ABSTRACT
Enterococcus faecalis is a commensal bacterium of the gastrointestinal tract that can cause nosocomial infections in immunocompromised humans. The hallmarks of this organism are its ability to survive in a variety of stressful habitats and, in particular, its ability to withstand membrane damage. One strategy used by E. faecalis to protect itself from membrane-damaging agents, including the antibiotic daptomycin, involves incorporation of exogenous fatty acids from bile or serum into the cell membrane. Additionally, the response regulator LiaR (a member of the LiaFSR [lipid II-interacting antibiotic response regulator and sensor] system associated with cell envelope stress responses) is required for the basal level of resistance E. faecalis has to daptomycin-induced membrane damage. This study aimed to determine if membrane fatty acid changes could provide protection against membrane stressors in a LiaR-deficient strain of E. faecalis. We noted that despite the loss of LiaR, the organism readily incorporated exogenous fatty acids into its membrane, and indeed growth in the presence of exogenous fatty acids increased the survival of LiaR-deficient cells when challenged with a variety of membrane stressors, including daptomycin. Combined, our results suggest that E. faecalis can utilize both LiaR-dependent and -independent mechanisms to protect itself from membrane damage.
IMPORTANCE Enterococcus faecalis is responsible for a significant number of nosocomial infections. Worse, many of the antibiotics used to treat E. faecalis infection are no longer effective, as this organism has developed resistance to them. The drug daptomycin has been successfully used to treat some of these resistant strains; however, daptomycin-resistant isolates have been identified in hospitals. Many daptomycin-resistant isolates are found to harbor mutations in the genetic locus liaFSR, which is involved in membrane stress responses. Another mechanism shown to increase tolerance to daptomycin involves the incorporation of exogenous fatty acids from host fluids like serum or bile. This improved tolerance was found to be independent of liaFSR and suggests that there are additional ways to impact sensitivity to daptomycin. Thus, further studies are needed to understand how host fatty acid sources can influence antibiotic susceptibility.
INTRODUCTION
Enterococcus faecalis is a Gram-positive, facultative anaerobe that resides in the gastrointestinal tract of humans and many other mammalian species (1). Additionally, the organism is known to persist in the external environment for significant periods of time, demonstrating its ability to withstand a variety of changing conditions. Despite the commensal nature of E. faecalis, it is a significant contributor to nosocomial infections, including bloodstream, skin and soft tissue, and urinary tract infections, endocarditis, and meningitis in immunocompromised patients (2, 3). Eradication of E. faecalis, especially in regard to such infections, is challenging as the organism is inherently resistant to a variety of classes of antibiotics and has the ability to acquire additional resistance mechanisms via horizontal gene transfer (2, 4–6). Given this, enterococci are considered serious public health threats, and calls for new antibiotic therapies and surveillance are ongoing (7).
Although resistant to many antibiotics, infections caused by E. faecalis have successfully been treated with the antibiotic daptomycin. Daptomycin is naturally synthesized by Streptomyces roseosporus (8, 9) and is FDA approved for the treatment of skin and soft tissue infections caused by susceptible Gram-positive bacteria. The antibiotic targets the cell membranes of Gram-positive bacteria, leading to membrane depolarization and eventual cell death (10, 11). More detailed studies on the mechanism of daptomycin action suggest that the antibiotic inserts into bacterial cell membranes in a calcium-dependent manner, which then allows monomers of daptomycin to oligomerize in the outer leaflet and finally translocate to the inner leaflet, forming pore-like structures (12). This sequence of events leads to a loss of membrane homeostasis, including leakage of ions from the cytoplasm (13, 14). Despite the success of this antibiotic, daptomycin-resistant strains of enterococci have been isolated during patient treatment (6, 15). Characterization of daptomycin-resistant isolates by whole-genome sequencing indicates that resistance develops by chromosomal mutations in genes related to cell membrane and envelope homeostasis (16, 17) rather than by acquisition of horizontally acquired elements.
The ability to adapt and respond to environmental changes is essential for the survival of bacterial cells. Given that the cell envelope is constantly exposed to the environment, adaptive responses must be maintained or cell viability will be lost (18, 19). Across many bacterial species, the regulatory process surrounding the cell envelope stress response consists of extracytoplasmic function (ECF) σ factors and two-component systems (TCS) (20, 21, 22). In the Firmicutes (low G+C Gram-positive bacteria), numerous two- and three-component systems respond to envelope-damaging agents, including antimicrobial peptides and antibiotics (18, 23). One such example is the LiaFSR (lipid II-interacting antibiotic response regulator and sensor) system, which was first identified in Bacillus subtilis (24). In this system, LiaS is a membrane-bound sensor histidine kinase, LiaR is the response regulator, and LiaF (25) is a membrane-anchored negative regulator thought to affect the function of LiaS (25–27). LiaR was shown to regulate the expression of the liaIHGFSR locus, which, using an unknown mechanism, aids in the cellular response against cell envelope-targeting antibiotics and antimicrobial peptides (25).
Genomic analysis of a daptomycin-susceptible and -resistant clinical strain pair of E. faecalis revealed that a codon deletion in liaF was responsible for the resistance phenotype (6, 15, 28). It is thought that this mutation increased the expression of LiaSR, activating the damage response pathway and effectively abolishing the bactericidal activity of the antibiotic (28, 29). Moreover, deletion of liaR, encoding the response regulator of the system, can render both E. faecalis and Enterococcus faecium hypersusceptible to daptomycin, independent of the strain background (30, 31).
Our lab has recently discovered a previously unknown mechanism of environmentally induced tolerance to membrane-damaging agents (32). Specifically, we found that supplementing E. faecalis with bile or serum reduced susceptibility to high bile levels, sodium dodecyl sulfate (SDS), and daptomycin. Further analysis confirmed that E. faecalis was able to incorporate exogenous fatty acids from these supplements into its membrane, thus altering the fatty acid composition of the membrane. Supplementation with specific fatty acids, such as oleic acid, a dominant fatty acid found in bile and serum, confirmed that growth in the presence of fatty acids provided tolerance to these stressors (32).
Given these observations, we sought to address the hypothesis that the presence of exogenous fatty acids triggers an LiaFSR-mediated envelope stress response in E. faecalis, improving the organism's survival from membrane-damaging agents. Herein, however, we present data showing that supplementation of E. faecalis with exogenous sources of fatty acids can reduce susceptibility to membrane stressors, including daptomycin, in the absence of liaR. These data suggest that the contribution of exogenous fatty acid incorporation to cell membrane protection is independent of the LiaFSR system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Enterococcus faecalis strains OG1RF, OG1RFΔliaR (31), OG1RFΔliaR::liaR (31), and S613 and R712 (6, 15) were grown statically in brain heart infusion (BHI) medium at 37°C unless otherwise stated. Overnight cultures were used to inoculate medium to an optical density at 600 nm (OD600) of 0.01. Cultures were supplemented as indicated in the text with bovine bile (Sigma-Aldrich), pooled human serum (ICN Biomedicals), fatty acids (Sigma-Aldrich), or the solvent control (ethanol).
GC-FAME preparation and analysis.
Strains were grown as above with the supplements indicated in Table 1. At exponential phase (OD600 of ≈0.4), 10 to 12 ml of culture was centrifuged at 3,500 rpm for 10 min. Cell pellets were washed extensively twice with 1× phosphate-buffered saline (PBS). Pellets were subsequently stored at −80°C and shipped on dry ice to Microbial ID, Inc. (Newark, DE) for gas chromatography-fatty acid methyl ester (GC-FAME) analysis. Cells underwent saponification using a sodium hydroxide-methanol mixture and a hexane extraction before GC-FAME analysis as previously described (33). Results show averages and standard deviations for three independent cultures.
TABLE 1.
Exponential phase generation times
| Strain | Generation times in medium constituent (min) |
||||
|---|---|---|---|---|---|
| BHIa | Serumb | Bilec | C18:1 cis9d | Ethanole | |
| WTf | 32.2 ± 0.5 | 36.5 ± 0.8 | 32.7 ± 1.9 | 39.6 ± 0.7 | 42.1 ± 1.0 |
| ΔliaR | 28.7 ± 1.3 | 36.2 ± 3.4 | 33.2 ± 3.4 | 35.2 ± 3.3 | 38.0 ± 4.1 |
| ΔliaR::liaR | 31.4 ± 2.9 | 34.1 ± 2.8 | 27.5 ± 3.4 | 35.2 ± 2.4 | 35.0 ± 1.9 |
| S613 | 29.7 ± 2.7 | 31.0 ± 3.4 | 48.1 ± 3.5 | 27.1 ± 1.5 | 31.1 ± 2.6 |
| R712 | 29.4 ± 2.5 | 36.0 ± 1.2 | 40.8 ± 3.8 | 30.0 ± 2.2 | 38.0 ± 1.0 |
BHI medium was used in all cultures with supplements as indicated.
Pooled human serum was supplemented to a final concentration of 15%.
Bovine bile was supplemented to a final concentration of 0.2%.
Oleic acid was added to a final concentration of 20 μg/ml.
Ethanol solvent control was added to a final concentration of 0.2%.
WT, wild type.
Membrane challenge assays.
Cells were harvested at mid-log phase (OD600 of ≈0.4), washed with 1× PBS, centrifuged, and resuspended in the appropriate challenge medium, as performed previously (32). For antibiotic treatment, cells were resuspended in BHI medium containing 100 mM CaCl2 and either 10 μg/ml or 40 μg/ml daptomycin as indicated in the text. For bile treatment, cells were resuspended in an equivalent volume of 20% bovine bile. For SDS treatment, cells were resuspended in an equivalent volume of fresh BHI containing 0.05% SDS. Serial dilutions were plated onto BHI agar at 0, 15, 30, and 60 min after resuspension in the challenge medium. The log ratio of survivors over time was calculated for three biological replicates; the averages and standard deviations for each experiment are shown.
Statistical analysis.
Comparisons between the growth conditions, membrane content, and log ratio of survivors were determined using two-tailed, unpaired Student's t tests as indicated in the text.
1H NMR analyses.
Stock solutions at 1.23 mM, 50 mM, and 100 mM of daptomycin (1.0 mg, 617 nmol in 500 μl methanol-d4), oleic acid (7.0 mg, 24.8 μmol in 500 μl methanol-d4), and calcium chloride (5.5 mg, 49.6 μmol in 500 μl methanol-d4), respectively, were prepared. The 1H NMR spectra were recorded on a VNMRS 500 MHz NMR spectrometer (Varian NMR Systems, Palo Alto, CA). The proton nuclear magnetic resonance (1H NMR) experiments consisted of 128 scans using PRESAT solvent suppression peak selection. To examine a potential interaction between daptomycin and oleic acid, we looked at the spectra of mixtures containing either an equivalent concentration of daptomycin and oleic acid (1:1, daptomycin/oleic acid) or a mixture containing an excess of oleic acid (1:5, daptomycin/oleic acid). Each mixture was homogenized by vortexing, and the resulting solution was allowed to incubate at room temperature for 30 min. Following incubation, the 1H NMR experiment was repeated.
To discover if calcium could induce an interaction between daptomycin and oleic acid, we also determined the spectra when calcium chloride was added. For all experiments, we used an overall molar ratio of 5:4 (calcium/daptomycin). The mixture was vortexed and allowed to incubate at room temperature for 30 min prior to the 1H NMR experiment to determine the baseline spectra of a calcium-daptomycin complex. For those experiments examining how this mixture may interact with oleic acid, the fatty acid was added following the incubation of calcium chloride and daptomycin. Oleic acid was added into the mixture at either a 1:1 or 1:5 molar ratio, the mixture was homogenized by vortexing and incubated at room temperature for 30 min, and then the 1H NMR experiment was repeated.
RESULTS
Incorporation of exogenous fatty acids is similar in the presence or absence of liaR in E. faecalis.
Previously, we demonstrated that growth in the presence of fatty acid sources impacted the generation time of E. faecalis OG1RF (32). Most notably, growth in the presence of saturated fatty acids significantly increased the generation time of OG1RF compared to that in unsupplemented cultures. The organism also readily incorporated exogenous fatty acids into its membrane, even if those fatty acids negatively impacted growth. As the LiaFSR system has been shown to be important to the cell membrane stress response in enterococci, we wondered whether it contributed to our past observations. Thus, we examined the growth rates and membrane fatty acid contents of the parental OG1RF (control), the ΔliaR, and the genetically complemented ΔliaR::liaR strains (31) in the presence and absence of exogenous fatty acid sources. It is important to note that E. faecalis does not possess genes for β-oxidation; therefore, the organism either incorporates exogenous fatty acids into its membrane or, in the case of exogenous short-chain fatty acids, potentially elongates such fatty acids (34, 35).
In general, the growth rates and the membrane contents were similar for the three strains grown in BHI with a few notable differences. The generation times for all in unsupplemented medium was approximately 30 min (Table 1), which was increased to about 40 min when the strains were grown in the presence of ethanol (solvent control; final concentration of 0.2%). As shown in Table 2, the dominant fatty acids for all strains grown in BHI were cis-vaccenic acid (C18:1 cis 11, approximately 40%) and palmitic acid (C16:0, about 37%). While the overall membrane content was similar between the strains, we did note that the genetic complement (ΔliaR::liaR) had significantly more palmitoleic acid (C16:1 cis 9) (P < 0.05) and less stearic acid (C18:0) (P < 0.05) than the wild-type and ΔliaR strains. However, these differences did not influence the overall saturated/unsaturated ratio, which was close to 1 for all three strains (Table 2).
TABLE 2.
Membrane analysis of wild-type and mutant strains during log phase growth
| Fatty acid | % of total membrane content for indicated supplement and strain (avg ± SD)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BHI |
Serumb |
Bilec |
C18:1
cis9d |
|||||||||
| WTe | ΔliaR | ΔliaR::liaR | WT | ΔliaR | ΔliaR::liaR | WT | ΔliaR | ΔliaR::liaR | WT | ΔliaR | ΔliaR::liaR | |
| C12:0 | 0.7 ± 0.1 | 0.6 ± 0.1 | 0.9 ± 0.1 | ND | 0.3 ± 0.0 | 0.3 ± 0.0 | ND | ND | 0.2 ± 0.2 | 0.7 ± 0.1 | 1.1 ± 0.3 | 1.1 ± 0.1 |
| C14:0 | 4.6 ± 0.1 | 4.4 ± 0.0 | 6.4 ± 0.1 | 2.4 ± 0.1 | 2.6 ± 0.1 | 2.9 ± 0.1 | 1.1 ± 0.0 | 1.0 ± 0.0 | 1.6 ± 0.1 | 0.7 ± 0.1 | 0.9 ± 0.3 | 0.5 ± 0.1 |
| C16:1 | 7.0 ± 0.1 | 6.5 ± 0.4 | 9.7 ± 0.1 | 4.9 ± 0.1 | 4.9 ± 0.1 | 5.1 ± 0.1 | 1.5 ± 0.5 | 1.5 ± 0.0 | 1.7 ± 0.1 | 1.8 ± 0.2 | 1.6 ± 0.1 | 1.2 ± 0.1 |
| C16:0 | 37.6 ± 0.6 | 37.4 ± 0.2 | 34.9 ± 0.3 | 35.9 ± 0.6 | 36.1 ± 0.4 | 37.2 ± 0.3 | 42.6 ± 0.1 | 42.6 ± 0.2 | 42.4 ± 0.2 | 3.4 ± 0.5 | 3.1 ± 0.7 | 1.7 ± 0.3 |
| C17:1 | ND | 1.5 ± 0.1 | ND | 0.6 ± 0.0 | 0.6 ± 0.0 | 0.5 ± 0.0 | 1.7 ± 0.2 | 1.0 ± 0.0 | 1.7 ± 0.1 | 0.3 ± 0.3 | 0.4 ± 0.3 | 0.7 ± 0.2 |
| C17:0 2OH | 5.2 ± 0.7 | 5.2 ± 0.8 | 5.1 ± 0.4 | 0.2 ± 0.3 | 0.6 ± 0.1 | 0.5 ± 0.0 | 0.3 ± 0.2 | 0.3 ± 0.0 | 0.3 ± 0.0 | ND | ND | ND |
| C18:1 cis9 | 0.7 ± 0.7 | 0.2 ± 0.4 | ND | 21.2 ± 0.3 | 20.3 ± 0.9 | 18.4 ± 0.2 | 42.3 ± 0.2 | 38.4 ± 1.8 | 40.8 ± 0.5 | 76.3 ± 1.0 | 68.5 ± 5.0 | 75.1 ± 0.7 |
| C18:1 cis11 | 39.0 ± 0.5 | 38.5 ± 1.4 | 38.9 ± 0.5 | 4.7 ± 0.1 | 4.9 ± 0.2 | 4.4 ± 0.2 | 3.7 ± 0.1 | 3.8 ± 0.1 | 3.6 ± 0.0 | ND | ND | ND |
| C18:0 | 4.7 ± 0.1 | 5.0 ± 0.2 | 2.9 ± 0.1 | 9.2 ± 0.2 | 9.1 ± 0.1 | 7.8 ± 0.1 | 2.8 ± 0.1 | 6.9 ± 0.8 | 3.0 ± 0.3 | 0.8 ± 0.1 | 0.8 ± 0.1 | 0.5 ± 0.1 |
| C20:0 | ND | ND | ND | ND | ND | ND | 0.6 ± 0.1 | 0.5 ± 0.0 | 0.4 ± 0.1 | 15.9 ± 0.7 | 23.7 ± 4.1 | 19.2 ± 0.9 |
| C18:2 | ND | ND | ND | 18.5 ± 0.5 | 18.1 ± 0.3 | 20.7 ± 0.2 | ND | ND | ND | ND | ND | ND |
| Othersf | 0.2 ± 0.4 | 0.3 ± 0.4 | 1.2 ± 1.1 | 2.5 ± 0.4 | 2.5 ± 1.6 | 2.3 ± 0.5 | 3.5 ± 0.3 | 3.9 ± 0.9 | 4.3 ± 0.8 | ND | ND | ND |
| Saturated/unsaturated | 1.0 ± 0.6 | 1.0 ± 0.2 | 0.9 ± 0.5 | 0.9 ± 0.9 | 1.0 ± 0.4 | 1.0 ± 0.7 | 0.9 ± 0.5 | 1.1 ± 0.5 | 1.0 ± 1.1 | 0.3 ± 1.0 | 0.4 ± 1.0 | 0.3 ± 1.4 |
| C10-C17/C18-C20g | 1.2 ± 0.03 | 1.3 ± 0.4 | 1.4 ± 0.03 | 0.8 ± 0.02 | 0.9 ± 0.03 | 0.9 ± 0.01 | 1.0 ± 0.005 | 1.0 ± 0.02 | 1.0 ± 0.01 | 0.1 ± 0.01 | 0.1 ± 0.01 | 0.1 ± 0.01 |
Membrane contents were determined by GC-FAME analysis by Microbial ID, Inc. Values represent averages and SD from for three independent cultures. ND indicates that fatty acid was not detected.
Pooled human serum was supplemented to a final concentration of 15%.
Bovine bile was supplemented to a final concentration of 0.2%.
Oleic acid was added to a final concentration of 20 μg/ml.
WT, wild type.
Others indicates fatty acids that comprised <1% of the total membrane content.
Total fatty acid length ratios including both saturated and unsaturated fatty acids.
As E. faecalis can readily cause wound infections and bacteremia, we examined both the growth rate and the membrane composition upon supplementation with 15% pooled human serum. The generation times were similar for the three strains with no statistical significance observed (Table 1). As with growth in unsupplemented medium, the dominant saturated fatty acid was palmitic acid (C16:0) for cultures grown in the presence of serum. Although not the major saturated fatty acid, stearic acid (C18:0) was approximately 2-fold higher (P < 0.001) in all strains in comparison to that for growth without serum. The greatest differences, however, were in the unsaturated fatty acid profiles upon supplementation. For cells grown with serum, the dominant unsaturated fatty acids were the eukaryote-derived oleic acid (C18:1 cis 9) and linoleic acid (C18:2 cis 9,12), which together constituted 40% of the total membrane content. There was a concomitant decrease in the amount of cis-vaccenic acid from approximately 40% in unsupplemented medium to less than 5% of the total membrane content in the presence of serum. These alterations did not alter the saturated/unsaturated ratios compared with those of unsupplemented cultures (Table 2). The lengths of the fatty acid tails were significantly longer for cells grown with serum (P < 0.001) (Table 2) as indicated by the ratio C10-C17/C18-C20. These findings are consistent with the composition of fatty acids in serum (32) and indicative of their incorporation by E. faecalis.
As E. faecalis naturally inhabits the intestine, we wanted to examine the effects of physiological levels of bile (0.2% bovine bile) upon growth and membrane content, as it too is a source of fatty acids that can be utilized by the organism (32). Growth with bile did not alter the generation times of any of the strains in comparison to growth in the absence of bile (Table 1). However, bile supplementation did impact the membrane contents of all strains examined. In all cases, palmitic acid (C16:0) remained the dominant saturated fatty acid and comprised approximately 42% of the membrane, which was a modest, but significant (P < 0.005), increase from growth in BHI alone. As was noted with serum supplementation, oleic acid (C18:1 cis 9) was the dominant unsaturated fatty acid, at approximately 40% of the total membrane content, and there was a concomitant reduction in the amount of cis-vaccenic acid (C18:1 cis 11). There was also an overall decrease in the total amounts of shorter-chain fatty acids, as indicated by the C10-C17/C18-C20 ratios (P < 0.005) (Table 2) when all strains were grown with bile versus without bile. Despite these changes, the saturated/unsaturated fatty acid ratios were essentially unaltered among the wild-type, ΔliaR, and ΔliaR::liaR strains.
We previously noted that E. faecalis can tolerate high levels of oleic acid (C18:1 cis 9) in culture and that this single fatty acid comprises the majority of the membrane content when supplemented at a final concentration of 20 μg/ml (32); indeed, this finding holds true even in a strain in which liaR is absent (Tables 1 and 2). For OG1RF and its derivatives examined here, oleic acid comprised approximately 70% of the membrane (Table 2). Essentially, cis-vaccenic acid (C18:1 cis 11), the native unsaturated C18 fatty acid, was replaced entirely in the membrane by oleic acid. Oleic acid supplementation also markedly influenced the membrane saturated fatty acid composition. For all strains grown with exogenous oleic acid, the dominant saturated fatty acid was arachidic acid (C20:0) and not palmitic acid (C16:0) (Table 2), which is the dominant fatty acid found in cells grown without this supplement. This presence of arachidic acid was surprising, as it was not detected in unsupplemented cultures (see Discussion). Overall, the contributions of oleic acid and arachidic acid led to a membrane composition dominated by long-chain fatty acids, far different from what was observed in unsupplemented cultures (Table 2).
Combined, these results suggests that induction of the LiaFSR response is not required for E. faecalis to incorporate exogenous fatty acids. Furthermore, the membrane content of the ΔliaR strain is not markedly altered from that of the parental strain under the conditions examined.
Supplementation of growth medium with fatty acids can protect an OG1RF ΔliaR strain from membrane stress.
Although the above results suggest that liaR is not needed for incorporation of exogenous fatty acids by E. faecalis, we decided to determine whether the lack of liaR impacted the ability of exogenous fatty acids to protect from membrane-damaging agents.
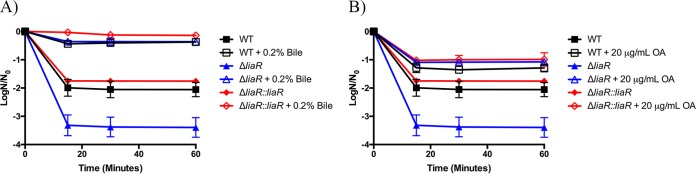
To assess the impact of LiaR and the LiaFSR system on membrane stress responses, E. faecalis was grown in the presence or absence of fatty acid sources and then exposed to 20% bovine bile. Figure 1A shows that all strains were susceptible to 20% bovine bile and that the ΔliaR strain was by far the most sensitive at all time points analyzed (15, 30, and 60 min). When strains were supplemented with 0.2% bile prior to challenge (providing a source of exogenous fatty acids), we observed an increase in survival for all strains. Importantly, supplementation of the medium with low levels of bile improved the survival of the deletion strain to the levels observed for wild-type OG1RF (Fig. 1A).
FIG 1.
Fatty acid supplementation protects liaR-deficient Enterococcus faecalis from high bile challenge. Shown are the averages ± standard deviations for n = 3. (A) Bile supplementation and challenge with 20% bile. All strains supplemented with 0.2% bile had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.001). (B) Oleic acid (OA) supplementation and challenge with 20% bile. All strains supplemented with 20 μg/ml OA had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.023). WT, wild type.
One of the main exogenous fatty acids incorporated into the membranes of these strains upon bile or serum supplementation was oleic acid (C18:1 cis 9) (Table 2). Our previous investigations demonstrated that supplementation solely with this fatty acid protected wild-type E. faecalis from membrane stress (32). We sought to determine if supplementation with oleic acid alone could protect the ΔliaR strain from bile-induced stress. We examined this phenomenon by comparing cultures supplemented with 20 μg/ml oleic acid to those without oleic acid prior to challenge with 20% bovine bile. The addition of oleic acid to the growth medium did provide tolerance to this membrane stress (Fig. 1B). Nonetheless, the overall survival for all strains was best when they were supplemented with 0.2% bile than with 20 μg/ml oleic acid. As we observed with bile supplementation, growth with oleic acid was able to protect the ΔliaR strain from bile at a level equivalent to that observed in the wild-type or the complemented strains; thus, the inherent sensitivity of the mutant strain could be overcome.
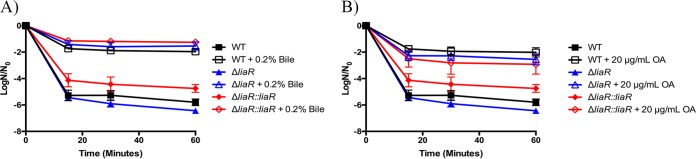
Given the improved survival of liaR-deficient E. faecalis when grown in medium supplemented with bile or oleic acid, we subsequently wanted to determine if such supplementation could protect from a different source of membrane damage. We grew strains in the presence of 0.2% bovine bile or 20 μg/ml oleic acid to exponential phase and then challenged the cells with 0.05% SDS (Fig. 2A and B, respectively). Similar to the results for the 20% bile challenge, the cellular viability for all strains was increased in the presence of 0.05% SDS when cultures were supplemented with 0.2% bile or 20 μg/ml oleic acid. In the case of SDS treatment, however, the presence or absence of liaR had no impact on survival compared to that of wild-type or complemented strains, although modification of the membrane composition did indeed rescue all strains from SDS damage.
FIG 2.
Fatty acid supplementation protects liaR-deficient Enterococcus faecalis from sodium dodecyl sulfate challenge. Shown are the averages ± standard deviations for n = 3. (A) Bile supplementation and challenge with 0.05% sodium dodecyl sulfate (SDS). All strains supplemented with 0.2% bile had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.0001). (B) Oleic acid (OA) supplementation and challenge with 0.05% SDS. All strains supplemented with 20 μg/ml OA had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P < 0.05).
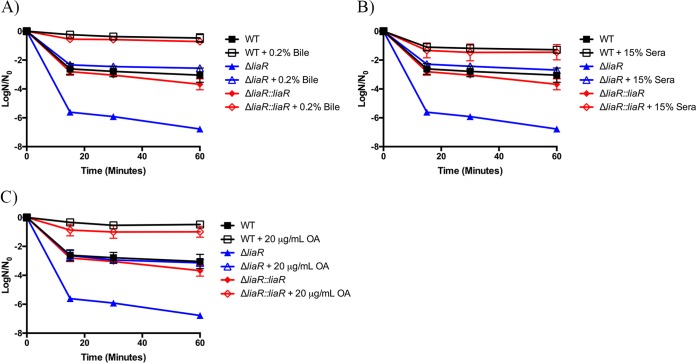
Sensitivity to daptomycin of liaR-deficient E. faecalis is decreased upon supplementation with exogenous sources of fatty acids.
To test whether the reduced daptomycin susceptibility mediated by exogenous sources of fatty acids (32) occurred through activation of the LiaFSR response, we examined daptomycin sensitivity in E. faecalis OG1RFΔliaR (31) grown in the presence or absence of fatty acid sources. We grew the parental, deletion, and genetically complemented strains to mid-log phase (OD600 of ≈0.4) in the presence of either 0.2% bile, 15% pooled human serum, or 20 μg/ml oleic acid and then exposed the cells to 10 μg/ml daptomycin. Figure 3 shows that the liaR deletion mutant was extremely susceptible to this concentration of daptomycin compared to the wild-type or ΔliaR::liaR strain. Survival against daptomycin challenge was significantly improved in the ΔliaR strain by supplementation with either bile, serum, or oleic acid. However, supplementation of the deletion strain did not yield as many survivors as supplementation of the wild-type or genetically complemented strains. Taken together, these data suggest that exogenous fatty acids can reduce daptomycin susceptibility using a mechanism independent of the LiaFSR response, but modification of the fatty acid membrane composition does not completely overcome the need for liaR.
FIG 3.
Fatty acid sources reduce daptomycin susceptibility in liaR-deficient Enterococcus faecalis. Shown are the averages ± standard deviations for n = 3. (A) Bile supplementation and challenge with 10 μg/ml daptomycin. All strains supplemented with 0.2% bile had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.002). (B) Serum supplementation and challenge with 10 μg/ml daptomycin. All strains supplemented with 15% sera had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.011). (C) Oleic acid (OA) supplementation and challenge with 10 μg/ml daptomycin. All strains supplemented with 20 μg/ml OA had a statistically increased number of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.002).
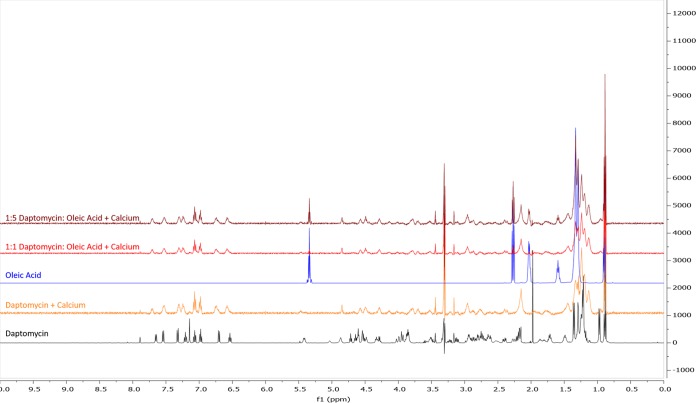
Increased tolerance to daptomycin is not due to interaction with free fatty acids.
Our data support the notion that the ΔliaR strain can incorporate exogenous fatty acids to a level similar to that of the wild-type strain, leading to better survival against membrane-damaging agents, including the antibiotic daptomycin. As daptomycin is known to insert into membranes and has a fatty acid tail (decanoic acid [C10:0]) within its structure, we wanted to verify that our observations were not due to an interaction between daptomycin and free fatty acids. Additionally, since studies have demonstrated that the presence of calcium can alter the structure of daptomycin (13) and that the activity of the antibiotic is dependent upon calcium (36), we wanted to examine if calcium could potentially mediate an interaction between free fatty acids and daptomycin. To do this, we employed proton nuclear magnetic resonance (1H NMR) to observe the interactions of daptomycin, calcium, and oleic acid.
Line broadening was observed in the spectrum containing calcium and daptomycin, which can be attributed to daptomycin aggregation, as previously reported (37). While the presence of calcium did impact the spectra of daptomycin (see Fig. S1 and S2 in the supplemental material), we noted no additional line broadening or chemical shifts in the spectra if oleic acid was added (Fig. 4).
FIG 4.
The addition of calcium does not direct an interaction between daptomycin and oleic acid. Shown is a superimposed image of five individual 1H NMR spectra, between 0.0 to 10.0 ppm. The spectra are organized as follows, from top to bottom, a 1:5 mixture of daptomycin/oleic acid plus excess calcium (maroon), a 1:1 mixture of daptomycin/oleic acid plus excess calcium (red), 50 mM solution of oleic acid (blue), 1.2 mM daptomycin plus excess calcium (orange), and 1.23 mM daptomycin solution (black). All solutions were made using methanol-d4, and spectra were generated using a VNMRS 500 MHz instrument. Spectra were superimposed using MestReNova software.
Thus, these data show that the lack of interaction between daptomycin and oleic acid indicates a role for altered cellular membranes and physiology in enhancing tolerance to daptomycin.
Clinically isolated E. faecalis strains can incorporate exogenous fatty acids.
Given the breadth of diversity of E. faecalis isolates (34), we wanted to ensure that our observations were not limited to OG1RF. Thus, we expanded our studies to include a clinical strain pair of daptomycin-susceptible and -resistant E. faecalis that were obtained from the bloodstream of a patient before and after daptomycin therapy (6). E. faecalis S613 is daptomycin susceptible (MIC of 0.5 to 1 μg/ml), and R712 is a daptomycin-resistant derivative of S613 (MIC of 8 μg/ml) (15, 28). Previous analyses of these strains showed that the sole contributing factor for their differences in daptomycin susceptibility was a mutation in the negative regulator liaF (28). Given that these strains are true clinical isolates, we sought to examine their abilities to both incorporate exogenous fatty acids and respond to membrane stressors.
Similar to OG1RF and its derivatives that were examined (Table 2), both clinical isolates had membranes dominated by palmitic acid (C16:0) and cis-vaccenic acid (C18:1 cis 11) when grown in BHI (Table 3). However, the clinical isolates had significantly larger amounts of cis-vaccenic acid (P < 0.001) than OG1RF and its derivatives (5 to 10% increase) and, consequently, statistically lower levels of palmitic acid (P < 0.005); this was particularly true for R712. Despite these differences, the saturated/unsaturated ratio was not significantly different from that for OG1RF or its derivatives examined here.
TABLE 3.
Membrane fatty acid analysis of clinical isolates during log-phase growth
| Fatty acid | % of total membrane content for indicated supplement and strain (avg ± SD)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| BHI |
Serumb |
Bilec |
C18:1
cis9d |
|||||
| S613 | R712 | S613 | R712 | S613 | R712 | S613 | R712 | |
| C12:0 | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.2 | 0.3 ± 0.3 | 0.9 ± 0.0 | 1.1 ± 0.0 |
| C14:0 | 4.8 ± 0.1 | 3.7 ± 0.1 | 3.6 ± 0.1 | 2.4 ± 0.0 | 1.3 ± 0.1 | 1.6 ± 0.0 | 0.4 ± 0.1 | 0.6 ± 0.0 |
| C16:1 | 7.9 ± 0.2 | 7.8 ± 0.2 | 5.0 ± 0.2 | 4.7 ± 0.0 | 1.1 ± 0.1 | 1.7 ± 0.1 | 1.0 ± 0.2 | 1.4 ± 0.1 |
| C16:0 | 33.2 ± 0.2 | 28.2 ± 0.6 | 37.4 ± 0.6 | 36.6 ± 0.9 | 43.4 ± 0.7 | 43.0 ± 0.9 | 1.5 ± 0.2 | 1.9 ± 0.1 |
| C17:0 2OH | 4.8 ± 0.2 | 6.1 ± 0.1 | 0.5 ± 0.1 | 0.6 ± 0.1 | ND | ND | ND | ND |
| C18:1 cis9 | ND | ND | 15.8 ± 0.8 | 18.6 ± 0.4 | 32.0 ± 1.6 | 38.1 ± 1.7 | 56.1 ± 3.4 | 57.8 ± 2.9 |
| C18:1 cis11 | 44.1 ± 0.3 | 49.1 ± 0.2 | 7.5 ± 0.4 | 4.8 ± 0.2 | 3.1 ± 0.2 | 3.9 ± 0.2 | 0.5 ± 0.8 | 0.5 ± 0.8 |
| C18:0 | 3.4 ± 0.2 | 3.4 ± 0.2 | 7.4 ± 0.1 | 7.3 ± 0.1 | 10.8 ± 1.7 | 6.2 ± 1.4 | 0.5 ± 0.0 | 0.4 ± 0.0 |
| C20:0 | ND | ND | 0.1 ± 0.1 | ND | 2.6 ± 0.4 | 0.4 ± 0.4 | 38.6 ± 4.6 | 36.4 ± 3.6 |
| C18:2 | ND | ND | 18.9 ± 1.2 | 21.3 ± 1.4 | ND | ND | ND | ND |
| C20:4 | ND | ND | 1.1 ± 0.1 | 1.3 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.1 | ND | ND |
| Otherse | 0.6 ± 1.0 | 0.7 ± 1.1 | 2.4 ± 1.4 | 2.3 ± 1.6 | 5.1 ± 0.7 | 4.4 ± 0.8 | 0.4 ± 0.1 | ND |
| Saturated/unsaturated | 0.8 ± 0.5 | 0.6 ± 0.8 | 1.0 ± 0.3 | 0.9 ± 0.5 | 1.6 ± 1.4 | 1.2 ± 1.1 | 0.7 ± 1.1 | 0.7 ± 1.0 |
| C10-C17/C18-C20f | 1.1 ± 0.01 | 0.9 ± 0.01 | 1.0 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.04 | 0.04 ± 0.0 | 0.05 ± 0.0 |
Membrane contents were determined by GC-FAME analysis by Microbial ID, Inc. Values represent averages and SD from three independent cultures. ND indicates that fatty acid was not detected.
Pooled human serum was supplemented to a final concentration of 15%.
Bovine bile was supplemented to a final concentration of 0.2%.
Oleic acid was added to a final concentration of 20 μg/ml.
Others indicates fatty acids that comprised <1% of the total membrane content.
Total fatty acid length ratios including both saturated and unsaturated fatty acids.
Upon supplementation with 15% serum, the clinical strains did not show major changes in generation times (Table 1). Additionally, their membrane contents, while altered from growth in unsupplemented medium, were similar to each other's and to those of OG1RF. We again noted that the proportion of stearic acid (C18:0), while not dominant, did double for both strains when grown in serum, and we saw similar, if not higher, increases in OG1RF as well (Table 2). The same decrease in cis-vaccenic acid (C18:1 cis 11), and concomitant increases in oleic acid (C18:1 cis 9) and linoleic acid (C18:2 cis 9, 12) observed in the OG1RF-derived strains were also present in the clinical isolates supplemented with serum.
When supplemented with 0.2% bile, the clinical strains showed increases in generation times that were not observed in OG1RF or its derivatives (Table 1). However, this was statistically significant only for S613 (P < 0.001). The overall trends in membrane incorporation remained constant between the clinical isolates and OG1RF derivatives. In particular, we observed high levels of palmitic acid (C16:0) and also increases in stearic acid (C18:0) (Table 3) for the clinical isolates that were similar to those observed with the OG1RF derivatives (Table 2).
The most dramatic difference between the clinical isolates and the OG1RF-derived strains can be seen in the membrane content of cultures supplemented with oleic acid (C18:1 cis 9). For the clinical strains, the membrane consisted of 57% oleic acid (Table 3), whereas for the OG1RF derivatives it was closer to 70% (Table 2). Again, for the clinical isolates, the dominant saturated fatty acid was C20:0 (arachidic acid), which made up more than 36% of the total membrane content. Interestingly, for OG1RF and its derivatives, as well as for the clinical isolates, this fatty acid was detected at significant levels only upon supplementation with oleic acid (C18:1 cis 9). It should be noted that the levels of arachidic acid in the clinical isolates (Table 3) were nearly double those observed for OG1RF and its derivatives (Table 2).
In summary, similar to OG1RF and its derivatives, the clinical isolates S613 and R712 also readily incorporate exogenous sources of fatty acids into their membranes. Thus, despite genetic differences between the strains, the ability to incorporate fatty acids appears to be consistent.
Specific fatty acid sources can alter sensitivity to membrane stress agents in clinical isolates.
Our analysis of the membrane content of clinical isolates upon supplementation with fatty acid sources demonstrated that OG1RF is not unique in its ability to incorporate exogenous fatty acids (Table 3). Given these data, we sought to understand how these clinical isolates responded to membrane stress after exogenous fatty acid supplementation using the experimental design outlined before for OG1RF and its derivatives.
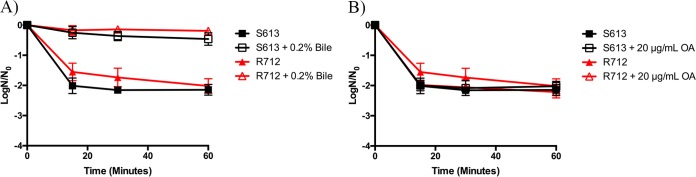
We exposed the clinical strain pair, S613 and R712, to high levels of bovine bile (20%) when grown in the presence or absence of 0.2% bile or 20 μg/ml oleic acid. We noted increased survival across all time points assessed (15, 30, and 60 min) when cultures were supplemented with low levels of bile (Fig. 5A). Surprisingly, supplementation with 20 μg/ml oleic acid was unable to protect either clinical isolate from the high bile challenge (Fig. 5B), in stark contrast to what we observed with OG1RF and its derivatives (Fig. 1B).
FIG 5.
Fatty acid supplementation shows variable protection in daptomycin-sensitive (S613) and daptomycin-resistant (R712) clinical pair isolates versus high bile challenge. Shown are the averages ± standard deviations for n = 3. (A) Bile supplementation and challenge with 20% bovine bile. All strains supplemented with 0.2% bile had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all analyzed time points (P ≤ 0.001). (B) Oleic acid (OA) supplementation and challenge with 20% bovine bile. All strains supplemented with 20 μg/ml OA versus those of their unsupplemented counterparts were not statistically different at all time points analyzed (P value > 0.05).
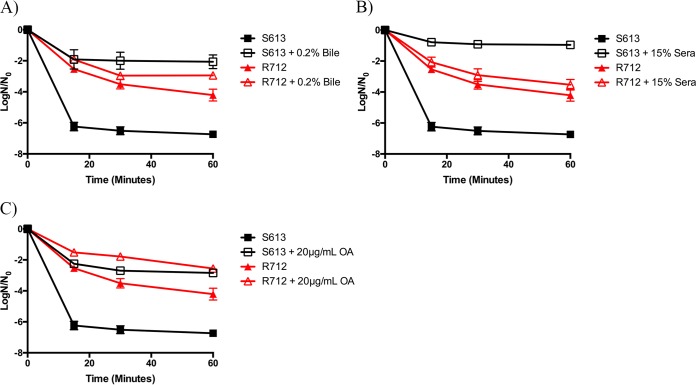
Given these findings, we attempted to determine if supplementing this clinical pair with exogenous fatty acids could alter the daptomycin susceptibility. We first supplemented these strains with 0.2% bile until exponential phase and then exposed S613 or R712 to 10 μg/ml or 40 μg/ml daptomycin, respectively. The rationale for this range of concentrations is the intrinsic daptomycin resistance of R712 (15). Interestingly, supplementation with 0.2% bile caused a significant increase in the ratio of S613 survivors for the entire time course when the strain was exposed to daptomycin (P < 0.001). Conversely, R712 had a moderate increase in survivors only after 60 min of exposure (P = 0.005) (Fig. 6A). When the clinical isolates were supplemented with 15% pooled human serum, a result similar to that observed with 0.2% bile was documented for S613 (Fig. 6B). However, R712 did not appear to benefit from the fatty acids in serum.
FIG 6.
Fatty acid sources demonstrate variable protection from daptomycin in daptomycin-sensitive (S613) and daptomycin-resistant (R712) clinical pair isolates. Shown are the averages ± standard deviations for n = 3. (A) Bile-supplemented S613 challenged with 10 μg/ml daptomycin or R712 challenged with 40 μg/ml daptomycin. S613 supplemented with 0.2% bile had statistically increased numbers of survivors versus those of its unsupplemented counterparts at all time points analyzed (P ≤ 0.037). (B) Serum supplementation and challenge with 10 μg/ml or 40 μg/ml daptomycin. S613 supplemented with 15% sera had a statistically increased number of survivors versus that of its unsupplemented counterpart at all time points analyzed (P values ≤ 0.0001), while the R712 supplemented cultures were not significantly different (P > 0.05). (C) Oleic acid (OA) supplementation and challenge with 10 μg/ml or 40 μg/ml daptomycin. All strains supplemented with 20 μg/ml OA had statistically increased numbers of survivors versus those of their unsupplemented counterparts at all time points analyzed (P ≤ 0.002).
Given that growth in both bile and serum reduced the daptomycin sensitivity of S613, we wanted to investigate whether oleic acid alone also altered daptomycin susceptibility, as was observed for the OG1RF derivatives. As shown in Fig. 6C, supplementation with oleic acid greatly reduced the sensitivity to daptomycin in S613 compared to that in unsupplemented cultures. We again examined the resistant isolate R712 under the same conditions and noted that growth in the presence of oleic acid decreased daptomycin susceptibility at a concentration of 40 μg/ml (Fig. 6C). This effect was far greater than that observed by supplementation with bile or serum. Taken together, these data suggest that exogenous sources of fatty acids can indeed be taken up and incorporated and subsequently alter the susceptibility of E. faecalis clinical strains to membrane-damaging agents.
DISCUSSION
Our previous data showed that E. faecalis OG1RF is able to incorporate exogenous fatty acids, which provide increased tolerance to membrane stressors such as bile, SDS, and daptomycin (32). These observations provided us with insights into how E. faecalis can utilize exogenous fatty acids from the host to reduce sensitivity to membrane stressors or membrane-damaging antibiotics. Moreover, the data described here suggest that increased tolerance to membrane stress was not a result of exogenous fatty acids activating the LiaFSR system. Using a clinical strain pair of E. faecalis clinical isolates, we also demonstrated that the ability to incorporate exogenous fatty acids, as well as the ability of such fatty acids to induce protection against membrane damage, is not limited to laboratory strains of E. faecalis such as OG1RF.
For all strains examined in this study, incorporation of exogenous fatty acids was conserved and fairly consistent across the genetic backgrounds. One interesting distinction, however, was the increased levels of arachidic acid (C20:0) in the membranes of the clinical isolate strains S613 and R712 compared to those of the OG1RF strains upon supplementation with oleic acid (Tables 2 and 3). The observation of arachidic acid in any of the strains was surprising: none of the strains produced detectable levels of this fatty acid when grown without supplementation. How then does supplementation with oleic acid lead to arachidic acid in the membrane? If E. faecalis were to elongate oleic acid, one would expect to see C20:1 cis 11 (38) and not arachidic acid (C20:0). It is possible that the cell is producing longer fatty acids through its de novo fatty acid biosynthetic pathway. The length of fatty acid tails during de novo biosynthesis is controlled via competition between the fatty acid acyltransferase and the fatty acid condensation (elongation) enzyme (reviewed in reference 39). Perhaps oleic acid supplementation directly or indirectly impacts the activity of one or both enzymes, leading to the observed increased fatty acid tail length. It is possible that the clinical isolates are more sensitive to these enzymatic changes, which might explain why we observe higher levels in these strains. Ongoing studies are geared to determine the source of this fatty acid.
Although the membrane composition of OG1RFΔliaR is similar to that of the wild-type strain, the deletion strain is far more sensitive to 20% bile and daptomycin (Fig. 1 and 3). These data support the critical role of the LiaR-mediated membrane stress responses seen in other bacterial species (24, 27). However, supplementation with specific sources of exogenous fatty acids can increase survival of the ΔliaR strain when challenged with 20% bile or daptomycin (Fig. 1A and C and 3A and C). Thus, while liaR is required for the basal level of tolerance to high bile, the cell can circumvent this need if exogenous fatty acid sources are provided (Fig. 1). In the case of daptomycin challenge, protection induced by fatty acids is independent of LiaFSR, but liaR is absolutely required for the optimal membrane adaptive response (Fig. 3). These observations indicate that there are different cellular responses, depending on the type of membrane damage. The combined data also support previous findings that the host-derived fatty acid, oleic acid, can reduce membrane damage and even have a role in cell growth and survival (32, 40).
Surprisingly, we did not observe increased sensitivity of the ΔliaR strain to SDS compared to that of the wild-type (Fig. 2). This suggests that liaR is dispensable for the basal level of tolerance to SDS and again suggests that E. faecalis responds to different membrane stressors in unique ways. Previous work has shown that E. faecalis has an altered transcriptional response when exposed to bile versus SDS (41). It is likely that other components within bile, such as bile salts, may contribute to these altered responses, but further analysis is needed.
Another interesting aspect of our study is that the supplementation of exogenous fatty acids to clinical isolates of E. faecalis may impact their tolerance to membrane damage (Fig. 5 and 6). However, the ability of fatty acids to induce protection in the clinical isolates was not necessarily consistent with that in the OG1RF derivatives. For example, growth in oleic acid was unable to protect either R712 or S613 from high bile damage (Fig. 5B), unlike what we observed for the derivatives of OG1RF (Fig. 1B). It is not clear what differences may contribute to this observation. One possibility is differences in the amounts of arachidic acid (C20:0) between the strains (Tables 2 and 3). For the clinical isolates, this fatty acid comprised >35% of the membrane content when the culture was supplemented with oleic acid, nearly double what was seen in the OG1RF derivatives This is reflected in the reduced ratio of C10-C17/C18-C20 fatty acids. This alteration might impact the expression or activity of membrane proteins (for example, efflux pumps) that may contribute to the overall sensitivity or resistance of the strains. More work is needed to elucidate the mechanism contributing to these observations. However, growth in oleic acid protects both S613 and R712 from daptomycin-induced damage (Fig. 6C), mirroring what is seen in OG1RF derivatives. This indicates that damage induced by daptomycin and bile is not equivalent and that there are genetic or physiological differences between enterococcal strains in how they handle membrane-damaging agents.
Overall, our results show that exogenous fatty acids impact membrane composition and the ability to survive a variety of membrane stressors. However, the mechanism by which fatty acids confer this protection is unclear. An altered membrane fatty acid profile would likely impact the level, distribution, and potential activity of membrane-associated proteins that may contribute to survival. Additionally, it is not clear what other metabolic processes may be impacted by shifting from de novo fatty acid biosynthesis for the generation of membranes to the use of exogenous fatty acids. These observations and the underlying mechanism(s) of fatty acid-induced membrane protection are critically important for understanding the host-pathogen interaction and bacterial response to antimicrobial peptides due to the abundance of free fatty acids. E. faecalis is a commensal organism that has access to fatty acids located in bile and serum and is naturally tolerant to these compounds. In the human host, E. faecalis from the gut may enter different compartments and alter their membranes in order to succeed in a hostile environment. Fatty acids in serum and tissues might help the bacterium survive membrane stressors driven by the innate immune system (i.e., antimicrobial peptides).
A growing number of studies are demonstrating that the microbes within the host, both commensal organisms and pathogens, are capable of utilizing host metabolites, including fatty acids. Utilization of these sources significantly impacts the microbes, leading to altered physiology, gene expression, and possibly virulence (42). These studies, in conjunction with our previous findings, demonstrate that host fatty acids can induce protection from membrane stressors, including antibiotics. It is worth noting, however, that measurements of MICs of antimicrobials are not often performed in the presence of host fatty acid sources (43). It is worth considering further how the host environment may lead to an altered sensitivity to such damaging agents and to take into account the host environment when such analyses are performed.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Reynolds for helpful discussions. We are grateful to J. Wells and J. Wen for editorial comments.
Funding for this work was through startup funds provided by the University of Tennessee and through NIH/NIAID grant R01AI116571, both to E.M.F. C.A.A. is funded by NIH/NIAID grants R01-AI093749, R21-AI114961, and R21/R33-AI121519.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00933-16.
REFERENCES
- 1.Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology 155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garsin DA, Frank KL, Silanpaa J, Ausubel FM, Hartke A, Shankar N, Murray BE. 2014. Pathogenesis and models of enterococcal infection. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 4.Kristich CJ, Rice LB, Arias CA. 2014. Enterococcal infection-treatment and antibiotic resistance. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 5.Munita JM, Murray BE, Arias CA. 2014. Daptomycin for the treatment of bacteraemia due to vancomycin-resistant enterococci. Int J Antimicrob Agents 44:387–395. doi: 10.1016/j.ijantimicag.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin Infect Dis 41:565–566. doi: 10.1086/432121. [DOI] [PubMed] [Google Scholar]
- 7.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J Jr, Infectious Diseases Society of America. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 8.Debono M, Abbott BJ, Molloy RM, Fukuda DS, Hunt AH, Daupert VM, Counter FT, Ott JL, Carrell CB, Howard LC, Boeck LVD, Hamill R. 1988. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: the synthesis and evaluation of daptomycin (LY146032). J Antibiot (Tokyo) 41:1093–1105. doi: 10.7164/antibiotics.41.1093. [DOI] [PubMed] [Google Scholar]
- 9.Debono M, Barnhart M, Carrell CB, Hoffmann JA, Occolowitz JL, Abbott BJ, Fukuda DS, Hamill RL, Biemann K, Herlihy WC. 1987. A21978C, a complex of new acidic peptide antibiotics: isolation, chemistry, and mass spectral structure elucidation. J Antibiot (Tokyo) 40:761–777. doi: 10.7164/antibiotics.40.761. [DOI] [PubMed] [Google Scholar]
- 10.Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother 47:2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, Muraih JK, Tishbi N, Herskowitz J, Victor RL, Silverman J, Uwumarenogie S, Taylor SD, Palmer M, Mintzer E. 2014. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem 289:11584–11591. doi: 10.1074/jbc.M114.554444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung D, Rozek A, Okon M, Hancock RE. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol 11:949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Tran TT, Munita JM, Arias CA. 2015. Mechanisms of drug resistance: daptomycin resistance. Ann N Y Acad Sci 1354:32–53. doi: 10.1111/nyas.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365:892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. 2013. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother 57:5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother 55:3345–3356. doi: 10.1128/AAC.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores-Kim J, Darwin AJ. 2014. Regulation of bacterial virulence gene expression by cell envelope stress responses. Virulence 5:835–851. doi: 10.4161/21505594.2014.965580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev 32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- 20.Flores-Kim J, Darwin AJ. 2015. Activity of a bacterial cell envelope stress response is controlled by the interaction of a protein binding domain with different partners. J Biol Chem 290:11417–11430. doi: 10.1074/jbc.M114.614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110. doi: 10.1016/S0065-2911(02)46002-X. [DOI] [PubMed] [Google Scholar]
- 22.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 23.Fabret C, Feher VA, Hoch JA. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol 181:1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol 188:5153–5166. doi: 10.1128/JB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrecke K, Jordan S, Mascher T. 2013. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol Microbiol 87:769–788. doi: 10.1111/mmi.12130. [DOI] [PubMed] [Google Scholar]
- 27.Shankar M, Mohapatra SS, Biswas S, Biswas I. 2015. Gene regulation by the LiaSR two-component system in Streptococcus mutans. PLoS One 10:e0128083. doi: 10.1371/journal.pone.0128083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, Shamoo Y, Dowhan W, Bayer AS, Arias CA. 2013. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4:e00281-13. doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. 2013. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 57:2831–2833. doi: 10.1128/AAC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panesso D, Reyes J, Gaston EP, Deal M, Londono A, Nigo M, Munita JM, Miller WR, Shamoo Y, Tran TT, Arias CA. 2015. Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob Agents Chemother 59:7327–7334. doi: 10.1128/AAC.01073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, Singh KV, Yeaman MR, Murray BE, Shamoo Y, Garsin D, Bayer AS, Arias CA. 2015. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis 211:1317–1325. doi: 10.1093/infdis/jiu602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito HE, Harp JR, Fozo EM. 2014. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol 80:6527–6538. doi: 10.1128/AEM.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasser M, Kunitsky C, Jackoway G, Ezzell JW, Teska JD, Harper B, Parker S, Barden D, Blair H, Breezee J, Carpenter J, Cheek WV, DeMartino M, Evans B, Ezzell JW, Francesconi S, Franko E, Gardner W, Glazier M, Greth K, Harper B, Hart T, Hodel M, Holmes-Talbot K, Hopkins KL, Iqbal A, Johnson D, Krader P, Madonna A, McDowell M, McKee ML, Park M, Parker S, Pentella M, Radosevic J, Robison RA, Rotzoll B, Scott K, Smith M, Syed N, Tang J, Teska JD, Trinh H, Williams LI, Wolcott M, AOAC. 2005. Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int 88:178–181. [PubMed] [Google Scholar]
- 34.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 36.Jones RN, Barry AL. 1987. Antimicrobial activity and spectrum of LY146032, a lipopeptide antibiotic, including susceptibility testing recommendations. Antimicrob Agents Chemother 31:625–629. doi: 10.1128/AAC.31.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ball LJ, Goult CM, Donarski JA, Micklefield J, Ramesh V. 2004. NMR structure determination and calcium binding effects of lipopeptide antibiotic daptomycin. Org Biomol Chem 2:1872–1878. doi: 10.1039/b402722a. [DOI] [PubMed] [Google Scholar]
- 38.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. 2014. Incorporation of extracellular fatty acids by a fatty-acid kinase dependent pathway in Staphylococcus aureus. Mol Microbiol 92:234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubota N, Kuzumoto K, Hidaka E, Yoshizawa K, Yumoto K, Oana K, Ogiso Y, Nakamura T, Kawakami Y. 2013. First isolation of oleate-dependent Enterococcus faecalis small-colony variants from the umbilical exudate of a paediatric patient with omphalitis. J Med Microbiol 62:1883–1890. doi: 10.1099/jmm.0.062752-0. [DOI] [PubMed] [Google Scholar]
- 41.Solheim M, Aakra A, Vebo H, Snipen L, Nes IF. 2007. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl Environ Microbiol 73:5767–5774. doi: 10.1128/AEM.00651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao J, Rock CO. 2015. How bacterial pathogens eat host lipids: implications for the development of fatty acid synthesis therapeutics. J Biol Chem 290:5940–5946. doi: 10.1074/jbc.R114.636241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.