ABSTRACT
The ability to sense and adapt to temperature fluctuation is critical to the aquatic survival, transmission, and infectivity of Vibrio cholerae, the causative agent of the disease cholera. Little information is available on the physiological changes that occur when V. cholerae experiences temperature shifts. The genome-wide transcriptional profile of V. cholerae upon a shift in human body temperature (37°C) to lower temperatures, 15°C and 25°C, which mimic those found in the aquatic environment, was determined. Differentially expressed genes included those involved in the cold shock response, biofilm formation, type VI secretion, and virulence. Analysis of a mutant lacking the cold shock gene cspV, which was upregulated >50-fold upon a low-temperature shift, revealed that it regulates genes involved in biofilm formation and type VI secretion. CspV controls biofilm formation through modulation of the second messenger cyclic diguanylate and regulates type VI-mediated interspecies killing in a temperature-dependent manner. Furthermore, a strain lacking cspV had significant defects for attachment and type VI-mediated killing on the surface of the aquatic crustacean Daphnia magna. Collectively, these studies reveal that cspV is a major regulator of the temperature downshift response and plays an important role in controlling cellular processes crucial to the infectious cycle of V. cholerae.
IMPORTANCE Little is known about how human pathogens respond and adapt to ever-changing parameters of natural habitats outside the human host and how environmental adaptation alters dissemination. Vibrio cholerae, the causative agent of the severe diarrheal disease cholera, experiences fluctuations in temperature in its natural aquatic habitats and during the infection process. Furthermore, temperature is a critical environmental signal governing the occurrence of V. cholerae and cholera outbreaks. In this study, we showed that V. cholerae reprograms its transcriptome in response to fluctuations in temperature, which results in changes to biofilm formation and type VI secretion system activation. These processes in turn impact environmental survival and the virulence potential of this pathogen.
INTRODUCTION
The facultative human pathogen Vibrio cholerae is found in aquatic reservoirs, such as estuaries, coastal waters, and freshwater lakes and rivers, where it is exposed to seasonal and interannual fluctuations in temperature (1–3). Studies have demonstrated a positive correlation between the occurrence of V. cholerae and surface water temperature in a variety of aquatic environments (4–10) and between cases of the disease cholera and elevated sea surface temperatures in areas where cholera is endemic (5, 6, 11). These aquatic habitats exhibit a temperature range of about 12°C to 30°C (5, 6), whereas the human host environment is a constant 37°C. Temperature is hypothesized to be a key signal to differentiate between host and environmental reservoirs, facilitating virulence factor expression in the human host and genes associated with environmental survival upon expulsion. Studies aimed at understanding the importance of temperature upshift in V. cholerae have shed light on mechanisms crucial to the survival of this pathogen inside the human host (12–14). However, less information is available regarding how V. cholerae survives and adapts to the downshift in temperature the pathogen experiences upon exiting the human host and entering the environment, which is crucial to the perpetuation of its infectious cycle.
Low-temperature environments impose numerous challenges to bacterial cell physiology. As temperatures decrease, the lipid composition of the cell membrane transitions from a liquid crystalline state to a rigid gel state (15, 16). Translation is impeded, as low temperatures cause poor ribosome assembly and the formation of extensive RNA secondary structures (17, 18). In Escherichia coli, a shift to low temperatures causes the upregulation of genes that encode proteins involved in ribosome function, helicase activity, and exoribonuclease activity, as well as four cold shock proteins (CSPs) (16). CSPs are predicted to counteract the widespread effects of RNA secondary-structure stabilization as chaperones (19, 20). The V. cholerae genome encodes four predicted cold shock genes, cspA, cspV, VCA0184, and VC1142; however, a previous study determined that only CspA and CspV were highly induced when exponentially growing V. cholerae cells were shifted from 37°C to lower temperatures (21). Currently, little is known about how these proteins and low-temperature shifts affect the physiology and cellular processes of this pathogen.
Biofilm formation is a cellular process that is critical to the infectious cycle and environmental survival of V. cholerae and is regulated by temperature (22). Biofilms are cell aggregates or surface-attached bacterial communities enclosed in an extracellular matrix. In the aquatic environment, V. cholerae forms biofilms on the surfaces of plankton, which greatly increases its survival and facilitates dissemination of the pathogen (23–26). Additionally, biofilm-like aggregates of V. cholerae have been found in the surface waters of Bangladesh in a partially dormant “conditionally viable” state (27). The extracellular matrix that surrounds the biofilm of V. cholerae is composed of Vibrio polysaccharides (VPS) (28, 29), matrix proteins RbmA, RbmC, and Bap1 (30, 31), and extracellular DNA (32). The vps-I (vpsU-vpsK) and vps-II (vpsL-vpsQ) operons are responsible for the biosynthesis of VPS and are required for mature biofilm formation (28, 29). Transcriptional regulation of biofilm genes is controlled by the activators AphA, VpsR, and VpsT and the repressor HapR. The response regulators VpsR and VpsT bind the vps promoters directly (33), while AphA affects biofilm formation through vpsT (34). Transcription of the genes that encode VPS and the matrix proteins is increased when levels of the intracellular messenger cyclic diguanylate (c-di-GMP) are high (35, 36). HapR is the master quorum-sensing regulator that represses biofilm formation at high cell density through direct binding of the vps regulatory region and vpsT (37).
The type VI secretion system (T6SS) mediates interspecies competition (38), virulence (39, 40), and natural transformation (41) in V. cholerae. The T6SS apparatus includes a base that spans the cell envelope, an inner tube composed of hemolysin-coregulated protein (Hcp) polymers, and an outer contractile sheath that is formed by VipA and VipB (42, 43). The genes encoding the T6SS components are organized into one large cluster (VCA0105 to VCA0124) and two auxiliary clusters (VCA0017 to VCA0022 and VC1415 to VC1421) (39, 44). Transcription of T6SS genes is positively regulated by HapR, the global regulator cyclic AMP (cAMP) receptor protein (CRP), the transcriptional regulator VasH, and the competence regulators Tfox and QstR and negatively regulated by the quorum-sensing regulator LuxO and global regulator TsrA (40, 41, 45–47). It has been reported that T6SS production is affected by temperature (48); however, it remains unclear if temperature impacts T6SS interspecies killing.
In this study, we identified V. cholerae genes that are temperature regulated by examination of the genome-wide transcriptional profile of V. cholerae upon a shift from 37°C to 15°C or 25°C. We found that the expression of genes encoding cold shock proteins, biofilm formation, T6SS, and virulence were differentially regulated upon a cold shift. We demonstrated that biofilm formation and the T6SS were affected by temperature. Mutational analysis and phenotypic characterization of the most highly induced gene, cspV, revealed that this cold shock gene affects biofilm formation and type VI secretion in V. cholerae in vitro and on the surface of the chitinous zooplankton Daphnia magna, revealing the mechanism by which V. cholerae responds to varying temperatures and how this response impacts V. cholerae environmental survival.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
V. cholerae O1 El Tor A1552 was used as our wild-type strain. E. coli CC118λpir and S17-1λpir were used for cloning and conjugation, respectively. Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.2 M NaCl [pH 7.5]) was used to grow all strains. Temperature shift experiments were performed by inoculating overnight-grown cultures of V. cholerae in a 1:200 dilution in LB medium and incubating them at 37°C until an optical density at 600 nm (OD600) of 0.4 was reached; cultures were then shifted to 15°C and 25°C for 1 h. All cultures were aerated by shaking at 200 rpm. When needed, ampicillin and rifampin were used at 100 μg/ml, and streptomycin was used at 50 μg/ml. Deletion constructs were generated using splicing by overlap extension PCR and cloned into pGP704-sacB28 suicide plasmids. Mutants were generated using previously published protocols (30) and verified by PCR.
Gene expression profiling.
Microarrays used were composed of spotted 70-mer oligonucleotides representing the open reading frames of the V. cholerae strain N16961 genome and were printed at the University of California, Santa Cruz, CA (discussed in greater detail in reference 49). Experimental RNA samples were isolated from V. cholerae cells that were exposed to a cold shift. Overnight-grown cultures of V. cholerae were diluted 1:200 in LB medium and incubated at 37°C until cells reached to an OD600 of 0.4; cultures were shifted to 15°C, 25°C, or maintained at 37°C for 1 h. RNA preparation and microarray hybridization and scanning were performed as described previously (49) Normalized signal ratios were obtained with locally weighted scatterplot smoothing (LOWESS) print tip normalization, using the Bioconductor packages (50) in the R environment. Differentially regulated genes were determined using three biological replicates and two technical replicates for each treatment (six data points for each spot), using the Significance Analysis of Microarrays (SAM) program (51), with a 2-fold difference in gene expression and a 3% false-discovery rate (FDR) as cutoff values.
Expression analysis: real-time PCR.
RNA was harvested as described previously (22). Results are from two independent experiments performed in triplicate. All samples were normalized to the expression of the 16S rRNA housekeeping gene using the Pfaffl method (52). Relative expression was calculated by normalizing expression at 15°C or 25°C to that at 37°C. Statistical analysis was performed using two-tailed Student's t test.
Analysis of biofilm formation.
Green fluorescent protein (GFP)-expressing V. cholerae cells from overnight-grown cultures were diluted into LB broth to an OD600 of about 0.02, and then 3 ml was inoculated into glass chambers (Thermo Fisher Scientific, Waltham, MA, USA) and incubated statically at 15°C, 25°C, or 37°C. After 24 h, biofilm formation was visualized using confocal laser scanning microscopy (CLSM) with a 5-Pa laser scanning microscope (LSM) (Zeiss, Oberkochen, Germany). Three-dimensional images were reconstructed using Imaris 7.6 and analyzed using COMSTAT (53). Each experiment included three independent biological replicates, and three images were taken for each replicate.
Determination of intracellular c-di-GMP levels.
Overnight-grown V. cholerae strains were diluted 1:200 and grown to an OD600 of 4.0, at which time cultures were harvested, and the remaining culture was shifted to 15°C for 1 h and then sampled. c-di-GMP extraction was performed as described previously (22). Each c-di-GMP quantification experiment was performed with four biological replicates. Levels of c-di-GMP in cells grown at different temperatures were compared using a two-tailed Student t test.
Analysis of Hcp production.
Overnight-grown V. cholerae strains were diluted 1:200 and grown to an OD600 of 4.0, at which time cultures were sampled, and the remaining culture was shifted to 15°C, 25°C, or maintained at 37°C for 1 h and then sampled. Sample preparation and Western blotting were performed as previously described (40). These experiments were conducted with three biological replicates.
Bacterial killing assay.
Killing assays were performed as described previously (44), with modifications. Bacterial strains were grown overnight on LB plates and resuspended in LB broth. V. cholerae and streptomycin-resistant E. coli MC4100 or Aeromonas sp. were mixed at a 10:1 ratio, and 25 μl was spotted onto LB agar plates and incubated for 4 h. E. coli alone was plated and incubated as described above for comparison. All spots were harvested, serially diluted in LB agar, and plated onto LB agar plates containing 50 μg/ml of streptomycin to enumerate surviving prey cells.
Preparation of bacterium-free and gnotobiotic D. magna.
Bacterium-free and gnotobiotic D. magna containing only one bacterial strain, Aeromonas (Xinb3-6, GenBank accession no. KF924766), were produced according to methods previously described. Bacterium-free animals were transferred in 80-ml sterile Aachener Daphnien medium (ADaM) and fed axenic or autoclaved Scenedesmus obliquus green algae. For gnotobiotic D. magna, the strain Aeromonas previously cultured in LB medium and resuspended in sterile ADaM was introduced into bacterium-free hatchlings and grown under the same environmental condition as the bacterium-free Daphnia animals until further experimentation.
D. magna attachment and T6SS killing assays.
Overnight-grown V. cholerae was washed twice using 1× phosphate-buffered saline (PBS). Cultures were diluted to an OD600 of 0.02 in 5 ml ADaM and gently vortexed, and then a single D. magna animal was added to each culture tube and incubated statically at 25°C for 24 h. For scanning electron microscopy (SEM), D. magna animals were fixed in 3% glutaraldehyde in 0.1 M phosphate buffer for 2 h, washed twice in distilled water for 10 s, and dehydrated in a graded ethanol series (30, 50, 70, and 90%). Samples were critical-point dried, sputtered with approximately 20-nm gold particles, and imaged with an FEI Quanta three-dimensional (3D) dual-beam microscope. For CFU counts, cultures were plated to enumerate the planktonic population, while the D. magna animals were removed, added to fresh ADaM, and then homogenized and plated to enumerate the attached population. A two-tailed Student t test was used to determine statistical significance. For live-D. magna T6SS-mediated killing assays, overnight-grown cultures of V. cholerae and Aeromonas sp. were washed twice using 1× PBS, at which time cultures were normalized to the same OD600, mixed at a 10:1 ratio, and then diluted and added to the D. magna as described above. After 4 h, the D. magna animals were removed, added to fresh ADaM, and then homogenized and plated onto LB medium plates containing 50 μg/ml streptomycin to enumerate the surviving prey cells.
RESULTS
Changes to V. cholerae transcriptional profile upon a cold shift.
Whole-genome expression profiling revealed that a total of 595 and 254 genes were significantly differentially regulated upon being shifted from 37°C to 15°C and 25°C, respectively (see Table S1 in the supplemental material). There were 112 genes differentially regulated at both 15°C and 25°C (Table S1). Fifty genes were upregulated in similar manners upon shifts to both 15°C and 25°C, suggesting a more general role in low-temperature adaptation (Table S1). These included genes predicted to encode proteins involved in amino acid biosynthesis, energy metabolism andtransport, and binding (Table S1). Additionally, three cold shock proteins were upregulated at low temperatures (discussed in further detail below) (Table S1). Forty-eight genes were downregulated in similar manners upon shifts to both 15°C and 25°C. These genes are predicted to encode proteins involved in cellular processes, including heat shock, carbohydrate transport and binding, and transcriptional regulation (Table S1). Additionally, genes involved in pathogenesis were downregulated at low temperatures (Fig. 1). Genes differentially regulated specifically at either 15°C or 25°C postshift included those involved in biofilm formation (Fig. 1), which were upregulated at 15°C but not at 25°C (Fig. 1), and the T6SS, which were downregulated at 15°C and upregulated at 25°C. Because temperature is an important signal that can be used by V. cholerae to distinguish between the human host and the environment, we were interested in genes that might affect the behavior of V. cholerae upon its transition from the human host into the aquatic environment. To this end, we further examined the effect that temperature has on biofilm formation and the T6SS and tested if the CSPs affect these processes.
FIG 1.

The V. cholerae transcriptome is altered by temperature downshifts. Expression profiles of a selected set of genes in V. cholerae cells grown at 37°C compared to those shifted to 15°C or 25°C for 1 h. Induced expression is represented in yellow, and repressed expression is represented in blue. Differential expression of genes involved in virulence (I), biofilm formation (II), T6SS (III), and cold shock (IV) was observed upon a cold shift. The color scale is shown at the bottom. The details of the expression data are provided in Table S1 in the supplemental material.
Regulation of biofilm formation by low temperature.
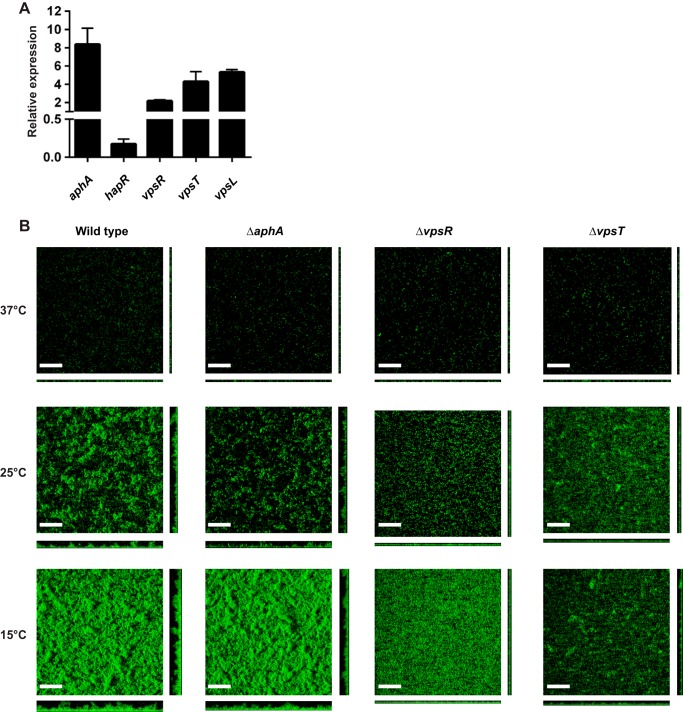
The expression of genes encoding major biofilm regulators, aphA, vpsR, and vpsT, and structural components, vps-I, vps-II, and rbmA, were upregulated upon a shift from 37°C to 15°C, while the gene encoding a negative regulator of biofilm formation, hapR, was downregulated (Fig. 1; see also Table S1 in the supplemental material). In agreement with the whole-genome expression analysis, reverse transcription-quantitative PCR (qRT-PCR) analysis revealed that aphA, vpsL, vpsR, and vpsT mRNA message abundance was increased 8.3-, 2.2-, 4.3-, and 5.3-fold, respectively, while hapR message abundance was decreased 7.1-fold 1 h after a shift from 37°C to 15°C (Fig. 2A). To determine if the major biofilm regulators were responsible for modulating biofilm gene expression at low temperatures, ΔaphA, ΔvpsR, and ΔvpsT mutants were grown at 15°C, 25°C, and 37°C in static chambers for 24 h, and biofilms were imaged and compared to wild-type V. cholerae. As we showed previously (22), biofilm formation is enhanced at 15°C and 25°C compared to that at 37°C (Fig. 2B and Table 1). However, the ΔvpsR and ΔvpsT mutants exhibit markedly lower biomass and thickness than the wild type at both 15°C and 25°C (Fig. 2 and Table 1). The ΔaphA mutant exhibited a small but reproducible decrease in biomass and thickness (Fig. 2 and Table 1) at low temperatures. These results suggest that temperature affects biofilm formation through modulation of the known biofilm regulators VpsR, VpsT, and AphA.
FIG 2.
Biofilm formation is modulated by temperature. (A) Analysis of biofilm gene expression in response to a low-temperature shift by qRT-PCR. Error bars indicate standard deviations of the results from three biological replicates. All genes exhibited a statistically significant difference between 15°C and 37°C (P < 0.05). (B) Three-dimensional biofilm structures of wild-type V. cholerae and the ΔaphA, ΔvpsR, and ΔvpsT mutants formed at 37°C, 25°C, and 15°C after 24 h of incubation. Images shown are from one representative experiment of three independent experiments. The scale bars represent 40 μm.
TABLE 1.
COMSTAT analysis of biofilms formed by mutants lacking major biofilm regulatorsa
| Strain/genotype | Temp (°C) | Biomass (μm3/μm2) | Thickness (μm) |
Roughness coefficient | |
|---|---|---|---|---|---|
| Avg | Maximum | ||||
| WT | 37 | 0.22 (0.02) | 0.30 (0.09) | 5.57 (1.02) | 1.83 (0.04) |
| 25 | 1.64 (0.25) | 1.89 (0.15) | 15.55 (1.02) | 1.19 (0.05) | |
| 15 | 6.64 (0.23) | 7.33 (0.24) | 22.00 (2.33) | 0.43 (0.02) | |
| ΔaphA | 37 | 0.12 (0.02) | 0.13 (0.02) | 7.63 (4.06) | 1.92 (0.02) |
| 25 | 2.71 (0.28) | 3.35 (0.12) | 18.77 (3.09) | 0.78 (0.06) | |
| 15 | 7.41 (0.37) | 7.73 (0.42) | 23.17 (1.02) | 0.40 (0.01) | |
| ΔvpsR | 37 | 0.17 (0.03) | 0.19 (0.02) | 5.87 (0.51) | 1.89 (0.02) |
| 25 | 1.17 (0.18) | 1.22 (0.07) | 6.45 (1.02) | 1.30 (0.05) | |
| 15 | 3.25 (0.28) | 3.09 (0.31) | 6.16 (0.88) | 0.30 (0.05) | |
| ΔvpsT | 37 | 0.16 (0.01) | 0.17 (0.03) | 5.28 (0.88) | 1.89 (0.01) |
| 25 | 0.87 (0.47) | 1.17 (0.47) | 7.33 (0.51) | 1.40 (0.25) | |
| 15 | 2.37 (0.36) | 2.62 (0.27) | 9.68 (0.88) | 0.54 (0.06) | |
Biofilms were grown at 15°C, 25°C, and 37°C in static chambers in LB medium for 24 h. Values in parentheses are standard deviations.
Regulation of the T6SS by low temperature.
The expression of genes involved in the biogenesis of type VI secretion was downregulated at 15°C and upregulated at 25°C. qRT-PCR analysis confirmed that hcp mRNA message abundance was decreased nearly 1.5-fold 1 h after the shift to 15°C and increased 6.0-fold 1 h after the shift to 25°C (Fig. 3A). We compared levels of Hcp protein production and secretion in cells before (37°C) and 1 h after (15°C or 25°C) the low-temperature shift. We found that Hcp production was very low, and no secretion was observed in cells grown at 15°C; however, Hcp production and secretion were increased in cells grown at 25°C (Fig. 3B). Next, we performed T6SS-mediated killing assays at 15, 25, and 37°C using E. coli as a prey strain. We observed a 5-fold reduction in the number of CFU of E. coli recovered after incubation with wild-type V. cholerae at 37°C (Fig. 3C) and a 16-fold reduction when incubated with wild-type V. cholerae at 25°C (Fig. 3C). However, we saw no significant decrease (1.1-fold) in the number of E. coli CFU recovered when cells were incubated with V. cholerae at 15°C, indicating that under the conditions tested, wild-type V. cholerae does not use its T6SS to kill E. coli when incubated at 15°C (Fig. 3C). Collectively, these results showed that T6SS production and type VI-mediated killing are strongly modulated by temperature.
FIG 3.
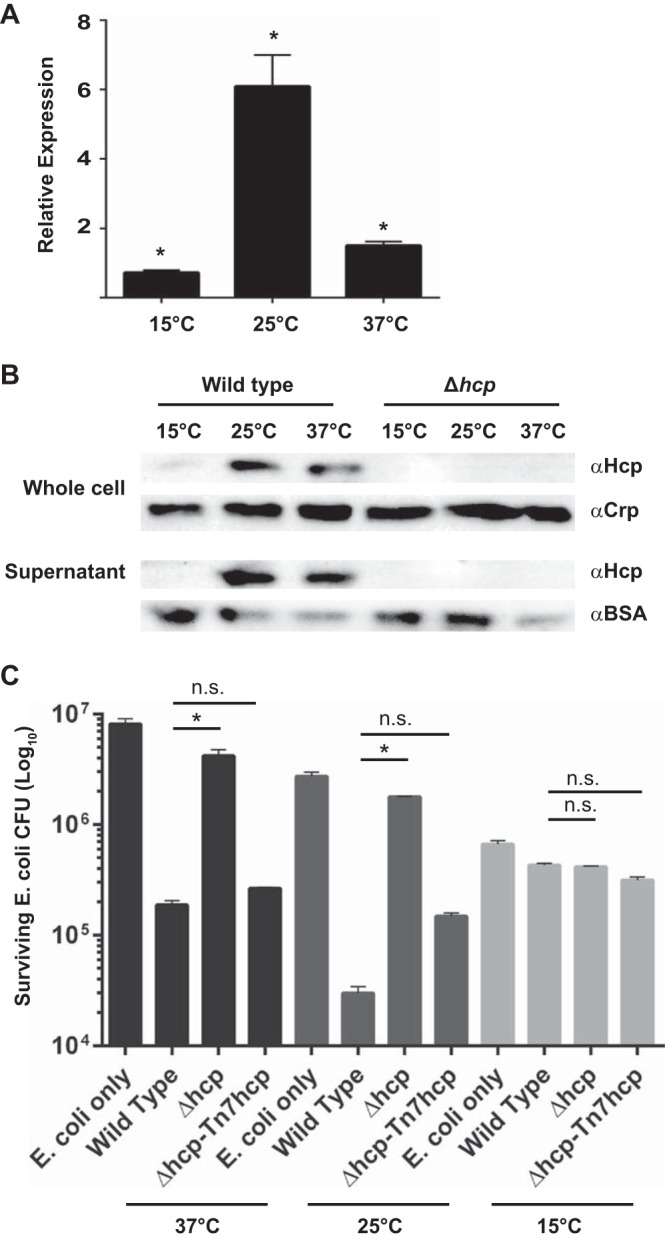
The T6SS is modulated by temperature. (A) Analysis of hcp gene expression by qRT-PCR. Error bars indicate standard deviations of the results from three biological replicates (P < 0.05). (B) Hcp production levels by Western blot analysis before (at 37°C) and 1 h after (15°C or 25°C) a low-temperature shift. BSA, bovine serum albumin control; α, anti. (C) T6SS-mediated killing of E. coli at 37°C, 25°C, and 15°C. Error bars indicate standard deviations of the results from three biological replicates. *, P < 0.05; n.s., not significantly different (P > 0.05).
Expression of cold shock genes.
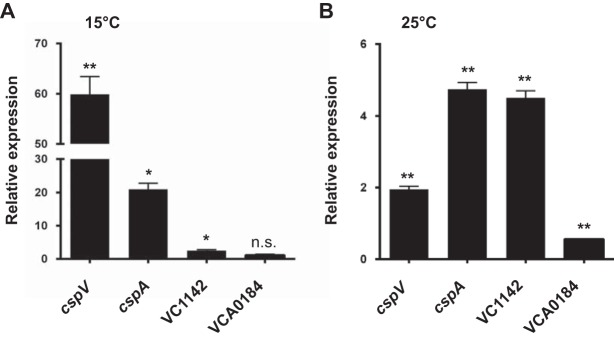
Because low temperatures dramatically affect both biofilm formation and the T6SS, we hypothesized that the CSPs may be important for these cellular processes. To test this hypothesis, we first determined which cold shock genes were upregulated upon the shift to low temperature in V. cholerae. At 15°C, the message abundance of cspV, cspA, and VC1142 was increased 60.9-, 21.9-, and 2.5-fold, respectively (Fig. 4A); however, there was no significant difference for VCA0184 (Fig. 4A). At 25°C, the message abundance of cspV, cspA, and VC1142 was increased 2.0-, 4.7-, and 4.5-fold, respectively (Fig. 4B), while VCA0184 was decreased 1.8-fold (Fig. 4B). Because cspV was the most highly expressed gene at 15°C, the temperature at which we observed the most dramatic effect on biofilm formation and the T6SS, we further studied cspV to see if it plays a role in these processes.
FIG 4.
Expression of three cold shock genes is differentially regulated by temperature. Analysis of cold shock gene expression in response to a shift from 37°C to 15°C (A) or 25°C (B) by qRT-PCR. Error bars indicate standard deviations of the results from three biological replicates. *, P < 0.05; **, P < 0.005; n.s., not significantly different (P > 0.05).
CspV regulates biofilm gene expression, biofilm formation, and cellular c-di-GMP levels.
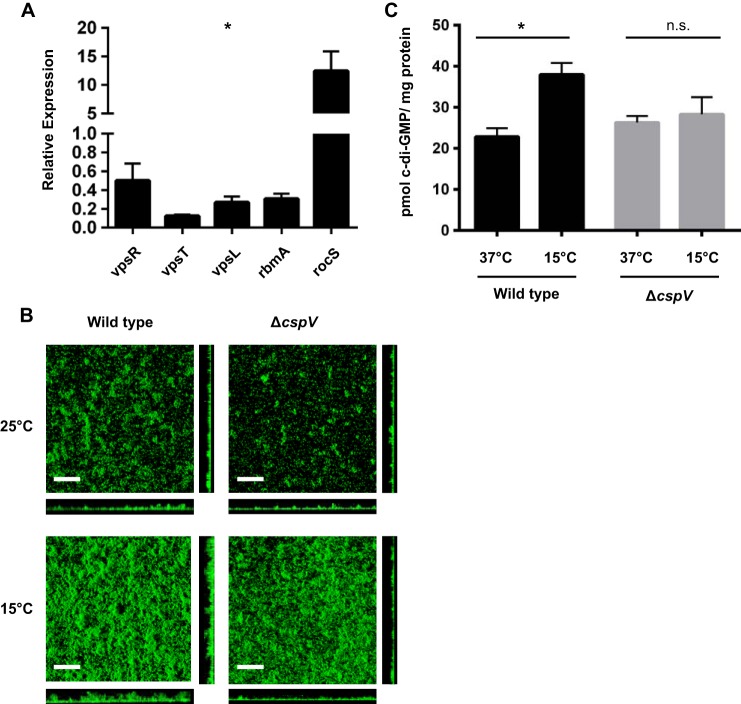
To determine if cspV plays a role in biofilm formation at low temperatures, we first used qRT-PCR to compare the mRNA message abundances of biofilm genes in the ΔcspV mutant and the wild type. The ΔcspV mutant exhibited a significant decrease in mRNA message abundance of the positive biofilm regulators vpsR and vpsT and biofilm matrix components vpsL and rbmA, as well as an increase in that of the phosphodiesterase gene rocS, which negatively regulates biofilm formation through the degradation of c-di-GMP (Fig. 5A). Next, we examined biofilm formation directly in a strain lacking cspV during growth at 15°C, 25°C, and 37°C. When grown at 15°C, the cspV mutant showed 1.8-, 2.4-, and 3.7-fold decreases in biomass, average thickness, and maximum thickness, respectively, compared with the wild type (Fig. 5B and Table 2). At 25°C, the ΔcspV mutant showed 1.1-, 1.3-, and 1.3-fold decreases in biofilm biomass, average thickness, and maximum thickness, respectively, compared with the wild type (Fig. 5B and Table 2). This indicates that cspV affects biofilm formation more dramatically at 15°C but also shows a small but reproducible defect at 25°C.
FIG 5.
cspV regulates biofilm formation. (A) Biofilm gene expression in the ΔcspV mutant compared with wild-type V. cholerae 1 h after a shift from 37°C to 15°C by qRT-PCR. All genes exhibited a statistically significant difference between the ΔcspV mutant and the wild type at 15°C (*,P < 0.05). Error bars indicate standard deviations of the results from four biological replicates. (B) Three-dimensional biofilm structures of wild-type V. cholerae and the strain lacking cspV formed at 25 and 15°C after 24 h of incubation. The images shown are from one representative experiment of three independent experiments. Scale bars represent 40 μm. (C) c-di-GMP levels in the ΔcspV mutant and wild-type V. cholerae grown at 37°C and then shifted to 15°C for 1 h. (A and C) Error bars indicate standard deviations of the results from four biological replicates. *, P < 0.05; n.s., not significantly different (P > 0.05).
TABLE 2.
COMSTAT analysis of biofilms formed by WT and a strain lacking cspV
| Strain/genotype | Temp (°C) | Biomass (μm3/μm2) | Thickness (μm) |
Roughness coefficient | |
|---|---|---|---|---|---|
| Avg | Maximum | ||||
| WT | 25 | 1.84 (0.76) | 1.78 (0.64) | 10.07 (3.22) | 1.05 (0.33) |
| 15 | 4.61 (1.24) | 5.10 (1.55) | 15.64 (3.01) | 0.53 (0.10) | |
| ΔcspV | 25 | 1.40 (0.92) | 1.17 (0.74) | 7.33 (2.69) | 1.16 (0.50) |
| 15 | 2.26 (0.89) | 1.90 (0.53) | 5.67 (2.85) | 0.51 (0.35) | |
a Biofilms were grown at 15°C and 25°C in static chambers in LB medium for 24 h. Values in parentheses are standard deviations.
In the ΔcspV mutant, we observed an upregulation of rocS, which encodes an important phosphodiesterase responsible for degrading c-di-GMP in V. cholerae; therefore, we hypothesized that cspV might regulate the expression of vps genes through modulation of c-di-GMP levels. Thus, cellular levels of c-di-GMP were quantified in the ΔcspV mutant before and 1 h after the shift from 37°C to 15°C and compared to those of the wild type. In agreement with previously published findings (22), we found that c-di-GMP levels were >1.5-fold higher (P < 0.005) at 15°C (postshift) than at 37°C (preshift) in wild-type V. cholerae (Fig. 5C). Yet, the ΔcspV mutant showed no significant increase in cellular levels of c-di-GMP at 15°C (postshift) compared to 37°C (preshift) (P > 0.05) (Fig. 5C). This suggests that cspV affects biofilm formation through the modulation of c-di-GMP, possibly through upregulation of the phosphodiesterase gene rocS.
Regulation of the T6SS in the ΔcspV mutant.
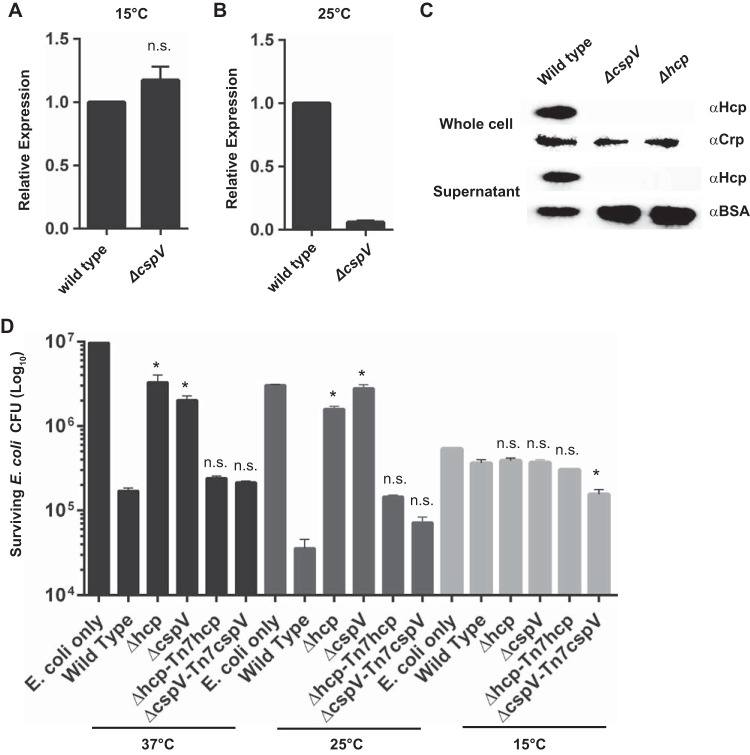
To determine if CspV is involved in the regulation of the T6SS, we first compared hcp mRNA message abundance 1 h after a shift from 37°C to 15°C or 25°C using qRT-PCR of the ΔcspV mutant and wild type. We observed no difference (1.04-fold) in hcp mRNA message abundance between the ΔcspV mutant and the wild type at 15°C (Fig. 6A). However, at 25°C, there was a 14.0-fold decrease in hcp levels in the ΔcspV mutant compared to those in the wild type (Fig. 6B). Next, we analyzed Hcp production and secretion in the ΔcspV mutant grown at 25°C, the temperature at which we observed a difference in hcp mRNA abundance. We found that the ΔcspV mutant did not produce Hcp, similar to what occurred with the Δhcp control strain (Fig. 6C). Finally, we compared levels of T6SS-mediated killing of E. coli between the ΔcspV mutant and wild-type V. cholerae. The ΔcspV strain exhibited a significant decrease in T6SS-mediated killing of E. coli, comparable to that of the Δhcp mutant, at 37°C and 25°C, while no killing was observed at 15°C in any of the strains (Fig. 6D). These results show that cspV is required for T6SS-mediated killing under the conditions tested.
FIG 6.
CspV impacts the T6SS. Analysis of hcp expression by qRT-PCR in the ΔcspV mutant and wild-type V. cholerae upon a shift from 37°C to 15°C (A) or to 25°C (B). Error bars indicate standard deviations of the results from four biological replicates. (C) Hcp production levels by Western blot analysis in the ΔcspV mutant at 25°C. (D) T6SS-mediated killing of E. coli wild-type and mutant strains at 37°C, 25°C, and 15°C. Error bars indicate standard deviations of the results from three biological replicates. *, P < 0.05; n.s., not significantly different (P > 0.05).
Analysis of D. magna colonization by the ΔcspV mutant.
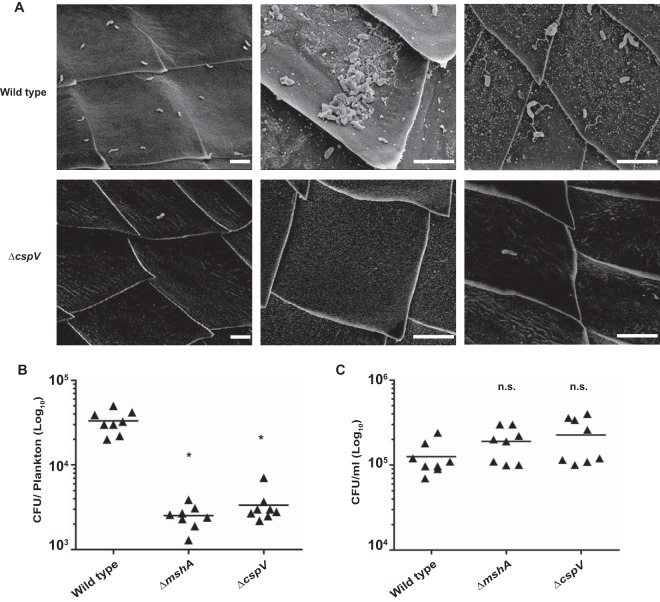
Because we determined that cspV is important for biofilm formation at temperatures that V. cholerae would experience in aquatic reservoirs, we hypothesized that cspV is required for the attachment of V. cholerae to zooplankton. To test this, we used D. magna as a model for zooplankton colonization, since V. cholerae has been shown to attach to the chitinous surface of D. magna (54), and Daphnia spp. have been found to coexist with V. cholerae in areas were cholera is endemic (55, 56). To determine if a strain lacking cspV has a colonization defect, D. magna was incubated with either wild-type V. cholerae or the cspV deletion mutant for 24 h and then imaged using scanning electron microscopy (SEM). We used D. magna incubated with a V. cholerae strain lacking the type IV pilus gene, mshA, as a control, since it was previously established that this mutant has an attachment defect on the surface of D. magna (54). We found single bacterial cells and aggregates of wild-type V. cholerae attached to the carapace of D. magna, primarily near the dorsal and ventral spinules (Fig. 7A). The ΔcspV mutant was found attached as single cells in the same regions, although at drastically reduced numbers, and no aggregates were observed (Fig. 7A), similar to what was seen with an mshA mutant (see Fig. S1 in the supplemental material).
FIG 7.
CspV impacts attachment to live D. magna. (A) SEM images showing the wild type and ΔcspV mutant attached to D. magna. Scale bars represent 5 μM. (B and C) Analysis of the abilities of wild-type V. cholerae, the ΔmshA mutant, and the ΔcspV mutant to colonize D. magna (B) and survive in ADaM (C). Error bars indicate standard deviations of results from eight biological replicates. *, P < 0.05; n.s., P > 0.05.
To further investigate the impact of cspV on plankton association, we analyzed the ability of both wild-type V. cholerae and the cspV deletion mutant to colonize zooplankton by individually infecting live D. magna cells with each strain and enumerating the surviving bacteria after 24 h. The mutant lacking cspV attached to Daphnia 9.8-fold less than the wild type (Fig. 7B), similar to the ΔmshA strain control, which attached 13-fold less than the wild type (Fig. 7B). In contrast, we did not see any difference in V. cholerae levels in planktonic cells, indicating that the ΔcspV mutant survives but does not attach to Daphnia (Fig. 7C). Collectively, these results show that cspV is important for attachment to chitinous surfaces of live zooplankton.
Type VI secretion-mediated killing on the surface of D. magna.
As shown above, the T6SS was highly active at 25°C (Fig. 3) and requires cspV (Fig. 6), suggesting that type VI-mediated competition may be important for surface colonization in aquatic environments. The chitinous surface of D. magna harbors a diverse microbial community (57). We hypothesized that V. cholerae would use its T6SS to kill the microbiota of D. magna. To address this, we used an Aeromonas sp. isolated from the D. magna microbiome as a prey strain to conduct T6SS in vitro killing assays. We observed a nearly 44-fold reduction in the number of CFU of Aeromonas sp. after incubation with wild-type V. cholerae at 25°C (Fig. 8A) and a much less dramatic reduction in the number of Aeromonas sp. CFU when this organism was incubated with the ΔcspV strain or the Δhcp control strain (3.2- or 1.8-fold, respectively) (Fig. 8A). We then performed killing assays in vivo using live D. magna as the attachment surface. We found that there was a 23-fold decrease in the number of Aeromonas sp. cells when they were incubated with wild-type V. cholerae on the surfaces of live D. magna (Fig. 8B) and only a 1.2- or 1.4-fold change when they were incubated with the ΔcspV strain or the Δhcp control strain, respectively (Fig. 8B). These results indicate that the V. cholerae T6SS can be used to kill zooplankton-associated bacteria and that cspV is crucial to this process.
FIG 8.
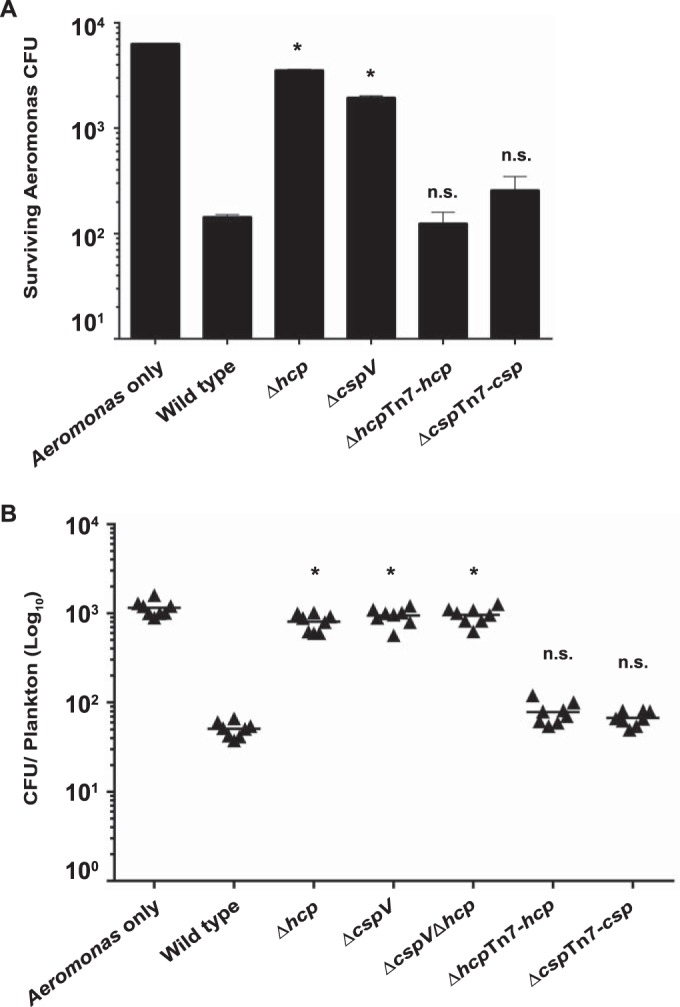
CspV contributes to interspecies killing on the surface of live D. magna. T6SS-mediated killing of Aeromonas sp. in vitro (A) and on the surface of D. magna (B). Error bars indicate standard deviations of the results from eight biological replicates. *, P < 0.05; n.s., not significantly different (P > 0.05).
DISCUSSION
Very little is known about the mechanism by which V. cholerae senses and responds to varying temperatures or how this response impacts V. cholerae environmental survival. Like Datta and Bhadra (21), we observed a lag in growth after a low-temperature shift (see Fig. S2 in the supplemental material). We determined that V. cholerae exhibits significant alterations in its transcriptome in response to low-temperature shifts. Many of these changes in gene expression are presumably oriented toward overcoming the challenges imposed by cold shock. Studies aimed at characterizing the cold shock responses of other organisms have commonly identified a number of genes involved in DNA modulation, transport and binding functions, membrane fluidity, cell envelope modification, and energy metabolism (16, 58–63). We found similar genes that exhibited differential expression upon a low-temperature shift in V. cholerae as well (Table S1).
Little is known about the regulation and function of CSPs in V. cholerae. In E. coli, there are nine cold shock proteins; however, only four are cold inducible (16). Our study revealed that three of the four predicted cold shock genes, cspV, cspA, and, to a lesser extent, VC1142, were induced in V. cholerae upon a shift to low temperatures. Previous work on cspV in V. cholerae has indicated that it contains a cold-inducible promoter that lacks a long 5′-untranslated region in its mRNA transcript, an unusual exception among cold shock genes that are traditionally posttranscriptionally regulated (21). Transcriptional profiling of human-shed V. cholerae found cspV to be highly induced (64), further supporting the hypothesis that cold shock genes may play an important role in the infectious cycle of V. cholerae by promoting survival and persistence in the environment after dissemination from the human host. Other transcriptional profiling studies have found that cspV transcript levels are elevated in the presence of bile (65) and high c-di-GMP (35). We found that the deletion of this gene causes a significant defect in biofilm formation and T6SS in V. cholerae (Fig. 5 and 7). Further studies are needed to determine the molecular underpinnings of CspV-mediated regulation of mRNA abundance.
This study also revealed that the expression of biofilm genes and major regulators of biofilm formation is controlled by temperature fluctuations. Low-temperature growth has previously been determined to affect biofilm formation in multiple bacteria (66–69), and in V. cholerae, temperature affects biofilm formation though the modulation of c-di-GMP signaling by a set of six diguanylate cyclases in V. cholerae (22). Here, we have demonstrated that low-temperature biofilm gene expression is controlled by transcriptional modulation of known biofilm regulators of aphA, hapR, vpsR, and vpsT (Fig. 2A). Both VpsR and VpsT have been demonstrated to be receptors of c-di-GMP (70, 71); therefore, they may be the receptors responsible for relaying increased c-di-GMP signals to downstream processes at low temperatures; however, future studies will be needed to address this. We found that the expression of aphA, a major regulator of virulence (72), is significantly enhanced at a low temperature. To our knowledge, this is the first report of the modulation of this virulence regulator at 15°C, suggesting that AphA may play an important role outside the human host.
Our study also determined that temperature has a dramatic impact on the T6SS. We found that hcp message abundance, protein production and secretion, and activity were highest at 25°C (Fig. 3A and B). It was previously reported that Hcp is increased in cells grown at 23°C compared to levels at 37°C (48), further corroborating our results. A molecular mechanism by which hcp transcription is decreased at 15°C is yet to be determined. However, we surmise that decreased levels of hapR at lower temperatures (Fig. 2A) might in part be responsible, as HapR positively regulates hcp expression (45, 73).
Water temperature and abundance of zooplankton covary with V. cholerae prevalence in the aquatic environment (5, 10, 11). Attachment to zooplankton is advantageous to V. cholerae in a multitude of ways. First, attached V. cholerae cells are able to utilize chitin as a source of carbon and nitrogen (74, 75). Second, contact with chitin induces type IV pili required for competence (76, 77), allowing V. cholerae to potentially increase its fitness by taking up and incorporating foreign DNA when it is closely associated with other bacteria on the zooplankton surface. Finally, attachment to zooplankton allows V. cholerae to disseminate, thereby increasing the chance of exposure to human hosts, and it has been shown to be important in transmission (23, 24, 26, 78). We found that CspV is important for the attachment to live zooplankton through direct microscopic observation and numbers of CFU recovered from colonization experiments (Fig. 7). In addition, we determined that CspV is required for type VI-mediated killing of an Aeromonas sp., the microbiome member isolated from D. magna, and has been demonstrated to affect the growth, survival, and reproduction of D. magna (57). This also may indicate that the presence of V. cholerae has implications on the health of zooplankton through alterations in microbiome composition.
V. cholerae is a human pathogen with an aquatic life cycle. It therefore offers a great opportunity to study the effects of environmental variables, such as temperature, on the environmental survival and transmission of an infectious agent. Our studies provide new insights into the adaptation of V. cholerae to fluctuating environmental parameters and reveal how this adaptation impacts the environmental survival of this important human pathogen.
Supplementary Material
ACKNOWLEDGMENTS
c-di-GMP quantification was performed at the UCSC Mass Spectrometry Facility. We kindly thank Q. Zhang for aid with the high-performance-tandem mass spectrometry (HPLC-MS/MS), B. Abrams at the UCSC Microscopy center for help with confocal microscopy, T. Yuzvinsky for assistance with sample preparation and electron microscopy, and the W. M. Keck Center for Nanoscale Optofluidics for use of the FEI Quanta 3D Dualbeam microscope. We also thank S. Beyhan and N. J. Shikuma for generously providing the csp and aphA deletion plasmids (respectively) and K. Ottemann, A. Rogers, P. Wadell, and members of the Yildiz lab for their input on this paper.
We declare no conflicts of interest.
This work was supported by NIH/NIAID grant AI102584 to F.H.Y. L.T. was supported in part by the Research Mentoring Institute (RMI) funded by the National Human Genome Research Institute (NHGRI) and the Eugene Cota-Robles (ECR) Fellowship. c-di-GMP quantification was performed at the UCSC Mass Spectrometry Facility, which is funded by NIH grant S10-RR20939 (MS equipment grant).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00807-16.
REFERENCES
- 1.Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique AK, Ali A, Huq A, Colwell RR. 2006. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ Microbiol 72:2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipp EK, Huq A, Colwell RR. 2002. Effects of global climate on infectious disease: the cholera model. Clin Microbiol Rev 15:757–770. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baffone W, Tarsi R, Pane L, Campana R, Repetto B, Mariottini GL, Pruzzo C. 2006. Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea (Italy) and study of their pathogenicity-associated properties. Environ Microbiol 8:1299–1305. doi: 10.1111/j.1462-2920.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- 5.Gil AI, Louis VR, Rivera ING, Lipp E, Huq A, Lanata CF, Taylor DN, Russek-Cohen E, Choopun N, Sack RB, Colwell RR. 2004. Occurrence and distribution of Vibrio cholerae in the coastal environment of Peru. Environ Microbiol 6:699–706. doi: 10.1111/j.1462-2920.2004.00601.x. [DOI] [PubMed] [Google Scholar]
- 6.Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, Morris JG Jr, Khan MN, Siddique AK, Yunus M, Albert MJ, Sack DA, Colwell RR. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol 71:4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipp EK, Rivera IN, Gil AI, Espeland EM, Choopun N, Louis VR, Russek-Cohen E, Huq A, Colwell RR. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl Environ Microbiol 69:3676–3680. doi: 10.1128/AEM.69.6.3676-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis VR, Russek-Cohen E, Choopun N, Rivera ING, Gangle B, Jiang SC, Rubin A, Patz JA, Huq A, Colwell RR. 2003. Predictability of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol 69:2773–2785. doi: 10.1128/AEM.69.5.2773-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner JW, Malayil L, Guadagnoli D, Cole D, Lipp EK. 2014. Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with respect to seasonal fluctuations in temperature and plankton abundance. Environ Microbiol 16:1019–1028. doi: 10.1111/1462-2920.12246. [DOI] [PubMed] [Google Scholar]
- 10.Vezzulli L, Brettar I, Pezzati E, Reid PC, Colwell RR, Höfle MG, Pruzzo C. 2012. Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J 6:21–30. doi: 10.1038/ismej.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner JW, Good B, Cole D, Lipp EK. 2009. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J 3:1082–1092. doi: 10.1038/ismej.2009.50. [DOI] [PubMed] [Google Scholar]
- 12.Parsot C, Mekalanos JJ. 1990. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc Nat Acad Sci U S A 87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slamti L, Livny J, Waldor MK. 2007. Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J Bacteriol 189:351–362. doi: 10.1128/JB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber GG, Kortmann J, Narberhaus F, Klose KE. 2014. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc Nat Acad Sci U S A 111:14241–14246. doi: 10.1073/pnas.1411570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hébraud M, Potier P. 1999. Cold shock response and low temperature adaptation in psychrotrophic bacteria. J Mol Microbiol Biotechnol 1:211–219. [PubMed] [Google Scholar]
- 16.Phadtare S, Inouye M. 2004. Genome-wide transcriptional analysis of the cold shock response in in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol 186:7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Shakhnovich EI. 2010. Thermal adaptation of viruses and bacteria. Biophys J 98:1109–1118. doi: 10.1016/j.bpj.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6:125–136. [PubMed] [Google Scholar]
- 19.Phadtare S, Severinov K. 2010. RNA remodeling and gene regulation by cold shock proteins. RNA Biol 7:788–795. doi: 10.4161/rna.7.6.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues DF, Tiedje JM. 2008. Coping with our cold planet. Appl Environ Microbiol 74:1677–1686. doi: 10.1128/AEM.02000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datta PP, Bhadra RK. 2003. Cold shock response and major cold shock proteins of Vibrio cholerae. Appl Environ Microbiol 69:6361–6369. doi: 10.1128/AEM.69.11.6361-6369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsley L, Yildiz FH. 2015. Temperature affects c-di-GMP signaling and biofilm formation in Vibrio cholerae. Environ Microbiol 17:4290–4305. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broza M, Gancz H, Kashi Y. 2008. The association between non-biting midges and Vibrio cholerae. Environ Microbiol 10:3193–3200. doi: 10.1111/j.1462-2920.2008.01714.x. [DOI] [PubMed] [Google Scholar]
- 24.Colwell RR, Huq A, Islam MS, Aziz KMA, Yunus M, Khan NH, Mahmud A, Sack RB, Nair GB, Chakraborty J, Sack DA, Russek-Cohen E. 2003. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci U S A 100:1051–1055. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2012. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front Microbiol 2:2601–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci U S A 103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fong JC, Syed KA, Klose KE, Yildiz FH. 2010. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A 96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol 188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong JCN, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol 189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seper A, Fengler VH, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. 2011. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol 82:1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zamorano-Sánchez D, Fong JCN, Kilic S, Erill I, Yildiz FH. 2015. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J Bacteriol 197:1221–1235. doi: 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Frey EM, Liu Z, Bishar R, Zhu J. 2010. The virulence transcriptional activator aphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect Immun 78:697–703. doi: 10.1128/IAI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol 188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 42.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J, Ho B, Mekalanos JJ. 2011. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. 2011. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol 193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, Uhlin BE, Wai SN. 2012. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun 80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect Immun 74:3633–3642. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Nat Acad Sci U S A 98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW. 2004. Quantification strategies in real-time PCR, p 87–112. In Bustin SA. (ed), A–Z of quantitative PCR. International University Line, La Jolla, CA. [Google Scholar]
- 53.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 54.Chiavelli DA, Marsh JW, Taylor RK. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol 67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huq A, Colwell RR, Rahman R, Ali A, Chowdhury MA, Parveen S, Sack DA, Russek-Cohen E. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl Environ Microbiol 56:2370–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miah MF, Roy S, Jinnat E, Khan ZK. 2013. Assessment of Daphnia, Moina and Cylops in freshwater ecosystems and the evaluation of mixed culture in laboratory. Am Int J Res Form Appl Nat Sci 3:1–7. [Google Scholar]
- 57.Sison-Mangus MP, Mushegian AA, Ebert D. 2014. Water fleas require microbiota for survival, growth and reproduction. ISME J 9:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beckering CL, Steil L, Weber MHW, Volker U, Marahiel MA. 2002. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J Bacteriol 184:6395–6402. doi: 10.1128/JB.184.22.6395-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fonseca P, Moreno R, Rojo F. 2011. Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ Microbiol Rep 3:329–339. doi: 10.1111/j.1758-2229.2010.00229.x. [DOI] [PubMed] [Google Scholar]
- 60.Gao H, Yang ZK, Wu L, Thompson DK, Zhou J. 2006. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood RR, Arias CR. 2011. Evaluation of global gene expression during cold shock in the human pathogen Vibrio vulnificus. Mar Biotechnol (NY) 13:942–954. doi: 10.1007/s10126-010-9356-1. [DOI] [PubMed] [Google Scholar]
- 62.Yang L, Zhou D, Liu X, Han H, Zhan L, Guo Z, Zhang L, Qin C, Wong HC, Yang R. 2009. Cold-induced gene expression profiles of Vibrio parahaemolyticus: a time-course analysis. FEMS Microbiol Lett 291:50–58. doi: 10.1111/j.1574-6968.2008.01434.x. [DOI] [PubMed] [Google Scholar]
- 63.Han Y, Zhou D, Pang X, Zhang L, Song Y, Tong Z, Bao J, Dai E, Wang J, Guo Z, Zhai J, Du Z, Wang X, Wang J, Huang P, Yang R. 2005. DNA microarray analysis of the heat- and cold-shock stimulons in Yersinia pestis. Microbes Infect 7:335–348. doi: 10.1016/j.micinf.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerda-Maira FA, Ringelberg CS, Taylor RK. 2008. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J Bacteriol 190:7441–7452. doi: 10.1128/JB.00584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fitzpatrick F, Humphreys H, O'gara JP. 2005. Evidence for low temperature regulation of biofilm formation in Staphylococcus epidermidis. J Med Microbiol 54:509–510. doi: 10.1099/jmm.0.45990-0. [DOI] [PubMed] [Google Scholar]
- 67.Piao Z, Sze CC, Barysheva O, Iida K, Yoshida S. 2006. Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl Environ Microbiol 72:1613–1622. doi: 10.1128/AEM.72.2.1613-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramli NS, Eng Guan C, Nathan S, Vadivelu J. 2012. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS One 7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White-Ziegler CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 70.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 73.Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. 2009. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res 37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colwell RR. 1970. Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J Bacteriol 104:410–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc Nat Acad Sci U S A 101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 77.Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, Dal Peraro M, Blokesch M. 2014. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet 10:e1004066. doi: 10.1371/journal.pgen.1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huq A, Yunus M, Sohel S, Bhuiya A, Emch M, Luby SP, Russek-Cohen E, Nair GB, Sack RB, Colwell RR. 2010. Simple sari cloth filtration of water is sustainable and continues to protect villagers from cholera in Matlab, Bangladesh. mBio 1(1):e00034-10. doi: 10.1128/mBio.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.