Abstract
The CB2 cannabinoid receptor (CB2) remains a tantalizing, but unrealized therapeutic target. CB2 receptor ligands belong to varied structural classes and display extreme functional selectivity. Here, we have screened diverse CB2 receptor ligands at canonical (inhibition of adenylyl cyclase) and noncanonical (arrestin recruitment) pathways. The nonclassic cannabinoid (−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol (CP55940) was the most potent agonist for both pathways, while the classic cannabinoid ligand (6aR,10aR)-3-(1,1-Dimethylbutyl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran JWH133) was the most efficacious agonist among all the ligands profiled in cyclase assays. In the cyclase assay, other classic cannabinoids showed little [(−)-trans-Δ9-tetrahydrocannabinol and (−)-(6aR,7,10,10aR)-tetrahydro-6,6,9-trimethyl-3-(1-methyl-1-phenylethyl)-6H-dibenzo[b,d]pyran-1-ol] (KM233) to no efficacy [(6aR,10aR)-1-methoxy-6,6,9-trimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromene(L759633) and (6aR,10aR)-3-(1,1-dimethylheptyl)-6a,7,8,9,10,10a-hexahydro-1-methoxy-6,6-dimethyl-9-methylene-6H-dibenzo[b,d]pyran]L759656. Most aminoalkylindoles, including [(3R)-2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenyl-methanone, monomethanesulfonate (WIN55212-2), were moderate efficacy agonists. The cannabilactone 3-(1,1-dimethyl-heptyl)-1-hydroxy-9-methoxy-benzo(c)chromen-6-one (AM1710) was equiefficacious to CP55940 to inhibit adenylyl cyclase, albeit with lower potency. In the arrestin recruitment assays, all classic cannabinoid ligands failed to recruit arrestins, indicating a bias toward G-protein coupling for this class of compound. All aminoalkylindoles tested, except for WIN55212-2 and (1-pentyl-1H-indol-3-yl)(2,2,3,3-tetramethylcyclopropyl)-methanone (UR144), failed to recruit arrestin. WIN55212-2 was a low efficacy agonist for arrestin recruitment, while UR144 was arrestin biased with no significant inhibition of cyclase. Endocannabinoids were G-protein biased with no arrestin recruitment. The diarylpyrazole antagonist 5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]hept-2-yl]-1H-pyrazole-3-carboxamide (SR144258) was an inverse agonist in cyclase and arrestin recruitment assays while the aminoalkylindole 6-iodo-2-methyl-1-[2-(4-morpholinyl)ethyl]-1H-indol-3-yl](4-methoxyphenyl)methanone (AM630) and carboxamide N-(1,3-benzodioxol-5-ylmethyl)-1,2-dihydro-7-methoxy-2-oxo-8-(pentyloxy)-3-quinolinecarboxamide (JTE907) were inverse agonists in cyclase but low efficacy agonists in arrestin recruitment assays. Thus, CB2 receptor ligands display strong and varied functional selectivity at both pathways. Therefore, extreme care must be exercised when using these compounds to infer the role of CB2 receptors in vivo.
Introduction
The endocannabinoid system is a highly conserved lipid signaling system whose core components include its two cognate receptors, CB1 and CB2 receptors, endogenous arachidonic acid–derived lipids that engage these receptors (endocannabinoids) and enzymes that synthesize and degrade the endocannabinoids (Pertwee et al., 2010; Console-Bram et al., 2012; Alexander et al., 2013) The CB1 and CB2 receptors belong to the G-protein-coupled receptor (GPCR) superfamily. These receptors have seven α-helices traversing the plasma membrane with an extracellular N- terminal domain and an intracellular C-terminal domain. Both CB1 and CB2 receptors preferentially couple to inhibitory G (Gi) proteins (Turu and Hunyady, 2010), although coupling of CB1R to Gs and Gq has also been observed (Glass and Felder, 1997; Abadji et al., 1999; Calandra et al., 1999; Lauckner et al., 2005).
Although the CB1 receptor is fairly well studied and characterized, lately substantial drug development efforts have been directed toward CB2 receptors since their activation is devoid of major psychotropic side effects and preclinical studies suggest efficacy in treating chronic pain, inflammation (Deng et al., 2015; Dhopeshwarkar and Mackie, 2014), fibrosis (Guillot et al., 2014), numerous neurodegenerative maladies such as multiple sclerosis (Pertwee, 2007), amyotrophic lateral sclerosis (Shoemaker et al., 2007), stroke (Pacher and Haskó, 2008), drug abuse, and depression (Onaivi et al., 2008; Xi et al., 2011; Ignatowska-Jankowska et al., 2013). Similar to CB1 receptors, CB2 receptor signaling involves canonical (G-protein-dependent, pertussis toxin–sensitive) and noncanonical (G-protein-independent, pertussis toxin–insensitive) components (McGuinness et al., 2009; van der Lee et al., 2009; Franklin et al., 2013).
Ligand engagement of a GPCR such as the CB2 receptor favors certain receptor conformations, some of which lead to the exchange of active GTP for inactive GDP on Gα subunits of Gαβγ heterotrimeric G proteins, leading to modulation of a variety of signaling pathways (Gudermann et al., 1997). For example, Gαi-GTP subunits inhibit adenylyl cyclase, decreasing cAMP production and thus suppressing cAMP-dependent protein kinase A activity (Bayewitch et al., 1995; Gonsiorek et al., 2000; Sugiura et al., 2000; Shoemaker et al., 2005; Cabral and Griffin-Thomas, 2009). Additionally, Gβγ subunits modulate certain calcium and potassium ion channels (Smrcka, 2008), ceramide biosynthesis (Herrera et al., 2006), and also promote phosphorylation and activation of a family of mitogen-activated protein kinases including ERK1/2, p38, and c-Jun N-terminal kinase as well as Akt kinase/protein kinase B and a range of second-messenger proteins and transcription factors (Bouaboula et al., 1996; Pertwee, 1997; McAllister et al., 1999; Sugiura et al., 2000; Howlett et al., 2002; Molina-Holgado et al., 2002; Ehrhart et al., 2005; Herrera et al., 2005, 2006; Pertwee et al., 2010; Atwood et al., 2012a).
The best described noncanonical CB2 receptor signaling is arrestin signaling. Arrestins are versatile monomeric cytosolic proteins that play a key role in receptor desensitization and internalization following phosphorylation of a GPCR by GPCR kinases (DeWire et al., 2007). They also act as adaptor proteins to direct GPCRs to clathrin-coated pits for endocytosis. Initially thought to have a role limited to receptor desensitization and internalization, arrestins are now appreciated for their role as multiscaffolding proteins that couple to various signaling proteins and form complexes with various signaling proteins, thus acting both as receptor signal transducers and terminators (Luttrell and Lefkowitz, 2002).
CB2 receptor ligands are structurally diverse, and similar to other GPCR ligands can be broadly classified into those that behave as agonists (positive efficacy), antagonists (neutral efficacy), and inverse agonists (negative efficacy) for a particular signaling pathway (Kenakin, 1987, 2002; Pertwee et al., 2010). These ligands stabilize different suites of CB2 receptor conformations, and hence have the ability to activate or inhibit varied subsets of signaling pathways with differing potencies and efficacies, a property known as functional selectivity or biased agonism (Kenakin, 2011; Atwood et al., 2012b). Functional selectivity is an important pharmacological concept that substantially increases the diversity of GPCR signaling. This ability of ligands to selectively engage specific cellular signaling pathways offers the theoretical possibility to design drugs/ligands that activate therapeutically relevant pathways, while avoiding those that lead to untoward side effects. Functional selectivity is also a tool that can be used to determine signaling pathways involved in specific biologic processes (Valant et al., 2014). Studies examining the functional selectivity at CB2 receptors were initially performed by Shoemaker et al. (2005) and more recently extended by Atwood et al. (2012b) and Schuehly et al. (2011) to additional classes of CB2 receptor ligands. The aim of the present investigation was to thoroughly explore/examine the functional selectivity elicited by structurally diverse CB2 receptor ligands at canonical and noncanonical CB2 receptor pathways.
Materials and Methods
(−)-Cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol (CP55940); 5-(4-Chlorophenyl)-1-(2,4-dichloro-phenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide (SR141716A); 5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]hept-2-yl]-1H-pyrazole-3-carboxamide (SR144528); and (−)-trans-Δ9-tetrahydrocannabinol (THC) were obtained from National institute of Drug Abuse Drug Supply Service (Bethesda, MD). Abbott Laboratories (Abbott Park, IL) generously provided N-[3-(2-methoxyethyl)-4,5-dimethyl-1,3-thiazol-2-ylidene]-2,2,3,3-tetramethylcyclopropane-1-carboxamide (A836339), while 3-(1,1-dimethyl-heptyl)-1-hydroxy-9-methoxy-benzo(c)chromen-6-one (AM1710); (1-(4-cyanobutyl)-3-(naphthalen-1-oyl)indole) (AM2232); (2-iodophenyl)[1-[(1-methyl-2-piperidinyl)methyl]-1H-indol-3-yl]-methanone (AM2233); and [1-(5-fluoropentyl)-1H-indol-3-yl](4-methyl-1-naphthalenyl)-methanone (MAM2201) were obtained from Dr. Alex Makriyannis (Northeastern University, Boston). 4-O-methylhonokiol and 4,11,11-trimethyl-8-methylene-bicyclo[7.2.0]undec-4-ene (E) β caryophyllene [(E) BCP] were gifts from Dr. Juerg Gertsch (University of Bern, Bern, Switzerland). Other reagents and drugs were purchased from Cayman Chemical (Ann Arbor, MI), Invitrogen (Carlsbad, CA), Sigma-Aldrich (St. Louis, MO), Tocris Cookson (Ellisville, MO), Thermo Fischer Scientific (Waltham, WA), Clontech (Mountain View, CA), or LI-COR Biosciences (Lincoln, NE).
Cell Culture.
Cyclase and internalization experiments were performed using human embryonic kidney (HEK) 293 cells stably expressing mouse CB2 receptors generated, expanded, and maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and penicillin/streptomycin (GIBCO, Carlsbad, CA) at 37°C in 5% CO2 (Atwood et al., 2012b). For ease of immunostaining, an amino-terminal hemagglutinin (HA) epitope tag was introduced into the CB2 receptor, a modification that does not appear to affect the extent or kinetics of CB2 internalization (Atwood et al., 2012b). To determine arrestin recruitment PathHunter Chinese hamster ovary (CHO) K1 CNR2 and CHO K1 CNR1 cells were purchased from DiscoveRx (Fremont, CA). These cells were grown and maintained in PathHunter AssayComplete media.
Cyclase Assays.
Cyclase assays were performed using the LANCE Ultra cAMP assay kit (Perkin Elmer, Boston). All cAMP assays were performed at room temperature with HEK293 cells transfected and stably expressing mouse CB2 receptors (mCB2) (HEK-mCB2) and were harvested from 50% confluent cell plates (log phase). Cells were centrifuged at 1000 rpm for 5 minutes and the pellet was resuspended in stimulation buffer [1X HEPES buffered saline (HBS), 5 mM HEPES, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX) 0.1% bovine serum albumin (BSA), pH 7.4, prepared fresh on the day of experiment] and incubated for 60 minutes at 37°C. About 500 cells/well (in 10 µl) were seeded in 384-well optiplate (Perkin Elmer) and stimulated with 5 µl of drugs (4X concentrated) and 5µl 4X forskolin (1 µM final concentration) for 5 minutes at room temperature. The reaction was terminated by addition of 5 µl cell lysis (1 × lysis buffer supplied by the vendor) followed by the addition of 10 µl Eu-cAMP tracer working solution (4X) and 10 µl Ulight anti-cAMP working solution (4X). After 60-minute incubation at room temperature, time-resolved fluorescence was measured on an Enspire multiplate reader (Perkin Elmer).
Arrestin Recruitment Assays.
Arrestin recruitment was measured using a cell-expressing proprietary enzyme complementation system (PathHunter, DiscoverX). These cells are engineered wherein an N-terminal deletion mutant of β-galactosidase enzyme (enzyme acceptor fragment) is fused with arrestin while a complimentary smaller fragment (ProLink) is fused with the C-terminal domain of a mouse cannabinoid receptor. Upon cannabinoid receptor activation, recruitment of arrestin leads to formation of an active β galactosidase enzyme, which then acts on the substrate to emit light that can be detected by luminescence. Briefly, CHO K1 CNR2 or CHO K1 CNR1 cells were seeded at a density of 20,000 cells/well in poly-D-lysine–coated 96-well plates (Costar 3596, Thermo Fisher Scientific). Cells were grown overnight in 90 µl of AssayComplete media under humidified conditions and 5% CO2 at 37°C. On the following day, medium was replaced by 90 µl of HBS/BSA (BSA: 0.2 mg/ml) and cells were challenged by adding 10 µl of compound/drug (10X) followed by incubation for 90 minutes at 37°C. (Compounds were dissolved in dimethylsulfoxide or ethanol as 10 mM stock solutions and were formulated in HBS/BSA) After the initial 90-minute drug incubation, 25 µl of PathHunter detection reagent was added to each well and the plate was further incubated for 60 minutes at room temperature. The extent of enzyme fragment complementation was monitored by the intensity of the chemiluminescent signal on an Enspire multiplate reader (Perkin Elmer). In preliminary experiments the assay was optimized for maximum chemiluminiscent signal by lysing cells at different times following addition of agonist. Optimum/maximum signal was achieved after 90-minute agonist incubation, which aligns well with the assay time suggested by DiscoverX in their product literature.
Internalization Assays.
On-cell internalization assays were performed using mouse HA-CB2 stably expressed in HEK cells grown to 95% confluency in Dulbecco’s modified Eagle’s medium (containing 10% fetal bovine serum, 0.5% penicillin/streptomycin) (Atwood et al., 2012b). Cells were gently washed once with HBS/BSA (BSA concentration, 0.08 mg/ml) 100 µl per well. Drugs dissolved in dimethylsulfoxide or ethanol were formulated in HBS/BSA and applied at EC90 concentrations (the EC90 concentration obtained from cyclase assay results of the selected compounds) and incubated further for 1 hour at 37°C. Cells were then fixed with 4% paraformaldehyde for 30 minutes and washed 5 times with HBS/BSA (300 µl per well) with Tris-buffered saline. Blocking buffer (Odyssey blocking buffer, LI-COR Biosciences) was applied at 150 μl per well for 1 hour at room temperature. Anti-HA antibody (mouse monoclonal, 1:200, Covance, Princeton, NJ) diluted in blocking buffer was then applied for 1 hour with gentle shaking at room temperature. Next, the plate was washed 5 times (300 µl per well) with Tris-buffered saline with 0.05% Tween 20. Secondary antibody (anti-mouse conjugated with an IR800 dye, 1:800) was then applied again for 1 hour at room temperature. Following this, the plate was washed 5 times with Tris-buffered saline with 0.05% Tween 20 (300 µl per well). The plate was then patted dry and scanned using LI-COR Biosciences Odyssey. Receptor internalization was calculated as the average integrated intensities of the drug-treated wells divided by the average integrated intensities of the untreated wells and the results are expressed as percentages.
Quantification of Signaling Bias.
The relatively unbiased (cyclase versus arrestin) mixed CB1/CB2 agonist, CP55940, was employed as the reference compound for these studies. Transduction coefficients (τ/KA) were determined by fitting concentration-response curves to the operational modal of bias (van der Westhuizen et al., 2014). The log R values [the logarithm of transduction coefficient (τ/KA)] obtained were normalized to the reference agonist, CP55940, to determine the relative effectiveness of select ligands to activate a signaling pathway [Δ log (τ/KA)] using the following equation:
| (1) |
To quantify the bias of selected ligands at the two pathways, relative to CP55940, the Δ log(τ/KA) values were normalized to the arrestin pathway to yield the log bias factor (BF) [ΔΔ log(τ/KA)] using the following equation:
| (2) |
Thus, bias was defined from the BF as follows:
| (3) |
The relative effectiveness of select ligands at the cyclase and arrestin pathways was calculated using the following equation:
| (4) |
All BFs reported are expressed as cyclase versus the arrestin pathway (bias of a ligand for adenylyl cyclase inhibition favored over arrestin recruitment).
Statistical Analysis.
Data are reported as mean ± S.E.M. or mean ± 95% confidence intervals. Nonlinear regression was employed to fit concentration-response curves. All graphs and statistical analyses were performed using GraphPad Prism version 4.0 (GraphPad Software, San Diego, CA). Unless otherwise mentioned, all assays were performed in triplicates.
Results
CB2 receptor ligands are highly structurally diverse (Supplemental Table 1; Table 1). The substantial heterogeneity of signaling by CB2 receptor ligands observed in earlier studies (Shoemaker et al., 2005; Atwood et al., 2012b) suggests the potential to design CB2 agonists that selectively activate specific signaling pathways, potentially with therapeutic value/utility, and may serve to explain the failure of CB2 agonists in clinical trials to date. Thus, in this study we have compared and contrasted how structurally diverse CB2 ligands signal via a G-protein and arrestin pathway. The mixed CB1/CB2 agonist CP55940 is generally regarded as a balanced, highly efficacious and potent agonist at CB2 receptors (Howlett et al., 2002). Hence, CP55940 was employed as a reference ligand in the present investigation. Activity thresholds were chosen as follows: compounds having an efficacy (% Emax) below 20%, with respect to the reference ligand CP55940, in either the cyclase or arrestin assays, were considered as inactive in that assay.
TABLE 1.
List of CB2 receptor ligand families and their structure-based classification (for agonists) examined in this study
| Class | Representative Members |
|---|---|
| Classic cannabinoids | Δ9THC, L759633, L759656, JWH133, KM233 |
| Nonclassic cannabinoids | CP55940, HU308 |
| Aminoalkylindoles | WIN55212-2, AM1241,STS135, JWH015, GW405833, UR144, MAM2201, AM2232, AM2233, AM1248 |
| Thiazoles | A836339 |
| Tricyclic pyrazole | GP1a |
| Cannabilactone | AM1710 |
| Carboxamides | SER601, 4Q3C |
| Pyrimidine analog | GW833972A |
| Eicosanoids | 2AG, methanandamide |
| Plant products | 4-Methylhonokiol, (E) β-caryophyllene (BCP) |
| Antagonists | AM630, JTE907, SR144528 |
AM1241, (2-iodo-5-nitrophenyl)-(1-(1-methylpiperidin-2-ylmethyl)-1H-indol-3-yl)methanone; AM1248, 1-[(N-methylpiperidin-2-yl)methyl]-3-(adamant-1-oyl)indole; AM2232, (1-(4-cyanobutyl)-3-(naphthalen-1-oyl)indole); A836339, N-[3-(2-Methoxyethyl)-4,5-dimethyl-1,3-thiazol-2-ylidene]-2,2,3,3-tetramethylcyclopropane-1-carboxamide; BCP, (1R,4E,9S)-4,11,11-trimethyl-8-methylidenebicyclo[7.2.0]undec-4-ene; GP1a, N-(piperidin-1-yl)-1-(2,4-dichlorophenyl)-1,4-dihydro-6-methylindeno[1,2-c]pyrazole-3-carboxamide; GW405833, 1-(2,3-dichlorobenzoyl)-5-methoxy-2-methyl-3- [2-(4-morpholinyl)ethyl]-1H-indole; GW833972A, 2-[(3-chlorophenyl)amino]-N-(4-pyridinylmethyl)-4-(trifluoromethyl)-5-pyrimidinecarboxamide hydrochloride; HU308, 4-[4-(1,1-dimethylheptyl)-2,6-dimethoxyphenyl]-6,6-dimethylbicyclo[3.1.1]hept-2-ene-2-methanol; JTE907, N-(1,3-benzodioxol-5-ylmethyl)-1,2-dihydro-7-methoxy-2-oxo-8-(pentyloxy)-3-quinolinecarboxamide; JWH133, (6aR,10aR)-3-(1,1-Dimethylbutyl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran; JWH015, (2-methyl-1-propyl-1H-indol-3-yl)-1-naphthalenylmethanone; KM233, (−)-(6aR,7,10,10aR)-tetrahydro-6,6,9-trimethyl-3-(1-methyl-1-phenylethyl)-6H-dibenzo[b,d]pyran-1-ol; L759633, (6aR,10aR)-1-methoxy-6,6,9-trimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromene; L759656, (6aR,10aR)-3-(1,1-dimethylheptyl)-6a,7,8,9,10,10a-hexahydro-1-methoxy-6,6-dimethyl-9-methylene-6H-dibenzo[b,d]pyran; MAM2201, [1-(5-fluoropentyl)-1H-indol-3-yl](4-methyl-1-naphthalenyl)-methanone; SER601, N-(Adamant-1-yl)-6-isopropyl-4-oxo-1-pentyl-1,4-dihydroquinoline-3-carboxamide Δ9THC, (−)-trans-Δ9-tetrahydrocannabinol; 4Q3C, 1,4-dihydro-8-methoxy-4-oxo-1-pentyl-N-tricyclo[3.3.1.13,7]dec-1-yl-3-quinolinecarboxamide; WIN55212-2, [(3R)-2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenyl-methanone, monomethanesulfonate; UR144, (1-pentyl-1H-indol-3-yl)(2,2,3,3-tetramethylcyclopropyl)-methanone.
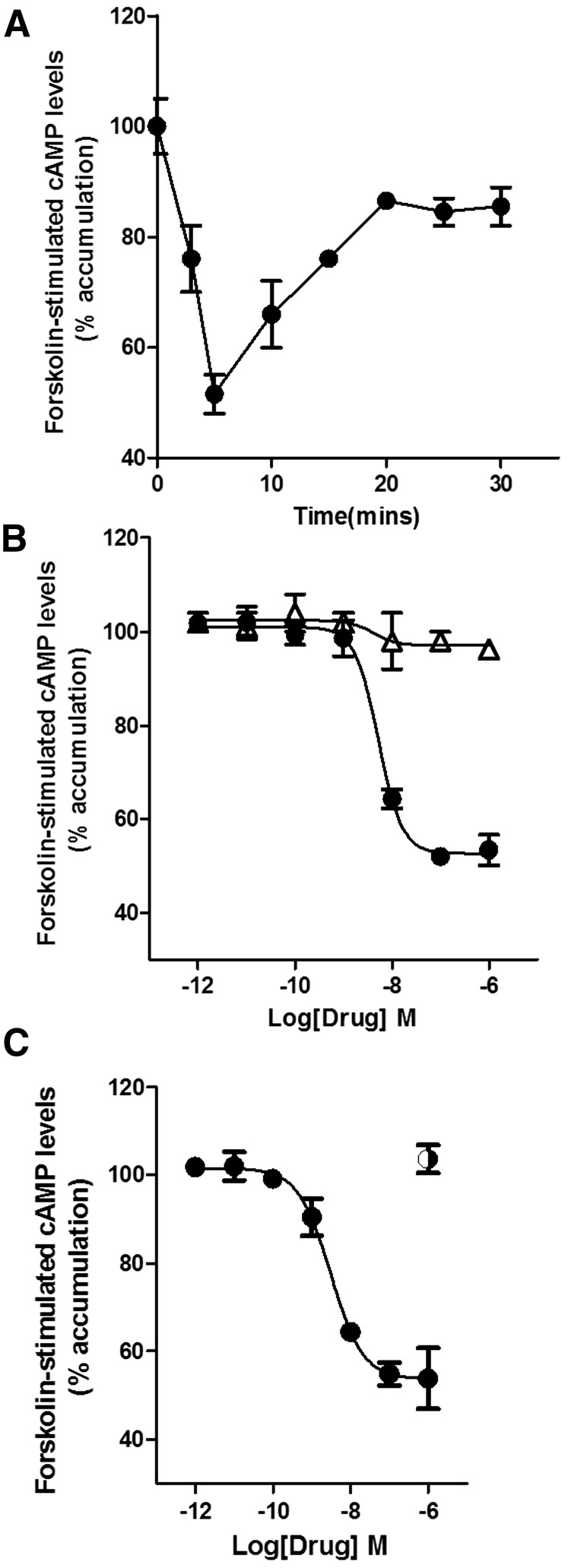
CB2 receptor ligands from different chemical scaffolds were first screened for adenylyl cyclase inhibition. The effects of these ligands on forskolin-stimulated cAMP levels (% accumulation) were performed using HEK293 cells stably transfected with HA-tagged mCB2 (HEK-mCB2). Figure 1A shows the time course for inhibition of forskolin-stimulated adenylyl cyclase by 1 µM CP55940. Inhibition of forskolin-stimulated cAMP levels was maximal at 5 minutes and then leveled off/plateaued to 90% of the forskolin-alone value at 20 minutes (Fig. 1A). CP55940 (1 µM) induced 50% ± 4.2% inhibition of forskolin-stimulated cAMP in HEK-mCB2 cells after a 5-minute treatment. Thus, subsequent experiments examined the inhibition of forskolin-stimulated cAMP production at 5 minutes of treatment. This inhibition was completely prevented by the potent and efficacious CB2 receptor antagonist, SR144528 (1 µM) (pretreated for 5 minutes) (Fig. 1B). CP55940-induced inhibition of forskolin-stimulated increases in cAMP levels was also pertussis toxin sensitive, indicating involvement of Gi/o proteins (Fig. 1C).
Fig. 1.
(A) Time course of CP55940 (●) (1 µM) inhibition of forskolin-stimulated adenylyl cyclase. Peak inhibition occurs after 5 minutes of treatment with CP55940 of HEK cells stably transfected with mCB2 receptors. (B) Effect of increasing concentrations of CP55940 was blocked by SR144528 (1 µM) (Δ). (C) CP55940 effects were pertussis toxin (◑) sensitive, indicating involvement of Gi/o G proteins in the inhibition of adenylyl cyclase. CP55940 EC50 and Emax were obtained by fitting the dose-response curve using nonlinear regression with GraphPad Prism 4.0. Data represent mean ± S.E.M. of at least three experiments.
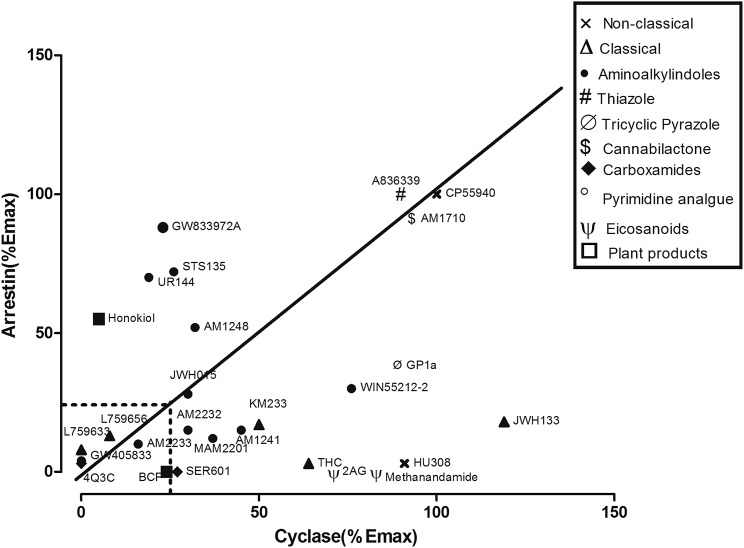
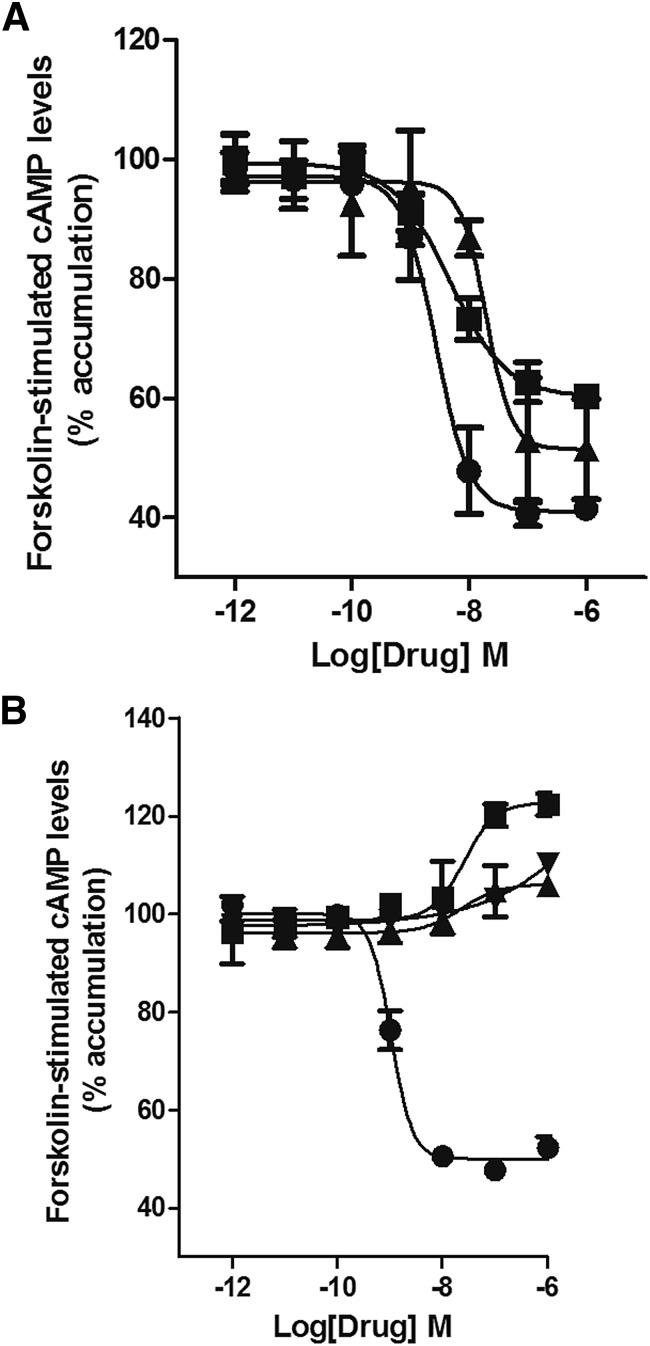
CB2 ligands affected cAMP levels with varying efficacies (as summarized in Fig. 2), even within the same chemical class. For example, among the classic cannabinoids, (6aR,10aR)-3-(1,1-Dimethylbutyl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran (JWH133) was slightly more efficacious than CP55940 in inhibiting the forskolin-stimulated increase in cAMP (Fig. 2; Supplemental Fig. 1B), while THC and KM233 were intermediate efficacy agonists (Fig. 2; Supplemental Fig. 2C) and L759656 and were inactive (Fig. 2; Table 2). All aminoalkylindoles were intermediate efficacy agonists in inhibiting adenylyl cyclase with the exception of GW405833 and UR144, which were inactive (Fig. 2; Supplemental Fig. 2; Table 2). The cannabilactone, AM1710, was equally efficacious but less potent than CP55940 in inhibiting adenylyl cyclase (Fig. 2; Supplemental Fig. 3A). The endocannabinoid, 2-arachidonoyl glycerol (2AG), and anandamide analog, methanandamide, were low efficacy agonists in the cyclase assays (Fig. 2; Fig. 3A). The three CB2 antagonists tested, 6-iodo-2-methyl-1-[2-(4-morpholinyl)ethyl]-1H-indol-3-yl](4-methoxyphenyl)methanone (AM630); SR144258; and N-(1,3-benzodioxol-5-ylmethyl)-1,2-dihydro-7-methoxy-2-oxo-8-(pentyloxy)-3-quinolinecarboxamide (JTE907) all behaved as inverse agonists in the cyclase assays (Fig. 3B).
Fig. 2.
Efficacy-based clustering of the CB2 receptor ligands based on their efficacy in inhibiting adenylyl cyclase and recruiting arrestin. CP55940, AM1710, and A836339 were efficacious and unbiased agonists. 4-O-methylhonokiol, STS135, UR144, and GW833972A were more arrestin biased with the remainder of the compounds either cyclase biased, weakly efficacious, or ineffective at either pathway.
TABLE 2.
EC50 and Emax values for CB2 ligands in the cyclase and arrestin assays
The EC50 values presented with 95% confidence intervals, while the Emax data are presented as mean ± S.E.M. and P values (*P < 0.05; **P < 0.01, ***P < 0.001). The P values were obtained use the Student’s t test and by comparing the efficacy of the test compound at 1 µM with the effect of 1 µM CP55940. For antagonists, the comparisons were to basal signaling. The *P and **P values represent compounds meeting >20% activation or inhibition thresholds (compared with CP55940); the ***P values represent compounds that failed to meet the 20% activation or inhibition thresholds (compared with CP55940).
| Compound |
Cyclase Assay |
Arrestin Recruitment Assay |
||||||
|---|---|---|---|---|---|---|---|---|
| EC50 |
95% CI |
Emaxa |
P value |
EC50 |
95% CI |
Emax |
P value |
|
| nM | nM | % | nM | |||||
| CP55940 | 3 | 1.1–8.5 | 57 ± 0.5 | NA | 3.2 | 0.2–5.5 | 100 | NA |
| THC | 7.3 | 2.3–20.4 | 52 ± 1.2 | * | NA | NA | 3 ± 1.3 | *** |
| L759633 | NA | NA | NA | NA | NA | NA | 10 ± 1.5 | *** |
| L759656 | NA | NA | NA | NA | NA | NA | 11 ± 1.1 | *** |
| JWH133 | 20 | 15.5–28.1 | 61 ± 1.1 | * | NA | NA | 18 ± 2.1 | *** |
| KM233 | 1.6 | 0.2–7.7 | 50 ± 1.3 | ** | NA | NA | 17 ± 3.4 | *** |
| HU308 | 30 | 27.6–35.1 | 60 ± 3.4 | NS | NA | NA | 3 ± 1.6 | *** |
| WIN55212-2 | 16 | 10.5–21.7 | 40 ± 1.2 | ** | 7.2 | 3.4–11.0 | 30 ± 1.2 | * |
| AM1241 | 20 | 18.1–22.5 | 27 ± 5.2 | * | NA | NA | 17 ± 1.9 | *** |
| STS135 | 52 | 40.9–65.11 | 31 ± 2.3 | * | 1.5 | 1–3.9 | 62 ± 1.1 | * |
| JWH015 | 30 | 24.4–38.1 | 23 ± 0.72 | * | 160 | 101–225 | 28 ± 1.5 | ** |
| GW405833 | NA | NA | 0 | *** | NA | NA | 4 ± 2.6 | *** |
| UR144 | NA | NA | 12 ± 0.7 | 95 | 78.2–135 | 70 ± 1.2 | ** | |
| MAM2201 | 7 | 1.1–11.5 | 26 ± 2.1 | * | NA | NA | 12 ± 3.4 | *** |
| AM2232 | 22 | 17.3–24.3 | 24 ± 1.3 | * | NA | NA | 15 ± 2.7 | *** |
| AM2233 | NA | NA | 7 ± 1.6 | *** | NA | NA | 10 ± 1.5 | *** |
| AM1248 | 15 | 12.3–17.3 | 24 ± 3.1 | * | 71 | 62.1–99 | 52 ± 2.3 | * |
| A836339 | 43 | 39.2-46 | 54 ± 1.1 | NS | 0.7 | 0.04–2.4 | 100 (NS) | NS |
| GP1a | 14 | 10.1–25.2 | 51 ± 7.6 | NS | 17 | 9.1–27.3 | 38 ± 1.2 | * |
| AM1710 | 11 | 5.5–15.6 | 48 ± 4.3 | NS | 4 | 1.6–7.1 | 91 ± 3.6 | NS |
| SER601 | 40 | 19.2–76.1 | 25 ± 1.1 | ** | NA | NA | NA | NA |
| 4Q3C | NA | NA | NA | NA | NA | NA | NA | NA |
| GW833972A | 28 | 20–56.1 | 22 ± 7.5 | * | 90 | 56.1–135 | 88 ± 2.5 | ** |
| 2AG | 4.1 | 0.4–7.8 | 39 ± 0.7 | * | NA | NA | NA | NA |
| Methanandamide | 20 | 17.9–25.3 | 55 ± 0.5 | * | NA | NA | NA | NA |
| 4 Methylhonokiol | NA | NA | NA | NA | 51 | 42.1–62 | 55 ± 4.6 | ** |
| β Caryophyllene | 30 | 22.1–36.6 | 5 ± 1.1 | *** | NA | NA | NA | NA |
| AM630 | 25 | 21.6–29.3 | 19 ± 2.4b | ** | 9 | 3.8–14.4 | 43 ± 4.2 | ** |
| JTE907 | 22 | 14.1–24.5 | 8 ± 0.8b | ** | 0.1 | 0.01–3.6 | 47 ± 1.5 | ** |
| SR144528 | NA | NA | 11 ± 0.7b | ** | 15 | 11–18.3 | 43 ± 3.1c | ** |
AM1241, (2-iodo-5-nitrophenyl)-(1-(1-methylpiperidin-2-ylmethyl)-1H-indol-3-yl)methanone; AM1248, 1-[(N-methylpiperidin-2-yl)methyl]-3-(adamant-1-oyl)indole; AM2232, (1-(4-cyanobutyl)-3-(naphthalen-1-oyl)indole); A836339, N-[3-(2-methoxyethyl)-4,5-dimethyl-1,3-thiazol-2-ylidene]-2,2,3,3-tetramethylcyclopropane-1-carboxamide; CI, confidence interval; Emax, maximal inhibition of forskolin-stimulated cAMP production; GP1a, N-(piperidin-1-yl)-1-(2,4-dichlorophenyl)-1,4-dihydro-6-methylindeno[1,2-c]pyrazole-3-carboxamide; GW833972A, 2-[(3-chlorophenyl)amino]-N-(4-pyridinylmethyl)-4-(trifluoromethyl)-5-pyrimidinecarboxamide hydrochloride; HU308, 4-[4-(1,1-dimethylheptyl)-2,6-dimethoxyphenyl]-6,6-dimethylbicyclo[3.1.1]hept-2-ene-2-methanol; JTE907, N-(1,3-benzodioxol-5-ylmethyl)-1,2-dihydro-7-methoxy-2-oxo-8-(pentyloxy)-3-quinolinecarboxamide; JWH015, (2-methyl-1-propyl-1H-indol-3-yl)-1-naphthalenylmethanone; JWH133, (6aR,10aR)-3-(1,1-Dimethylbutyl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran; KM233, (−)-(6aR,7,10,10aR)-tetrahydro-6,6,9-trimethyl-3-(1-methyl-1-phenylethyl)-6H-dibenzo[b,d]pyran-1-ol; L759633, (6aR,10aR)-1-methoxy-6,6,9-trimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromene; L759656, (6aR,10aR)-3-(1,1-dimethylheptyl)-6a,7,8,9,10,10a-hexahydro-1-methoxy-6,6-dimethyl-9-methylene-6H-dibenzo[b,d]pyran; MAM2201, [1-(5-fluoropentyl)-1H-indol-3-yl](4-methyl-1-naphthalenyl)-methanone; NA, not applicable/cannot be determined (inhibition or activation <20% threshold); NS, non statistically significant difference; SER601, N-(Adamant-1-yl)-6-isopropyl-4-oxo-1-pentyl-1,4-dihydroquinoline-3-carboxamide THC, tetrahydrocannabinol; 4Q3C, 1,4-dihydro-8-methoxy-4-oxo-1-pentyl-N-tricyclo[3.3.1.13,7]dec-1-yl-3-quinolinecarboxamide; WIN55212-2, [(3R)-2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenyl-methanone, monomethanesulfonate.
Percentage of inhibition of forskolin-stimulated cAMP production.
Above basal.
Below basal.
Fig. 3.
Cyclase assay: (A) 2AG (▪) (EC50 4.1 nM) and the stable analog of anandamide, methanandamide (▲) (EC50 20 nM), were intermediate efficacy agonists of the cyclase pathway, with 2AG being more potent. CP55940 (●) inhibition of adenylyl cyclase is shown for reference. (B) AM630 (▪), JTE907 (▲), and SR144528 (▼) behaved as inverse agonists by increasing the accumulation of cAMP above that seen with forskolin alone. EC50 and Emax were obtained by fitting the dose-response curve using nonlinear regression with GraphPad Prism 4.0. Data represent mean ± S.E.M. of at least three experiments.
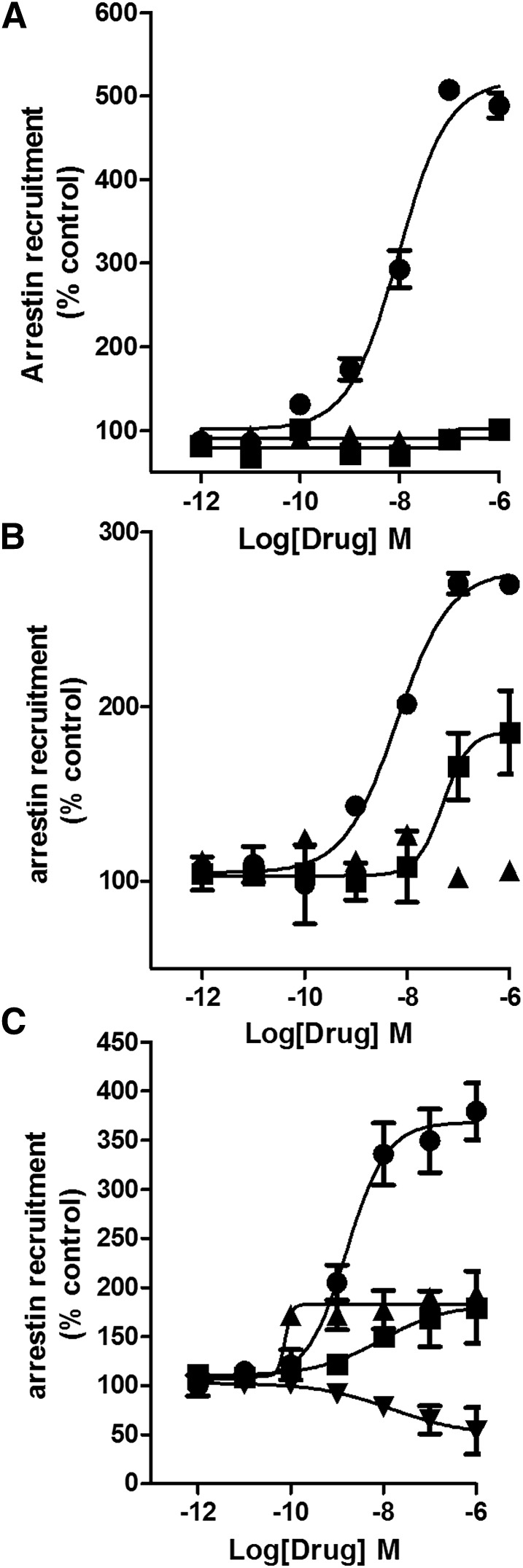
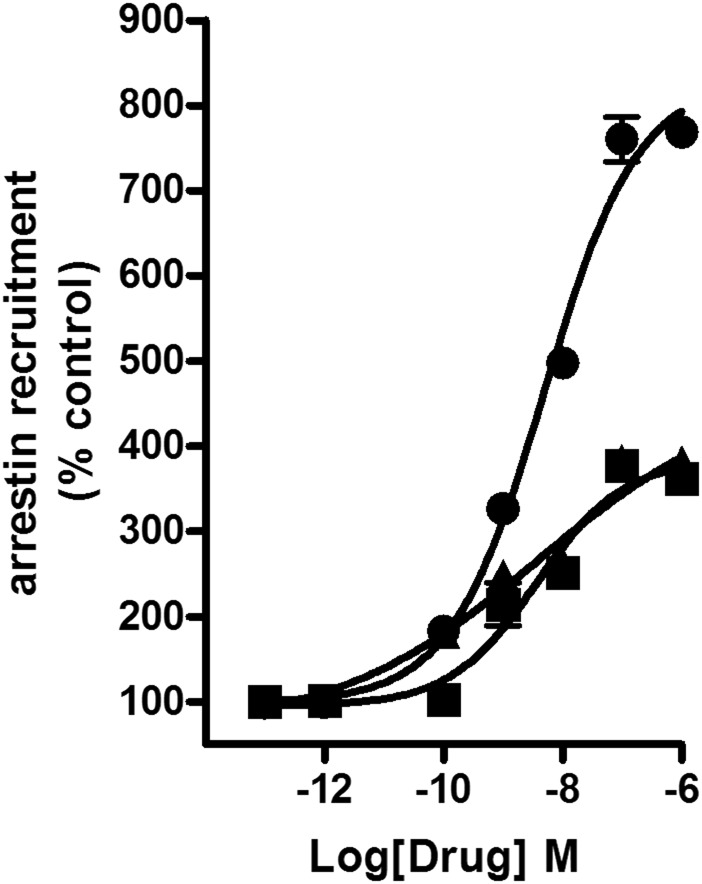
Next, arrestin recruitment was evaluated using an enzyme complementation assay (see Materials and Methods). The nonclassic cannabinoid, CP55940, was potent and efficacious in recruiting arrestin with an EC50 value of 3.2 nM (Emax 100%, set as the reference ligand) (Fig. 2; Fig. 4A). Interestingly, all classic cannabinoids failed to substantially recruit arrestin (P > 0.05 at 1 µM), even JWH133, which efficaciously inhibited adenylyl cyclase. This result indicates a potential for strong G-protein bias for this structural family at CB2 receptors. Interestingly, the nonclassic cannabinoid, 4-[4-(1,1-dimethylheptyl)-2,6-dimethoxyphenyl]-6,6-dimethylbicyclo[3.1.1]hept-2-ene-2-methanol (HU308, a congener of CP55940), also failed to recruit arrestin. The ability of aminoalkylindoles to recruit arrestin varied the most among the CB2 agonist families investigated: GW405833; MAM2201; AM2232; AM2233; and AM1241 weakly recruited arrestins (Table 2). WIN55,212-2 had intermediate efficacy, which aligns well with our earlier data (Atwood et al., 2012b), as did AM1421 and JWH015. The designer drug STS135 was efficacious and potent in recruiting arrestin, and had moderate efficacy and low potency to inhibit adenylyl cyclase. STS135 and UR144, an important constituent of some Spice/K2 preparations, were the only aminoalkylindoles that were arrestin biased (Supplemental Fig. 4, B and C). Interestingly, UR144 and STS135 minimally inhibited cAMP production. Thus, aminoalkylindoles—exceptions being WIN55,212-2; JWH015; AM1248; STS135; and UR144—displayed strikingly varied functional selectivity, with the majority of them being low efficacy agonists in cyclase assays with little activation of the arrestin pathway (Table 2). The thiazole, A836339, and cannabilactone, AM1710, both had efficacy similar to CP55940 in the arrestin recruitment assay (Supplemental Fig. 5, A and B). 4-O-Methylhonokiol recruited arrestin with intermediate potency and efficacy (Fig. 4B); the other natural product tested, (E) β-caryophyllene, was inactive (Fig. 4B). Two of the three CB2 antagonists tested, AM630 and JTE907, recruited arrestin with low efficacy, while SR144258 showed inverse agonism in this assay (Fig. 4C).
Fig. 4.
Arrestin recruitment assay: (A) 2AG (▪) and the stable anandamide analog, methanandamide (▲), failed to recruit arrestins. (B) The natural product, 4-O-methylhonokiol (▪) (EC50 51 nM), was a low efficacy agonist, while β caryophyllene (▲) failed to recruit arrestins. (C) SR144528 (▼) (EC50 2.5 nM) was an inverse agonist, while AM630 (▪) and JTE907 (▲) were low efficacy agonists in recruiting arrestins. EC50 and Emax values were obtained by fitting the dose-response curve using nonlinear regression with GraphPad Prism 4.0. Data represent mean ± S.E.M. of at least three experiments.
The endocannabinoid, 2AG and the anandamide analog, methanandamide, failed to recruit arrestins via CB2, indicating a strong bias toward G-protein signaling (Fig. 4A). It is intriguing to observe that 2AG and methanandamide do recruit arrestins via CB1 receptors (CHO cells transfected with mCB1 receptors) (Fig. 5). Thus, these agonists differentially activate arrestin signaling through CB1 and CB2 receptors.
Fig. 5.
Concentration-response curves for mCB1-mediated arrestin recruitment by CP55940 (●), 2AG (▪), and the stable anandamide analog methanandamide (▲). 2AG and methanandamide were low efficacy agonists in arrestin recruitment to CB1 receptor. EC50 and Emax values were obtained by fitting the dose-response curve using nonlinear regression with GraphPad Prism 4.0. Data represent mean ± S.E.M. of at least three experiments.
To sum up briefly, CP55940; AM1710; A836339, AM1248, and JWH015 were the most balanced compounds evaluated, although the latter two had lower efficacy. The majority of the other compounds screened were G-protein biased, while a few were more arrestin biased: STS135; UR144; 4-O-methylhonokiol; and GW833972A (Table 3). Thus, few of the compounds evaluated showed bias toward recruitment of arrestin (see Supplemental Fig. 6). Several compounds failed to show activity in our model system based on our thresholds: L759633, L759656; AM2233; GW405833; and 4Q3C showed very low activity for either inhibition of adenylyl cyclase or stimulation of arrestin recruitment (see Table 2 for a comprehensive list of EC50 and Emax values for these ligands).
TABLE 3.
Transduction ratios and BFs of CB2 receptor ligands (cyclase over arrestin pathway)
| Compound | cAMP |
Arrestin |
|||||
|---|---|---|---|---|---|---|---|
| Log R | Δ Log(τ/KA) | Log R | Δ Log(τ/KA) | ΔΔ Log (τ/KA) | BF | 95% CI | |
| CP55940 | 8.10 | 0.00 | 8.27 | 0.00 | 0.00 | 1.00 | 0.95–1.08 |
| THC | 8.23 | 0.13 | 6.88 | −1.39 | 1.52 | 33.11 | 28.3–35.6 |
| L759633 | NA | NA | NA | NA | NA | NA | NA |
| L759656 | NA | NA | NA | NA | NA | NA | NA |
| JWH133 | 7.1 | −1 | 7 | −1.62 | 0.62 | 4.16 | 3.99–4.27 |
| KM233 | 8.73 | −0.04 | 7.1 | −1.52 | 1.48 | 30.19 | 28.19–33.56 |
| HU308 | 10.5 | 2.00 | 9.1 | 0.4 | 1.6 | 39.8 | 38.1–42.3 |
| WIN55212-2 | 8.9 | 0.867 | 8.22 | −0.007 | 0.874 | 7.41 | 5.8–8.7 |
| AM1241 | 7.73 | −0.46 | 6.8 | −1.9 | 1.44 | 27.54 | 24.9–31.6 |
| STS135 | 7.88 | −0.62 | 8.79 | 0.05 | −0.67 | 0.2 | 0.14–0.29 |
| JWH015 | 8.56 | 0.363 | 7.65 | −1.09 | 1.45 | 28.13 | 26.1–32.9 |
| GW405833 | NA | NA | NA | NA | NA | NA | NA |
| UR144 | 7.9 | −0.8 | 10.5 | 2 | −2.8 | 0.0015 | 0.0007–0.002 |
| MAM2201 | 8.25 | −0.32 | 10.1 | 1.36 | 1.68 | 47.86 | 41.4–56.1 |
| AM2232 | 8.34 | 0.09 | 7.0 | −1.23 | 1.32 | 20.89 | 16.2-28.7 |
| AM2233 | NA | NA | NA | NA | NA | NA | NA |
| AM1248 | 7.39 | −1.2 | 8.78 | 0.04 | −1.16 | 0.079 | 0.043–0.12 |
| A836339 | 9.1 | 0.4 | 9.1 | 0.32 | 0.08 | 1.2 | 0.91–1.7 |
| GP1a | 7.8 | −0.7 | 7.95 | −0.75 | 0.05 | 1.12 | 0.4–1.9 |
| AM1710 | 7.92 | −0.33 | 7.919 | −0.358 | −0.028 | 0.95 | 0.5–1.2 |
| SER601 | 8.85 | 0.35 | 7.04 | −1.23 | 1.58 | 38.01 | 30.2–45.7 |
| 4Q3C | NA | NA | NA | NA | NA | NA | NA |
| GW833972A | 7.65 | −0.85 | 7.37 | −1.25 | 0.4 | 2.5 | 1.7–2.9 |
| 2AG | 8.393 | 0.290 | 6.9 | −1.37 | 1.66 | 47.8 | 45.2–51.1 |
| Meth-anandamide | 9.67 | 1.1 | 7.51 | −0.381 | 1.5 | 32 | 30.51–35.78 |
| 4 Methylhonokiol | 7 | −1.103 | 10.010 | 1.733 | −2.836 | 0.002 | 0.0014–0.032 |
| β Caryophyllene | NA | NA | NA | NA | NA | NA | NA |
| AM630 | NA | NA | NA | NA | NA | NA | NA |
| JTE907 | NA | NA | NA | NA | NA | NA | NA |
| SR144528 | NA | NA | NA | NA | NA | NA | NA |
AM1241, (2-iodo-5-nitrophenyl)-(1-(1-methylpiperidin-2-ylmethyl)-1H-indol-3-yl)methanone; AM1248, 1-[(N-methylpiperidin-2-yl)methyl]-3-(adamant-1-oyl)indole; AM2232, (1-(4-cyanobutyl)-3-(naphthalen-1-oyl)indole); A836339, N-[3-(2-methoxyethyl)-4,5-dimethyl-1,3-thiazol-2-ylidene]-2,2,3,3-tetramethylcyclopropane-1-carboxamide; GP1a, N-(piperidin-1-yl)-1-(2,4-dichlorophenyl)-1,4-dihydro-6-methylindeno[1,2-c]pyrazole-3-carboxamide; GW833972A, 2-[(3-chlorophenyl)amino]-N-(4-pyridinylmethyl)-4-(trifluoromethyl)-5-pyrimidinecarboxamide hydrochloride; HU308, 4-[4-(1,1-dimethylheptyl)-2,6-dimethoxyphenyl]-6,6-dimethylbicyclo[3.1.1]hept-2-ene-2-methanol; JTE907, N-(1,3-benzodioxol-5-ylmethyl)-1,2-dihydro-7-methoxy-2-oxo-8-(pentyloxy)-3-quinolinecarboxamide; JWH015, (2-methyl-1-propyl-1H-indol-3-yl)-1-naphthalenylmethanone; JWH133, (6aR,10aR)-3-(1,1-Dimethylbutyl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran; KM233, (−)-(6aR,7,10,10aR)-tetrahydro-6,6,9-trimethyl-3-(1-methyl-1-phenylethyl)-6H-dibenzo[b,d]pyran-1-ol; L759633, (6aR,10aR)-1-methoxy-6,6,9-trimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromene; L759656, (6aR,10aR)-3-(1,1-dimethylheptyl)-6a,7,8,9,10,10a-hexahydro-1-methoxy-6,6-dimethyl-9-methylene-6H-dibenzo[b,d]pyran; MAM2201, [1-(5-fluoropentyl)-1H-indol-3-yl](4-methyl-1-naphthalenyl)-methanone; NA, not applicable/cannot be determined; SER601, N-(Adamant-1-yl)-6-isopropyl-4-oxo-1-pentyl-1,4-dihydroquinoline-3-carboxamide; THC, tetrahydrocannabinol; 4Q3C, 1,4-dihydro-8-methoxy-4-oxo-1-pentyl-N-tricyclo[3.3.1.13,7]dec-1-yl-3-quinolinecarboxamide; WIN55212-2, [(3R)-2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenyl-methanone, monomethanesulfonate.
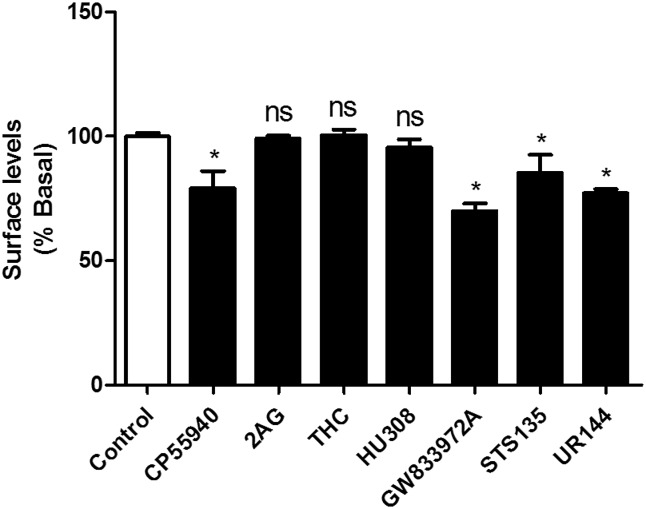
Ligand-induced activation of CB2 receptors often leads to receptor internalization. Internalization is independent of Gi/o protein signaling since it is not blocked by pertussis toxin but requires arrestin. Thus, a ligand biased toward the arrestin pathway may display higher efficacy in receptor internalization while a ligand biased toward G-protein coupling may fail to cause internalization or induce only modest internalization. Using on-cell western internalization assays, we tested this hypothesis by evaluating select ligands that were either balanced or biased to either the cyclase or arrestin pathway (see Table 3). Figure 6 depicts the extent of internalization induced by these agonists at a concentration that produced 90% inhibition of adenylyl cyclase. We found that adenylyl cyclase–biased agonists (Table 3) for cyclase inhibition versus arrestin recruitment, 2AG (BF, 47.8), tetrahydrocannabinol (BF, 33.1), and HU308 (BF 39.8) failed to internalize CB2 receptors (P > 0.05 at a concentration that resulted in 90% inhibition of adenylyl cyclase inhibition; EC90 concentration), while arrestin-biased agonists, UR144 (BF, 0.0015) and STS135 (BF, 0.2), efficaciously internalized CB2 receptors (P < 0.05 at the EC90 for arrestin recruitment). CP55940 (BF, 1) and GW833972A (BF, 2.5), which displayed similar arrestin and cyclase efficacy, also internalized CB2 receptors (P < 0.05, EC90) (see Table 4 for details).
Fig. 6.
Bar graph depicting the efficacy of select agonists in internalizing the CB2 receptor at their EC90 concentrations for activating their preferred signaling pathway. Values are shown as mean ± 95% confidence intervals. EC90 concentrations were obtained by fitting the dose-response curve (nonlinear regression) from cyclase assays for cyclase biased and unbiased agonists. For arrestin-biased agonists, EC90 concentrations were obtained by fitting the dose-response curve (nonlinear regression) from arrestin recruitment assays. Data were analyzed using one-way analysis of variance with the post hoc Bonferroni test (*P < 0.05).
TABLE 4.
Percent basal surface levels and 95% confidence intervals of CB2 receptors
The results represent data obtained after treatment with select ligands (at their EC90 concentrations for inhibition of adenylyl cyclase or stimulation of arrestin recruitment).
| Treatment |
Basal Level (Mean ± S.E.M.) |
95% CI |
|---|---|---|
| % | ||
| Control | 99 ± 1.2 | 97–101 |
| CP55940 | 78 ± 2.3 | 74– 80 |
| 2AG | 99 ± 1.1 | 96–103 |
| THC | 100 ± 2.4 | 92–105 |
| HU308 | 95 ± 3.3 | 90–103 |
| GW833972A | 69 ± 3.0 | 66–72 |
| STS135 | 85 ± 3.3 | 76–89 |
| UR144 | 76 ± 2.1 | 73–80 |
CI, confidence interval; GW833972A, 2-[(3-chlorophenyl)amino]-N-(4-pyridinylmethyl)-4-(trifluoromethyl)-5-pyrimidinecarboxamide hydrochloride; HU308, 4-[4-(1,1-dimethylheptyl)-2,6-dimethoxyphenyl]-6,6-dimethylbicyclo[3.1.1]hept-2-ene-2-methanol; THC, tetrahydrocannabinol.
Discussion
CB2 receptors display rich and pleiotropic signaling and are activated by ligands with diverse chemical structures (Atwood et al., 2012b), whose unique fingerprints of receptor activation lead to biased agonism (van der Westhuizen et al., 2014). The relative extent to which a particular pathway is activated by a ligand can be characterized by two parameters, KA and τ, where KA is the functional equilibrium dissociation constant for a particular signaling pathway/cell environment and serves as a measure of potency of a ligand for that pathway and τ is the intrinsic efficacy or the ability of an agonist to efficaciously recruit a particular pathway/evoke a biologic response compared with a reference ligand. The ratio of these two parameters on a logarithmic scale yields a transduction coefficient [log (τ/KA)] and serves as a measure of agonist interaction with its receptor for a specific signaling pathway. Furthermore, the BFs (the ratio of transduction coefficients) help estimate the differences in molecular efficacies of ligands for one pathway over the other (Evans et al., 2011; Rajagopal et al., 2011; Kenakin et al., 2012; Kenakin and Christopoulos, 2013; van der Westhuizen et al., 2014). The advantage of calculating transduction coefficients and BFs is that these values are independent of receptor density or tissue component (Kenakin et al., 2012). In the present study transduction coefficients and BFs were computed using an operational model of bias (van der Westhuizen et al., 2014). We have attempted to minimize the influence of observation and system bias by comparing ligand activity with reference to the unbiased agonist, CP55940. It should be kept in mind that these assays were performed with overexpressed receptors in cell lines and signaling in this context may vary from the signaling profiles observed in native cells. However, these results do indicate that these compounds are capable of eliciting differential signaling from CB2 receptors, at least under the conditions studied here.
SR144528 and AM630 are generally regarded as CB2 receptor inverse agonists (Bouaboula et al., 1999; Bolognini et al., 2012). However, our results indicate the inverse agonism of CB2 ligands is also subject to functional selectivity. We found that while SR144528 was an inverse agonist in both the cyclase and arrestin assays, AM630 and JTE907 were inverse agonists only in the cyclase assays and behaved as very low efficacy agonists in the arrestin recruitment assays, thus again highlighting the importance of evaluating inverse agonists/antagonists in different signaling pathways. The inverse agonism of SR144528, but not AM630, in recruiting arrestin may explain why the former, but not the latter, antagonist causes externalization of CB2 receptors (Atwood et al., 2012b).
Generally, since agonists of other GPCRs that induce high levels of receptor phosphorylation by GPCR kinases preferentially activate the arrestin pathway compared with agonists that stimulate low levels of phosphorylation, which preferentially stimulate G-protein signaling (Zheng et al., 2008, 2011), it will be interesting to determine if STS135; GW833972A; UR144; or 4-O-methylhonokiol induce higher levels of CB2 receptor phosphorylation when compared with other ligands and also if they display increased endocytic dwell times (Flores-Otero et al., 2014).
Many of the ligands screened in the current study have been widely used in CB2 receptor research. A lack of precise knowledge of the repertoire of signaling pathways engaged by these ligands may lead to misleading conclusions and incorrect inferences when using these compounds to ascertain the biologic roles of CB2 receptors. Another interesting aspect of this study was that the classic cannabinoids (L759633 and L759656), the aminoalkylindoles (AM2233 and GW405833), and the carboxamide derivative 4Q3C were inactive in both assays employed in our studies. There are a number of possible reasons why these compounds failed to stimulate either of the pathways. These include a potential role for allosterism, the type of assay employed (Ross et al., 1999) or that activation of different sets of signaling pathways are relevant for the CB2-mediated behavioral response, independent of arrestin and Gαi subunits (for example, mitogen-activated protein kinases, ceramide, Akt kinase/protein kinase B, or G-protein-coupled inwardly rectifying potassium channels) (see Blättermann et al., 2012). Finally, the cell line employed and receptor expression levels in that particular cell line may impact signaling.
Functional selectivity increases the scope and diversity of CB2 receptor signaling. Indeed, arrestin-biased signaling has been reported to have useful therapeutic effects for other GPCRs (Wisler et al., 2007). Biased ligands may offer an advantage over unbiased or less-biased ligands in achieving desired therapeutic efficacy. There is a growing interest and appreciation in the therapeutic utility of CB2 receptor ligands, especially in the treatment of neuropathic and inflammatory pain. However, several ligands that displayed promising activity in preclinical models have failed in clinical trials (Dhopeshwarkar and Mackie, 2014). While there are many reasons for a ligand that appears effective in preclinical models to fail in the clinic, one important factor, which may be overlooked, is functional selectivity (Dhopeshwarkar and Mackie, 2014). Our data have important implications when extrapolating from preclinical studies to humans. We have found that certain ligands, which are widely used in in vitro (cell-based studies) and in vivo (animal studies) are strongly biased toward one or another of these two pathways. (Of course, this bias may extend to other pathways, as well.) To the best of our knowledge, a broad signaling profiling of the CB2 agonists that have been clinically tested has not been published. Exploring the signaling biases of these ligands may help to explain why these CB2 agonists have failed in clinical trials.
Supplementary Material
Acknowledgments
The authors thank Dr. Jürg Gertsch (University of Bern) for the gift of 4-O-methylhonokiol and (E) β-caryophyllene and Dr. Alex Makriyannis (Northeastern University) for AM1710, AM2232, AM2233, and MAM2201.
Abbreviations
- AM1710
3-(1,1-dimethyl-heptyl)-1-hydroxy-9-methoxy-benzo(c)chromen-6-one
- AM2233
(2-iodophenyl)[1-[(1-methyl-2-piperidinyl)methyl]-1H-indol-3-yl]-methanone
- AM630
6-iodo-2-methyl-1-[2-(4-morpholinyl)ethyl]-1H-indol-3-yl](4-methoxyphenyl)methanone
- BF
bias factor
- BSA
bovine serum albumin
- CB1
cannabinoid receptor type 1
- CB2
cannabinoid receptor type 2
- CHO
Chinese hamster ovary
- CP55940
(−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol
- Emax, maximal effect Gi
inhibitory G proteins
- GPCR
G-protein-coupled receptor
- GW405833
1-(2,3-dichlorobenzoyl)-5-methoxy-2-methyl-3-[2-(4-morpholinyl)ethyl]-1H-indole
- HA
hemagglutinin
- HBS
HEPES buffered saline
- HEK
human embryonic kidney
- SR144528
5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2S,4R)-1,3,3-trimethylbicyclo[2.2.1]hept-2-yl]-1H-pyrazole-3-carboxamide
- STS135
N-(adamantan-1-yl)-1-(5-fluoropentyl)-1H-indole-3-carboxamide
- 2AG
2-arachidonoyl glycerol
- UR144
(1-pentyl-1H-indol-3-yl)(2,2,3,3-tetramethylcyclopropyl)-methanone
Authorship Contributions
Participated in research design: Dhopeshwarkar, Mackie.
Conducted experiments: Dhopeshwarkar.
Performed data analysis: Dhopeshwarkar, Mackie.
Wrote or contributed to the writing of the manuscript: Dhopeshwarkar, Mackie.
Footnotes
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grants DA021696, DA009158, and DA035068].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Abadji V, Lucas-Lenard JM, Chin C, Kendall DA. (1999) Involvement of the carboxyl terminus of the third intracellular loop of the cannabinoid CB1 receptor in constitutive activation of Gs. J Neurochem 72:2032–2038. [DOI] [PubMed] [Google Scholar]
- Alexander SP, Benson HE, Faccenda E, Pawson AJ, Sharman JL, McGrath JC, Catterall WA, Spedding M, Peters JA, Harmar AJ, et al. CGTP Collaborators (2013) The Concise Guide to Pharmacology 2013/14: overview. Br J Pharmacol 170:1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood BK, Straiker A, Mackie K. (2012a) CB₂ cannabinoid receptors inhibit synaptic transmission when expressed in cultured autaptic neurons. Neuropharmacology 63:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood BK, Wager-Miller J, Haskins C, Straiker A, Mackie K. (2012b) Functional selectivity in CB2 cannabinoid receptor signaling and regulation: implications for the therapeutic potential of CB2 ligands. Mol Pharmacol 81:250–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayewitch M, Avidor-Reiss T, Levy R, Barg J, Mechoulam R, Vogel Z. (1995) The peripheral cannabinoid receptor: adenylate cyclase inhibition and G protein coupling. FEBS Lett 375:143–147. [DOI] [PubMed] [Google Scholar]
- Blättermann S, Peters L, Ottersbach PA, Bock A, Konya V, Weaver CD, Gonzalez A, Schroder R, Tyagi R, Luschnig P, et al. (2012) A biased ligand for OXE-R uncouples Gα and Gβγ signaling within a heterotrimer. Nat Chem Biol 8:631–638. [DOI] [PubMed] [Google Scholar]
- Bolognini D, Cascio MG, Parolaro D, Pertwee RG. (2012) AM630 behaves as a protean ligand at the human cannabinoid CB2 receptor. Br J Pharmacol 165:2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaboula M, Dussossoy D, Casellas P. (1999) Regulation of peripheral cannabinoid receptor CB2 phosphorylation by the inverse agonist SR 144528. Implications for receptor biological responses. J Biol Chem 274:20397–20405. [DOI] [PubMed] [Google Scholar]
- Bouaboula M, Poinot-Chazel C, Marchand J, Canat X, Bourrié B, Rinaldi-Carmona M, Calandra B, Le Fur G, Casellas P. (1996) Signaling pathway associated with stimulation of CB2 peripheral cannabinoid receptor. Involvement of both mitogen-activated protein kinase and induction of Krox-24 expression. Eur J Biochem 237:704–711. [DOI] [PubMed] [Google Scholar]
- Cabral GA, Griffin-Thomas L. (2009) Emerging role of the CB2 cannabinoid receptor in immune regulation and therapeutic prospects. Expert Rev Mol Med 11:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra B, Portier M, Kernéis A, Delpech M, Carillon C, Le Fur G, Ferrara P, Shire D. (1999) Dual intracellular signaling pathways mediated by the human cannabinoid CB1 receptor. Eur J Pharmacol 374:445–455. [DOI] [PubMed] [Google Scholar]
- Console-Bram L, Marcu J, Abood ME. (2012) Cannabinoid receptors: nomenclature and pharmacological principles. Prog Neuropsychopharmacol Biol Psychiatry 38:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Guindon J, Cornett BL, Makriyannis A, Mackie K, Hohmann AG. (2015) Chronic cannabinoid receptor 2 activation reverses paclitaxel neuropathy without tolerance or cannabinoid receptor 1-dependent withdrawal. Biol Psychiatry 77:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. (2007) Beta-arrestins and cell signaling. Annu Rev Physiol 69:483–510. [DOI] [PubMed] [Google Scholar]
- Dhopeshwarkar A, Mackie K. (2014) CB2 Cannabinoid receptors as a therapeutic target—what does the future hold? Mol Pharmacol 86:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhart J, Obregon D, Mori T, Hou H, Sun N, Bai Y, Klein T, Fernandez F, Tan J, Shytle RD. (2005) Stimulation of cannabinoid receptor 2 (CB2) suppresses microglial activation. J Neuroinflammation 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BA, Broxton N, Merlin J, Sato M, Hutchinson DS, Christopoulos A, Summers RJ. (2011) Quantification of functional selectivity at the human α1A-adrenoceptor. Mol Pharmacol 79:298–307. [DOI] [PubMed] [Google Scholar]
- Flores-Otero J, Ahn KH, Delgado-Peraza F, Mackie K, Kendall DA, Yudowski GA. (2014) Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat Commun 5:4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin JM, Vasiljevik T, Prisinzano TE, Carrasco GA. (2013) Cannabinoid agonists increase the interaction between β-Arrestin 2 and ERK1/2 and upregulate β-Arrestin 2 and 5-HT2A receptors. Pharmacol Res 68:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M, Felder CC. (1997) Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. J Neurosci 17:5327–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsiorek W, Lunn C, Fan X, Narula S, Lundell D, Hipkin RW. (2000) Endocannabinoid 2-arachidonyl glycerol is a full agonist through human type 2 cannabinoid receptor: antagonism by anandamide. Mol Pharmacol 57:1045–1050. [PubMed] [Google Scholar]
- Gudermann T, Schöneberg T, Schultz G. (1997) Functional and structural complexity of signal transduction via G-protein-coupled receptors. Annu Rev Neurosci 20:399–427. [DOI] [PubMed] [Google Scholar]
- Guillot A, Hamdaoui N, Bizy A, Zoltani K, Souktani R, Zafrani ES, Mallat A, Lotersztajn S, Lafdil F. (2014) Cannabinoid receptor 2 counteracts interleukin-17-induced immune and fibrogenic responses in mouse liver. Hepatology 59:296–306. [DOI] [PubMed] [Google Scholar]
- Herrera B, Carracedo A, Diez-Zaera M, Gómez del Pulgar T, Guzmán M, Velasco G. (2006) The CB2 cannabinoid receptor signals apoptosis via ceramide-dependent activation of the mitochondrial intrinsic pathway. Exp Cell Res 312:2121–2131. [DOI] [PubMed] [Google Scholar]
- Herrera B, Carracedo A, Diez-Zaera M, Guzmán M, Velasco G. (2005) p38 MAPK is involved in CB2 receptor-induced apoptosis of human leukaemia cells. FEBS Lett 579:5084–5088. [DOI] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, et al. (2002) International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev 54:161–202. [DOI] [PubMed] [Google Scholar]
- Ignatowska-Jankowska BM, Muldoon PP, Lichtman AH, Damaj MI. (2013) The cannabinoid CB2 receptor is necessary for nicotine-conditioned place preference, but not other behavioral effects of nicotine in mice. Psychopharmacology (Berl) 229:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T. (1987) Agonists, partial agonists, antagonists, inverse agonists and agonist/antagonists? Trends Pharmacol Sci 8:423–426. [Google Scholar]
- Kenakin T. (2002) Efficacy at G-protein-coupled receptors. Nat Rev Drug Discov 1:103–110. [DOI] [PubMed] [Google Scholar]
- Kenakin T. (2011) Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther 336:296–302. [DOI] [PubMed] [Google Scholar]
- Kenakin T, Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov 12:205–216. [DOI] [PubMed] [Google Scholar]
- Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. (2012) A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci 3:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauckner JE, Hille B, Mackie K. (2005) The cannabinoid agonist WIN55,212-2 increases intracellular calcium via CB1 receptor coupling to Gq/11 G proteins. Proc Natl Acad Sci USA 102:19144–19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Lefkowitz RJ. (2002) The role of β-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci 115:455–465. [DOI] [PubMed] [Google Scholar]
- McAllister SD, Griffin G, Satin LS, Abood ME. (1999) Cannabinoid receptors can activate and inhibit G protein-coupled inwardly rectifying potassium channels in a Xenopus oocyte expression system. J Pharmacol Exp Ther 291:618–626. [PubMed] [Google Scholar]
- McGuinness D, Malikzay A, Visconti R, Lin K, Bayne M, Monsma F, Lunn CA. (2009) Characterizing cannabinoid CB2 receptor ligands using DiscoveRx PathHunterTM β-arrestin assay. J Biomol Screen 14:49–58. [DOI] [PubMed] [Google Scholar]
- Molina-Holgado E, Vela JM, Arévalo-Martín A, Almazán G, Molina-Holgado F, Borrell J, Guaza C. (2002) Cannabinoids promote oligodendrocyte progenitor survival: involvement of cannabinoid receptors and phosphatidylinositol-3 kinase/Akt signaling. J Neurosci 22:9742–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Meozzi PA, Myers L, Perchuk A, Mora Z, Tagliaferro PA, Gardner E, et al. (2008) Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PLoS One 3:e1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Haskó G. (2008) Endocannabinoids and cannabinoid receptors in ischaemia-reperfusion injury and preconditioning. Br J Pharmacol 153:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG. (1997) Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacol Ther 74:129–180. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. (2007) Cannabinoids and multiple sclerosis. Mol Neurobiol 36:45–59. [DOI] [PubMed] [Google Scholar]
- Pertwee RG, Howlett AC, Abood ME, Alexander SP, Di Marzo V, Elphick MR, Greasley PJ, Hansen HS, Kunos G, Mackie K, et al. (2010) International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB₁ and CB₂. Pharmacol Rev 62:588–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S, Ahn S, Rominger DH, Gowen-MacDonald W, Lam CM, Dewire SM, Violin JD, Lefkowitz RJ. (2011) Quantifying ligand bias at seven-transmembrane receptors. Mol Pharmacol 80:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RA, Brockie HC, Stevenson LA, Murphy VL, Templeton F, Makriyannis A, Pertwee RG. (1999) Agonist-inverse agonist characterization at CB1 and CB2 cannabinoid receptors of L759633, L759656 and AM630. Br J Pharmacol 126:665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuehly W, Paredes JM, Kleyer J, Huefner A, Anavi-Goffer S, Raduner S, Altmann KH, Gertsch J. (2011) Mechanisms of osteoclastogenesis inhibition by a novel class of biphenyl-type cannabinoid CB2 receptor inverse agonists. Chem Biol 18:1053–1064. [DOI] [PubMed] [Google Scholar]
- Shoemaker JL, Ruckle MB, Mayeux PR, Prather PL. (2005) Agonist-directed trafficking of response by endocannabinoids acting at CB2 receptors. J Pharmacol Exp Ther 315:828–838. [DOI] [PubMed] [Google Scholar]
- Shoemaker JL, Seely KA, Reed RL, Crow JP, Prather PL. (2007) The CB2 cannabinoid agonist AM-1241 prolongs survival in a transgenic mouse model of amyotrophic lateral sclerosis when initiated at symptom onset. J Neurochem 101:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrcka AV. (2008) G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci 65:2191–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T, Kondo S, Kishimoto S, Miyashita T, Nakane S, Kodaka T, Suhara Y, Takayama H, Waku K. (2000) Evidence that 2-arachidonoylglycerol but not N-palmitoylethanolamine or anandamide is the physiological ligand for the cannabinoid CB2 receptor. Comparison of the agonistic activities of various cannabinoid receptor ligands in HL-60 cells. J Biol Chem 275:605–612. [DOI] [PubMed] [Google Scholar]
- Turu G, Hunyady L. (2010) Signal transduction of the CB1 cannabinoid receptor. J Mol Endocrinol 44:75–85. [DOI] [PubMed] [Google Scholar]
- Valant C, May LT, Aurelio L, Chuo CH, White PJ, Baltos JA, Sexton PM, Scammells PJ, Christopoulos A. (2014) Separation of on-target efficacy from adverse effects through rational design of a bitopic adenosine receptor agonist. Proc Natl Acad Sci USA 111:4614–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee MM, Blomenröhr M, van der Doelen AA, Wat JW, Smits N, Hanson BJ, van Koppen CJ, Zaman GJ. (2009) Pharmacological characterization of receptor redistribution and β-arrestin recruitment assays for the cannabinoid receptor 1. J Biomol Screen 14:811–823. [DOI] [PubMed] [Google Scholar]
- van der Westhuizen ET, Breton B, Christopoulos A, Bouvier M. (2014) Quantification of ligand bias for clinically relevant β2-adrenergic receptor ligands: implications for drug taxonomy. Mol Pharmacol 85:492–509. [DOI] [PubMed] [Google Scholar]
- Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, Shenoy SK, Lefkowitz RJ. (2007) A unique mechanism of β-blocker action: carvedilol stimulates β-arrestin signaling. Proc Natl Acad Sci USA 104:16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi ZX, Peng XQ, Li X, Song R, Zhang HY, Liu QR, Yang HJ, Bi GH, Li J, Gardner EL. (2011) Brain cannabinoid CB₂ receptors modulate cocaine’s actions in mice. Nat Neurosci 14:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chu J, Qiu Y, Loh HH, Law PY. (2008) Agonist-selective signaling is determined by the receptor location within the membrane domains. Proc Natl Acad Sci USA 105:9421–9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zhang Y, Loh HH, Law PY. (2011) Modulating μ-opioid receptor phosphorylation switches agonist-dependent signaling as reflected in PKCε activation and dendritic spine stability. J Biol Chem 286:12724–12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.