ABSTRACT
The increasing prevalence of carbapenem-resistant Acinetobacter baumannii (CRAB) strains in intensive care units has caused major problems in public health worldwide. Our aim was to determine whether this phage could be used as an alternative therapeutic agent against multidrug-resistant bacterial strains, specifically CRAB clinical isolates, using a mouse model. Ten bacteriophages that caused lysis in CRAB strains, including blaOXA-66-like genes, were isolated. YMC13/01/C62 ABA BP (phage Bϕ-C62), which showed the strongest lysis activity, was chosen for further study by transmission electron microscopy (TEM), host range test, one-step growth and phage adsorption rate, thermal and pH stability, bacteriolytic activity test, genome sequencing and bioinformatics analysis, and therapeutic effect of phage using a mouse intranasal infection model. The phage Bϕ-C62 displayed high stability at various temperatures and pH values and strong cell lysis activity in vitro. The phage Bϕ-C62 genome has a double-stranded linear DNA with a length of 44,844 bp, and known virulence genes were not identified in silico. In vivo study showed that all mice treated with phage Bϕ-C62 survived after intranasal bacterial challenge. Bacterial clearance in the lung was observed within 3 days after bacterial challenge, and histologic damage also improved significantly; moreover, no side effects were observed.
IMPORTANCE In our study, the novel A. baumannii phage Bϕ-C62 was characterized and evaluated in vitro, in silico, and in vivo. These results, including strong lytic activities and the improvement of survival rates, showed the therapeutic potential of the phage Bϕ-C62 as an antimicrobial agent. This study reports the potential of a novel phage as a therapeutic candidate or nontoxic disinfectant against CRAB clinical isolates in vitro and in vivo.
INTRODUCTION
Bacteriophages (phages) are viruses that infect bacteria and are ubiquitous in the biosphere (1). Recently, phages have been reported as therapeutic agents to treat bacterial infectious diseases in humans and animals (2–5). Phages possess several advantages as therapeutic agents over conventional antibiotics, in that they have precise effective bacteriolytic activity against target bacteria and have shown no critical side effects to date (6–8).
Antimicrobials have been crucial for the prevention and treatment of infectious diseases in humans, animals, and plants since the 1940s (9). Today, however, certain isolated strains have exhibited resistance to all currently available commercial antimicrobial agents (9, 10). Many scientists and medical professionals have emphasized the urgent need to prevent the emergence and spread of drug-resistant bacteria (11). Despite efforts to reduce the dissemination of drug-resistant pathogens in health care settings, antimicrobial resistance has been constantly increasing and becoming a serious global clinical challenge with little accompanying novel antibiotic discovery (12). Hence, phages are currently being viewed as alternative therapeutic agents (6).
Acinetobacter spp. are Gram-negative bacilli that are ubiquitous and important infectious pathogens in clinical settings, and they are responsible for various infections, including pneumonia, meningitis, septicemia, wound infection, and urinary tract infection (13). In particular, A. baumannii is one of the major bacterial species causing serious nosocomial infections in intensive care units (ICUs). They exhibit a high rate of resistance to most commercial drugs, leading to higher mortality and morbidity (14, 15). Carbapenems are extended-spectrum β-lactam antibiotics exhibiting potent and excellent efficacy, particularly in the treatment of serious infections caused by multidrug-resistant Gram-negative bacteria (16). However, the current emergence and prevalence of A. baumannii expressing resistance to carbapenems have been increasingly reported in many countries (17). These carbapenem-resistant A. baumannii (CRAB) strains lead to community- and hospital-acquired infections that are difficult to control and treat, and these problems have caused a serious medical threat worldwide (18, 19).
In this study, we isolated and characterized the lytic bacteriophage Bϕ-C62, which is able to infect CRAB clinical isolates. Our aim was to determine whether this phage could be used as an alternative therapeutic agent against multidrug-resistant bacterial strains, specifically CRAB strains, using a mouse model. This study reports on the safety and therapeutic efficacy of a novel phage against CRAB isolated from clinical samples, using the mouse model as a surrogate host.
MATERIALS AND METHODS
Bacterial strains.
A total of 45 clinical carbapenem-resistant Acinetobacter species isolates were selected from clinical samples, including respiratory, urine, and pus samples, at a university-affiliated hospital in 2013. The identification and antimicrobial susceptibility of the clinical isolates were determined using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Vitek MS system; bioMérieux Inc., Marcy l'Etoile, France) and the VITEKN132 system (bioMérieux). Collected CRAB isolates were used for initial isolation and evaluation of the phage host spectrum. Clonal differences of the A. baumannii isolates that showed clear zones on a plate, i.e., plaques, based on the phage host spectrum test were confirmed using pulsed-field gel electrophoresis (PFGE) with the contour-clamped homogeneous electric field (CHEF) DR-II system (Bio-Rad Laboratories, Hercules, CA). Phylogenetic analyses were performed using InfoQuest FP software (version 4.50; Bio-Rad Laboratories, Inc.). To determine the epidemiological relationships of these strains, multilocus sequence typing (MLST) was performed, and results were analyzed using the MLST database (http://pubmlst.org/abaumannii/). Detection of the OXA carbapenemase genes in A. baumannii strains was performed by multiplex PCR (20). The modified Hodge test (MHT) was performed for all isolates as previously described by Lee et al. (21). The carbapenem-resistant YMC13/01/C62 strain was specifically used as the host bacterial species for characterization and in vivo testing in order to estimate the therapeutic potential of phage Bϕ-C62.
Isolation and propagation of bacteriophage.
Ten bacteriophages capable of lysing carbapenem-resistant Acinetobacter spp. were isolated from sewage water at a hospital in South Korea. The isolation and purification of phages were performed using polyethylene glycol (PEG; Sigma, St. Louis, MO, USA) treatment and the double layer method (22). The sewage sample was treated with NaCl (1 M; Merck) and PEG 8000 (final concentration of 10%) and was incubated at 4°C for 24 h. The sample solution was centrifuged and filtered using 0.22-μm membranes (Millipore Corporation, Bedford, MA, USA). Phages were harvested by ultracentrifugation (12,000 × g for 1 h at 4°C) and resuspended in sterilized sodium chloride-magnesium sulfate (SM) buffer (100 mM NaCl, 8 mM MgSO4, 2% gelatin, 50 mM Tris-HCl, pH 7.5).
To amplify phages against collected clinical strains, phage samples (40 μl) and all strains were mixed in 4 ml of Luria-Bertani (LB) broth medium (Difco, Detroit, MI, USA) and incubated overnight at 37°C. The cultures next were centrifuged (12,000 × g for 10 min at 4°C) and filtered (0.22-μm membrane; Millipore Corporation, Bedford, MA, USA) to remove bacterial debris. The purification steps of single plaques using plaque assays were repeated three times. Host bacterial strains (optical density at 600 nm [OD600] of 0.5) in 4 ml of LB broth medium were mixed with 10 μl of purified phage solution and incubated at 37°C for 12 h with shaking. Culture samples were centrifuged (12,000 × g for 10 min at 4°C) and filtered to remove cell debris. After PEG 8000 (a final concentration of 10%) treatment, the phage solutions were incubated for 12 h at 4°C and centrifuged (12,000 × g, at 4°C for 10 min), followed by resuspension in SM buffer. The number of PFU of the concentrated phage solutions was confirmed through plaque assays using the double layer method.
Transmission electron microscopy (TEM).
Ten purified phage particles were mounted on copper grids and negatively stained with 2% uranyl acetate for 15 s. Phage morphology was then observed using a transmission electron microscope (JEM-1011; JEOL, Tokyo, Japan) at an operating voltage of 80 kV.
Host range test.
A total of 45 clinical carbapenem-resistant Acinetobacter species isolates were used for purified phage host range testing using spot tests as described previously, with some modifications (23). Briefly, the phage solution was spotted directly onto a bacterial lawn on the LB agar plate and incubated at 37°C overnight. According to the degree of clarity, the plaque-forming level of the phage was measured as a clear zone (++) or turbid zone (+).
Host cell lysis activity.
A carbapenem-resistant A. baumannii YMC13/01/C62 strain culture (early exponential phase; OD600 of 0.2) was infected with the phage at a multiplicity of infection (MOI) of 0.1, 1, or 10 while shaking at 37°C. Samples were collected at 1-h intervals for 6 h, and microbial growth was measured according to the OD600 using spectrophotometry. This experiment was performed in triplicate.
Genome sequencing and bioinformatics analysis.
The extraction of phage genomic DNA was carried out using the phenol-chloroform precipitation method as described previously (24). The complete genome of phage Bϕ-C62 was sequenced using a 454 GS Junior genome analyzer (Roche, Branford, CT, USA). Gap filling was performed using standard PCR. The entire genome sequence was determined using the Roche GS Assembler, version 2.6 (Roche), and CLC Genomics Workbench 4.8 (CLCbio USA, Cambridge, MA). The NCBI open reading frame (ORF) finder and GenMark.hmm software (25) were used for the prediction of ORFs. The comparison of genome sequences with those of other phages was performed using the NCBI (http://www.ncbi.nlm.nih.gov/) database, MAUVE software (version 2.3.1) (26), and Easyfig software (version 2.1) (27). BLASTP and PSI-BLAST searches (http://www.ebi.ac.uk/Tools/sss/fasta/) were used to determine the similarity of all putative proteins in the genome sequence. The tRNA genes were analyzed using the tRNAscan-SE program (28).
Therapeutic effect of phage in the mouse model.
To evaluate mortality due to phage Bϕ-C62, mice were divided into six groups: (i) phosphate-buffered saline (PBS) and SM buffer treated (n = 6), (ii) A. baumannii and SM buffer treated (n = 6), (iii) PBS and phage Bϕ-C62 treated (n = 6), (iv) A. baumannii and phage Bϕ-C62 treated (MOI = 10, n = 6), (v) A. baumannii and phage Bϕ-C62 treated (MOI = 1; n = 6), and (vi) A. baumannii and phage Bϕ-C62 treated (MOI = 0.1; n = 6). Briefly, female C57BL/6 mice, 7 to 8 weeks of age, first were immunized with 200 mg/kg of body weight cyclophosphamide (Sigma, MO, USA) by intraperitoneal (i.p.) injection, followed by a second i.p. injection after 48 h. Mice were anesthetized with a Zoletil-Rompun mixture by i.p. injection (29). The mice were inoculated with bacteriophage (1 × 1010 PFU/ml; 30 μl) through the same route after 30 min of intranasal bacterial (1 × 109 CFU/ml; 30 μl) infection. Survival rate and body weight were monitored for 12 days. For histological analysis, mice were divided into four groups: (i) PBS and SM buffer treated (n = 15), (ii) PBS and phage Bϕ-C62 treated (n = 15), (iii) A. baumannii and SM buffer treated (n = 15), and (iv) A. baumannii and phage Bϕ-C62 treated (MOI = 10; n = 15). Briefly, mice (n = 5 mice per group) were sacrificed at day 1, 3, or 5 after bacterial infection, and lung tissues and serum samples were collected for analysis. Lung tissues were homogenized and centrifuged at 8,000 rpm for 1 min. The collected blood samples were centrifuged at 12,000 rpm for 20 min, and the sera were collected. All of the samples were stored at −80°C until tested.
Histological analysis.
The left lobe of one lung was removed from each experimental group (n = 5 mice per group) at day 1 or 3, fixed in 10% formalin, and embedded in paraffin. Slides of hematoxylin-eosin (H&E)-stained tissues were observed for histopathology analysis using an optical microscope. Histopathology was evaluated for severity level in a blinded manner. Lung tissue inflammation was evaluated as a severity score of inflammation: 0, no inflammatory lesions; 1, mild; 2, mild to moderate; 3, moderate; 4, moderate to severe; 5, severe.
Bacterial clearance and phage counting.
The collected lung samples were weighed and homogenized at each time point (day 1, 3, or 5) and then serially diluted in PBS. Lung lysates and sera were plated on LB agar plates containing ampicillin (50 μg/ml) to determine viable bacterial counts. Bacteriophage counts in lung lysates and sera were determined using the double layer agar method.
Ethics statement.
All animal studies were approved by and followed the guidelines and regulations of the Institutional Animal Care and Use Committee, Yonsei University College of Medicine, Seoul, South Korea.
Statistical analysis.
Statistical calculations for survival rate were performed using the log-rank (Mantel-Cox) test, and comparisons of bacteria, phage count, and cytokine levels were performed using a one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test (GraphPad Prism software, version 6; GraphPad Software, San Diego, CA, USA).
Nucleotide sequence accession number.
The complete genomic sequence of phage Bϕ-C62 was deposited in the GenBank database under accession number KJ817802.
RESULTS
Characterization of bacteria.
All 45 clinical A. baumannii strains used in this study were multidrug-resistant A. baumannii strains resistant to carbapenems (Table 1; also see Table S1 in the supplemental material). Sixteen of 45 CRAB strains, which showed a clear zone by phage Bϕ-C62 in the spot test, were clonally different when reviewed by PFGE (see Fig. S1). Based on multiplex PCR, 16 CRAB isolates contained the blaoxa-66-like genes encoding the class D carbapenem-hydrolyzing oxacillinases, and these strains showed positive results on the Hodge test. These isolates also were identified as sequence type 357, belonging to European clone II based on MLST analysis (Table 1).
TABLE 1.
Antibiotic resistance profiles of A. baumannii strains used in this studya and host spectrum of A. baumannii phage Bϕ-C62
| Host strain | MLSTb | OTCc | MHTd | MIC (μg/ml) forf: |
Infectivity of Bϕ-C62e | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin | Ampicillin/sulbactam | Ceftazidime | Colistin | Cefepime | Cefotaxime | Gentamicin | Imipenem | Levofloxacin | Meropenem | Minocycline | Piperacillin/tazobactam | Co-trimoxazole | Tigecycline | |||||
| YMC13/01/C62 | 357 | OXA-66-like | + | ≥64 (R) | ≤16 (I) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R129 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤1 (S) | ++ |
| YMC13/01/R317 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤1 (S) | ++ |
| YMC13/01/U249 | 357 | OXA-66-like | + | ≤8 (S) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≤2 (S) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R280 | 357 | OXA-66-like | + | ≥64 (R) | ≤8 (S) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≤4 (I) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤1(S) | ++ |
| YMC13/01/R224 | 357 | OXA-66-like | + | ≥64 (R) | ≤16 (I) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R656 | 357 | OXA-66-like | + | ≤8 (S) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≤4 (S) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤1(S) | ++ |
| YMC13/01/P187 | 357 | OXA-66-like | + | ≤8 (S) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≤1 (S) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R1400 | 357 | OXA-66-like | + | ≥64 (R) | ≤8 (S) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≤4 (I) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤1(S) | ++ |
| YMC13/01/R1224 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R1919 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤1 (S) | ++ |
| YMC13/01/R2058 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R1049 | 357 | OXA-66-like | + | ≥64 (R) | ≤8 (S) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
| YMC13/01/R3197 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤4 (S) | ≥128 (R) | ≥320 (R) | ≤1(S) | + |
| YMC13/03/R1238 | 357 | OXA-66-like | + | ≥64 (R) | ≤8 (S) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤0.5 (S) | + |
| YMC13/04/B720 | 357 | OXA-66-like | + | ≥64 (R) | ≥32 (R) | ≥64 (R) | ≤0.5 (S) | ≥64 (R) | ≥64 (R) | ≥16 (R) | ≥16 (R) | ≥8 (R) | ≥16 (R) | ≤8 (I) | ≥128 (R) | ≥320 (R) | ≤2 (S) | ++ |
Antibiotic resistance was determined by the disk diffusion test method.
MLST, multilocus sequence type.
OTC, OXA-type carbapenemase.
MHT, modified Hodge test; +, positive (distinct distortion of ≥3 mm).
Phage activity against collected bacteria: ++, clear plaque; +, turbid plaque.
S, susceptible; I, intermediate; R, resistant.
Of the 16 isolates, one A. baumannii YMC13/01/C62 strain was used as the host for phage Bϕ-C62. This strain was resistant to amikacin, ceftazidime, cefepime, cefotaxime, gentamicin, imipenem, levofloxacin, meropenem, piperacillin-tazobactam, and co-trimoxazole but was susceptible to colistin, minocycline, and tigecycline and was intermediate to ampicillin-sulbactam.
Phage characterization.
Ten Acinetobacter phages, including phage Bϕ-C62, were isolated in this study. All of the phages belonged to the Myoviridae family and the order Caudovirales (see Fig. 6; see also Fig. S2 in the supplemental material). As shown in Table 1, the infection rate of phage Bϕ-C62 was approximately 35% (16 of 45) against CRAB clinical isolates based on spot test analyses. Moreover, phage Bϕ-C62 formed plaques on two colistin-resistant A. baumannii strains. However, no other tested Gram-negative bacterial strains were lysed by phage Bϕ-C62, including Pseudomonas aeruginosa and Escherichia coli strains (data not shown).
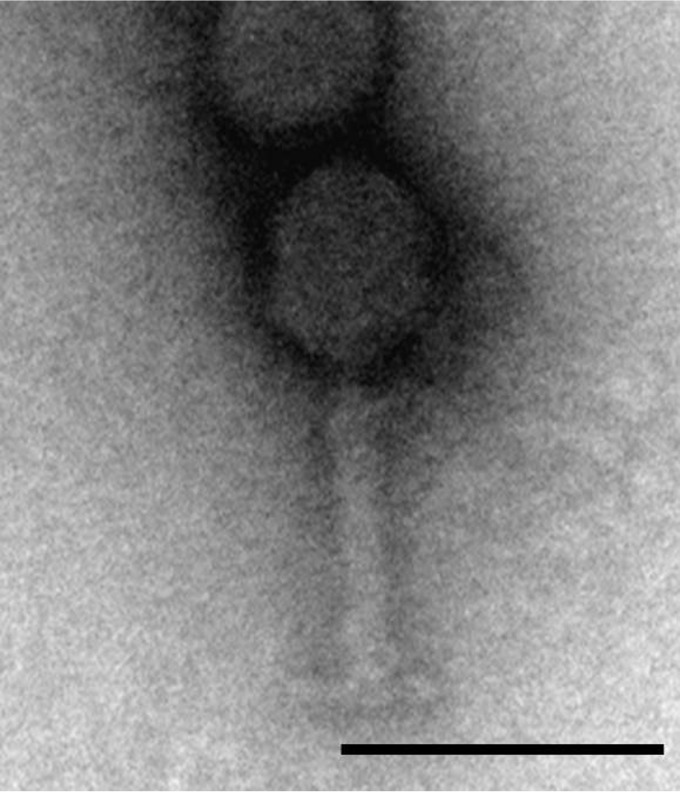
FIG 6.
Transmission electron microscopy images of A. baumannii phage Bϕ-C62. A. baumannii phage Bϕ-C62 has an icosahedral head and contractile tail and belongs to the Myoviridae family. Bar length, 100 nm.
According to the one-step growth curve analysis, phage Bϕ-C62 showed a latency period of approximately 20 min with a burst size of 76 PFU/cell (see Fig. S3a in the supplemental material) and an absorption rate of approximately 80% within 1 min (see Fig. S3b). Phage Bϕ-C62 showed high temperature stability at 25°C (99%) and 37°C (97%) for up to 9 h and maintained activity at 62%, 55%, and 46% at 40°C, 50°C, and 60°C, respectively. No activity was detected at 70°C (see Fig. S4a). The optimal pH for phage Bϕ-C62 was at pH 7 (91%) and pH 7.5 (92%) for up to 10 months. High stability (>90%) was retained at pH 4 to pH 10 at day 1, with significant stability (>60%) noted over 10 months (see Fig. S4b).
The lysis activity of phage Bϕ-C62 against A. baumannii YMC13/01/C62 was evaluated in cultured bacteria in the early exponential phase with phage at MOIs of 0.1, 1, and 10. Phage inhibited bacterial growth at all MOIs (OD600 of <0.3 at 6 h), while the bacterial culture without phage grew rapidly (OD600 of 1.3 at 6 h) according to culture time points (Fig. 1).
FIG 1.
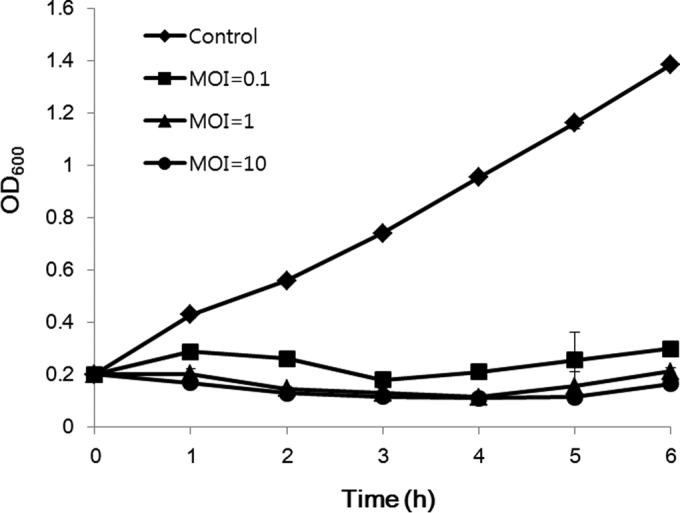
Bacteriolytic effect of phage Bϕ-C62 against carbapenem-resistant A. baumannii YMC13/01/C62. Early-exponential-phase cultures (OD600 of 0.2) of A. baumannii YMC13/01/C62 were infected by phage Bϕ-C62 at an MOI of 0.1, 1, or 10 and cultured for up to 6 h. The results shown are the means ± standard deviations from triplicate experiments.
Genome sequencing and bioinformatics analysis.
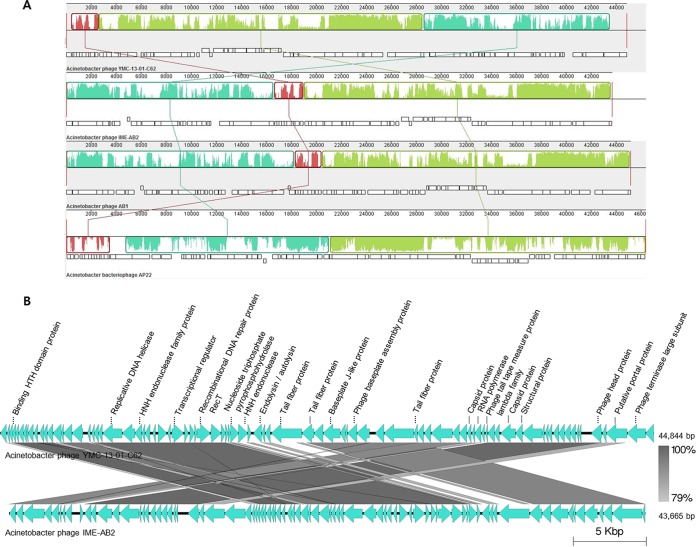
The complete genome sequence of phage Bϕ-C62 was determined to contain 44,844 bp, with read lengths of 90,862 and coverage of 937 with a GC content of 37.6%. Among the 84 ORFs identified, 71 were predicted on the negative strands while only 13 ORFs were predicted on the positive strand. Of the 84 ORFs, 75 had a predicted ATG initiation codon, 8 were predicted to contain GTG, and 1 was posited to harbor TTG. Only 21 (25%) of the 84 ORFs in the genome were assigned putative functions by BLASTP and PSI-BLAST, and no predicted tRNAs were found in the genome (see Table S2 in the supplemental material). Compared with other phage genomes, the phage Bϕ-C62 genome showed highest similarity of 80%, 57%, and 50% with phage IME-AB2 (GenBank accession number JX976549), phage AB1 (GenBank accession number HM368260), and phage AP22 (GenBank accession number HE806280), respectively (Fig. 2A). Nucleotide alignment of the four A. baumannii phages showed that some functional regions were highly homologous, with significant rearrangements observed. In the genome map of phage Bϕ-C62, each of the annotated genes was divided into three groups: (i) phage head morphology and structure (orf45, orf46, orf48, orf51, orf55, orf61, orf65, orf68, orf69, orf80, orf82, and orf83), (ii) phage DNA replication and modification (orf22, orf23, orf30, orf34, orf40, and orf63), and (iii) lysis (orf42) (Fig. 2B; also see Fig. S5 and Table S2).
FIG 2.
Genome comparison of A. baumannii phage Bϕ-C62, A. baumannii phage IME-AB2, phage AB1, and phage AP22 at the DNA level. This analysis was carried out using MAUVE software 2.3.1 with the default parameters. The connecting lines indicate similar regions between Bϕ-C62 ORFs and the three Acinetobacter phages. (A) White blocks indicate annotated genes with the reverse strand shifted downward. (B) The genome map of Bϕ-C62 and its comparison with ORFs from phage IME-AB2 as determined by Easyfig software, which indicates a putative function of each ORF, with the direction of transcription shown by arrows.
Effect of phage treatment against bacterial infection in the mouse model.
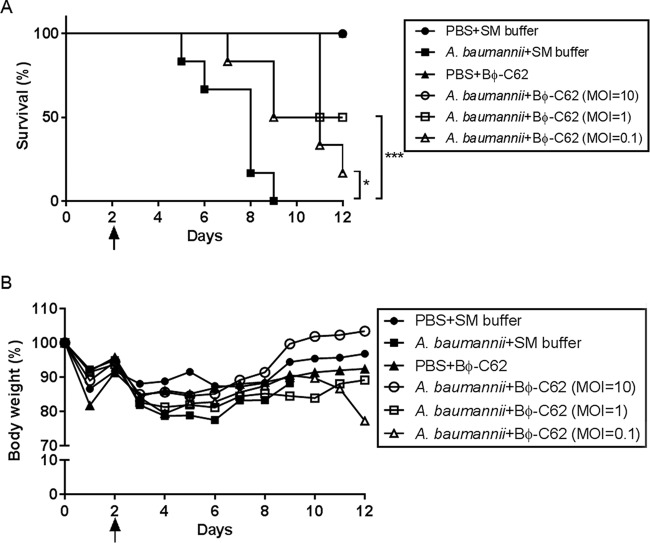
To evaluate the therapeutic effects of Bϕ-C62 against bacterial infection and the safety of Bϕ-C62 in the mouse model, we evaluated the survival rate, histological features of the lung, the number of bacteria and bacteriophage in the lung, and immunogenicity through intranasal administration of phage against a carbapenem-resistant Acinetobacter sp. strain. The minimum lethal dose (MLD) against bacterial infection through the intranasal route was confirmed as 2 × 109 CFU per mouse prior to the experimentation (data not shown). As shown in Fig. 5A, after the injection of cyclophosphamide (CP), the intranasal bacterium-infected mouse group without phage administration died by day 3 postinfection, and all mortality was noted within day 7 postinfection. After bacterial infection, phage-treated mouse groups exhibited a 100% survival rate at an MOI of 10, and at MOIs of 1 and 0.1 the mice showed survival rates of 50% and ∼16%, respectively. Therefore, survival rates differed according to phage MOI, and only mice that were phage treated had reduced or nonexistent mortality or morbidity (Fig. 3).
FIG 5.
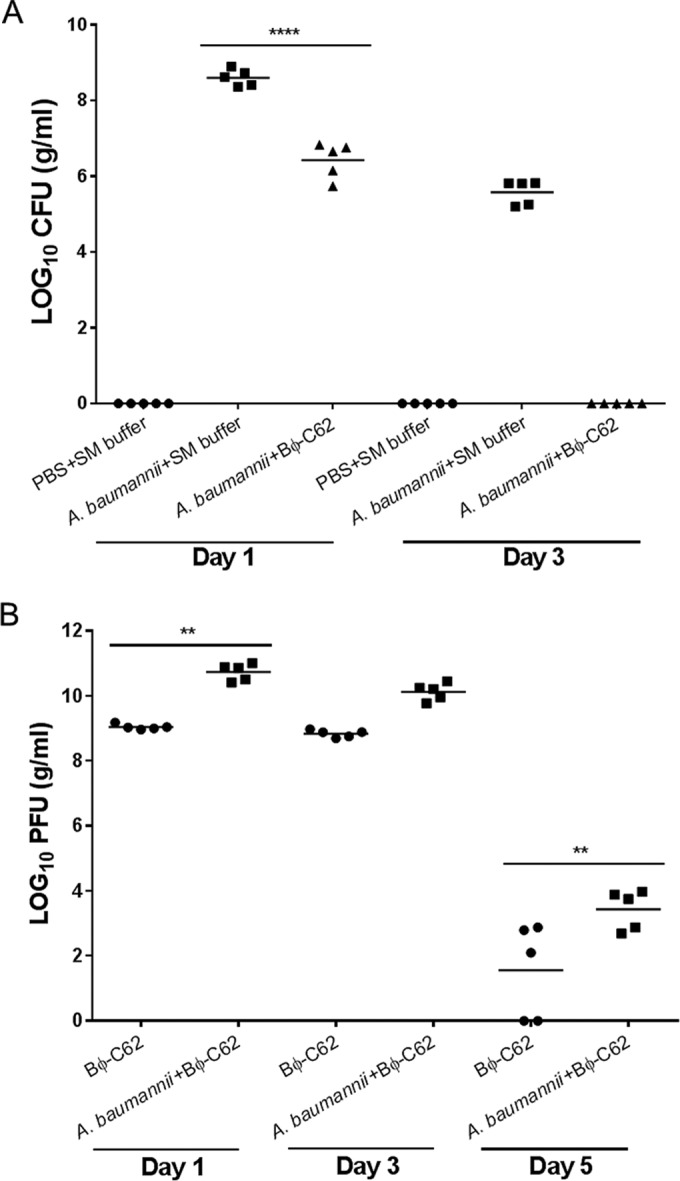
Bacterial CFU (A) and phage PFU (B) in the lungs of phage Bϕ-C62-treated and untreated mice after bacterial infection (A. baumannii YMC13/01/C62). Phage were administered 30 min postinfection. Five mice were sacrificed from each mouse group at days 1, 3, and 5. The horizontal bar indicates the mean value for each group. The counts were compared using one-way ANOVA with Tukey's multiple-comparison test (****, P < 0.0001; **, P < 0.05).
FIG 3.
Survival kinetics (A) and body weights (B) of phage Bϕ-C62-administered mice infected with A. baumannii YMC13/01/C62. (A) Female C57BL/6 mice were intranasally inoculated with phage and/or bacteria. •, buffer treated (PBS and SM); ■, bacterium infected; ▲, treated with phage only; ○, bacterium infected and phage treated 30 min postinfection (MOI = 10); □, bacterium infected and phage treated 30 min postinfection (MOI = 1); △, bacterium infected and phage treated 30 min postinfection (MOI = 0.1). (B) Body weights of C57BL/6 mice were measured daily. The black arrow represents a second cyclophosphamide (CP) administration and the first bacterial infection time point. Values given are averages (n = 6 mice per group). Statistical analysis was performed using the log-rank (Mantel-Cox) test. ***, P < 0.0001; *, P = 0.0236.
Histological changes and cytokine analysis.
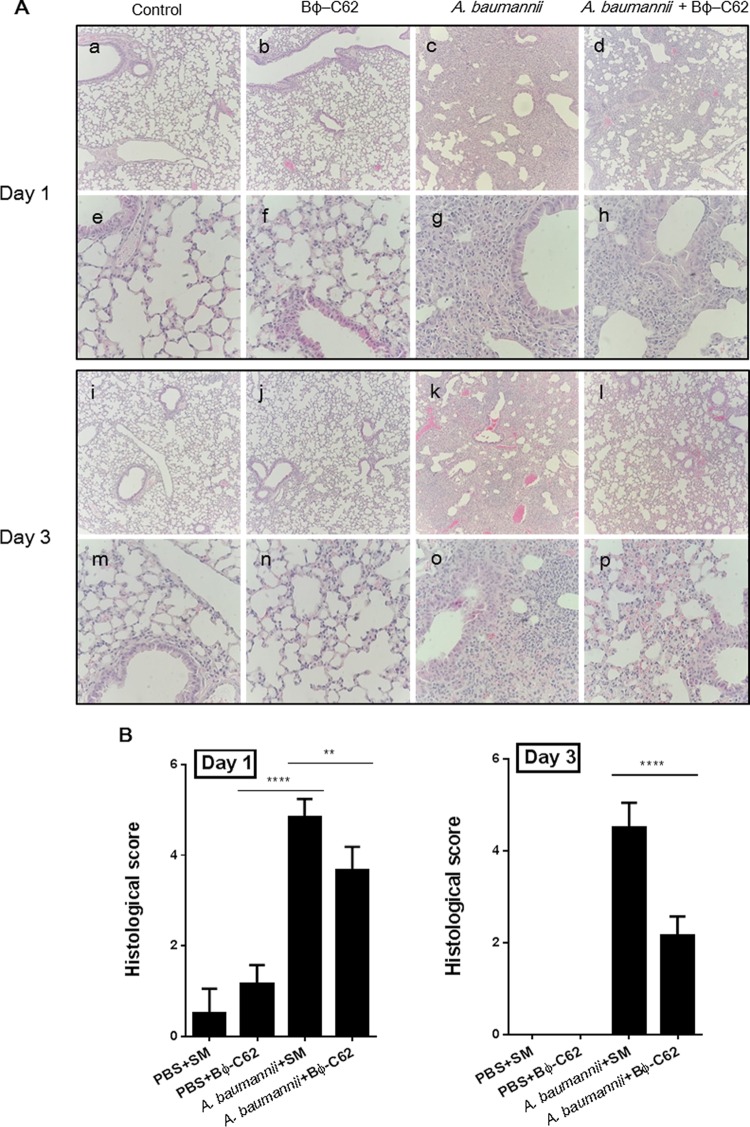
To assess the therapeutic effect of phage in intranasally bacterium-infected mice, the histological changes of lung samples were observed using H&E staining. The bacterium-infected group showed severe thickening of the alveolar walls and an infiltration of neutrophils in the alveolar space at days 1 and 3 (Fig. 4A, c and k). In contrast, most histological sections of the phage-treated group after bacterial infection exhibited moderate alveolar wall thickening and neutrophils in the alveolar space at day 1. The sections of lung at day 3 showed mild to moderate alveolar wall thickening and a number of neutrophils in the alveolar space (Fig. 4A, d and l). The buffer-treated control group showed slightly increased neutrophil numbers in response to PBS and SM buffer treatment at day 1 (Fig. 4A, a and i); however, no histological changes were seen in the lung (Fig. 4A, e and m). Moreover, the phage-administered group revealed little damage in the lung compared to the control group (Fig. 4A, b and j), with no difference identified on day 3 (Fig. 4A, f and n).
FIG 4.
Histopathological changes in mouse lungs of bacterium (A. baumannii YMC13/01/C62)-infected mice or phage Bϕ-C62-treated mice according to H&E staining (magnification with a 10× objective [a to d and i to l] and a 40× objective [e to h and m to p]) (A) and histological severity of mouse lungs (B). We tested a control (PBS and SM buffer) group, phage Bϕ-C62-treated group, A. baumannii-infected group, and phage Bϕ-C62-treated group (30 min postinfection) on day1 (a to h) and day 3 (i to p). One-way ANOVA with Tukey's multiple-comparison test was used to compare histological severity (****, P < 0.0001; **, P < 0.05).
To evaluate the immunogenicity of phage administration and bacterial infection in the lung, we measured the concentrations of cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-6 [IL-6]) in the lungs and blood of five mice from each mouse group at day 1 or 3 after bacterial infection or phage administration (see the supplemental material). The level of TNF-α in the lungs of the phage-treated postinfection group was slightly reduced relative to that in the bacterium-infected group at day 1; however, the difference was not statistically significant (see Fig. S6a in the supplemental material). On the other hand, a significant reduction in IL-6 was seen in the bacterium-infected group (****, P < 0.0001; see Fig. S6b). The levels of TNF-α and IL-6 in the phage-treated group were similar to those of the control group. Levels of TNF-α and IL-6 were not detected in serum collected at day 1 or day 3 from any experimental group (data not shown). Also, the quantification of the histologic severity of infection in lung tissues showed a significant reduction in the phage-treated group postinfection (day 1, score of 3.6; day 3, score of 2.1) relative to the bacterium-infected group (day 1, score of 4.8; day 3, score of 4.5). The phage-administered group (day 1, score of 1; day 3, score of 0) presented a mild or zero severity score (Fig. 4B).
Bacterial and phage counts in the mouse lungs of each group were measured at days 1, 3, and 5. Bacteria counts in the phage-treated group were reduced from 8.6 log10 to 6.4 log10 compared with those of the bacterium-infected group at day 1, and most of the bacteria were eliminated from the lungs at day 3. Bacteria were not detected in the buffer-treated group or phage-administered group at any of the time points (Fig. 5A). The PFU value of the phage-treated postinfection group (10.8 log10) was increased by more than 1 log unit relative to that of the phage-treated group (9.1 log10) at day 1, which was maintained until day 3. At day 5, the phage count sharply decreased from 10.8 log10 to 3.4 log10 in the phage-treated group postinfection and 9.1 log10 to 1.5 log10 in the phage-administered group (Fig. 5B). Phage was not detected in the buffer-treated group or in the bacterium-infected group at any time point (data not shown). These results indicate that phage Bϕ-C62 has significant therapeutic potential against CRAB.
DISCUSSION
In recent decades, the rise of multidrug-resistant pathogens and the decrease of newly approved antibiotics have led to the use of bacteriophages as a novel alternative strategy for control of antibiotic-resistant bacteria (30, 31). Lately, some phage products, such as ListShield and ListexP100, have been approved by the U.S. Food and Drug Administration (USFDA) for use as food biopreservatives (32).
Most phages tend to have high host specificity and a narrow host range, leading to the rapid emergence of phage-resistant bacteria on lytic phages. This resistance is a major obstacle to phage applications as therapeutic agents (6). However, the combination of phages and antibiotics has been researched as a successful strategy to deal with the above-described drawbacks (32). Therefore, the isolation and characterization of many novel and strong lytic phages are very important for their application in phage therapy for treatment of multidrug-resistant pathogens.
Colistin, as last-resort therapy, is the most effective agent for treatment of several infectious diseases caused by multidrug-resistant (MDR) Gram-negative pathogens, including CRAB strains (33). However, it has a risk of renal toxicity. Moreover, the emergence of colistin-resistant A. baumannii in a clinical setting has been reported recently (33, 34). In spite of several global incidents with MDR A. baumannii, the isolation and detailed characterization of lytic phages against CRAB strains have been rarely reported (35).
To date, approximately 20 A. baumannii phages have been isolated from sewage, the hospital environment, and marine sediment samples (35). Based on morphology as determined by TEM, these phages were classified as members of the Myoviridae, Siphoviridae, and Podoviridae families of the Caudovirales order (35). To date, most of these phages have been characterized for phage stability, antibacterial activity, or genome sequence and have been suggested as therapeutic applications against MDR A. baumannii (23, 35–41). In this study, we isolated and characterized the lytic phage Bϕ-C62, which is specific to clinical A. baumannii isolates, and evaluated its potential as an alternative therapeutic agent against CRAB isolated from patients using the mouse lung infection model. Although Soothill (42) demonstrated that phage BS46 effectively protected mice against highly virulent A. baumannii peritoneal cavity injection, the efficacy of phages for the therapeutic treatment of infectious diseases caused by MDR A. baumannii resistant to carbapenems, including imipenem and meropenem, has not been studied using an animal lung infection model.
In this study, 16 of 45 CRAB clinical strains, which were strains susceptible to the phage Bϕ-C62, were found to contain a blaOXA-66-like gene that is one of the known carbapenemase genes reducing susceptibility to carbapenems (43). It is worth noting that all of these isolates were the ST357 A. baumannii strains that were first isolated and reported in South Korea in 2013 (Table 1) (44). Also, the 16S rRNA methylase gene armA (45), which is one of the aminoglycoside resistance genes, was detected in 10 CRAB strains (YMC13/01/R317, R280, R224, R1400, R1224, R1919, R2058, R1049, R3197, and B720) (data not shown).
The phage Bϕ-C62 belongs to the Myoviridae family, which is composed of an ∼62-nm-diameter head and a contractile tail ∼89 nm in length (n = 10) (Fig. 6) and is similar in morphology to A. baumannii phage AP22 (41). The phage Bϕ-C62 showed a relatively longer latency period and a smaller burst size than most known Acinetobacter phages belonging to Myoviridae (40, 46). Phage Bϕ-C62 exhibited significant host cell lytic activities against the host bacterial strain at all MOIs (Fig. 1). In addition, phage Bϕ-C62 maintained significant antibacterial activity and stability at various pH and temperature levels compared to other Acinetobacter phages, such as phage AB1 and phage ZZ1, belonging to the Myoviridae (see Fig. S4 in the supplemental material) (46, 47). Among the known A. baumannii phages, phage Bϕ-C62 has substantial similarity in genome sequence, genome size, and GC content to phage IME-AB2 and phage AP22, which also belong to the Myoviridae family of tailed phages (Fig. 2). The phage Bϕ-C62 genome did not possess known virulence genes such as toxin-related or antibiotic resistance-related genes. These results suggest that Bϕ-C62 is a lytic phage that is specific to A. baumannii and could be used to effectively eliminate CRAB strains from clinical environments.
In this research, we observed that a single dose of phage Bϕ-C62 at an MOI of 10 showed effective therapeutic potential in the mouse model. This phage increased the survival rate of mice with CRAB infections. Moreover, a complete clearance of CRAB YMC13/01/C62 from the lung was observed in all surviving mice at day 3. Also, phage Bϕ-C62 was not harmful to the mice at the highest-titer phage injections, and histological results corresponded with the above-described findings. These studies demonstrate that the A. baumannii phage Bϕ-C62 can be used to treat mice with intranasal CRAB infection; therefore, phage Bϕ-C62 is a viable candidate for phage therapy against CRAB infections. Our results also support that bacteriophages can be an effective alternative strategy to conventional antibiotic treatment for therapy against MDR pathogens.
In this study, the novel Acinetobacter phage Bϕ-C62, which lyses OXA-66-like carbapenemase-producing A. baumannii strains, was characterized in vitro and in silico and was evaluated in vivo for its therapeutic potential to treat lung infection. The current results describe fundamental research into bacteriophage therapy for lung infection in the mouse model.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2009064); by the BioNano Health-Guard Research Center, funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (grant H-GUARD_2014M3A6B2060509); and by a grant from the Brain Korea 21 PLUS Project for Medical Science, Yonsei University.
We are grateful to Sori Jong (The Research Institute of Antimicrobial Resistance, Yonsei University College of Medicine, Seoul, Republic of Korea) for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00526-16.
REFERENCES
- 1.Ackermann HW. 1996. Frequency of morphological phage descriptions in 1995. Arch Virol 141:209–218. doi: 10.1007/BF01718394. [DOI] [PubMed] [Google Scholar]
- 2.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. 2011. Phage treatment of human infections. Bacteriophage 1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahony J, McAuliffe O, Ross RP, van Sinderen D. 2011. Bacteriophages as biocontrol agents of food pathogens. Curr Opin Biotechnol 22:157–163. doi: 10.1016/j.copbio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Merril CR, Scholl D, Adhya SL. 2003. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2:489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 5.Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wittebole X, De Roock S, Opal SM. 2014. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsson AS. 2014. Phage therapy-constraints and possibilities. Ups J Med Sci 119:192–198. doi: 10.3109/03009734.2014.902878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 28:591–595. doi: 10.1016/j.tibtech.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Cohen ML. 1992. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science 257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 10.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 11.Norrby SR, Nord CE, Finch R, European Society of Clinical Microbiology and Infectious Diseases. 2005. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect Dis 5:115–119. doi: 10.1016/S1473-3099(05)70086-4. [DOI] [PubMed] [Google Scholar]
- 12.Tenover FC. 2001. Development and spread of bacterial resistance to antimicrobial agents: an overview. Clin Infect Dis 33(Suppl 3):S108–S115. doi: 10.1086/321834. [DOI] [PubMed] [Google Scholar]
- 13.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Munoz-Price LS, Weinstein RA. 2008. Acinetobacter infection. N Engl J Med 358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 15.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann P, Poirel L. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect 8:321–331. doi: 10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 17.Durante-Mangoni E, Zarrilli R. 2011. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol 6:407–422. doi: 10.2217/fmb.11.23. [DOI] [PubMed] [Google Scholar]
- 18.Vila J, Pachon J. 2012. Therapeutic options for Acinetobacter baumannii infections: an update. Expert Opin Pharmacother 13:2319–2336. doi: 10.1517/14656566.2012.729820. [DOI] [PubMed] [Google Scholar]
- 19.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Lee K, Kim CK, Yong D, Jeong SH, Yum JH, Seo YH, Docquier JD, Chong Y. 2010. Improved performance of the modified Hodge test with MacConkey agar for screening carbapenemase-producing Gram-negative bacilli. J Microbiol Methods 83:149–152. doi: 10.1016/j.mimet.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 23.Lin NT, Chiou PY, Chang KC, Chen LK, Lai MJ. 2010. Isolation and characterization of phi AB2: a novel bacteriophage of Acinetobacter baumannii. Res Microbiol 161:308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Wilcox SA, Toder R, Foster JW. 1996. Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res 4:397–398. doi: 10.1007/BF02257276. [DOI] [PubMed] [Google Scholar]
- 25.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manepalli S, Gandhi JA, Ekhar VV, Asplund MB, Coelho C, Martinez LR. 2013. Characterization of a cyclophosphamide-induced murine model of immunosuppression to study Acinetobacter baumannii pathogenesis. J Med Microbiol 62:1747–1754. doi: 10.1099/jmm.0.060004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry M, Lavigne R, Debarbieux L. 2013. Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob Agents Chemother 57:5961–5968. doi: 10.1128/AAC.01596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann NH. 2008. The potential of phages to prevent MRSA infections. Res Microbiol 159:400–405. doi: 10.1016/j.resmic.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 33.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 34.Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother 60:1163–1167. doi: 10.1093/jac/dkm305. [DOI] [PubMed] [Google Scholar]
- 35.Peng F, Mi Z, Huang Y, Yuan X, Niu W, Wang Y, Hua Y, Fan H, Bai C, Tong Y. 2014. Characterization, sequencing and comparative genomic analysis of vB_AbaM-IME-AB2, a novel lytic bacteriophage that infects multidrug-resistant Acinetobacter baumannii clinical isolates. BMC Microbiol 14:181. doi: 10.1186/1471-2180-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thawal ND, Yele AB, Sahu PK, Chopade BA. 2012. Effect of a novel podophage AB7-IBB2 on Acinetobacter baumannii biofilm. Curr Microbiol 65:66–72. doi: 10.1007/s00284-012-0127-2. [DOI] [PubMed] [Google Scholar]
- 37.Merabishvili M, Vandenheuvel D, Kropinski AM, Mast J, De Vos D, Verbeken G, Noben JP, Lavigne R, Vaneechoutte M, Pirnay JP. 2014. Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS One 9:e104853. doi: 10.1371/journal.pone.0104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Chen B, Song Z, Song Y, Yang Y, Ma P, Wang H, Ying J, Ren P, Yang L, Gao G, Jin S, Bao Q, Yang H. 2012. Bioinformatic analysis of the Acinetobacter baumannii phage AB1 genome. Gene 507:125–134. doi: 10.1016/j.gene.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 39.Jeon J, Kim JW, Yong D, Lee K, Chong Y. 2012. Complete genome sequence of the podoviral bacteriophage YMC/09/02/B1251 ABA BP, which causes the lysis of an OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolate from a septic patient. J Virol 86:12437–12438. doi: 10.1128/JVI.02132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CN, Tseng TT, Lin JW, Fu YC, Weng SF, Tseng YH. 2011. Lytic myophage Abp53 encodes several proteins similar to those encoded by host Acinetobacter baumannii and phage phiKO2. Appl Environ Microbiol 77:6755–6762. doi: 10.1128/AEM.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popova AV, Zhilenkov EL, Myakinina VP, Krasilnikova VM, Volozhantsev NV. 2012. Isolation and characterization of wide host range lytic bacteriophage AP22 infecting Acinetobacter baumannii. FEMS Microbiol Lett 332:40–46. doi: 10.1111/j.1574-6968.2012.02573.x. [DOI] [PubMed] [Google Scholar]
- 42.Soothill JS. 1992. Treatment of experimental infections of mice with bacteriophages. J Med Microbiol 37:258–261. doi: 10.1099/00222615-37-4-258. [DOI] [PubMed] [Google Scholar]
- 43.Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob Agents Chemother 53:2657–2659. doi: 10.1128/AAC.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung JY, Koo SH, Cho HH, Kwon KC. 2013. Nosocomial infection by sequence type 357 multidrug-resistant Acinetobacter baumannii isolates in a neonatal intensive care unit in Daejeon, Korea. Ann Lab Med 33:279–282. doi: 10.3343/alm.2013.33.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oshiro S, Tada T, Kameoka Y, Suzuki K, Ohmagari N, Miyoshi-Akiyama T, Kirikae T. 2015. Development and evaluation of immunochromatography to detect Gram-negative bacteria producing ArmA 16S rRNA methylase responsible for aminoglycoside resistance. J Microbiol Methods 118:159–163. doi: 10.1016/j.mimet.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Jin J, Li ZJ, Wang SW, Wang SM, Huang DH, Li YH, Ma YY, Wang J, Liu F, Chen XD, Li GX, Wang XT, Wang ZQ, Zhao GQ. 2012. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol 12:156. doi: 10.1186/1471-2180-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H, Liang L, Lin S, Jia S. 2010. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol 10:131. doi: 10.1186/1471-2180-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.