Abstract
Endohyphal bacteria (EHB) can influence fungal phenotypes and shape the outcomes of plant-fungal interactions. Previous work has suggested that EHB form facultative associations with many foliar fungi in the Ascomycota. These bacteria can be isolated in culture, and fungi can be cured of EHB using antibiotics. Here, we present methods for successfully introducing EHB into axenic mycelia of strains representing two classes of Ascomycota. We first establish in vitro conditions favoring reintroduction of two strains of EHB (Luteibacter sp.) into axenic cultures of their original fungal hosts, focusing on fungi isolated from healthy plant tissue as endophytes: Microdiplodia sp. (Dothideomycetes) and Pestalotiopsis sp. (Sordariomycetes). We then demonstrate that these EHB can be introduced into a novel fungal host under the same conditions, successfully transferring EHB between fungi representing different classes. Finally, we manipulate conditions to optimize reintroduction in a focal EHB-fungal association. We show that EHB infections were initiated and maintained more often under low-nutrient culture conditions and when EHB and fungal hyphae were washed with MgCl2 prior to reassociation. Our study provides new methods for experimental assessment of the effects of EHB on fungal phenotypes and shows how the identity of the fungal host and growth conditions can define the establishment of these widespread and important symbioses.
INTRODUCTION
As appreciation for their diversity and importance grows, plant microbiomes are increasingly of interest for diverse medical, industrial, and agricultural applications (1, 2). However, many plant-associated microbes remain undescribed (3, 4) and/or are found in association with, or as symbionts of, other microorganisms (5–14, 49–52). One example of these microbial symbioses is that between plant-associated fungi and their bacterial endosymbionts (endobacteria, endofungal bacteria, or endohyphal bacteria [EHB]). Recent studies have indicated that EHB are widespread in rhizosphere fungi from diverse fungal phyla (e.g., mycorrhizal and pathogenic fungi from the Basidiomycota, Glomeromycota, and Mucoromycotina [6–13]) and in the highly diverse Ascomycota that infect roots, stems, and leaves as endophytes (14) (class 3 endophytes, sensu [15]). However, functional relationships have been studied for only a few associations, limiting inferences regarding the scope and potential importance of EHB-fungal associations in ecological interactions and human applications (but see references 16 and 17).
Most studies aimed at understanding functional relationships between EHB and fungi have focused on root-associated fungi, especially arbuscular mycorrhizal fungi in the Glomeromycota and plant-pathogenic Rhizopus in the Mucoromycotina (6–11, 18–23). EHB in rhizosphere fungi are often vertically transmitted and host specific, and they frequently maintain obligate relationships with their hosts (18–20). Although EHB can alter the phenotypes of root-associated fungi and profoundly influence the establishment and outcomes of plant-fungal interactions (21–23), these often obligate and closely coevolved relationships can make it difficult to isolate the symbionts and to determine their individual contributions to plant health or other applications.
In contrast, relationships between EHB and many species of Ascomycota appear to be facultative (14, 24). Previous studies focusing on foliar endophytes in several of the most species-rich clades of Ascomycota have documented a lack of phylogenetic concordance between these fungi and their EHB (14), consistent with facultative symbioses and frequent transfer of EHB among fungal strains. Moreover, these fungi can be cured of their EHB via antibiotic treatment, and both partners often can be cultivated in isolation on standard nutrient media (14, 24, 25). Much like their fungal hosts, most EHB of foliar endophytes appear to be horizontally transmitted (14, 15, 24–29), but the factors determining the dynamics of these associations are poorly understood.
EHB of foliar endophytes can strongly influence fungal phenotypes, with consequences for plant-fungal interactions (24; K. R. Arendt, S. J. Araldi-Brondolo, D. A. Baltrus, and A. E. Arnold, unpublished data). Curing fungi of EHB provides one important tool for empirical assessment of the effects of these bacteria and has been used in a variety of studies (14, 22, 24, 30). However, to date, few studies have successfully reintroduced axenic EHB into fungal mycelia (see references 22 and 30), and to our knowledge, the capacity of EHB to be transferred among fungal strains has not been demonstrated experimentally.
Here, we examine methods for successfully introducing EHB into axenic fungal mycelia. We focus on two fungal species representing distantly related classes of Ascomycota that were originally isolated as foliar endophytes from a woody plant. We first establish in vitro conditions favoring reintroduction of two strains of axenic EHB (Luteibacter sp. [Gammaproteobacteria]) into axenic strains of their original fungal hosts. We then demonstrate that these EHB can be introduced into novel fungal hosts under the same conditions, successfully transferring EHB between members of the Dothideomycetes and Sordariomycetes. Finally, we manipulate conditions to optimize reintroduction in a focal EHB-fungal association, examining the importance of the nutrient content for the coculture medium, the mycelium/bacterium ratio in coculture, the age of the bacterial culture at the time of coculturing, the treatment of the axenic cultures prior to coculturing, and the nutrient content of the solid medium onto which the coculture is plated.
Our study provides a new suite of methods for assessing the effects of cultivable EHB on the phenotypes of cultivable fungi and indicates that both the fungal host and culture conditions can influence the establishment of these widespread and important symbioses. By understanding how these symbioses are initiated and maintained, we can gain new insights into the cryptic ecological interactions that shape ubiquitous associations between plants and the Ascomycota, the largest and most economically important phylum of fungi.
MATERIALS AND METHODS
As part of a previous study, endophytes were isolated from healthy, surface-sterilized foliage of Platycladus orientalis (Cupressaceae) in Durham, NC (14). This collection included Pestalotiopsis sp. strain 9143 (Xylariales, Sordariomycetes) with its naturally occurring bacterial symbiont, Luteibacter sp. strain 9143, and Microdiplodia sp. strain 9145 (Botryosphaeriales, Dothideomycetes) with its naturally occurring symbiont, Luteibacter sp. strain 9145. Although the fungi represent distinct classes of Ascomycota, the bacteria are closely related: their 16S rRNA sequences are 100% identical, and their whole-genome sequences are nearly invariant (D. A. Baltrus, K. Dougherty, K. R. Arendt, M. Huntemann, A. Clum, M. Pillay, K. Palaniappan, N. Varghese, N. Mikhailova, D. Stamatis, T. B. K. Reddy, C. Y. Ngan, C. Daum, N. Shapiro, V. Markowitz, N. Ivanova, N. Kyrpides, T. Woyke, and A. E. Arnold, unpublished data). Both associations are accessioned as living cultures at the Robert L. Gilbertson Mycological Herbarium at the University of Arizona (accession numbers MYCO-ARIZ 9143 and 9145).
Preparation of axenic cultures.
Each fungal strain was cured of EHB by cultivation on 2% malt extract agar (MEA) amended with four antibiotics: ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (10 μg/ml), and ciprofloxacin (40 μg/ml) (14, 24, 26–29). We confirmed that fungal cultures were free of EHB using the molecular and visualization methods described below. Unless otherwise stated, axenic fungal strains were maintained on 2% MEA at 25°C.
EHB were isolated from naturally infected fungal cultures on 2% MEA that were incubated for 72 h at 36°C (14, 24, 25). At this temperature, bacteria emerged from hyphae and were isolated by streaking onto Luria broth (LB) agar (31). Stocks were prepared in LB by transfer under sterile conditions. To prevent contamination by other bacteria, the master stock of each EHB was passaged once from liquid culture to LB agar plates amended with rifampin (50 μg/ml). One growing colony was obtained per EHB strain and used as the source for further experiments. Unless otherwise stated, axenic bacterial strains were maintained in LB at 25°C.
Introduction of EHB into axenic fungi.
We introduced EHB into axenic mycelia of their original host species (i.e., Luteibacter sp. 9143 into Pestalotiopsis sp. 9143 and Luteibacter sp. 9145 into Microdiplodia sp. 9145) and the alternate host species (i.e., Luteibacter sp. 9145 into Pestalotiopsis sp. 9143 and Luteibacter sp. 9143 into Microdiplodia sp. 9145). Prior to reassociation, the fungal and bacterial strains were prepared as follows.
For each axenic fungus, a plug of mycelium (1.25-cm diameter) was collected under sterile conditions from inside the edge of an actively growing colony on 1× potato dextrose agar (PDA) (2.4%). Each plug was separately blended in three 5-s, high-speed pulses in a sterile blender (Waring 51BL31) in 100 ml of 1× potato dextrose broth (PDB) and then transferred to a sterile flask and incubated on a rotary shaker at 27°C and 100 rpm for 7 days. Mycelium was collected via vacuum filtration onto sterile 8-μm Whatman filter papers, washed twice with sterile 10 mM MgCl2 (32, 33), removed from the filter papers with forceps under sterile conditions, resuspended in 100 ml of 1× PDB, and blended as before.
Bacterial cultures were inoculated into 5 ml of LB and incubated on a rotary shaker at 36°C and 200 rpm for 3 days. The cultures were then centrifuged at a relative centrifugal force (RCF) of 300 for 3 min, and the supernatant was discarded. The pelleted cells were washed twice with 4 ml of sterile 10 mM MgCl2 and resuspended in 4 ml of 1× PDB. We used MgCl2 for the washing step because this has been a common step for other systems involving plant-associated bacteria: it is used to limit changes to bacterial cultures due to osmotic shock compared to washing with water alone (34).
Fungal and bacterial suspensions were combined in a ratio of 5:1 (mycelium/bacterium ratio) based on absorbance (ABS; i.e., optical density at 600 nm [OD600]) values for the respective suspensions. The absorbances of axenic cultures after washing and resuspension were 0.10 ABS for Microdiplodia sp. 9145, 0.15 ABS for Pestalotiopsis sp. 9143, and 1.10 and 0.90 for both Luteibacter strains after 3 days and 1 day (see below). Bacterial suspensions (0.9 ml and 1.4 ml) were added to 50-ml suspensions of Microdiplodia and Pestalotiopsis, respectively, and 1× PDB was added to each coculture to bring the yield to a total volume of 100 ml. These quantities were chosen because at higher concentrations of bacteria, we often observed bacteria growing on the external surfaces of fungal hyphae. Each coculture mixture was cultured for 7 days at 27°C in full darkness with agitation on a rotary shaker at 100 rpm.
Each coculture was prepared twice. After incubation, 20 μl of each coculture was transferred to six petri plates containing nutrient media: three plates contained 1× PDA, and three contained water agar. The plates were incubated at 27°C for 14 days. The bacterial infection status was verified as described below. The success of establishing symbioses was consistent across all replicates on each medium for each EHB-fungal association. After successful infection, fungi were subcultured three times on 2% MEA to confirm the stability of the association.
Molecular analysis and identification of EHB.
The presence and identity of EHB were confirmed using molecular analysis. For the former, total genomic DNA was extracted directly from fresh fungal mycelium collected from inside the growing edge of a fungal colony using a modified protocol from the Extract-N-Amp tissue PCR kit (Sigma-Aldrich). Genomic DNA was screened for the presence of bacteria by 16S rRNA PCR using RedTaq (Sigma) with primers 27F/1492R (35). The PCR conditions followed those described previously (14), but with an annealing temperature of 50°C and 40 cycles.
Positive 16S rRNA PCR amplicons were cleaned using ExoSap-It (Affymetrix) and sequenced bidirectionally with the primers used in PCR at the University of Arizona Genetics Core. Sequences were assembled automatically, bases were called, and quality scores were assigned by Phred (36) and Phrap (37) with orchestration by Mesquite v. 1.06 (38). Consensus sequences were edited manually in Sequencher 5.1 (Gene Codes Corp.).
In all cases, the sequences of bacteria obtained here were 100% identical to those reported previously from these cultures (14). BLAST searches of GenBank were conducted using BLASTn and highly similar sequences (38). Taxonomic placement within Luteibacter, validated previously by phylogenetic analysis (24), was confirmed using a ≥99% match over the full sequence length. As needed, the same methods were used to confirm the identity of EHB growing axenically. Because Luteibacter sp. 9143 and 9145 have identical 16S rRNA sequences, the strains were distinguished based on colony traits and associated phenotypes (Arendt et al., unpublished).
We did not observe any additional EHB or free-living bacteria in the fungal cultures used in this study. Visual and PCR-based evidence that fungi were free of EHB was confirmed by cloning from negative PCR products (i.e., PCR products generated as described above for which no bands were evident after 16S rRNA PCR). Cloning methods followed the manufacturer's instructions (Agilent; StrataClone) for reactions using half volumes. No positive clones were recovered from fungi after antibiotic treatment or from negative controls, whereas clones from EHB-infected strains consistently have provided evidence of EHB presence (14).
Visual confirmation of successful introduction.
We used a strain of Luteibacter sp. 9143 expressing a fluorescent protein (tdTomato) in Pestalotiopsis sp. 9143 to visually confirm that EHB were successfully introduced and that they were present within hyphae. Luteibacter sp. 9143 (strain DBL564) was mated with Escherichia coli strains containing the plasmids pRK2013 (39), pTNS2 (40), and pBT276 (41). A single Luteibacter colony was picked following selection for gentamicin, rifampin, and nitrofurantoin resistance (39–41). The resultant strain (DBL920) contained tdTomato integrated at its Tn7 site, with expression driven by Plac.
DBL920 was introduced into Pestalotiopsis sp. 9143 as described above. Fluorescence microscopy was used to confirm that the introduction was successful. Fresh mycelium was harvested under sterile conditions from cultures grown on 1× PDA, wet mounted on a glass microscope slide with 20 μl double-sterilized Milli-Q water, prepared with a coverslip, and secured with clear nail polish. The sample was examined using a Leica BX61 compound microscope with a 100-W mercury arc lamp, a Chroma Technology U-MWG filter (510- to 550-nm excitation/590-nm long pass emission), a direct-fluorescence (DF) stage filter, a 40× objective, and Leica software (LAS-AF v.1.8.2). This method was repeated using antibiotic-cured isolates of Pestalotiopsis sp. 9143, which showed no evidence of endohyphal bacteria, and axenic DBL920.
Confirmation of partner viability.
Visual assessments were used to confirm that EHB and fungi were viable throughout the experimental manipulations described above. Hyphae with EHB, hyphae without EHB, and axenic bacteria were evaluated using the LIVE/DEAD BacLight Bacterial Viability kit (Invitrogen) following the method of Hoffman and Arnold (14).
To prepare fungal samples for visualization, fresh hyphae were scraped from the surface of the growing edge of each fungal colony on 2% MEA. Axenic bacteria were prepared by scraping a single colony from LB agar. Hyphae or bacterial cells were placed on a glass slide with 15 μl of 1:1:18 LIVE/DEAD stain (component A:component B:sterile deionized water), covered with a coverslip, and incubated in darkness for 20 min. Sterile distilled water then was pulled through the slide mount with blotting paper. The slides were sealed with clear acrylic nail polish, which was allowed to dry before viewing. A Leica 4000MB compound microscope with a 100-W mercury arc lamp was used for fluorescent imaging at room temperature with a Chroma Technology 35002 filter set (480-nm excitation/520-nm emission) and a 100× APO oil objective. Three replicate slides were prepared per fungal culture, and in all cases, replicates from the same material were consistent.
Manipulation of reintroduction conditions.
We focused on the pairing of Pestalotiopsis sp. 9143 with Luteibacter sp. 9143 to further examine the culture conditions under which an EHB could be successfully reintroduced to axenic mycelium of a host fungus. Using conditions that permitted successful reassociation as a baseline protocol (see above), we altered the nutrient content for the coculture medium (1×, 0.1×, 0.01×, 0.001×, and 0.0001× PDB) and the mycelium/bacterium ratio in the coculture (10:1, 7:1, and 5:1). We also examined the role of the age of the bacterial culture at the time of coculturing (1 day old versus 3 days old), whether axenic cultures were washed with MgCl2 before coculturing, and the nutrient content of the solid medium onto which the coculture was plated (water agar versus 1× PDA). For this experiment, we adjusted the coculture volume to 5 ml to include more replicates. The cocultures were then incubated for 3 days in culture tubes. Each treatment was replicated twice, and one fungal colony from each replicate was screened for EHB. The treatments are shown in detail in Table S1 in the supplemental material.
The role of these treatments was quantified using nominal logistic analysis, with success of association as the response variable (yes or no) and the above-mentioned treatments as explanatory variables. Successful association (i.e., yes) was defined by detection of the bacterium using molecular analysis and confirmation that the bacteria were viable, were present in viable fungal tissue, and occurred within hyphae (rather than epihyphally).
RESULTS
Both Pestalotiopsis sp. 9143 and Microdiplodia sp. 9145 were viable on 1× PDA and water agar in the presence and absence of EHB. Each strain of Luteibacter was viable on LB agar and in LB under the conditions described above, and it could be isolated reliably following heat treatment of infected mycelia. EHB could be detected reliably from the original cultures using the above-described molecular and visualization methods, and they were confirmed to be absent following antibiotic treatment (see Fig. S1 in the supplemental material).
Reintroduction of EHB.
EHB were successfully reintroduced to their original host strains after the fungi were treated with antibiotics (Fig. 1; see Fig. S1 in the supplemental material). In each case, the bacteria were confirmed to be endohyphal (Fig. 1) and viable (green fluorescence with LIVE/DEAD) (results not shown). Reinfected strains resembled naturally infected strains with regard to hyphal morphology on 2% MEA.
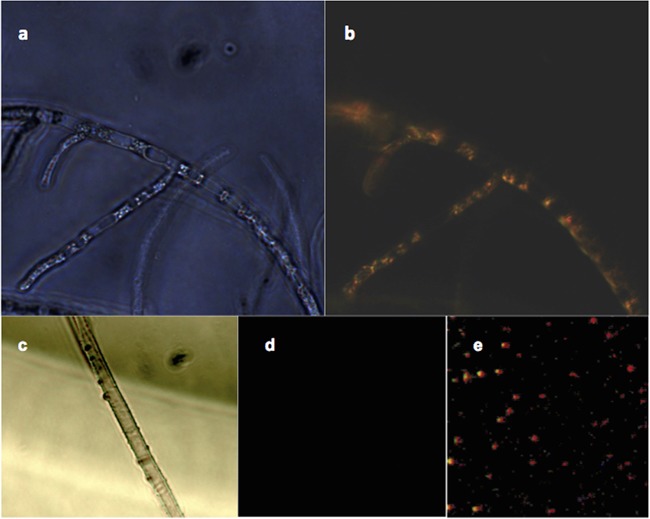
FIG 1.
(a and b) Successful reintroduction of Luteibacter sp. 9143 (tdTomato construct) into hyphae of Pestalotiopsis sp. 9143, illustrated with phase-contrast (a) and dark-field/fluorescence (b) microscopy. (c and d) Absence of Luteibacter sp. 9143 in cured hyphae of Pestalotiopsis sp. 9143, illustrated with phase-contrast (c) and dark-field/fluorescence (d) microscopy. (e) Free-living Luteibacter sp. 9143 (tdTomato construct) in pure culture, seen with dark-field/fluorescence microscopy. Magnification, 400× (a, b, and e) or 1,000× (c and d).
Reintroduction of Luteibacter sp. 9145 into Microdiplodia sp. 9145 was successful when the coculture was plated on PDA or water agar. However, reintroduction of Luteibacter sp. 9143 into Pestalotiopsis sp. 9143 was successful only on water agar. In each case, reinfected fungal strains maintained these associations through at least three subculturing events on 2% MEA. No bacterial growth was observed on these subculture plates. The identities of EHB were confirmed by PCR and sequencing.
Cross-inoculation of EHB.
Each bacterial strain was successfully introduced into the nonhost fungal strain (see Fig. S1 in the supplemental material). Luteibacter sp. 9143 was successfully introduced into Microdiplodia sp. 9145 when the coculture was plated on either PDA or water agar. Luteibacter sp. 9145 was successfully introduced into Pestalotiopsis sp. 9143 when the coculture was plated on water agar. In each case, the bacteria were confirmed to be endohyphal and viable and were maintained in their novel fungal hosts through at least three subculturing events on 2% MEA. The reinfected strains resembled the naturally infected strains with regard to hyphal morphology and the density of bacteria within hyphae.
Manipulation of reassociation conditions.
We examined the effects of particular culture conditions on reintroduction of EHB by focusing on the association between Pestalotiopsis sp. 9143 and Luteibacter sp. 9143. In total, 120 trials were conducted, each representing a bacterial culture of a given age (1 day old or 3 days old). Treatments included washing of axenic cultures with MgCl2 or not, various concentrations of the medium in which the coculture was grown (1×, 0.1×, 0.01×, 0.001×, and 0.0001× PDB), various mycelium/bacterium ratios in the coculture (10:1, 7:1, and 5:1), and final cultivation of the coculture on 1× PDA or water agar. All treatment combinations and their outcomes are shown in Table S1 in the supplemental material.
Nominal logistic analysis revealed that when all culture conditions were considered, those most relevant to successful resynthesis were (i) whether the axenic cultures were washed in MgCl2, (ii) the concentration of PDB in which the coculture was grown, and (iii) the solid medium on which the coculture was plated (Table 1). Culture age and the mycelium/bacterium ratio were less important (Table 1).
TABLE 1.
Results of nominal logistic regression assessing the importance of cultivation variables to successful in vitro reintroduction of an endohyphal bacterium (Luteibacter sp. 9143) into living mycelium of Pestalotiopsis sp. 9143 (a foliar fungus; Sordariomycetes, Ascomycota)a
| Factor | n | df | χ2 | P |
|---|---|---|---|---|
| Age of bacterial culture | 1 | 1 | 2.44 | 0.1186 |
| MgCl2 wash | 1 | 1 | 47.38 | <0.0001 |
| Concn of PDB | 4 | 4 | 25.25 | <0.0001 |
| Mycelium/bacterium ratio | 2 | 2 | 3.72 | 0.1559 |
| Medium | 1 | 1 | 38.22 | <0.0001 |
Success of reintroduction (yes or no) was used as the response variable. The explanatory variables are shown in all the other columns in Table S1 in the supplemental material (age of the bacterial culture, whether cultures were washed with MgCl2, the concentration of PDB, the mycelium/bacterium ratio, and the medium on which cocultures were plated). The whole model was significant (chi-square = 83.87; df = 9; P < 0.0001), and the results of effect tests, determined by likelihood ratios, are shown here. The results of this analysis were used to prune the total data set to identify the factors associated with positive reintroduction, which are shown in Table 2.
Examination of the data revealed that resynthesis failed in all 60 trials in which axenic cultures were not washed with MgCl2 (see Table S1 in the supplemental material). We therefore excluded those 60 trials from further analysis. Among the remaining 60 trials, resynthesis was more often successful when the coculture was plated on water agar rather than on 1× PDA: only 1 of 30 resynthesis attempts using PDA was successful (3-day-old bacterial culture, 0.01× PDB, and 7:1 mycelium/bacterium ratio), whereas 18 of 30 resynthesis attempts were successful using water agar (see Table S1 in the supplemental material). We therefore excluded the 30 trials on PDA from further analysis. These findings are consistent with the results of our initial assessment of the influence of the culture medium on the success of reintroducing Luteibacter sp. 9143 into Pestalotiopsis sp. 9143.
Among the remaining 30 trials, resynthesis attempts failed when the coculture was grown in 0.0001× PDB, but some resynthesis attempts were successful on each of the remaining concentrations of PDB (see Table S1 in the supplemental material). We therefore excluded the trials on 0.0001× PDB from further analysis.
Finally, we analyzed the remaining data set to more precisely evaluate the importance of the age of the bacterial culture, the concentration of PDB (1×, 0.1×, 0.01×, or 0.001×), and the mycelium/bacterium ratio in coculture establishment. Nominal regression of this reduced data set did not reveal significant differences among the suites of treatments listed here (simplified whole model, χ2 = 8.57, df = 6, P = 0.1993; no significant effects of any factor, P = 0.2659, P = 0.1009, and P = 0.4009, respectively). However, resynthesis was always successful when 0.01× PDB was used as the coculture medium, whereas success was more variable on higher concentrations of PDB (Table 2; see Table S1 in the supplemental material).
TABLE 2.
In vitro reintroduction of Luteibacter sp. 9143 into Pestalotiopsis sp. 9143 always failed when axenic cultures were not washed with MgCl2 and when cocultures were cultivated in 0.0001× PDB and failed in 29 of 30 trials when the cocultures were plated on 1× PDAa
| Coculture medium | Mycelium/bacterium ratio | Resynthesis |
|
|---|---|---|---|
| 1 day old | 3 days old | ||
| 1× PDB | 5:1 | No | Yes |
| 7:1 | No | Yes | |
| 10:1 | Yes | No | |
| 0.1× PDB | 5:1 | No | Yes |
| 7:1 | Yes | Yes | |
| 10:1 | Yes | No | |
| 0.01× PDB | 5:1 | Yes | Yes |
| 7:1 | Yes | Yes | |
| 10:1 | Yes | Yes | |
| 0.001× PDB | 5:1 | No | Yes |
| 7:1 | Yes | Yes | |
| 10:1 | Yes | Yes | |
Shown are qualitative outcomes of resynthesis attempts when cultures were washed with MgCl2, PDB concentrations were >0.0001×, and cocultures were plated on water agar for resynthesis attempts started with 1- and 3-day-old bacterial cultures. Nominal regression of this reduced data set did not reveal significant differences among the suites of treatments listed here; however, we note that resynthesis was always successful when 0. 0.001× PDB was used as the coculture medium, whereas success was more variable on other concentrations of PDB (see Table S1 in the supplemental material).
DISCUSSION
Previous work has suggested that endohyphal bacteria are facultative symbionts in many fungi (14, 20, 23, 24). Several studies have successfully cured fungal strains of their EHB for experimental use (21, 22, 24, 30). To our knowledge, in vitro reestablishment of the symbiosis between EHB and fungi has been achieved in one association previously: that of the root pathogen Rhizopus microsporus (Mucoromycotina) and its bacterial endosymbiont, Burkholderia rhizoxinica (Betaproteobacteria) (22, 30). In these studies, bacteria were removed via antibiotic treatments and then were reintroduced into their fungal host through coculturing on agar medium or microinjected into the fungal cytoplasm via a laser microbeam (22, 30). The production of bacterial chitinolytic enzymes may play an important role in the invasion of bacteria into fungal hyphae in this system (42).
The present study demonstrates resynthesis of associations between EHB and foliar fungal endophytes in the Ascomycota, the phylum that includes the vast majority of endophytic and plant-pathogenic fungi with relevance to agriculture and natural systems (17). We also demonstrate resynthesis involving Gammaproteobacteria, which are common in foliar endophytes studied to date (14) and can have profound effects on fungal traits (24; Arendt et al., unpublished). We show that resynthesis can be achieved readily in the laboratory using methods described here for two strains of EHB and two distantly related Ascomycota. Finally, we show that EHB can be moved readily between fungal strains, providing an important basis for future studies examining the degrees to which, and mechanisms by which, EHB influence fungal phenotypes. Importantly, the present study focused on two closely related bacteria; ongoing work will establish whether these conditions are appropriate for reintroduction of other EHB and for symbioses involving other fungal partners.
Consistent with previous work (14, 24), we found that the EHB association was facultative in these foliar fungi: both fungi and bacteria could be grown axenically on standard media. We were able to isolate bacteria from EHB-infected fungi by incubating cultures on 2% MEA at high temperature (36°C). Our observations of other fungal strains suggest that treatment at 36°C is not always successful in causing EHB to emerge in culture (Arendt et al., unpublished). In some cases, we observed bacteria emerging when mycelia were stored as vouchers in sterile water. In both thermal and water treatments, fungi appeared to be experiencing stress; whether this stress encourages bacterial proliferation or instead leads to fungal cell lysis and bacterial release remains to be explored.
Successful reassociation and cross-inoculation.
Context dependency is common in both the establishment and maintenance of many facultative fungal symbioses and in the functional outcomes of such associations (43–47) and likely shapes the success of EHB-fungal encounters in nature. Our results reveal that nutrient conditions can be important for successful reassociations. Reassociation was successful for both Pestalotiopsis sp. 9143-Luteibacter sp. 9143 and Microdiplodia sp. 9145-Luteibacter sp. 9145 on low-nutrient medium (water agar). When grown on water agar, hyphae of both fungal species were sparse, transparent, and thin; in contrast, both strains had robust, dense, and pigmented or opaque hyphae on PDA. Resynthesis of Microdiplodia sp. 9145 with Luteibacter sp. 9145 was also successful on high-nutrient medium. Cross-infection of both fungal hosts with novel EHB displayed the same patterns that were observed with their native EHB. In general, we observed more successful reassociations when the coculture step was performed under relatively dilute nutrient conditions, followed by plating onto low-nutrient medium. These results, combined with previous work indicating the facultative nature of EHB infections (14), might indicate that such associations are promoted by starvation in one or both partners. In future work, we will examine the potential contributions of substrates to the establishment of EHB infections in foliar endophytes.
In our experiments, we blended fungal mycelium at two points. This allowed us to standardize mycelial quantities in our treatments. Mycelial blending can break fungal cell walls but did not inhibit the viability of the fungal strains studied here, all of which were viable after treatment and grew as expected (data not shown). Traversing fungal cell walls may be important in establishing EHB in fungi, echoing previous studies that introduced EHB into fungal cytoplasm directly (22, 30). We speculate that horizontally transmitted and facultative EHB may enter foliar Ascomycota, such as endophytes, during the saprotrophic life phase of these fungi, during which hyphae may be damaged by agents such as microarthropods and the process of leaf fragmentation (48). We could not attempt reassociations or cross-inoculations on plates with solid media because the capacity of these EHB to grow readily on MEA meant that fungal and bacterial cells were intermixed with one another, making it difficult to determine whether the bacteria were epihyphal or endohyphal in such settings.
Our results suggest that washing cells with MgCl2 enhances the success of reassociations and cross-inoculations. Washing with MgCl2 is commonly used to limit osmotic shock in experimental manipulations of diverse bacteria (e.g., Gammaproteobacteria [34]). The widespread use of this method in bacterial studies encouraged our approach, but the efficacy of other salts could be explored.
When the MgCl2 wash was used, PDB concentrations were ca. 0.001× or greater, and cocultures were plated on water agar, resynthesis of Pestalotiopsis sp. 9143-Luteibacter sp. 9143 was achieved at various mycelium/bacterium ratios (5:1, 7:1, 10:1) and with 1-day-old or 3-day-old bacterial cultures. Qualitative examination of the results (Table 2; see Table S1 in the supplemental material) indicated that the most consistent success was obtained using 0.01× PDB, but the different suites of conditions did not differ statistically (for details, see Table S1 in the supplemental material). As such, the methods outlined in Table 2 were generally comparable, but for convenience, we suggest using 0.01× PDB.
When comparing resynthesis success with 1- versus 3-day-old bacterial cultures, we observed the trend that trials started with 3-day-old bacterial cultures reestablished symbiosis when the mycelium/bacterium ratio was lower and when the nutrient content of the coculturing medium was higher than in trials started with the 1-day-old culture. This could be due to a difference in the physiological state of the bacterial cells at the time of coculturing. Analysis of axenic cultures suggests that Luteibacter sp. 9143 reached stationary phase by 24 h after inoculation (see Fig. S2 in the supplemental material). We also noted that when cocultures started with the 1-day-old bacterial culture were plated on 1× PDA after cultivation in 1× PDB, viable bacteria often were observed living outside fungal cells (see Table S1 in the supplemental material). As such, resynthesis per se could not be verified. We therefore were conservative in our analyses and treated these as unsuccessful reintroductions.
Conclusions.
Here, we successfully cured two fungal endophytes of their endohyphal bacteria, reintroduced the bacteria into their original hosts, cross-inoculated to introduce each bacterium into a novel host from a different fungal class, and evaluated experimental methods to facilitate these processes in vitro. Together, the results show that fungal-host and culture conditions can define the outcome of symbiosis establishment between facultative, horizontally transmitted EHB and members of two classes of Ascomycota. Our findings, coupled with the emergence of EHB from fungal hyphae under stress, are consistent with the overarching hypothesis that EHB associations with these fungi are likely context dependent. Future work will aim to address the conditions in nature under which EHB are acquired or lost and the relevance for fungal phenotypes in natural and applied settings.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jana U'Ren, Naupaka Zimmerman, Kevin Dougherty, Anyangatia Ndobegang, Justin Shaffer, Ramiro Garza, and Julian Gonzales III for helpful discussions and technical assistance. We thank Marc Orbach for comments on early drafts of the manuscript.
Funding Statement
This work, including the efforts of A. Elizabeth Arnold, was funded by the National Science Foundation (NSF) (grants DEB-104766 and DEB-0702825). Work including the efforts of both A. Elizabeth Arnold and David A. Baltrus was also funded by NSF (IOS-1354219, to D.A.B., R. E. Gallery, and A.E.A). Further funding from NSF (award 0934013, to K. Lowe, University of Texas Pan American) supported R. Garza and J. Gonzales III through the University of Arizona Summer Research Institute. Additional support was provided by the College of Agriculture and Life Sciences and the School of Plant Sciences at the University of Arizona. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00452-16.
REFERENCES
- 1.Turner TR, James EK, Poole PS. 2013. The plant microbiome. Genome Biol 14:209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg G, Grube M, Schloter M, Smalla K. 2014. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol 5:148. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Putten WH, Klironomos JN, Wardle DA. 2007. Microbial ecology of biological invasions. ISME J 1:28–37. doi: 10.1038/ismej.2007.9. [DOI] [PubMed] [Google Scholar]
- 5.Márquez LM, Redman RS, Rodriguez RJ, Roossnick MJ. 2007. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513–515. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- 6.Fujimura R, Nishimura A, Ohshima S, Sato Y, Nishizawa T, Oshima K, Hattori M, Narisawa K, Ohta H. 2014. Draft genome sequence of the betaproteobacterial endosymbiont associated with the fungus Mortierella elongata FMR23-6. Genome Announc 2:e01272-14. doi: 10.1128/genomeA.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianciotto V, Lumini E, Bonfante P, Vandamme P. 2003. ‘Candidatus Glomeribacter gigasporarum’ gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int J Syst Evol Microbiol 53:121–124. doi: 10.1099/ijs.0.02382-0. [DOI] [PubMed] [Google Scholar]
- 8.Bertaux J, Schmid M, Hutzler P, Hartmann A, Garbaye J, Frey-Klett P. 2005. Occurrence and distribution of endobacteria in the plant-associated mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Environ Microbiol 7:1786–1795. doi: 10.1111/j.1462-2920.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- 9.Izumi H, Anderson IC, Alexander IJ, Killham K, Moore E. 2006. Endobacteria in some ectomycorrhiza of Scots pine (Pinus sylvestris). FEMS Microbiol Ecol 56:34–43. doi: 10.1111/j.1574-6941.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J, Kämpfer P, Domann E, Schäfer P, Hartmann A, Kogel K-H. 2008. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol 10:2235–2246. doi: 10.1111/j.1462-5822.2008.01202.x. [DOI] [PubMed] [Google Scholar]
- 11.Sato Y, Narisawa K, Tsuruta K, Umezu M, Nishizawa T, Tanaka K, Yamaguchi K, Komatsuzaki M, Ohta H. 2010. Detection of Betaproteobacteria inside the mycelium of the fungus Mortierella elongata. Microbes Environ 25:321–324. doi: 10.1264/jsme2.ME10134. [DOI] [PubMed] [Google Scholar]
- 12.Desiro A, Faccio A, Kaech A, Bidartondo MI, Bonfante P. 2015. Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytol 205:1464–1472. doi: 10.1111/nph.13136. [DOI] [PubMed] [Google Scholar]
- 13.Desiro A, Salvioli A, Ngonkeu EL, Mondo SJ, Epis S, Faccio A, Kaech A, Pawlowska TE, Bonfante P. 2014. Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J 8:257–270. doi: 10.1038/ismej.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman M, Arnold AE. 2010. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76:4063–4075. doi: 10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez RJ, White JF Jr, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 16.Lackner G, Hertweck C. 2011. Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog 7:e1002096. doi: 10.1371/journal.ppat.1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. 2011. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75:583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianciotto V, Genre A, Jargeat P, Lumini E, Becard G, Bonfante P. 2004. Vertical transmission of endobacteria in the arbuscular mycorrhizal fungus Gigaspora margarita through generation of vegetative spores. Appl Environ Microbiol 70:3600–3608. doi: 10.1128/AEM.70.6.3600-3608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naumann M, Schuessler A, Bonfante P. 2010. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J 4:862–871. doi: 10.1038/ismej.2010.21. [DOI] [PubMed] [Google Scholar]
- 20.Mondo SJ, Toomer KH, Morton JB, Lekberg Y, Pawlowska TE. 2012. Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution 66:2564–2576. doi: 10.1111/j.1558-5646.2012.01611.x. [DOI] [PubMed] [Google Scholar]
- 21.Lumini E, Bianciotto V, Jargeat P, Novero M, Salvioli A, Faccio A, Becard G, Bonfante P. 2007. Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria. Cell Microbiol 9:1716–1729. doi: 10.1111/j.1462-5822.2007.00907.x. [DOI] [PubMed] [Google Scholar]
- 22.Partida-Martinez LP, Monajembashi S, Greulich K-O, Hertweck C. 2007. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol 17:773–777. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 23.Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L, Cruveiller S, Bianciotto V, Piffanelli P, Lanfranco L, Bonfante P. 2012. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6:136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman MT, Gunatilaka MK, Wijeratne K, Gunatilaka L, Arnold AE. 2013. Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte. PLoS One 8:e73132. doi: 10.1371/journal.pone.0073132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman M. 2010. Bacterial endosymbionts of endophytic fungi: diversity, phylogenetic structure, and biotic interactions. Ph.D. thesis University of Arizona, Tucson, AZ. [Google Scholar]
- 26.Fisher PJ. 1996. Survival and spread of the endophyte Stagonospora pteridiicola in Pteridium aquilinum, other ferns and some flowering plants. New Phytol 132:119–122. doi: 10.1111/j.1469-8137.1996.tb04516.x. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues FK. 1994. The foliar fungal endophytes of the Amazonian palm Euterpe oleracea. Mycologia 86:376–385. doi: 10.2307/3760568. [DOI] [Google Scholar]
- 28.Lodge DJ, Fisher PJ, Sutton BC. 1996. Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia 88:733–738. doi: 10.2307/3760967. [DOI] [Google Scholar]
- 29.Gamboa MA, Bayman P. 2001. Communities of endophytic fungi in leaves of a tropical timber tree (Guarea guidonia: Meliaceae). Biotropica 33:352–360. [Google Scholar]
- 30.Lackner G, Moebius N, Hertweck C. 2011. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J 5:252–261. doi: 10.1038/ismej.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertani G. 1951. Studies on lysogenesis: the mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138:1315–1341. [Google Scholar]
- 33.Jankowska A, Laubitz D, Antushevich H, Zabielski R, Grzesiuk E. 2008. Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco-2 cells. J Biomed Biotechnol 2008:357964. doi: 10.1155/2008/357964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar S, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY. [Google Scholar]
- 36.Ewing B, Green P. 1998. Basecalling of automated sequencer traces using Phred: error probabilities. Genome Res 8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 37.Ewing B, Hiller L, Wendl M, Green P. 1998. Basecalling of automated sequencer traces using Phred: accuracy assessment. Genome Res 8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 41.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luitjen E, Parsek MR, Wong GC. 2013. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moebius N, Üzüm Z, Dijksterhuis J, Lackner G, Hertweck C. 2014. Active invasion of bacteria into living fungal cells. eLife 3:e03007. doi: 10.7554/eLife.03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCreadie JW, Beard CE, Adler PH. 2005. Context-dependent symbiosis between black flies (Diptera: Simuliidae) and trichomycete fungi (Harpellales: Legeriomycetaceae). Oikos 108:362–370. doi: 10.1111/j.0030-1299.2005.13417.x. [DOI] [Google Scholar]
- 44.Daskin JH, Alford RA. 2012. Context-dependent symbioses and their potential roles in wildlife diseases. Proc R Soc B Biol Sci 279:1457–1465. doi: 10.1098/rspb.2011.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith FA, Grace EJ, Smith SE. 2009. More than a carbon economy: nutrient trade and ecological sustainability in facultative arbuscular mycorrhizal symbioses. New Phytol 182:347–358. doi: 10.1111/j.1469-8137.2008.02753.x. [DOI] [PubMed] [Google Scholar]
- 46.Denison RF, Kiers ET. 2011. Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr Biol 21:R775–R785. doi: 10.1016/j.cub.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Picard KT, Letcher PM, Powell MJ. 2013. Evidence for a facultative mutualist nutritional relationship between the green coccoid alga Bracteacoccus sp. (Chlorophyceae) and the zoosporic fungus Rhizidium phycophilum (Chytridiomycota). Fungal Biol 117:319–328. doi: 10.1016/j.funbio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Crowther TW, Boddy L, Jones TH. 2012. Functional and ecological consequences of saprotrophic fungus-grazer interactions. ISME J 6:1992–2001. doi: 10.1038/ismej.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Partida-Martinez LP, Groth I, Schmitt I, Richter W, Roth M, Hertweck C. 2007. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int J Syst Evol Microbiol 57:2583–2590. doi: 10.1099/ijs.0.64660-0. [DOI] [PubMed] [Google Scholar]
- 50.Tarkka MT, Sarniguet A, Frey-Klett P. 2009. Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr Genet 55:233–243. doi: 10.1007/s00294-009-0241-2. [DOI] [PubMed] [Google Scholar]
- 51.Torres-Cortés G, Ghignone S, Bonfante P, Schüßler A. 2015. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci U S A 112:7785–7790. doi: 10.1073/pnas.1501540112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naito M, Morton JB, Pawlowska TE. 2015. Minimal genomes of mycoplasa-related endobacteria are plastic and contain host-derived genes for sustained life within Glomeromycota. Proc Natl Acad Sci U S A 112:7791–7796. doi: 10.1073/pnas.1501676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.