iron levels of the ccc1Δ strain transformed with soybean ferritins (SFerH1, SFerH2, and SFerH1 plus SFerH2) were compared to the iron content in ccc1Δ cells that lacked ferritin (vector) grown in SC medium with 10 μM or 100 μM FAS. The statistical significance of the endogenous iron levels of wild-type cells shown in Fig. 6B was determined by two-way analysis of variance (ANOVA) using GraphPad Prism 6 software. The ferritin and iron concentrations in the growth medium were considered the ANOVA variables. In all cases, the P value was <0.01.
FIG 5.

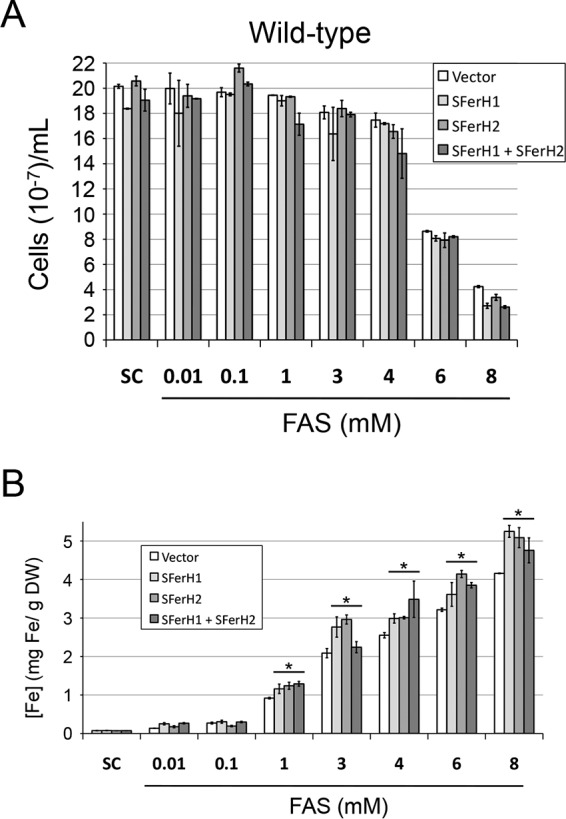
Iron accumulation in ccc1Δ cells that express soybean ferritin. Yeast ccc1Δ cells were transformed with plasmids that express soybean seed ferritin as detailed in the legend of Fig. 2. The yeast transformants were then inoculated at an A600 of 0.2 in SC−Ura−His medium (SC) alone or with increasing Fe(NH4)2(SO4)2 (FAS) concentrations. Numbers of cells per milliliter (A) and endogenous iron contents displayed as weight of iron per yeast dry weight (DW) (B and C) were determined after 24 h of incubation at 30°C. Data and error bars represent the averages and standard deviations of data from at least three independent biological samples. Asterisks indicate statistically significant differences compared to ccc1Δ cells that lacked soybean ferritin (P < 0.01).
FIG 6.
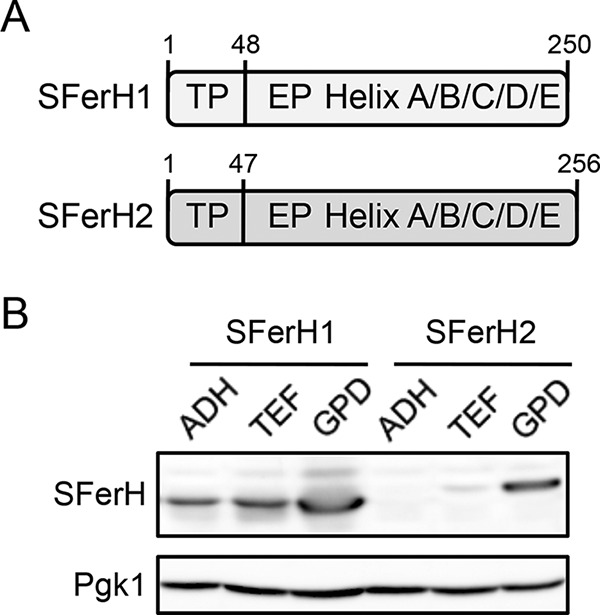
Iron accumulation in wild-type cells that express soybean ferritin. Wild-type BY4741 yeast cells were transformed with plasmids that express soybean seed ferritin as detailed in the legend of Fig. 2. Yeast cells were then inoculated at an A600 of 0.2 in SC−Ura−His medium (SC) alone or with increasing Fe(NH4)2(SO4)2 (FAS) concentrations. Numbers of cells per milliliter (A) and endogenous iron contents displayed as weight of iron per yeast dry weight (DW) (B) were determined after 24 h of incubation at 30°C. Data and error bars represent the averages and standard deviations of data from at least three independent biological samples. Asterisks indicate statistically significant differences compared to wild-type cells that lacked soybean ferritin (P < 0.006).
RESULTS
Cloning and expression of soybean seed ferritin in Saccharomyces cerevisiae.
Unlike mammalian ferritins, which reside in the cytoplasm, phytoferritins localize to plastids. Therefore, in order to express SFerH1 and SFerH2 in baker's yeast, both ferritin-coding sequences lacking their transit peptide were cloned into vectors that drove expression under the control of the yeast gene promoter ADH1, TEF2, or TDH3 (Fig. 1A) (see Materials and Methods for more details). Wild-type cells that expressed soybean ferritins were grown in synthetic medium (SC−Ura medium for SFerH1 or SC−His medium for SFerH2) to the exponential phase, and proteins were extracted. To determine ferritin levels, Western blot analysis was performed with an anti-SFerH1 antibody, which cross-reacted with SFerH2. As expected, maximal expression in both cases was achieved for the p413GPD and p416GPD plasmids that drove gene expression with TDH3 (the strongest expression promoter), and minimum ferritin levels were obtained with ADH1 (the weakest promoter) (Fig. 1B). Similar expression results were observed when yeast cells were incubated in the presence of iron in the growth medium or for longer time periods (data not shown). Surprisingly, the expression of the multicopy 2μ plasmids did not lead to higher ferritin levels (data not shown). Thereafter, SFerH1 and SFerH2 genes were expressed in the centromeric plasmids under the control of the constitutive TDH3 promoter.
Soybean ferritin binds iron when expressed in yeast.
To address whether soybean ferritin expressed in yeast that assembled into functional complexes was capable of capturing intracellular iron, Prussian blue staining was performed, which detects iron bound to ferritin (49). Wild-type cells that lacked ferritin, and which expressed either SFerH1, SFerH2, or both genes, were grown to the exponential phase in medium with 5 mM ferric citrate. Proteins were extracted, resolved by PAGE under nondenaturing conditions, and stained with ferrocyanide (Prussian blue). Characteristic blue bands were observed in cells that expressed SFerH1 or SFerH2, which was consistent with the multimeric ferritin complexes that bound iron in vivo (Fig. 2A). Prussian blue staining was dramatically decreased when yeast cells that expressed ferritin were grown in medium with no added iron (data not shown). As expected, SFerH2 assembled into a complex larger than that of SFerH1, whereas a heteropolymer of an intermediate size formed when both SFerH1 and SFerH2 were coexpressed in the same cells. A similar result was obtained for ccc1Δ mutant cells (data not shown). These results demonstrated that soybean seed ferritin expressed in yeast assembled into a multimeric complex that bound iron in vivo.
FIG 2.

Soybean ferritin accumulates iron when expressed in yeast. The wild-type BY4741 and ccc1Δ yeast strains were cotransformed with p416GPD plus p413GPD (vector), p416GPD-SFerH1 plus p413GPD (SFerH1), p416GPD plus p413GPD-SFerH2 (SFerH2), or p416GPD-SFerH1 plus p413GPD-SFerH2 (SFerH1 plus SFerH2). (A) Prussian blue staining of soybean seed ferritin expressed in wild-type yeast cells. Yeast transformants were grown in SC−Ura−His medium with 5 mM ferric citrate for 8 h to the exponential phase. Heat-labile proteins were removed from cell extracts via a 15-min heat treatment at 75°C. Protein samples were resolved in two 7% nondenaturing polyacrylamide gels. Samples were stained for protein with Coomassie blue and for iron with Prussian blue. (B and C) mRNA levels of iron-regulated genes in wild-type (B) or ccc1Δ (C) cells that expressed soybean seed ferritin. Yeast transformants were cultivated to the exponential growth phase in SC−Ura−His medium without additional iron; total RNA was extracted; and CTH2, FET3, and FIT3 mRNA levels were determined by qPCR. ACT1 was used to normalize the mRNA values. The averages and standard deviations of data from at least three independent biological experiments are represented.
To further address how soybean ferritin influenced iron homeostasis when expressed in yeast, wild-type cells transformed with the vectors, SFerH1, SFerH2, or both ferritins were grown, and the expression levels of specific iron-regulated genes, such as CTH2, FET3, and FIT3, were determined by qPCR. As shown in Fig. 2B, the expression levels of all these three genes were increased in ferritin-expressing cells, especially in SFerH1-expressing cells. When the experiment was performed with cells that lacked the Ccc1 vacuolar iron transporter, both SFerH1 and SFerH2 also brought about an increase in the CTH2, FET3, and FIT3 mRNA levels. Interestingly, coexpression of SFerH1 and SFerH2 in ccc1Δ cells caused a further induction of these iron-regulated genes. Since the CTH2, FET3, and FIT3 genes are members of the iron regulon, which are upregulated by the Aft1 transcription factor when cells sense iron deficiency, these results indicated that soybean ferritin reduced intracellular iron availability. Taken together, our results strongly suggested that soybean ferritin expressed in yeast bound iron in vivo, and cells perceived it by responding accordingly.
Soybean ferritin increases resistance of ccc1Δ cells to elevated iron concentrations.
As soybean seed ferritin bound iron when expressed in yeast cells, a decision was made to determine whether ferritin conferred resistance to toxic levels of extracellular iron. For this purpose, wild-type and ccc1Δ cells were transformed with plasmids that expressed SFerH1 and SFerH2, and growth on solid medium that contained increasing FeCl3 concentrations was assayed. Spot assays were performed, which consisted of 10-fold dilutions of yeast drops starting at an A600 of 0.1 on plates containing SC−Ura−His medium, to maintain selection for both the SFerH1 and SFerH2 plasmids or for empty vectors. When expressed in wild-type cells, soybean ferritin did not alter growth on iron-containing plates (data not shown). However, ccc1Δ cells that expressed either SFerH1 or SFerH2 grew better at 4 mM FeCl3 than did ccc1Δ cells without ferritin (Fig. 3A). SFerH1 and SFerH2 allowed growth at 5 mM FeCl3, an inhibitory concentration for cells that lacked the Ccc1 detoxifying factor (Fig. 3A). No growth was observed at higher FeCl3 concentrations (data not shown). To further characterize the effect of soybean ferritin on iron resistance of ccc1Δ cells, growth with synthetic liquid medium that contained 5 mM another iron salt, ferrous ammonium sulfate (FAS), in 96-well plates was assayed. Soybean ferritin expression did not alter yeast growth in liquid SC−Ura−His medium (Fig. 3B). It is noteworthy that the expression of SFerH1 and SFerH2, either separately or simultaneously, markedly increased the growth rate when 5 mM FAS was added to the medium (Fig. 3C). The coexpression of SFerH1 and SFerH2 yielded maximal growth at high iron concentrations (Fig. 3C). Similar conclusions were drawn after calculating the doubling time in the exponential growth phase. Whereas no difference was observed in synthetic medium, the doubling time was longer when cells were grown in 5 mM FAS (Fig. 3D). Yeast cells that lacked ferritin obtained the longest doubling time, whereas yeast cells that coexpressed both phytoferritins showed the most rapid growth in the exponential phase (Fig. 3D). These results indicated that soybean ferritin increased the resistance of ccc1Δ cells to high iron concentrations by probably detoxifying intracellular iron. An additive effect was observed when both the H1 and H2 phytoferritin forms were coexpressed.
FIG 3.
Growth of the ccc1Δ strain that expresses soybean ferritin in medium with high iron concentrations. Yeast ccc1Δ cells were transformed with plasmids that express soybean seed ferritin, as detailed in the legend of Fig. 2. (A) After overnight growth in SC−Ura−His medium, yeast cells were spotted in 1:10 dilutions starting at an A600 of 0.1 onto plates containing SC−Ura−His solid medium with increasing FeCl3 concentrations. Plates were incubated at 30°C for 3 days and then photographed. (B and C) ccc1Δ transformants were grown to the exponential phase in SC−Ura−His medium and then reinoculated in 96-well plates at an A600 of 0.2 in liquid SC−Ura−His medium without (B) or with (C) 5 mM Fe(NH4)2(SO4)2 (FAS). Cells were incubated for 3 days at 28°C, and the A600 was determined every 30 min with a SPECTROstar Nano microplate reader. ccc1Δ yeast cells transformed with p416GPD plus p413GPD are shown in blue, those transformed with p416GPD-SFerH1 plus p413GPD are shown in red, those transformed with p416GPD plus p413GPD-SFerH2 are shown in green, and those transformed with p416GPD-SFerH1 plus p413GPD-SFerH2 are shown in purple. Data from a representative experiment of at least three independent biological replicates are shown. (D) Doubling time in the exponential phase of the growth curves shown in panels B and C. Data and error bars represent averages and standard deviations of data from three independent biological samples.
Soybean ferritin reduces yeast oxidative stress produced by elevated iron concentrations.
Iron at high concentrations is harmful to cells because it catalyzes redox reactions that generate ROS. As soybean ferritin captures iron in yeast, a decision was made to determine the cellular redox state in ccc1Δ cells that expressed soybean seed ferritin compared to control ccc1Δ yeast cells. For this purpose, growth on plates that contained the biocompatible redox indicator methylene blue, which gains a blue color when oxidized, was assayed (57). As shown in Fig. 4, the expression of SFerH1 or SFerH2 separately, but especially the coexpression of both phytoferritins, markedly reduced the blue color induced by increasing FeCl3 concentrations. This indicated that soybean ferritin reduced the cellular redox state. Therefore, these results suggested that soybean ferritin expression protected yeast cells from oxidative stress produced by excess iron.
FIG 4.
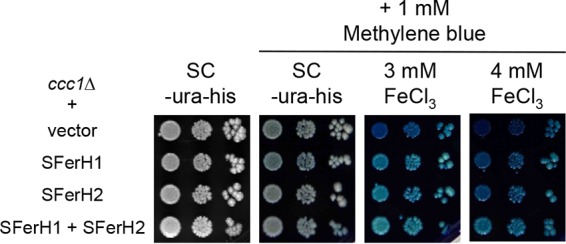
Growth of ccc1Δ yeast cells that express soybean ferritin on plates containing the redox state indicator methylene blue. Yeast ccc1Δ cells were transformed with plasmids that express soybean seed ferritin as detailed in the legend of Fig. 2 and then spotted onto plates as indicated in the legend of Fig. 3A. Methylene blue was added at 1 mM to plates containing SC−Ura−His medium with or without FeCl3 at the indicated concentrations. Plates were incubated at 30°C for 3 days and then photographed.
Soybean ferritin expression increases yeast iron accumulation.
Here, we showed that soybean seed ferritin expressed in yeast that bound iron in vivo increased the resistance of ccc1Δ cells to high iron concentrations and reduced the redox state of ccc1Δ cells grown at toxic iron levels. Therefore, whether soybean ferritin expression allowed yeast cells to increase iron accumulation was ascertained. As shown in Fig. 5A, ccc1Δ cells that expressed soybean ferritin reached a slightly higher maximal cell density than that reached by cells that lacked CCC1 when incubated in synthetic medium with no additional iron. The maximal cell density was not altered when 10 μM or 100 μM FAS was added to the growth medium. However, yeast growth was negatively affected at FAS concentrations of 1 mM or higher (Fig. 5A). Consistent with the data shown in Fig. 3, soybean ferritin expression increased the resistance of ccc1Δ cells to high iron concentrations. Whereas the ccc1Δ strain grew with only up to 3 mM FAS, ccc1Δ cells that expressed either SFerH1 or SFerH2 were able to grow with 6 mM FAS, and ccc1Δ cells that coexpressed both phytoferritin forms were able to grow with up to 8 mM FAS (Fig. 5A).
In order to determine yeast cellular iron concentrations as the weight of iron per yeast dry weight (grams of Fe per gram of dry yeasts), the endogenous iron levels and yeast cell dry weights of each transformant were measured after 1 day of incubation in each tested iron-containing medium (see Materials and Methods) (51). No significant differences were found for iron accumulation by ccc1Δ cells incubated in synthetic medium with no additional iron (Fig. 5B). Although growth was not affected, the addition of 10 μM or 100 μM FAS to synthetic medium allowed ccc1Δ ferritin-expressing cells to significantly increase their iron accumulation (Fig. 5B). Iron levels were maximal for the SFerH1/SFerH2-coexpressing cells, which reached 3-fold-higher endogenous iron levels than those of ccc1Δ cells that lacked ferritin when incubated with 0.1 mM FAS (∼400 μg Fe/g dry yeast) (Fig. 5B). Yeast cells gradually increased their iron content as the environmental FAS concentration rose (Fig. 5C). It is noteworthy that ccc1Δ cells that expressed ferritin did not accumulate more iron than the control cells with 1 and 3 mM FAS (Fig. 5C). Yeast ccc1Δ cells that expressed soybean ferritin reached maximal endogenous iron levels (∼7 mg Fe/g dry yeast) when incubated with 6 or 8 mM FAS in the medium (Fig. 5C).
Although no iron resistance alteration took place when wild-type cells that expressed soybean ferritin were assayed for growth on solid medium with increasing iron concentrations (data not shown), the results obtained for ccc1Δ cells under liquid conditions encouraged further characterization of the effects of soybean ferritin expression on wild-type cells. Similarly to ccc1Δ cells, wild-type cells transformed with empty vectors, SFerH1, SFerH2, or both phytoferritin forms were cultivated for 1 day in iron-containing medium, and both cell densities and endogenous iron levels were determined. As expected, wild-type cells were more resistant to iron than were ccc1Δ cells, even when grown in medium with 8 mM FAS (Fig. 6A), whereas ccc1Δ cells tolerated only up to 3 mM FAS (Fig. 5A). Soybean ferritin expression did not alter wild-type cell growth. The only exception was with 8 mM FAS-containing medium, which caused slightly reduced growth (Fig. 6A). Soybean ferritins significantly increased the iron accumulation (up to 26%) of wild-type cells grown in medium with iron concentrations of 1 mM or higher (Fig. 6B). Taken together, these results suggested that at certain environmental iron concentrations, soybean seed ferritin expression facilitates iron accumulation in both wild-type and ccc1Δ yeast cells.
DISCUSSION
In this study, we explored how soybean seed ferritin expression influenced yeast iron metabolism. By using Prussian blue staining, we first demonstrated that soybean seed ferritin bound iron in vivo when expressed in yeast cells. Interestingly, iron accumulation in phytoferritin complexes led to the transcriptional activation of Aft1 target genes, probably due to a reduction of intracellular iron availability for iron-sulfur cluster biosynthesis. The induction of iron-responsive genes was also observed when human ferritin was expressed in yeast cells (58).
Plant studies have shown that EP-mediated ferritin autodegradation facilitates iron release from soybean ferritin and its utilization during seed germination (25). Although yeast-expressed phytoferritin stores iron in a nontoxic state, it is unclear whether iron bound to soybean ferritin can be recycled to be utilized in yeast metabolism. Previous studies demonstrated that human mitochondrial ferritin protects yeast frataxin mutants from death induced by oxidative stress and excess iron or copper, extends life span, and improves respiratory function (59–61). However, human mitochondrial ferritin is not a functional homolog of yeast frataxin (61). Instead, yeast expression of human ferritin makes mitochondrial iron more available for heme and cytochrome synthesis in mutants that accumulate insoluble and biologically unavailable iron in mitochondria, such as those with impaired Fe-S cluster biogenesis (61, 62). Although the mechanism is not known, it has been proposed that ferritin can bind a small proportion of mitochondrial precipitated iron and release it as Fe2+ to restore heme synthesis (61).
In animals, excess iron is stored in the form of ferritin, whereas plant cells primarily keep iron supplies in vacuoles. Studies of Arabidopsis have suggested that phytoferritin functions in iron buffering by protecting cells against oxidative stress (26). However, most iron in pea seeds is stored in the form of phytoferritin, which is critical for germination and early growth (27, 28). Here, it was observed that soybean ferritin exerted both iron storage and oxidative protection functions when expressed in yeast. First, soybean ferritin expression increased yeast iron accumulation; second, ferritin enhanced yeast resistance to elevated iron concentrations; and third, ferritin reduced the yeast redox state under high-iron conditions. All these observations became even more evident in ccc1Δ mutants than in wild-type cells, probably because the mutant strain lacked the vacuolar iron detoxification pathway, which protects wild-type cells from excess iron and partially masks the ferritin effect. The present work actually showed that soybean ferritins protected ccc1Δ yeast cells from iron toxicity, despite previous studies that used human ferritins not being able to show a protective effect when expressed in a ccc1Δ strain (63). Previous in vitro studies reported that SFerH1/SFerH2 heteropolymers display greater iron oxidation activity and DNA protection from oxidative damage during iron oxidative deposition than soybean ferritin homopolymers (30, 64). Here, it was observed that simultaneously expressed SFerH1 and SFerH2 assembled into heteropolymers in yeast. Moreover, heteropolymers displayed greater iron accumulation and resistance and lower oxidative stress than did ferritin homopolymers when expressed in ccc1Δ yeast cells. This finding indicated that ferritins H1 and H2 positively interacted in yeast.
Plant iron biofortification seems the most appropriate strategy to combat IDA worldwide (65). Biotechnological approaches, which have consisted of soybean seed ferritin ectopic expression in cereals, have resulted in only moderately enriched iron contents (31–33). Since plants are multicellular organisms, genetic engineering strategies need to improve various aspects of iron homeostasis, including root iron uptake, transport to edible parts, resistance to excess iron, and bioavailability, in order to obtain appropriate fortified vegetable foods (34, 35). By using a unicellular organism such as S. cerevisiae, the present work bypassed several obstacles. Soybean seed ferritin upregulated the expression of the yeast iron uptake machinery, which probably contributed to iron accumulation. Previous studies with human ferritins reported increases in yeast endogenous iron accumulation, which ranged from 2- to 5-fold when cells were incubated with 20 mM ferric citrate (49). However, our study obtained a modest increase in the iron level when wild-type cells were grown with 8 mM FAS (Fig. 6B) as well as a 3-fold increase when ccc1Δ cells were grown with 100 μM FAS (Fig. 5B). In our case, the main problem that limited iron accumulation was that soybean ferritin expression levels were too low. Since it was not possible to increase ferritin expression by using multicopy plasmids, additional approaches need to be explored. For instance, previous studies substantially enhanced the expression of animal or human ferritin in yeast by using specific yeast strains defective in the vacuolar proteases Pep4 and Prb1, by modifying codon translation efficiency in the amino-terminal region, and by optimizing incubation times and iron concentrations (48–50). Additional approaches could include the use of yeast for the high-throughput genetic selection of engineered phytoferritin variants with an enhanced iron accumulation capacity, which was recently done with bacterial ferritin (66), or by identifying and expressing in yeast a phytoferritin chaperone that increases iron loading, which has been described for the human PCBP1 ferritin chaperone (67).
In summary, this report demonstrates that plant ferritins assemble into functional complexes that capture iron and protect yeast cells from oxidative stress derived from high environmental iron concentrations. Although further work is necessary to improve soybean ferritin expression and, consequently, iron accumulation in yeast, this approach represents a promising novel dietary iron source to treat iron deficiency.
ACKNOWLEDGMENTS
We are indebted to the students from the University of Valencia who contributed to this work. We are grateful to the Biotechnology Department at the Instituto de Agroquímica y Tecnología de Alimentos (IATA) and the Department of Biochemistry and Molecular Biology of the University of Valencia for their technical and scientific assistance. We thank Taro Masuda and Martin Funk for the gift of biological material used in this work.
Funding Statement
This work has been funded by grants AGL2011-29099 and BIO2014-56298-P from the Spanish Ministry of Economy and Competitiveness to S.P., a predoctoral fellowship from the Spanish Ministry of Economy and Competitiveness to A.M.R., and a postdoctoral JAE-Doc contract from the Spanish Research Council (CSIC) and the European Social Fund to R.D.L.
REFERENCES
- 1.Zimmermann MB, Hurrell RF. 2007. Nutritional iron deficiency. Lancet 370:511–520. doi: 10.1016/S0140-6736(07)61235-5. [DOI] [PubMed] [Google Scholar]
- 2.Huma N, Salim-Ur-Rehman, Anjum FM, Murtaza MA, Sheikh MA. 2007. Food fortification strategy—preventing iron deficiency anemia: a review. Crit Rev Food Sci Nutr 47:259–265. doi: 10.1080/10408390600698262. [DOI] [PubMed] [Google Scholar]
- 3.Beard JL, Burton JW, Theil EC. 1996. Purified ferritin and soybean meal can be sources of iron for treating iron deficiency in rats. J Nutr 126:154–160. [DOI] [PubMed] [Google Scholar]
- 4.Murray-Kolb LE, Takaiwa F, Goto F, Yoshihara T, Theil EC, Beard JL. 2002. Transgenic rice is a source of iron for iron-depleted rats. J Nutr 132:957–960. [DOI] [PubMed] [Google Scholar]
- 5.Lonnerdal B, Bryant A, Liu X, Theil EC. 2006. Iron absorption from soybean ferritin in nonanemic women. Am J Clin Nutr 83:103–107. [DOI] [PubMed] [Google Scholar]
- 6.Lonnerdal B. 2009. Soybean ferritin: implications for iron status of vegetarians. Am J Clin Nutr 89:1680S–1685S. doi: 10.3945/ajcn.2009.26736W. [DOI] [PubMed] [Google Scholar]
- 7.Lv C, Zhao G, Lonnerdal B. 2015. Bioavailability of iron from plant and animal ferritins. J Nutr Biochem 26:532–540. doi: 10.1016/j.jnutbio.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Liao X, Yun S, Zhao G. 2014. Structure, function, and nutrition of phytoferritin: a newly functional factor for iron supplement. Crit Rev Food Sci Nutr 54:1342–1352. doi: 10.1080/10408398.2011.635914. [DOI] [PubMed] [Google Scholar]
- 9.Zhao G. 2010. Phytoferritin and its implications for human health and nutrition. Biochim Biophys Acta 1800:815–823. doi: 10.1016/j.bbagen.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Zielinska-Dawidziak M. 2015. Plant ferritin: a source of iron to prevent its deficiency. Nutrients 7:1184–1201. doi: 10.3390/nu7021184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arosio P, Ingrassia R, Cavadini P. 2009. Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta 1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Theil EC. 2011. Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr Opin Chem Biol 15:304–311. doi: 10.1016/j.cbpa.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surguladze N, Patton S, Cozzi A, Fried MG, Connor JR. 2005. Characterization of nuclear ferritin and mechanism of translocation. Biochem J 388:731–740. doi: 10.1042/BJ20041853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, Sanford D, Arosio P, Drysdale J. 2001. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem 276:24437–24440. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 15.Proudhon D, Wei J, Briat J, Theil EC. 1996. Ferritin gene organization: differences between plants and animals suggest possible kingdom-specific selective constraints. J Mol Evol 42:325–336. doi: 10.1007/BF02337543. [DOI] [PubMed] [Google Scholar]
- 16.Briat JF, Duc C, Ravet K, Gaymard F. 2010. Ferritins and iron storage in plants. Biochim Biophys Acta 1800:806–814. doi: 10.1016/j.bbagen.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Briat JF, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, Cellier F, Gaymard F. 2010. New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann Bot 105:811–822. doi: 10.1093/aob/mcp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ragland M, Briat JF, Gagnon J, Laulhere JP, Massenet O, Theil EC. 1990. Evidence for conservation of ferritin sequences among plants and animals and for a transit peptide in soybean. J Biol Chem 265:18339–18344. [PubMed] [Google Scholar]
- 19.Lescure AM, Proudhon D, Pesey H, Ragland M, Theil EC, Briat JF. 1991. Ferritin gene transcription is regulated by iron in soybean cell cultures. Proc Natl Acad Sci U S A 88:8222–8226. doi: 10.1073/pnas.88.18.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laulhere JP, Laboure AM, Briat JF. 1989. Mechanism of the transition from plant ferritin to phytosiderin. J Biol Chem 264:3629–3635. [PubMed] [Google Scholar]
- 21.van Wuytswinkel O, Briat JF. 1995. Conformational changes and in vitro core-formation modifications induced by site-directed mutagenesis of the specific N-terminus of pea seed ferritin. Biochem J 305(Part 3):959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Wuytswinkel O, Savino G, Briat JF. 1995. Purification and characterization of recombinant pea-seed ferritins expressed in Escherichia coli: influence of N-terminus deletions on protein solubility and core formation in vitro. Biochem J 305(Part 1):253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Fu X, Qi X, Hu X, Chasteen ND, Zhao G. 2009. Protein association and dissociation regulated by ferric ion: a novel pathway for oxidative deposition of iron in pea seed ferritin. J Biol Chem 284:16743–16751. doi: 10.1074/jbc.M109.011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Yun S, Yang X, Zhao G. 2013. Stability and iron oxidation properties of a novel homopolymeric plant ferritin from adzuki bean seeds: a comparative analysis with recombinant soybean seed H-1 chain ferritin. Biochim Biophys Acta 1830:2946–2953. doi: 10.1016/j.bbagen.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Fu X, Deng J, Yang H, Masuda T, Goto F, Yoshihara T, Zhao G. 2010. A novel EP-involved pathway for iron release from soya bean seed ferritin. Biochem J 427:313–321. doi: 10.1042/BJ20100015. [DOI] [PubMed] [Google Scholar]
- 26.Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. 2009. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57:400–412. doi: 10.1111/j.1365-313X.2008.03698.x. [DOI] [PubMed] [Google Scholar]
- 27.Lobreaux S, Briat JF. 1991. Ferritin accumulation and degradation in different organs of pea (Pisum sativum) during development. Biochem J 274(Part 2):601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marentes E, Grusak M. 1998. Iron transport and storage within the seed coat and embryo of developing seeds of pea (Pisum sativum L.). Seed Sci Res 8:575–582. [Google Scholar]
- 29.Masuda T, Goto F, Yoshihara T. 2001. A novel plant ferritin subunit from soybean that is related to a mechanism in iron release. J Biol Chem 276:19575–19579. doi: 10.1074/jbc.M011399200. [DOI] [PubMed] [Google Scholar]
- 30.Deng J, Liao X, Yang H, Zhang X, Hua Z, Masuda T, Goto F, Yoshihara T, Zhao G. 2010. Role of H-1 and H-2 subunits of soybean seed ferritin in oxidative deposition of iron in protein. J Biol Chem 285:32075–32086. doi: 10.1074/jbc.M110.130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. 1999. Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- 32.Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishman S, Oliveira M, Goto F, Datta SK. 2003. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci 164:371–378. doi: 10.1016/S0168-9452(02)00421-1. [DOI] [Google Scholar]
- 33.Drakakaki G, Marcel S, Glahn RP, Lund EK, Pariagh S, Fischer R, Christou P, Stoger E. 2005. Endosperm-specific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bioavailable iron. Plant Mol Biol 59:869–880. doi: 10.1007/s11103-005-1537-3. [DOI] [PubMed] [Google Scholar]
- 34.Lucca P, Hurrell R, Potrykus I. 2002. Fighting iron deficiency anemia with iron-rich rice. J Am Coll Nutr 21:184S–190S. doi: 10.1080/07315724.2002.10719264. [DOI] [PubMed] [Google Scholar]
- 35.Wirth J, Poletti S, Aeschlimann B, Yakandawala N, Drosse B, Osorio S, Tohge T, Fernie AR, Gunther D, Gruissem W, Sautter C. 2009. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol J 7:631–644. doi: 10.1111/j.1467-7652.2009.00430.x. [DOI] [PubMed] [Google Scholar]
- 36.Sanvisens N, Puig S. 2011. Causes and consequences of nutritional iron deficiency in living organisms, p 245–276. In Merkin TC. (ed), Biology of starvation in humans and other organisms. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 37.Kaplan CD, Kaplan J. 2009. Iron acquisition and transcriptional regulation. Chem Rev 109:4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- 38.Philpott CC, Leidgens S, Frey AG. 2012. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta 1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 40.Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, Botstein D, Philpott CC. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J Biol Chem 276:49244–49250. doi: 10.1074/jbc.M109220200. [DOI] [PubMed] [Google Scholar]
- 41.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Chen OS, McVey Ward D, Kaplan J. 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Bagley D, Ward DM, Kaplan J. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H, Li L, Jia X, Ward DM, Kaplan J. 2011. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem 286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pas M, Piskur B, Sustaric M, Raspor P. 2007. Iron enriched yeast biomass: a promising mineral feed supplement. Bioresour Technol 98:1622–1628. doi: 10.1016/j.biortech.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Gaensly F, Wille G, Brand D, Bonfim T. 2011. Iron enriched Saccharomyces cerevisiae maintains its fermenting power and bakery properties. Food Sci Technol Int 31:980–983. doi: 10.1590/S0101-20612011000400025. [DOI] [Google Scholar]
- 47.Kyyaly MA, Powell C, Ramadan E. 2015. Preparation of iron-enriched baker's yeast and its efficiency in recovery of rats from dietary iron deficiency. Nutrition 31:1155–1164. doi: 10.1016/j.nut.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 48.Shin YM, Kwon TH, Kim KS, Chae KS, Kim DH, Kim JH, Yang MS. 2001. Enhanced iron uptake of Saccharomyces cerevisiae by heterologous expression of a tadpole ferritin gene. Appl Environ Microbiol 67:1280–1283. doi: 10.1128/AEM.67.3.1280-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HJ, Kim HM, Kim JH, Ryu KS, Park SM, Jahng KY, Yang MS, Kim DH. 2003. Expression of heteropolymeric ferritin improves iron storage in Saccharomyces cerevisiae. Appl Environ Microbiol 69:1999–2005. doi: 10.1128/AEM.69.4.1999-2005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo HY, Chung YJ, Kim SJ, Park CU, Kim KS. 2003. Enhanced expression and functional characterization of the human ferritin H- and L-chain genes in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 63:57–63. doi: 10.1007/s00253-003-1350-3. [DOI] [PubMed] [Google Scholar]
- 51.Martínez-Garay CA, de Llanos R, Romero AM, Martínez-Pastor MT, Puig S. 2016. Responses of Saccharomyces cerevisiae strains from different origins to elevated iron concentrations. Appl Environ Microbiol 82:1906–1916. doi: 10.1128/AEM.03464-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mumberg D, Muller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 53.Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 55.Tamarit J, Irazusta V, Moreno-Cermeno A, Ros J. 2006. Colorimetric assay for the quantitation of iron in yeast. Anal Biochem 351:149–151. doi: 10.1016/j.ab.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Sanvisens N, Romero AM, An X, Zhang C, de Llanos R, Martinez-Pastor MT, Bano MC, Huang M, Puig S. 2014. Yeast Dun1 kinase regulates ribonucleotide reductase inhibitor Sml1 in response to iron deficiency. Mol Cell Biol 34:3259–3271. doi: 10.1128/MCB.00472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishida K, Silver PA. 2012. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol 10:e1001269. doi: 10.1371/journal.pbio.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim KS, Chang YJ, Chung YJ, Park CU, Seo HY. 2007. Enhanced expression of high-affinity iron transporters via H-ferritin production in yeast. J Biochem Mol Biol 40:82–87. doi: 10.5483/BMBRep.2007.40.1.082. [DOI] [PubMed] [Google Scholar]
- 59.Desmyter L, Dewaele S, Reekmans R, Nystrom T, Contreras R, Chen C. 2004. Expression of the human ferritin light chain in a frataxin mutant yeast affects ageing and cell death. Exp Gerontol 39:707–715. doi: 10.1016/j.exger.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 60.Campanella A, Isaya G, O'Neill HA, Santambrogio P, Cozi A, Arosio P, Levi S. 2004. The expression of human mitochondrial ferritin rescues respiratory function in frataxin-deficient yeast. Hum Mol Genet 13:2279–2288. doi: 10.1093/hmg/ddh232. [DOI] [PubMed] [Google Scholar]
- 61.Sutak R, Seguin A, Garcia-Serres R, Oddou JL, Dancis A, Tachezy J, Latour JM, Camadro JM, Lesuisse E. 2012. Human mitochondrial ferritin improves respiratory function in yeast mutants deficient in iron-sulfur cluster biogenesis, but is not a functional homologue of yeast frataxin. Microbiologyopen 1:95–104. doi: 10.1002/mbo3.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seguin A, Sutak R, Bulteau A-L, Garcia-Serres R, Oddou J-L, Lefevre S, Santos R, Dancis A, Camadro J-M, Latour J-M, Lesuisse E. 2010. Evidence that yeast frataxin is not an iron storage protein in vivo. Biochim Biophys Acta 1802:531–538. doi: 10.1016/j.bbadis.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Leidgens S, Bullough KZ, Shi H, Li F, Shakoury-Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A, Stemmler TL, Philpott CC. 2013. Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J Biol Chem 288:17791–17802. doi: 10.1074/jbc.M113.460253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao X, Lv C, Zhang X, Masuda T, Li M, Zhao G. 2012. A novel strategy of natural plant ferritin to protect DNA from oxidative damage during iron oxidation. Free Radic Biol Med 53:375–382. doi: 10.1016/j.freeradbiomed.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Bouis HE. 2003. Micronutrient fortification of plants through plant breeding: can it improve nutrition in man at low cost? Proc Nutr Soc 62:403–411. doi: 10.1079/PNS2003262. [DOI] [PubMed] [Google Scholar]
- 66.Matsumoto Y, Chen R, Anikeeva P, Jasanoff A. 2015. Engineering intracellular biomineralization and biosensing by a magnetic protein. Nat Commun 6:8721. doi: 10.1038/ncomms9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi H, Bencze KZ, Stemmler TL, Philpott CC. 2008. A cytosolic iron chaperone that delivers iron to ferritin. Science 320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]